Highlights
-
•
We estimated the time to the most recent common ancestor (TMRCA) and evolutionary rate of SARS-CoV-2 to be 12 November 2019 (95% BCI: 11 October 2019 and 09 December 2019) and 9.90 × 10-4 substitutions per site per year (95% BCI: 6.29 × 10-4–1.35 × 10-3), respectively.
-
•
Our results emphasize the importance of using phylogenetic and phylodynamic analyses to provide insights into the roles of various interventions to limit the spread of SARS-CoV-2 in China and beyond.
-
•
Understanding epidemic dynamics of SARS-CoV-2 in real time is increasingly important for guiding prevention efforts.
Keywords: COVID-19, SARS-CoV-2, TMRCA, Evolutionary rate, Re, Lockdown
Abstract
To investigate the evolutionary and epidemiological dynamics of the current COVID-19 outbreak, a total of 112 genomes of SARS-CoV-2 strains sampled from China and 12 other countries with sampling dates between 24 December 2019 and 9 February 2020 were analyzed. We performed phylogenetic, split network, likelihood-mapping, model comparison, and phylodynamic analyses of the genomes. Based on Bayesian time-scaled phylogenetic analysis with the best-fitting combination models, we estimated the time to the most recent common ancestor (TMRCA) and evolutionary rate of SARS-CoV-2 to be 12 November 2019 (95 % BCI: 11 October 2019 and 09 December 2019) and 9.90 × 10−4 substitutions per site per year (95 % BCI: 6.29 × 10−4–1.35 × 10−3), respectively. Notably, the very low Re estimates of SARS-CoV-2 during the recent sampling period may be the result of the successful control of the pandemic in China due to extreme societal lockdown efforts. Our results emphasize the importance of using phylodynamic analyses to provide insights into the roles of various interventions to limit the spread of SARS-CoV-2 in China and beyond.
1. Introduction
On December 31, 2019, the World Health Organization (WHO) was informed of an outbreak of respiratory illnesses, including atypical pneumonia, which seriously threatened the global public health, detected around Wuhan Huanan Seafood Wholesale Market in the Chinese city of Wuhan, Hubei Province–the seventh-largest city in China with 11 million city residents. Of note, some of the first reported infected individuals from the wet market showed symptoms as early as December 8, 2019. Subsequently, the wet market was closed on January 1, 2020. The virus causing the outbreak of mysterious pneumonia cases was quickly determined to be a novel coronavirus, and this novel coronavirus was further named 2019-nCoV by WHO (Zhu et al., 2020; Zhou et al., 2020a; Wu et al., 2020a). On 23 January, 2020, Chinese authorities introduced unprecedented measures to contain the virus, stopping movement in and out of Wuhan and 15 other cities in Hubei Province. Consequently, WHO declared the 2019-nCoV outbreak to be a Public Health Emergency of International Concern (PHEIC) under International Health Regulations on 30 January 2020. The newly emerged coronavirus (SARS-CoV-2) is similar to betacoronaviruses detected in bats, reportedly sharing ∼96 % sequence identity to the BetaCoV/bat/Yunnan/RaTG13/2013 (EPI_ISL_402131) genome, a coronavirus isolated from an intermediate horseshoe bat (Rhinolophus affinis) in Yunnan Province, China (Zhou et al., 2020b). SARS-CoV-2, a member of the betacoronavirus genus of the Coronaviridae family, is a single, positive-strand RNA, approximately 30 kb in length, however, the mortality and transmissibility of SARS-CoV-2 are still unknown. On 11 February 2020, the International Committee on Taxonomy of Viruses officially renamed 2019-nCoV, which is responsible for the current outbreak of coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus belongs to the same family as the SARS-CoV-1 pathogen, which was responsible for >8 000 cases and 774 deaths in 37 countries during the 2002–2003 SARS outbreak (Drosten et al., 2003; Ksiazek et al., 2003; Zhong et al., 2003), and the MERS-CoV pathogen, which was responsible for 2 494 cases and 858 deaths in 27 countries during the 2012 MERS outbreak (Zaki et al., 2012; de Groot et al., 2013). Notably, the current COVID-19 outbreak is characterized by its significant dispersal into many major urban centers in China and beyond, further facilitating its continued spread from person to person (Chan et al., 2020; Li et al., 2020a), and has caused considerable morbidity and mortality in China and elsewhere. As of 14 July 2020, a total of 12 964 809 confirmed cases including 570 288 deaths in 216 countries, areas or territories, have been reported globally by WHO (https://www.who.int/emergencies/diseases/novel-coronavirus-2019), with USA, Brazil, India, and Russia especially hard hit. Although the number of confirmed cases of COVID-19 worldwide has excessed 12 million, it showed that countries had only discovered on average about 6 % of coronavirus infections and the true number of infected people worldwide may already have reached several tens of millions(https://medicalxpress.com/news/2020-04-covid-average-actual-infections-worldwide.html). Notably, there are many asymptomatic carriers remaining in humans and the nucleic acid of SARS-CoV-2 from some convalescent patients could be tested positive again which means that the virus cannot be eradicated and can replicate again. These factors will contribute the subsequent COVID-19 outbreaks, and many scientists believe that COVID-19 outbreaks will be recurrence.
Over the past three and half decades at least 30 new infectious agents affecting humans have emerged including SARS-CoV-2, and most of them are zoonotic. It was also reported that 61 % infectious organisms affecting humans are zoonotic diseases which can infect both human and animals (Nii-Trebi, 2017; McArthur, 2019). Previous studies have revealed that both SARS-CoV-1 and MERS-CoV originated in bats (Lau et al., 2010; Guan et al., 2003; Lau et al., 2005; Li et al., 2005), with SARS-CoV-1 jumping to humans from palm civets (Song et al., 2005; Chinese, 2004; Wang et al., 2005) and MERS-CoV jumping to humans from camels (Muller et al., 2014; Chu et al., 2014) following intermediate transmission from bats (Lau et al., 2010; Guan et al., 2003; Lau et al., 2005; Li et al., 2005). Research has also revealed that SARS-CoV-2 likely originated in bats, either directly or through an as-yet unidentified animal host (Zhou et al., 2020b). Initial cases have been linked to Wuhan Huanan Seafood Wholesale Market; however, the specific animal source is yet to be determined (Li et al., 2020a). The detection of SARS-CoV-2 in humans without knowing the animal source of infection has heightened concerns not only in China, but also internationally. Therefore, identifying the animal source of SARS-CoV-2 is still a top research priority for controlling the COVID-19 outbreak. The deadly pandemic has prompted a high-speed race to understand how the coronavirus is evolving and spreading. But doing so requires an unprecedented collaboration among scientists, across the globe, to decode the virus and its path.
Since the first whole-genome sequence (Wuhan-Hu-1; GenBank accession number MN908947, also named hCoV-19/Wuhan/Hu-1/2019 with accession ID EPI_ISL_402125 in GISAID) of the novel coronavirus, SARS-CoV-2, which was isolated from a 41-year old man who worked at Wuhan Huanan Seafood Wholesale Market, was shared online on 11 January, 2020, that first genome became the baseline for researchers to track the SARS-CoV-2 virus as it spreads around the world (Wu et al., 2020b). Since the start of the COVID-19 outbreak and the identification of the pandemic virus, laboratories around the world are generating viral genome sequence data with unprecedented speed, researchers have sequenced and shared some 66 000 viral genomes from around the world on 14 July, 2020. Such a vast amount of available genetic data presents a unique opportunity for researchers to trace the origin and spread of COVID-19 outbreaks in different countries and gain real-time insights into the pandemic, enabling real-time progress in the understanding of the new disease and in the research and development of candidate medical countermeasures. Sequence data are essential to design and evaluate diagnostic tests, to track and trace the ongoing outbreak, and to identify potential intervention options. Therefore, tracking the accumulating nucleotide mutations in SARS-CoV-2 virus’s genome as the pandemic progresses will help us better understand the pandemic and could help improve antiviral drug and vaccine effectiveness.
In the present study, we employed state-of-the-art methods to investigate the evolutionary and epidemiological dynamics of the virus based on 112 genomes of SARS-CoV-2 strains sampled from China and 12 other countries with sampling dates between 24 December 2019 and 9 February 2020. Rapid evolutionary and epidemiological analyses have become ever more important in response to the ongoing public health crisis in order to understand pathogenic origins, transmission dynamics, and subsequent host adaptations, and to investigate effective prevention measures for controlling pathogenic outbreaks. Our study should provide insights into the evolutionary and epidemiological histories of SARS-CoV-2 in China and elsewhere.
2. Materials and methods
2.1. Collation of SARS-CoV-2 genome-wide dataset
As of 19 February 2020, more than 100 genomes of human-obtained SARS-CoV-2 strains have been released on GISAID (http://gisaid.org/) (Elbe and Buckland-Merrett, 2017). No statistical methods were used to predetermine the number of genomes in the present study, we downloaded all available genomes of human-obtained SARS-CoV-2 strains. The dataset used in present study was also not randomized. Notably, due to the difficulty of sequencing samples with low virus concentrations, certain sequences were excluded from this study in order to avoid potential biases, e.g., sequences that were too short, re-sequences of the same sample, sequences with insufficient associated information, and sequences that showed evidence of artefacts due to the appearance of nucleotide variation. The final dataset (“dataset_112”) included 112 genomes of SARS-CoV-2 from Australia (n = 8), Belgium (n = 1), China (n = 53), Finland (n = 1), France (n = 10), Germany (n = 1), Japan (n = 7), Korea (n = 1), Nepal (n = 1), Singapore (n = 11), Thailand (n = 2), UK (n = 2), and USA (n = 14) with sampling dates between 24 December 2019 and 9 February 2020. Of the 53 genomes collected from China, three were from Chongqing, two were from Fujian Province, 16 were from Guangdong Province, 21 were from Hubei Province, one was from Jiangsu Province, one was from Jiangxi Province, one was from Sichuan Province, three were from Taiwan, one was from Yunnan Province, and four were from Zhejiang Province (Supplementary Table 1). We first aligned the collected dataset (“dataset_112”) using MAFFT v7.222 (Katoh and Standley, 2013) and subsequently edited the alignment manually using BioEdit v7.2.5 (Hall, 1999).
2.2. Recombination screening and maximum-likelihood analysis
Recombination may impact evolutionary estimates and is known to occur in coronaviruses (Graham and Baric, 2010). To assess recombination of our dataset (“dataset_112”), we employed the pairwise homoplasy index (PHI) to measure similarity between closely linked sites using SplitsTree v4.15.1 (Huson and Bryant, 2006) and the default recombination detection methods using the Recombination Detection Program (RDP) v4.100 (Martin et al., 2015). The best-fit nucleotide substitution model for “dataset_112” was identified according to the Bayesian information criterion (BIC) method with three (24 candidate models) or 11 (88 candidate models) substitution schemes in jModelTest v2.1.10 (Darriba et al., 2012). To evaluate the phylogenetic signals of “dataset_112”, we performed likelihood-mapping analysis (Schmidt and von Haeseler, 2007) using TREE-PUZZLE v5.3 (Schmidt et al., 2002), with 280 000 randomly chosen quartets for the dataset. Split network analysis was performed for “dataset_112” using Kishino-Yano-85 (Kimura, 1980) distance transformation with the NeighborNet method, which can be loosely thought of as a “hybrid” between the neighbor-joining (NJ) and split decomposition methods, implemented in TREE-PUZZLE v5.3 (Schmidt et al., 2002). Maximum-likelihood (ML) phylogenetic trees for the dataset were estimated using PhyML v3.1 (Guindon et al., 2010) under a Hasegawa-Kishino-Yano (HKY) (Kimura, 1980) nucleotide substitution model with a proportion of invariable sites, which was identified as the best fitting model for ML inference by jModelTest v2.1.10 (Darriba et al., 2012). Branch support was inferred using 1 000 bootstrap replicates (Felsenstein, 1985) and trees were midpoint rooted. Analysis of temporal molecular evolutionary signals for the dataset was conducted using TempEst v1.5 (Rambaut et al., 2016). In brief, regression analyses were used to determine the relationship between sampling dates and root-to-tip genetic divergence obtained from the ML phylogeny. The slope of the regression line provides an estimate of the rate of evolution in substitutions per site per year, and the intercept with the time-axis constitutes an estimate of the age of the root. We also estimated the evolutionary rate and time to the most recent common ancestor (TMRCA) for “dataset_112” using ML dating in the TreeTime package (Sagulenko et al., 2018).
2.3. Molecular clock phylogenetics
To estimate the Bayesian molecular clock phylogenies of SARS-CoV-2, Bayesian inference analyses were performed for “dataset_112” through a Markov chain Monte Carlo (MCMC) (Yang and Rannala, 1997) framework implemented in BEAST v1.8.4 (Drummond et al., 2012), with the BEAGLE v2.1.2 library program (Suchard and Rambaut, 2009) used for computational enhancement. For model selection, we tested five coalescent tree priors for our dataset: a constant-size population (Kingman, 1982), an exponential growth population with growth rate parameterization (Griffiths and Tavare, 1994), another exponential growth population with doubling time parameterization (Griffiths and Tavare, 1994), a Bayesian skyline tree prior (five groups, piecewise-constant model) (Drummond et al., 2005), and a Bayesian Skygrid tree prior (five population sizes across our 0.2 year interval, allowing a different population size to be estimated for 14.6 days (d)) (Gill et al., 2013). We kept the default option of a ‘Random starting tree’ to start the inference process. For each tree prior, we tested two clock models: a strict clock and an uncorrelated relaxed clock with log-normal distribution (UCLN) (Drummond et al., 2006). In each case, we set an uninformative continuous-time Markov chain (CTMC) reference prior (Ferreira and Suchard, 2008) on the molecular clock rate. For all 10 model combinations, we selected the best fitting model by marginal likelihood comparison using path-sampling (PS) and stepping-stone sampling (SS) estimations (Gelman and Meng, 2020; Baele et al., 2012; Baele et al., 2013). We sampled for 100 path steps with a chain length of one million, with power posteriors determined from evenly spaced quantiles of a beta (0.3, 1.0) distribution (Xie et al., 2011). All Bayesian analyses were run for 100 million MCMC steps with sampling parameters and trees every 10 000 generations. Convergence of MCMC chains was evaluated by calculating the effective sample sizes of parameters using Tracer v1.7.1 (Rambaut et al., 2018). All parameters had an effective sample size >200, indicative of sufficient sampling. We extracted clock rate and TMRCA estimates using Tracer v1.7.1 (Rambaut et al., 2018) and identified the maximum clade credibility (MCC) tree using TreeAnnotator v1.8.4 after discarding the first 10 % as burn-in, followed by tree visualization using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
2.4. Estimation of Re for SARS-CoV-2
We used the Bayesian birth-death skyline (BDSKY) model (Stadler et al., 2013) to estimate time-varying rates of epidemic spread, measured as changes in Re, denoted as Re (t) (Stadler et al., 2013), and implemented in BEAST v2.6.1 (Bouckaert et al., 2019). The nucleotide substitution process was modeled under HKY (Kimura, 1980) with a proportion of invariable sites, and evolutionary rates were estimated using an UCLN model (Drummond et al., 2006). We employed lognormal distribution with a mean of 0 and standard deviation of 1.0 for Re, which placed most weight below 5.18 (95 % quantile). The selected number of intervals for Re was 5 with equidistant intervals per step. We used a normal distribution with a mean of 48.7 and standard deviation of 15 (corresponding to a 95 % credible interval from 19.3–78.1) for the rate of becoming uninfectious (denoted as δ), which placed most weight below 73.4 (95 % quantile). These values are expressed as units per year and reflect the inverse of the time of infectiousness (mean = 7.49 d, 95 % credible interval: 4.67–18.91 d) according to previous study (Li et al., 2020a). We used a beta distribution with parameters α = 1.0 and β = 9 999 for the sampling proportion (denoted as s), corresponding to a minority of sampled cases (95 % credible interval: 2.53 × 10−6–3.69 × 10−4). The origin of the epidemic was estimated using a normal distribution with a mean of 0.25 and standard deviation of 0.05 units per year. Bayesian analysis was run for 500 million MCMC steps and sampled every 50 000 steps. Mixing of the MCMC chains was visually inspected using Tracer v1.7.1 (Rambaut et al., 2018), with an effective sample size of >200 for each parameter. We used the bdskytools package in R (https://github.com/laduplessis/bdskytools) to plot the BDSKY results.
3. Results
3.1. Demographic characteristics of SARS-CoV-2
“Dataset_112” included 112 genomes of SARS-CoV-2 strains sampled from Australia (n = 8), Belgium (n = 1), China (Chongqing, n = 3; Fujian Province, n = 2; Guangdong Province, n = 16; Hubei Province, n = 21; Jiangsu Province, n = 1; Jiangxi Province, n = 1; Sichuan Province, n = 1; Taiwan, n = 3; Yunnan Province, n = 1; and Zhejiang Province, n = 4), Finland (n = 1), France (n = 10), Germany (n = 1), Japan (n = 7), Korea (n = 1), Nepal (n = 1), Singapore (n = 11), Thailand (n = 2), UK (n = 2), and USA (n = 14) with sampling dates between 24 December 2019 and 9 February 2020 (Supplementary Table 1). The samples were primarily from China (53/112, 47.32 %) and Hubei Province (21/112, 18.75 %), the Chinese Province acknowledged as the original epicenter of the SARS-CoV-2 outbreak.
3.2. Tree-like signals and phylogenetic analyses
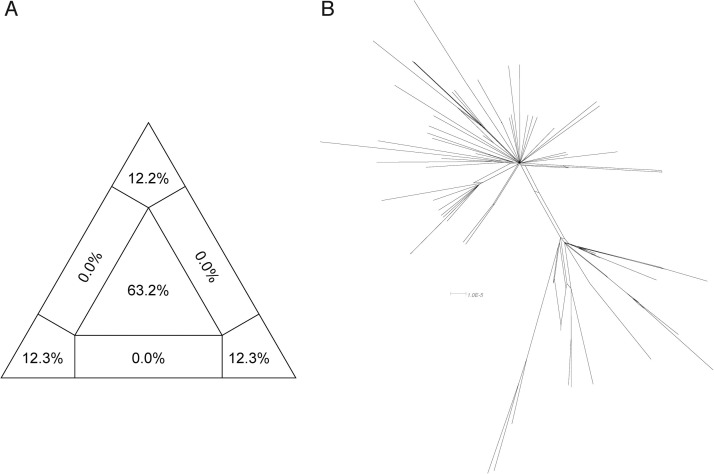
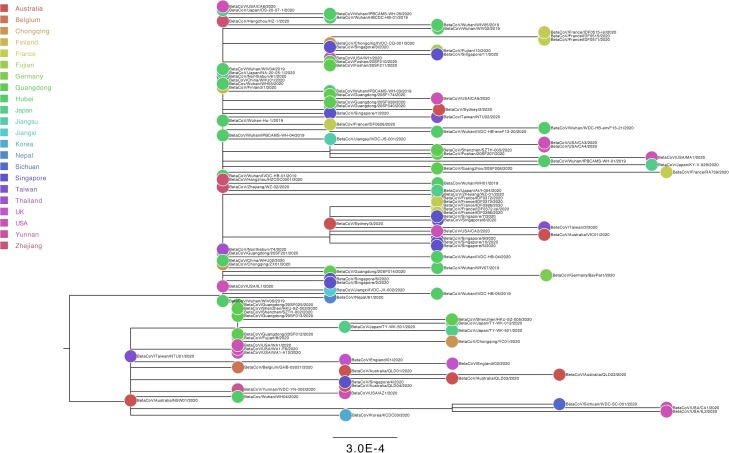
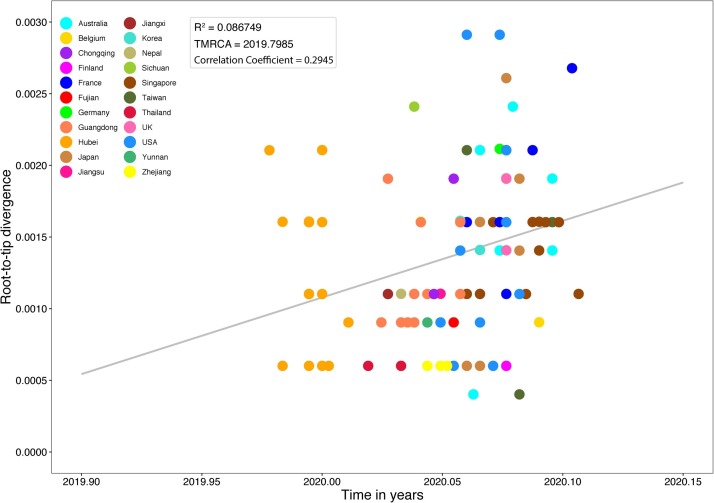
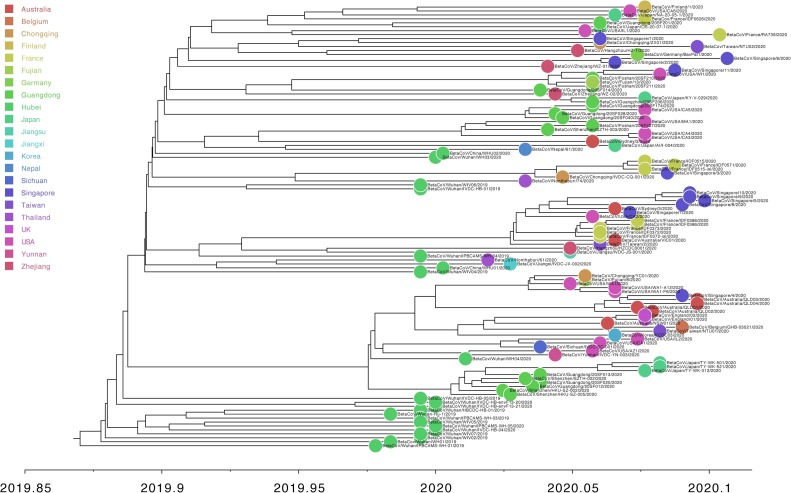
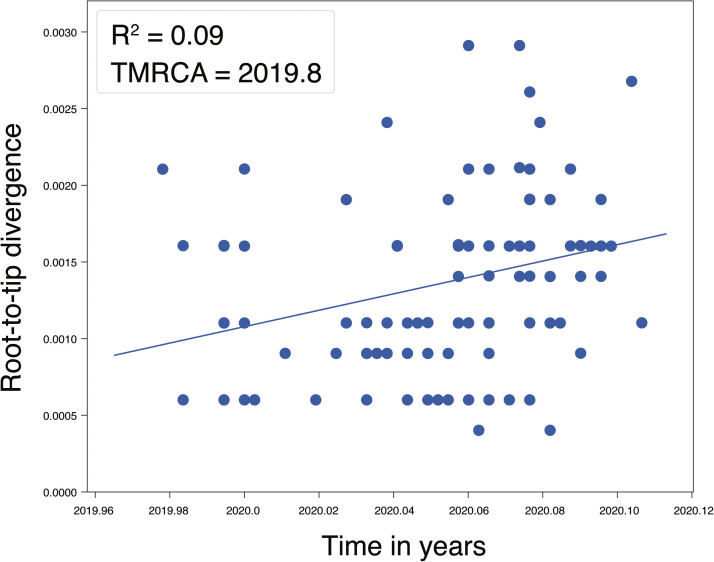
For “dataset_112”, a HKY (Kimura, 1980) nucleotide substitution model with a proportion of invariable sites was the model of best fit across the two different substitution schemes (i.e., 24 and 88 candidate models) according to the BIC method, and was thus used in subsequent likelihood-mapping and phylogenetic analyses. The PHI test of “dataset_112” did not find statistically significant evidence of recombination (p = 1.0). In addition, no evidence of recombination was found using RDP v4.100 (Martin et al., 2015). Our likelihood-mapping analysis revealed that the quartets from “dataset_112” were primarily distributed in the center (63.2 %) rather than the corners (36.8 %) or sides (0%) of the triangle, indicating a strong star-like topology signal, which may be due to exponential epidemic spread (Fig. 1 A), in accordance with previous studies (Li et al., 2020b; Li et al., 2020c; Li et al., 2020d). The split network generated for “dataset_112” using the NeighborNet method revealed the existence of polytomies, and thus was highly unresolved. This indicated that the phylogenetic relationship of our dataset was probably best represented by a star-like phylogenetic tree rather than a strictly bifurcating tree (Fig. 1B), suggesting possible rapid early spread of SARS-CoV-2, in accordance with the likelihood-mapping results. ML phylogenetic analysis of “dataset_112” also showed star-like topology (Fig. 2 ), indicating the introduction of a new virus to an immunologically naive population, in accordance with the likelihood-mapping and split network results. Root-to-tip linear regression analyses between genetic divergence and sampling date using the best-fitting root, which minimizes the mean of the squares of the residuals, showed that “dataset_112” had a minor positive temporal signal (R2 = 0.087; correlation coefficient = 0.2945), thus suggesting a minor clocklike pattern of molecular evolution (Fig. 3 ). We estimated the whole-genome evolutionary rate of SARS-CoV-2 to be 5.3504 × 10−3 substitutions per site per year and the TMRCA of SARS-CoV-2 to be 19 October 2019. The ML dating analyses between root-to-tip genetic divergence and sampling date also showed that our dataset had a minor strong positive temporal signal (R2 = 0.09) (Supplementary Fig. 1). The evolutionary rate and TMRCA date estimates of SARS-CoV-2 for “dataset_112” were 5.35 × 10−3 substitutions per site per year and 19 October 2019, respectively, in accordance with the root-to-tip regression results using TempEst v1.5 (Rambaut et al., 2016). Based on Bayesian time-scaled phylogenetic analysis using the tip-dating method, the estimated TMRCA dates and evolutionary rates of SARS-CoV-2 for “dataset_112” ranged from 12 November 2019 to 7 December 2019 (95 % BCI: 11 October 2019 and 21 December 2019) and from 8.37 × 10−4 to 1.12 × 10−3 substitutions per site per year (95 % BCI: 5.06 × 10−4–1.53 × 10−3), respectively (Table 1 ). Notably, the estimated TMRCA dates and evolutionary rates of SARS-CoV-2 were consistent across different molecular clock models but were distinct across different coalescent tree prior models. The best-fitting combination was an UCLN relaxed molecular clock along with an exponential growth tree prior model with growth rate parameterization, as shown by the marginal likelihood estimates for “dataset_112” when comparing the two clock models and five tree prior models. Thus, the TMRCA date and evolutionary rate estimates of SARS-CoV-2 for “dataset_112” with the best-fitting combination were 12 November 2019 (95 % BCI: 11 October 2019 and 09 December 2019) and 9.90 × 10−4 substitutions per site per year (95 % BCI: 6.29 × 10−4–1.35 × 10−3), respectively (Table 1). The estimates of the MCC phylogenetic relationships among the SARS-CoV-2 genomes for our dataset from the Bayesian coalescent framework using the tip-dating method, as well as the exponential coalescent tree prior with doubling time parameterization and UCLN relaxed molecular clock, are displayed in Fig. 4 . As shown, “dataset_112” exhibited more genetic diversity than our previous datasets (Li et al., 2020b; Li et al., 2020c; Li et al., 2020d).
Fig. 1.
Likelihood-mapping and split network analyses of SARS-CoV-2.
Likelihood-mapping (A) and split network (B) analyses of SARS-CoV-2 for “dataset_112” are shown. For likelihood-mapping analysis, corners represent tree-like phylogenetic signals and those at sides represent network-like signals. Central area of likelihood map represents star-like signals of unresolved phylogenetic information.
Fig. 2.
Estimated maximum-likelihood phylogenetic tree of SARS-CoV-2.
Maximum-likelihood phylogenetic tree of SARS-CoV-2 for “dataset_112” is shown. Tree is midpoint rooted. Colors indicate different sampling locations. Scale bar at bottom indicates 0.0003 nucleotide substitutions per site.
Fig. 3.
Root-to-tip genetic divergence plot of SARS-CoV-2.
Root-to-tip plot shows regression of genetic divergence against sampling dates. Colors indicate different sampling locations. Gray color indicates linear regression line.
Table 1.
Bayesian phylogenetic estimates of evolutionary parameters and model comparison for genome sequences of SARS-CoV-2 under different clock models and coalescent tree priors.
| Clock model | Coalescent tree prior | Substitution rate (substitutions/site/year) |
TMRCA |
PS | Rank | SS | Rank | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Lower 95 % HPD | Upper 95 % HPD | Mean | Lower 95 % HPD | Upper 95 % HPD | ||||||
| Strict | Constant | 1.02E-03 | 6.83E-04 | 1.38E-03 | 2019-11-14 | 2019-10-16 | 2019-12-07 | −42140.6 | 10 | −42141 | 9 |
| Exponentiala | 9.97E-04 | 6.56E-04 | 1.39E-03 | 2019-11-14 | 2019-10-16 | 2019-12-09 | −42117.4 | 2 | −42117 | 2 | |
| Exponentialb | 1.11E-03 | 7.38E-04 | 1.53E-03 | 2019-11-29 | 2019-11-09 | 2019-12-15 | −42123.5 | 3 | −42124 | 3 | |
| Skyline | 9.90E-04 | 5.43E-04 | 1.44E-03 | 2019-12-06 | 2019-11-15 | 2019-12-21 | −42124.6 | 5 | −42125.3 | 5 | |
| Skygrid | 8.37E-04 | 5.06E-04 | 1.23E-03 | 2019-12-05 | 2019-11-19 | 2019-12-16 | −42129.6 | 7 | −42130.5 | 7 | |
| Relaxed | Constant | 1.03E-03 | 6.94E-04 | 1.40E-03 | 2019-11-13 | 2019-10-14 | 2019-12-09 | −42140.5 | 9 | −42141 | 10 |
| Exponentiala | 9.90E-04 | 6.29E-04 | 1.35E-03 | 2019-11-12 | 2019-10-11 | 2019-12-09 | −42116.1 | 1 | −42117 | 1 | |
| Exponentialb | 1.12E-03 | 7.40E-04 | 1.53E-03 | 2019-11-30 | 2019-11-10 | 2019-12-16 | −42127.8 | 6 | −42129 | 6 | |
| Skyline | 1.00E-03 | 5.09E-04 | 1.47E-03 | 2019- | 2019-11-16 | 2019-12-21 | −42123.7 | 4 | −42125 | 4 | |
| Skygrid | 8.57E-04 | 5.32E-04 | 1.24E-03 | 2019-12-06 | 2019-11-19 | 2019-12-16 | −42130.1 | 8 | −42131 | 8 | |
PS: path sampling, SS: stepping-stone sampling.
exponential growth population with growth rate parameterization.
exponential growth population with doubling time parameterization.
Fig. 4.
Estimated maximum-clade-credibility tree of SARS-CoV-2.
Circle at tip is colored according to sampling location.
3.3. Phylodynamic analyses of SARS-CoV-2
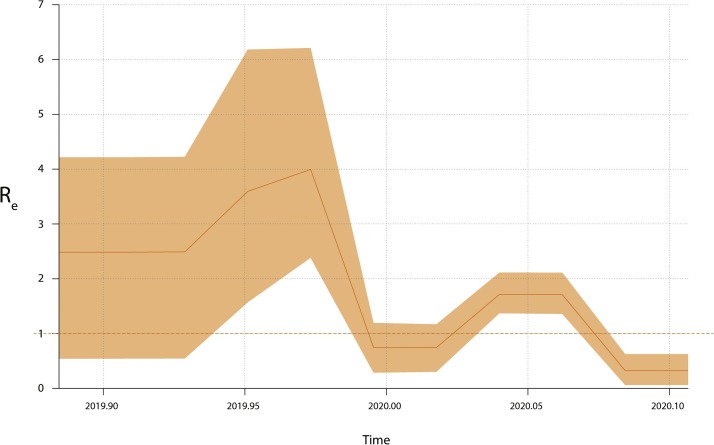
Analysis showed that the Re estimates of SARS-CoV-2 for “dataset_112” experienced complex phylodynamics characterized by at least two growing and two declining phases (Fig. 5 ). The mean Re estimates of SARS-CoV-2 for our dataset ranged from 0.336 to 4.137, and the first growth phase had more uncertainty compared to the remaining phases due to the wider 95 % highest posterior density (HPD) interval of Re estimates. Notably, we found very low Re estimates of SARS-CoV-2 for “dataset_112″ during the recent sampling time period. The low Re estimates suggest that China’s extreme lockdowns may be responsible for the successful control of SARS-CoV-2 in China.
Fig. 5.
Re estimates over time of SARS-CoV-2.
Re estimates obtained using Bayesian birth-death skyline model over five equidistant intervals. Horizontal red dotted line represents epidemiological threshold (Re = 1). Shaded area represents 95 % BCI.
4. Discussion
To investigate the global epidemic spread of SARS-CoV-2, we performed comprehensive evolutionary analyses of 112 genomes from “dataset_112”. Our likelihood-mapping analysis confirmed increasing tree-like phylogenetic signals over time as more genome sequences of SARS-CoV-2 strains were added to our study compared with previous results (Li et al., 2020c; Li et al., 2020d; Li et al., 2020e). This indicates more complex genetic divergence of SARS-CoV-2 in humans and greater adaptation to humans (Fig. 1A), consistent with our earlier studies (Li et al., 2020c; Li et al., 2020d; Li et al., 2020e). Split network analysis of SARS-CoV-2 based on “dataset_112” using the NeighborNet method was more resolved over time as more genome sequences were added to our study compared with our previous results (Li et al., 2020d). This indicates increasing tree-like evolution of SARS-CoV-2, consistent with our likelihood-mapping analysis (Fig. 1B). These results are also consistent with our ML phylogenetic analyses, which showed a more bifurcating tree topology from “dataset_112” compared to our previous results (Li et al., 2020c; Li et al., 2020d; Li et al., 2020e), (Fig. 2). Our dataset still had a minor positive temporal signal based on regression analysis using TempEst v1.5 (Rambaut et al., 2016) and ML dating analysis using TreeTime package (Sagulenko et al., 2018) compared to our previous results (Li et al., 2020d). Furthermore, the estimated TMRCA dates and evolutionary rates of SARS-CoV-2 for “dataset_112” were found to be nearly identical using both analyses (Fig. 3 and Supplementary Fig. 1), consistent with earlier results (Li et al., 2020d). The estimated TMRCA dates of SARS-CoV-2 based on “dataset_112” using TempEst v1.5 (Rambaut et al., 2016) (19 October 2019) and ML dating analysis using TreeTime (Sagulenko et al., 2018) (19 October 2019) were also identical to our previous results (Li et al., 2020d). However, the estimated evolutionary rates of SARS-CoV-2 for “dataset_112” using TempEst v1.5 (Rambaut et al., 2016) (5.3504 × 10−3 substitutions per site per year) and ML dating analysis using TreeTime (Sagulenko et al., 2018) (5.35 × 10−3 substitutions per site per year) were very distinct to our prior results (Li et al., 2020d) using TempEst v1.5 (Rambaut et al., 2016) (3.3452 × 10−4 substitutions per site per year) and ML dating analysis using TreeTime (Sagulenko et al., 2018) (3.34 × 10−4 substitutions per site per year). The estimated TMRCA dates and evolutionary rates of SARS-CoV-2 for “dataset_112” were very similar across different clocks using the tip-dating method, but very distinct across different coalescent tree priors (e.g., parametric coalescent and nonparametric coalescent models) (Table 1). Notably, the estimated TMRCA dates and evolutionary rates of SARS-CoV-2 were more similar between exponential growth population with growth rate parameterization and constant population size models than those determined using exponential growth population with growth rate parameterization and another exponential growth population with doubling time parameterization models. Bayesian analyses with the tip-dating method using an UCLN relaxed molecular clock as well as an exponential growth coalescent tree prior with doubling time parameterization model suggested that SARS-CoV-2 is evolving at a rate of 9.90 × 10−4 substitutions per site per year (Table 1). This in accordance with our prior studies (Li et al., 2020c; Li et al., 2020d; Li et al., 2020e), but very distinct to results based on regression analysis using TempEst v1.5 (Rambaut et al., 2016) and ML dating analysis using TreeTime (Sagulenko et al., 2018), which showed that SARS-CoV-2 is evolving at a rate of 5.3504 × 10−3 and 5.35 × 10−3 substitutions per site per year, respectively. These findings suggest that the virus originated on 12 November 2019, in agreement with our previous studies (Li et al., 2020c; Li et al., 2020d; Li et al., 2020e), but distinct from earlier regression analysis (Rambaut et al., 2016) and ML dating analysis results (Sagulenko et al., 2018), which both showed that the virus originated on 19 October 2019. In summary, the TMRCA date and evolutionary rate estimates of SARS-CoV-2 for “dataset_112” are still sensitive to the tree prior, and additional genomes should make these estimates more robust towards the tree prior choice. We found Bayesian approaches to be more powerful than regression analysis and ML dating analysis. We employed the BDSKY model (Stadler et al., 2013) and found that the Re estimates of SARS-CoV-2 for “dataset_112” experienced a complex phylodynamic history (Fig. 5). However, the epidemic spread of SARS-CoV-2 had very low Re estimates during the recent sampling time period, suggesting that the introduction of effective prevention measures (e.g., joint defense and control strategies in China, particularly the extreme lockdown of Wuhan) limited viral spread within the sampled populations. If performed in real time, such analyses could provide actionable targets for prevention. The limitations of these evolutionary analyses are discussed in our previous studies (Li et al., 2020c; Li et al., 2020d; Li et al., 2020e), and can also be applied here. Both Bayesian coalescent and BDSKY models assume that the population is well-mixed. That is, they assume that there is no significant population structure and that the sequences are a random sample from the population. For the epidemiological analysis of SARS-CoV-2 from “dataset_112”, the non-random and non-well-mixing of the sampled population, and the non-constant sampling effort may be potential strong sources of bias in the estimates of SARS-CoV-2 epidemic parameters, which is an issue for all molecular evolutionary studies using real world data. It is important to note that there is currently not enough genomic data from the early COVID-19 outbreak period to interpret the early history of global transmissions of COVID-19 from few genomes in detail. Links of all paired genomic sequences that seem directly connected now from our phylogenetic trees in the present study are likely to be connected more closed with other genomic sequences from other countries not sampled and sometimes can be connected differently later with more genomic sequences becoming available. The phylogenetic relationships of genomic sequences of SARS-CoV-2 in the future will be much more complex than the early incomplete picture presented in this study. Therefore, our results and conclusions should be explained with caution due to the limited number of SARS-CoV-2 genomes presented in this study over a short time period. The 95 % BCI estimates for the evolutionary rates and TMRCA dates are averaged over many plausible phylogenetic reconstructions of the genome data; thus, as more patients with COVID-19 are sampled and more SARS-CoV-2 genomes become available, we expect these estimates will become narrower.
In conclusion, this study characterized the epidemic spread patterns of SARS-CoV-2 in China (including 10 provinces) and beyond (including 12 other countries) based on genome data generated from patients with COVID-19 between 24 December 2019 and 9 February 2020. Our results shed light on the evolutionary and epidemiological histories of SARS-CoV-2 over time, and suggest that a strategy of ‘suppression’ (e.g., social distancing of the entire population, case isolation, household quarantine, and school and university closure) is needed to reduce deaths and prevent healthcare systems being overwhelmed. Our results also emphasize the importance of using phylogenetic and phylodynamic analyses to provide insights into the roles of various interventions to limit the spread of SARS-CoV-2 in China and beyond. Understanding epidemic dynamics of SARS-CoV-2 in real time is increasingly important for guiding prevention efforts.
Author contributions
X.L. conceived and designed the study and drafted the manuscript. X.L analyzed the data. X.L., Q.N., W.C., D.L., Y.C., H.L., D.L., M.T., J.Z., and W.T. interpreted the data and provided critical comments. All authors reviewed and approved the final manuscript.
Author statement
All authors have reviewed and confirmed the revised manuscript.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
This work was supported by a grant from the Special Project for Prevention and Control of Novel Coronavirus Pneumonia in Weifang Science and Technology Development Plan in 2020 (No. 2020YQFK015) to Associate Senior Technologist Qing Nie and Dr. Xingguang Li. We gratefully acknowledge the Authors and Originating and Submitting Laboratories for their sequences and meta-data shared through GISAID (Elbe and Buckland-Merrett, 2017), on which this research is based.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198098.
Contributor Information
Qing Nie, Email: nieqing0454@163.com.
Xingguang Li, Email: xingguanglee@hotmail.com.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Baele G. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012;29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G., Li W.L., Drummond A.J., Suchard M.A., Lemey P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol. Biol. Evol. 2013;30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese S.M.E.C. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Chu D.K. MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 2014;20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A., Shapiro B., Pybus O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Drummond A.J., Ho S.Y., Phillips M.J., Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenge. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ferreira M.A.R., Suchard M.A. Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 2008;36:355–368. doi: 10.1002/cjs.5550360302. [DOI] [Google Scholar]
- Gelman A., Meng X.-L. Simulating normalizing constants: from importance sampling to bridge sampling to path sampling. Stat. Sci. 2020;13:163–185. [Google Scholar]
- Gill M.S. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol. Biol. Evol. 2013;30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R.C., Tavare S. Sampling theory for neutral alleles in a varying environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994;344:403–410. doi: 10.1098/rstb.1994.0079. [DOI] [PubMed] [Google Scholar]
- Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guindon S. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. doi:citeulike-article-id:691774. [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/bf01731581. [DOI] [PubMed] [Google Scholar]
- Kingman J.F.C. The coalescent. Stoch. Process. Their Appl. 1982;13:235–248. doi: 10.1016/0304-4149(82)90011-4. [DOI] [Google Scholar]
- Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lau S.K. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Q. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zai J., Wang X., Li Y. Potential of large “first generation” human-to-human transmission of 2019-nCoV. J. Med. Virol. 2020;92:448–454. doi: 10.1002/jmv.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Transmission dynamics and evolutionary history of 2019-nCoV. J. Med. Virol. 2020 doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zai J., Wang X., Li Y. Potential of large’ first generation’ human-to-human transmission of 2019-nCoV. J. Med. Virol. 2020 doi: 10.1002/jmv.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1 doi: 10.1093/ve/vev003. vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur D.B. Emerging infectious diseases. Nurs. Clin. North Am. 2019;54:297–311. doi: 10.1016/j.cnur.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nii-Trebi N.I. Emerging and neglected infectious diseases: insights, advances, and challenges. Biomed Res. Int. 2017;(2017) doi: 10.1155/2017/5245021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Lam T.T., Max Carvalho L., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2 doi: 10.1093/ve/vew007. vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko P., Puller V., Neher R.A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4 doi: 10.1093/ve/vex042. vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.A., von Haeseler A. Maximum-likelihood analysis using TREE-PUZZLE. Curr. Protoc. Bioinf. 2007 doi: 10.1002/0471250953.bi0606s17. Chapter 6, Unit 6 6. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Song H.D. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler T., Kuhnert D., Bonhoeffer S., Drummond A.J. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proc. Natl. Acad. Sci. U. S. A. 2013;110:228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M.A., Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. Author correction: a new coronavirus associated with human respiratory disease in China. Nature. 2020;580:E7. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Lewis P.O., Fan Y., Kuo L., Chen M.H. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst. Biol. 2011;60:150–160. doi: 10.1093/sysbio/syq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhong N.S. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/s0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.