Abstract
Sophisticated analysis and simplified visualization are crucial for understanding complicated structures of biomacromolecules. DSSR (Dissecting the Spatial Structure of RNA) is an integrated computational tool that has streamlined the analysis and annotation of 3D nucleic acid structures. The program creates schematic block representations in diverse styles that can be seamlessly integrated into PyMOL and complement its other popular visualization options. In addition to portraying individual base blocks, DSSR can draw Watson-Crick pairs as long blocks and highlight the minor-groove edges. Notably, DSSR can dramatically simplify the depiction of G-quadruplexes by automatically detecting G-tetrads and treating them as large square blocks. The DSSR-enabled innovative schematics with PyMOL are aesthetically pleasing and highly informative: the base identity, pairing geometry, stacking interactions, double-helical stems, and G-quadruplexes are immediately obvious. These features can be accessed via four interfaces: the command-line interface, the DSSR plugin for PyMOL, the web application, and the web application programming interface. The supplemental PDF serves as a practical guide, with complete and reproducible examples. Thus, even beginners or occasional users can get started quickly, especially via the web application at http://skmatic.x3dna.org.
INTRODUCTION
Sophisticated analysis and simplified visualization are crucial for interpreting complicated 3D structures of biomacromolecules, such as those deposited in the PDB (1). Unlike proteins and carbohydrates, nucleic acids possess unique structural features that require dedicated software tools for their in-depth analysis and effective depiction. The aromatic, planar bases are a defining feature of RNA and DNA, and they can pair and stack with each other in various ways. Together with the sugar-phosphate backbone, the bases participate in many tertiary motifs (2,3). Thus, bases have been simplified as slabs to emphasize base pairing and stacking interactions (4), or as sticks in ribbon representations (5).
With the exception of Jmol/JSmol (6), however, available visualization tools lack a sophisticated analysis engine for nucleic acid structures. They cannot select features such as hairpin/junction loops or noncanonical pairs, render minor grooves as an attribute, or simplify RNA/DNA representations beyond the level of individual bases or Watson-Crick (WC) pairs. As a salient example, none of them can automatically identify and highlight G-quadruplexes, an increasingly important class of noncanonical nucleic acid structures (7–9).
DSSR (Dissecting the Spatial Structure of RNA) (10) is an integrated computational tool that has been designed from the bottom up to streamline the analysis and annotation of 3D nucleic acid structures. Starting from an atomic-coordinate file in PDB or PDBx/mmCIF format, the program automatically identifies and characterizes numerous structural features, including modified nucleotides, arrays of stacked bases, (noncanonical) base pairs, higher-order base associations (multiplets), pseudoknots of arbitrary complexity, various types of ‘closed’ loops, canonical stems, coaxially stacked helices, and G-quadruplexes. DSSR has been extensively tested using all nucleic-acid-containing structures in the PDB and is continuously developed following user feedback. Over the years, DSSR has been widely cited in scientific literature (11–18) and adopted into many other structural bioinformatics resources (19–26).
The DSSR-Jmol integration (6) introduces a novel and powerful SQL-like selection mechanism for DSSR-derived structural features, and provides simplified representations of nucleic acids as step diagrams and base blocks in Jmol/JSmol. This work fills a gap in RNA/DNA structural bioinformatics, and serves as an example for incorporating DSSR-derived features into other molecular viewers. PyMOL (Schrödinger, Inc.) is an industry leader in 3D molecular visualization, and it is especially well-developed for protein structures. For nucleic acids, however, PyMOL is deficient in two main aspects. Firstly, it lacks a selection mechanism for RNA/DNA-specific features (including WC pairs or G-quadruplexes). Secondly, its representation styles of nucleic acids, most notably the planar nitrogenous bases, are limited. DSSR has unique features that can significantly enhance the selection and visualization of nucleic acid structures in PyMOL. Specifically, DSSR creates schematic block representations in diverse styles that can be seamlessly integrated into and complemented with other popular visualization options of PyMOL. DSSR dramatically simplifies the depiction of G-quadruplexes by automatically detecting G-tetrads and depicting them as large square blocks. The DSSR-PyMOL schematics are simple, informative, and appealing. They are especially effective for RNA and DNA structures with up to dozens of nucleotides (Supplementary Figure S1).
The idea of representing bases and WC-pairs as rectangular blocks came from the pioneering work of Calladine et al. (27,28) The block schematics were first implemented in the pair of SCHNAaP/SCHNArP programs (29,30) for rigorous analysis and reversible rebuilding of double-helical nucleic acid structures. The algorithms that underpinned SCHNAaP/SCHNArP laid the foundation of ‘analyze’ and ‘rebuild’, two core components of the 3DNA suite of programs (31–33). 3DNA also takes advantage of the standard base reference frame (34), and comprises quite a few other related programs. One of them is ‘blocview’, a script which calls several 3DNA utility programs to generate individual base blocks and set the view, MolScript (35) to produce backbone ribbons, and Raster3D (36) to render the composite image. The 3DNA ‘blocview’ schematics catch characteristic attributes of nucleic acid structures. They have gradually become popular and been adopted into the RCSB PDB (1) and the NDB (37), and then propagated into other bioinformatics resources (e.g., the ‘RNA Structure Atlas’ website hosted by the Leontis-Zirbel RNA group).
DSSR supersedes ‘blocview’ by eliminating all the internal and external dependencies of the 3DNA utility program. DSSR produces block representations, not only of individual bases but also WC-pairs and G-tetrads, that can be fed directly into PyMOL. The DSSR-PyMOL integration is easier to use, has more features, and produces better schematics than the original 3DNA-blocview approach. Users can easily create cartoon-block images via the command-line interface (CLI), the DSSR plugin for PyMOL, the web application, or the web application programming interface (API). Together, these four interfaces cover virtually all conceivable cases of usage. Moreover, the supplemental PDF has been diligently written to serve as a practical guide, with complete and reproducible examples. Thus, even beginners or occasional users can get started quickly, particularly via the web application at http://skmatic.x3dna.org.
MATERIALS AND METHODS
There are four ways to produce DSSR-enabled schematics with PyMOL: the CLI, the dssr_block PyMOL plugin, the web application, and the web API. The CLI interface requires the installation of DSSR, as detailed in the supplemental PDF. In essence, users simply need to download one binary executable that is <2MB in size and available for macOS, Linux, or Windows. DSSR is self-contained, with zero runtime dependencies on third-party libraries. The dssr_block plugin is available from the PyMOL Wiki page. The script can be directly executed, imported as a Python module, or installed with the ‘Plugin Manager’ of PyMOL. The simplest user interface is via the web application at http://skmatic.x3dna.org: it only requires a modern web browser with internet connection. Software developers can take advantage of the web API (http://skmatic.x3dna.org/api) programmatically, without installation of DSSR or the plugin.
In this section, we first define DSSR blocks (Figure 1), and then outline the major aspects of each of the four interfaces for creating DSSR-PyMOL cartoon-block images. See the supplemental PDF for complete and reproducible examples presented in a tutorial style.
Figure 1.
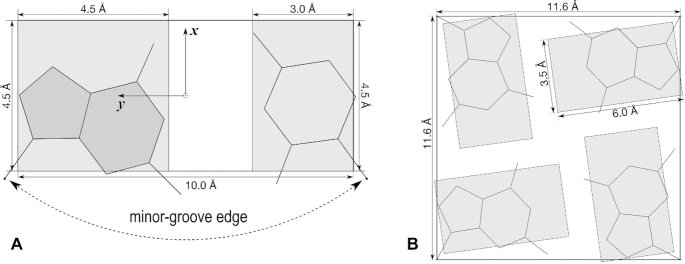
Definition of various types of DSSR blocks, illustrated using idealized, planar Watson-Crick (WC) base-pair and G-tetrad geometries. (A) Purine (A, G) and pyrimidine (C, T, U) base blocks and the WC-pair block in default dimensions. The standard base reference frame is attached, and the minor-groove edge is marked. (B) The slim purine (guanine) block and the square G-tetrad block for simplified visualizations of G-quadruplexes. The slim pyrimidine block is of dimension 4 Å-by-3.5 Å. All blocks have a thickness of 0.5 Å by default. This figure and the following ones were annotated using Inkscape (https://inkscape.org).
Definition of blocks
Currently available DSSR blocks are defined in Figure 1. The default purine (A, G) and pyrimidine (C, T, U) base blocks (Figure 1A) have already been used in the 3DNA-blocview program (31–33). The long WC-pair block, however, is new to DSSR. The standard base reference frame (34) has its positive x-axis pointing from the minor-groove edge to the major groove. The combined usage of WC-pair blocks and minor-groove edges reveals double-helical stems (10), their right- or left-handedness, and the alternations of minor- and major-grooves in unprecedented clarity (see Figure 2).
Figure 2.
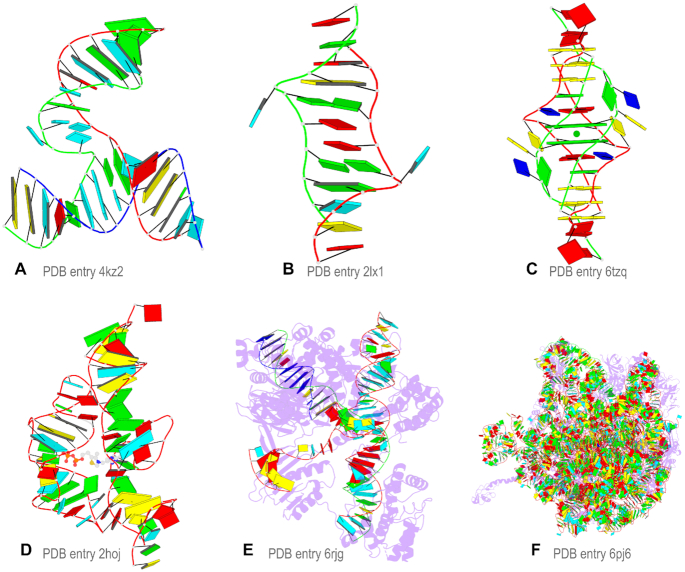
Cartoon-block schematics enabled by the DSSR-PyMOL integration for six representative PDB entries. Note WC-pairs as long blocks and minor-groove edges rendered in black (A, B), G-tetrads as square blocks and the metal ion as sphere (C), the ligand as balls-and-sticks (D), and proteins as purple cartoons (E, F). Color code: A, red; C, yellow; G, green; T, blue; U, cyan; G-tetrad, green; WC-pairs, per base in the leading strand.
A brand-new feature of DSSR is the automatic identification and comprehensive characterization of G-quadruplexes (Figure 3, see also http://g4.x3dna.org). Details of the underlying algorithms and survey results on all G-quadruplexes available in the PDB will be reported elsewhere. Here we introduce the slim-guanine block that unambiguously highlights the H-bonding directionality in G-tetrads, and the large square G-tetrad block that greatly simplifies the visualization of G-quadruplexes (Figure 1B, see also Figure 3).
Figure 3.
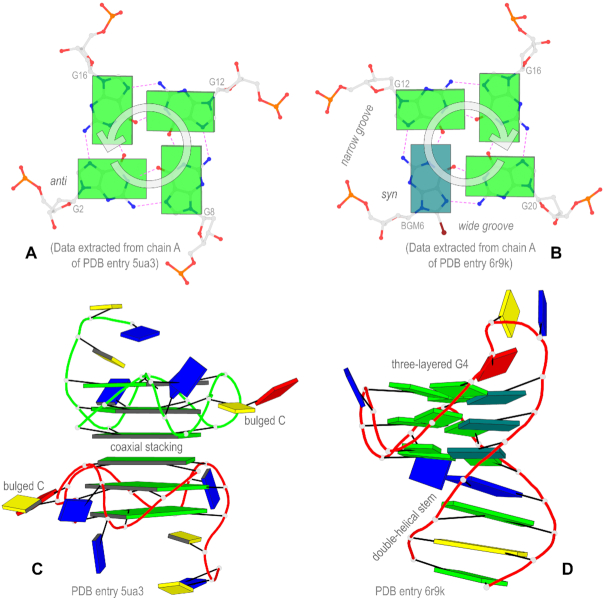
Innovative block representations introduced in DSSR to simplify the visualization of G-quadruplexes. (A) The anti guanine (lower-left) leads to a counter-clockwise orientation of H-bonding interactions (dashed lines in magenta) from the WC edge to the major-groove (Hoogsteen) edge. (B) The syn guanine (colored teal) reverses the direction of the H-bonds to clockwise, and it also creates a wide groove and a narrow groove. (C) A parallel G-quadruplex (G4) illustrated with square blocks for G-tetrads, highlighting a six-layered G4-helix composed of two three-layered G4-stems via coaxial stacking interactions. The two bulged cytosines are marked. (D) Another G4 with a (1+3) hybrid conformation, emphasizing the duplex-quadruplex transition and the three guanines in syn conformation.
The command-line interface
The CLI is the most efficient and versatile way to explore the numerous features that DSSR has to offer. It also provides the basis for the other three interfaces that make DSSR-PyMOL schematics easily accessible to a wider audience. There are five options to set DSSR block types and PyMOL rendering parameters: (a) The block-file option turns on the block representation, with features specified as a list of keywords (Supplementary Figure S2A–C). For example, the list ‘wc-minor-g4’ (i.e., block-file=wc-minor-g4) leads to WC-pairs being rendered as long blocks, minor-grooves colored in black, and G-tetrads as square blocks (Figure 1, see also examples in Figures 2 and 3). The order of the keywords in a list does not matter. For instance, ‘g4-wc-minor’ would have the same effect as ‘wc-minor-g4’. If a structure does not possess a feature, the corresponding keyword will be ignored. For example, if ‘wc’ is specified for a structure without WC pairs (e.g., Figures 2C and 3C), nothing will occur. (b) The block-color option is used to customize the colors of blocks or DSSR-derived features (Supplementary Figure S2D). (c) The block-depth option sets the thickness of blocks (Supplementary Figure S2D). (d) The cartoon-block option combines PyMOL cartoon representations and DSSR block schematics with a selected set of style settings (Supplementary Figure S3A). (e) The blocview option is like cartoon-block but it also sets the structure into the most extended view and draws solid-colored blocks with black outlines (Figures 2 and 3, and Supplementary Figure S3B). The last two options can be used to automate the process of creating DSSR-PyMOL schematics. See Supplementary Figures S4–S6 and the corresponding commands listings for worked examples.
The dssr_block PyMOL plugin
The dssr_block plugin makes DSSR transparent to PyMOL users (Supplementary Figure S7). Essentially, it adds the dssr_block command to PyMOL for interactive creation of DSSR blocks in various styles (Figure 1, see also examples in Figures 2 and 3). Instead of working directly with coordinate inputs and block outputs on the command line, the plugin takes advantage of PyMOL’s atom selections and displays the results instantaneously. This way, it seamlessly integrates and complements the other visualization options in PyMOL.
The web application
The http://skmatic.x3dna.org site provides a cross-platform, web-based application for easy creation of DSSR-PyMOL schematics (Supplementary Figure S8). Implemented with standard web technologies (see the supplemental PDF), the client-side user interface works in all modern web browsers across a wide spectrum of platforms. The web application is intuitive, allowing even novices or occasional users to get started quickly.
The web application consists of two components: (a) pre-calculated schematics and summary information of PDB entries, a common use case (Figure 4, and Supplementary Figure S9); (b) the general application, necessitating the input of an atomic-coordinate file together with optional customizations.
Figure 4.
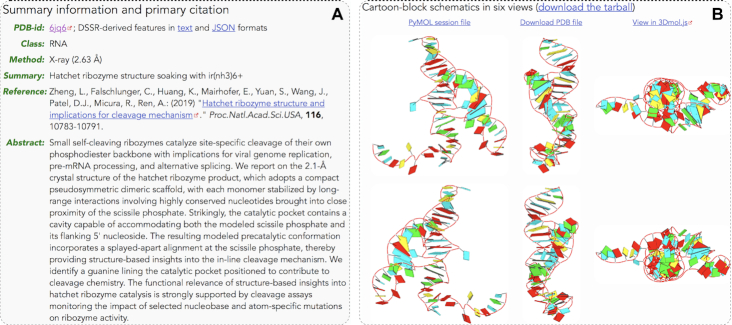
Screenshots of a sample pre-calculated PDB entry (6jq6) from the web application at http://skmatic.x3dna.org. (A) Structure summary and primary citation compiled from the RCSB PDB and the PubMed. (B) DSSR-PyMOL schematics in six orthogonal perspectives. The images are based on the asymmetric unit of the PDB entry.
The web API
The web API offers a programmatic means to generate DSSR-enabled schematics with PyMOL, without the need of any DSSR-related installations. It has been implemented in Ruby, using the Roda routing-tree web toolkit (https://roda.jeremyevans.net/). The main access endpoint is http://skmatic.x3dna.org/api. See the supplemental PDF for more information.
RESULTS AND DISCUSSIONS
Cartoon-block schematics
Figure 2 showcases representative cartoon-block schematics enabled by the DSSR-PyMOL integration. The six PDB entries illustrated there cover different types of RNA, DNA, and their complexes with ligands or proteins: (a) 4kz2, a crystal structure of the phi29 pRNA three-way junction core (38); (b) 2lx1, the major conformation of the internal loop 5′GAGU/3′UGAG (39); (c) 6tzq, a DNA G-quadruplex/i-motif hybrid (40); (d) 2hoj, the crystal structure of an Escherichia coli thi-box riboswitch bound to thiamine pyrophosphate (41); (e) 6rjg, a cryo-EM structure of the St1Cas9-sgRNA-AcrIIA6-tDNA59-ntPAM complex (42); and (f) 6pj6, a high resolution cryo-EM structure of the E. coli 50S subunit (43). See the supplemental PDF for detailed instructions to reproduce these images.
The DSSR block-file=wc-minor-g4 settings works extremely well with the blocview option in the majority of cases (see also below for pre-calculated PDB entries). Additional features can be turned on: Figure 2C shows the Ba2+ metal ion between the two G-tetrads, and Figure 2D highlights the thiamine diphosphate (a small-molecule metabolite). As depicted in Figure 2A–E and Supplementary Figure S5, the cartoon-block schematics are simple and aesthetic, highly effective for nucleic acid structures with dozens of nucleotides. For gigantic ribosomal structures (available only in PDBx/mmCIF format in the PDB) such as 6pj6 (Figure 2F), the image becomes a colorful blob, yet it remains easily recognizable.
By default, DSSR reads in the first model and therefore only creates a representative schematic of an NMR ensemble. With the dssr_block PyMOL plugin, it is straightforward to produce an aggregate image with all models to show conformational variations (Supplementary Figure S7B). Biological units of X-ray crystal structures may contain multiple models formatted in an NMR-like ensemble in the PDB. This is confusing since the different models in a biological unit are symmetry related, whereas in an authentic NMR ensemble, they are fully independent. In such cases (e.g., Figure 2C for 6tzq), the DSSR symm option is required to process the entire structure of a biological unit composed of multiple models. DSSR also auto-detects polypeptide chains in proteins (18), and outputs PyMOL commands to render them as purple cartoons with 70% transparency (Figure 2E, F).
Simplified representations of G-quadruplexes
G-quadruplexes are noncanonical nucleic acid structures formed from guanine-rich base sequences. The building block of G-quadruplexes is G-tetrad, a special type of higher-order, planar base associations (termed ‘multiplets’ in DSSR) (10). The four guanines of a G-tetrad are arranged in a cyclic manner via four consecutive G+G pairs (10,31), each held together by two H-bonds (Figure 3A-B, and Supplementary Figure S6). G-quadruplexes arise from stacking of G-tetrads and are stabilized by cations (Figure 3C, D, and also Figure 2C). The guanine base can be in anti or syn glycosidic torsion angle conformation, leading to polymorphic topologies and groove-width variations in the 3D structures of G-quadruplexes (Figure 3).
DSSR can detect and characterize G-quadruplexes, simply starting from an atomic-coordinate file. Please see http://g4.x3dna.org for experimentally determined G-quadruplexes that are auto-annotated by DSSR from the PDB. A detailed description of this new DSSR module on G-quadruplexes will be reported in a separate article. Figure 3 showcases some of the structural features of G-quadruplexes that are made clear by the DSSR-PyMOL integration: panels A and B show the molecular images of two G-tetrads, overlaid with guanines as slim blocks. Switching a guanine from anti to syn conformation reverses the H-bonding directionality of the G-tetrads from counter-clockwise to clockwise. The syn conformation (of the modified nucleotide BGM, 8-bromo-2′-deoxyguanosine) stands out by being colored teal instead of green for the anti guanines. Panel C illustrates the distinction between helix and stem and the definition of coaxial stacking interactions (10), in the context of G-quadruplexes. The meaning of the three terms is explicit in the simplified representation, using large square blocks for G-tetrads and different colorings (red and green) for the two DNA chains. Panel D demonstrates the duplex-quadruplex transition using a combination of long blocks for WC-pairs and slim guanine blocks for G-tetrads. The solid-colored blocks make the three-layered G-quadruplex, including the syn guanines, easily discernible. In Figure 3, panels A and C are based on the crystal structure of a parallel DNA G-quadruplex with bulged cytosines (PDB entry 5ua3) (13), whilst panels B and D pertain to the (1+3) hybrid G-quadruplex structure with lateral-propeller-propeller loops and three syn guanines (PDB entry 6r9k) (44).
As shown in Figures 1B and 3A, B, the square-block representation is defined specifically for the G-tetrad of G-quadruplexes. By design, the square block for the G-tetrad parallels the long-block for WC pairs in canonical DNA and RNA double helices. Just as there are numerous types of noncanonical base pairs, many forms of non-G tetrads have been reported (9,45,46), including A-, C-, T- and U-tetrads, or mixed tetrads. Also, there are cases of pseudo-G-tetrads (47,48), where some of the G·G pairs are distinct from the one in the G-tetrad of G-quadruplexes (see above and Figure 3A, B). A recent survey of the PDB reveals that over 80% of all tetrads are the G-tetrad in G-quadruplexes (46). As noted at the beginning of this section, a tetrad (G-tetrad or otherwise) is a special type of ‘multiplets’ in DSSR (10), and it may belong to a larger context with more than four bases (pentad, hexad, heptad, and octad etc.) The WC-pair block schematics not only make the canonical duplexes stand out, but also highlight single-stranded regions and noncanonical pairs (e.g., Figure 2A, B). In a similar fashion, the G-tetrad block schematics introduced here allow the G-quadruplexes and nearby components (including other tetrads) to be immediately recognizable (e.g., Figures 2C and 3C).
The advantages enabled by DSSR for the visualization of G-quadruplexes in PyMOL are obvious (Figure 3). The DSSR-PyMOL integration, like the DSSR-Jmol case (6), exemplifies the critical roles that a domain-specific analysis engine may play in an otherwise general-purpose molecular visualization tool.
Pre-calculated PDB entries
A common usage of the DSSR-enabled schematics with PyMOL is for nucleic-acid-containing structures in the PDB. The web application (http://skmatic.x3dna.org) allows users to conveniently access pre-calculated PDB entries via a clean and clutter-free interface. The default image (main view) of a PDB entry was created with the two DSSR options block-file=wc-minor-g4 and blocview (see above), for the first model of an NMR-ensemble or the asymmetric unit of a crystal or cryo-EM structure.
As an example, Figure 4 shows screenshots for PDB entry 6jq6, a hatchet ribozyme structure solved by X-ray crystallography (49). The results include two parts: (a) Summary information and the primary citation (including abstract, if any) compiled automatically from the RCSB PDB (1) and the PubMed. DSSR-derived features in human-readable text and machine-friendly JSON formats are available. (b) Cartoon-block schematics in six orthogonal views (also accessible as a tarball): the main view (top-left) and the other five orientations derived from it via simple rotations. The PyMOL session file for the main view can be downloaded for further customizations. The corresponding PDB coordinate file for this view can also be downloaded or visualized directly online using 3Dmol.js (50). Furthermore, the main-view image is accessible programmatically via the web API (see the supplemental PDF for details).
CONCLUSIONS
DSSR-enabled block schematics bring innovative, simplified visualizations of 3D nucleic acid structures into PyMOL. DSSR implements long blocks for WC-pairs, minor-grooves highlighting, and most notably, large square blocks for G-tetrads in G-quadruplexes. These features can be seamlessly integrated into PyMOL and complement its other popular visualization options. As demonstrated in Figures 2–4, the DSSR-PyMOL schematics are informative, aesthetically pleasing, and highly characteristic of nucleic acids. For small to mid-sized RNA- or DNA-containing structures, the base identity, pairing geometry, stacking interactions, double-helical stems, and G-quadruplexes are clear. The four interfaces allow users to easily create novel, publication quality schematics of 3D nucleic acid structures. Finally, all results reported here are completely reproduceable (see the supplemental PDF). Any questions related to this work are welcome and will be openly addressed on the 3DNA Forum (http://forum.x3dna.org).
Supplementary Material
ACKNOWLEDGEMENTS
I would like to thank Christopher A. Hunter, Christopher R. Calladine, Helen M. Berman, Catherine L. Lawson, Zukang Feng, Wilma K. Olson and Harmen J. Bussemaker for their helpful input on the block schematic during its continuous evolution for over two decades. I appreciate Thomas Holder (PyMOL Principal Developer, Schrödinger, Inc.) for writing the DSSR plugin for PyMOL, and for providing insightful comments on the manuscript and the web application interface. I also thank Jessalyn Lu and Yin Yin Lu for proofreading the manuscript, and the user community for feedback.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM096889]. Funding for open access charge: National Institutes of Health (NIH) [R01GM096889].
Conflict of interest statement. None declared.
REFERENCES
- 1. Burley S.K., Berman H.M., Christie C., Duarte J.M., Feng Z., Westbrook J., Young J., Zardecki C.. RCSB Protein Data Bank: sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education. Protein Sci. 2018; 27:316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore P.B. Structural motifs in RNA. Annu. Rev. Biochem. 1999; 68:287–300. [DOI] [PubMed] [Google Scholar]
- 3. Butcher S.E., Pyle A.M.. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011; 44:1302–1311. [DOI] [PubMed] [Google Scholar]
- 4. Couch G.S. Nucleic acid visualization with UCSF Chimera. Nucleic Acids Res. 2006; 34:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lindow N., Baum D., Leborgne M., Hege H.-C.. Interactive visualization of RNA and DNA structures. IEEE Trans. Vis. Comput. Graph. 2019; 25:967–976. [DOI] [PubMed] [Google Scholar]
- 6. Hanson R.M., Lu X.-J.. DSSR-enhanced visualization of nucleic acid structures in Jmol. Nucleic Acids Res. 2017; 45:W528–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lightfoot H.L., Hagen T., Tatum N.J., Hall J.. The diverse structural landscape of quadruplexes. FEBS Lett. 2019; 593:2083–2102. [DOI] [PubMed] [Google Scholar]
- 10. Lu X.-J., Bussemaker H.J., Olson W.K.. DSSR: An integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015; 43:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desai N., Brown A., Amunts A., Ramakrishnan V.. The structure of the yeast mitochondrial ribosome. Science. 2017; 355:528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bayrak C.S., Kim N., Schlick T.. Using sequence signatures and kink-turn motifs in knowledge-based statistical potentials for RNA structure prediction. Nucleic Acids Res. 2017; 45:5414–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meier M., Moya-Torres A., Krahn N.J., McDougall M.D., Orriss G.L., McRae E.K.S., Booy E.P., McEleney K., Patel T.R., McKenna S.A. et al.. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018; 46:5319–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan D., Piana S., Dirks R.M., Shaw D.E.. RNA force field with accuracy comparable to state-of-the-art protein force fields. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E1346–E1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berger K.D., Kennedy S.D., Turner D.H.. Nuclear magnetic resonance reveals that GU base pairs flanking internal loops can adopt diverse structures. Biochemistry. 2019; 58:1094–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giambaşu G.M., Case D.A., York D.M.. Predicting site-binding modes of ions and water to nucleic acids using molecular solvation theory. J. Am. Chem. Soc. 2019; 141:2435–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuturello F., Tiana G., Bussi G.. Assessing the accuracy of direct-coupling analysis for RNA contact prediction. RNA. 2020; 26:637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kribelbauer J.F., Lu X.-J., Rohs R., Mann R.S., Bussemaker H.J.. Toward a mechanistic understanding of DNA methylation readout by transcription factors. J. Mol. Biol. 2020; 432:1801–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baulin E., Yacovlev V., Khachko D., Spirin S., Roytberg M.. URS DataBase: Universe of RNA structures and their motifs. Database. 2016; 2016:baw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zok T., Antczak M., Zurkowski M., Popenda M., Blazewicz J., Adamiak R.W., Szachniuk M.. RNApdbee 2.0: multifunctional tool for RNA structure annotation. Nucleic Acids Res. 2018; 46:W30–W35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Antczak M., Zablocki M., Zok T., Rybarczyk A., Blazewicz J., Szachniuk M.. RNAvista: a webserver to assess RNA secondary structures with non-canonical base pairs. Bioinformatics. 2019; 35:152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallego D., Darré L., Dans P.D., Orozco M.. VeriNA3d: An R package for nucleic acids data mining. Bioinformatics. 2019; 35:5334–5336. [DOI] [PubMed] [Google Scholar]
- 23. Thiel B.C., Beckmann I.K., Kerpedjiev P., Hofacker I.L.. 3D based on 2D: Calculating helix angles and stacking patterns using forgi 2.0, an RNA Python library centered on secondary structure elements. F1000Research. 2019; 8:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yesselman J.D., Eiler D., Carlson E.D., Gotrik M.R., d’Aquino A.E., Ooms A.N., Kladwang W., Carlson P.D., Shi X., Costantino D.A. et al.. Computational design of three-dimensional RNA structure and function. Nat. Nanotechnol. 2019; 14:866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sagendorf J.M., Markarian N., Berman H.M., Rohs R.. DNAproDB: An expanded database and web-based tool for structural analysis of DNA-protein complexes. Nucleic Acids Res. 2020; 48:D277–D287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zok T., Popenda M., Szachniuk M.. ElTetrado: a tool for identification and classification of tetrads and quadruplexes. BMC Bioinformatics. 2020; 21:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Calladine C.R., Drew H.R.. A base-centred explanation of the B-to-A transition in DNA. J. Mol. Biol. 1984; 178:773–782. [DOI] [PubMed] [Google Scholar]
- 28. Calladine C.R., Drew H.R., Luisi B.F., Travers A.A.. Understanding DNA: The molecule & how it works (third edition). 2004; Amsterdam: Elsevier. [Google Scholar]
- 29. Lu X.-J., El Hassan M.A., Hunter C.A.. Structure and conformation of helical nucleic acids: Analysis program (SCHNAaP). J. Mol. Biol. 1997; 273:668–680. [DOI] [PubMed] [Google Scholar]
- 30. Lu X.-J., El Hassan M.A., Hunter C.A.. Structure and conformation of helical nucleic acids: Rebuilding program (SCHNArP). J. Mol. Biol. 1997; 273:681–691. [DOI] [PubMed] [Google Scholar]
- 31. Lu X.-J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu X.-J., Olson W.K.. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008; 3:1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li S., Olson W.K., Lu X.-J.. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019; 47:W26–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olson W.K., Bansal M., Burley S.K., Dickerson R.E., Gerstein M., Harvey S.C., Heinemann U., Lu X.-J., Neidle S., Shakked Z. et al.. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001; 313:229–237. [DOI] [PubMed] [Google Scholar]
- 35. Kraulis P.J. MolScript: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991; 24:946–950. [Google Scholar]
- 36. Merritt E.A., Bacon D.J.. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997; 277:505–524. [DOI] [PubMed] [Google Scholar]
- 37. Narayanan B.C., Westbrook J., Ghosh S., Petrov A.I., Sweeney B., Zirbel C.L., Leontis N.B., Berman H.M.. The Nucleic Acid Database: New features and capabilities. Nucleic Acids Res. 2014; 42:D114–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang H., Endrizzi J.A., Shu Y., Haque F., Sauter C., Shlyakhtenko L.S., Lyubchenko Y., Guo P., Chi Y.-I.. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the phi29 hexameric motor pRNA. RNA. 2013; 19:1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kennedy S.D., Kierzek R., Turner D.H.. Novel conformation of an RNA structural switch. Biochemistry. 2012; 51:9257–9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu B., Zhang D., Paukstelis P.J.. A DNA G-quadruplex/i-motif hybrid. Nucleic Acids Res. 2019; 47:11921–11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edwards T.E., Ferré-D’Amaré A.R.. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006; 14:1459–1468. [DOI] [PubMed] [Google Scholar]
- 42. Fuchsbauer O., Swuec P., Zimberger C., Amigues B., Levesque S., Agudelo D., Duringer A., Chaves-Sanjuan A., Spinelli S., Rousseau G.M. et al.. Cas9 allosteric inhibition by the anti-CRISPR protein AcrIIA6. Mol. Cell. 2019; 76:922–937. [DOI] [PubMed] [Google Scholar]
- 43. Stojković V., Myasnikov A.G., Young I.D., Frost A., Fraser J.S., Fujimori D.G.. Assessment of the nucleotide modifications in the high-resolution cryo-electron microscopy structure of the Escherichia coli 50S subunit. Nucleic Acids Res. 2020; 48:2723–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karg B., Mohr S., Weisz K.. Duplex‐guided refolding into novel G‐quadruplex (3+1) hybrid conformations. Angew. Chem. Int. Ed. 2019; 58:11068–11071. [DOI] [PubMed] [Google Scholar]
- 45. Liu H., Wang R., Yu X., Shen F., Lan W., Haruehanroengra P., Yao Q., Zhang J., Chen Y., Li S. et al.. High-resolution DNA quadruplex structure containing all the A-, G-, C-, T-tetrads. Nucleic Acids Res. 2018; 46:11627–11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Popenda M., Miskiewicz J., Sarzynska J., Zok T., Szachniuk M.. Topology-based classification of tetrads and quadruplex structures. Bioinformatics. 2020; 36:1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Russo Krauss I., Pica A., Merlino A., Mazzarella L., Sica F.. Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. Acta Crystallogr. D Biol. Crystallogr. 2013; 69:2403–2411. [DOI] [PubMed] [Google Scholar]
- 48. Pica A., Russo Krauss I., Parente V., Tateishi-Karimata H., Nagatoishi S., Tsumoto K., Sugimoto N., Sica F.. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res. 2017; 45:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng L., Falschlunger C., Huang K., Mairhofer E., Yuan S., Wang J., Patel D.J., Micura R., Ren A.. Hatchet ribozyme structure and implications for cleavage mechanism. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:10783–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rego N., Koes D.. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2015; 31:1322–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.