Abstract
The spike protein of SARS-CoV-2 has been undergoing mutations and is highly glycosylated. It is critically important to investigate the biological significance of these mutations. Here, we investigated 80 variants and 26 glycosylation site modifications for the infectivity and reactivity to a panel of neutralizing antibodies and sera from convalescent patients. D614G, along with several variants containing both D614G and another amino acid change, were significantly more infectious. Most variants with amino acid change at receptor binding domain were less infectious, but variants including A475V, L452R, V483A, and F490L became resistant to some neutralizing antibodies. Moreover, the majority of glycosylation deletions were less infectious, whereas deletion of both N331 and N343 glycosylation drastically reduced infectivity, revealing the importance of glycosylation for viral infectivity. Interestingly, N234Q was markedly resistant to neutralizing antibodies, whereas N165Q became more sensitive. These findings could be of value in the development of vaccine and therapeutic antibodies.
Keywords: COVID-19, SARS-CoV-2, spike, variants, mutants, glycosylation, pseudotyped virus
Graphical Abstract
Eighty natural variants and 26 glycosylation spike mutants of SARS-CoV-2 were analyzed in terms of infectivity and antigenicity using high throughput pseudovirus assay in conjunction with neutralizing antibodies.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic is a tremendous threat globally. As of July 3, 2020, 216 countries have reported COVID-19 cases, with more than 10 million confirmed cases and approximately 518,000 deaths (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/). The causative agent of COVID-19, SARS-CoV-2 causes a lower respiratory tract infection that can progress to severe acute respiratory syndrome and even multiple organ failure (Lv et al., 2020a; Yang et al., 2020).
SARS-CoV-2 is a single-stranded positive-strand RNA virus whose genome encodes four structural proteins: spike (S), small protein (E), matrix (M), and nucleocapsid (N) (Chan et al., 2020). The S protein is a type I fusion protein that forms trimers on the surface of the virion. It is composed of two subunits, with S1 responsible for receptor binding and S2 for membrane fusion (Walls et al., 2020; Wrapp et al., 2020). The SARS-CoV-2 utilizes angiotensin-converting enzyme 2 (ACE2) as the receptor for entry into target cells (Letko et al., 2020). Therefore, the S protein determines the infectivity of the virus and its transmissibility in the host (Hulswit et al., 2016). As this protein is the major antigen inducing protective immune responses (Du et al., 2009; He et al., 2004; Li, 2016; Walls et al., 2020), all vaccines under development are directed against it. Clearly, it is pivotal to closely monitor antigenic evolution of the spike in the circulating viruses. As it is a heavily glycosylated protein, investigation of the effects of the site-specific glycans on infectivity and immune escape is also of unquestionable importance.
RNA viruses are known to have higher mutation rates than DNA viruses (Duffy, 2018; Lauring and Andino, 2010). Amino acid changes in the surface protein can significantly alter viral function and/or interactions with neutralizing antibodies. For example, A226V of Chikungunya virus E1 protein facilitated its adaptability in the vector Aedesalbopictus, resulting in an increased transmissibility (Tsetsarkin et al., 2007). Similarly, A82V of Ebola virus GP protein led to increased viral infectivity and mortality (Diehl et al., 2016; Urbanowicz et al., 2016). Moreover, the highly pathogenic avian influenza H5N1 with 4 amino acid changes demonstrated enhanced transmission (Herfst et al., 2012), whereas in H7N9, the combined amino acid change A143V/R148K of hemagglutinin decreased the sensitivity of the virus to neutralizing antibodies by more than 10 times (Ning et al., 2019; Petrie and Lauring, 2019).
Although SARS-CoV-2 was only discovered in humans recently, mutations in the gene encoding Spike (S) protein are being continuously reported (Dawood, 2020; Korber et al., 2020; Saha et al., 2020; Sheikh et al., 2020; van Dorp et al., 2020). As of May 6, 2020, 329 naturally occurring variants in S protein have been reported in public domain. Notably, there were only 13 amino acid sites with a rate of more than 0.1%. Preliminary study suggested that the increased fatality rate may be associated with the most dominant variant D614G. Presumably, this change may have induced a conformational change in the S protein, thereby resulting in the increased infectivity (Becerra-Flores and Cardozo, 2020). However, it remains largely unclear as to whether these reported variants could influence viral infectivity, transmissibility, or reactivity with neutralizing antibodies.
It is also well documented that mutations affecting glycosylation of viral proteins could also profoundly affect viral life cycle and its interaction with the host. For example, N-glycosylation at specific sites of the HIV-1 Env protein is critical for Env expression and assembly (François and Balzarini, 2011; Kong et al., 2015; Wang et al., 2013). Deletion of certain glycosylation sites could decrease the binding of Env protein to the CD4 receptor, abolishing the infectivity of the resulting viral particles (François and Balzarini, 2011). Moreover, glycosylation site mutations are also known to render the virus resistant to neutralization by the antibodies (Wang et al., 2013; Wang et al., 2015), whereas deletion of certain glycosylation sites in the H5N1 HA protein has been found to affect HA cleavage, replication, stability, and antigenicity (Zhang et al., 2015). It is of note that although the S protein of SARS-CoV-2 is much more heavily glycosylated, with 22 potential N-glycosylation sites (Kumar et al., 2020; Vankadari and Wilce, 2020), how these glycosylation sites could affect viral infectivity and antibody-mediated neutralization remains unknown.
In this study, we investigated the biological significance of natural variants with amino acid change(s) as well as mutants at the putative N-linked glycosylation sites in the SARS-CoV-2 S protein. To achieve these, we generated 106 S mutants reported in the public domain or mutants at putative N-linked glycosylation sites and analyzed their infectivity and reactivity to neutralizing antibodies using the high-throughput pseudotyped virus system (Nie et al., 2020). We report that some natural variants and N-linked glycan deleted mutants have evolved to possess remarkable alterations in their infectivity and antigenicity.
Results
Construction of Pseudotyped Viruses with Natural Variants and Deletions of the Glycosylation Sites
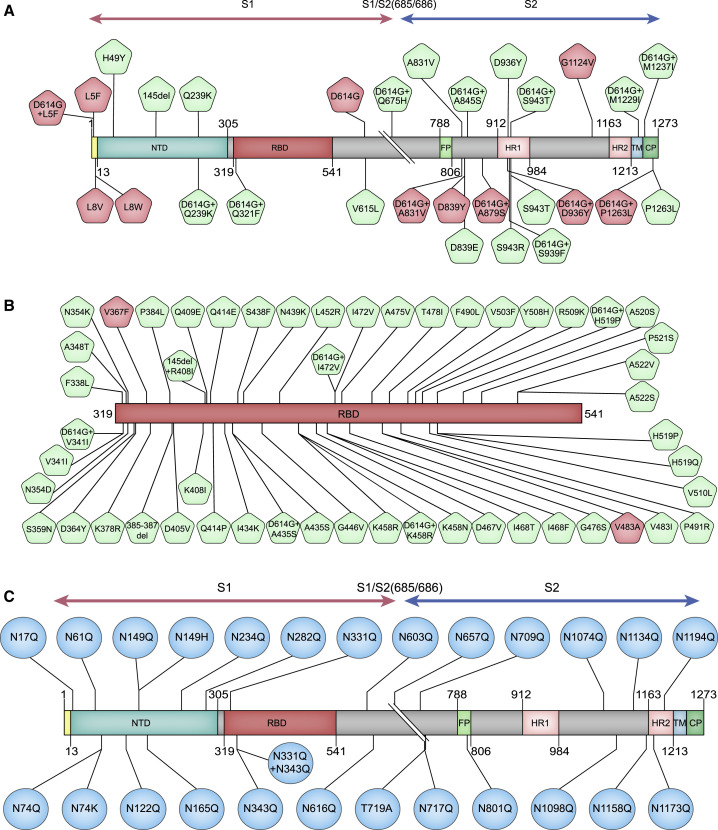
For ease of description in this communication, variants are referred to as those that occurred naturally (reported in GISAID) and mutants (investigational or experimental mutants) for those we introduced experimentally. For the analyses of variants, we first retrieved all S sequences reported in GISAID database up to May 6, 2020. Following removal of incomplete, redundant, and ambiguous sequences, a total of 13,406 sequences of the S protein were selected for alignment analyses. Using Wuhan-1 strain (GenBank: MN_908947) as a template, we have selected three groups of variants and mutants to construct the pseudotyped viruses. As shown in Figure 1 , group A represents all high-frequency variants and combined variants with D614G across the entire S gene (29 strains) excluding receptor-binding domain (RBD) region. Group B includes variants in RBD (51 strains). Although the eight single mutations (i.e., Q239K, V341I, A435S, K458R, I472V, H519P, A831V, and S943T) in both groups A and B do not exist by themselves, they were found to occur in combination with D614G. Therefore, the pseudotyped viruses for the eight single mutants were also constructed to compare with the double mutants with D614G. Group C is comprised of 26 mutants at the putative glycosylation sites (22 sites). This group includes both variants (N74K, N149H, and T719A) and investigational mutants that we made for the analyses of the effects of glycosylation. Specifically, all 22 sites (N to Q) were made in the lab to generate 22 individual mutants; we also made a combination by deleting the two glycosylation sites in RBD.
Figure 1.
Illustration of Amino Acid Changes Selected for This Study
(A) Variants and combined variants with D614G across the entire S gene excluding the RBD region.
(B) Variants in RBD.
(C) Mutants at the putative glycosylation sites (22 sites). This group include mutations we introduced at all 22 putative glycosylation sites (N to Q), a combination of two glycosylation site mutations in RBD, and three naturally occurring variants, N74K, N149H, and T719A, with ablated glycosylation sites.
High-frequency amino acid change sites (frequency > 0.1%) are highlighted in red. See also Table S1.
In total, we have generated 106 pseudotyped viruses (i.e., 80 variants and 26 glycosylation mutants) (Figure 1). These viruses were prepared as described previously (Nie et al., 2020; see STAR Methods).
Significantly Altered Infectivity of Variants and Deletions of the Glycosylation Site Mutants
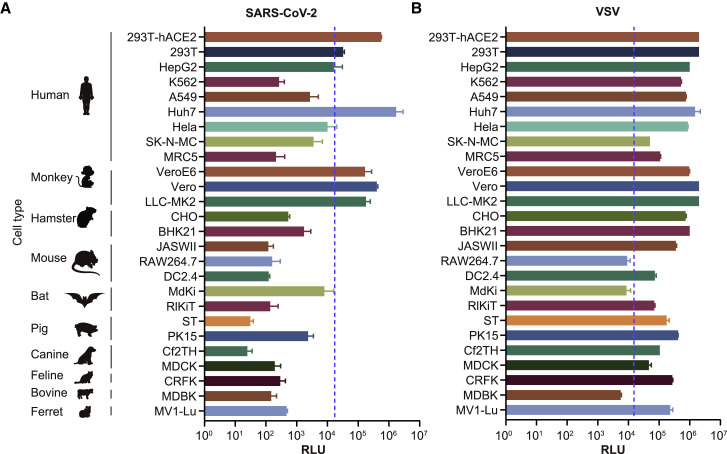
To determine the infectivity of these variants and mutants, we first infected 26 cell lines with pseudotyped viruses with either SARS-CoV-2 S protein or VSV G protein (see STAR Methods). As expected, the two types of pseudotyped viruses are different in the infection efficiency in the 26 cell lines (Figure 2 ). Although almost all cell lines were generally susceptible to infection by VSV G pseudotyped virus, SARS-CoV-2 pseudotyped virus could efficiently infect certain cell lines including three human cell lines (293T-hACE2, 293T, and Huh-7) and three non-human primate cell lines (Vero, VeroE6, and LLC-MK2). As such, we selected these four out of the six cell lines in subsequent experiments, including 293T-hACE2, Huh-7, Vero, and LLC-MK2.
Figure 2.
Selection of Susceptible Cell Lines
A total of 26 different cell lines were infected with pseudotyped viruses with either SARS-CoV-2 S (A) or VSV G (B). The infected cell lysates were diluted by 10× and analyzed for luminescence activities (RLU). All results were obtained from three independent experiments (mean ± SEM). When the RLU for the tested cell reached 1% of that for Huh-7, it was deemed as a permissive cell line. The dash line indicates the 1% RLU value of Huh-7 cell.
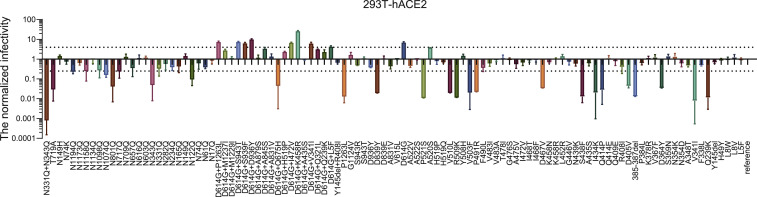
We first tested the infectivity of 106 pseudotyped viruses (80 natural variants and 26 glycosylation mutants) in 293T-hACE2 cells, where a difference by 4-fold in RLU compared with the reference Wuhan-1 strain (GenBank: MN_908947) was deemed as being significant (Figure S1 ). Of all 106 pseudotyped viruses, 24 were determined as low-infectivity (17 natural mutants and 7 glycosylation mutants), with RLU reading decreased by 4- to 100-fold (Figure S1). Among them, 13 were located in the RBD region. Variant V341I and investigational glycosylation mutant (N331Q+N343Q) were deemed as no-infectivity as demonstrated by over 100-fold decrease in RLU values compared with the reference strain. Both of them were located in RBD. It is worth noting that double glycosylation deletions at N331 and N343 resulted in a drastic reduction in viral infectivity (1,200-fold), whereas single deletion at each site caused modest reduction in viral infectivity, with the infectivity of N331Q reduced by only 3-fold and N343Q by 20-fold. Moreover, the non-natural double glycosylation mutations in RBD (N331Q and N343Q) resulted in significantly reduced infectivity, suggesting that the two glycosylation sites in the RBD region may participate in the binding of the receptor or maintain the conformation of the RBD region.
Figure S1.
Infectivity Analysis of Variants and Mutants in 293T-ACE2 Cell, Related to Figure 3
RLU values resulting from infection with variant pseudotyped viruses were quantified by luminescence meter and normalized to the reference strain (Wuhan-1) produced in parallel, with the difference by 4-fold being considered as significant; all experiments were conducted three times (mean ± SEM) unless specified. The horizontal dashed lines indicate the threshold of 4-fold difference.
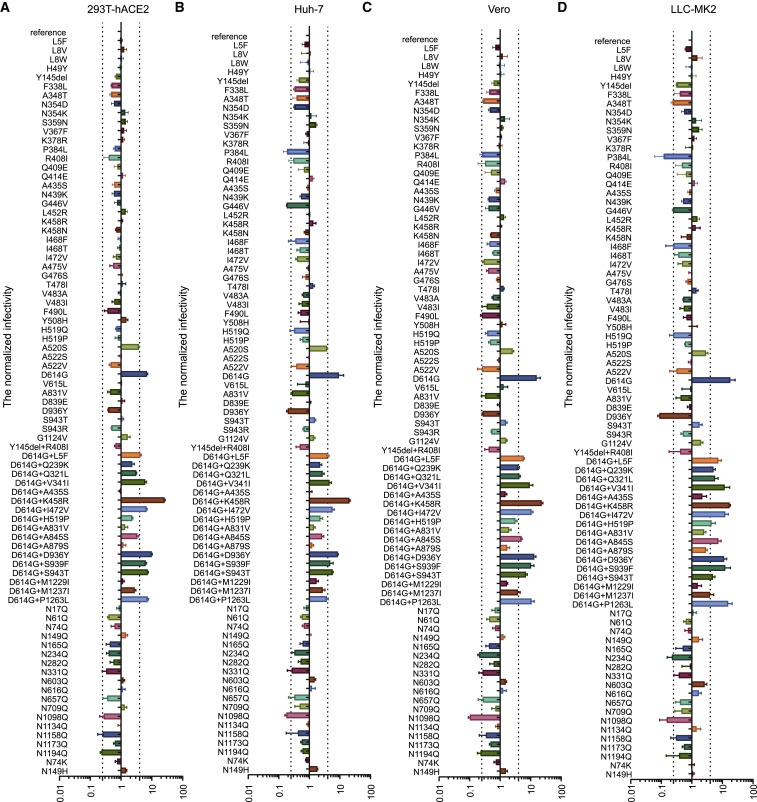
The remaining 63 variants were tested further with other three cell lines for infectivity. Notably, single D614G and combined variants with D614G (D614G+L5F, D614G+V341I, D614G+K458R, D614G+I472V, D614G+D936Y, D614G+S939F, and D614G+S943T) demonstrated increased infectivity compared to the reference strain in all the four cell lines (Figures 3A–3D), whereas no difference was found between single D614G and D614G combined variants, suggesting that the enhanced infectivity was more likely ascribed to D614G itself.
Figure 3.
Infectivity Analysis of Variants and Deletions of the Glycosylation Site Mutants
Infectivity of natural variants and experimental mutants conducted in 293T-ACE2 (A), Huh-7 (B), Vero (C), and LLC-MK2 (D). RLU values generated with the infection of the variants or mutants, as measured by luminescence meter, were compared with the reference strain (Wuhan-1). A difference by 4-fold is considered significant; all experiments were conducted three times (mean ± SEM) unless specified. See also Figure S1.
Differential Antigenicity of Natural Variants and Experimental Mutants with Monoclonal Antibodies
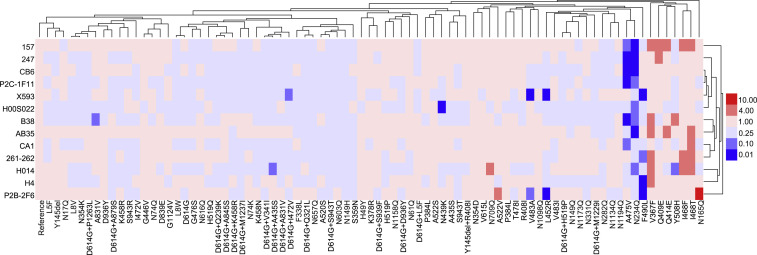
Having identified the variants with altered infectivity, we next set out to investigate the antigenicity of the infectious mutants using 13 neutralizing monoclonal antibodies (mAbs) (see STAR Methods). It was noted that some changes in RBD region demonstrated altered sensitivity to neutralizing mAbs (Figures 4 and S2 ). Specifically, A475V reduced the sensitivity to mAbs 157, 247, CB6, P2C-1F11, B38, and CA1, whereas F490L reduced the sensitivity to mAbs X593, 261-262, H4, and P2B-2F6. Moreover, V483A became resistant to mAbs X593 and P2B-2F6, and L452R to mAbs X593 and P2B-2F6. Finally, Y508H reduced the sensitivity to mAb H014, N439K to mAb H00S022, A831V to mAb B38, D614G+I472V to mAb X593, and D614G+A435S to mAb H014 by more than 4 times. In addition, some changes in the RBD region, including V367F, Q409E, Q414E, I468F, I468T, Y508H, and A522V, were observed to be more susceptible to neutralization mediated by mAbs.
Figure 4.
Analyses of Antigenicity of the Natural Variants and Experimental Mutants Using a Panel of Neutralizing mAbs
Serial dilutions of mAb preparations were pre-incubated with the pseudotyped viruses at 37°C for 1 h before they were added to Huh-7 cells. Luciferase activity was measured 24 h later to calculate EC50 of each antibody. The ratio of EC50 between the variant or mutant strains and the reference strain (Wuhan-1) was calculated and analyzed to generate heatmap using Hem I (Deng et al., 2014). The data were the results from 3–5 replicates. The red and blue boxes indicate the increase or decrease of the neutralization activity as shown in the scale bar. See also Figure S2.
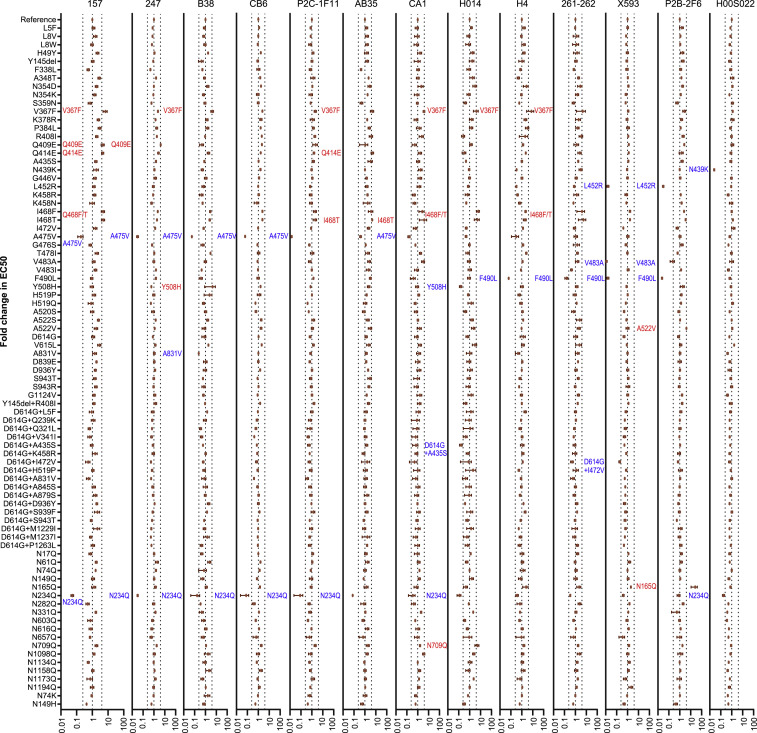
Figure S2.
Analyses of Antigenicity of Natural Variants and Experimental Mutants Using a Panel of Neutralizing mAbs, Related to Figure 4
Serial dilutions of mAb preparations were pre-incubated with the virus at 37°C for one hour before they were added to Huh-7 cells. Luciferase activity was measured 24 hours later to calculate EC50 of each antibody. The y axis represents the ratio of EC50 between the variant/mutant strain and the reference strain (Wuhan-1). The data (mean ± SEM) were the results from 3-5 replicates. The vertical dashed lines indicate the threshold of 4-fold difference. The significant changes were marked with colored symbols, blue for decreased, red for increased.
We next determine how infectious glycosylation mutants reacted to the same panel of mAbs. Mutant N165Q actually became more sensitive to mAb P2B-2F6, whereas N234Q reduced the neutralization sensitivity to different set of mAbs including 157, 247, CB6, P2C-1F11, H00S022, B38, AB35, and H014. These results confirmed that these two glycosylation sites are important for receptor binding.
These mAbs have proven to be valuable in our analyses of the amino acid changes. As shown in Figure 4, five mAbs (i.e., 157, 247, CB6, P2C-1F11 and B38) were unable to effectively neutralize both A475V and N234Q. Neither X593 nor P2B-2F6 was effective in neutralizing L452R, V483A, and F490L, whereas P2B-2F6 was more effective in neutralizing N165Q. In addition, mAb H014 was incapable of neutralizing N234Q, Y508H, and D614G+A435S, whereas mAbs H4 and 261-262 were found not to neutralize F490L. Furthermore, H00S022 was unable to neutralizing N439K and N234Q.
Altered Reactivity of Natural Variants and Experimental Mutants to Human Convalescent Sera
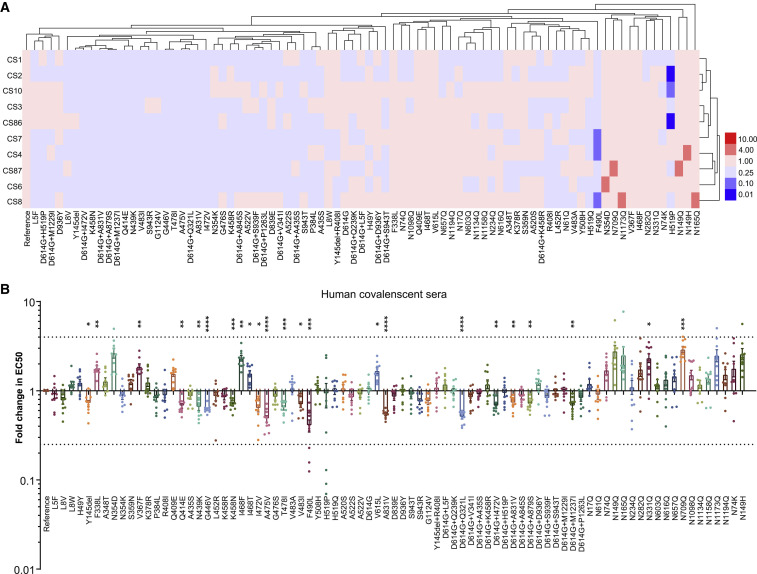
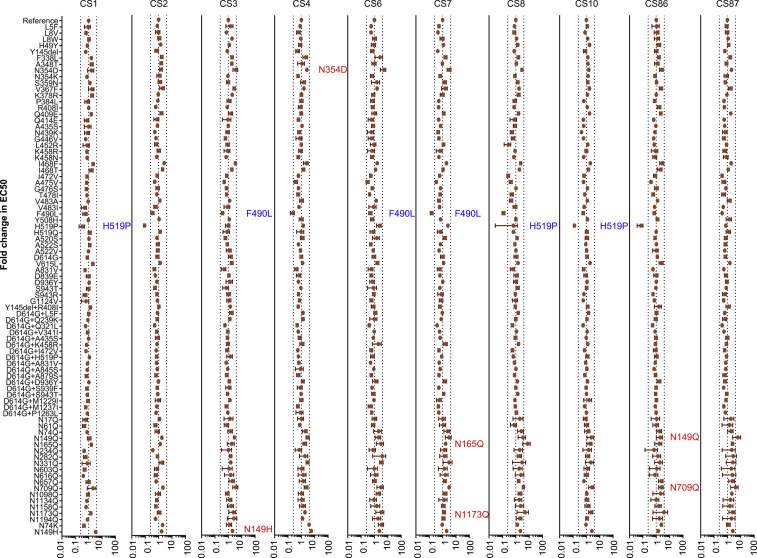
Finally, we determined the sensitivity of the strains with amino acid changes to ten COVID-19 convalescent sera (see STAR Methods). None of the variants and mutants demonstrated significantly altered sensitivity to all 10 convalescent sera, i.e., the EC50 values were not altered by more than 4-fold, irrespective of an increase or decrease, when compared with the reference strain (Figures 5 A and S3 ). However, the neutralization sensitivity of both F490L and H519P to three of ten patient sera were found to have decreased by more than 4 times, while six variants and mutants (N149H, N149Q, N165Q, N354D, N709Q, and N1173Q) became over 4-fold sensitive to one or two of the ten tested sera. Notably, five out of the six were glycan deletion mutants.
Figure 5.
Differential Sensitivity of the Natural Variants and Experimental Mutants to a Panel of Convalescent Serum Samples
(A) Serial dilutions of 10 patient serum samples were individually mixed with the pseudotyped viruses at 37°C for 1 h before added to Huh-7 cells for incubation of 24 h to determine the EC50. The experiments were repeated at least 3 times. Hem I software was used to analyze the data and draw the heatmap. The red and blue boxes indicate the increase or decrease of the neutralization activity as shown in the scale bar.
(B) Summary of the data from ten serum samples, with the values presented as mean ± SEM. The horizontal dashed lines indicate the threshold of 4-fold difference. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
See also Figure S3.
Figure S3.
Analyses of Antigenicity of the Natural Variants and Experimental Mutants Using Ten Convalescent Serum Samples, Related to Figure 5
The data (mean ± SEM) were the results from 3-5 replicates. The vertical dashed lines indicate the threshold of 4-fold difference. The significant changes were marked with colored symbols, blue for decreased, red for increased.
As shown in Figure 5B, when the data of individual convalescent sera were pooled together to analyze the sensitivity of all variants, no marked difference was observed (>4-fold). However, modest differences between some variants and reference strain (within 4-fold) were observed in their reactivity to grouped convalescent sera. These differences were statistically significant (p < 0.05). It is worth mentioning that some variants including F338L, V367F, I468F, I468T, and V615L (Figure 5B) were even more sensitive to the convalescent sera compared with reference strain, whereas more variants were found to be resistant to the convalescent sera. These variants include single amino acid change such as Y145del, Q414E, N439K, G446V, K458N, I472V, A475V, T478I, V483I, F490L, and A831V, as well as the double amino acid changes including D614G+Q321L, D614G+I472V, D614G+A831V, D614G+A879S and D614G+M1237I.
Similar to natural variants, although the magnitude of some glycosylation deletions in sensitivity to the sera is less than 4-fold, the differences between mutants and the reference strain (Wuhan-1) were found to be still several-fold and statistically significant, i.e., glycosylation mutants N331Q and N709Q significantly increased the sensitivity to convalescent sera (Figure 5B).
Discussion
As an RNA virus, SARS-CoV-2 virus is expected to continue evolving over time in human populations. Close monitoring of circulating virus strains is of unquestionable importance to inform research and development of vaccines and therapeutics. Here, we analyzed all spike sequences (over 13,000) reported to GISAID database. After filtering out incomplete, redundant, and ambiguous sequences, we narrowed down to 80 variants. Moreover, as glycosylation of viral protein is well documented to affect viral replication and immune response and SARS-CoV-2 S protein is heavily glycosylated, we also made 26 substitutional mutations at all 22 putative glycosylation sites. In total, we made 106 pseudotyped viruses, allowing us to characterize them using the established method (Nie et al., 2020; see STAR Methods).
Table 1 summarizes the characteristics of variants and investigational mutants. Of all variants, D614G is of particular note. This variant has been shown to rapidly accumulating since its emergence and linked to more clinical presentations (Korber et al., 2020). At the beginning of this study (May 6, 2020), it accounted for 62.8% (Table S1) of all circulating strains, but by July 3, it had reached 75.7%. This dominant strain could effectively infect the four cell lines tested, being 10-fold more infectious than the original Wuhan-1 strain (Figure 3).
Table 1.
Characteristics of Variants and Mutants
| Group A | Group B | Group C | |
|---|---|---|---|
| Number of variants or mutants | 29 | 51 | 26 |
| Increased infectivity | D614G, D614G+L5F, D614G+D936Y, D614G+S939F, D614G+S943T | D614G+V341I, D614G+K458R, D614G+I472Va | none |
| Decreased infectivity | Q239K, D839Y, P1263L, D614G+Q675H | V341I, D364Y, 385-387del, D405V, Q414P, I434K, S438F, D467V, P491R, V503F, R509K, V510L, P521S | N122Q, N343Q, N717Q, T719A, N801Q, N1074Q, N331Q+N343Q |
| Increased sensitivity to neutralizing mAbs | none | V367F, Q409E, Q414E, I468F, I468T, Y508H, A522V | N165Q, N709Q |
| Decreased sensitivity to neutralizing mAbs | A831V | N439K, L452R, A475V, V483A, F490L, Y508H, D614G+A435S, D614G+I472Va | N234Q |
| Increased sensitivity to convalescent sera | V615L | F338L, V367F, I468F, I468T | N149H, N149Q, N165Q, N331Q, N354D, N709Q, N1173Q |
| Decreased sensitivity to convalescent sera | Y145del, A831V, D614G+A831V, D614G+A879S, D614G+M1237I | Q414E, N439K, G446V, K458N, I472V, A475V, T478I, V483I, F490L, H519P, D614G+Q321L, D614G+I472Va | none |
D614G+I472V is the only variant with increased infectivity and decreased sensitivity to neutralizing mAb and convalescent sera. It is of note only one sequence is recorded in GISAID.
Another important finding is that natural variants capable of affecting the reactivity to neutralizing mAbs were almost all located in the RBD region (except A831V) because all antibodies used in this study were targeting the RBD (Cao et al., 2020; Ju et al., 2020; Lv et al., 2020b; Shi et al., 2020). Specifically, P2B-2F6 and P2C-1F11 were obtained using RBD as a bait to isolate RBD-specific B cells in peripheral blood mononuclear cells (PBMCs) of SARS-CoV-2 patients (Ju et al., 2020). P2C-1F11 and P2B-2F6 actually bind to overlapping epitope, with the latter being better characterized. Specifically, P2B-2F6 is involved in three hydrophobic interaction sites on RBD (Y449, L452, and F490) (Figures 6A and 6B). Indeed, both L452R and F490L were natural variants, with decreased sensitivity to neutralization by P2B-2F6 mAb; because both L452R and F490L remain sensitive to P2C-1F11, suggesting this mAb is not derived from the same clone for P2B-2F6. Moreover, both mutants displayed decreased sensitivity to another neutralizing mAb X593 by 10-fold compared with the reference strain (Figure 4).
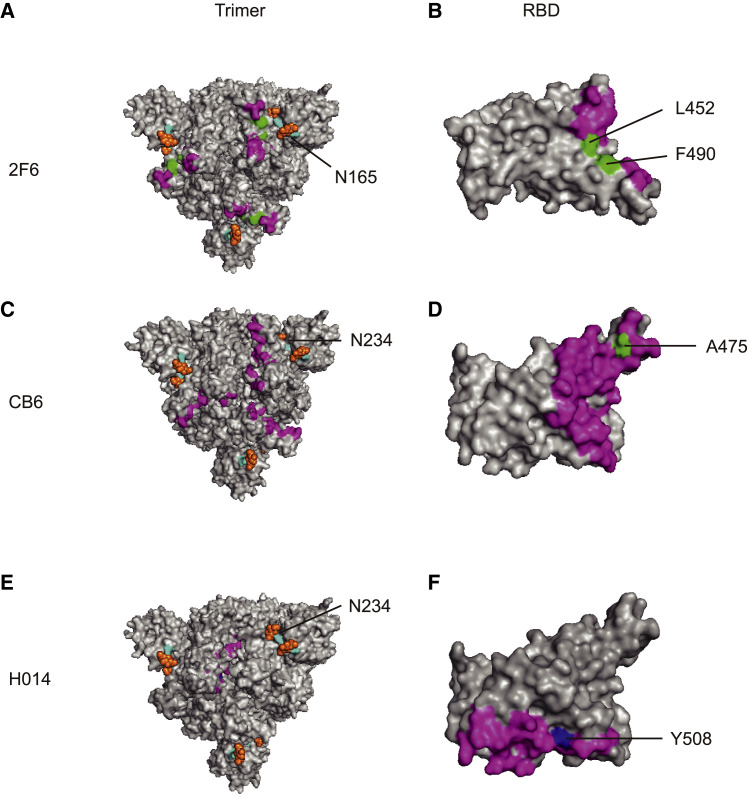
Figure 6.
mAb Epitopes and Antigenicity-Related Sites
(A and B) Epitope of the mAb P2B-2F6 on the trimer (A) and RBD (B).
(C and D) Epitope of the mAb CB6 on the trimer (C) and RBD (D).
(E and F) Epitope of the mAb H014 on the trimer (E) and RBD (F).
The monoclonal antibody epitope is indicated in purple. The glycosylation sites are indicated in light blue. The sugar chain is shown in orange. The amino acid sites that coincide with the epitopes of monoclonal antibodies are indicated in green. Amino acid sites that do not coincide with the monoclonal antibody epitope are indicated in dark blue.
Although we identified multiple variants with decreased sensitivity to neutralizing mAbs, we need to look at how frequent these variants are in the field. V483A in RBD is one of the two variants with a mutation frequency of over 0.1%. It showed decreased reactivity to the two mAbs (P2B-2F6 and X593) (Figures 6A and 6B; Ju et al., 2020). Another RBD variant A475V sits in the binding epitope of RBD. It is significantly resistant to several neutralizing mAbs including P2C-1F11, CA1, 247, and CB6. It is noteworthy that CB6 mAb targets the receptor binding epitope (Figures 6C and 6D; Shi et al., 2020). Specifically, Y508 was buried in the epitope targeted by mAb H014 (Figures 6E and 6F; Lv et al., 2020b). Indeed, the Y508H was found to be resistant to this mAb.
It is worth mentioning that D614G+I472V has shown increased infectivity and more resistance to neutralizing antibodies (Table 1), but only one sequence (originated from Canada) was reported in GISAID. Moreover, some variants, including N439K, L452R, A475V, V483A, F490L, and Y508H, do have decreased sensitivity to neutralizing mAbs. However, only V483A exceeded 0.1% in frequency at the beginning of the study, all of which were found in the United States, with 28 sequences reported as of May 6, 2020, and 36 up to July 3, 2020. Variants containing N439K showed a significant increase in circulation, i.e., with 5 cases reported as of May 6, 2020 (all in the United Kingdom) to 47 by July 3, 2020 (45 in the United Kingdom and 2 in Romania). In addition, only one sequence from France containing Y508H was deposited in GIRSAID as of May 6, whereas four sequences reported as of July 3, 2020, of which two originated from Netherlands, one from Sweden, and one from France. Only one or two isolates were reported for other variants, which have not been observed to have increased during the time frame we have been monitoring. Nevertheless, as RNA viruses mutate all the time and some variants may only appears during certain period of time, whereas others could emerge in an unpredictable fashion, continued analyses of the circulating strains in terms of the mutation frequency and temporal pattern are warranted.
Our results suggest that the 13 mAbs used in this study could be divided into seven groups, because they appear to be different in the inhibitory effects on the variants. As such, it would be interesting to formulate a therapeutic regimen comprised of at least two mAbs. For example, a combination of P2C-1F11 and X593 should be effective to inhibit all variants in this study. It would be of interest to test more neutralizing antibodies that could be targeting epitopes outside RBD.
With regard to the glycosylation mutants analyzed in this study, N165Q increased the sensitivity to mAb P2b-2F6 whereas N234Q displayed resistance to neutralizing mAbs such CA1, CB6, 157, and others. Although neither of them is found in circulation, the reactivity of these two mutants to neutralizing mAb is still worth noting. As N165 and N234 are located near the RBD region (Watanabe et al., 2020), these mutants may affect some epitopes targeted by neutralizing mAbs. Specifically, N165 glycosylation site is involved in the binding of mAb to the RBD region of S protein (Cao et al., 2020). It is likely that the sugar chain can mask the epitope targeted by the antibody. This type of glycan shield has been observed in other viruses such as HIV-1. Specifically, the deletion of the N197 sugar chain of HIV-1 gp120 could enhance neutralization by targeting this epitope (Li et al., 2008; Utachee et al., 2010; Wang et al., 2013). In this study, the other glycosylation mutant N234Q, which is also close to the RBD, is resistant to several mAbs. Nevertheless, we cannot exclude the possibility that the sugar chains themselves are the targets of the neutralizing mAbs, similar to observations made in other viruses. Specifically, the sugar chain at residues 142 and 144 in influenza virus H1N1 is essential for the induction of neutralizing antibodies (Huang et al., 2020), while the broadly neutralizing antibodies against the epitopes of HIV-1 gp120, 2G12, PGT121, and PG9 have been found to target the sugar chains (Doores, 2015). Because COVID-19 vaccines are being developed in different platform such as bacteria, mammalian cells, and plant expression systems, it is important to consider that different glycosylation profiles could affect the immunogenicity of the vaccines.
The use of sera from 10 convalescent patients in neutralizing assay largely confirmed the results obtained with the well-characterized neutralizing mAbs. It is understood that the magnitude of altered reactivity is slightly smaller with human sera than that with mAbs, given that polyclonal antibodies from convalescent patents are directed against multi-epitopes on the full-length S protein; as a result, these polyclonal antibodies could complement one another. However, the differences in their reactivity to the human antibodies were found to be by several fold in most cases and all determined as statistically significant. Notably, some RBD variants such as A475V and F490L have been confirmed to have decreased sensitivity to both human sera and multiple neutralizing mAbs. A475V reduced the sensitivity to 6 mAb out of the 13 mAb used in this study, whereas F490L reduced the sensitivity to neutralization by 3 mAbs. It is possible that antibodies in convalescent sera are able to neutralize these critical epitopes targeted by these mAbs that are known to disrupt the binding of the S protein to hACE2 receptor (Ju et al., 2020; Shi et al., 2020; Walls et al., 2020; Wang et al., 2020). The A475V could weaken the hydrogen bond and hydrophobic interaction (Shi et al., 2020), whereas F490L may erode the hydrophobic interaction between molecules (Ju et al., 2020).
In summary, we have analyzed over 100 pseudotyped viruses in terms of their infectivity and sensitivity to neutralization by well-characterized mAbs or human sera from convalescent patients. Amino acid changes have been observed across the entire spike protein. An amino acid change (D614G) outside the RBD was found to be more infectious, but no evidence of being resistant to neutralizing antibodies has been demonstrated. However, the increasing dominance of D614G particularly deserves attention. Although some strains with amino acid changes at RBD studied here lost their infectivity, suggesting they may not likely become widespread, the RBD natural variants with increased resistance to antibody-mediated neutralization should be closely watched. Moreover, ablation of glycosylation sites affected their reactivity to neutralizing antibodies, along with their infectivity, enforcing the notion that the glycan could substantially affect SARS-CoV-2 viral replication and vaccine-induced immune responses. Collectively, our findings help shed light on the implications of some evolving strains in circulating viruses with respect to enhanced infectivity and altered antigenicity.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| X593 | Laboratory of X. Sunney Xie | N/A |
| CA1 | Laboratory of Jinghua Yan; Shi et al., 2020 | N/A |
| CB6 | Laboratory of Jinghua Yan; Shi et al., 2020 | N/A |
| H014 | Laboratory of Dr. Liangzhi Xie | N/A |
| H00S022 | Laboratory of Dr. Liangzhi Xie | N/A |
| P2C-1F11 | Laboratory of Linqi Zhang; Ju et al., 2020 | N/A |
| P2B-2F6 | Laboratory of Linqi Zhang; Ju et al., 2020 | N/A |
| AB35 | Laboratory of Linqi Zhang | N/A |
| 261-262 | Laboratory of Linqi Zhang | N/A |
| 157 | Laboratory of Linqi Zhang | N/A |
| 247 | Laboratory of Linqi Zhang | N/A |
| H4 | Laboratory of Fu Gao; Wu et al., 2020 | N/A |
| B38 | Laboratory of Fu Gao; Wu et al., 2020 | N/A |
| Bacterial and Virus Strains | ||
| DH5α Chemically Competent Cell | Invitrogen | Cat#12034013 |
| G∗ΔG-VSV | Kerafast | Cat#EH1020-PM |
| Biological Samples | ||
| Convalescent patient serum, CS1 | This paper | N/A |
| Convalescent patient serum, CS2 | This paper | N/A |
| Convalescent patient serum, CS3 | This paper | N/A |
| Convalescent patient serum, CS4 | This paper | N/A |
| Convalescent patient serum, CS6 | This paper | N/A |
| Convalescent patient serum, CS7 | This paper | N/A |
| Convalescent patient serum, CS8 | This paper | N/A |
| Convalescent patient serum, CS10 | This paper | N/A |
| Convalescent patient serum, CS86 | This paper | N/A |
| Convalescent patient serum, CS87 | This paper | N/A |
| Critical Commercial Assays | ||
| Britelite plus reporter gene assay system | PerkinElmer | Cat#6066769 |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
| 293T-hACE2 | Sino Biological Company | N/A |
| A549 | ATCC | Cat#CCL-185; RRID: CVCL_0023 |
| BHK21 | ATCC | Cat#CCL-10; RRID: CVCL_1915 |
| Cf2TH | ATCC | Cat#CRL-1430; RRID: CVCL_3363 |
| CHO | ATCC | Cat#CCL-61; RRID: CVCL_0214 |
| CRFK | ATCC | Cat#CCL-94; RRID: CVCL_2426 |
| DC2.4 | Millipore | Cat#SCC142; RRID: CVCL_J409 |
| HeLa | ATCC | Cat#CCL-2; RRID: CVCL_0030 |
| HepG2 | ATCC | Cat#HB-8065; RRID: CVCL_0027 |
| Huh-7 | JCRB | Cat#0403; RRID: CVCL_0336 |
| JASWII | ATCC | Cat#CRL-11904; RRID: CVCL_3727_ |
| K562 | ATCC | Cat#CCL-243; RRID: CVCL_0004 |
| LLC-MK2 | ATCC | Cat#CCL-7; RRID: CVCL_3009 |
| MDBK | ATCC | Cat#CCL-22;; RRID: CVCL_0421 |
| MDCK | ATCC | Cat#CCL-34; RRID: CVCL_0422 |
| MdKi | Laboratory of Dr. Zhengli Shi; Yang et al., 2019 | N/A |
| MRC-5 | ATCC | Cat#CCL-171; RRID: CVCL_0440 |
| Mv 1 Lu | ATCC | Cat#CCL-64; RRID: CVCL_0593 |
| PK15 | ATCC | Cat#CCL-33; RRID: CVCL_2160 |
| RAW264.7 | ATCC | Cat#TIB-71; RRID: CVCL_0493 |
| RlKiT | Laboratory of Dr. Zhengli Shi; Yang et al., 2019 | N/A |
| SK-N-MC | ATCC | Cat#HTB-10; RRID: CVCL_0530 |
| ST | ATCC | Cat#CRL-1746; RRID: CVCL_2204 |
| Vero | ATCC | Cat#CCL-81; RRID: CVCL_0059 |
| VeroE6 | ATCC | Cat#CRL-1586; RRID: CVCL_0574 |
| Oligonucleotides | ||
| VSV (P protein)-F:TCTCGTCTGGATCAGGCGG | GENEWIZ | N/A |
| VSV (P protein)-R: TGCTCTTCCACTCCA TCCTCTTGG |
GENEWIZ | N/A |
| Primers design for mutagenesis were located in Table S2 | N/A | N/A |
| Recombinant DNA | ||
| Plasmid:pcDNA3.1.S2 (codon-optimized S gene of SARS-CoV-2, GenBank: MN_908947) | Nie et al., 2020 | Addgene ID: 149457 |
| Plasmid:pcDNA3.1.S2-L5F | This paper | N/A |
| Plasmid:pcDNA3.1.S2-L8V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-L8W | This paper | N/A |
| Plasmid:pcDNA3.1.S2-H49Y | This paper | N/A |
| Plasmid:pcDNA3.1.S2-Y145del | This paper | N/A |
| Plasmid:pcDNA3.1.S2-Q239K | This paper | N/A |
| Plasmid:pcDNA3.1.S2-F338L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-V341I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-A348T | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N354D | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N354K | This paper | N/A |
| Plasmid:pcDNA3.1.S2-S359N | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D364Y | This paper | N/A |
| Plasmid:pcDNA3.1.S2-V367F | This paper | N/A |
| Plasmid:pcDNA3.1.S2-K378R | This paper | N/A |
| Plasmid:pcDNA3.1.S2-P384L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-385-387del | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D405V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-R408I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-Q409E | This paper | N/A |
| Plasmid:pcDNA3.1.S2-Q414E | This paper | N/A |
| Plasmid:pcDNA3.1.S2-Q414P | This paper | N/A |
| Plasmid:pcDNA3.1.S2-I434K | This paper | N/A |
| Plasmid:pcDNA3.1.S2-A435S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-S438F | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N439K | This paper | N/A |
| Plasmid:pcDNA3.1.S2-G446V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-L452R | This paper | N/A |
| Plasmid:pcDNA3.1.S2-K458R | This paper | N/A |
| Plasmid:pcDNA3.1.S2-K458N | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D467V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-I468F | This paper | N/A |
| Plasmid:pcDNA3.1.S2-I468T | This paper | N/A |
| Plasmid:pcDNA3.1.S2-I472V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-A475V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-G476S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-T478I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-V483A | This paper | N/A |
| Plasmid:pcDNA3.1.S2-V483I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-F490L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-P491R | This paper | N/A |
| Plasmid:pcDNA3.1.S2-V503F | This paper | N/A |
| Plasmid:pcDNA3.1.S2-Y508H | This paper | N/A |
| Plasmid:pcDNA3.1.S2-R509K | This paper | N/A |
| Plasmid:pcDNA3.1.S2-V510L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-H519Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-H519P | This paper | N/A |
| Plasmid:pcDNA3.1.S2-A520S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-P521S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-A522S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-A522V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G | This paper | N/A |
| Plasmid:pcDNA3.1.S2-V615L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-A831V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D839E | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D839Y | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D936Y | This paper | N/A |
| Plasmid:pcDNA3.1.S2-S943T | This paper | N/A |
| Plasmid:pcDNA3.1.S2-S943R | This paper | N/A |
| Plasmid:pcDNA3.1.S2-G1124V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-P1263L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-Y145del+R408I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+L5F | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+Q239K | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+Q321L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+V341I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+A435S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+K458R | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+I472V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+H519P | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+Q675H | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+A831V | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+A845S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+A879S | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+D936Y | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+S939F | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+S943T | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+M1229I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+M1237I | This paper | N/A |
| Plasmid:pcDNA3.1.S2-D614G+P1263L | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N17Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N61Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N74Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N122Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N149Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N165Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N234Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N282Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N331Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N343Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N603Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N616Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N657Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N709Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N717Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N801Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N1074Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N1098Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N1134Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N1158Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N1173Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N1194Q | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N74K | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N149H | This paper | N/A |
| Plasmid:pcDNA3.1.S2-T719A | This paper | N/A |
| Plasmid:pcDNA3.1.S2-N331Q+N343Q | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism version 8.0.1(244) | GraphPad Software | https://www.graphpad.com |
| Microsoft Office Home and Student 2019 | Microsoft Corporation | https://www.microsoft.com/microsoft-365 |
| Heatmap Illustrator (HemI) version 1.0.3.7 | Deng et al., 2014 | http://ccd.biocuckoo.org |
| BioEdit version 7.2 | BioEidt Software | https://bioedit.software.informer.com |
| Adobe Illustrator CC 2018 | Adobe | https://www.adobe.com |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Youchun Wang (wangyc@nifdc.org.cn).
Materials Availability
All the unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Models and Subject Details
Cell lines
Most cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM, high glucose; Hyclone, Cat#SH30243.01). They include 293T(Homo sapiens, embryonic kidney), 293T-hACE2 (293T cells stably expressed hACE2), A549 (Homo sapiens, liver), BHK21 (Mesocricetus auratus, kidney), Cf2TH (Canis familiaris, thymus), CHO (Cricetulus griseus, ovary), CRFK (Felis catus, kidney), HeLa (Homo sapiens, cervix), HepG2 (Homo sapiens, liver), Huh-7 (Homo sapiens, liver, gallbladder), LLC-MK2 (Macaca mulatta, kidney), MDBK (Bos taurus, kidney), MDCK (Canis familiaris, kidney), MRC-5 (Homo sapiens, lung), MV1-Lu (Neovison vison, lung), PK15 (Sus scrofa, kidney), RAW264.7 (Mus musculus, leukemia, ascites), SK-N-MC (Homo sapiens, brain), ST (Sus scrofa, testis), Vero (Cercopithecus aethiops, kidney), and VeroE6 (Cercopithecus aethiops, kidney) cells. DC2.4 (Mus musculus, bone marrow derived dendritic cells), K562 (Homo sapiens, bone marrow) and MdKi (Myotis davidii, kidney) and cells were incubated in RPMI medium modified (Hyclone, Cat#SH30809.01). RlKiT(R. leschenaultii, kidney) were cultured in DMEM/F-12, GlutaMAX (GIBCO, Cat#10565-018). JASWII (Mus musculus, bone marrow) were cultured in MEMα, nucleosides (GIBCO, Cat#12571063) with 5 ng/ml Recombinant Murine GM-CSF (Peprotech, Cat#315-03). All the cells were cultured in media supplemented with 100 U/mL of Penicillin-Streptomycin solution (GIBCO, Cat#15140163), 20mM N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid (HEPES, GIBCO, Cat#15630080) and 10% fetal bovine serum (FBS, Pansera ES, PAN-Biotech GmbH, Cat#ST30-2602), a humidified atmosphere with 5% CO2 at 37°C, with the exception of JASWII cells which were cultured in medium with 20% FBS. 0.25% Trypsin-EDTA (GIBCO, Cat#25200056) were used to detach cells for subculture.
Human sera
Human serum samples from 10 convalescent patients in Wuhan (CS1, CS2, CS3, CS4, CS6, CS7, CS8 and CS10) and Shandong (CS86 and CS87) were collected on Feb 18, 2020 and Mar 5, 2020. All volunteers signed informed consent forms.
Method Details
Site-directed mutagenesis
pcDNA3.1.S2 recombinant plasmid (GenBank: MT_613044), constructed by inserted the codon-optimized S gene of SARS-CoV-2 (GenBank: MN_908947) into pcDNA3.1 (Nie et al., 2020), was used as the template to generate the plasmid with mutagenesises in S gene. Following procedure of circular PCR, 15 to 20 nucleotides before and after the target mutation site were selected as forward primers, while the reverse complementary sequences were selected as reverse primers. Following site-directed mutagenesis PCR, the template chain was digested using DpnI restriction endonuclease (NEB, USA). Afterward, the PCR product was directly used to transform E. coli DH5α competent cells; single clones were selected and then sequenced. The primers designed for the specific mutation sites are listed in Table S2, and the frequency of different variants in the epidemic population is listed in Table S1.
Production and titration of pseudotyped viruses
Pseudotyped viruses incorporated with spike protein from either SARS-CoV-2, variants or mutants were constructed using a procedure described by us recently (Nie et al., 2020). On day before transfection, 293T cells were prepared and adjusted to the concentration of 5 −7 × 105 cell/ml, 15 mL of which were transferred into a T75 cell culture flask and incubated overnight at 37°C in an incubator conditioned with 5% CO2. The cells generally reach 70%–90% confluence after overnight incubation. Thirty microgram of DNA plasmid expressing the spike protein was transfected according to the user’s instruction manual. The transfected cells were subsequently infected with G∗ΔG-VSV (VSV G pseudotyped virus) at concentration of 7.0 × 104TCID50/ml. These cells were incubated at 37°C for 6-8 hours in the presence of in 5% CO2. Afterward, cell supernatant was discarded, followed by rinsing the cells gently with PBS +1% FBS. Next, 15ml fresh complete DMEM was added to the flask and cultured for 24 h. Twenty-four hours post infection, SARS-CoV-2 pseudotyped viruses containing culture supernatants were harvested, filtered (0.45-μm pore size, Millipore, Cat#SLHP033RB) and stored at −70°C in 2 mL aliquots until use.
The 50% tissue culture infectious dose (TCID50) of SARS-CoV-2 pseudovirus was determined using a single-use aliquot from the pseudovirus bank to avoid inconsistencies resulted from repeated freezing-thawing cycles.
For titration of the pseudotyped virus, a 2-fold initial dilution with six replicates was made in 96-well culture plates followed by serial 3-fold dilutions. The last column was employed as the cells control without pseudotyped virus. Subsequently, the 96-well plates were seeded with Huh-7 cells adjusted to 2 × 105cells/ml. After 24 h incubation at 37°C in a humidified atmosphere with 5% CO2, the supernatant was aspirated and discarded gently to leave 100 μL in each well; next, 100 μL of luciferase substrate (Perkinelmer, Cat#6066769) was added to each well. After 2-min incubation at room temperature in the dark, 150 μL of lysate was transferred to white 96-well plates for the detection of luminescence using a luminometer (PerkinElmer, Ensight). Positive was determined to be ten-fold higher than the negative (cells only) in terms of relative luminescence unit (RLU) values. The 50% tissue culture infectious dose (TCID50) was calculated using the Reed–Muench method (Nie et al., 2020).
Quantification of pseudotyped virus particles using RT-PCR
Before quantification, all the pseudotyped viruses were purified through a 25% sucrose cushion by ultra-centrifugation at 100,000 × g for 3 h (Nie et al., 2020). Viral RNA was extracted from 140 μL of purified pseudotyped viruses using the QIAamp Viral RNA Mini Kit (QIAGEN, Cat#52906), and served as template for reverse transcription using the SuperScript III First-Strand Synthesis System for RT-PCR kit reagent (Invitrogen, Cat#18080-051). Virus quantification by real-time PCR was performed using the TB Green Premix Ex TaqII (TaKaRa, Cat#RR820A), following the supplier’s instruction. The P protein gene of VSV virus was cloned into the vector pCDNA3.1(+) as a plasmid standard, with the viral copy number calculated accordingly. See primers in the Key Resources Table.
Infection assay
Using the quantitative RT-PCR, we normalized the pseudotyped virus particles to the same amount. After normalization, 100 μL of the pseudotyped virus with 10-fold dilution was added to wells in 96-well cell culture plate. After the cells were trypsin-digested, 2 × 104/100 μL cells were added to each well in the 96-well plates. The plates were then incubated at 37°C in a humidified atmosphere with 5% CO2. After incubation for 24 hours, chemiluminescence detection was performed as described in the titration of pseudotyped viruses. Each group contained 3-5 replicates.
Neutralization assay
The virus neutralization assay was conducted as described previously (Nie et al., 2020). Briefly, 100 μL serial dilutions of human sera or monoclonal antibody preparations were added into 96-well plates. After that, 50 μL pseudoviruses with concentration of 1300 TCID50/ml were added into the plates, followed by incubation at 37°C for 1 hour. Afterward, Huh-7 cells were added into the plates (2 × 104 cells/100 μL cells per well), followed by incubation at 37°C in a humidified atmosphere with 5% CO2. Chemiluminescence detection was performed after 24 hours incubation. The Reed-Muench method was used to calculate the virus neutralization titer. The results are based on 3-5 replicates unless specified. In order to validate the test operation process, the Coefficient of Variance (CV) control of replicates is set within 30% of six wells, so is the CV for the duplicate sample wells.
Quantification and Statistical Analysis
GraphPad Prism 8 was used for plotting and statistical analysis; the values were expressed as mean ± SEM. One-way ANOVA and Holm-Sidak’s multiple comparisons test was used to analyze the differences between groups. A p value of less than 0.05 was considered to be significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001, ns represents no significant difference.
Acknowledgments
We would like to thank Prof. X. Sunney Xie of Peking University for providing mAb X593; Prof. JinghuaYan of the Institute of Microbiology, Chinese Academy of Sciences for mAbs CA1 and CB6; Dr. Liangzhi Xie from Sino Biological Company for mAbs H014 and H00S022 and the 293T-hACE2 cell line; Prof. Linqi Zhang of Tsinghua University for mAbs P2C-1F11, P2B-2F6, AB35, 261-262, 157, and 247; and Prof. Yan Wu of the Institute of Microbiology, Chinese Academy of Sciences for H4 and B38. We would like to thank Dr. Xiaoming Yang from China National Biotec Group Company Ltd. for providing convalescent sera (CS1, CS2, CS3, CS4, CS6, CS7, CS8, and CS10) and Ms. Fangyu Dong from China Biologic Products Holdings, Inc. for convalescent sera CS86 and CS87. This work was supported by the National Science and Technology Major Projects of Drug Discovery (2018ZX09101001), the National Science and Technology Major Projects of Infectious Disease (2017ZX10304402), and the Bill and Melinda Gates Foundation (INV-006379).
Author Contributions
Y.W. and W.H. conceived, designed, and supervised the experiments. J.N., Li Zhang, Y.W., Xuguang Li, and W.H. wrote the manuscript. Q. Li, J.W., H.H., S.L., C.Z., H.L., H.Q., L.N., J.L., M.W., Q.S., Q. Lu, and Xiaoyu Li performed the experiments. Linqi Zhang and Q.Z. provided some mAbs and analyzed them. All of the authors have read and approved the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: July 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.07.012.
Supplemental Information
References
- Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020:e13525. doi: 10.1111/ijcp.13525. Published online May 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood A.A. Mutated COVID-19, May Foretells Mankind in a Great Risk in the Future. New Microbes New Infect. 2020;35:100673. doi: 10.1016/j.nmni.2020.100673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H., Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS ONE. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl W.E., Lin A.E., Grubaugh N.D., Carvalho L.M., Kim K., Kyawe P.P., McCauley S.M., Donnard E., Kucukural A., McDonel P. Ebola Virus Glycoprotein with Increased Infectivity Dominated the 2013-2016 Epidemic. Cell. 2016;167:1088–1098. doi: 10.1016/j.cell.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015;282:4679–4691. doi: 10.1111/febs.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16:e3000003. doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François K.O., Balzarini J. The highly conserved glycan at asparagine 260 of HIV-1 gp120 is indispensable for viral entry. J. Biol. Chem. 2011;286:42900–42910. doi: 10.1074/jbc.M111.274456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- Herfst S., Schrauwen E.J., Linster M., Chutinimitkul S., de Wit E., Munster V.J., Sorrell E.M., Bestebroer T.M., Burke D.F., Smith D.J. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Owino S.O., Crevar C.J., Carter D.M., Ross T.M. N-Linked Glycans and K147 Residue on Hemagglutinin Synergize To Elicit Broadly Reactive H1N1 Influenza Virus Antibodies. J. Virol. 2020;94:e01432-19. doi: 10.1128/JVI.01432-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 doi: 10.1038/s41586-020-2380-z. Published online May 26, 2020. [DOI] [PubMed] [Google Scholar]
- Kong L., Wilson I.A., Kwong P.D. Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262. Proteins. 2015;83:590–596. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 doi: 10.1016/j.cell.2020.06.043. Published online July 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Maurya V.K., Prasad A.K., Bhatt M.L.B., Saxena S.K. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virusdisease. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cleveland B., Klots I., Travis B., Richardson B.A., Anderson D., Montefiori D., Polacino P., Hu S.L. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 2008;82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M., Luo X., Estill J., Liu Y., Ren M., Wang J., Wang Q., Zhao S., Wang X., Yang S., On Behalf Of The Covid-Evidence And Recommendations Working Group Coronavirus disease (COVID-19): a scoping review. Euro Surveill. 2020;25:2000125. doi: 10.2807/1560-7917.ES.2020.25.15.2000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. bioRxiv. 2020 doi: 10.1101/2020.06.02.129098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning T., Nie J., Huang W., Li C., Li X., Liu Q., Zhao H., Wang Y. Antigenic Drift of Influenza A(H7N9) Virus Hemagglutinin. J. Infect. Dis. 2019;219:19–25. doi: 10.1093/infdis/jiy408. [DOI] [PubMed] [Google Scholar]
- Petrie J.G., Lauring A.S. Influenza A(H7N9) Virus Evolution: Which Genetic Mutations Are Antigenically Important? J. Infect. Dis. 2019;219:3–5. doi: 10.1093/infdis/jiy409. [DOI] [PubMed] [Google Scholar]
- Saha P., Banerjee A.K., Tripathi P.P., Srivastava A.K., Ray U. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201312. BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J.A., Singh J., Singh H., Jamal S., Khubaib M., Kohli S., Dobrindt U., Rahman S.A., Ehtesham N.Z., Hasnain S.E. Emerging genetic diversity among clinical isolates of SARS-CoV-2: Lessons for today. Infect. Genet. Evol. 2020;84:104330. doi: 10.1016/j.meegid.2020.104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2381-y. Published online May 26, 2020. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin K.A., Vanlandingham D.L., McGee C.E., Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz R.A., McClure C.P., Sakuntabhai A., Sall A.A., Kobinger G., Muller M.A., Holmes E.C., Rey F.A., Simon-Loriere E., Ball J.K. Human Adaptation of Ebola Virus during the West African Outbreak. Cell. 2016;167:1079–1087. doi: 10.1016/j.cell.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utachee P., Nakamura S., Isarangkura-Na-Ayuthaya P., Tokunaga K., Sawanpanyalert P., Ikuta K., Auwanit W., Kameoka M. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. J. Virol. 2010;84:4311–4320. doi: 10.1128/JVI.02619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Nie J., Prochnow C., Truong C., Jia Z., Wang S., Chen X.S., Wang Y. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology. 2013;10:14. doi: 10.1186/1742-4690-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zirkle B., Nie J., Ma J., Gao K., Chen X.S., Huang W., Kong W., Wang Y. N463 Glycosylation Site on V5 Loop of a Mutant gp120 Regulates the Sensitivity of HIV-1 to Neutralizing Monoclonal Antibodies VRC01/03. J. Acquir. Immune Defic. Syndr. 2015;69:270–277. doi: 10.1097/QAI.0000000000000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020 doi: 10.1126/science.abb9983. eabb9983. Published online May 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.L., Tan C.W., Anderson D.E., Jiang R.D., Li B., Zhang W., Zhu Y., Lim X.F., Zhou P., Liu X.L. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat. Microbiol. 2019;4:390–395. doi: 10.1038/s41564-018-0328-y. [DOI] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen S., Yang D., Wang X., Zhu J., Peng D., Liu X. Role of stem glycans attached to haemagglutinin in the biological characteristics of H5N1 avian influenza virus. J. Gen. Virol. 2015;96:1248–1257. doi: 10.1099/vir.0.000082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.