Abstract
COVID-19 can result in severe lung injury. It remained to be determined why diabetic individuals with uncontrolled glucose levels are more prone to develop the severe form of COVID-19. The molecular mechanism underlying SARS-CoV-2 infection and what determines the onset of the cytokine storm found in severe COVID-19 patients are unknown. Monocytes and macrophages are the most enriched immune cell types in the lungs of COVID-19 patients and appear to have a central role in the pathogenicity of the disease. These cells adapt their metabolism upon infection and become highly glycolytic, which facilitates SARS-CoV-2 replication. The infection triggers mitochondrial ROS production, which induces stabilization of hypoxia-inducible factor-1α (HIF-1α) and consequently promotes glycolysis. HIF-1α-induced changes in monocyte metabolism by SARS-CoV-2 infection directly inhibit T cell response and reduce epithelial cell survival. Targeting HIF-1ɑ may have great therapeutic potential for the development of novel drugs to treat COVID-19.
Keywords: Covid-19, SARS-CoV-2, metabolism, monocyte, inflammation, interferon, HIF-1alpha, glycolysis, mitochondria, diabetes
Graphical Abstract
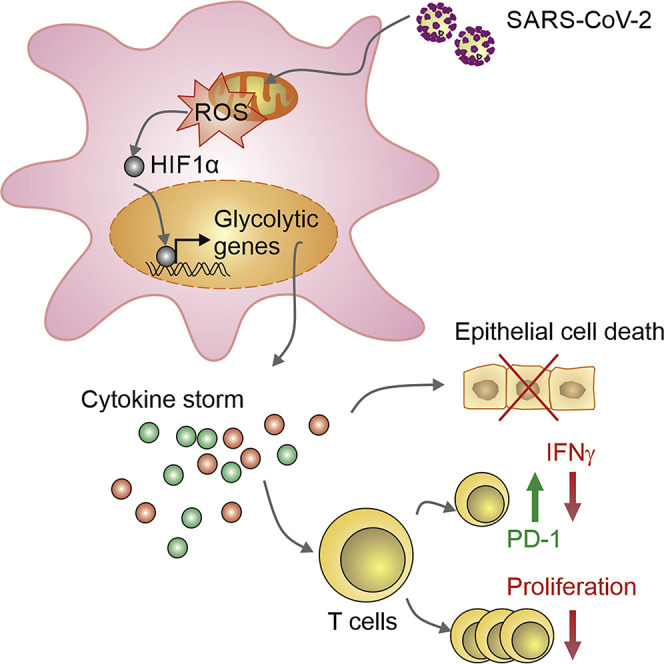
Highlights
-
•
Elevated glucose levels regulate viral replication and cytokine production in monocytes
-
•
Glycolysis sustains CoV-2-induced monocyte response and viral replication
-
•
mtROS/HIF-1α is necessary for CoV-2 replication and monocyte cytokine production
-
•
Monocyte-derived cytokines drive T cell dysfunction and epithelial cell death
Diabetic people with uncontrolled blood glucose levels have a greater risk to develop severe COVID-19 disease. Codo et al. show that elevated glucose levels and glycolysis promote SARS-CoV-2 (CoV-2) replication and cytokine production in monocytes through a mitochondrial ROS/hypoxia-inducible factor-1α dependent pathway, resulting in T cell dysfunction and epithelial cell death.
Context and Significance
COVID-19 is a highly infectious disease. There is an urgent need for novel therapeutic approaches to treat COVID-19 disease. Why certain individuals are more prone to develop the severe form of COVID-19 remains elusive. Obese and diabetic people with uncontrolled blood glucose levels have a greater risk to develop the severe form of COVID-19. The cytokine storm observed in severe COVID-19 patients is thought to be associated with dysfunctional adaptive immune response and epithelial cell death. Here we show that elevated glucose and sustained aerobic glycolysis in monocytes directly promote viral replication, cytokine production, and the subsequent T cell dysfunction and lung epithelial cell death. Thus, targeting metabolic pathways may provide new therapeutic strategies to treat COVID-19 disease.
Introduction
COVID-19 is a highly infectious disease caused by the acute respiratory syndrome coronavirus 2 (SARS-CoV-2). As of July 3, 2020, the pandemic of COVID-19 had already affected over 10 million people and taken over 500,000 lives according to the Coronavirus Resource Center of John Hopkins University. There is an urgent need for treatments and vaccines; however, little is known about the cellular mechanisms that underlie SARS-CoV-2 (CoV-2) infection. The reasons why certain individuals are more prone to develop the severe form of COVID-19 are also elusive, although it is clear that age, cardiovascular diseases, and diabetes are among the main risk factors for severe COVID-19 symptoms (Guo et al., 2020; Jordan et al., 2020; Zhou et al., 2020; Zhu et al., 2020). In a cohort of more than 7,000 COVID-19 patients with or without diabetes, it was shown that uncontrolled blood glucose levels correlate with worse prognosis and higher mortality (Zhu et al., 2020). Dysregulated innate immune response with exaggerated inflammatory cytokine production is also a defining feature driving severe COVID-19 disease (Tay et al., 2020; Blanco-Melo et al., 2020). Myeloid cells, such as monocytes and macrophages, are the most enriched immune cell types in the lungs of COVID-19 patients and play a major role in the pathogenicity of the disease (Bost et al., 2020). It was also described that inhibition of glucose flux with 2-deoxy-D-glucose (2-DG) prevented CoV-2 replication in a colon adenocarcinoma cell line (Bojkova et al., 2020). Thus, the link between elevated glucose levels and uncontrolled inflammatory response in the lungs with the severity of COVID-19 disease prompted us to investigate the mechanism by which monocytes respond to SARS-CoV-2 infection in a context of elevated glucose supply.
Results and Discussion
Elevated Glucose Levels Enhance SARS-CoV-2 Replication and Cytokine Expression in Monocytes
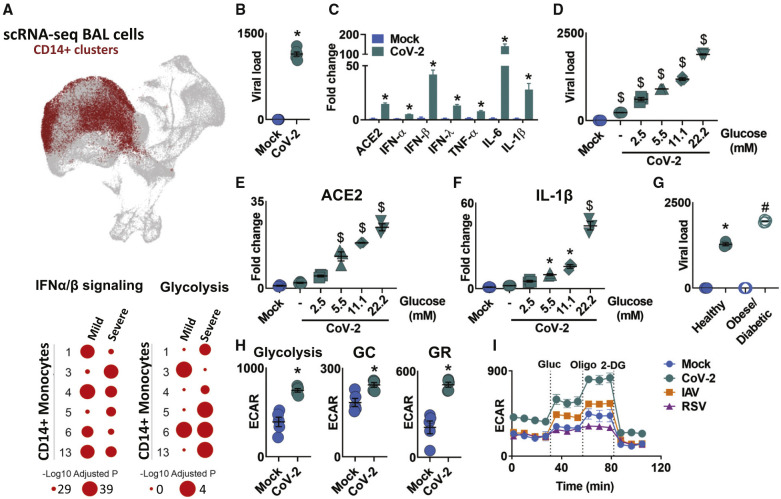
Once in the airway, CoV-2 infects host cells and starts to replicate. Upon infection, there is an accumulation of immune cells in the lungs, especially macrophages and monocytes (Merad and Martin, 2020). Using publicly available single-cell RNA sequencing (RNA-seq) data from bronchoalveolar lavage (BAL) of mild and severe COVID-19 patients and controls (Bost et al., 2020), we identified that several genes associated with “interferon α/β signaling pathway” are upregulated in mild and severe COVID-19 patients compared to controls in all 6 clusters of monocytes (Figure 1 A). These monocytes are likely recruited by chemokines produced by infected epithelial cells, while interferons (IFNs) induce angiotensin-converting enzyme 2 (ACE2), one of the proteins used by the virus to enter the cell (Hoffmann et al., 2020; Wrapp et al., 2020). We showed that CoV-2 effectively infects peripheral blood monocytes and enhances the expression of ACE2 (Figures 1B and 1C). Monocytes infected with CoV-2 expressed higher levels of IFN α, β, and λ, and higher levels of the proinflammatory cytokines TNF-α, IL-1β, and IL-6, which are associated with the COVID-19 “cytokine storm” (Figure 1C).
Figure 1.
SARS-CoV-2 Infection Induces Glycolysis in Human Monocytes
(A) Single-cell RNA-seq data showing CD14+ monocyte clusters expressing IFN-α and IFN-β in both mild and severe COVID-19 patients compared to healthy individuals. Enrichment of glycolysis in monocytes of severe COVID-19 patients.
(B and C) Human monocytes were infected with mock control or SARS-CoV-2 (CoV-2) (MOI 0.1) for 1 h under continuous agitation and incubated for 24 h.
(B) CoV-2 viral load in human monocytes.
(C) Relative mRNA expression of ACE2, IFN-α, IFN-β, IFN-λ, TNF-α, IL-6, and IL-1β by qPCR.
(D–F) Human monocytes were infected with mock control or CoV-2 (MOI 0.1) for 1 h under continuous agitation and rested for 24 h in media containing different glucose concentrations (0, 2.5, 5.5, 11.1, and 22.2 mM).
(D) Analysis of viral load by qPCR.
(E and F) Relative mRNA expression of (E) ACE2 and (F) IL-1β.
(G) Peripheral blood mononuclear cells from healthy controls (Healthy) and obese/diabetic patients were infected with mock control or SARS-CoV-2 (CoV-2) (MOI 0.1) for 1 h under continuous agitation and incubated for 24 h; viral load was determined by qPCR.
(H) Extracellular acidification rate (ECAR) analysis of glycolysis (following glucose injection), glycolytic capacity (GC; following oligomycin injection), and glycolytic reserve (GR; glycolytic capacity; glycolysis) in mock control or CoV-2-infected human monocytes.
(I) ECAR of human monocytes upon glycolytic stress (following injections of glucose, oligomycin, and 2-DG) 24 h post-infection with CoV-2, RSV, and IAV.
Data represent mean ± SEM of at least two independent experiments performed in triplicate. ∗p < 0.05 compared to mock. $p < 0.05 compared to all other groups. #p < 0.05 compared to CoV-2 from healthy patients (one-way ANOVA and Tukey post hoc tests).
Uncontrolled blood glucose levels observed in diabetic patients are a major risk factor for the severity of COVID-19 (Zhu et al., 2020). Thus, we hypothesized that glucose availability may affect viral replication capacity. We used increasing glucose concentrations to mimic human physiological and pathological conditions. Monocytes were cultured in low (2.5 mM, equivalent to 40 mg/dL), normal (5.5 mM, equivalent to 100 ng/dL), and high glucose conditions (11.1 mM and 22.2 mM, equivalent to 200 and 400 mg/dL, respectively). Glucose directly increased viral load, ACE2, and IL-1β expression in CoV-2-infected monocytes in a dose-dependent manner (Figures 1D–1F). Moreover, increased glucose levels further augmented TNF-α, IL-6, and IFN α, β, and λ expression (Figure S1A). These data suggest that individuals with elevated circulating glucose levels may be more susceptible to CoV-2 infection. To test this, we infected monocytes isolated from obese/diabetic patients and observed increased viral load compared to healthy controls (Figure 1G). Altogether, these data indicate that elevated glucose levels directly promote viral replication and cytokine expression.
Once inside the cells, glucose can be oxidized by glycolysis to produce ATP and a number of other metabolites (Breda et al., 2019). We observed that several glycolysis-associated genes were upregulated in BAL monocytes of COVID-19 patients (Figure 1A). Consistent with widespread metabolic remodeling, carbon metabolism pathways were enriched among the 144 differentially expressed proteins associated with energy metabolism in the proteomic analysis of CoV-2-infected monocytes cultured under 11.1 mM glucose (used in all subsequent experiments unless stated otherwise). In total, 510 differentially expressed proteins were identified, playing roles in a range of biological processes (Figures S2A–S2C). In agreement with the metabolic remodeling observed in our proteomics study, CoV-2 stimulates glycolysis and increases the glycolytic capacity and glycolytic reserve in monocytes (Figure 1H). To test whether increased glycolysis is a trait specific to CoV-2, we infected human monocytes with other RNA respiratory viruses that also may cause acute respiratory syndrome (human influenza A H1N1 virus [IAV] and respiratory syncytial virus, subgroup A [RSV]) (Hammer et al., 1997; Ribeiro et al., 2010). We observed that monocytes infected with either IAV or RSV did not display increased glycolysis or glycolytic reserve (Figures 1I and S1B). In summary, our results show that, at least in monocytes, increased glycolysis is a trait specifically elicited by CoV-2.
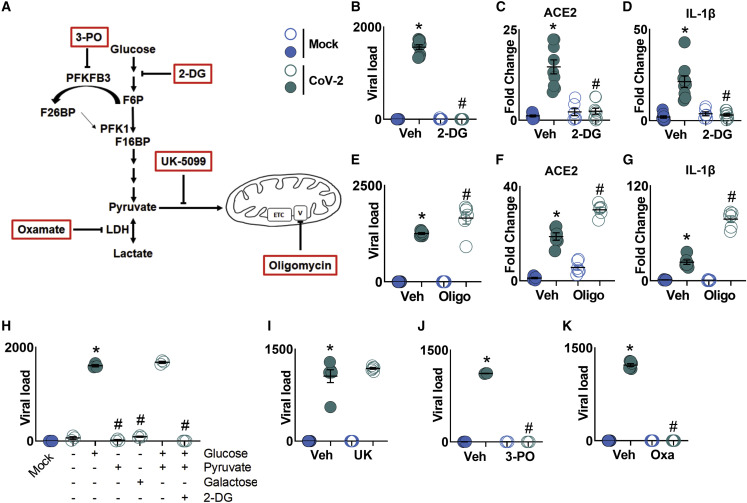
Glycolysis Is Necessary for SARS-CoV-2 Replication and Monocyte Response
Next, we assessed the impact of CoV-2 infection on the metabolism and function of human monocytes. We used 2-DG to inhibit glucose flux and oligomycin to inhibit ATP synthase (Figure 2 A). Treatment with 2-DG completely blocked viral replication in CoV-2-infected monocytes, as well as the CoV-2-induced increase of ACE2 and IL-1β expression (Figures 2B–2D). 2-DG treatment also inhibited CoV-2-induced TNF-α, IL-6, and IFN α, β, and λ expression in monocytes (Figure S3A). In contrast, oligomycin treatment led to an increase in the viral load, ACE2 and IL-1β expression, and TNF-α, IL-6, α, β, and λ IFN expression in monocytes (Figures 2E–2G and S3B). These data indicate that carbon flux through glycolysis, but not respiration coupled to ATP synthesis, is necessary for CoV-2 replication and for CoV-2-induced monocyte immune response.
Figure 2.
Glycolysis Sustains SARS-CoV-2 Replication in Human Monocytes
Human monocytes were pre-treated for 2 h with metabolic modulators (PFKFB3 inhibitor 3-PO, glycolysis inhibitor 2-DG, mitochondrial pyruvate carrier inhibitor UK-5099 [UK], LDH-A inhibitor oxamate [OXA], and ATP synthase inhibitor oligomycin [Oligo]) prior to SARS-CoV-2 (CoV-2) infection (MOI 0.1) for 1 h under continuous agitation and incubation for 24 h.
(A) Schematic of metabolism modulator mechanism of action.
(B) Viral load by qPCR of infected monocytes pre-treated with 2-DG.
(C and D) Relative mRNA expression of (C) ACE2 and (D) of IL-1β in infected monocytes pre-treated with 2-DG by qPCR.
(E) Viral load by qPCR of infected monocytes pre-treated with Oligo.
(F and G) Relative mRNA expression of (F) ACE2 and (G) IL-1β in monocytes treated with Oligo.
(H) Human monocytes were given glucose, pyruvate, or galactose as a carbon source for ATP. 2-DG was used as a glucose metabolism inhibitor. These cells were infected for 1 h with CoV-2 (MOI 0.1), followed by incubation for 3 h; viral load was analyzed by qPCR.
(I–K) Viral load in CoV-2-infected monocytes treated with (I) UK, (J) 3-PO, and (K) OXA by qPCR.
All data represent mean ± SEM of at least two independent experiments performed in triplicate. ∗p < 0.05 compared to mock. #p < 0.05 compared to CoV-2 (one-way ANOVA and Tukey post hoc tests).
One glucose molecule yields two net ATP molecules through glycolysis. Galactose blunts aerobic glycolysis because galactose enters the glycolytic pathway at a lower rate (Leloir pathway) and yields no glycolytic-derived ATP. This obligates the cell to rely exclusively on oxidative phosphorylation (OXPHOS) for ATP generation (Aguer et al., 2011; Buck et al., 2016). We infected human monocytes in glucose-free media and added glucose, pyruvate, or galactose as the main source of carbon for oxidation and subsequent ATP production. We observed that viral replication occurred when glucose or glucose + pyruvate were available and that the glucose + pyruvate effect was abolished in the presence of 2-DG (Figure 2H). When given only pyruvate or galactose, CoV-2 was unable to replicate (Figure 2H). We infected monocytes with IAV and RSV and cultured those cells in media containing normal glucose levels (5.5 mM), high glucose levels (11.1 mM), galactose, or glucose + 2-DG to assess whether changes in glycolytic flux could affect the replication of those viruses. Elevated glucose levels resulted in increased viral load for CoV-2, IAV, and RSV (Figure S3C). In addition, unlike CoV-2, both IAV and RSV were still able to replicate in the presence of galactose and glucose + 2-DG (Figure S3C). This indicates that glycolysis is essential for CoV-2 replication, but not IAV or RSV. Consistent with this notion, we inhibited pyruvate mitochondrial carrier with UK-5099 and determined that pyruvate-derived TCA cycle intermediates did not affect CoV-2 replication and ACE2 and IL-1β expression in CoV-2-infected monocytes (Figures 2I and S3D). These data suggest that the glycolytic flux is not only necessary but also sufficient for CoV-2 replication. To reduce the glycolytic rate of monocytes, we inhibited the enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3), an allosteric activator of phosphofructokinase-1 (Breda et al., 2019), with 3-PO. This also resulted in decreased viral replication and ACE2 and IL-1β expression (Figures 2J and S3E). Finally, to evaluate the importance of aerobic glycolysis for CoV-2 infection, we blocked lactate dehydrogenase A (LDH-A) activity with oxamate. This also completely abrogated viral replication and reduced ACE2 and IL-1β expression in CoV-2-infected cells (Figures 2K and S3F). We cannot exclude that other metabolites originated from glycolysis also provide substrates necessary for the observed effect (Baardman et al., 2018; Rodriguez et al., 2019). Taken together, our results indicate that CoV-2 replication in monocytes and CoV-2-induced monocyte response are sustained by a switch to aerobic glycolysis (the Warburg effect).
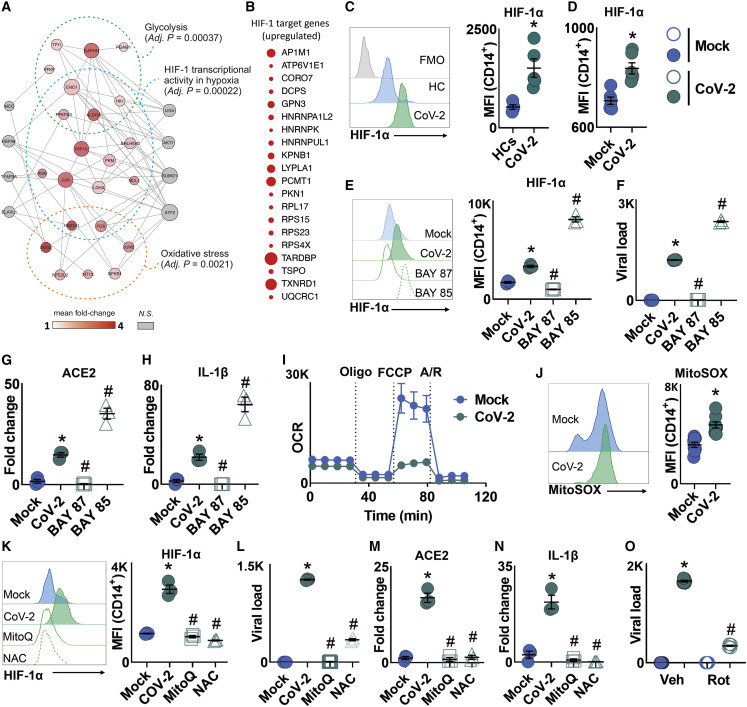
Mitochondrial ROS-Mediated HIF-1α Stabilization Sustains SARS-CoV-2 Replication and Cytokine Production in Monocytes
When compared to healthy controls, BAL monocytes from severe COVID-19 patients had higher expression of HIF-1 target genes (Figure 3 A). Our proteomic analysis further supports that CoV-2-infected monocytes have increased HIF-1α function given that HIF-1α targets are enriched among the upregulated proteins (Figure 3B). HIF-1α is a strong inducer of glycolysis and IL-1β transcription (Tannahill et al., 2013). Blood monocytes from severe COVID-19 patients present high expression of HIF-1α in comparison to healthy donors (Figure 3C). We observed that HIF-1α protein levels and its transcriptional activity measured by the target genes GLUT-1, PFKFB3, PKM2, and LDH-A, which are involved in the glucose transport and glycolytic pathway, were increased in CoV-2-infected monocytes (Figures 3D and S4A). IAV and RSV also increased HIF-1α levels, but to a lesser extent than CoV-2 (Figure S4B). To investigate the role of HIF-1α in CoV-2-mediated monocyte response, we treated cells with either the HIF-1α stabilizer BAY85-3934 (BAY85) or the HIF-1α inhibitor BAY87-2243 (BAY87). BAY87 decreased HIF-1α levels and completely abrogated CoV-2 replication (Figures 3E and 3F). Moreover, the expression of HIF-1α target genes was reduced upon BAY87 treatment in CoV-2-infected monocytes (Figure S4C). In contrast, HIF-1α stabilization exacerbated the effects of CoV-2 infection, as shown by increased viral load and expression of HIF-1α target genes (Figures 3E, 3F, and S4C). To determine whether this behavior is common to other viruses, we treated monocytes with either BAY85 or BAY87 and infected these cells with IAV and RSV. HIF-1α modulators did not affect viral load in monocytes infected with IAV (Figure S4D). In addition, inhibition of HIF-1α had no effect on RSV replication, whereas BAY85-induced HIF-1α stabilization reduced RSV viral load (Figure S4D). These data demonstrate that HIF-1α plays an essential role in CoV-2 infection in monocytes.
Figure 3.
mtROS/HIF-1α Axis Is Necessary for SARS-CoV-2 Replication in Human Monocytes
(A) Single-cell RNA-seq data showing enrichment of HIF-1, glycolysis, and oxidative stress pathways in SARS-CoV-2 (CoV-2)-infected human monocytes.
(B) Human monocytes were infected with mock control or CoV-2 (MOI 0.1) for 1 h under continuous agitation and incubated for 24 h. Enrichment of HIF-1α target genes in CoV-2-infected monocytes.
(C) Peripheral blood mononuclear cells from healthy controls (HCs) and COVID-19 patients (CoV-2) were stained with CD14 plus HIF-1α, and HIF-1α was determined by flow cytometry.
(D) Human monocytes were infected with mock control or CoV-2 (MOI 0.1) for 1 h under continuous agitation and incubated for 24 h. HIF-1α expression was assessed by flow cytometry on CD14+ monocytes.
(E–H) Human monocytes were pre-treated for 2 h with HIF-1α stabilizer BAY85-3934 (BAY85) and inhibitor BAY87-2243 (BAY87) prior to CoV-2 infection (MOI 0.1) for 1 h under continuous agitation.
(E and F) After 24 h, (E) HIF-1α expression was assessed by flow cytometry on CD14+ monocytes and (F) viral load was determined by qPCR.
(G and H) Relative gene expression of (G) ACE2 and (H) IL-1β in CoV-2-infected human monocytes pre-treated with vehicle, BAY85, or BAY87.
(I and J) Human monocytes were infected with mock control or CoV-2 (MOI 0.1) for 1 h under continuous agitation.
(I) Real-time oxygen consumption rate (OCR) by human monocytes upon mitochondrial stress (MitoStress test: injections of oligomycin, FCCP, rotenone, and antimycin A) 24 h post-infection.
(J) Mitochondrial superoxide (MitoSOX) production was assessed by flow cytometry on CD14+ monocytes 24 h post-infection.
(K–N) Human monocytes were pre-treated for 2 h with antioxidant mitoquinol (MitoQ) or N-acetyl cysteine (NAC) prior to SARS-CoV-2 infection (MOI 0.1) for 1 h under continuous agitation.
(K and L) After 24 h, (K) HIF-1α expression was assessed on CD14+ monocytes and (L) viral load was determined by qPCR.
(M and N) Relative mRNA expression of (M) ACE2 and (N) IL-1β in mock/SARS-CoV-2-infected monocytes pre-treated with vehicle, MitoQ, or NAC.
(O) Human monocytes were pre-treated for 2 h with rotenone (Rot) prior to CoV-2 infection (MOI 0.1) for 1 h under continuous agitation. After 24 h, viral load was determined by qPCR.
All data are representative of at least two independent experiments performed in triplicate. Error bars represent mean ± SEM. ∗p < 0.05 compared to mock. #p < 0.05 compared to CoV-2 (one-way ANOVA and Tukey post hoc tests).
Next, we investigated the effect of HIF-1α stabilization or inhibition over ACE2 and cytokine expression. Inhibition of HIF-1α prevented the CoV-2-mediated increase in the expression of ACE2, IL-1β, TNF-α, IL-6, and IFN α, β, and λ (Figures 3G, 3H, and S4E). In contrast, stabilization of HIF-1α increased the expression of ACE2, IL-1β, TNF-α, IL-6, and IFN α, β, and λ in CoV-2-infected monocytes (Figures 3G, 3H, and S4E). Our findings demonstrated that HIF-1α is necessary for the induction of glycolysis and the consequent proinflammatory state of CoV-2-infected monocytes.
Next, we aimed to determine the mechanism by which CoV-2 increases HIF-1α levels and promotes a proinflammatory state. It is known that succinate accumulation drives HIF-1α stabilization (Tannahill et al., 2013). We used cell-permeable diethyl succinate and observed that succinate boosted CoV-2 viral load (Figure S4F). In contrast, dimethyl malonate, which has been shown to inhibit succinate oxidation through its action on succinate dehydrogenase (SDH) (Mills et al., 2016), partially decreased CoV-2 replication in monocytes (Figure S4G). Thus, succinate oxidation is important for CoV-2 replication.
Reactive oxygen species (ROS) are strong inducers of HIF-1α (Mills et al., 2016). During metabolic reprogramming, the mitochondria are an important source of intracellular ROS (Mills et al., 2016). We observed that oxidative stress-associated genes are enriched in BAL of severe COVID-19 patients (Figure 3A). Proteomic analysis performed in CoV-2-infected monocytes confirmed alterations on oxidative metabolism with an overall downregulation of proteins involved in the TCA cycle (Figure S2D). Moreover, several components of respiratory electron chain complexes are downregulated in CoV-2-infected monocytes, such as NDUFV1 (complex I), SDHA (complex II), cytochrome c reductase COR1 and UQCRC2 (complex III), ATP5PF, ATP5F1A, ATP5PD, F-type ATPase α, and PPA2 (ATP synthase) (Figure S2D). In agreement with our proteomic data, the spare respiratory capacity was reduced in CoV-2-infected monocytes (Figure 3I). Reduced respiration in LPS-activated macrophages is known to increase mitochondrial ROS (mtROS) production (Mills et al., 2016). We observed that CoV-2-infected monocytes have increased mtROS production as measured by mitoSOX (Figure 3J). Treatment of CoV-2-infected monocytes with antioxidants, such as mitoquinol (MitoQ) or thiol reductant N-acetyl cystein (NAC), was sufficient to inhibit viral replication and CoV-2-induced HIF-1α stabilization and expression of ACE2 and IL-1β (Figures 3K–3N). These antioxidants also prevented the upregulation of TNF-α; IL-6; IFN α, β, and λ; and HIF-1α target genes (Figure S4H).
Succinate is known to drive reverse electron transport (RET) (Chouchani et al., 2014; Mills et al., 2016). Furthermore, inhibition of complex I with rotenone only reduces complex I-derived mtROS when RET is taking place (Mills et al., 2016; Votyakova and Reynolds, 2001). We investigated whether CoV-2 drives ROS production via RET, as described in LPS-activated macrophages (Mills et al., 2016). CoV-2-infected monocytes treated with rotenone have reduced viral replication (Figure 3O). These data indicate that RET is an important source of mtROS in CoV-2-infected monocytes. Altogether, these results highlight the metabolic alterations that take place in CoV-2-infected monocytes under high glucose conditions. These metabolic adaptations favor mtROS-induced HIF-1α stabilization to promote CoV-2 replication and monocyte inflammatory response.
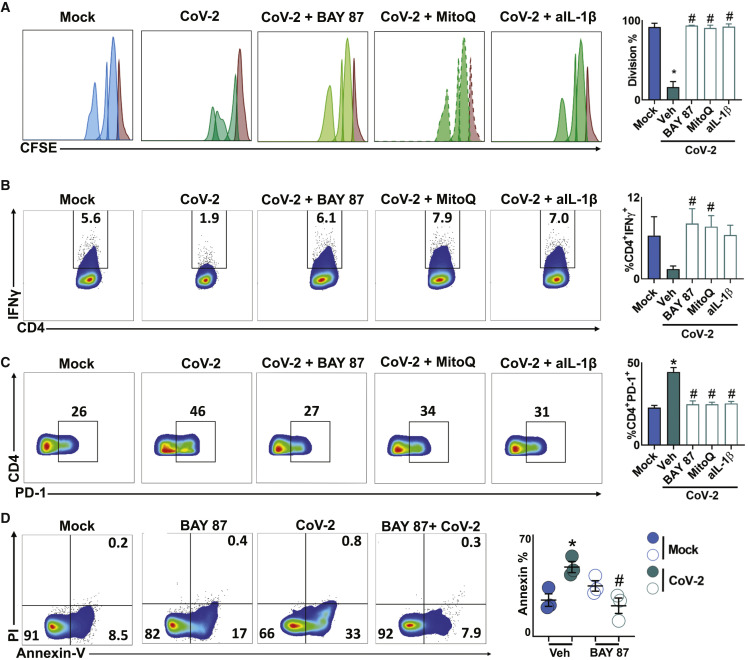
SARS-CoV-2-Induced Metabolic Reprogramming of Monocytes Directly Affects T Cell Response and Lung Epithelial Cell Death
Exacerbated proinflammatory stimuli can negatively affect T cell response (de Marcken et al., 2019). The cytokines TNF-α, IL-1β, and IL-6, which were highly expressed in CoV-2-infected monocytes (Figure 1C), resemble the cytokine storm proposed in severe COVID-19 patients (Tay et al., 2020). The cytokine storm is thought to be associated with lymphopenia in COVID-19 patients (Zheng et al., 2020). We tested whether immunometabolic reprogramming of monocytes under high glucose concentrations during CoV-2 infection inhibits T cell proliferation. We co-cultured lymphocytes with allogeneic peripheral blood mononuclear cells in the presence of UV-inactivated conditioned media (CM) from mock (CM-mock) or CoV-2-infected monocytes treated with vehicle (CM-CoV-2), BAY87 (CM-BAY87), or MitoQ (CM-MitoQ). We found that CM-CoV-2 impaired CD4 and CD8 T cell proliferation (Figures 4 A and S4I), whereas CM-BAY87 or CM-MitoQ restored T cell proliferation to the same extent as CM-Mock (Figures 4A and S4I). Because HIF-1α directly promotes IL-1β, but not TNF-α and IL-6 expression (Tannahill et al., 2013), we tested whether IL-1β secretion affects T cell response. We observed that neutralization of IL-1β with anti-IL-1β (CM-aIL-1β) from UV-inactivated CM of CoV-2-infected monocytes restored T cell proliferation (Figures 4A and S4I). The effect of CM-aIL-1β on T cell proliferation was similar to that observed with CM-BAY87 or CM-MitoQ (Figures 4A and S4I). Published data showed that IFN-γ expression was reduced in CD4 T cells co-cultured with RNA virus-infected monocytes (de Marcken et al., 2019). Similarly, CM-CoV-2 decreased IFN-γ production by CD4 T cells, and this was reversed in CM-BAY87 and CM-MitoQ and appeared to be reversed in CM-aIL-1β (Figures 4B and S4J). Persistent viral infections are associated with increased PD-1 expression, a marker of T cell exhaustion (Diao et al., 2020). CM-CoV-2 increased PD-1 expression by CD4 T cells, and this was reversed in CM-BAY87, CM-MitoQ, and CM-IL-1β (Figure 4C). These results provide evidence to explain why elevated glucose levels during CoV-2 infection may lead to T cell dysfunction and lymphopenia.
Figure 4.
Monocyte Metabolism Modulates T Cell and Epithelial Cell Response to SARS-CoV-2
(A–C) Human lymphocytes were isolated from peripheral blood, stained with CFSE, and cocultured with allogeneic peripheral blood mononuclear cells for 72 h in the presence of conditioned media from mock or SARS-CoV-2 (CoV-2) monocytes pre-treated with vehicle, BAY87-2243 (BAY87), and Mitoquinol (MitoQ). For the experiment with anti-IL-1β (aIL-1β), lymphocytes were given conditioned media from CoV-2-infected monocytes with aIL-1β. (A) Proliferation, (B) percentage of viable CD4+IFN-γ+ T cells, and (C) percentage of viable CD4+PD-1+ T cells were determined by flow cytometry and are shown by representative dot plots. Brown peak in the histograms represents non-proliferative population. Data represent mean ± SEM of at least two independent experiments performed in quadruplicate. ∗p < 0.05 compared to mock. #p < 0.05 compared to CoV-2. (A–C) One-way ANOVA and Tukey post hoc tests. (B) Unpaired Student’s t test, CoV-2 versus BAY87 and CoV-2 versus MitoQ.
(D) Human lung epithelial cell line A549 was cultured for 24 h in the presence of conditioned media from mock or CoV-2-infected monocytes pre-treated with vehicle or BAY87. Representative dot plots of cellular apoptosis by Annexin V/PI staining. Data represent mean ± SEM of at least two independent experiments performed in triplicate. ∗p < 0.05 compared to mock. #p < 0.05 compared to CoV-2 (one-way ANOVA and Tukey post hoc tests).
Cytokine storm also plays a critical role in lung injury of patients with severe COVID-19 (Shi et al., 2020). However, whether this contributes to epithelial cell dysfunction is not clear. We treated the human pulmonary epithelial cell line A549 with CM-CoV-2 or CM-BAY87 and measured cell death. CM-CoV-2 induced apoptosis of A549 cells (Figure 4D), which was prevented when CoV-2-infected monocytes were treated with a HIF-1α inhibitor (Figure 4D). This suggests that under high glucose conditions, CoV-2-infected monocytes can promote epithelial cell death in an mtROS/HIF-1α-dependent manner.
In conclusion, we showed that CoV-2-infected monocytes express large amounts of proinflammatory cytokines and IFNs. Elevated glucose levels directly induce viral replication and proinflammatory cytokine expression. Glycolytic flux is required for CoV-2 replication. CoV-2-induced mtROS production stabilizes HIF-1α, which in turn upregulates glycolytic genes and IL-1β expression. Finally, we demonstrate that CoV-2-infected monocytes promote T cell dysfunction and lung epithelial cell death. These data may explain why uncontrolled diabetes may lead to compromised adaptive immune response and lung dysfunction in patients with severe COVID-19 symptoms. Our results also provide mechanistic evidence to suggest that the mtROS/HIF-1α/glycolysis axis could be targeted to treat COVID-19 disease.
Limitations of the Study
Our study focuses on metabolic changes promoted by CoV-2 infection in isolated monocytes. Besides ATP, substrates derived from other branches of glycolysis might also be important for CoV-2 infection in these cells. Some of the compounds used in this study, such as MitoQ and NAC, are commercially available for personal consumption. We would like to highlight that our experiments were performed ex vivo in a controlled environment and that the findings described above are context dependent. The inhibitors used in this study have well-established targets and characterization. However, we cannot exclude the possibility of unknown off-target effects, and genetic models of HIF-1α deletion are a non-pharmacological way of assessing the interaction of HIF-1α and mitochondrial ROS production.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD14-FITC (clone M5E2) | BD Biosciences | Cat#555397; RRID: AB_395798 |
| anti-HIF-1α-PE (clone 241812) | R&D Systems | Cat.IC1935P; RRID: AB_2232941 |
| anti-IFN-γ-PECy7 (clone B27) | BD Biosciences | Cat#557643; RRID: AB_396760 |
| anti-CD4-APC (clone RPA-T4) | BD Biosciences | Cat#555349; RRID: AB_398593 |
| anti-CD3-PERCPCy5.5 (clone SP34-2) | BD Biosciences | Cat#552852; RRID: AB_394493 |
| anti-CD8-APCCy7 (clone SK1) | BD Biosciences | Cat#557834;RRID: AB_396892 |
| anti-CD279-PE (clone MIH4) | BD Biosciences | Cat#557946; RRID: AB_647199 |
| anti-IL-1β (clone MAB601) | R&D System | Cat.DY201; RRID: AB_2848158 |
| Bacterial and Virus Strains | ||
| HIAE-02 SARS-CoV-2/SP02/human/2020/BRA | Isolated from patient | N/A |
| A/California/04/2009 (pH1N1) virus | Neumann et al., 1999 | N/A |
| Respiratory syncytial virus, subgroup A – RSV A2 strain | ATCC | ATCC VR-1540 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FICOLL - PAQUE PLUS | Sigma-Aldrich | Cat.GE17-1440-03 |
| RPMI 1640 | Thermo Fisher Scientific | Cat#11875119 |
| Penicillin-Streptomycin | Sigma-Aldrich | Cat.P4333 |
| Minimum Essential Medium (MEM) | Sigma-Aldrich | Cat. M0325 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat#12657029 |
| Dulbecco’s Modified Eagle Medium (DMEM) | Thermo Fisher Scientific | Cat#11885084 |
| 2-DG (5mM) | Cayman Chemicals | Cat#14325 |
| 3-PO (10μM) | Cayman Chemicals | Cat#19276 |
| UK-5099 (10μM) | Cayman Chemicals | Cat#16980 |
| Oligomycin D (1μM) | Cayman Chemicals | Cat#20184 |
| BAY 85-3934 (10 μM) | Cayman Chemicals | Cat#15297 |
| MitoQ (500nM) | Cayman Chemicals | Cat#89950 |
| N-acetyl-L-Cysteine (1mM) | Cayman Chemicals | Cat#20261 |
| Rotenone (500nM) | Sigma-Aldrich | Cat.R8875 |
| Diethyl succinate 98% (5mM) | Alfa Aesar | Cat.A15777 |
| Dimethyl malonate 98% (10mM) | Alfa Aesar | Cat.A11007 |
| Oxamate (40mM) | Biocompare | Cat#2580 |
| BAY 87-2243 (10 μM) | Selleckchem | Cat.S7309 |
| TRIzol Reagent | Sigma-Aldrich | Cat#564406 |
| BD Horizon Fixable Viability Stain 510 | BD Biosciences | Cat#564406 |
| MitoSOX Red Mitochondrial ROS Indicator | Thermo Fisher Scientific | Cat.M36008 |
| phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat. P1585 |
| calcium ionophore (A23187) | Sigma-Aldrich | Cat. C7522 |
| Brefeldin A | BioLegend | Cat#420601 |
| p-formaldehyde | Sigma-Aldrich | Cat.F1635 |
| Galactose (500μM) | Sigma-Aldrich | Cat.G0750 |
| carboxyfluorescein succinimidyl ester (CellTrace CFSE) | BioLegend | Cat#423801 |
| Critical Commercial Assays | ||
| GoScript Reverse Transcriptase cDNA synthesis kit | Promega | Cat. A2791 |
| SybrGreen Supermix | QIAGEN | Cat. 208156 |
| Deposited Data | ||
| Raw and analyzed data | This paper | https://doi.org/10.6019/PXD019157 |
| Experimental Models: Cell Lines | ||
| Vero CCL-81 | ATCC | ATCC CCL-81 |
| A549 CCL-185 | ATCC | ATCC CCL-185 |
| Software and Algorithms | ||
| Prism8 | GaphPad | N/A |
| FlowJo software v10 | BD Biosciences | N/A |
| Progenesis software | Waters | N/A |
Resource Availability
Lead Contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Pedro Manoel M. Moraes-Vieira (pmvieira@unicamp.br).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019157 and https://doi.org/10.6019/PXD019157. All other supporting data in this study are available from the Lead Contact on request.
Experimental Model and Subject Details
Sample Acquisition from Obese/Diabetic Patients
Human blood samples from obese/diabetic patients used in this study were obtained from individuals admitted at the Clinics Hospital, University of Campinas. All obese/diabetic patients (2 males and 2 females, age between 43-63 years old) presented blood glucose 144 mg/dL ± 24.5 mg/dL or body mass index 32.6 kg/m2 ± 2.72 kg/m2. Detailed parameters of obese/diabetc patients recruited for this study are described on Table S1. All individuals in this study provided a signed statement of consent. This study was approved by the Brazilian Committee for Ethics in Human Studies (CAEE 30227920 9 0000 5404).
Sample Acquisition from COVID-19 Patients
Human blood samples from severe COVID-19 patients analyzed in this study were obtained from individuals admitted at the Clinics Hospital, University of Campinas and included in a clinical trial (UTN: U1111-1250-1843). All individuals in this study provided a signed statement of consent. All samples were collected before intervention. SARS-CoV-2 infection was diagnosed using real-time PCR and severe COVID-19 was defined by the presence of typical lung CT-scan images and low resting blood O2 saturation. Lung CT-scans were analyzed by two lung-specialized radiologists and ranked according to a previous publication (Wang et al., 2020). COVID-19 patients (all male, age between 30-57 years old) presented body mass index 28.0 kg/m2 ± 5.44 kg/m2. All parameters of COVID-19 patients analyzed in this study are described on Table S1. All study was approved by the Brazilian Committee for Ethics in Human Studies (CAEE: 30227920.9.0000.5404).
PBMCs Isolation
Peripheral blood from healthy donors was obtained from buffy coats provided by the Hematology and Hemotherapy Center of the University of Campinas (SP-Campinas, Brazil). The study was approved by the Brazilian Committee for Ethics in Human Studies (CAEE: 31622420.0.0000.5404). Peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient centrifugation using 1.077 g × mL-1 Ficoll gradient. Briefly, buffy coats were gently mixed and then diluted (1:1) with Phosphate Buffer Saline (PBS) and carefully transferred to 50-mL tube containing 10 mL of Ficoll, which was centrifuged at 2700 rpm for 20 min at room temperature. Total PBMCs were cultured as adherent monolayers (1,5 × 106 cell × mL-1) in RPMI 1640. After 2-3 h of adhesion, cells were washed with PBS and incubated until infection with RPMI 1640 containing 10% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin at 37°C with 5% CO2 atmosphere.
Method Details
Viruses and Cell Lines
HIAE-02 SARS-CoV-2/SP02/human/2020/BRA (GenBank: MT126808.1) virus was isolated from the second confirmed case in Brazil and kindly donated by Professor Dr. Edson Luiz Durigon. Respiratory syncytial virus, subgroup A – RSV A2 strain was kindly donated by Dr. Fernando Polack. Human influenza A H1N1 virus – IAV (A/California/04/2019; H1N1) was kindly donated by Dr. Eurico Arruda. All Sars-CoV-2 virus stocks were propagated in the Vero cell line and supernatant was harvested at 2–3 dpi. The viral titers were determined by plaque assays on Vero cells. Vero CCL-81 cells were cultivated in MEM supplemented with 10% heat inactivated fetal bovine serum (FBS) and 1% Penicillin-Streptomycin, and incubated at 37°C with 5% CO2 atmosphere. A549 cell line was cultivated in DMEM supplemented with 10% FBS and 1% Penicillin-Streptomycin and incubated at 37°C with 5% CO2 atmosphere.
Reagents and Infection
PBMCs were treated with inhibitors for 2 h prior to viral infection. Cells were infected with SARS-CoV-2, IAV or RSV at MOI 0.1 under continuous agitation at 15 rpm for 1 h for virus absorption. After infection, cells were washed with pre-warmed PBS twice and incubated in RPMI containing 10% FBS and 1% Penicillin-Streptomycin for 24 h at 37°C with 5% CO2 atmosphere.
RNA Extraction, Viral Load and Gene Expression Analyses
Total RNA extractions were performed using TRIzol Reagent according to manufacturer’s instructions. RNA concentration was determined by NanoDrop 2000 spectrophotometer (Thermo Scientific). Extracted total RNA was reverse-transcribed using GoScript Reverse Transcriptase cDNA synthesis kit according to the manufacturer’s instructions. For viral load detection, specific SARS-CoV-2 N1 primers targeting the N1 region were used as previously described (Won et al., 2020). Standard curve was generated using serial dilutions of SARS-CoV-2. Viral load and gene expression qRT-PCR were performed using SybrGreen Supermix. All qRT-PCR reactions were performed using BIO-RAD CFX394 Touch Real-Time PCR Detection System on 384-well plates. Gene expression fold change was calculated with the ΔΔCt method using Microsoft Excel. Briefly, ΔΔCt = ΔCt (SARS-CoV-2-infected) – ΔCt (mock control) with ΔCt = Ct(gene-of-interest) – Ct(housekeeping-gene-18S). The fold change for each gene is calculated as 2ˆ-ΔΔCt. Primer sequences used: 18S (Forward: 5′-CCCAACTTCTTAGAGGGACAAG-3′; Reverse: 5′-CATCTAAGGGCATCACAGACC-3′); IFN-α (Forward: 5′-GACTCCATCTTGGCTGTGA-3′; Reverse: 5′-TGATTTCTGCTCTGACAACCT-3′); IFN-β (Forward: 5′-AAACTCATGAGCAGTCTGCA-3′; Reverse: 5′-AGGAGATCTTCAGTTTCGGAGG-3′); IFN-λ (Forward: 5′-ACCTATTTTGTGGCCTATCAGAGCT-3′; Reverse: 5′-CGGCTCCACTTCAAAAAGGTAAT-3′); ACE2 (Forward: 5′-GGACCCAGGAAATGTTCAGA-3′; Reverse: 5′-GGCTGCAGAAAGTGACATGA-3′); SLC2A1 – GLUT1 (Forward: 5′-CTGCTCATCAACCGCAAC-3′; Reverse: 5′-CTTCTTCTCCCGCATCATCT-3′); LDH-A (Forward: 5′-AGCCCGATTCCGTTACCT-3′; Reverse: 5′-CACCAGCAACATTCATTCCA-3′); PKM2 (Forward: 5′-ATCGTCCTCACCAAGTCTGG-3′; Reverse: 5′-GAAGATGCCACGGTACAGGT-3′); TNF-α (Forward: 5′-CCGAGGCAGTCAGATCATCTT-3′; Reverse: 5′- AGCTGCCCCTCAGCTTGA-3′); IL-1β (Forward: 5′-AAGCTGATGGCCCTAAACAG-3′; Reverse: 5′-AGGTGCATCGTGCACATAAG-3′); IL-6 (Forward: 5′-CCAGCTATGAACTCCTTCTC-3′; Reverse: 5′-GCTTGTTCCTCACATCTCTC-3′); and PFKFB3 (Forward: 5′-CAGTTGTGGCCTCCAATATC-3′; Reverse: 5′-GGCTTCATAGCAACTGATCC-3′).
Real-Time Metabolic Assays
An XFe24 Extracellular Flux analyzer (Agilent) was used to determine the bioenergetic profile of PBMCs. PBMCs were plated at 4,000,000 cells per well in XFe24 plates 3 h before infection with SARS-CoV-2. Glycolytic Stress and Mito Stress Tests were performed on XFe24 Bioanalyzer at 24 h post infection, all assays were performed following manufacturer’s protocols. Results were normalized to cell number.
mtROS Production
After 24 h post infection, PBMCs were collected and stained with BD Horizon Fixable Viability Stain 510, MitoSOX Red Mitochondrial ROS Indicator and anti-CD14-FITC for 15 min at 37°C. Cells were washed with PBS, fixed with 4% p-formaldehyde for 30 min at 4°C and transferred to polypropylene FACS tubes. Cells were then analyzed using a FACSVerse (Becton & Dickinson, San Diego, CA, USA) flow cytometer, and data were analyzed using FlowJo software.
HIF-1α Staining
After 24 h post infection, PBMCs were fixed with 4% p-formaldehyde for 30 min at 4°C. Cells were washed with PBS, permeabilized, and incubated with anti-CD14-FITC and anti-HIF-1α-PE for 1 h at 4°C. Cells were then acquired by flow cytometry (FACSVerse – Becton & Dickinson, San Diego, CA, USA) and analyzed using FlowJo software.
Generation of Conditioned Medium
Conditioned medium was generated by incubating PBMCs with mock control or SARS-CoV-2 as described previously. Supernatant from each condition was collected and placed under UV light for 30 min for residual virus inactivation. Supernatants were stored in −80°C until used in human T cell and epithelial cell assays.
Human Lymphocyte Isolation and Mixed Lymphocyte Reaction
PBMCs isolated from buffy coats from healthy volunteers were incubated at 37°C and 5% CO2 for 2 h to allow monocytes adherence to the plate surface. Non-adherent cells in suspension were collected and stained with carboxyfluorescein succinimidyl ester (CellTrace CFSE). Briefly, cells were incubated with CFSE diluted in pre-warmed PBS to the desired concentrations at 37° for 15 min and then resuspended in a fresh pre-warmed medium for another 30 min and then washed with PBS, according to the manufacturer’s recommendation. 5 × 105 lymphocytes were cocultured with 5 × 104 allogenic PBMCs in the presence of different conditioned medium for 72 h. After 72 h, cells were stimulated with 0.1 μg × mL-1 1 phorbol 12-myristate 13-acetate (PMA) and 0.5 μg × mL−1 calcium ionophore (A23187) in the presence of brefeldin A (10 μg × mL−1) for 4 h, labeled with fixable viability stain, permeabilized, and incubated with anti-IFN-γ-PECy7, anti-CD4-APC, anti-CD3-PERCPCy5.5, anti-CD8-APCCy7 and anti-CD279-PE. Cells were then acquired by flow cytometry (FACSymphony -Becton & Dickinson, San Diego, CA, USA) and analyzed using FlowJo software.
Epithelium Apoptosis Assay
Epithelial cells from cell line A549 were incubated in the presence of different conditioned medium for 24 h. The percentage of live (AnnexinV-/PI-), early (AnnexinV+/PI-) and late (AnnexinV+/PI+) apoptotic cells was determined by flow cytometry (FACSVerse – Becton & Dickinson, San Diego, CA, USA) after labeling with Annexin V/PI, as previously described (Medeiros et al., 2002).
Proteomics Sample Preparation, Mass Spectrometry, and Data Processing
Macrophages and monocytes isolated from 5 healthy controls were chemically (Lysis Buffer: 100 mM TrisHCL, 1 mM EDTA, 150 mM NaCL, 1% triton-x, protease and phosphatase inhibitors), and mechanically lysed (Ultrasonication probe) then the total protein extract was quantified by Bradford assay, according to manufacturer’s instruction (Bio-Rad). Aliquots of 60 μg of total protein extract were transferred on Microcon-10 Centrifugal Filter, with 10 kDa cut off, for filter-aided sample preparation (FASP) protein digestion, as described (Distler et al., 2016). Proteins were reduced by 10 mM (dithiothreitol) DTT and alchilated by 50 mM iodoacetamide (IAA). Each protein aliquot was digested over-night by trypsin. In detail, trypsin digestions were carried out at 37°C, 16-18 h in 50 mM ammonium bicarbonate (AMBIC) pH = 8.0. The day after, all digestions were blocked by adding formic acid (FA) to a final concentration of 0.5% (v/v) and the peptides were recovered from the filter in 50 mM AMBIC, concentrated in a speedvac and stored at −80°C until use.
Digested peptides from each sample were resuspended in 0.1% FA. The separation of tryptic peptides was performed on an ACQUITY MClass System (Waters Corporation). 0.50 μg of each digested sample was loaded onto a Symmetry C18 5 μm, 180 μm × 20 mm precolumn (Waters Corp.) used as trapping column and subsequently separated by a 120 min reversed phase gradient at 300 nL × min-1 (linear gradient, 3%–55% ACN over 90 min) using an HSS T3 C18 1.8 μm, 75 μm × 150 mm nanoscale and LC column (Waters Corp.) maintained at 40°C. For the gradient elution water-Formic Acid (99.9/0.1, v/v) was used as eluent A and Acetonitrile Formic Acid (99.9/0.1, v/v) as eluent B. The Separated peptides have been analyzed by High Definition Synapt G2-Si Mass spectrometer directly coupled to the chromatographic system. Differential protein expression was evaluated with a data-independent acquisition (DIA) of shotgun proteomics analysis by Expression configuration mode (MSe). The mass spectrometer operated in “Expression Mode” switching between low (4 eV) and high (25–60 eV) collision energies on the gas cell, using a scan time of 1.0 s per function over 50–2000 m/z. All spectra were acquired in Ion Mobility Mode by applying a wave velocity for the ion separation of 1.000 m/s and a transfer wave velocity of 175 m/s. The processing of low and elevated energy, added to the data of the reference lock mass ([Glu1]-Fibrinopeptide B Standard, Waters Corp.) provides a time-aligned inventory of accurate mass retention time components for both the low and elevated-energy (EMRT, exact mass retention time) Each sample was run in three technical replicates.
Continuum LC-MS data from three replicate experiments for each sample were processed for qualitative and quantitative analysis using the software Progenesis (Waters Corp.). The qualitative identification of proteins was obtained by searching in Homo sapiens database (UNIPROT Protein reviewed release 2020-04). Search parameters were set as: automatic tolerance for precursor ions and for product ions, minimum 2 fragment ions matched per peptide, minimum 3 fragment ions matched per protein, minimum 1 peptide matched per protein, 2 missed cleavage, carbamydomethylation of cysteines as fixed modification and oxidation of methionines as variable modifications, false discovery rate (FDR) of the identification algorithm < 1%. Label free quantitative analysis was obtained using the relative abundance intensity integrated in Progenesis software, using all peptides identified for normalization. The expression analysis was performed considering technical replicates available for each experimental condition following the hypothesis that each group is an independent variable. Filtered tables were generated to include proteins found at least in two out of three technical replicates and to exclude proteins showing less than 20% change and those showing no statistical significance according to ANOVA ≥ 0.05.
Protein Ontologies and Network Analysis
To identify biologically relevant molecular pathways, the proteomic dataset was analyzed by a bioinformatic analysis tool based on QIAGEN’S Ingenuity Pathways Analysis (QIAGEN’S Ingenuity pathway analysis, Ingenuity Systems, http://www.qiagen.com/ingenuity). This allows us to explore functional associations relevant to the experimental results. The analysis parameters were set as: Direct and indirect relationship; endogenous chemical substances included; all molecules and/or relationships considered as Summary Filter. The most significant categories associated with the loaded datasets were identified by calculating their significance (p value, Fischer test). A p value threshold of 0.05 has been set as significant for an association between gene/proteins present in the datasets and each pathway (Canonical pathway, Upstream regulators and Biological Function). For the Gene Ontology analysis, we made use of the PANTHER 14.0 (Protein ANalysis THrough Evolutionary Relationships) Classification System (GENEONTOLOGY Unifying Biology) containing 15.552 protein families, and STRING: functional protein association networks, v.11.0.
The heatmap was built with the heatmapper (http://www.heatmapper.ca/expression), using the median value of the 3 normalized log2 technical replicates of the sum of the Intensity Peaks for each protein identified in each patient. We used an Average Linkage clustering method and the Pearson correlation method for the measurement of the linear correlation between variable samples.
The visualization of proteomic data was performed in the Python programming language (v. 3.7.3). The pathway enrichment analyses were carried out in ClusterProfiler (Yu et al., 2012) and were visualized in R (v. 4.0) (5) and Cytoscape environments (v. 3.8) (Otasek et al., 2019; Shannon et al., 2003).
Re-analysis of scRNA-seq Data
The scRNA-seq data from bronchoalveolar lavage fluid (BALF) from patients with varying severity of COVID-19 disease and from healthy was acquired from the Gene Expression Omnibus (GEO) database under the series number GEO: GSE145926, which contains 12 data of lung tissue generated using 3′ V2 chemistry kit on Chromium Single cell controller (10xGenomics). Filtered feature-barcode matrix was used in the following analysis. All additional analysis was performed using Seurat v3. Cells with less than 200 genes detected or greater than 10% mitochondrial RNA content were excluded from analysis, with 90,046/94,583 cells passing filter (95%).
For clustering of all cell types in the BALF of patients, raw UMI counts were log normalized and variable genes called on each dataset independently based on average expression greater than 0.1 and average dispersion greater than 1. To remove batch effects between samples associated with a housekeeping gene expression signature, we assigned a housekeeping score using the AddModuleScore function based on C1orf43 CHMP2A, EMC7, GPI, PSMB2, PSMB4, RAB7A, REEP5, SNRPD3, VCP, VPS29 genes. Additionally, we assigned scores for S and G2/M cell cycle phases based on previously defined gene sets using the CellCycleScoring function. Scaled z-scores for each gene were calculated using the ScaleData function and regressed against the number of UMIs per cell, mitochondrial RNA content, S phase score, G2/M phase score and housekeeping score. Scaled data was used as an input into PCA based on variable genes. Clusters were identified using shared nearest neighbor (SNN) based clustering based on the first 20 PCs with k = 30 and resolution = 0.4. The same principal components were used to generate the UMAP projections, which were generated with a minimum distance of 1 and 20 neighbors.
Differentially expressed genes between mild/severe COVID-19 patients and controls for each cluster were identified using FDR < 0.05 and |avg_logFC| > 0.25. Functional enrichment analysis was performed using Enrichr tool (Chen et al., 2013). As input, we utilized the genes that were upregulated in mild/severe COVID-19 patients compared to controls in each one of the 6 CD14+ monocyte clusters. Corrplot R package (Simko, 2017) was used to display the enrichment score.
Network analysis was performed with NetworkAnalyst tool (Zhou et al., 2019). As input, we utilized the genes that were up-rgulated in severe COVID-19 patients compared to controls in at least 3 out 6 CD14+ monocyte clusters (parameters: IMEx Interactome as protein-protein interaction and Minimum Network). Data was visualized by Cytoscape tool (Shannon et al., 2003).
Quantification and Statistical Analysis
Proteomic and RNA-seq were analyzed as described above. Additional data were plotted and analyzed using the GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA). For analyses between 2 groups, Student’s t test was used. For comparisons among more than 2 groups, either one-way ANOVA, followed by Tukey post hoc tests was used. Differences were considered statistically significant when P value was < 0.05.
Acknowledgments
We thank the Hematology and Hemotherapy Center of Unicamp (Campinas, São Paulo, Brazil) and Nelson Cabral Junior and Regina Amelia Moraes for providing the buffy coats used for peripheral blood mononuclear cell isolation in this study. We thank Edson Durigon for providing the SARS-CoV-2 and Eurico Arruda for providing RSV and IAV. We thank Elzira E. Saviani for technical support. We would like to thank Aline Mika Matsuguma for designing our schematic figures. We gratefully acknowledge Dr. Catherine A. Reardon from the Committee on Molecular Metabolism and Nutrition, University of Chicago and from Ben May Department for Cancer Research, University of Chicago, Chicago, IL, USA, for revising the manuscript. We thank the São Paulo Research Foundation – FAPESP (grant numbers 2015/15626-8, 2016/23328-0, 2016/18031-8, 2018/22505-0, 20/04579-7, 2020/04746-0, 2019/06372-3, 2020/04558-0, 2020/04919-2, 2020/04583-4, 2017/01184-9, 2020/04522-5, and 2019/00098-7); Fundo de Apoio ao Ensino, Pesquisa e Extensão (FAEPEX), Unicamp (grant number 2274/20); the Brazilian National Council for Scientific and Technological Development – CNPq; and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) (Finance Code 001) for funding this project.
Author Contributions
Conceptualization, A.C.C., G.G.D., L.d.B.M., and P.M.M.-V.; Methodology, A.C.C., G.G.D., L.d.B.M., P.M.M.-V., and A.S.F.; Software, H.I.N., R.F.C., C.A.O.d.B.J., J.L.J.R., D.M.-d.-S., V.C.C., F.C., P.H.V., and G.R.-d.-O.; Validation, A.S.V.; Formal Analysis, A.C.C., G.G.D., A.S.F., D.M.-d.-S., R.F.C., and H.I.N.; Investigation, A.C.C., G.G.D., L.d.B.M., A.S.F., G.F.d.S., S.P.M., J.V.V.-d.-S., J.S.P., M.C.M., P.L.P., D.A.T.-T., K.B.d.S., R.E.M., L.D.C., A.B., V.O.B., N.S.B., D.M.-d.-S., V.C.C., F.C., P.H.V., G.R.-d.-O., E.M., R.G.U., A.F.B., T.A.N., L.C.R., A.C.P., M.V.A., M.L.M., and P.M.M.-V.; Resources, P.M.M.-V., M.A.M., J.L.P.-M., and L.A.V.; Data Curation, A.C.C., G.G.D., L.d.B.M., P.M.M.-V., A.S.F., D.M.-d.-S., H.I.N., and R.F.C.; Writing – Original Draft, A.C.C., G.G.D., L.d.B.M., and P.M.M.-V.; Writing – Review & Editing, A.C.C., G.G.D., L.d.B.M., H.I.N., A.D., A.S.F., J.L.P.-M., M.A.M., M.A.R.V., and P.M.M.-V.; Visualization, A.C.C., G.G.D., L.d.B.M., and A.S.F.; Project Administration, A.C.C., G.G.D., L.d.B.M., and P.M.M.-V.; Supervision, P.M.M.-V.
Declaration of Interests
Authors declare no competing interests.
Published: July 17, 2020; corrected online: August 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cmet.2020.07.007.
Supplemental Information
References
- Aguer C., Gambarotta D., Mailloux R.J., Moffat C., Dent R., McPherson R., Harper M.E. Galactose enhances oxidative metabolism and reveals mitochondrial dysfunction in human primary muscle cells. PLoS One. 2011;6:e28536. doi: 10.1371/journal.pone.0028536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baardman J., Verberk S.G.S., Prange K.H.M., van Weeghel M., van der Velden S., Ryan D.G., Wüst R.C.I., Neele A.E., Speijer D., Denis S.W. A defective pentose phosphate pathway reduces inflammatory macrophage responses during hypercholesterolemia. Cell Rep. 2018;25:2044–2052.e5. doi: 10.1016/j.celrep.2018.10.092. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020 doi: 10.1038/s41586-020-2332-7. Published online May 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breda C.N.S., Davanzo G.G., Basso P.J., Saraiva Câmara N.O., Moraes-Vieira P.M.M. Mitochondria as central hub of the immune system. Redox Biol. 2019;26:101255. doi: 10.1016/j.redox.2019.101255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., O’Sullivan D., Klein Geltink R.I., Curtis J.D., Chang C.H., Sanin D.E., Qiu J., Kretz O., Braas D., van der Windt G.J. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani E.T., Pell V.R., Gaude E., Aksentijević D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Ord E.N.J., Smith A.C. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marcken M., Dhaliwal K., Danielsen A.C., Gautron A.S., Dominguez-Villar M. TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci. Signal. 2019;12:eaaw1347. doi: 10.1126/scisignal.aaw1347. [DOI] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler U., Kuharev J., Navarro P., Tenzer S. Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 2016;11:795–812. doi: 10.1038/nprot.2016.042. [DOI] [PubMed] [Google Scholar]
- Guo W., Li M., Dong Y., Zhou H., Zhang Z., Tian C., Qin R., Wang H., Shen Y., Du K. Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab. Res. Rev. 2020 doi: 10.1002/dmrr.3319. Published online March 31, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J., Numa A., Newth C.J. Acute respiratory distress syndrome caused by respiratory syncytial virus. Pediatr. Pulmonol. 1997;23:176–183. doi: 10.1002/(sici)1099-0496(199703)23:3<176::aid-ppul2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan R.E., Adab P., Cheng K.K. Covid-19: risk factors for severe disease and death. BMJ. 2020;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- Medeiros A.I., Bonato V.L., Malheiro A., Dias A.R., Silva C.L., Faccioli L.H. Histoplasma capsulatum inhibits apoptosis and Mac-1 expression in leucocytes. Scand. J. Immunol. 2002;56:392–398. doi: 10.1046/j.1365-3083.2002.01142.x. [DOI] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Däbritz J.H.M., Gottlieb E., Latorre I. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Watanabe T., Ito H., Watanabe S., Goto H., Gao P., Hughes M., Perez D.R., Donis R., Hoffmann E. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otasek D., Morris J.H., Bouças J., Pico A.R., Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20:185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Brasileiro G.S., Soleiman L.N., Silva C.C., Kavaguti C.S. Severe acute respiratory syndrome caused by the influenza A (H1N1) virus. J. Bras. Pneumol. 2010;36:386–389. doi: 10.1590/s1806-37132010000300017. [DOI] [PubMed] [Google Scholar]
- Rodriguez A.E., Ducker G.S., Billingham L.K., Martinez C.A., Mainolfi N., Suri V., Friedman A., Manfredi M.G., Weinberg S.E., Rabinowitz J.D., Chandel N.S. Serine metabolism supports macrophage IL-1β production. Cell Metab. 2019;29:1003–1011.e4. doi: 10.1016/j.cmet.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko, T.W.a.V. (2017). R package “corrplot”: visualization of a correlation matrix (Version 0.84).
- Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votyakova T.V., Reynolds I.J. DeltaPsi(m)-Dependent and -independent production of reactive oxygen species by rat brain mitochondria. J. Neurochem. 2001;79:266–277. doi: 10.1046/j.1471-4159.2001.00548.x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X., Shi H., Zhou M. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020 doi: 10.1148/radiol.2020200843. Published online March 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J., Lee S., Park M., Kim T.Y., Park M.G., Choi B.Y., Kim D., Chang H., Kim V.N., Lee C.J. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19) Exp. Neurobiol. 2020;29:107–119. doi: 10.5607/en20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Soufan O., Ewald J., Hancock R.E.W., Basu N., Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., She Z.G., Cheng X., Qin J.J., Zhang X.J., Cai J., Lei F., Wang H., Xie J., Wang W. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020;31:1068–1077.e3. doi: 10.1016/j.cmet.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019157 and https://doi.org/10.6019/PXD019157. All other supporting data in this study are available from the Lead Contact on request.