Abstract
Skeletal muscle constitutes more than 30% of total body mass using substrates such as glycogen, glucose, free fatty acids, and creatinine phosphate to generate energy. Consequently, multinucleated myofibers and resident mononucleated stem cells (satellite cells) generate several metabolites, which enter into circulation affecting the function of other organs, especially during exercise and atrophy. The present study was aimed at building a comprehensive profile of metabolites in primary human skeletal muscle cells during myogenic progression in an untargeted metabolomics approach using a high resolution Orbitrap Fusion Tribrid Mass Spectrometer. Identification of metabolites with multivariate statistical analyses showed a global shift in metabolomic profiles between myoblasts undergoing proliferation and differentiation along with distinctly separable profiles between early and late differentiating cultures. Pathway analyses of 71 unique metabolites revealed that Pantothenate metabolism and Coenzyme A biosynthesis and Arginine Proline metabolism play dominant roles in proliferating myoblasts, while metabolites involved in vitamin B6, Glyoxylate and Dicarboxylate, Nitrogen, Glutathione, and Tryptophan metabolism were upregulated during differentiation. We found that early and late differentiating cultures displayed differences in Phenylalanine, Tyrosine, Glycine, Serine and Threonine metabolism. Our results identify metabolites during maturation of muscle from progenitor myoblasts that have implications in muscle regeneration and pathophysiology.
Subject terms: Biological techniques, Cell biology
Introduction
Skeletal muscle physiology is critically dependent on the functionality of a progenitor population called, “satellite cells” that proliferate and differentiate to form multinucleated myofibers, thereby contributing to muscle regeneration during injury or exercise1–4. Conversely, defects in satellite cell function can result in loss of muscle mass and decline in performance that is frequently observed in chronic illnesses and aging5. In this aspect, the field of metabolomics has emerged with the intention of providing a comprehensive profile of metabolites and low molecular weight molecules in specific organ tissues, thereby enabling precision medicine and biomarker discovery in various disease states6. Thus, identification of metabolic pathways during myogenic progression may provide information on energy requirements of cells during various physiological states of the muscle tissue.
Recent studies have characterised the skeletal muscle metabolome during strenuous exercise in humans, neuromuscular diseases such as Pompe disease and Duchenne’s muscular dystrophy, daily variations in tissue metabolites vis-à-vis nutritional challenges, overexpression of metabolic regulators, and aging7–12. Metabolic profiles of young, post-mortem, and aging murine satellite cells have also been evaluated using measurement of mitochondrial function and analysis of metabolic gene signatures associated with different myogenic cell cycle states 13,14. Thus, studies from murine myogenic cells suggest that metabolic requirements are critically linked to myogenic cell fates and therefore the quality of regeneration. While the use of animal models has hugely aided in developing personalized metabolic profiles in each condition, few studies have used human subjects or cell culture models to achieve similar objectives. For such studies, muscle biopsies have been extracted from control and test subjects including serum samples to detect circulating metabolites and expand the repertoire of metabolites associated with a specific physiological state9,10 . Myogenic cell culture studies, on the other hand provide direct evidence of the presence of muscle-specific metabolites without confounding results arising from inter-tissue communication and endocrine regulators targeting skeletal muscle7,15. The objective of our study, therefore, was to develop a standard metabolomic profile of human primary myoblasts using ultra-high performance liquid chromatography tandem mass spectrometry (UHPLC-MS/MS) coupled to multivariate statistical analyses during different stages of myogenic progression, which can then be used for identifying signature metabolic profiles of skeletal muscles in altered physiological states.
Results
Metabolomic analysis
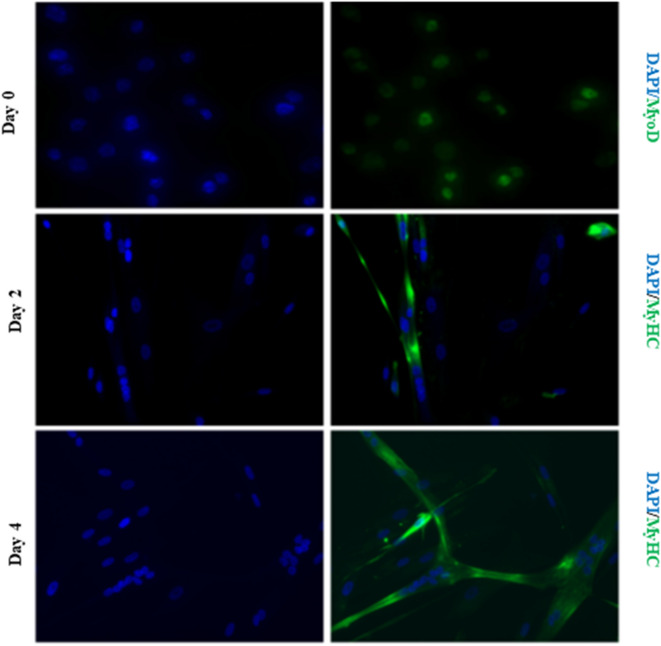
Cells lysates were derived from proliferating primary hSkmc at (Day 0) and differentiating HSMM cultures at two different days after induction of differentiation (Day 2 and Day 4) and were analyzed using UHPLC-MS/MS. Metabolites were identified and then analyzed using PCA analysis to obtain general information of the datasets, relationships between the different groups, and the detection of outliers (located outside the 95% confidence region of the model) (Supplementary Fig. 1B). Of the total 71 metabolites that were detected, principal component PC2 effectively and distinctly separated the Day 0 and Day 2/4 cultures suggesting that the switch from proliferation to differentiation causes a significant change in global metabolite profiles (Supplementary Fig. 1B). Proliferating myogenic cultures (Day 0) displayed greater variation as evidenced by the spread along the PC1 axis, possibly due to heterogeneity arising from asynchronous cell cycle times amongst cells. Early differentiating cultures (Day 2) while distinct from proliferating myoblasts, still showed considerable variation probably due to cells transiting from proliferation to differentiation. This is evident by the presence of mononucleated and multinucleated cells in the culture (Fig. 1, middle panel). Late differentiating cultures (Day 4), on the other hand, were represented by a tight cluster indicating culture homogeneity and increased presence of multinucleated cells (Fig. 1, Supplementary Fig. 1B). Gene expression analysis of known myogenic genes, MyoD, Myogenin, and Myosin Heavy Chain (MyHC) were activated as reported before during proliferation, early and late differentiation respectively (Supplementary Fig. 1A). Visualization of data using heat map analysis corroborated the distinction in PCA analyses between metabolite patterns of proliferating, early and late differentiating cultures (Supplementary Fig. 2).
Figure 1.
hSKmc cultured in growth media for 3 days were fixed and stained with MyoD (top panel), or switched to differentiation media for 2 days (middle panel) and 4 days (last panel) and stained for MyHC.
Of the 71 identified unique metabolites, approximately 60% of the metabolites fell in the category of amino acids or derivatives and metabolites of amino acids. Of the rest, 18% of the metabolites were represented by carboxylic acids, 8% by nucleosides, 4% by esters, 3% by aldehyde and their derivatives, and less than 2% by carbohydrate polymers, alkaloids, and purine derivatives (Table 1).
Table 1.
Global and differential metabolite profiles during myogenic progression of primary human myoblasts.
| S. no | Metabolite name | Mode | Day-0/ versus Day-2 | Day-0/ versus Day-4 | Day-2/ versus Day-4 | |||
|---|---|---|---|---|---|---|---|---|
| Ratio | p value | Ratio | p value | Ratio | P value | |||
| 1 | 3-Methylhistidine | Positive Hilic | 1.957483761 | 0.017058732 | 1.969849069 | 0.01706663 | 1.482804653 | 0.956364752 |
| 2 | 5-Hydroxytryptophan | Positive Hilic | 0.892634278 | 0.501064879 | 0.990409135 | 0.857467453 | 0.539920534 | 0.391557516 |
| 3 | Arginine | Positive Hilic | 2.063405284 | 0.021845753 | 3.019269217 | 0.129708384 | 64.10815532 | 0.510461226 |
| 4 | Carnosine | Positive Hilic | 1.381165782 | 0.231663886 | 0.441840836 | 0.388254784 | 0.925133238 | 0.326132666 |
| 5 | Galactosamine | Positive Hilic | 2.400296514 | 0.011032868 | 3.063427592 | 0.014300249 | 0.063095683 | 0.055447395 |
| 6 | Glutathione (oxidized) | Positive Hilic | 0.155939966 | 0.008695074 | 0.140495964 | 0.040022369 | 18.45594754 | 0.451109586 |
| 7 | Glutathione (reduced) | Positive Hilic | 0.219887217 | 0.478180308 | 0.508236024 | 0.371266079 | 47.07290561 | 0.653463664 |
| 8 | Gly-Val | Positive Hilic | 2.618389502 | 0.265807903 | 2.610573581 | 0.239896174 | 0.06066872 | 0.980335803 |
| 9 | Histidine | Positive Hilic | 0.897460993 | 0.824289642 | 0.68752904 | 0.041119919 | 5.333627958 | 0.590077911 |
| 10 | Kynurenine | Positive Hilic | 1.403423107 | 0.206203297 | 1.877591002 | 0.045644309 | 0.313956092 | 0.206462409 |
| 11 | Lysine | Positive Hilic | 1.07404302 | 0.69878706 | 1.262601926 | 0.439139483 | 41.26193431 | 0.370038949 |
| 12 | N-Acetylhistidine | Positive Hilic | 8.663894138 | 0.010640991 | 12.71642777 | 0.011309978 | 0.043353965 | 0.166758226 |
| 13 | l-Methylhistidine | Positive Hilic | 1.146680443 | 0.006762015 | 0.909198318 | 0.088270644 | 1.191416337 | 0.015088396 |
| 14 | N-Methylisoleucine | Positive Hilic | 0.883831782 | 0.656091485 | 1.693482061 | 0.733860783 | 2.87311408 | 0.690666886 |
| 15 | N-Methylproline | Positive Hilic | 1.434443191 | 0.356711878 | 2.093768751 | 0.288915143 | 1.250040637 | 0.208222283 |
| 16 | N-Methylserine | Positive Hilic | 0.630955667 | 0.050240636 | 0.51163564 | 0.019520417 | 21.3565562 | 0.250600044 |
| 17 | Octopine | Positive Hilic | 5.287217325 | 0.197774148 | 12.78205406 | 0.172678835 | 0.738545396 | 0.019932737 |
| 18 | Proline–hydroxyproline | Positive Hilic | 1.699381081 | 0.015190354 | 2.008944192 | 0.033992985 | 1.515058751 | 0.174178233 |
| 19 | Propionylcarnitine | Positive Hilic | 1.178580994 | 0.131295737 | 0.804876763 | 0.7130271 | 11.61778227 | 0.530650712 |
| 20 | SDMA | Positive Hilic | 2.317179658 | 0.429051246 | 3.662975851 | 0.304548183 | 0.005772778 | 0.300783066 |
| 21 | Taurine | Positive Hilic | 1.152807858 | 0.378443043 | 1.135080404 | 0.532004759 | 0.724821458 | 0.939454594 |
| 22 | Uridine-5-diphosphoacetylglucosamine | Positive Hilic | 0.236797735 | 0.285847398 | 0.320233968 | 0.203784286 | 332.6518156 | 0.70071203 |
| 23 | Beta-alanine | Positive Hilic | 2.343509429 | 0.009954229 | 2.496581273 | 0.056740483 | 0.100688392 | 0.820501657 |
| 24 | Glycine | Positive Hilic | 0.46210005 | 0.16294956 | 0.487130168 | 0.056597298 | 2.098289127 | 0.766787981 |
| 25 | Palmitate | Positive Hilic | 0.614986594 | 0.464993079 | 1.090262794 | 0.880114504 | 0.002434426 | 0.600636379 |
| 26 | Cholate | Positive Hilic | 1.098018515 | 0.157506917 | 1.094544916 | 0.364194308 | 13.14097994 | 0.977100429 |
| 27 | Glycerophosphocholine | Positive Hilic | 0.410966821 | 0.028671262 | 0.361175125 | 0.173575736 | 9.097481347 | 0.726250188 |
| 28 | Glutamate | Positive Hilic | 1.221537042 | 0.207605635 | 1.025410416 | 0.690595173 | 2.45540182 | 0.176857341 |
| 29 | Leucine | Positive Hilic | 1.444908173 | 0.176917283 | 1.728757289 | 0.275269829 | 10.29942336 | 0.483717136 |
| 30 | 4-Quinolinecarboxylic acid | Negative Hilic | 1.418438368 | 0.513853945 | 2.159369267 | 0.218075199 | 0.103449101 | 0.086485699 |
| 31 | Aspartic acid | Negative Hilic | 0.482476444 | 0.065825708 | 0.602504147 | 0.113049977 | 0.127367313 | 0.516568913 |
| 32 | Glutamic acid | Negative Hilic | 1.046514892 | 0.780023849 | 0.827811878 | 0.242537341 | 0.150792733 | 0.415386587 |
| 33 | Ornithine | Negative Hilic | 1.850187949 | 0.391006337 | 5.755406751 | 0.145940321 | 0.078217893 | 0.165848257 |
| 34 | Pyroglutamic acid | Negative Hilic | 1.367259238 | 0.093669826 | 1.891645605 | 0.043315005 | 2.369792082 | 0.366558893 |
| 35 | Valine | Negative Hilic | 1.753550671 | 0.174813102 | 2.384865691 | 0.113296857 | 61.62403609 | 0.040640995 |
| 36 | Phenylalanine | Negative Hilic | 0.935194029 | 0.86993207 | 0.409014557 | 0.042444283 | 0.791986697 | 0.090863383 |
| 37 | Tryptophan | Negative Hilic | 0.371330134 | 0.081424087 | 0.192753035 | 0.042524748 | 0.01204981 | 0.032135949 |
| 38 | 4-Hydroxybenzaldehyde | Positive RP | 0.038566578 | 0.338542448 | 0.08710944 | 0.030486282 | 2.748006594 | 0.55634682 |
| 39 | 2-Quinolinecarboxylic acid | Positive RP | 1.284244421 | 0.178306336 | 1.296689552 | 0.039303244 | 1.799999525 | 0.963141641 |
| 40 | 4-Guanidinobutanoate | Positive RP | 0.542825386 | 0.391616229 | 1.237852105 | 0.544874186 | 1.46736347 | 0.207158492 |
| 41 | 4-Pyridoxate | Positive RP | 0.906943765 | 0.438937202 | 0.87509599 | 0.296192773 | 0.067630149 | 0.857936761 |
| 42 | Creatinine | Positive RP | 1.253400463 | 0.16561792 | 1.26221635 | 0.080361469 | 2.527037031 | 0.902938632 |
| 43 | Deoxycarnitine | Positive RP | 1.26442431 | 0.070202975 | 2.712227268 | 0.414190457 | 0.165367546 | 0.531546157 |
| 44 | d-pantothenic acid | Positive RP | 2.54453839 | 0.010420678 | 1.630641604 | 0.003710638 | 1.16289204 | 0.095092403 |
| 45 | Guanosine | Positive RP | 1.145849982 | 0.355689984 | 1.129363317 | 0.50751644 | 5.768618975 | 0.89840141 |
| 46 | Hippurate | Positive RP | 1.127259102 | 0.136289877 | 1.406303815 | 0.016533704 | 0.219676668 | 0.131795401 |
| 47 | Hypoxanthine | Positive RP | 1.359667365 | 0.457112959 | 1.608746416 | 0.099129029 | 7.05463603 | 0.809897126 |
| 48 | Carnitine | Positive RP | 1.90728994 | 0.143435009 | 1.564029523 | 0.172039547 | 0.018010572 | 0.589277748 |
| 49 | Proline | Positive RP | 1.1825476 | 0.10029429 | 1.364221698 | 0.019455593 | 1.09422421 | 0.087632586 |
| 50 | N-Acetyl-d-Tryptophan | Positive RP | 0.513159962 | 0.129235482 | 0.496551741 | 0.022394724 | 0.019329765 | 0.799179294 |
| 51 | Nicotinamide | Positive RP | 1.854686932 | 0.213557125 | 3.014205827 | 0.199623561 | 294.7130137 | 0.171017445 |
| 52 | Phenethylamine | Positive RP | 0.947278292 | 0.830253378 | 1.539430825 | 0.158597412 | 0.158329201 | 0.080856795 |
| 53 | Pipecolate | Positive RP | 1.25016381 | 0.293213253 | 1.427971372 | 0.207201663 | 1.071570022 | 0.09885495 |
| 54 | Pyridoxal | Positive RP | 1.217768919 | 0.031096573 | 1.27808282 | 0.111845597 | 0.112332187 | 0.515255364 |
| 55 | Pyridoxine | Positive RP | 1.087248254 | 0.317809478 | 1.096173651 | 0.700340773 | 4.820930313 | 0.965070128 |
| 56 | Cytidine | Positive RP | 2.322579759 | 0.180942623 | 2.507571618 | 0.258966415 | 29.54734542 | 0.869145933 |
| 57 | Citrulline | Positive RP | 1.24280421 | 0.682704165 | 0.603931311 | 0.531030346 | 0.000524472 | 0.53161591 |
| 58 | Inosine | Positive RP | 1.899386369 | 0.182257102 | 1.390470066 | 0.310097325 | 50.39782881 | 0.198914094 |
| 59 | Cytosine | Positive RP | 2.198331093 | 0.138106621 | 2.197735973 | 0.21587362 | 0.096448908 | 0.999444736 |
| 60 | Uracil | Positive RP | 0.944308633 | 0.227098922 | 1.026581624 | 0.238988701 | 0.184148495 | 0.131452792 |
| 61 | Guanine | Positive RP | 1.262679439 | 0.538371956 | 1.28191787 | 0.421569109 | 97.45431759 | 0.983003715 |
| 62 | Folate | Positive RP | 1.330385823 | 0.627752558 | 0.447595415 | 0.203196636 | 0.015697847 | 0.219888549 |
| 63 | Betaine | Positive RP | 2.045353528 | 0.181453617 | 2.995576467 | 0.1168387 | 2.012165194 | 0.007590226 |
| 64 | Pyroglutamate | Positive RP | 0.755668365 | 0.123082312 | 0.807394884 | 0.226135842 | 1566.855327 | 0.704018417 |
| 65 | Adenosine | Positive RP | 1.371651672 | 0.378510887 | 0.894426306 | 0.530775604 | 41.39432776 | 0.044975022 |
| 66 | 2-Hydroxyphenylacetate | Positive RP | 1.100243522 | 0.374777295 | 1.16996763 | 0.295039165 | 0.890312864 | 0.493264835 |
| 67 | Lipoamide | Positive RP | 1.23055559 | 0.104304526 | 1.21925258 | 0.072138847 | 0.442513586 | 0.929302085 |
| 68 | Suberate | Positive RP | 0.959311235 | 0.696818973 | 0.964122724 | 0.863292691 | 5.735613668 | 0.985533593 |
| 69 | 4-Methylcatechol | Positive RP | 0.816546234 | 0.152010857 | 0.621372018 | 0.037113323 | 0.807864486 | 0.0491021 |
| 70 | Uridine | Negative RP | 1.337277003 | 0.490020692 | 1.13709077 | 0.724444642 | 5.622283342 | 0.382538807 |
| 71 | 4-Acetamidobutanoate | Negative RP | 4.059047216 | 0.120056303 | 5.167043856 | 0.096549799 | 0.000822875 | 0.158797722 |
Italics values represent metabolites with significant values p < 0.05.
Significant changes in metabolites during myogenic progression
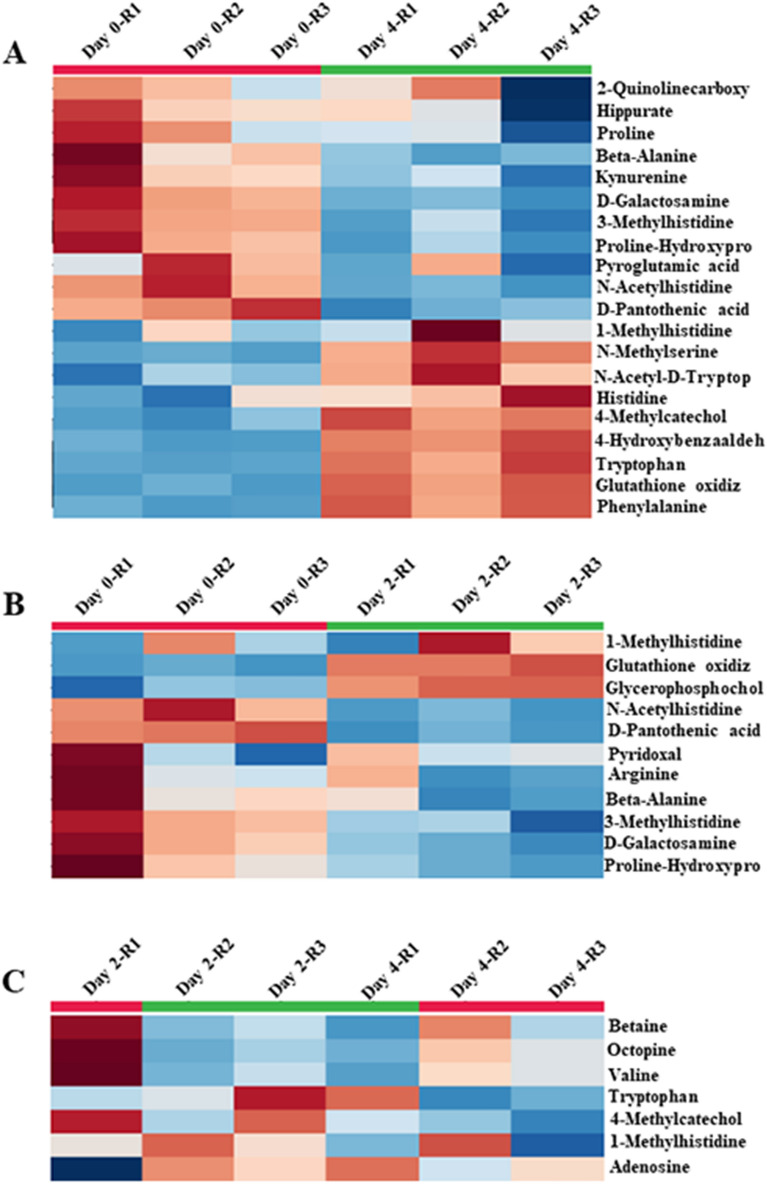
Using the MetaboAnalyst 4.0 software package, a total of 23 unique metabolites were identified that displayed significantly altered levels between proliferating and differentiation cultures, including 8 common between early and late differentiating cultures (Fig. 2A,B). Of these, 20 metabolites were observed to be significantly altered between proliferating and late differentiating cultures (Day 0 and Day 4) and 11 metabolites between proliferating and early differentiating cultures (Day 0 and Day 2), suggesting a global shift in metabolism when human myogenic cell progress from proliferation to a terminally differentiated state. Expectedly, numbers of metabolites showing differential expression between early and late differentiating cultures were smaller with 7 metabolites displaying significant differences in the levels of Betaine, Octopine, Valine, Tryptophan, 4-Methylcatechol, 1 Methylhistidine, and adenosine (Fig. 2C). Table 1 lists the changes in ratios of metabolites from proliferating to early differentiation (Day 0 to Day 2), early to late differentiation (Day 2 to Day 4), and from proliferation to late differentiation (Day 0 to Day 4).
Figure 2.
Heat map displaying metabolites that were significantly altered over the progression of myogenic culture. (A) 20 metabolites that were differentially altered from proliferating myogenic culture (Day 0) to late differentiating culture (Day 4). (B) 11 metabolites that were differentially altered from proliferating (Day 0) to early differentiating myogenic culture (Day 2). (C) 7 metabolites that were differentially altered from early (Day 2) to late differentiating culture (Day 4).
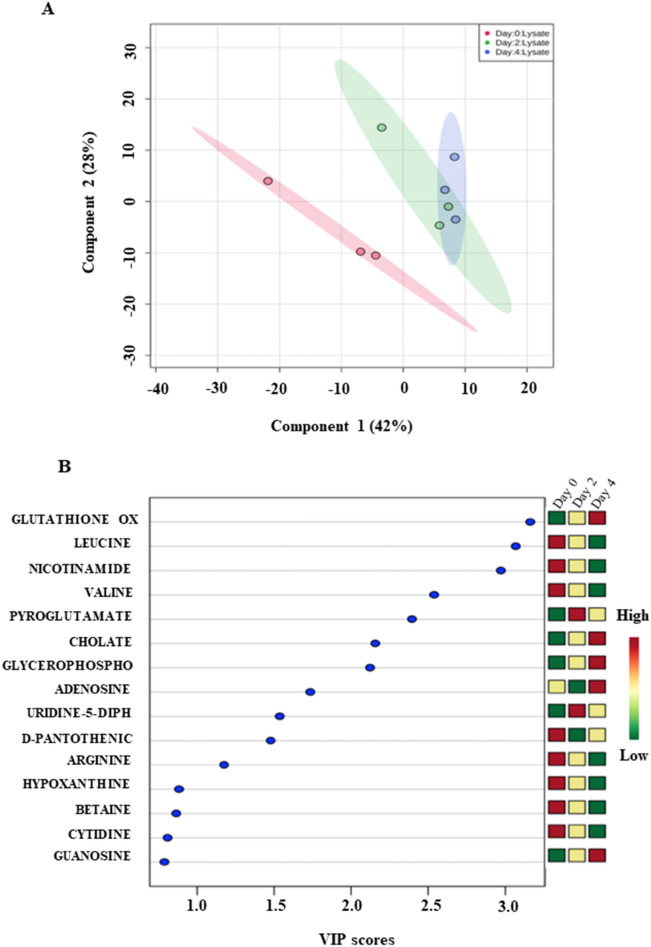
To identify those metabolites with the most important features that contributed to differences between the 3 groups, we performed a PLS-DA plot (Fig. 3A). The PLS-DA score plot with component one accounting for 42% of the variation and component two explaining 28% of the variation, shows a clear separation between proliferation and differentiation, as well as a distinction between early and late differentiation (Fig. 3A). The PLS-DA analysis showed a distinct separation (R2Y = 0.9) and good predictability (Q2 = 0.56) with an accuracy of 45% (Supplementary Fig. 1C). The variable importance in projection (VIP) score plot was used to identify the top 11 most important metabolite features between the 3 different groups with scores greater than 1 and that contributed to the classification (Fig. 3B). In order to confirm that the variation in concentration of the identified metabolites was statistically significant, a t test was done to identify the important discriminants with a threshold of p < 0.05 (Table 1). We identified 6 such significant metabolites including Glutathione oxidized, valine, Glycerophosphocholine, Adenosine, d-pantothenic acid, and Arginine (Fig. 3B, Table 1). Of these metabolites, d-pantothenic acid, Arginine, and Valine were significantly upregulated in proliferating myoblasts, while, Glutathione oxidized, Glycerophosphocholine, and Adenosine were upregulated in late differentiating cultures (Fig. 3B).
Figure 3.
(A) Partial least square discriminate analysis (PLS-DA) scores plot of proliferating (Day 0), early differentiating (Day 2) and late differentiating myogenic culture (Day 4). First component versus second component showing the separation of myogenic culture (Day 0) to early (Day 2) and late differentiating myogenic culture (Day 4). Component 1 and 2 explaining about 70% of the data. (B) VIP scores with the corresponding expression heat map. Green and red indicates the low and high metabolites levels respectively.
Metabolomic pathway analysis in myogenic progression
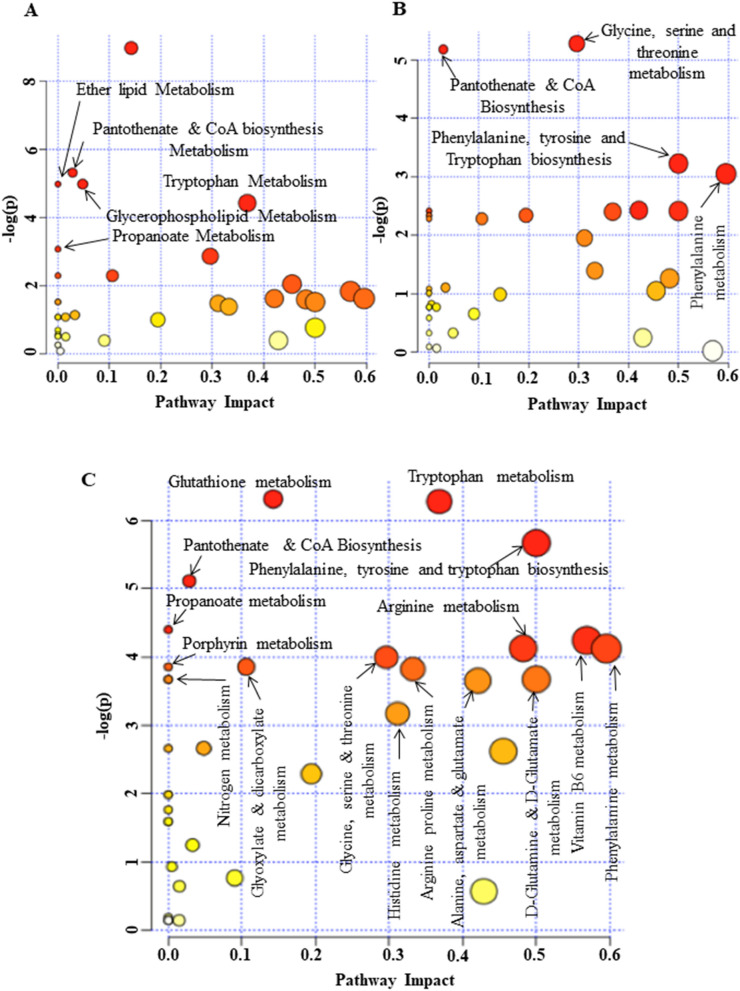
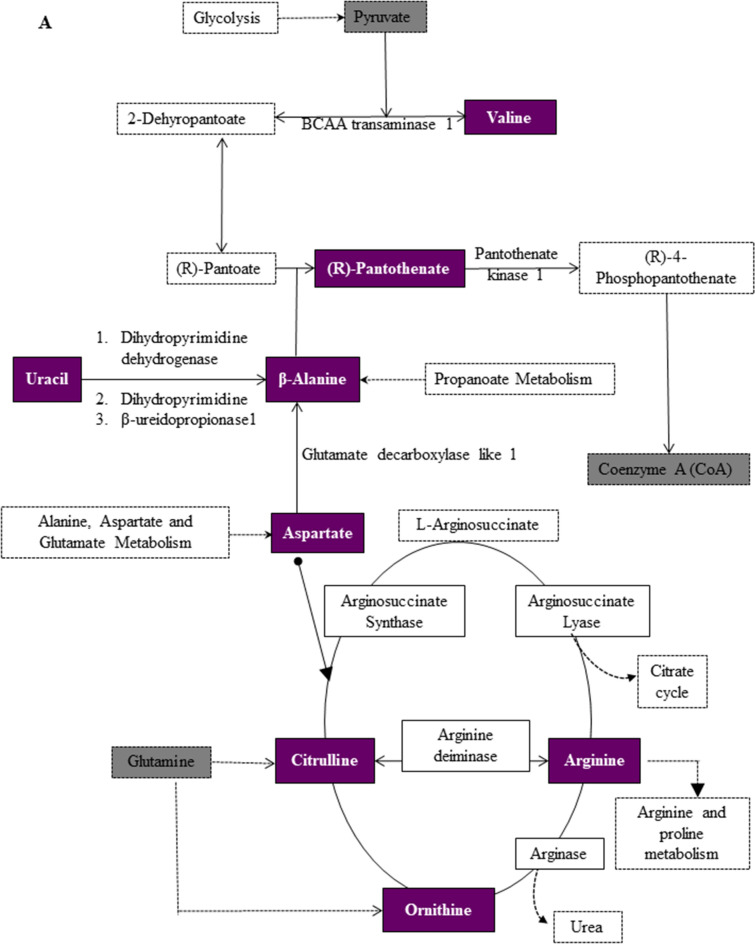
To investigate which of the metabolic pathways were significantly altered during myogenic progression, we performed Pathways analysis with MetaboAnalyst 4.0 and identified the Pantothenate metabolic pathway and Coenzyme A (CoA) biosynthesis to be significantly regulated throughout myogenic progression (Fig. 4, Table 2). Specifically, Pantothenate and CoA biosynthesis was upregulated in proliferating cultures compared to differentiating myogenic cultures (Fig. 4, Table 2). Additionally, we detected several metabolites indicating a dominant role for Arginine and Proline metabolism in proliferating myoblasts (Fig. 4, Tables 1, 2). Metabolic pathway maps displayed key metabolites related to the Pantothenate and CoA biosynthesis pathway that were altered during proliferation including Pantothenate, β-alanine, Uracil, Valine, and Aspartate, while those related to Arginine and Proline metabolism included Arginine, Ornithine, Citrulline and Aspartate (Fig. 5A).
Figure 4.
Summary of pathway analysis with Ingenuity pathway analysis based on KEGG database. All the significantly match (p value < 0.05) were labeled. The color and size of each circle was based on p value and pathway impact value, respectively. Pathways that were significantly altered between (A) proliferating myoblasts (Day 0) and early differentiating culture (Day 2), (B) proliferating (Day 0) and late differentiating culture (Day 4), and (C) early and late differentiating cultures.
Table 2.
Metabolic pathways displaying significant changes in the primary human myoblasts during the course of myogenic progression.
| S. no | Pathways displayed significant changes during Day 0 to Day 2 transformation | Match status | p value | FDR |
|---|---|---|---|---|
| 1 | Pantothenate and CoA Biosynthesis | 5/19 | 0.0049 | 0.062407 |
| 2 | Glycerophospholipid Metabolism | 1/36 | 0.0069 | 0.062407 |
| 3 | Ether lipid metabolism | 1/20 | 0.0069 | 0.062407 |
| 4 | Tryptophan Metabolism | 3/41 | 0.0120 | 0.086932 |
| 5 | Pantothenate Metabolism | 1/23 | 0.0466 | 0.27999 |
| Pathways displayed significant changes during Day 2 to Day 4) transformation | ||||
| 1 | Glycine, Serine and Threonine Metabolism | 2/33 | 0.0051 | 0.10159 |
| 2 | Pantothenate and CoA Biosynthesis | 5/19 | 0.0056 | 0.10159 |
| 3 | Phenylalanine, Tyrosine and Tryptophan Biosynthesis | 1/4 | 0.0396 | 0.26212 |
| 4 | Phenylalanine Metabolism | 4/10 | 0.0472 | 0.26212 |
| Pathways displayed significant changes during Day 0 to Day 4 transformation | ||||
| 1 | Glutathione metabolism | 5/28 | 0.0018 | 0.033886 |
| 2 | Tryptophan Metabolism | 3/41 | 0.0018 | 0.033886 |
| 3 | Phenylalanine, Tyrosine and Tryptophan Biosynthesis | 1/4 | 0.0034 | 0.041434 |
| 4 | Pantothenate and CoA Biosynthesis | 5/19 | 0.0060 | 0.054196 |
| 5 | Pantothenate Metabolism | 1/23 | 0.0122 | 0.058348 |
| 6 | Vitamin B6 Metabolism | 3/9 | 0.0143 | 0.058348 |
| 7 | Arginine Biosynthesis | 5/14 | 0.0161 | 0.058348 |
| 8 | Phenylalanine Metabolism | 4/10 | 0.0161 | 0.058348 |
| 9 | Glycine, Serine and Threonine Metabolism | 2/33 | 0.0184 | 0.058348 |
| 10 | Glyoxylate and Dicarboxylate Metabolism | 2/32 | 0.0211 | 0.058348 |
| 11 | Porphyrin Metabolism | 2/30 | 0.0211 | 0.058348 |
| 12 | Arginine and Proline Metabolism | 6/38 | 0.0211 | 0.058348 |
| 13 | d-Glutamine and d-Glutamate Metabolism | 1/6 | 0.0254 | 0.058348 |
| 14 | Butanoate Metabolism | 1/15 | 0.0254 | 0.058348 |
| 15 | Nitrogen Metabolism | 1/6 | 0.0254 | 0.058348 |
| 16 | Alanine, Aspartate and Glutamate Metabolism | 2/28 | 0.0259 | 0.058348 |
| 17 | Histidine Metabolism | 5/16 | 0.0419 | 0.088755 |
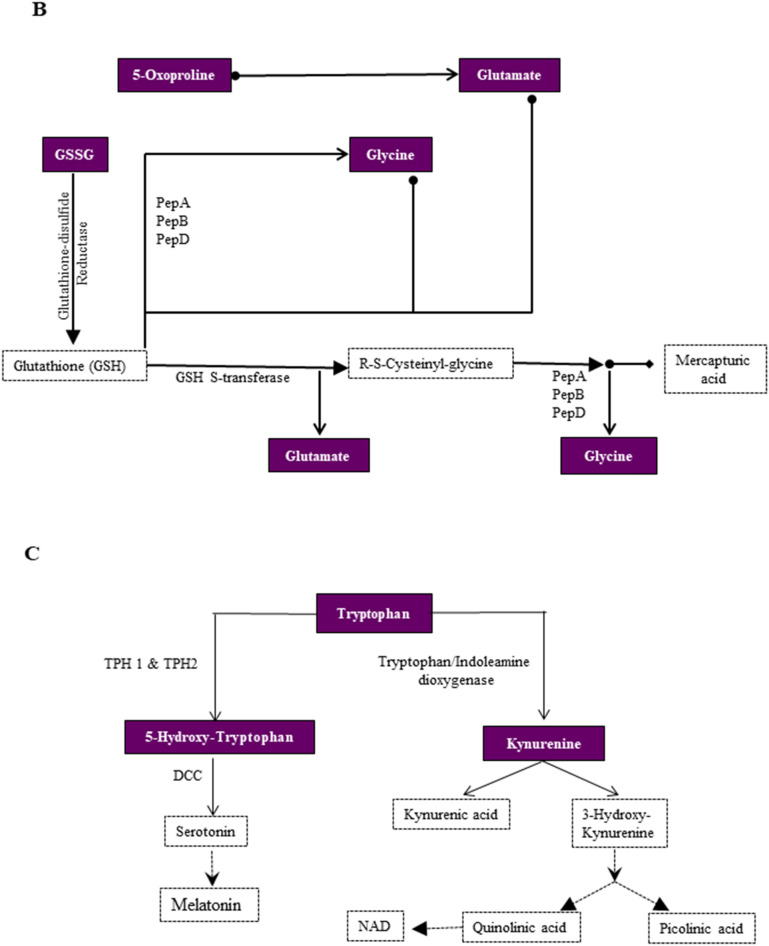
Figure 5.
Representative metabolic pathway maps of significantly altered metabolites during proliferation and differentiation. Key metabolites related to (A) Pantothenate and CoA biosynthesis and Arginine and Proline metabolism that display significant alteration during proliferation and (B) Glutathione metabolism, (C) Tryptophan metabolism during differentiation. Colored boxes represent identified metabolites. Images were modified from map00770 and map00220 respectively for (A), map00480 for (B), and map00380 for (C) from the KEGG database (https://www.kegg.jp).
Of the 12 unique pathways upregulated during differentiation, vitamin B6, Glyoxylate and Dicarboxylate, Nitrogen, Glutathione, and Tryptophan metabolism were upregulated in late differentiation (Table 2). Pathway analyses between early and late differentiating cultures identified significant alterations primarily in specific amino acid metabolism such as Phenylalanine, Tyrosine, Glycine, Serine and Threonine metabolism (Fig. 4, Table 2). Metabolic pathway maps showed key metabolites related to the Glutathione pathway were altered during differentiation including Glutathione oxidised (GSSG), Glycine, Glutamate, and 5-Oxoproline, while metabolites related to the Tryptophan metabolism pathway were Kynurenine and 5-Hydroxy tryptophan (Fig. 5B,C).
Discussion
Skeletal muscle performance in the context of physical activity and endocrine regulation of other organ systems is critically dependent on energy demands of individual myogenic cells. These cells are characterised by specific metabolite profiles that signify as biomarkers of a particular stage of myogenic progression13,14. While gene expression and in vivo functional studies have indicated the importance of specific factors for muscle mass maintenance and physiology, less is known about measureable changes in metabolites corresponding to these pathways, the tissue source, and the temporal kinetics of these metabolites during the course of myogenic progression16–18. Using two multivariate statistical methods of PCA and PLS-DA analyses, our results identify the predominant metabolites and pathways important for proliferation and distinct differentiation stages during myogenic progression in human primary myoblasts.
From our results we found that a key pathway to be active in proliferation was Arginine and Proline metabolism with Arginine displaying a VIP score > 1 (Fig. 3B). Specifically, we found that proline–hydroxyproline was upregulated 1.7-fold in proliferating myoblasts over early differentiating cultures and 2-fold over later differentiating cultures (Fig. 3A). Both proline and hydroxyproline comprise 30% of the amino acids in collagen, a vital component in the extracellular matrix of skeletal muscle19. These amino acids contribute to the stability of the helices in collagen and are responsible for the stiffness of this polymer indicating that these are necessary components in creating bio-artificial muscles in tissue engineering19,20. The increase in metabolite levels of proline and hydroxyproline in the early stages suggest a preparatory step in myogenic progenitors for terminal differentiation. It has also been reported that metabolomic analysis of aged muscles has revealed a reduction in proline and hydroxyproline that might be responsible for the progressive loss of muscle mass and function in sarcopenia, in accordance with dysfunctional satellite cell activation in aging muscles11.
Our analysis revealed that one of the pathways dominating the proliferative phase is the Pantothenate metabolic pathway and CoA biosynthesis with d-pantothenic acid displaying a high VIP score (Fig. 3B). Pantothenic acid is the primary substrate for Pantothenate kinase, the rate limiting step in CoA biosynthesis, accounting for about 66% of CoA in skeletal muscle, the highest amount than in any other tissue21–23. In muscle energy metabolism, CoA contributes to the fatty acyl-CoA pool in mitochondria via the carnitine shuttle system, that is subsequently used for the carnitine palmitoyl-transferase 2 (CPT2) mediated transesterification of acyl-carnitine to carnitine and acyl CoA, for mitochondrial fatty acid β-oxidation, oxidative decarboxylation of pyruvate to acetyl CoA, and finally as a key substrate for α-ketoglutarate in the TCA cycle24–27. Importantly, CoA functions as a substrate for the enzyme, acyl CoA synthetase (ACS) that enables a critical fuel selection switch from glucose to fatty acids (FA) during activity28. Indeed, impaired ACS function result in a significant decline in FA oxidation, heightened insulin sensitivity, depleted glucose reserves resulting in a form of hypoglycaemia that could not be compensated by hepatic gluconeogenesis, and reduction in muscle endurance capacity28. Physiological regulation of CoA became evident in a study examining the effects of altering CoA levels in skeletal muscle that resulted in altered mitochondrial morphology, lowered ATP levels, increased oxidative stress, reduced grip strength and endurance in exercise29 Thus, it may seem that Pantothenate and CoA profiles and their metabolites in muscle cells might potentially be altered during specific pathological situations.
Another key metabolite that was modestly, but significantly upregulated in differentiating human myoblasts was Pyridoxal, suggesting the importance of vitamin B6 and one carbon metabolism (OCM) in myogenic progression. Specifically, vitamin B6 is a source for pyridoxal phosphate (PLP), a coenzyme required for several phases of OCM, further highlighting the importance of this dietary nutrient in aiding normal muscle growth and function, especially during muscle development and postnatal regeneration. Additionally, recent reports suggest that PLP is essential for the synthesis of carnosine, a histidine containing dipeptide that is prevalent in Type ll glycolytic fibers, with increased PLP levels present in highly contractile muscle, suggesting a role for vitamin B6 metabolism in muscle regeneration30,31.
Of the metabolites present at increasing levels during differentiation, we found glutathione metabolism particularly GSSG displaying the highest VIP score of 3.2, to be significantly upregulated more than 2-fold above proliferating myoblasts, indicating the importance of regulation of oxidative stress in myogenic progression (Fig. 3B). These results are concordant with a recent report examining antioxidant transcription and myogenic differentiation in satellite cells, indicating that a pro-oxidative environment is essential for differentiation32. Despite the observation that GSSG is a notoriously difficult compound to quantify by mass spectrometry, it is highly unlikely that its presence is an artefact of the methodology, since we detect this compound at retention time 8.3 with very good peak shape and ion intensity using HILIC negative mode as is indicated by high resolution EIC analysis (Supplementary Fig. 4). Increasing concentrations of tryptophan, a precursor of serotonin and IGF-1 that is associated with increasing muscle mass, suggest the direct impact that this essential amino acid has on differentiation. Indeed, it has been shown that reduced levels of tryptophan are associated with sarcopenia, while its metabolite kynurenine has been observed to accumulate in the peripheral tissues of rats with advanced age33,34. Specifically, mice fed with tryptophan, but not its metabolite kynurenine showed increased expression of myogenic, as well Akt1 and Akt2, two factors essential for myogenic differentiation and myotube maturation35. This suggests that the increase in muscle mass on tryptophan administration arises because of its requirement during terminal myogenic differentiation.
Conclusion
Overall, our study aims to use high throughput metabolomics analysis as a foundational study to characterise skeletal muscle-specific metabolic changes during myogenic progression that includes proliferating, early differentiating and late differentiating human myogenic cultures. Using an untargeted metabolomics approach coupled to multivariate analyses, this is the first study to implicate specific pathways such as Pantothenate metabolism and CoA biosynthesis and arginine and proline metabolism during the proliferation step of myogenic progression. Several unique pathways including vitamin B6, Tryptophan, Glutathione, and Glyoxylate and Dicarboxylate metabolism were identified as the key pathways that are altered during the differentiation phase of myogenic progression. Although the study captures a wide unbiased landscape of metabolites, this could be a precursor for future studies pursuing individual metabolites that are perturbed during myogenic differentiation for specific muscle diseases.
Materials and methods
Cell culture
Human skeletal muscle myoblasts (hSKmc) (CC-2580), growth media, supplements, and serum (CC-3245) were purchased from Lonza (Walkersville, MD). HSMM differentiation medium (DM) was prepared by adding 2% horse serum to DMEM (Hyclone). HSMM were maintained in growth media for 3–4 days till cultures reached 70% confluence followed by three washes in phosphate buffered saline (PBS, pH 7) and switched to DM. Intracellular metabolites from cell lysates were collected from Day 0 cultures (before addition of DM), Day 2 and Day 4 cultures (DM added for 2 and 4 days respectively). For kinetic analysis of intracellular metabolites, hSKmc cultures for Day 0, 2, and 4 underwent three washes in PBS, scraped in RIPA lysis and extraction buffer (Cat # 89900, ThermoFisher Scientific), and stored at – 80 °C.
Sample preparation
Metabolites were extracted from the quenched cells using 100% methanol extraction method. Cells pellet frozen at − 80 °C was thawed for 10 min and resuspended in 500 μl of 100% chilled methanol (MS-grade, Sigma) and vortexed for 1 min and frozen at − 80 °C for 10 min. The freeze–thaw cycle was repeated twice followed by pelleting of cells by centrifugation at 15,000g for 10 min at 4 °C. After centrifugation each supernatant (~ 400 μl) was collected in a separate microfuge tube without disturbing the pellet. Each fraction (100 μl) was dried using a speed vacuum at room temperature for 20–25 min. Samples were stored at − 80 °C till further analysis. For sample injection, each sample was resuspended in 25 μl of methanol water mixture (3:17, methanol: water), vortexed briefly and centrifuged at 11,000 rpm for 10 min at 4 °C.
Measurement of metabolites
Orbitrap Fusion mass spectrometer (Thermo Scientific) coupled with heated electrospray ion source was used for data acquisition. Data acquisition methods have been followed as per published protocols36,37 with minor modifications. Briefly for MS1 mode, mass resolution was kept at 120,000 and for MS2 acquisition, mass resolution was 30,000. Mass range of data acquisition was 60–900 da. Extracted metabolites were separated on UPLC ultimate 3,000. Data were acquired on reverse phase and HILIC column and positive and negative ionization mode both. Reverse phase column was HSS T3 and HILIC column was XBridge BEH Amide (Waters Corporation). For polar compound separation, solvent A was 20 mM ammonium acetate in the water of PH 9.0 and mobile phase B was 100% acetonitrile. The elution gradient starts from 85% B to 10% B over 14 min with flow rate of 0.35 ml/min. For reverse phase, Solvent A was water and B was methanol with 0.1% formic acids added in both. The elution gradient starts with 1% B to 95% B over 10 min with flow rate 0.3 ml/min. sample injection volume was 5ul. Pool quality control (QC) sample was run after every five samples to monitor signal variation and drift in mass error. Data matrices have been provided in Supplementary Table 1 and representative spectra of one of the samples acquired in positive and negative modes of HILIC and RP have been provided in Supplementary Fig. 3.
Data processing
All LC/MS acquired data has been processed using the Progenesis QI for metabolomics (Water Corporation) software using the default setting. The untargeted workflow of Progenesis QI was used to perform retention time alignment, feature detection, deconvolution, and elemental composition prediction. Metascope plug of Progenesis QI has been used for the in-house library with accurate mass, fragmentation pattern and retention time for database search. We have also used online available spectral library for further confirmation of identification. Cut-off for retention time match was 0.5 min and spectral similarity was more than 30% fragmentation match in Progenesis QI. Peaks that had a coefficient of variation (CV) less than 30% in pool QC samples were kept for the further analysis of data. Additionally, manual verification of each detected feature has been done for the selection of right peaks.
Statistical analysis
For each of the stages of myogenic progression (Day 0, 2, and 4), three independent experiments with equal number of cells counts were performed. Processing of the raw data lead to identification of total 71 metabolites at all the three stages (Day 0, Day 2 and Day 4) of myogenic progression. All the further statistical and functional analysis including the PCA, heat map, molecular pathways identification, analysis of variance (ANOVA) was done based on the identified peaks intensity using the online freely available Metaboanalyst 4.0 software. For analysis of PCA, heatmaps, ANOVA data were put in matrix, with samples in rows and features in columns. Before final analysis data integrity check was performed and raw data were normalized by sum methods and scaling was done using the Pareto scaling.
Supplementary information
Acknowledgements
We are grateful to Dr. Satyajit Rath (Indian Institute of Science Education and Research, Pune) for discussions and comments on the design of the study. We are also extremely grateful to Drs. T Ramchandran (THSTI) and K. Desiraju (THSTI) for critical comments on the statistical analysis methods used in this study. This work was supported by grants awarded to Suchitra Gopinath from the Department of Biotechnology, India (BT/09/IYBA/2015), THSTI collaborative intramural grants Suchitra Gopinath and Niraj Kumar, and a collaborative Grant (BT/PR29599/PFN/20/1393/2018) awarded to Suchitra Gopinath from the Department of Biotechnology, India.
Author contributions
S.D.G. and N.K. conceived, designed, and coordinated the study. S.D.G. generated the study material. A.K. performed most of the experiments. S.K. helped in peak identification. A.K. and Y.K. performed data analysis. JKS performed immunocytochemistry. S.D.G., Y.K., and A.K. wrote the manuscript. All authors approved final submission of the manuscript.
Data availability
Data deposited to MetaboLights with ID-MTBLS1818 (https://www.ebi.ac.uk/metabolights/MTBLS1818).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yashwant Kumar, Email: y.kumar@thsti.res.in.
Niraj Kumar, Email: nkumar@thsti.res.in.
Suchitra Devi Gopinath, Email: sgopinath@thsti.res.in.
Supplementary information
is available for this paper at 10.1038/s41598-020-68796-4.
References
- 1.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sambasivan R, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 4.Schultz E. Satellite cell proliferative compartments in growing skeletal muscles. Dev. Biol. 1996;175:84–94. doi: 10.1006/dbio.1996.0097. [DOI] [PubMed] [Google Scholar]
- 5.Biressi S, Gopinath SD. The quasi-parallel lives of satellite cells and atrophying muscle. Front. Aging Neurosci. 2015;7:140. doi: 10.3389/fnagi.2015.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015;1:a000588. doi: 10.1101/mcs.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda R, Uchitomi R, Oyabu M, Hatazawa Y, Kamei Y. Metabolomic analysis of C2C12 myoblasts induced by the transcription factor FOXO1. FEBS Lett. 2019;593:1303–1312. doi: 10.1002/1873-3468.13445. [DOI] [PubMed] [Google Scholar]
- 8.Meinke P, Limmer S, Hintze S, Schoser B. Assessing metabolic profiles in human myoblasts from patients with late-onset Pompe disease. Ann. Transl. Med. 2019;7:277. doi: 10.21037/atm.2019.04.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saoi M, et al. Characterization of the human skeletal muscle metabolome for elucidating the mechanisms of bicarbonate ingestion on strenuous interval exercise. Anal. Chem. 2019;91:4709–4718. doi: 10.1021/acs.analchem.9b00149. [DOI] [PubMed] [Google Scholar]
- 10.Sato S, Parr EB, Devlin BL, Hawley JA, Sassone-Corsi P. Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Mol. Metab. 2018;16:1–11. doi: 10.1016/j.molmet.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchitomi R, et al. Metabolomic analysis of skeletal muscle in aged mice. Sci. Rep. 2019;9:10425. doi: 10.1038/s41598-019-46929-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph J, Cho DS, Doles JD. Metabolomic analyses reveal extensive progenitor cell deficiencies in a mouse model of duchenne muscular dystrophy. Metabolites. 2018;8:61. doi: 10.3390/metabo8040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pala F, et al. Distinct metabolic states govern skeletal muscle stem cell fates during prenatal and postnatal myogenesis. J. Cell Sci. 2018 doi: 10.1242/jcs.212977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryall JG. Simultaneous measurement of mitochondrial and glycolytic activity in quiescent muscle stem cells. Methods Mol. Biol. 2017;1556:245–253. doi: 10.1007/978-1-4939-6771-1_13. [DOI] [PubMed] [Google Scholar]
- 15.Seldin MM, et al. A strategy for discovery of endocrine interactions with application to whole-body metabolism. Cell Metab. 2018;27:1138–1155. doi: 10.1016/j.cmet.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015;14:58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 17.Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell. Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 18.Sakuma K, Aoi W, Yamaguchi A. The intriguing regulators of muscle mass in sarcopenia and muscular dystrophy. Front. Aging Neurosci. 2014;6:230. doi: 10.3389/fnagi.2014.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson DL, Lehninger AL, Cox MM. Lehninger Principles of Biochemistry. 4. New York City: W.H. Freeman and Co.; 2005. [Google Scholar]
- 20.Thorrez L, DiSano K, Shansky J, Vandenburgh H. Engineering of human skeletal muscle with an autologous deposited extracellular matrix. Front. Physiol. 2018;9:1076. doi: 10.3389/fphys.2018.01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith CM, Narrow CM, Kendrick ZV, Steffen C. The effect of pantothenate deficiency in mice on their metabolic response to fast and exercise. Metabolism. 1987;36:115–121. doi: 10.1016/0026-0495(87)90003-5. [DOI] [PubMed] [Google Scholar]
- 22.Zano SP, Pate C, Frank M, Rock CO, Jackowski S. Correction of a genetic deficiency in pantothenate kinase 1 using phosphopantothenate replacement therapy. Mol. Genet. Metab. 2015;116:281–288. doi: 10.1016/j.ymgme.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YM, Rock CO, Jackowski S. Feedback regulation of murine pantothenate kinase 3 by coenzyme A and coenzyme A thioesters. J. Biol. Chem. 2005;280:32594–32601. doi: 10.1074/jbc.M506275200. [DOI] [PubMed] [Google Scholar]
- 24.Bremer J, Wojtczak AB. Factors controlling the rate of fatty acid -oxidation in rat liver mitochondria. Biochim. Biophys. Acta. 1972;280:515–530. doi: 10.1016/0005-2760(72)90131-2. [DOI] [PubMed] [Google Scholar]
- 25.Cooper RH, Randle PJ, Denton RM. Stimulation of phosphorylation and inactivation of pyruvate dehydrogenase by physiological inhibitors of the pyruvate dehydrogenase reaction. Nature. 1975;257:808–809. doi: 10.1038/257808a0. [DOI] [PubMed] [Google Scholar]
- 26.Garland PB, Yates DW, Haddock BA. Spectrophotometric studies of acyl-coenzyme A synthetases of rat liver mitochondria. Biochem. J. 1970;119:553–564. doi: 10.1042/bj1190553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oram JF, Wenger JI, Neely JR. Regulation of long chain fatty acid activation in heart muscle. J. Biol. Chem. 1975;250:73–78. [PubMed] [Google Scholar]
- 28.Li LO, et al. Compartmentalized acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes. 2015;64:23–35. doi: 10.2337/db13-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corbin DR, et al. Excess coenzyme A reduces skeletal muscle performance and strength in mice overexpressing human PANK2. Mol. Genet. Metab. 2017;120:350–362. doi: 10.1016/j.ymgme.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crozier PG, Cordain L, Sampson DA. Exercise-induced changes in plasma vitamin B-6 concentrations do not vary with exercise intensity. Am. J. Clin. Nutr. 1994;60:552–558. doi: 10.1093/ajcn/60.4.552. [DOI] [PubMed] [Google Scholar]
- 31.Suidasari S, et al. Carnosine content in skeletal muscle is dependent on vitamin B6 status in rats. Front. Nutr. 2015;2:39. doi: 10.3389/fnut.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajasekaran NS, Shelar SB, Jones DP, Hoidal JR. Reductive stress impairs myogenic differentiation. Redox Biol. 2020 doi: 10.1016/j.redox.2020.101492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Grant R. Changes in kynurenine pathway metabolism in the brain, liver and kidney of aged female Wistar rats. FEBS J. 2011;278:4425–4434. doi: 10.1111/j.1742-4658.2011.08366.x. [DOI] [PubMed] [Google Scholar]
- 34.Caballero B, Gleason RE, Wurtman RJ. Plasma amino acid concentrations in healthy elderly men and women. Am. J. Clin. Nutr. 1991;53:1249–1252. doi: 10.1093/ajcn/53.5.1249. [DOI] [PubMed] [Google Scholar]
- 35.Dukes A, et al. The aromatic amino acid tryptophan stimulates skeletal muscle IGF1/p70s6k/mTor signaling in vivo and the expression of myogenic genes in vitro. Nutrition. 2015;31:1018–1024. doi: 10.1016/j.nut.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhary E, Sharma R, Kumar Y, Agarwal N. Conditional silencing by CRISPRi reveals the role of DNA gyrase in formation of drug-tolerant persister population in Mycobacterium tuberculosis. Front. Cell Infect. Microbiol. 2019;9:70. doi: 10.3389/fcimb.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naz S, et al. Development of a liquid chromatography-high resolution mass spectrometry metabolomics method with high specificity for metabolite identification using all ion fragmentation acquisition. Anal. Chem. 2017;89:7933–7942. doi: 10.1021/acs.analchem.7b00925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposited to MetaboLights with ID-MTBLS1818 (https://www.ebi.ac.uk/metabolights/MTBLS1818).