Highlights
-
•
Clinical performance of five different commercially available automated SARS-CoV-2 antibody tests.
-
•
No overlap of “false” positive samples between different serology assays was observed.
-
•
The ability to rule out acute SARS-CoV-2 infection at hospital admission with serology is limited.
Abbreviations: Ab, antibody; CLIA, chemiluminescence immunoassays; COVID-19, coronavirus disease 2019; Ct value, cycle threshold value; ECLIA, electrochemiluminescence immunoassay; ELISA, enzyme linked immunosorbent assay; Ig, Immunoglobulin; N-protein, nucleocapsid-protein; PPV, positive predictive value; RBD, receptor binding domain; SARS-CoV-2, sever acute respiratory syndrome coronavirus 2; S-protein, viral spike-protein; S1/S2, viral spike protein subdomains
Keywords: SARS-CoV-2, COVID-19, Serology
Abstract
Background
The global market for SARS-CoV-2-immunoassays is becoming ever more crowded with antibody-tests of various formats, targets and technologies, careful evaluation is crucial for understanding the implications of individual test results. Here, we evaluate the clinical performance of five automated immunoassays on a set of clinical samples.
Methods
Serum/plasma samples of 75 confirmed COVID-19 patients and 320 pre-pandemic serum samples of healthy blood donors were subjected to two IgG and three total antibody SARS-CoV-2-immunoassays. All test setups were automated workflows.
Results
Positivity of assays (onset of symptoms > 10 days) ranged between 68.4 % and 81.6 % (Diasorin 68.4 %, Euroimmun 70.3 %, Siemens 73.7 %, Roche 79.0 % and Wantai 81.6 %). All examined assays demonstrated high specificity of >99 % (Euroimmun, Diasorin: 99.1 %, Wantai: 99.4 %) but only two reached levels above 99.5 % (Roche: 99.7 %, Siemens 100 %). Interestingly, there was no overlap in false positive results between the assays. The strongest correlation of quantitative results was observed between the Diasorin and Euroimmun IgG tests (r2 = 0.76). Overall, we observed no difference in the distribution of test results between female and male patients (p-values: 0.18−0.87). A significant difference between severely versus critically ill patients was demonstrated for the Euroimmun, Diasorin, Wantai and Siemens assays (p-values: < 0.041).
Conclusion
All assays showed good clinical performance. Our data confirm that orthogonal test strategies as recommended by the CDC can enhance clinical specificity. However, the suboptimal rates of test positivity found at time of hospitalization in this cohort underline the importance of molecular diagnostics to rule out/confirm active infection with SARS-CoV-2.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody tests are now becoming increasingly available following vocal demand by clinicians and policy makers alike [1]. Given the rapid development and distribution of these assays, careful evaluation is required to avoid misinterpretation of results [[2], [3], [4]]. The majority of currently offered assays are based on detection of antibodies binding to the highly immunogenic viral spike(S)- or nucleocapsid(N)-proteins, the former of which holds the highest potential to convey neutralizing capacity, whereas the latter is the most abundant [5]. The semi-quantitative Euroimmun SARS-CoV-2 immunoglobin (Ig) G enzyme linked immunosorbent assay (ELISA) targeting the S1-Domain of the S-protein was one of the first assays to become commercially available, thus also featuring a comparably large body of available performance data in literature [[4], [5], [6]]. In contrast, the Wantai SARS-CoV-2 antibody (Ab) ELISA based on a double-antigen sandwich (S-protein, receptor binding domain (RBD)) potentially offers higher sensitivity due to its ability to also detect IgM and IgA [4]. More recently, high-throughput solutions such as the Diasorin LIAISON® SARS-CoV-2 S1/S2 IgG assay (spike subdomains RBD/S2, chemiluminescence immunoassays (CLIA)), the Roche Elecsys® Anti-SARS-CoV-2 (N-protein, electrochemiluminescence immunoassay (ECLIA)), and the Siemens Atellica® IM SARS-CoV-2 Total (RBD, CLIA) have entered the scene, allowing for faster turnaround times and easy expansion of testing capacity. The aim of this study was to evaluate and compare the clinical performance of five different commercially available automated SARS-CoV-2 antibody tests in an inpatient cohort with confirmed coronavirus disease 2019 (COVID-19) and archived blood donor samples from the pre-pandemic time acting as negative controls.
2. Materials and methods
2.1. Ethics, samples and RT-PCR
To assess positivity, the first serum/plasma sample available after hospitalization was analyzed (n = 75, clinical characteristics Table S1). The initial test consisted of a modified E-gene assay adapted as ‘cobas Omni Utility Channel’-protocol and performed on the cobas6800 system [7,8] (cycle threshold value (Ct value) < 34 was defined as positive). 66/75 patients showed positive PCR results in at least two independent samples, using the above-mentioned method or the Roche SARS-CoV-2 IVD-Test for the cobas6800 system. The remaining nine patients received positive PCR results from external (certified) diagnostic laboratories, these patients are marked by an arrow in Fig. 1 F. To analyse specificity, a set of anonymized retained samples of a pre-pandemic blood donor cohort (n = 320, equally distributed between the age of 18–70; m/f ratio 1:1, collected 01.03.17 – 09.04.17) was used. All samples were stored at −20 °C prior to analysis. The use of anonymized samples was approved by the local ethics committee. Assays were performed according to the manufacturers’ recommendations (Table 1 ). For details on study design and testing see supplementary materials.
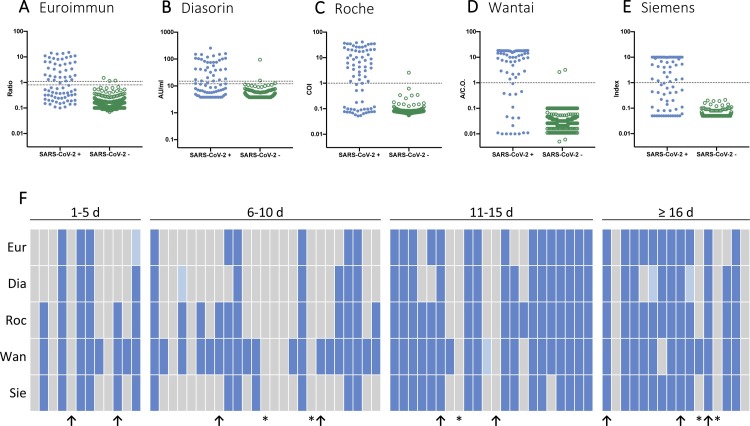
Fig. 1.
Test results of examined SARS-CoV-2 serology assays.
A-E The scatter plots with logarithmic scales display test results of SARS-CoV-2 PCR positive patients (blue dots, n = 75) and pre-pandemic blood donors (green dots, n = 320). Each dot represents one sample. Cut-off values for positivity are 1.1 for Euroimmun, 15 for Diasorin and 1 for Roche, Wantai and Siemens (upper dotted line). Cut-off values for borderline test results are 0.8 for Euroimmun and 12 for Diasorin (lower dotted line). F Test results of SARS-CoV-2 PCR positive patients (n = 75). Each column represents one patient. Samples are sorted by time since onset of symptoms in days. Negative test results are displayed in grey, positive test results in blue and borderline test results in light blue. Immunosuppressed patients are marked by a * in the bottom row and a ↑ in the bottom row indicates that RT-PCR was performed by an external certified lab.
Abbreviations: A/C.O., absorbance/cut-off value; AU/mL, arbitrary units/mL; COI, cut-off index (signal sample/cut-off); d, days; Dia, Diasorin; Eur, Euroimmun; Roc, Roche; Ratio, ratio (extinction sample/extinction calibrator); Sie, Siemens; Wan, Wantai.
Table 1.
Detailed information on SARS-CoV-2 serology assays.
| name | provider | platform | method | detection of Ab isotypes | antigen targeted | unit | cut-off for positivity | border-line | duration of test/batch | tests per h/run* |
sensitivity according to manufacturer | specificity according to manufacturer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Euroimmun | ||||||||||||
| Anti-SARS-CoV-2 ELISA (IgG) | EUROIMMUN AG, Lübeck, Germany | Euroimmun Analyzer I-2 P (EUROIMMUN AG, Lübeck, Germany) |
ELISA | IgG | S1-domain, spike protein (including the RBD) | Ratio | > 1.1 | 0.8 - 1.1 | 4.5 h | 180/run | 93.8 % (n = 16, ≥ 21 days since onset of symptoms) |
99.3 % (n = 150, German blood donors) |
| Diasorin | ||||||||||||
| LIAISON® SARS-CoV-2 S1/S2 IgG | DiaSorin S.p.A., Saluggia, Italy | LIAISON® XL Analyzer (DiaSorin S.p.A., Saluggia, Italy) | CLIA | IgG | S1 and S2 protein | AU/mL | > 15 | 12 - 15 | 35 min. | 180/h | 97.4 % (n = 39, > 15 days since onset of symptoms) |
98.5 % (n = 985, blood donors) |
| Roche | ||||||||||||
| Elecsys® Anti-SARS-CoV-2 | Roche Diagnostics Deutschland Gmbh, Mannheim, Germany | cobas e 411 (Roche Diagnostics Deutschland Gmbh, Mannheim, Germany) | ECLIA | total Ab | nucleocapsid protein | COI | > 1 | / | 18 min. | 75/h | 99.5 % (n = 185, ≥ 14 days since onset of symptoms) |
99.8 % (n = 4148, blood donors) |
| Wantai | ||||||||||||
| WANTAI SARS-CoV-2 Ab ELISA | Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China | Euroimmun Analyzer I-2 P (EUROIMMUN AG, Lübeck, Germany) |
ELISA | total Ab | recombinant SARS-CoV-2 antigen (RBD) | A/C.O. | > 1 | 0.9 - 1.1 | 4 h | 180/run | 94.5 % (n = 310, 1−39 days since onset of symptoms) |
100 % (n = 333, healthy individuals) |
| Siemens | ||||||||||||
| Atellica® IM SARS-CoV-2 Total (COV2T) | Siemens Healthcare Gmbh, Erlangen, Germany | Atellica® IM Analyzer (Siemens Healthcare Gmbh, Erlangen, Germany) | CLIA | total Ab | spike protein | Index | > 1 | / | 10 min. | 440/h | 100 % (n = 42, ≥14 days since onset of symptoms) | 99.8 % (n = 993, healthy individuals) |
* on the respective platform.
Abbreviations: Ab, antibody; A/C.0., absorbance/cut-off; AU/mL, arbitrary units/mL; CLIA, chemiluminescence immunoassay; COI, Cut-off index (sample signal/cut-off); ECLIA, electroluminescence assays; ELISA, enzyme-linked immunosorbent assay; h, hours; IgG, immunoglobin G, Ratio, ratio (extinction sample/extinction calibrators); max., maximum; min., minutes; n/a, data not available; No., number.
3. Results
3.1. Positivity and specificity
Overall, the Wantai ELISA showed the highest clinical sensitivity with a positivity rate of 81.6 % (CI: 66.6−90.8%) in patient samples collected > 10 days from the onset of symptoms, followed in order by the Roche ECLIA 79.0 % (CI: 63.7−88.9%), the Siemens CLIA 73.7 % (CI: 58.0−85.0%), the Euroimmun ELISA 70.3 % (CI: 54.2−82.5%) and the Diasorin CLIA 68.4 % (CI: 52.5−80.9%; Table 2 ). Out of all five immunoassays, the Wantai total Ig ELISA had the highest positivity rate of 8/12 (66.7 %) collected at day 1–5, 19/25 (76 %) at day 6–10 and 19/22 (86.4 %) at day 11–15 after symptom onset (Fig. 1 E, Table S2). In specimens collected more than 15 days after symptom onset, the Roche, Wantai and Siemens assays demonstrated the highest positivity rate of 12/16 (75 %) positive samples. (Fig. 1 , Table S2). A concordant positive test result in all five tests was observed in 31/75 (41.3 %) of samples, whereas in 15/75 (20 %) of samples, none of the immunoassays generated a positive result (this includes 4/5 immunocompromised patients). For test results of patients who tested negative in at least one assay (onset of symptoms > 10 days) see Table S3 (supplements). Using pre-pandemic samples as negative controls, all assays demonstrated low false positive rates of 3/320 (0.9 %, Euroimmun and Diasorin), 2/320 (0.6 %, Wantai), 1/320 (0.3 %, Roche) and 0/320 (0%, Siemens), amounting to a specificity of 99.1 %, 99.4 %, 99.7 % and 100 %, respectively (Table 2, Fig. 1). False positive test values were 1.14, 1.51 and 1.18 for Euroimmun (median of true positive: 5; range: 1–15), 15.9, 12.5 and 94.7 for Diasorin (median of true positive: 56.6; range: 12–256), 3.2 and 2.6 for Wantai (median of true positive: 15.6; range: 1–18.4) and 2.59 for Roche (median of true positive: 12.3, range: 1.2–41.4).
Table 2.
Positivity, specificity, NPV and PPV of examined SARS-CoV-2 serology assays.
| specificity |
positivity |
NPV |
PPV |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (n = 320) | 1−10 d (n = 37) | > 10 d (n = 38) | prevalence 0.8 % |
||||||||
| 1−10 d | > 10 d | 1−10 d | > 10 d | ||||||||
| % | 95 % CI | % | 95 % CI | % | 95 % CI | % | % | % | % | ||
| Euroimmun | |||||||||||
| 99.1 | (97.3−99.7) | 27.0 | (15.4−43.0) | 70.3 | (54.2−82.5) | 99.4 | 99.8 | 19.5 | 38.6 | ||
| Diasorin | |||||||||||
| 99.1 | (97.3−99.7) | 29.7 | (17.5−45.8) | 68.4 | (52.5−80.9) | 99.4 | 99.7 | 21.0 | 38.0 | ||
| Roche | |||||||||||
| 99.7 | (98.3−100) | 46.0 | (31.0−61.6) | 79.0 | (63.7−88.9) | 99.6 | 99.8 | 55.3 | 68.0 | ||
| Wantai | |||||||||||
| 99.4 | (97.8−100) | 73.0 | (57.0−84.6) | 81.6 | (66.6−90.8) | 99.8 | 99.9 | 49.5 | 52.3 | ||
| Siemens | |||||||||||
| 100 | (98.8−100) | 35.1 | (21.8−51.2) | 73.7 | (58.0−85.0) | 99.5 | 99.8 | 100 | 100 | ||
To determine positivity rates and specificity, samples from SARS-CoV-2 PCR positive patients (n = 75) collected in March and April of 2020 and samples from pre-pandemic blood donations made in March of 2017 (n = 320) were tested with each assay. NPV and PPV were calculated for a prevalence of 0.8 % according to our local currently low prevalence setting (Hamburg, Germany). Patients were grouped according to the time between onset of symptoms and date of blood sampling in days (1−10 days and >10 days).
Abbreviations: CI, confidence interval; d, days; NPV, negative predictive value; PPV, positive predictive value.
3.2. Distribution and correlations
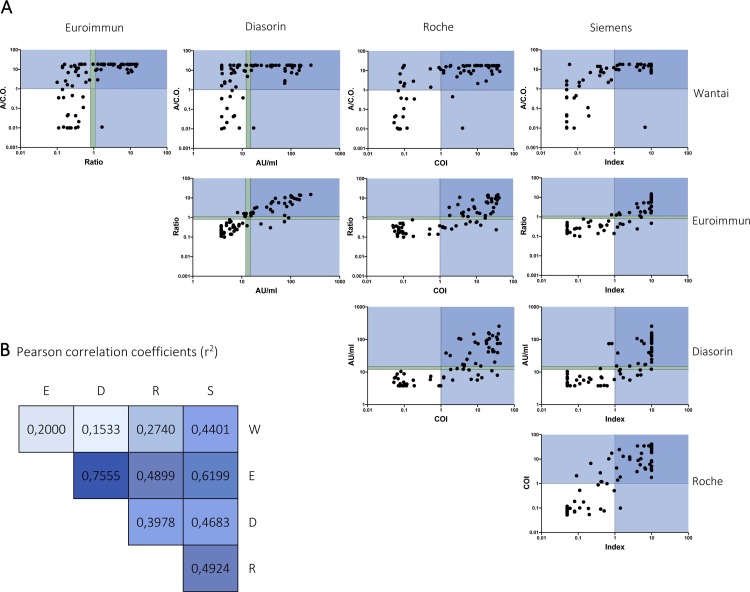
To analyze possible gender differences in antibody responses, we compared the distribution of test results between female and male COVID-19 patients for each assay, but no significant differences were observed (p-values ranging from 0.18 to 0.87). For comparison of quantitative responses, we conducted a side-by-side comparison of all test results in the SARS-CoV-2 PCR-positive patient cohort (Fig. 2 ). As expected, the strongest correlation was observed between the two IgG assays by Euroimmun and Diasorin with an r2 value of 0.76. The second strongest correlation was demonstrated between the Euroimmun and Siemens assays with a r2 value of 0.62. Neither assay correlated well with the results of the total antibody assay from Wantai (r2 values <0.45). Comparison between disease severity and test results showed a significant difference between severely and critically ill COVID-19 patients for the Euroimmun, Diasorin, Wantai and Siemens assays (p-values < 0.041; Figure S1 supplements).
Fig. 2.
Correlation between SARS-CoV-2 serology assays.
A Scatter plots with logarithmic scales, each displaying the correlation between two tests. Test results of SARS-CoV-2 PCR positive patients (n = 75) are plotted. Each dot represents one sample. Dotted lines indicate assay cut-offs for positivity (upper line) and borderline test results (lower line). Values in dark blue areas tested positive in both assays and values in light blue areas in one assay. B Person correlation coefficients (r2) arranged in the same format as in A. Values are color-coded, with values close to 1 displayed in a dark blue and values close to 0 in a light blue. All correlations are significant (p-value: ≤0.0003).
Abbreviations: A/C.O., absorbance/cut-off value; AU/ml, arbitrary units/mL; COI, cut-off index (signal sample/cut-off); D, Diasorin; E, Euroimmun; R, Roche; Ratio, ratio (extinction sample/extinction calibrator); S, Siemens; W, Wantai.
4. Discussion
Here, we present the performance characteristics of five commercial high-throughput SARS-CoV-2 immunoassays in samples from a COVID-19 inpatient cohort and negative controls consisting of pre-pandemic blood donor samples. All assays demonstrated good specificity above 99 % with the Roche and Siemens assay reaching 99.7 % and 100 % respectively. Nonetheless, for clinical use the prevalence in the respective patient’s population is critical. In a low prevalence environment such as Germany (seroprevalence 0.8 %–1 % (9)), the positive predictive values determined in our study ranged from 19.5 %–100 % and 38.0 %–100 % (for ≤ 10 days or >10 days after start of symptoms, respectively (see also Table 2). Interestingly we observe no overlap of false positive results between the different assays. Therefore, our data support the current recommendation of the CDC for low prevalence settings (<5% prevalence) to use a second serology assay to increase the PPV above 90 %. The overall assay positivity rates in this study are lower than reported by the respective manufacturers, but test results reported here fall in line with other clinical evaluation studies [4,5,9]. A major limitation of this study is that no longitudinal sera were analyzed so we cannot exclude that some patients might have seroconverted at later time points. According to currently available literature, emergence of S-protein antibodies is usually preceded by those against N-protein [10,11], however in our cohort the S-protein based ELISA by Wantai was the most sensitive assay followed by the N-protein based Elecsys assay (Roche) with 79 % vs. 81.6 % positivity (> 10 days after onset of symptoms), respectively.
As expected, assays detecting IgG, IgM and IgA had a higher sensitivity in our cohort than tests detecting only IgG [9]. Overall, there was good qualitative agreement between all assays, but moderate quantitative correlation (r2 = 0.76) was only observed between the two assays detecting specifically IgG (Euroimmun and Diasorin, Fig. 2). Regarding gender specific differences in immune response, we observed no disparity in number of positive samples or titers of SARS-CoV-2 antibodies in any of the five assays, but critically ill patients had significantly higher IgG antibody responses compared to patients with mild/severe disease course in accordance to results from other cohorts [12]. Overall, the suboptimal rate of test positivity found at time of hospitalization in this cohort underline the importance of molecular diagnostics to rule out/confirm active infection with SARS-CoV-2.
Funding
The research was partially funded by the BGV (Behörde für Gesundheit und Verbraucherschutz der Freien und Hansestadt Hamburg) of the city of Hamburg. ML, MMA, SuP, TTB, AWL and JSzW are funded by the German Center for Infection Research (DZIF). JSzW, ML and LSP are funded by German Research Foundation (DFG, SFB841).
CRediT authorship contribution statement
Lisa Sophie Pflüger: Conceptualization, Writing - original draft, Visualization, Investigation. Johannes H. Bannasch: Conceptualization, Writing - original draft, Visualization, Investigation. Thomas Theo Brehm: Data curation, Writing - original draft, Visualization, Investigation. Susanne Pfefferle: Writing - review & editing. Armin Hoffmann: Writing - review & editing. Dominik Nörz: Writing - review & editing. Marc van der Meirschen: Writing - review & editing, Resources. Stefan Kluge: Writing - review & editing, Resources. Munif Haddad: Writing - review & editing, Resources. Sven Pischke: Writing - review & editing. Jens Hiller: Writing - review & editing, Resources. Marylyn M. Addo: Writing - review & editing, Resources. Ansgar W. Lohse: Writing - review & editing, Resources. Julian Schulze zur Wiesch: Writing - review & editing, Resources. Sven Peine: Resources, Writing - review & editing. Martin Aepfelbacher: Writing - review & editing, Resources. Marc Lütgehetmann: Conceptualization, Data curation, Writing - original draft, Supervision.
Declaration of Competing Interest
Marc Lütgehetmann has received travel expenses and speakers’ honoraria (Roche Diagnostics, Diasorin, Biomerieux). The other authors declare that they have no conflict of interest.
Acknowledgements
None.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104549.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bryant J.E., Azman A.S., Ferrari M.J., Arnold B.F., Boni M.F., Boum Y. Serology for SARS-CoV-2: apprehensions, opportunities, and the path forward. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc6347. [DOI] [PubMed] [Google Scholar]
- 2.Theel E.S., Slev P., Wheeler S., Couturier M.R., Wong S.J., Kadkhoda K. The role of antibody testing for SARS-CoV-2: is there one? J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FDA . US Food & Drug Administration Website; 2020. EUA Authorized Serology Test Performance. [Google Scholar]
- 4.Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C., Fomsgaard A., Krogfelt K.A. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020;2020 04.09.20056325. [Google Scholar]
- 5.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beavis K.G., Matushek S.M., Abeleda A.P.F., Bethel C., Hunt C., Gillen S. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25(9) doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plebani M., Padoan A., Negrini D., Carpinteri B., Sciacovelli L. Diagnostic performances and thresholds: the key to harmonization in serological SARS-CoV-2 assays? Clin. Chim. Acta. 2020 doi: 10.1016/j.cca.2020.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. medRxiv. 2020 [Google Scholar]
- 11.Guo L., Ren L., Yang S., Xiao M., Chang Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long Q.-X.-X., Liu B.-Z.-Z., Deng H.-J.-J., Wu G.-C.-C., Deng K., Chen Y.-K.-K. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.