Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative virus of the coronavirus disease 2019 (COVID-19) pandemic. To establish a safe and convenient assay system for studying entry inhibitors and neutralizing antibodies against SARS-CoV-2, we constructed a codon-optimized, full-length C-terminal mutant spike (S) gene of SARS-CoV-2. We generated a luciferase (Luc)-expressing pseudovirus containing the wild-type or mutant S protein of SARS-CoV-2 in the envelope-defective HIV-1 backbone. The key parameters for this pseudovirus-based assay, including the S mutants and virus incubation time, were optimized. This pseudovirus contains a Luc reporter gene that enabled us to easily quantify virus entry into angiotensin-converting enzyme 2 (ACE2)-expressing 293T cells. Cathepsin (Cat)B/L inhibitor E−64d could significantly block SARS-CoV-2 pseudovirus infection in 293T-ACE2 cells. Furthermore, the SARS-CoV-2 spike pseudotyped virus could be neutralized by sera from convalescent COVID-19 patients or recombinant ACE2 with the fused Fc region of human IgG1. Thus, we developed a pseudovirus-based assay for SARS-CoV-2, which will be valuable for evaluating viral entry inhibitors and neutralizing antibodies against this highly pathogenic virus.
Keywords: Antiviral therapeutics, Coronavirus, Neutralizing antibodies, Pseudovirus, SARS-CoV-2, Spike protein
Introduction
Coronaviruses are membrane-enveloped, non-segmented positive-strand RNA viruses that are broadly distributed among humans, other mammals, and birds, causing acute and persistent infections.1 Three of these viruses can cause severe respiratory syndromes in humans; these include severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2, and Middle East respiratory syndrome coronavirus (MERS-CoV).2 SARS-CoV-2, closely related to SARS-CoV, is a novel coronavirus reported in 2019 that caused the recent outbreak of coronavirus disease-2019 (COVID-19). By May 25, 2020, the World Health Organization (WHO) reported that 5.2 million people worldwide had been infected with SARS-CoV-2 and 341,155 individuals died of COVID-19. Thus, there is an urgent need to develop effective viral inhibitors and antibody-based therapeutics against this highly pathogenic virus.
Similar to SARS-CoV, the cellular receptor of SARS-CoV-2 is angiotensin-converting enzyme 2 (ACE2)3; however, the SARS-CoV-2 spike (S) protein has a 10- to 20-fold higher affinity for ACE2 than the corresponding S protein of SARS-CoV.4 The S protein of SARS-CoV-2, which consists of two subunits S1 and S2, plays an important role in viral entry by mediating receptor binding and membrane fusion.5 The cellular transmembrane protease serine 2 (TMPRSS2) and cathepsin (Cat)B/L inhibitors are critical for priming the SARS-CoV-2 S protein to enhance ACE2-mediated viral entry.6 Thus, the S protein is a major target for vaccines, neutralizing antibodies, and SARS-CoV-2 entry inhibitors.6,7
In this study, we constructed a luciferase (Luc)-expressing pseudovirus bearing the S protein of SARS-CoV-2, based on the HIV-1 backbone to avoid handling of infectious coronaviruses. We established an efficient and quantitative pseudovirus-based inhibition assay for the evaluation of SARS-CoV-2 cell entry mediated by the viral S protein. This pseudovirus-based inhibition assay system is safe, versatile, and useful for studying the effects of viral entry inhibitors and neutralizing antibodies against SARS-CoV-2.
Materials and methods
Plasmids
The full length of the S gene from the SARS-CoV-2 (previously 2019-nCoV) strain Wuhan-Hu-1 (GenBank: MN908947) was codon-optimized (sequence shown in Supplementary Table 1), synthesized, and cloned into the KpnI and XbaI sites of the pCMV3 vector (pCMV3-SARS-CoV-2-S-FL, pS-FL for short) by Sino Biological Inc. (Beijing, China). The translated amino acid sequence was identical to QHD43416. The primers 5′-TGCTGAAAGGAGTGGCACTGGCCTACACCTGAATCT-3′ and 5′-AGATTCAGGTGTAGGCCAGTGCCACTCCTTTCAGCA-3′ were used to generate the plasmid pCMV3-SARS-CoV-2-S-Mut (pS-Mut) encoding a mutant S protein containing mutations K1269A and H1271A in the cytoplasmic tail, which destroyed the “KxHxx” motif (the last five amino acids were changed to ALAYT).The primers 5′-CGGGGTACCATGTTTGTGTTCCTGGTGCTGC-3′ and 5′-TGCTCTAGATCAACAACAGGAGCCACAGGAACAAC-3′ were used to generate the plasmid pCMV3-SARS-CoV-2-S-C19del (pS-C19del) encoding a mutant S protein with a C-terminal 19 amino acid deletion. The constructed recombinant SARS-CoV-2 plasmids containing the full-length (pS-FL) and mutant S variants (pS-Mut and pS-C19del, respectively) were confirmed by DNA sequencing. The expression plasmid for human ACE2 was obtained from Genecopoeia (EX-U1285-M02-B).
Cell lines
HEK293T cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA).Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM; Hyclone, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA), 100 mg/mL of streptomycin, and 100 unit/mL of penicillin at 37 °C in 5% CO2. HEK293T cells transfected with human ACE2 (293T-ACE2) were cultured under the same conditions with the addition of G418 (0.5 mg/mL) to the medium.
Inhibitors, antibodies, and sera
Camostat mesylate (Tokyo Chemical Industry, Tokyo, Japan) and aloxistatin (E−64d; MedChemExpress, Monmouth Junction, NJ, USA) were dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 50 mM. The anti-RBD monoclonal antibody against the SARS-CoV-2 S protein was kindly provided by Prof. Aishun Jin (College of Basic Medial, Chongqing Medical University). ACE2-Ig (Cat: 10108-H02H) was purchased from Sino Biological Inc.
Three convalescent COVID-19 patient sera were generously provided by Dr. Quan-Xin Long from Chongqing Medical University. All sera were tested positive using MCLIA kits supplied by Bioscience Co. (Tianjin, China).8 Patient sera were incubated at 56 °C for 30 min to inactivate the complement prior to experiments.
Analysis of SARS-CoV-2 S protein expression and subcellular localization
To analyze S protein expression and localization in cells, pS-FL, pS-Mut, and pS-C19del were transfected into HEK293T cells. Total protein was extracted from cells using RIPA Lysis Buffer (CWbiotech, Beijing, China) containing 1 mM phenyl methyl sulfonyl fluoride (PMSF; Beyotime, Shanghai, China). Cell lysates were denatured in 6 × sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, separated by 10% SDS-PAGE, and electro-transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The immunoblots were probed with the indicated antibodies (anti- SARS-CoV-2 S and actin). Protein bands were visualized using SuperSignal™ West Pico Chemiluminescent Substrate kits (Bio-Rad, Hercules, CA, USA) and quantified by densitometry using ImageJ software (National Center for Biotechnology Information [NCBI], Bethesda, MD, USA).
Confocal microscopy analysis was performed to study the localization of the S protein. Briefly, SARS-CoV-2 S proteins were detected using the mouse anti-SARS-CoV-2 S RBD monoclonal antibody. Rabbit anti-Calreticulin (12238#; Cell Signaling Technology, Danvers, MA, USA) as an endoplasmic reticulum marker. Specific signals were visualized using the Alexa Fluor 488 secondary antibody (Invitrogen, Carlsbad, CA, USA). For nuclear staining, cells were treated with 1 μg/mL 4′, 6-diamidino-2-phenylindole (DAPI, 10236276001; Roche Diagnostics GmbH, Mannheim, Germany) for 3 min. Stained sections were analyzed using a laser scanning confocal microscope (Leica TCS SP8, Solms, Germany).
Production and titration of SARS-CoV-2 S pseudoviruses
The HIV-1 NL4-3 ΔEnv Vpr Luc reporter vector (pNL4-3.Luc.R-E−) constructed by N. Landau9 was provided by Cheguo Cai from Wuhan University. To generate the SARS-CoV-2 S pseudotyped HIV-1 single-round luciferase virus, 5 × 106 HEK293T cells were co-transfected with 6 μg pNL4-3.Luc.R-E− and 6 μg recombinant SARS-CoV-2S plasmids (pS-FL, pS-Mut, or pS-C19del) using the Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer's instructions. The cells were transferred to fresh DMEM 12 h later. The supernatant containing SARS-CoV-2 pseudoviruses were harvested 48–72 h after transfection and filtered through a 0.45 μm filter. The vesicular stomatitis virus G (VSV-G)-expressing plasmid pMD2.G was co-transfected with the pNL4-3.Luc.R-E− plasmid to package the VSV-G pseudovirus.
Then, 293T-ACE2 cells (2 × 104 cells/well) were seeded into 96-well plates and infected with 50 μL of the pseudotyped viruses supplemented with polybrene (5 μg/mL). After incubation for 12 h, the pseudovirus-containing supernatant was removed and replaced with fresh DMEM containing 10% FBS. After 48–72 h post-infection, the 293T-ACE2 cells were lysed with 30 μL lysis buffer (Promega, Madison, WI, USA) to measure the pseudoviral transduction. Relative luminescence units (RLU) of Luc activity was detected using the Luciferase Assay Kit (Promega). All experiments were performed at least three times and expressed as means ± standard deviations (SDs).
Neutralization and inhibition assays
The 293T-ACE2 cells (2 × 104 cells/well) were seeded in 96-well plates. For the neutralization assay, 50 μL pseudoviruses (~2×104 RLU) were incubated with serial dilutions of sera samples (dilutions of 1:20, 40, 80, 160, 320, 640, 1280, 2560, 5120, 10240, and 20480) from patients and human normal serum as a negative control for 1 h at 37 °C, then added to the 96-well 293T-ACE2 cells. For the inhibition assay, the cells were pretreated with the protease inhibitors (camostat mesylate and E−64d) 2 h before infection. After 12 h of infection, fresh culture was added to each well. Luciferase activity was measured 72 h after infection and the percent neutralization was calculated using GraphPad Prism 6.0 software (GraphPad Software, San Diego, CA, USA).
Statistical analysis
All data are presented as means ± standard deviations (SDs). GraphPad Prism 6.0 software (GraphPad Software) was used to perform all statistical analyses and prepare graphs. Statistical significance was determined using a one-way ANOVA for multiple comparisons. Student's t-tests were applied to compare the two groups. Differences with p-values < 0.05 were deemed statistically significant.
Results
Expression and subcellular localization of SARS-CoV-2 S protein
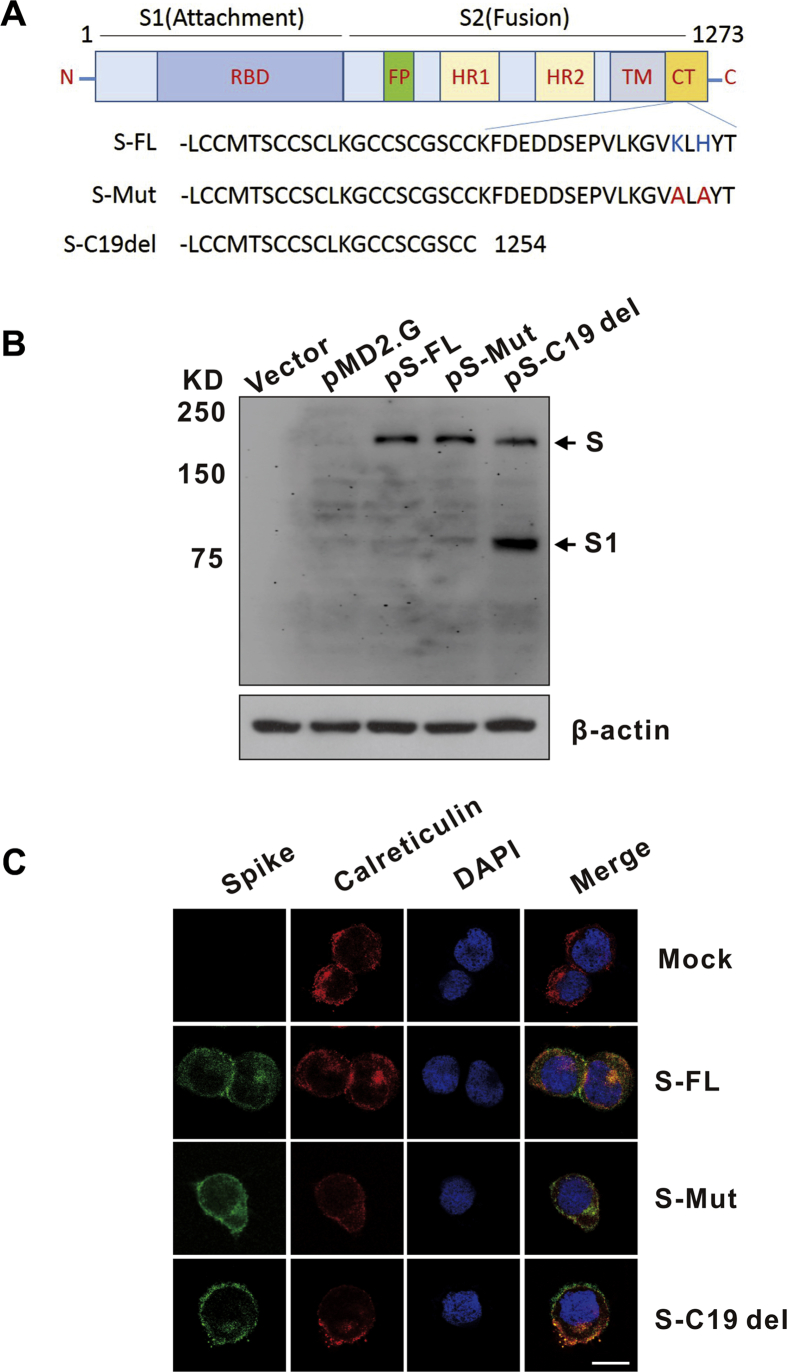
The S protein of SARS-CoV-2, containing 1273 amino acids, forms a trimeric spike on the virion surface that plays essential roles in viral entry. The cytoplasmic tail (CT) domain of several CoV S proteins contains intracellular retention signals, which are important for S incorporation into virions.10 To evaluate the expression and subcellular localization of SARS-CoV-2 S protein in the human cell line HEK293T, we synthesized and cloned a codon-optimized S gene (pS-FL). We also generated pS-Mut plasmid bearing two mutations that destroy the endoplasmic reticulum (ER) retrieval signal (“KxHxx” motif) in the CT domain of the S protein and pS-C19del plasmid lacking the C-terminal 19 amino acids (Fig. 1A). Immunoblot analysis of lysates from pS-FL-, pS-Mut-, and pS-C19del-transfected HEK293T cells revealed two major protein bands (unprocessed S and cleaved S1 subunit) that reacted with the monoclonal antibody targeting the RBD on the SARS-CoV-2 spike (Fig. 1B). Interestingly, the pS-C19del-transfected cells showed the strongest S1 signal. It is known that S proteins from different coronaviruses remain largely intracellular when they are expressed exogenously.11 Our confocal microscopy showed that S-FL mainly localized to the ER while mutagenesis of the C-terminal intracellular retention signal (S-Mut and S–C19del) promoted the transport of S to the cell surface (Fig. 1C).These data suggest that the ER retrieval signal (“KxHxx” motif) in the CT domain of SARS-CoV-2 S contributes to its subcellular localization.
Figure 1.
Detection of SARS-CoV-2 spike (S) protein expression and localization. (A) Schematic illustration of the SARS-CoV-2 full-length spike (S-FL) and mutant S variants. The RBD (receptor binding domain) is in subunit S1; the FP (fusion peptide), HR1 (heptad repeat 1), HR2 (heptad repeat 2), TM (transmembrane domain), and CT (cytoplasmic tail) are in subunit S2. The endoplasmic reticulum retrieval signals (“KxHxx” motif) in the CT domain of S-FL were destroyed in S-Mut protein. The C-terminal 19 amino acids were lacking in S-C19del. (B) Detection of SARS-CoV-2 S expression in HKE293T cells by Western blot using the anti-RBD monoclonal antibody. Cells were transfected with pS-FL, pS-Mut, and pS-C19del plasmids or with an empty vector. (C) Detection of SARS-CoV-2 S subcellular localization in HKE293T cells by confocal microscopy. Cells were grown on glass coverslips for 24 h preceding transfection of plasmids encoding S protein variants. The cells were harvested and labeled with the corresponding antibodies. Calreticulin, ER marker. Nuclei were counterstained with DAPI. Bar = 20 μm.
Production of SARS-CoV-2 S pseudotyped lentiviral particles
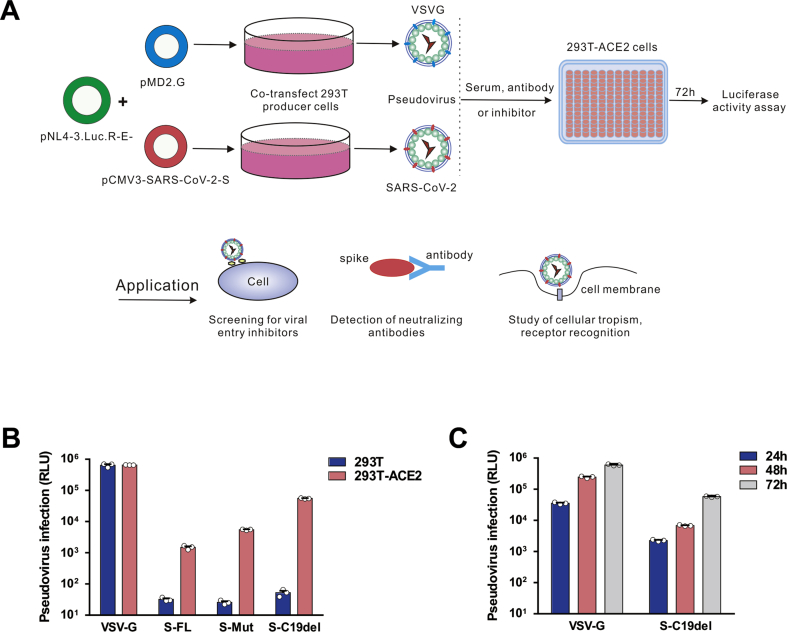
Due to the highly pathogenic nature of SARS-CoV-2, infectious SARS-CoV-2 must be handled in a biosafety level 3 (BSL-3) facility. We generated a SARS-CoV-2 pseudovirus to detect anti-SARS-CoV-2 entry inhibitors and neutralizing antibodies. The pseudoviruses were packaged and titrated using a method similar to SARS-CoV and MERS-CoV pseudoviruses.12,13 For this lentivirus-based pseudovirus system, the backbone was provided by HIV-1 (Fig. 2A). A firefly Luc gene was inserted into the pNL4-3 nef gene leading to frame shifts in env and vpr.9 Virus titers were determined by measuring RLU.
Figure 2.
Detection of SARS-CoV-2 S pseudotyped virus infectivity. (A) Schematic representation of the pseudovirus production and neutralization assay and the applications of the pseudovirus. (B) HEK293T and 293T-ACE2 cells were infected with lentiviruses pseudotyped with vesicular stomatitis virus G (VSV-G) and SARS-CoV-2 S protein variants. The y-axis shows the relative luminescence units (RLU) detected at 48 h post-pseudovirus inoculation. (C) Optimization of the incubation time for pseudovirus luciferase assay. Luciferase activities were measured 24–72 h post-virus infection. For this purpose, 72 h was chosen as the optimized incubation time. The data are presented as the means ± standard deviations (SDs) of three independent biological replicates.
pNL4-3.Luc.R-E− was co-transfected with pS-FL, pS-Mut, and pS-C19del respectively to package the SARS-CoV-2 S pseudotyped single-round Luc virus in HEK293T cells. We established HEK293T cells expressing human ACE2 (293T-ACE2) to test the correlation between ACE2 expression and SARS-CoV-2 pseudovirus susceptibility. VSV-G pseudovirus was used as a positive control. Virus infectivity was determined by a Luc assay expressed in RLU. As shown in Fig. 2B, both HEK293T and 293T-ACE2 cells could be effectively transduced by VSV-G pseudoviruses. However, when transduced by SARS-CoV-2 pseudoviruses, 293T-ACE2 cells showed an approximately 1200-fold increase in Luc activity compared to HEK293T cells, suggesting that the entry of SARS-CoV-2 pseudovirus is highly dependent on its cellular receptor ACE2 expression.
Next, we compared the virus titers between pseudoviruses with the full-length S protein and two S-protein variants. For the S-FL pseudotyped virus, the average infectivity was measured at approximately 1.6 × 103 RLU. The highest viral titer was observed in S–C19del pseudotyped virus (8 × 104 RLU), suggesting that the plasma membrane localization of this mutant S facilitates lentivirus packaging. Thus, we used this S–C19del pseudotyped virus in subsequent studies. We next determined the optimal time for Luc detection after SARS-CoV-2 pseudovirus infection. The highest transduction efficiency (approximately 1 × 105 RLU) was observed 72 h post-infection with SARS-CoV-2 pseudoviruses (Fig. 2C).
Evaluating viral entry inhibitors using a SARS-CoV-2 pseudovirus inhibition assay
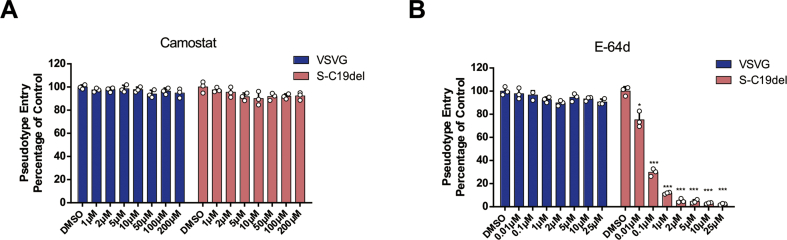
We next established a SARS-CoV-2 pseudovirus inhibition assay to evaluate therapeutics targeting the virus entry. Two clinically proven protease inhibitors, camostat mesylate and E−64d, which block host TMPRSS2 and CatB/L, respectively, were identified as potent inhibitors of S-mediated SARS-CoV-2 entry.6 We detected the inhibitory activities of these protease inhibitors on SARS-CoV-2 pseudovirus entry in 293T-ACE2 cells. As shown in Fig. 3A and B, camostat mesylate did not inhibit SARS-CoV-2 pseudovirus infection in 293T-ACE2 cells, but E−64d blocked >90% of SARS-CoV-2 pseudovirus entry at concentrations of 2 μM. As expected, neither of the compounds inhibited VSV-G pseudovirus infection, suggesting that SARS-CoV-2-S-driven entry into TMPRSS2 deficient 293T cells was CatB/L dependent. Together, these data demonstrate that this pseudovirus-based inhibition assay could be used as an effective and convenient method to screen SARS-CoV-2 entry inhibitors.
Figure 3.
Detection of entry inhibitors against SARS-CoV-2 pseudovirus infection. (A–B) Inhibitors camostat mesylate and E-64d blocked SARS-CoV-2 pseudovirus entry. 293T-ACE2 cells were pre-incubated with camostat mesylate (A) or E-64d (B), and subsequently inoculated with pseudovirions. The VSV-G pseudovirus was used as the control. RLU were detected at 72 h post-pseudovirus inoculation. Cell viability was examined by the methyl thiazolyl tetrazolium (MTT) assay. The data are presented as the means ± SDs of three independent biological replicates.
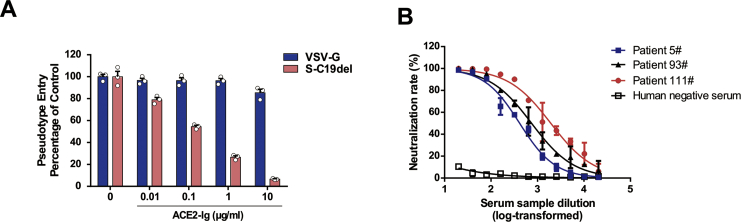
Detection neutralization effect of ACE2-Ig and convalescent COVID-19 patient sera
Next, we performed neutralization assays in 96-well plates. We detected the inhibitory ability of ACE2-Ig, a fusion protein consisting of the extracellular domain (Met 1-Ser 740) of human ACE2 linked to the Fc region of human IgG1 at the C-terminus.14 Our neutralization assay showed that SARS-CoV-2 was potently neutralized by ACE2-Ig and the IC50 (the concentration causing 50% inhibition of SARS-CoV-2 pseudovirus infection) was 0.13 μg/mL (Fig. 4A). We then evaluated the activity of serum samples from convalescent patients with COVID-19 against this SARS-CoV-2 pseudovirus. Three tested representative convalescent patient sera demonstrated neutralizing activity against SARS-CoV-2 pseudovirus infection while the control sera from healthy individuals showed no neutralizing activity. The half-maximal neutralizing titers (NT50) of the three serum samples were 412, 745, and 1930, respectively.
Figure 4.
Detection of neutralizing antibodies in sera against SARS-CoV-2 pseudovirus infection. (A) Inhibition of SARS-CoV-2 pseudovirus entry by ACE2-Ig. S-C19del or VSV-G pseudovirions were preincubated with ACE2-Ig and added to 293T-ACE2 cells. (B) Sera from convalescent COVID-19 patients neutralized the SARS-CoV-2 pseudovirus. Serum sample from healthy individual was tested as a negative control. RLU were detected at 72 h post-pseudovirus inoculation. The data are presented as the mean percentages of inhibition ± SDs of duplicate wells.
Collectively, our data indicate that a safe and convenient assay has been established to test the neutralizing activity of sera and entry inhibitors against SARS-CoV-2.
Discussion
Lentiviral vectors can be pseudotyped with various heterologous viral glycoproteins that modulate cellular tropism and cell-entry properties.15 Pseudovirus-based assays have been widely used for the study of cellular tropism, receptor recognition, and viral inhibitors as well as evaluation of neutralizing antibodies. The present study generated a pseudotyped SARS-CoV-2 based on the viral S protein using a lentiviral HIV-1 system, which incorporated a Luc reporter gene for the easy quantification of coronavirus S-mediated entry. We used a codon-optimized and CT-modified S protein to facilitate more efficient packaging into lentiviruses. This SARS-CoV-2 pseudovirus could effectively enter HEK293T cells transfected with the ACE2 receptor, providing a safe and convenient method to study SARS-CoV-2 entry without the requirement of BSL-3 laboratories.
Using this pseudotype system, we demonstrated the potential of the protease inhibitor E−64d and ACE2-Ig to block SARS-CoV-2 pseudovirus infection. We also showed that the entry of SARS-CoV-2 pseudovirus into 293T-ACE2 cells could be blocked by convalescent sera from COVID-19 patients. This convenient pseudovirus-based inhibition assay will simplify the detection of neutralizing antibodies against SARS-CoV-2. However, one limitation of this assay is that the pseudovirus only recapitulates viral entry events; therefore, additional assays are required to analyze other processes during the viral life cycle.
It is known that lentiviral particles bud at the cytoplasmic membrane of host cells.16 However, coronaviruses assemble and bud at membranes of the intermediate compartment between the ER and Golgi complex.17 In the lentivirus-based pseudovirus system, heterologous viral envelope glycoproteins, such as the coronavirus S protein, need to be transported to the cytoplasmic membrane for incorporation into pseudotyped progeny virions during packaging. Our results showed that the full-length S protein of SARS-CoV-2 was present in the ER and plasma membrane when expressed in HEK293T cells, which was in line with previous studies on other S glycoproteins of coronaviruses.11 Moreover, mutagenesis of the “KxHxx” motif on the CT domain or deletion of 19 amino acids on the CT promoted S transport to the plasma membrane, where it might be cleaved by the membrane-bound protease more efficiently. Therefore, the plasma membrane localization of this mutant S significantly improved the packaging efficiency of SARS-CoV-2 pseudotyped virus. Very recently, another group also generated SARS-CoV-2 pseudovirions with C-terminal-deleted S proteins using a similar lentiviral pseudotype system.18 However, Nie et al replaced the original N-terminal signal peptide of the SARS-CoV S protein with a tissue plasminogen activator signal (MDAMKRGLCCVLLLCGAVFVSA) to improve the packaging efficiency of SARS-CoV pseudovirions.12 These studies indicate that the subcellular localization of S is important for pseudotyped lentivirus packaging and so, modifying the CT or N-terminal signal peptide on the S protein of coronaviruses can increase the titers of S-pseudotyped particles.
In addition to the lentiviral HIV-1 based pseudotype system, there are several other pseudotype systems that have been developed to package highly pathogenic coronaviruses for studying S protein-mediated infection, including simian immunodeficiency virus (SIV) and murine leukemia virus (MLV) pseudotyped SARS-CoV,19 MLV pseudotyped MERS-CoV,20 and VSV pseudotyped SARS-CoV-2.21 The Luc reporter gene in the backbone enables easy and rapid quantification of the infectivity of viral pseudotyped particles in infected host cells through a simple Luc activity assay. Although each of these pseudotype systems have their advantages and disadvantages, future directions for the development and application of these pseudoviruses include: 1) establishment of high-titer viral vector production platforms, 2) development of self-inactivating vectors, and 3) analysis of individual viral entry and fusion events using single-particle tracking techniques.20
In summary, we generated a SARS-CoV-2 S pseudotyped lentivirus that can be used as a safe and convenient system for studying infection with the highly important emerging pathogen SARS-CoV-2. This pseudovirus assay will be useful for the screening of viral entry inhibitors, detection of neutralizing antibodies, development of receptor binding and entry assays, and characterization of the S protein function of SARS-CoV-2.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We would like to thank Prof. Cheguo Cai (Wuhan University, Wuhan, China) for providing the pNL4-3.Luc.R-E- plasmid. We also thank Dr. Hongbing Jiang (Washington University in St. Louis, St. Louis, MO, USA) for the suggestions. We would like to thank Editage (www.editage.cn) for English language editing. This work was supported by the Emergency Project from the Science & Technology Commission of Chongqing (cstc2020jscx-fyzx0053), a Major National Science & Technology Program grant (2017ZX10202203) from the Science & Technology Commission of China, the Leading Talent Program of CQ CSTC (CSTCCXLJRC201719), the Scientific Research Innovation Project for Postgraduate in Chongqing (CYB19168) and the Emergency Project for Novel Coronavirus Pneumonia from the Chongqing Medical University (CQMUNCP0302).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.07.006.
Contributor Information
Ni Tang, Email: nitang@cqmu.edu.cn.
Kai Wang, Email: wangkai@cqmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Masters P.S. Coronaviruses: an overview of their replication and pathogenesis. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.-L., Wang X.-G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281-292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long Q.-X., Liu B.-Z., Deng H.-J. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 9.Connor R.I., Chen B.K., Choe S., Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 10.Ujike M., Taguchi F. Ncorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lontok E., Corse E., Machamer C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nie Y., Wang P., Shi X. Highly infectious SARS-CoV pseudotyped virus reveals the cell tropism and its correlation with receptor expression. Biochem Biophys Res Commun. 2004;321:994–1000. doi: 10.1016/j.bbrc.2004.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao G., Du L., Ma C. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J. 2013;10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei C., Qian K., Li T. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat Commun. 2020;11:2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandrin V., Boson B., Salmon P. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 16.Milone M.C., O'Doherty U. Clinical use of lentiviral vectors. Leukemia. 2018;32:1529–1541. doi: 10.1038/s41375-018-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen V.P., Hogue B.G. Protein interactions during coronavirus assembly. J Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou X., Liu Y., Lei X. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore M.J., Dorfman T., Li W. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millet J.K., Tang T., Nathan L. Production of pseudotyped particles to study highly pathogenic coronaviruses in a biosafety level 2 setting. J Vis Exp JoVE. 2019;145 doi: 10.3791/59010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie J., Li Q., Wu J. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microb Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.