Abstract
Background
The rapid spread of novel coronavirus called SARS-CoV-2 or nCoV has caused countries all over the world to impose lockdowns and undertake stringent preventive measures. This new positive-sense single-stranded RNA strain of coronavirus spreads through droplets of saliva and nasal discharge.
Purpose
US FDA has authorized the emergency use of Remdesivir looking at the increasing number of cases of COVID-19, however there is still no drug approved to treat COVID-19. An alternative way of treatment could be the use of naturally derived molecules with known antiviral properties.
Method
We reviewed the antiviral activities of two polyphenols derived from tea, epigallocatechin-3-gallate (EGCG) from green tea and theaflavins from black tea. Both green tea and black tea polyphenols have been reported to exhibit antiviral activities against various viruses, especially positive-sense single-stranded RNA viruses.
Results
Recent studies have revealed the possible binding sites present on SARS-CoV-2 and studied their interactions with tea polyphenols. EGCG and theaflavins, especially theaflavin-3,3′-digallate (TF3) have shown a significant interaction with the receptors under consideration in this review. Some docking studies further emphasize on the activity of these polyphenols against COVID-19.
Conclusion
This review summarizes the available reports and evidences which support the use of tea polyphenols as potential candidates in prophylaxis and treatment of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Tea polyphenols, EGCG, Theaflavin
Abbreviations: 2′-O-MTase, 2′-O-methyltransferase; 3CLpro, Chymotrypsin-like protease; ACE2, Angiotensin-converting enzyme 2; CTD, C-terminal domain; E, Envelope protein; EGCG, Epigallocatechin Gallate; GRP78, Glucose-regulated Proteins; HCV, Hepatitis C virus; HE, Hemagglutinin Esterase; HIV, Human immunodeficiency virus; M, Membrane protein; MERS-CoV, Middle East Respiratory Syndrome Coronavirus; Mpro, Main protease; N, Nucleocapsid protein; Nsp, Non-structural proteins; NTD, N-terminal domain; PLpro, Papain like protease; PRRSV, Porcine reproductive and respiratory syndrome virus; RBD, Receptor-binding domain; RdRp, RNA dependant RNA polymerase; S, Spike protein; SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; TF, Theaflavin; TF2A, Theaflavin-3-gallate; TF2B, Theaflavin-3′-gallate; TF3, Theaflavin-3,3′-digallate; WNV, West Nile Virus
Graphical abstract
Introduction
COVID-19 is a viral disease that affects the epithelial cells of the respiratory system and causes inflammation of the mucosal membrane. This leads to alveolar damage and eventually pneumonia. It is caused by SARS-CoV-2, commonly known as novel coronavirus, a positive-sense single-stranded RNA virus. Earlier, coronaviruses have been reported to cause severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) (Xie and Chen, 2020). Amongst the drugs being tested in clinical trials, hydroxychloroquine, favipiravir, remdesivir, and lopinavir/ritonavir have gathered a lot of attention (Abd El-Aziz and Stockand, 2020). Several other antiviral drugs and new chemical molecules are also being tested for treatment against coronavirus. A few dietary molecules with previously established antiviral activities are also among the candidates evaluated for COVID-19 treatment.
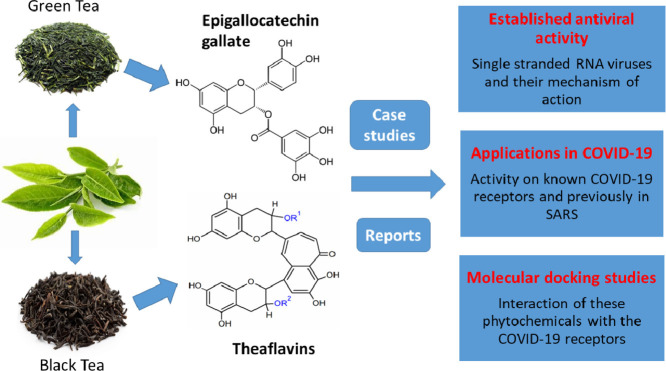
In this article, we will review the activity of phytochemicals derived from green tea and black tea. Irrespective of the type of tea, a typical procedure for production of green tea and black tea involves 5 general processes. Initially, the leaves are withered where the moisture content in the tea leaves is minimized. Then these leaves are fixed by enzymatically browning them and are allowed to get oxidized by exposing to oxygen. Finally, these leaves are rolled to give them a suitable shape and then dried to enhance the shelf-life (Tea Processing Steps, 2017). Epigallocatechin gallate (EGCG) is one of the most abundant polyphenolic catechin found in Camellia sinensis (L.) Kuntze (tea plant), especially in green tea. EGCG has been tested for its antiviral activity against several viruses and found to be a potential treatment option over synthetic chemical drugs. It is recognized as a multi-functional bioactive molecule exhibiting antitumorigenic, anti-inflammatory, antibacterial, antioxidative, and antiproliferative properties in addition to its antiviral effects (Chacko et al., 2010). Theaflavins (TFs) are another class of polyphenols found in abundance in black tea. The TF derivatives found majorly in black tea are theaflavin (TF1), theaflavin-3-gallate (TF2A), theaflavin-3′-gallate (TF2B), and theaflavin-3,3′-digallate (TF3). All these TFs are being researched for their bioactive properties and are known for their broad-spectrum biological properties, like anti-tumor, anti-viral, anti-inflammatory, anti-oxidative, and anti-bacterial properties (Higdon and Frei, 2003; Lambert and Yang, 2003; Yang and Landau, 2000). Since EGCG and TFs are of natural sources and consumed regularly by a majority of the population, investigating the anti-viral properties of these polyphenols against COVID-19 could be an advancement in finding a treatment to this latest pandemic.
Structure of SARS-CoV-2
SARS-CoV-2 has a significant resemblance with SARS-CoV and MERS-CoV with the percentage resemblance to the genome being 79% and 50%, respectively (Lu et al., 2020). There are also studies showing that SARS-CoV and SARS-CoV-2 have nucleotide similarity of 89.1% and nucleotide identity of 80% (Wu et al., 2020b; Zhou et al., 2020a). The virus has four main structural proteins, called spike protein (S), an envelope protein (E), membrane protein (M), and nucleocapsid protein (N), along with other non-structural proteins (Nsps) (Bosch et al., 2003). Another glycoprotein exists on the structure of β-strands known as hemagglutinin esterase (HE) (Hilgenfeld, 2014). The SARS-CoV-2 virus is known to infect the host cell via binding to the angiotensin-converting enzyme (ACE2) with the help of its S proteins (Zhou et al., 2020b). The S protein has two subunits, named as S1 and S2. The S1 subunit is known to attach to the receptor on the cell i.e. ACE2 with the help of its receptor-binding domain (RBD) (Du et al., 2009). The S2 subunit is involved in the membrane fusion of the virus and host cell. The E protein also has two domains, a hydrophobic domain and a charged cytoplasmic tail (Venkatagopalan et al., 2015). It is known to be functional in viral assembly by the formation of ion channels which help in coordinating with other viral proteins (Arbely et al., 2004; Venkatagopalan et al., 2015). The M protein has three transmembrane domains with long C-terminals and short N-terminals (Ruch and Machamer, 2012). Its major function is to maintain the viral envelope by interaction with other viral proteins by stabilizing the N protein (Schoeman and Fielding, 2019). The N protein plays an important role in RNA binding in the viral replication cycle. It forms a ribonucleoprotein complex with the help of its N-terminal domain (NTD) and performs dimerization using the C-terminal domain (CTD) (Fehr and Perlman, 2015). This protein has three domains, known as N-arm, C-tail, and the central linker region. It also performs roles like inhibition of host cell protein translation, alteration of host cell metabolism, and cell apoptosis (Hilgenfeld, 2014; McBride et al., 2014).
Active binding sites for SARS-CoV-2 and other targets
There are many binding sites present on the SARS-CoV-2 that might be the potential druggable targets. The major binding sites being investigated in COVID-19 are discussed in the following section.
Chymotrypsin-like protease (3CLpro, PDB ID: 6LU7)
3CLpro, also known as main protease (Mpro) or Nsp5 is an important enzyme found in SARS-CoV that is responsible for proteolytic function in the maturation stage of the virus. It is found to cleave at least 11 sites on polyprotein-1a and polyprotein-1ab found in the viral genome to cleave Nsp4-Nsp16 (Hegyi et al., 2002; Yang et al., 2005). These non-structural proteins namely RNA dependant RNA polymerase (RdRp), helicase, endonuclease, exonuclease, and 2′-O-methyltransferase (2′-O-MTase) are very important proteins in the viral genome (Benkert et al., 2011; Yang et al., 2003). Hence, 3CLpro is the most important target for drugs against coronaviruses (Chen et al., 2014a). Small molecules and peptide inhibitors are expected to inhibit 3CLpro activity (Lu et al., 2020).
RNA dependant RNA polymerase (RdRp, PDB ID: 6NUR)
RdRp (Nsp12) is the crucial enzyme that has an important role in the replication and transcription of the virus. With the help of its other co-factors Nsp7 and Nsp8, the interaction between Nsp12 and the RNA is strengthened, thereby enhancing the RdRp activity (Imbert et al., 2006). Hence, the Nsp12-RdRp is an important drug target for treatment against COVID-19. The RdRp domain of the protein is at the C-terminus with a Ser-Asp-Asp motif (Subissi et al., 2014).
Hemagglutinin esterase (HE, PDB ID: 3CL5)
HE is a structural protein present specifically in β-coronaviruses and is recognized as one of its markers. It acts both as a receptor-destroying enzyme and a lectin that attaches to the O-acetylated-sialic-acids (Zeng et al., 2008). HE-sialic acid complex (PDB ID: 3CL5) is thus another druggable target in COVID-19 (Prajapat et al., 2020).
Papain like protease (PLpro, PDB ID: 4OW0)
PLpro cleaves the N-terminus of the polyprotein to generate Nsps; Nsp1, Nsp2, and Nsp3 by cleaving them at their boundaries with the help of 3CLpro (Harcourt et al., 2004). PLpro is also crucial in alienating the innate immunity of the host cell (Chen et al., 2014b; Yuan et al., 2015). Due to its important function in the viral replication cycle, PLpro is another important druggable target (Wu et al., 2020a).
2′-O-methyltransferase (2′-O-MTase, PDB ID: 3R24)
2′-O-MTase (Nsp16) and S-adenosylmethionine (SAM) dependent nucleoside perform the role of methylating the ribose 2′-O position of the first two nucleosides of mRNA of the virus, thereby camouflaging and protecting the virus from the immunity system of the host cell (Menachery et al., 2014). 2′-O-MTase gets activated on binding with the Nsp10. Hence, Nsp10-Nsp16 complex (PDB: 3R24) is a druggable target owing to its indispensable role in viral replication (Khan et al., 2020).
Helicase (PDB ID: 6JYT)
Helicase (Nsp13) has the function of unwinding the double-stranded oligonucleotides in an NTP-dependant manner in the 5′−3′ direction (Harcourt et al., 2004). It has a metal-binding domain at N-terminal and a helicase domain (Wu et al., 2020a). Nsp 13 is another important target to develop a treatment method against coronaviruses (Kong et al., 2020).
Spike RBD (PDB ID: 6LXT)
RBD is the major binding domain of the S protein located on the S1 subunit. It complexes with the ACE2 receptor on the host cells. A total of 18 residues of ACE2 attach with 14 amino acids of RBD, the most important being K341 of ACE2 with the R453 of the RBD (Prabakaran et al., 2004). The binding activity of the RBD can be altered if there is a mutation on D454 or R441 residues of the RBD (Du et al., 2009). Any alteration in the structural proteins of RBD could be an effective prophylactic measure and inhibition of RBD-ACE2 complex can be another promising treatment strategy (Prajapat et al., 2020).
Structural proteins (S-PBD ID: 3SCI, E-PDB ID: 5X29, N-PDB ID: 1SSK, 2CJR)
Apart from RBD, there is another druggable target on the S protein present on the S2 subdomain. HR1/HR2 domains are observed in the post-fusional state of the virus which are typically 6-helices complex (PDB ID: 1WYY). This complex can be targeted for prophylactic action (S et al., 2005). The M protein needs to form a complex with S and N protein to perform various vital functions in the viral assembly (Arbely et al., 2004). Two pathways are activated by the M protein, namely the kappa pathway and IFN-β pathway. The other two major structural domains are NTD and CTD of the N protein (Chang et al., 2016). The NTD has multiple sites for RNA binding which can be targets for specifically designed inhibitors/proteins/peptides that compete with CTD to inhibit the oligomerization process (Lin et al., 2014; Roh, 2012).
ACE2 receptor (PDB ID: 6ACK)
ACE2 receptor is the primary binding site of the S protein which facilitates viral entry (Zhou et al., 2020b). The affinity of SARS-CoV-2 S protein to the ACE2 receptor is higher than that of SARS-CoV (Wrapp et al., 2020). The ACE2 receptor and the ACE2-RBD (PDB ID: 6VW1) complex are thus the two important sites for prophylaxis of COVID-19 (Wu et al., 2020a).
Glucose-regulated proteins (GRP78, PDB ID: 3LDL)
GRP78 also known as Binding immunoglobulin protein (BiP) or heat shock 70 kDa protein 5 (HSPA5), is a chaperone protein found in the lumen of the endoplasmic reticulum (ER). Its function is to prevent the unfolding of proteins that get translocated into the ER. In conditions where the unfolded proteins get accumulated above a threshold, GRP78 releases enzymes like activating transcription factor 6, inositol-requiring enzyme 1, and protein kinase RNA-like endoplasmic reticulum kinase, which enhance the protein folding and inhibit protein synthesis (Shen et al., 2002). GRP78 is overexpressed under cell stress and translocates from the ER to the plasma membrane, where it can be recognized as a receptor to mediate viral infection with the help Pep42 protein of the virus-cell (Ibrahim et al., 2019). Ibrahim et al. showed the interactions of the S protein of SARS-CoV-2 with the GRP78 and concluded that the S protein GRP78 binding site is a potential druggable target for treatment of COVID-19 (Ibrahim et al., 2020).
Antiviral properties of EGCG
Various studies have been done for discovering the antiviral activity of tea polyphenols, especially EGCG. Table 1 gives an overview of some studies performed in ssRNA.
Table 1.
Summary of known antiviral activity of EGCG.
| Virus | Genome | Conclusion | Reference |
| Porcine reproductive and respiratory syndrome virus (PRRSV) | ssRNA | EGCG effectively inhibits PRRSV infection and replication in porcine alveolar macrophages. It prevents MARC-145 cells from getting infected from PRRSV | Ge et al. (2018) |
| Hepatitis C virus (HCV) | ssRNA | EGCG prevents infection by inhibiting the entry of HCV into hepatoma cell lines and primary human hepatocytes thus | Ciesek et al. (2011) |
| Human immunodeficiency virus (HIV) | ssRNA | EGCG interferes with the interaction of host cell receptors and virus envelope and inhibits the entry of the virus into target cells | Liu et al. (2005) |
| Zika virus | ssRNA | Cells pre-treated with EGCG showed no virus infection | Carneiro et al. (2016) |
| Chikungunya | ssRNA | The entry, replication, and release were inhibited of CHIKV in vitro by EGCG | Lu et al. (2017) |
| West Nile Virus (WNV) | ssRNA | EGCG has a direct effect on the WNV when treated at early stages of the infection | Carneiro et al. (2016) |
| Dengue | ssRNA | EGCG directly interacts with the virus molecule causing virus deformation and thus preventing the virus from infecting further cells | Raekiansyah et al. (2018) |
| Influenza A/H1N1, A/H3N2, B | ssRNA | EGCG inhibits acidification of intracellular endosome compartments essential in the fusion of membranes of virus and host cell | Song et al. (2005) |
| Rotavirus | ssRNA | EGCG reduces the reactive oxygen species produced and prevents infection | Ho et al. (2009) |
| Ebola | ssRNA | EGCG inhibits HSPS5 protein of the host which is the target of Ebola virus treatments thus reducing the virus multiplication once infected | Reid et al. (2014) |
| Murine norovirus and feline calicivirus | ssRNA | EGCG at 100 μM was found out to be the most potent prophylactic agent when compared with other flavonoids | Seo et al. (2016) |
EGCG was found to inhibit Porcine reproductive and respiratory syndrome virus (PRRSV) infection irrespective of whether it was administered before or after infection; a concentration of 125 μM was enough to completely inhibit the infectivity of viral cells (Ge et al., 2018). In Hepatitis C virus (HCV) infection, EGCG was found out to inhibit the infection by attaching to the target cell and preventing the spread of the infection to different cells (Calland et al., 2012). It was also proposed that EGCG inhibits the entry of the ZIKA virus by interacting with the lipid bilayer. A significant point to be noted is that an in vivo study showed that ECGC can cross the placental barrier and spread to the brain, eyes and heart of the foetus, hence making its administration effective in pregnant women and possibly the foetus also (Chu et al., 2006). EGCG was found to inhibit not only the intracellular chikungunya virus replication but also the extracellular infection in the pre and post stages of viral infection (Lu et al., 2017). EGCG is expected to inhibit the intermediate stages of the influenza virus cycle apart from the known mechanism of affecting the virus attachment by inhibiting hemagglutination activity (Song et al., 2005). EGCG has also been found effective against HIV-1 and inhibits viral replication by acting at various stages. It blocks interaction of gp120 with CD4 by interfering with reverse transcriptase (Sodagari et al., 2016). A report suggested that EGCG inhibits the production of p24 antigen on isolated CD4 receptor cells, macrophages and CD4±T cells depending on its dose (Nance et al., 2009). Another report suggested that even at concentrations obtained by consumption of green tea, EGCG is seen effective in inhibiting gp120-CD4 attachment (Williamson et al., 2006).
As evident from the mechanisms of action of EGCG in various viruses, it is a wide spectrum antiviral agent with its mechanism differing from infection to infection. The difference in the antiviral activity can be attributed to the number of hydroxyl groups present on the benzene ring and galloyl group, together with the pyrogallol group which is responsible for exhibiting diverse mechanisms (Xu et al., 2017).
Antiviral properties of theaflavins
Table 2 gives a summary of few reports on antiviral activity of TFs
Table 2.
Summary of known antiviral activity of TFs.
| Virus | Genome | Conclusion | Reference |
| Sindbis Virus | ssRNA | TFs extract inhibited the viral infection by 99% at a concentration of 14.6mM | Villagomez (2017) |
| TMV | ssRNA | TFs interfered with the viral replication cycle by attachment to TMV-RNA complex | Okada et al. (1977) |
| Influenza A and B | ssRNA | TF1 had an IC50 value of 16.21 μg/ml against the virus, which is the best among 13 flavonoids studied using cytopathic effect inhibition assay. | Yang et al. (2014) |
| HSV | dsDNA | TF3 in combination with acyclovir showed an increased 21.8% inhibition than acyclovir alone in WST-1 assay | Berkefeld (2015) |
| Rotavirus and coronavirus | ssRNA | Synergistic activity when all TFs were administered in-vitro | Clark et al. (1998) |
| HCV | ssRNA | Act directly on the viral particles and inhibit the ability to bind to receptor surface | Chowdhury et al. (2018) |
| Caliciviruses | ssRNA | Best in-silico antiviral activity among 2080 small molecules screened. | Ohba et al. (2017) |
| HIV-1 | ssRNA | TF3 inhibits the entry of virus by targeting gp41. | Jie et al. (2011) |
Like EGCG, antiviral properties of theaflavin polyphenols and their derivatives have also been explored in several viral diseases. In a study on HSV-1, among all the theaflavins tested individually, TF1, TF2, and TF3 showed strong inhibition of the viral lytic cycle with TF3 being the most potent. It was shown that there was more than 99% inhibition of the infectivity of viral molecules when they were treated with TF3 at 50 μM for 1 h, thus proving its prophylactic antiviral activity (de Oliveira et al., 2015). In another study, TF2 was found to exhibit its anti-inflammatory effects in vivo by suppressing the levels of COX-2, TNF-A, 1CAM-1, and NFkB mRNA. Also, the radical quenching ability of TF3 was observed with the induction of mitochondria hyperpolarization which indicated activation of apoptotic signaling (Gosslau et al., 2011). A study on the influenza virus also showed the antiviral activity of TFs via neuraminidase inhibition; the activity of TF3 was better than other derivatives and almost comparable to the control oseltamivir carboxylate (Zu et al., 2012). TF3 was also seen to prevent the virus from adsorbing into the MDCK cells hence inhibiting the hemagglutinin of the virus (Nakayama et al., 1993). TF2B was found to be the most potent among all the other tea polyphenols in the inhibition of HIV at a concentration of 1 μM and a selectivity index greater than 200. The number of galloyl groups on the TF had a direct relation on its activity and it is estimated that these molecules interact with the gp41 six-helix bundle to prevent viral entry into the host cell (Liu et al., 2005). At a relatively higher concentration, TFs were also found to inhibit reverse transcriptase in HIV (Jie et al., 2011). TFs were found to have best in-silico antiviral activity among 2080 small molecules against caliciviruses. The study also proposed that hydroxyl groups in TFs are more essential in exhibiting the activity over the galloyl groups (Ohba et al., 2017). A study reported that TFs show activity directly on the HCV viral particles to inhibit their binding to the surface of the receptor. It highlighted that TF3 exhibited better anti-HCV activity than TF1 and TF2 (Chowdhury et al., 2018).
Application in COVID-19
Inhibition of 3CLpro
In a study investigating phytochemical mediated inhibition of SARS-CoV 3CLpro, molecular docking and in vitro studies were performed to screen different naturally obtained flavonoids. It was observed that EGCG with a docking score of −11.7, showed the best in silico activity among all the compounds tested. In vitro studies showed that EGCG exhibited 85% inhibition of 3CLpro at a concentration of 200 μM and had an IC50 value of 73 ± 2 μM (Thi Thanh Hanh et al., 2012). SARS-CoV 3CLpro was also studied by Singh and Konwar et al. with a remark that the inhibiting activity could be further improved by finding analogues of EGCG with higher bioavailability (Singh and Konwar, 2013). Tahir ul Qamar et al. showed that the 3CLpro structure of SARS-CoV-2 is highly similar to that of SARS-CoV, suggesting the validity of earlier studies in the present outbreak (Tahir Ul Qamar et al., 2020). Recently, another study recommended EGCG as one of the most potential inhibitor of 3CLpro among the phytochemicals found in nature on basis of molecular docking studies (Khaerunnisa et al., 2020). Hence on these foundations, we can expect EGCG to be a potential inhibitor of 3CLpro for the treatment of COVID-19. It was also found in a study on SARS-CoV that TF1, TF2a, TF2b, and TF3 showed higher potency in 3CLpro inhibition than other catechins including EGCG at IC50 less than 10 μM. This could be because of the unstable nature of EGCG and the other catechins. Among the TFs, TF2b and TF3 showed better 3CLpro inhibition activity because of the presence of the gallate group (Chen et al., 2014a). TF2b was found to form more hydrogen bonds with the 3CLpro receptor compared to drugs like Lopinavir, Darunavir and Atazanavir thus exhibiting the activity to maintain the interaction (Bhardwaj et al., 2020). Another similar study also showed that TF3 had better docking score (−10.574) than more than 20 antiviral drugs like lopinavir (−9.918), darunavir (−8.843), amprenavir (−8.655) as well as phytochemicals like hesperidin, biorobin, rosmarinic acid, etc. (Peele et al., 2020). A report described the use of TF in the prophylaxis of SARS-CoV-2. TF2 showed a docking score of −9.8 and TF3 showed a docking score of −10 on the 3CLpro receptor in a molecular docking study (Bhatia et al., 2020).
Inhibition of RDRP
In a study by Lung et al., it was found that out of 83 molecules already known in Chinese traditional medicine, TF was found to have the highest interaction with a binding energy of −9.11 kcal/mol. This interaction was based on hydrogen bonds, π-cation interaction and hydrophobic interactions (Lung et al., 2020b).
Inhibition of spike RBD
It was found that TF has a good binding with the RBD in SARS-CoV-2 with a high docking score. The driving forces of these interactions were majorly hydrophobic interactions along with hydrogen bonds at sites like ARG454, PHE456, ASN460, CYS480, GLN493, ASN501, and VAL503 of SARS-CoV-2 RBD, near the ACE2-S protein contact area. For the most favourable interaction of TF with the RBD in SARS-CoV-2, the binding energy (ΔG) was estimated to be −8.53 kcal/mol (Lung et al., 2020a).
Inhibition of structural proteins
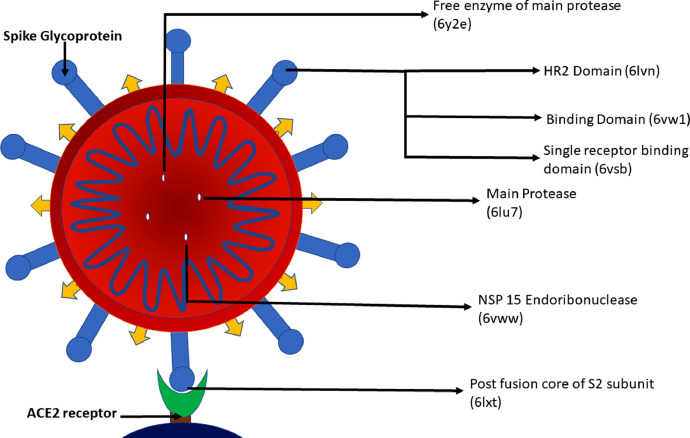
Khan et al. conducted a study to check the molecular docking of 18 phytoconstituents which have been already reported for their antiviral activity with 7 different proteins of the novel coronavirus. These docking results were compared with two drugs which are possibly the most potent against COVID-19, remdesivir and chloroquine. The protein data bank IDs of the main protease, HR2 domain, post-fusion core of the S2 subunit, S protein, RBD-ACE2 complex, NSP15 endoribonuclease, and free enzyme crystal structure main protease were 6lu7, 6lvn, 6lxt, 6vsb, 6vw1, 6vww, and 6y2e, respectively as shown in Fig. 1 . EGCG showed a very high binding affinity and a low inhibition constant among all the phytoconstituents screened, especially in the case of 6vw1, which is a potential target of SARS-CoV-2. Tables 3 and 4 show the comparative evaluation of the binding energies and the inhibition constant (μM) of EGCG with remdesivir and chloroquine. The highlight of this study was that EGCG exhibited better binding with the viral proteins and hence, is expected to show better antiviral activity than the reference drugs, remdesivir and chloroquine (Mf et al., 2020).
Fig. 1.
Sites on SARS-CoV-2 docked with therapeutic molecules.
Table 3.
Binding energies (μM) of EGCG, Remdesivir and Chloroquine with 6lu7, 6lvn, 6lxt, 6vsb (Mf et al., 2020).
| Compound | 6lu7 | 6lvn | 6lxt | 6vsb |
| EGCG | −6.99 | −4.90 | −7.57 | −7.26 |
| Remdesivir | −2.47 | −2.68 | −4.84 | −4.27 |
| Chloroquine | −3.62 | −3.26 | −4.35 | −4.79 |
Table 4.
Inhibition constants (μM) of EGCG, Remdesivir and Chloroquine with 6lu7, 6lvn, 6lxt, 6vsb (Mf et al., 2020).
| Compound | 6lu7 | 6lvn | 6lxt | 6vsb |
| EGCG | 7.57 | 255.95 | 2.84 | 4.75 |
| Remdesivir | 15.4 × 10−5 | 10.7 × 10−5 | 281.48 | 745.64 |
| Chloroquine | 2.2 × 10−5 | 4.0 × 10−5 | 651.5 | 309.3 |
Similar studies are being performed in SARS-CoV-2 to check the antiviral activity of TF. In one such study performed on 169 different phytoconstituents typically used as spices or flavours, TF3 was found to have a considerable affinity with M protease along with antioxidant activity (Zhang et al., 2020).
Inhibition of ACE2 receptor binding ability
Since the ACE2 receptor is the attaching site for the RBD of the viral S protein, direct interaction with ACE2 can prevent the infection of the cell by the virus. In the previously mentioned study by Zhang et al., TF3 was also found to directly bind to the ACE2 receptor, thus encouraging its use for prophylaxis (Zhang et al., 2020). The ability of TF3 to prevent the spike RBD from attaching to the ACE2 receptor was also shown by Maiti and Banerjee (2020).
Inhibition of glucose regulated protein-78 (GRP78)
While working on glioblastoma, Bhattacharjee et al. found out that EGCG forms many hydrogen bonds as well as non-bonded interaction with this druggable target. EGCG was found to inhibit the ATPase activity of GRP78, thus making it less flexible (Ibrahim et al., 2019). Inferring from the studies by Ibrahim et al. and Bhattacharjee et al., we can expect EGCG to be a potential inhibitor of COVID-19 S protein-GRP78 binding site (Bhattacharjee et al., 2015).
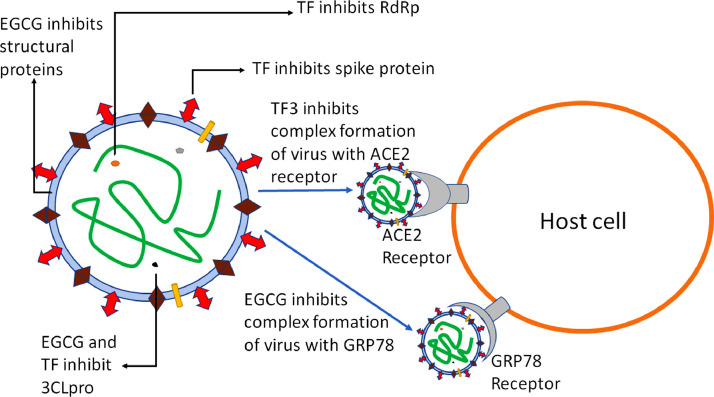
Fig. 2 shows possible mechanism of actions for tea polyphenols on different active sites. Table 5 summarizes the studies performed on SARS-CoV-2 using tea polyphenols
Fig. 2.
Depiction of role of tea polyphenols on druggable targets of COVID-19.
Table 5.
Summary of recent studies on in-silico activity of tea polyphenols on COVID-19 receptors.
| Sr. no | Polyphenol studied | Receptor targeted | Activity | Reference |
| 1 | EGCG | 3CLpro | Inhibition activity better than other phytochemicals | Khaerunnisa et al. (2020) |
| 2 | TF2b | 3CLpro | Better interaction as compared to repurposed drugs | Bhardwaj et al. (2020) |
| 3 | TF | RdRp | Best in-silico interaction among compounds from Chinese traditional medicine | Lung et al. (2020b) |
| 4 | TF | Spike RBD | Good molecular docking score with multiple hydrogen bonds | Lung et al. (2020a) |
| 5 | TF3 | M protease | Better affinity among 169 phytoconstituents used as spices and flavours | Zhang et al. (2020) |
| 6 | EGCG | Multiple structural proteins | Better interaction than Chloroquine and Remdesivir | Mf et al. (2020) |
| 7 | TF3 | ACE2 Receptor | Directly binds to the receptor and acts as prophylactic agent | Maiti and Banerjee (2020), Zhang et al. (2020) |
| 8 | TF2 and TF3 | 3CLpro | Molecular docking scores of −9.8 and −10 on the receptor repectively | Bhatia et al. (2020) |
| 9 | TF | 3CLpro | Better in-silico activity than 24 approved antiviral drugs and more than 20 screened phytochemicals | Peele et al. (2020) |
Challenges and future perspectives
COVID-19 is spreading at an alarming rate and the lack of an approved treatment is causing a major load on the healthcare systems. Several antiviral drugs are under clinical trials, however, owing to possible side-effects higher doses of these drugs cannot be administered. Comparing with the mechanism of action of possible drug candidates in previously known viral diseases, we can shortlist some potential viral targets and drugs that can act on these sites. EGCG and TFs are polyphenolic catechins found abundantly in green tea and black tea, respectively with a vast array of health benefits. Their antiviral activities also have been reported against various viral infections. In depth analysis of antiviral activities of EGCG and TFs reveals that both of them are wide spectrum antiviral molecules with no definite interaction sites. They act at different stages of the viral cycle. Some studies have also suggested that EGCG and TFs have prophylactic activity. 3CLpro is a vital enzyme found in SARS-CoV and SARS-CoV-2. Considering the genomic similarities between the two viruses, the nature of 3CLpro found in them is also similar. 3CLpro is also a druggable target owing to its important function in viral cell replication. EGCG and all the TFs are found to inhibit 3CLpro at lesser concentrations and also exhibit good binding to this target. TFs also exhibit a good binding with the RBD in SARS-CoV-2, by forming hydrophobic interactions as well as hydrogen bonds with some sites on viral S protein. Additionally, TFs are known to show RdRp inhibition and ACE2 binding activity. Hence, both EGCG and TFs are potential antiviral agents which should be explored as treatment and prophylactic alternatives for COVID-19. GRP78 is a protein found in ER and performs the role of preventing the unfolding of the proteins that are being synthesized. When the cell is under external stress, GRP78 moves to the cell membrane from the ER and becomes a target for viral infection. EGCG is known to interact with the COVID-19 S protein GRP78 binding site, which is a druggable target, via hydrogen and non-bonded interactions, thus exhibiting its antiviral activity. Apart from these, the broad-spectrum antiviral activity has already been established for these tea polyphenols. Hence, amongst all the suggested targets for COVID-19, tea polyphenols can potentially show inhibitory activity with few or all of them. Only further docking and modeling studies can help us analyze these interactions in detail.
However, EGCG is unstable and when consumed orally has low bioavailability. It tends to get oxidized quite easily before it reaches the target. Hence, many studies have suggested structural derivatives of EGCG to enhance its bioavailability. Ester derivatives of EGCG showed better antioxidant activity in scavenging reactive oxygen species like peroxyl radicals. The antiviral activity was also increased since the derivatives showed better inhibition of HCV protease as well as a-glucosidase in HIV treatment (Zhong et al., 2011). Some studies suggest the use of nanoparticles to encapsulate EGCG to attain better efficacy (Munin and Edwards-Lévy, 2011). Further, the administration of EGCG with other antiviral drugs can improve its bioavailability. In a study involving HCV, the combination of EGCG with monoclonal antibodies showed improved delivery of EGCG in vivo. Lipophilic derivatives of water-soluble EGCG like EGCG palmitate and EGCG stearate were evaluated for improvements in antimicrobial properties and EGCG palmitate was found to be 8.7 times more potent in HSV infection than EGCG (Hsu, 2015). The same author also suggested prophylactic use of EGCG palmitate by incorporating it into sanitizers. EGCG-monoesters were also synthesized with butanoyl, octanoyl, laurinol, palmitoyl and eicosanoyl modifications to test the effects of alkyl length on the anti-influenza activity of EGCG. It was observed that long acyl groups could significantly increase the anti-influenza activity of EGCG, and EGCG palmitate, the most potent among all the ester derivatives of EGCG, was about 24 times more potent than native EGCG (Mori et al., 2008). EGCG stearate was shown to inhibit HSV infection as well as treat its associated symptoms ( Zhao et al., 2012). By performing substitution like methylation, acylation, esterification, or glycosylation at different sites of EGCG, the pharmacokinetic and pharmacodynamic properties of EGCG can be improved (Xu et al., 2017). EGCG and its derivatives show some promising results in multiple viral infections and it will be worth studying the applications of these molecules in COVID-19 with the help of molecular docking.
To summarize, both the tea polyphenols need further detailed evaluation to validate their anti-COVID-19 applications. TFs, especially TF2b and TF3 can be used as good prophylactic agents owing to their ability to bind to RBD. EGCG and its stable lipophilic derivatives could also be potential prophylactic as well as therapeutic agents looking at their properties to dock to various active sites of SARS-CoV-2.
Tea is the most widely consumed beverage in the world, and developing antiviral polyphenolic molecules from the same is an exciting idea. The FDA has already assigned these polyphenols the coveted GRAS status, which further encourages their use over synthetic toxic antiviral drugs with respect to higher dosages. However, concluding EGCG and TFs as drug candidates on the basis of currently available literature would be an overstatement. Since these polyphenols have no specific activity, we cannot be sure of their targeted activity to the COVID-19 receptors discussed earlier. These might also bind to other proteins in the body and produce side-effects. Establishing compliance with the stringent regulatory affairs for molecules to be deemed drugs over the same molecule as nutraceutical is time consuming and strict. Hence, in the immediate future in terms of application in COVID-19, these molecules may not be used in treatment, but as dietary supplements or nutraceuticals. After extensive studies on these polyphenols regarding their specificity, activity, bioavailability and safety, there can be considerations on their use in treatment of viral infections including COVID-19.
Author contributions
Mr. Susmit Mhatre has written the original draft of the manuscript. He collected the data, analysed and interpreted it. He has also made all the tables and figures included in the manuscript. Ms. Tishya Srivastava assisted in writing the manuscript. Mr. Shivraj Naik designed the review work and revised the final draft. Dr. Vandana Patravale planned the manuscript as well as administered and supervised the entire work. All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy.
Funding
This work did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
We confirm that there are no conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
References
- Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2) - an update on the status. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbely E., Khattari Z., Brotons G., Akkawi M., Salditt T., Arkin I.T. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 2004;341:769–779. doi: 10.1016/j.jmb.2004.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkefeld C. Dissertations and Culminating Projects; 2015. In Vitro Synergistic Antiviral Activity of Black Tea Theaflavins and Acyclovir on Herpes Simplex Virus Types 1 and 2 in A549 Cells. [Google Scholar]
- Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomolecular Struct. Dyn. 2020;0:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia, S., Giri, S., Lal, A.F., Singh, S., 2020. Battle against coronavirus: repurposing old friends (food borne polyphenols) for new enemy (COVID-19).
- Bhattacharjee R., Devi A., Mishra S. Molecular docking and molecular dynamics studies reveal structural basis of inhibition and selectivity of inhibitors EGCG and OSU-03012 toward glucose regulated protein-78 (GRP78) overexpressed in glioblastoma. J. Mol. Model. 2015;21:272. doi: 10.1007/s00894-015-2801-3. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Van der Zee R., de Haan C., Rottier P. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calland N., Albecka A., Belouzard S., Wychowski C., Duverlie G., Descamps V., Hober D., Dubuisson J., Rouille Y., Seron K. (-)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology. 2012;55:720–729. doi: 10.1002/hep.24803. [DOI] [PubMed] [Google Scholar]
- Chacko S.M., Thambi P.T., Kuttan R., Nishigaki I. Beneficial effects of green tea: a literature review. Chin Med. 2010;5:13. doi: 10.1186/1749-8546-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Lo S.-.C., Wang Y.-.S., Hou M.-.H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today. 2016;21:562–572. doi: 10.1016/j.drudis.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-.N., Lin C.P.C., Huang K.-.K., Chen W.-.C., Hsieh H.-.P., Liang P.-.H., Hsu J.T.-A. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3’-digallate (TF3) Evidence-Based Complementary Alternat. Med. 2014;2 doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, P., Sahuc, M.-.E., Rouillé, Y., Vandeputte, A., Brodin, P., Goswami, M., Bandyopadhyay, T., Dubuisson, J., Séron, K., 2018. Theaflavins, polyphenols of black tea, inhibit entry of hepatitis C virus. bioRxiv 325126. [DOI] [PMC free article] [PubMed]
- Chu K.O., Wang C.C., Chu C.Y., Choy K.W., Pang C.P., Rogers M.S. Uptake and distribution of catechins in fetal organs following in utero exposure in rats. Hum. Reprod. 2006;22:280–287. doi: 10.1093/humrep/del353. [DOI] [PubMed] [Google Scholar]
- Clark K.J., Grant P.G., Sarr A.B., Belakere J.R, Swaggerty C.L., Phillips T.D., Woode G.N. An in vitro study of theaflavins extracted from black tea to neutralize bovine rotavirus and bovine coronavirus infections. Vet. Microbiol. 1998;63:147–157. doi: 10.1016/S0378-1135(98)00242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira A., Prince D., Lo C.-.Y., Lee L.H., Chu T.-.C. Antiviral activity of theaflavin digallate against herpes simplex virus type 1. Antiviral Res. 2015;118:56–67. doi: 10.1016/j.antiviral.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B., Jiang S. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. Clifton, N.J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge M., Xiao Y., Chen H., Fatao L., Du G., Zeng F. Multiple antiviral approaches of (–)-epigallocatechin-3-gallate (EGCG) against porcine reproductive and respiratory syndrome virus infection in vitro. Antiviral Res. 2018:158. doi: 10.1016/j.antiviral.2018.07.012. [DOI] [PubMed] [Google Scholar]
- Gosslau A., En Jao D.L., Huang M.-.T., Ho C.-.T., Evans D., Rawson N.E., Chen K.Y. Effects of the black tea polyphenol theaflavin-2 on apoptotic and inflammatory pathways in vitro and in vivo. Mol. Nutr. Food Res. 2011;55:198–208. doi: 10.1002/mnfr.201000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A., Friebe A., Gorbalenya A.E., Ziebuhr J. Mutational analysis of the active centre of coronavirus 3C-like proteases. J. Gen. Virol. 2002 doi: 10.1099/0022-1317-83-3-581. [DOI] [PubMed] [Google Scholar]
- Higdon J.V., Frei B. Tea Catechins and Polyphenols: health Effects, Metabolism, and Antioxidant Functions. Crit. Rev. Food Sci. Nutr. 2003;43:89–143. doi: 10.1080/10408690390826464. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. Compounds Derived from Epigallocatechin-3-Gallate (EGCG) as a Novel Approach to the Prevention of Viral Infections. Inflamm. Allergy - Drug Targets (Formerly Curr. Drug Targets - Inflamm. Allergy) 2015;14:13–18. doi: 10.2174/1871528114666151022150122. [DOI] [PubMed] [Google Scholar]
- Ibrahim I.M., Abdelmalek D.H., Elfiky A.A. GRP78: a cell's response to stress. Life Sci. 2019;226:156–163. doi: 10.1016/j.lfs.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 0. 2020 doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.-.C., Bourhis J.-.M., Bussetta C., Coutard B., Egloff M.-.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie Y., Li L., Tan S., Jin H., Qiu J., Mao Q., Li R., Xia C., Jiang Z.-.H., Jiang S., Liu S. A natural theaflavins preparation inhibits HIV-1 infection by targeting the entry step: potential applications for preventing HIV-1 infection. Fitoterapia. 2011;83:348–355. doi: 10.1016/j.fitote.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Suhartati, S., Soetjipto, S., 2020. Potential Inhibitor of COVID-19 Main Protease (Mpro) From Several Medicinal Plant Compounds by Molecular Docking Study.
- 32.Khan, R.J., Kumar Jha, R., Amera, G.M., Jain, M., Singh, E., Pathak, A., Singh, R.P., Muthukumaran, J., Singh, A.K., 2020. Targeting Novel Coronavirus 2019: a Systematic Drug Repurposing Approach to Identify Promising Inhibitors Against 3C-like Proteinase and 2’-O-Ribose Methyltransferase. [DOI] [PMC free article] [PubMed]
- Kong, R., Yang, G., Xue, R., Liu, M., Wang, F., Hu, J., Guo, X., Chang, S., 2020. COVID-19 Docking Server: an interactive server for docking small molecules, peptides and antibodies against potential targets of COVID-19. [DOI] [PMC free article] [PubMed]
- Lambert J.D., Yang C.S. Mechanisms of Cancer Prevention by Tea Constituents. J. Nutr. 2003;133:3262S–3267S. doi: 10.1093/jn/133.10.3262S. [DOI] [PubMed] [Google Scholar]
- Lin S.-.Y., Liu C.-.L., Chang Y.-.M., Zhao J., Perlman S., Hou M.-.H. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J. Med. Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Lu H., Zhao Q., He Y., Niu J., Debnath A.K., Wu S., Jiang S. Theaflavin derivatives in black tea and catechin derivatives in green tea inhibit HIV-1 entry by targeting gp41. Biochimica et Biophysica Acta (BBA) - General Subjects. 2005;1723:270–281. doi: 10.1016/j.bbagen.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Lu J.-.W., Hsieh P.S., Lin C.-.C., Hu M.-.K., Huang S.-.M., Wang Y.-.M., Liang C.-.Y., Gong Z., Ho Y.-.J. Synergistic effects of combination treatment using EGCG and suramin against the chikungunya virus. Biochem. Biophys. Res. Commun. 2017:491. doi: 10.1016/j.bbrc.2017.07.157. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, J., Lin, Y.-.S., Yang, Y.-.H., Chou, Y.-.L., Chang, G.-.H., Tsai, M.-.S., Hsu, C.-.M., Yeh, R.-.A., Shu, L.-.H., Cheng, Y.-.C., Liu, H.T., Wu, C.-.Y., 2020a. The potential SARS-CoV-2 entry inhibitor. bioRxiv 2020.03.26.009803.
- Lung J., Lin Y.-.S., Yang Y.-.H., Chou Y.-.L., Shu L.-.H., Cheng Y.-.C., Liu H.T., Wu C.-.Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020 doi: 10.1002/jmv.25761. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S., Banerjee A. Bioinformatics and Molecular Docking Study; 2020. Epigallocatechin-Gallate and Theaflavin-Gallate Interaction in SARS CoV-2 Spike-Protein Central-Channel With Reference to the Hydroxychloroquine Interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Man, Jiang Jinyan, Zheng Rongrong, Pearl Henna, Dickinson Douglas, Fu Baiping, Hsu Stephen. A Proprietary Topical Preparation Containing EGCG-Stearate and Glycerin with Inhibitory Effects on Herpes Simplex Virus: case Study. Inflamm. Allergy - Drug Targets (Discontinued) 2012;11:364–368. doi: 10.2174/187152812803251033. [DOI] [PubMed] [Google Scholar]
- McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-o-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mf, K., Ma, K., Za, K., T, A., Wa, A., 2020. Identification of Dietary Molecules as Therapeutic Agents to Combat COVID-19 Using Molecular Docking Studies.
- Mori S., Miyake S., Kobe T., Nakaya T., Fuller S.D., Kato N., Kaihatsu K. Enhanced anti-influenza A virus activity of (-)-epigallocatechin-3-O-gallate fatty acid monoester derivatives: effect of alkyl chain length. Bioorg. Med. Chem. Lett. 2008;18:4249–4252. doi: 10.1016/j.bmcl.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Munin A., Edwards-Lévy F. Encapsulation of natural polyphenolic compounds; a review. Pharmaceutics. 2011;3:793–829. doi: 10.3390/pharmaceutics3040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Suzuki K., Toda M., Okubo S., Hara Y., Shimamura T. Inhibition of the infectivity of influenza virus by tea polyphenols. Antiviral Res. 1993;21:289–299. doi: 10.1016/0166-3542(93)90008-7. [DOI] [PubMed] [Google Scholar]
- Nance C.L., Siwak E.B., Shearer W.T. Preclinical development of the green tea catechin, epigallocatechin gallate, as an HIV-1 therapy. J. Allergy Clin. Immunol. 2009;123:459–465. doi: 10.1016/j.jaci.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba M., Oka T., Ando T., Arahata S., Ikegaya A., Takagi H., Ogo N., Zhu C., Owada K., Kawamori F., Wang Q., Saif L.J., Asai A. Antiviral effect of theaflavins against caliciviruses. J. Antibiot. 2017;70:443–447. doi: 10.1038/ja.2016.128. [DOI] [PubMed] [Google Scholar]
- Okada F., Takeo T., Okada S., Tamemasa O. Antiviral effect of theaflavins on tobacco mosaic virus. Agric. Biol. Chem. 1977;41:791–794. [Google Scholar]
- Peele K.A., Potla Durthi C., Srihansa T., Krupanidhi S., Sai A.V., Babu D.J., Indira M., Reddy A.R., Venkateswarulu T.C. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inform. Med. Unlocked. 2020;19 doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Xiao X., Dimitrov D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004;314:235–241. doi: 10.1016/j.bbrc.2003.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomed. 2012;7:2173–2179. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The Coronavirus E Protein: assembly and Beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S D., A V., Pj R., Fa R., Bj B. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335:276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of golgi localization signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Singh, S.P., Konwar, B.K., 2013. Molecular Docking, DFT and ADME-Toxicity Studies on Analogues of Epigallocatechin Gallate as SARS Coronavirus 3CL Protease Inhibitors [WWW Document].
- Sodagari H., Bahramsoltani R., Farzaei M.H., Abdolghaffari A.H., Rezaei N., Taylor-Robinson A.W. Tea polyphenols as natural products for potential future management of HIV infection - an overview. J. Natl. Remedies. 2016;16:60–72. [Google Scholar]
- Song J.-.M., Lee K.-.H., Seong B.-.L. Antiviral effect of catechins in green tea on influenza virus. Antiviral Res. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir Ul Qamar M., Alqahtani S., Alamri M., Chen L.-.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tea Processing Steps: Tea Making And Manufacturing Process - Teabox, 2017. . Tea 101. URLhttps://tea101.teabox.com/tea-processed-classified/ (accessed 6.28.20).
- Thi Thanh Hanh N., Woo H.-.J., Kang H.-.K., Nguyen V., Kim Y.-.M., Kim D.-.W., Ahn S.-.A., Xia Y., Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34:831–838. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagomez J. Dissertations and Culminating Projects; 2017. In Vitro Antiviral Activity of Black Tea Polyphenols on Sindbis Virus in Vero Cells. [Google Scholar]
- Williamson M.P., McCormick T.G., Nance C.L., Shearer W.T. Epigallocatechin gallate, the main polyphenol in green tea, binds to the T-cell receptor, CD4: potential for HIV-1 therapy. J. Allergy Clin. Immunol. 2006;118:1369–1374. doi: 10.1016/j.jaci.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-.M., Wang W., Song Z.-.G., Hu Y., Tao Z.-.W., Tian J.-.H., Pei Y.-.Y., Yuan M.-.L., Zhang Y.-.L., Dai F.-.H., Liu Y., Wang Q.-.M., Zheng J.-.J., Xu L., Holmes E.C., Zhang Y.-.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Chen Q. Insight into 2019 novel coronavirus — an updated interim review and lessons from SARS-CoV and MERS-CoV. Int. J. Infect. Dis. 2020;94:119–124. doi: 10.1016/j.ijid.2020.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xu Z., Zheng W. A review of the antiviral role of green tea catechins. Molecules. 2017;22:1337. doi: 10.3390/molecules22081337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.S., Landau J.M. Effects of Tea Consumption on Nutrition and Health. J. Nutr. 2000;130:2409–2412. doi: 10.1093/jn/130.10.2409. [DOI] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U S A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-.F., Bai L.-.P., Huang W., Li X.-.Z., Zhao S.-.S., Zhong N.-.S., Jiang Z.-.H. Comparison of in vitro antiviral activity of tea polyphenols against influenza A and B viruses and structure-activity relationship analysis. Fitoterapia. 2014;93:47–53. doi: 10.1016/j.fitote.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G., Chen X., Xiao Z.-.X., He F., Zhang L. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290:3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L.W., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-.J., Shen X., Yan Y.-.M., Wang Y., Cheng Y.-.X. Discovery of anti-SARS-CoV-2 agents from commercially available flavor via docking screening (preprint) Open Sci. Framework. 2020 [Google Scholar]
- Zhong Y., Ma N., Shahidi C.M. Antioxidant and antiviral activities of lipophilic epigallocatechin gallate (EGCG) derivatives. J. Funct. Foods. 2011;4:87. doi: 10.1016/j.jff.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-.L., Wang, X.-.G., Hu, B., Zhang, L., Zhang, W., Si, H.-.R., Zhu, Y., Li, B., Huang, C.-.L., Chen, H.-.D., Chen, J., Luo, Y., Guo, H., Jiang, R.-.D., Liu, M.-.Q., Chen, Y., Shen, X.-.R., Wang, X., Zheng, X.-.S., Zhao, K., Chen, Q.-.J., Deng, F., Liu, L.-.L., Yan, B., Zhan, F.-.X., Wang, Y.-.Y., Xiao, G., Shi, Z.-.L., 2020a. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv 2020.01.22.914952.
- Zhou P., Yang X.-.L., Wang X.-.G., Hu B., Zhang L., Zhang W., Si H.-.R., Zhu Y., Li B., Huang C.-.L., Chen H.-.D., Chen J., Luo Y., Guo H., Jiang R.-.D., Liu M.-.Q., Chen Y., Shen X.-.R., Wang X., Zheng X.-.S., Zhao K., Chen Q.-.J., Deng F., Liu L.-.L., Yan B., Zhan F.-.X., Wang Y.-.Y., Xiao G.-.F., Shi Z.-.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu M., Yang F., Zhou W., Liu A., Du G.-.H., Zheng L. In vitro anti-influenza virus and anti-inflammatory activities of theaflavin derivatives. Antiviral Res. 2012;94:217–224. doi: 10.1016/j.antiviral.2012.04.001. [DOI] [PubMed] [Google Scholar]