Abstract
Tissue homeostasis and regeneration rely upon resident stem cells (SCs), whose behavior is regulated through niche-dependent crosstalk. The mechanisms underlying SC identity are still unfolding. Here, using spatiotemporal gene ablation in murine hair follicles (HFs), we uncover a critical role for transcription factors (TFs) NFIB and NFIX in maintaining SC identity. Without NFI-TFs, SCs lose hair-regenerating capability, and produce skin bearing striking resemblance to irreversible human alopecia, which also displays reduced NFIs. Through single cell transcriptomics, ATAC-seq and ChIP-seq profiling, we expose a key role for NFIB/NFIX in governing super-enhancer maintenance of the key HF-SC specific TF genes. When NFIB/NFIX are genetically removed, the stemness epigenetic landscape is lost. Super-enhancers driving SC identity are decommissioned, while unwanted lineages are de-repressed ectopically. Together, our findings expose NFIB/NFIX as crucial rheostats of tissue homeostasis, functioning to safeguard the SC epigenome from a breach in lineage confinement that otherwise triggers irreversible tissue degeneration.
Adult stem cells (SCs) are required to make and repair tissues. How SCs balance self-renewal and differentiation is critical for tissue maintenance and regeneration. During homeostasis, the concerted action of local niche signals and intrinsic epigenetic regulators establish stable gene expression patterns to maintain SC identity and function1,2. Disturbance of the niche environment, e.g. upon wounding, triggers rapid rewiring of SC regulatory programs allowing them to cope with stress and restore tissue homeostasis3,4. Thus, sensitive to their microenvironment, tissue SCs fine-tune gene expression to execute proper lineage, differentiation, developmental and wound-repair programs in vivo with remarkable precision.
How transcriptional circuits are established and maintained within adult SCs remains poorly understood. Even less clear is how transcriptional programs respond to perturbations in their environment and how they are restored following return to homeostasis. This becomes particularly relevant not only in wound-repair and aging, but also in disease states, where dysfunctions in SC balance can lead to tissue degeneration and/or tumorigenesis5,6.
Murine skin offers an excellent genetically tractable system to tackle these issues. Skin SCs reside at the epithelial-mesenchymal interface, where signals from their local environment determine when they will become activated and what kind of tissue they will make3 (Fig. 1a). The hair follicle (HF) is a particularly interesting model, since it transitions through synchronized programmed episodes of tissue regeneration. With each new hair cycle, quiescent SCs residing in a niche (bulge) located at the follicle base become transiently activated to self-renew and fuel HF regeneration and hair growth7,8. In response to injury, these SCs can also be mobilized to switch fates and re-epithelialize damaged epidermis9,10.
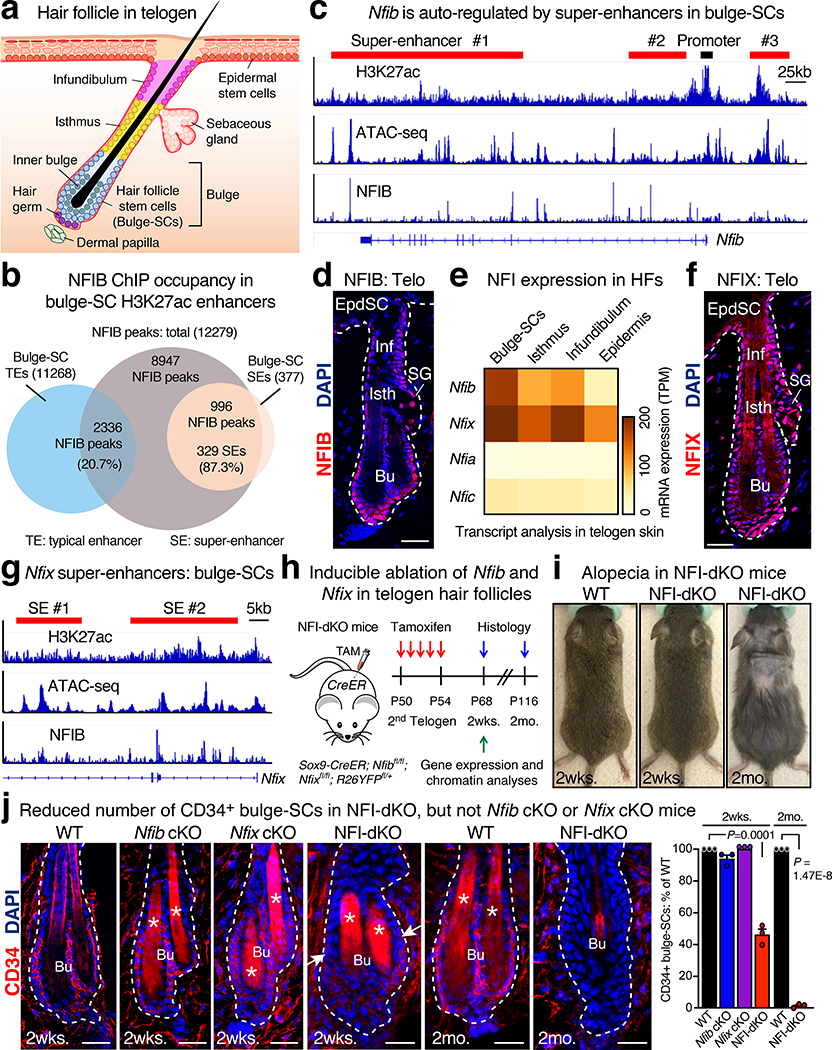
Fig. 1 |. Nfib and Nfix Redundantly Govern Bulge-SC Maintenance.
a, Schematic depicting the HF during quiescence (telogen) and relevant progenitor populations. b, Venn diagram showing enrichment of NFIB ChIP-seq peaks within bulge-SC super-enhancers (SEs) compared with typical enhancers (TEs). c, In vivo ATAC-seq and NFIB ChIP-seq tracks of the bulge-SC TF gene Nfib and its associated active super-enhancers marked by H3K27ac. Red bars denote location of super-enhancers. Exon/intron structure shown at bottom, with arrowheads indicating direction of transcription. d, NFIB immunofluorescence in 2nd telogen HFs. Newest bulge is always associated with the underlying dermal papilla, DAPI-stained. Representative image of three biological replicates. e, Gene expression (RNA-seq) profiles of NFI family members in skin epithelial progenitors. f, NFIX immunofluorescence in 2nd telogen HFs. Relative to NFIB, NFIX exhibits broader expression among skin epithelial progenitors. Representative image of three biological replicates. g, In vivo ATAC-seq and NFIB ChIP-seq tracks of the bulge-SC TF gene Nfix and its associated active super-enhancers marked by H3K27ac. Red bars denote location of super-enhancers. h, Strategy for inducible knockout of Nfib ± Nfix selectively in HFs. i, Images of NFI-dKO mice at 2wks and 2mo following targeting. Representative images of five biological replicates. j, Immunofluorescence comparing bulge-SC marker expression in telogen HFs of WT, Nfib cKO, Nfix cKO and Nfib/Nfix-dKO mice. Arrows point to decline of CD34+ bulge-SCs only in double Nfib/Nfix targeted (dKO) mice. Mean and standard deviation are shown (3–5 HFs analyzed/mouse). P values from one-way ANOVA (2 weeks timepoints; n = 3 mice) or unpaired two-tailed t-test (2 months timepoint; n = 3 mice). Asterisk denotes autofluorescence of hair shafts. All scale bars = 20μm. Bu, bulge. SG, Sebaceous gland. Inf, Infundibulum. Isth, Isthmus. Epi, Epidermis. Dashed lines, HF-dermal border. See also Source Data.
In embryonic SCs, key pluripotency genes are regulated by master transcription factors (TFs) that bind cooperatively to clustered recognition motifs within large open chromatin domains called ‘super-enhancers’11,12. Following this paradigm, quiescent bulge-SCs express a TF cohort which binds within super-enhancers to regulate critical genes, including TF genes involved in maintaining stemness13. Moreover, expression of these TF genes is highly sensitive to the niche microenvironment, rooted in additional TFs whose binding to super-enhancers is influenced by environmental signals14,15. Whereas super-enhancers represent only ~5% of active enhancers, their enrichment with signal-responsive TFs allows SCs to respond rapidly to perturbations.
Paradoxically, despite their seemingly shared role in regulating super-enhancers, bulge TFs exhibit distinct conditional loss-of-function phenotypes: Sox9 ablation transforms the bulge into an epidermal cyst16; Lhx2 ablation transforms the bulge into a sebaceous gland-like structure17; ablation of WNT effectors Tcf3/Tcf4 precociously activates the hair cycle18; and loss of Nfatc1 or Foxc1, downstream of BMP signaling, shortens quiescence and causes constitutive hair cycling19–22. These phenotypes often develop slowly, manifesting weeks following ablation.
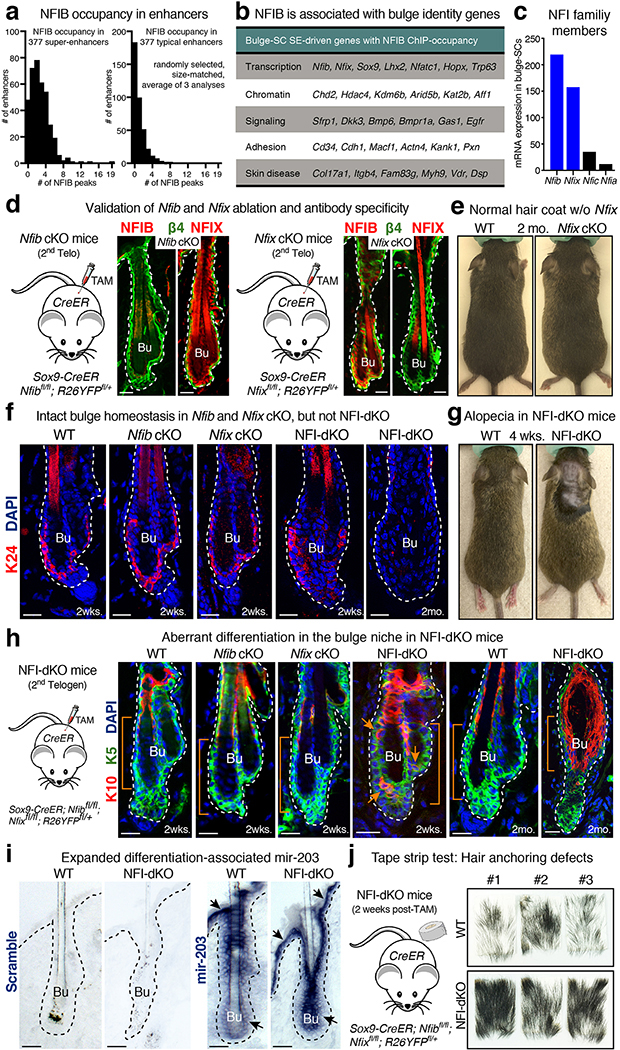
In searching for a unifying factor(s) that might govern stemness genes in bulge-SCs, we were intrigued by the NFI family of TFs. Like the aforementioned TFs, NFIB binds within most bulge-SC super-enhancers. Although NFIB loss was without consequence to bulge-SC function23, closely related NFIX is highly expressed in bulge-SCs, raising the possibility of redundancy. Indeed, despite widespread expression of NFI genes, strong phenotypes for loss of individual NFI factors are limited mostly to cortical, lung and bone development24–30. When we discovered that both Nfib and Nfix genes are themselves regulated by super-enhancers, NFI factors became strong candidates for chromatin regulators of stemness.
Most of our knowledge of NFI function at the chromatin level comes from cancers, where gain-of-function NFI fusions and locus amplifications have been reported31. In small cell lung cancer metastasis, the NFIB locus amplifications increase chromatin accessibility32. Less clear is how physiological levels of NFI factors regulate chromatin. In vitro, NFI factors bind nucleosomes33 and chromatin modifiers34, control chromatin loop boundaries35,36 and impact the histone modifications associated with differentiation genes37. Lacking is knowledge of how NFI proteins function in a physiological context, whether they share similar functions and how chromatin dynamics change upon NFI loss in a tissue.
Here, we use conditional gene targeting, histological analyses, single cell RNA-sequencing, ‘Assay for Transposase-Accessible Chromatin using sequencing’ (ATAC-seq38), and ‘Chromatin Immunoprecipitation sequencing’ (ChIP-seq) analyses to address these issues. In doing so, we unearth a striking parallel between NFI loss and human scarring alopecia, and a hitherto unrecognized role for NFI family members in regulating the super-enhancers that govern SC identity. Our discoveries provide new insights into why NFI factors may be more broadly and redundantly expressed than other TFs within tissues. Additionally, our work suggests why NFIs are often overexpressed in cancers, which in contrast to tissue SCs, are irreversibly locked into a permanent, undifferentiated and self-renewing state.
RESULTS
Nfib and Nfix Play Redundant Roles in Bulge-SCs and are Essential for Maintaining the Hair Coat
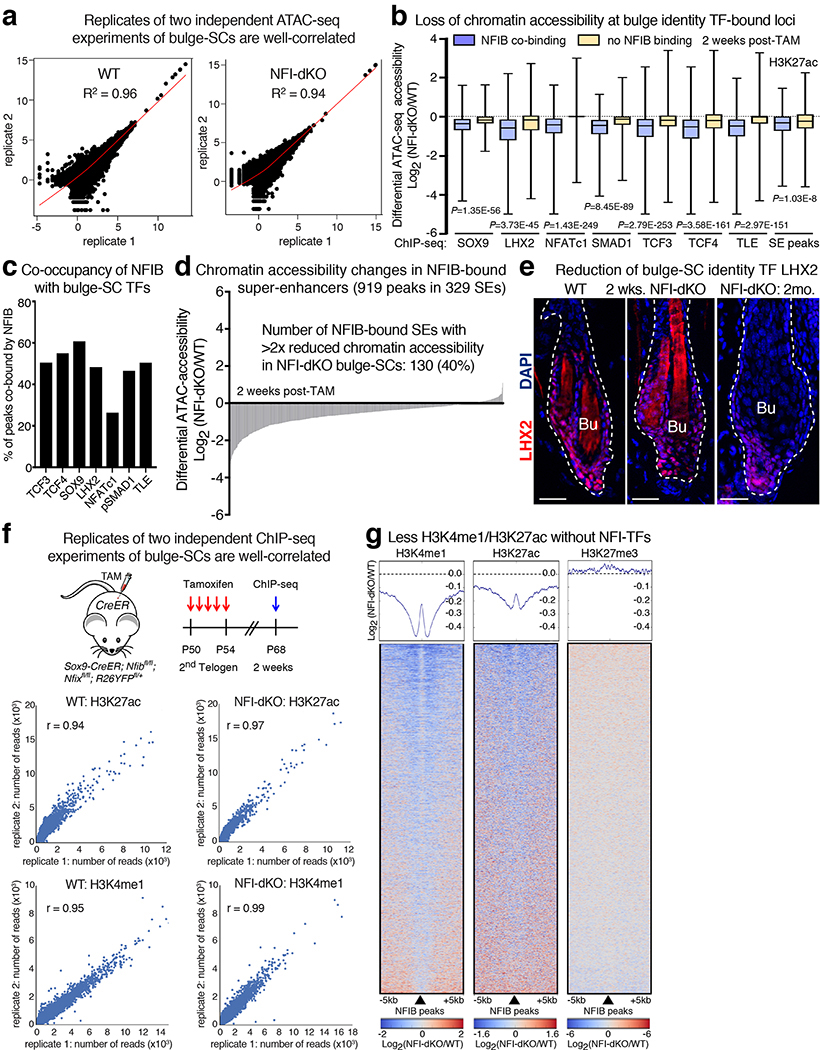
Super-enhancers are distinguished by the magnitude of open chromatin (>15 kb), the density of clustered binding motifs for key TFs and the robustness of H3K27 acetylation11. Super-enhancer regulated genes often include the super-enhancer regulating TFs themselves11–13. Pursuing this paradigm, we analyzed our prior bulge-SC super-enhancer13 and NFIB ChIP-seq data23. While NFIB-occupied sequence motifs were found in typical enhancers, they were enriched within the 377 bulge-SC super-enhancers (Fig. 1b), even after correcting for enhancer size differences (Extended Data Fig. 1a). 87% of super-enhancers harbored canonical NFIB sites within their short stretches (‘epicenters’) of clustered bulge TF-motifs. Moreover, Nfib was among the bulge super-enhancer regulated genes, as judged by H3K27ac and ATAC-seq38 (Fig. 1c, d and Extended Data Fig. 1b). Notably, auto-regulation is a typifying feature of TFs that possess a special role in super-enhancers.
Although Nfib single conditional targeting was without phenotypic consequence to the HF epithelium23, NFIX levels were comparable to NFIB in bulge-SCs (Fig. 1e,f and Extended Data Fig. 1c). Like NFIB, NFIX recognizes the palindromic consensus binding sequence 5’-TGGCA-(N3–5)-TGCCA-3’39,40. Interestingly, the Nfix locus also harbored two bulge-SC super-enhancers (Fig. 1g). We thus considered two possible scenarios: 1) NFIX is more relevant in bulge-SCs than NFIB; or 2) NFIB and NFIX are redundant, necessitating combined ablation to uncover their role in bulge-SCs.
To specifically target HFs, we used Sox9-CreER recombinase, whose robust activity can be spatially and temporally controlled by tamoxifen (TAM) administration7,41. Using Nfibfl/fl and Nfixfl/fl mice bred on a Rosa26-fl-stop-fl-YFP background to monitor Cre activity, we generated singly and doubly conditional knockout mice (Fig. 1h and Extended Data Fig. 1d). TAM was administered during second telogen (~P45-P80), and consequences to SC maintenance were analyzed during the subsequent hair cycle. Immunofluorescence confirmed efficient and selective targeting (Extended Data Fig. 1d).
Neither individual knockouts yielded discernible HF defects, and bulge-SC numbers remained normal (Fig. 1j and Extended Data Fig. 1e,f;23). By contrast, beginning at 4 weeks after combined Nfib/Nfix ablation, signs of hair loss emerged (Fig. 1i and Extended Data Fig. 1g). This was accompanied by a temporal decline in the CD34+/K24+ bulge-SC population (Fig. 1j and Extended Data Fig. 1f). Derived from bulge-SCs7, the “hair-anchoring inner bulge” layer of differentiated barrier cells (Fig. 1a) is also SOX9+, and they express NFI proteins (Extended Data Fig. 1d). Without NFIs, it differentiated aberrantly and became unable to anchor hair (Extended Data Fig. 1h–j).
Hair Loss Resembling Human Primary Cicatricial Alopecia
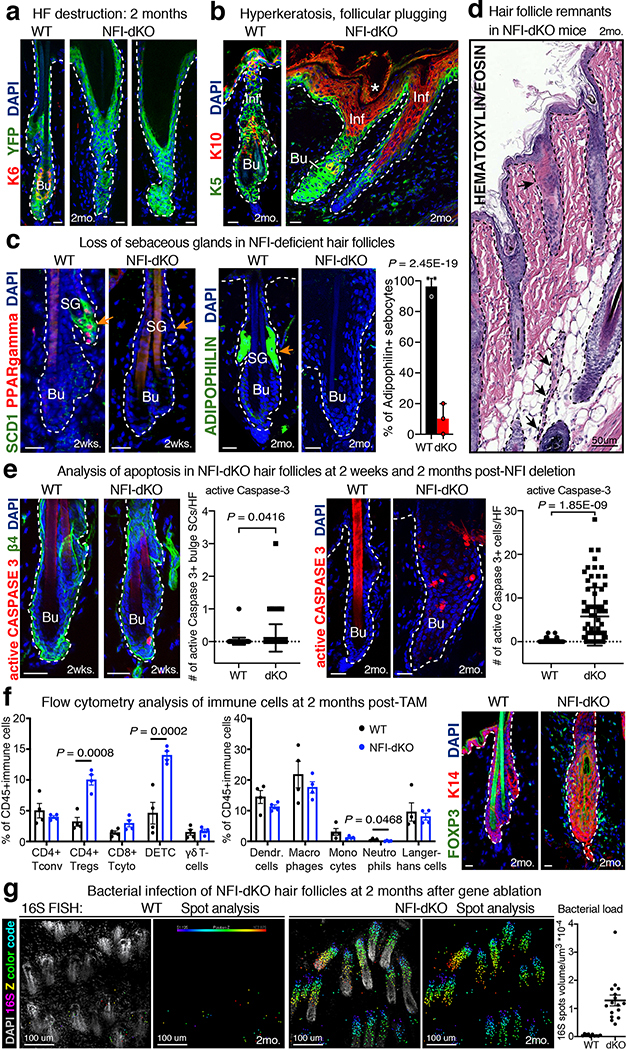
By 2 months post-TAM, NFI-dKO mice exhibited numerous skin aberrations (Fig. 2a, Extended Data Fig. 2a–d; Supplementary Table 1). Most HFs had degenerated, consistent with the inability to maintain the bulge-SC niche. Skin exhibited hyperkeratosis and follicular ostia, which appeared dilated and plugged with K10+ dead, keratinized deposits, and sebaceous glands were missing. In more advanced stages, follicular scarring was observed, in which fibrous strands ran perpendicular to the epidermis from remnants of dystrophic HFs.
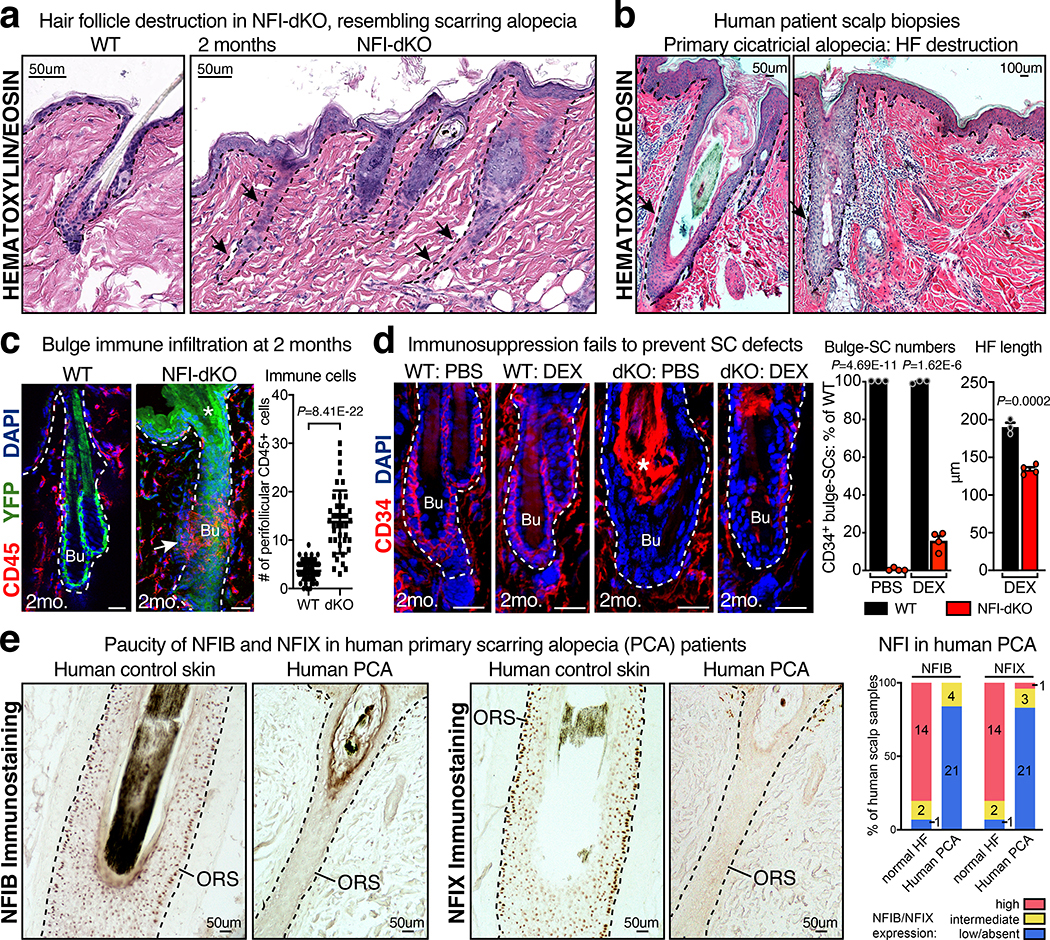
Fig. 2 |. NFI-Deficiency Leads to a Phenotype Resembling Primary Cicatricial (Scarring) Alopecia.
a, b, Hematoxylin & Eosin images of Nfib/Nfix-dKO skin at 2 months post-TAM (a) reveals similarities to human primary cicatricial alopecia (PCA) (b). Representative images of three biological replicates. c, Immunofluorescence and quantifications reveal infiltration of immune cells (CD45+) around the HFSC niche at 2 months post ablation of Nfib/Nfix relative to control. YFP labels Sox9-CreER-targeted HFs. Mean and standard deviation are shown. P value is from unpaired, two-tailed t-test based on n=69 WT HFs and n=41 NFI-dKO HFs (total, pooled from 3 mice). d, Experimental design, immunofluorescence and quantifications comparing 2nd telogen HFs of WT and NFI-dKO mice at 2 months post-TAM ± immunosuppression by dexamethasone (DEX). Note that DEX fails to prevent decline of CD34+ bulge-SCs in NFI-dKOs. DEX-treated NFI-dKO HFs were also shorter compared to DEX-treated WT HFs. These features were consistent with stem cell exhaustion. Mean and standard deviation are shown (3–6 HFs analyzed/mouse for CD34 quantifications, and 6–9 HFs analyzed/mouse for HF length measurements). P values comparing WT (n=3 mice) vs. dKO (n=4 mice) are from unpaired, two-tailed t-test. e, Immunohistochemistry of representative human scalp biopsies shows reduced expression of NFIB and NFIX in PCA compared to normal skin. ORS, outer root sheath. (right) Quantifications for 17 normal and 25 PCA replicates. Scale bars = 20μm, unless otherwise specified. Bu, bulge. Inf, Infundibulum. Dashed lines, HF-dermal border. Asterisk denotes autofluorescence. See also Source Data.
These features were reminiscent of irreversible hair loss in humans known as primary cicatricial alopecia (PCA) (Fig. 2b, Supplementary Table 1). Clinical features of PCA further include pigmentary incontinence, basal cell vacuolization and follicular apoptosis, which were also features of NFI-dKO mice (Extended Data Fig. 2e). While obvious, follicular scarring was milder in our mice than in human PCA, likely due to the relatively short timeframe of our analyses post-Nfi ablation. Overall, it was notable that of the 10 major features of human PCA, all but one was prominent in our mice.
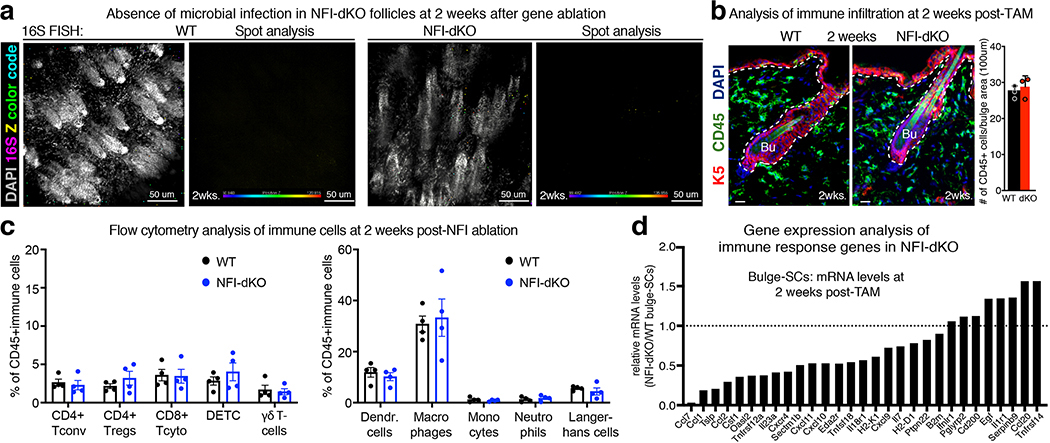
In the more common, sometimes self-resolving alopecia areata, inflammatory responses are directed towards the hair bulb42. By contrast, irreversible PCA is typified by perifollicular inflammatory infiltrates surrounding the non-cycling HF portion, including bulge43,44 (Fig. 2b). Akin to PCA, immune cells (CD45+) surrounded the Nfib/x-null bulges at 2 months post-TAM (Fig. 2c). Another feature of certain subtypes of human PCA skin is the increase in both regulatory T cells (Tregs) and γδ T cells45,46. Similarly, NFI-dKO mice displayed a marked increase in FOXP3+/CD4+ Tregs and also γδ T cells (in this case dendritic epithelial T cells, DETCs) (Extended Data Fig. 2f and Extended Data Fig. 3a,b). Follicles were also infiltrated with microbes, common in certain types of human PCA (e.g. Folliculitis decalvans) (Extended Data Fig. 2g).
Interestingly, neither bacterial nor immune infiltration were prominent early, at 2 weeks post NFI-ablation when bulge phenotypes were already apparent (Extended Data Fig. 4a–c, Fig. 1j). Additionally, transcriptional profiling of Nfib/x-null bulge-SCs at 2 weeks post-TAM revealed few differences compared to WT bulge-SCs in chemokines (e.g. Ccl1, Ccl2, Cxcl9/10/11), cytokine receptors (e.g. Il18r1) and immune-response genes (e.g. Oasl2, Pglyrp2), known to be induced upon barrier breach in murine HFs47 and in advanced human PCA43. Expression of Cd200, which endows the bulge with immune tolerance-promoting signals48, was also maintained at this stage (Extended Data Fig. 4d). Thus, the early bulge defects of NFI-dKO HFs appeared to occur prior to the immune response.
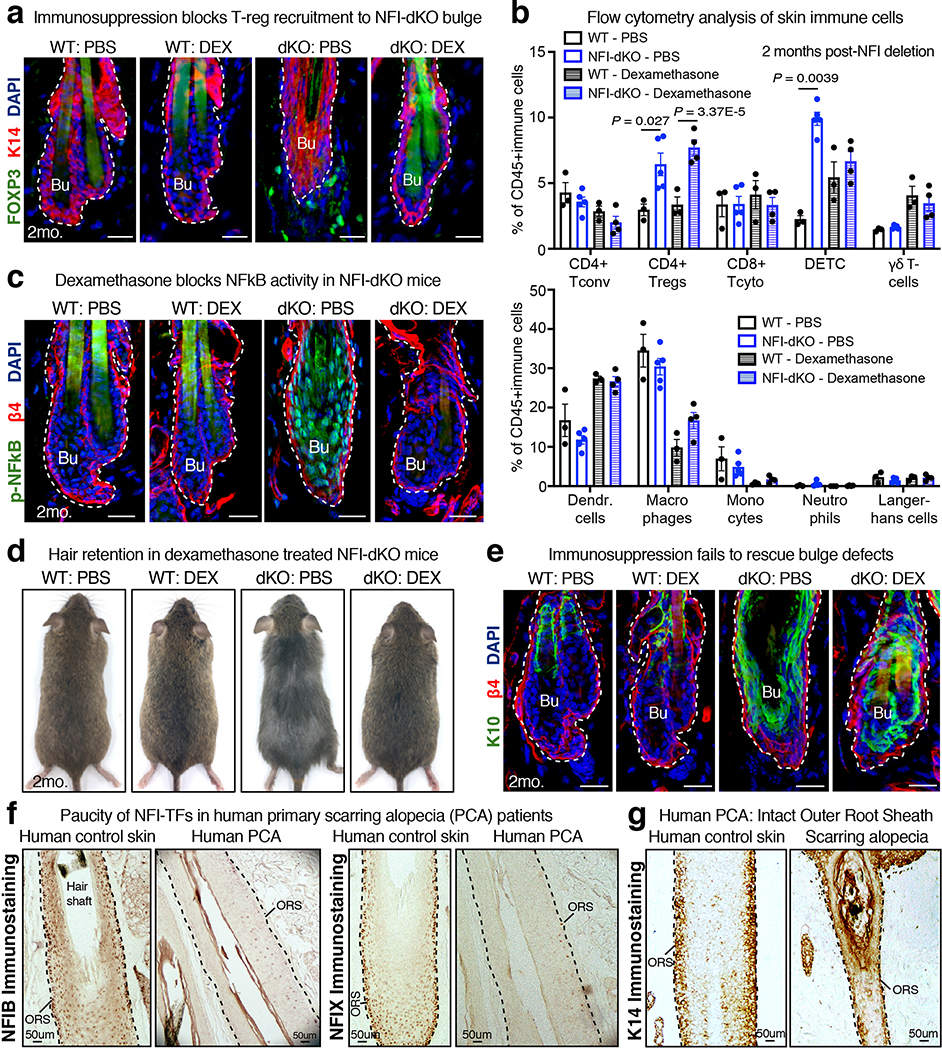
To assess secondary features of the phenotype, we treated mice with pan-immunosuppressant dexamethasone (DEX) for 2 months following NFI-deletion. DETC levels were comparable under DEX-treated conditions, and although DEX did not reduce total Treg numbers in NFI-dKO skin, it blocked their recruitment to the bulge niche and prevented NF-kB activation (Extended Data Fig. 5a–c). Most strikingly, features associated with inflammatory states, e.g. hair loss and HF degeneration were prevented in DEX-treated NFI-dKO mice (Extended Data Fig. 5d). However, immunosuppression failed to prevent the decline in bulge-SC numbers and ectopic differentiation, and follicles were shorter in length compared to WT (Fig. 2d and Extended Data Fig. 5e). These features are signs of SC exhaustion8. Collectively, whereas immune infiltration contributed to hair loss and HF degeneration, the perturbations in the bulge were linked to intrinsic bulge-SC defects, driven by the loss of NFI-TFs.
Given these parallels, we examined NFIB/NFIX’s expression status in human skin of PCA patients and healthy individuals (Supplementary Table 2). Unlike normal scalp skin, which displayed NFIB/NFIX immunolabeling enriched in the outer root sheath (ORS) of HFs, 84% (21/25) of PCAs showed weak or no immunolabeling (Fig. 2e and Extended Data Fig. 5f). This was not due to a loss of ORS cells, as K14+ ORS cells were still observed in human PCA patients (Extended Data Fig. 5g). Overall, although NFIB and NFIX mutations may not underlie the genetic basis of PCA49,50, our findings suggest that NFI downregulation may be an important early component of PCA in humans, as it is in our mice.
Nfib/Nfix Ablation Alters HFSC Identity
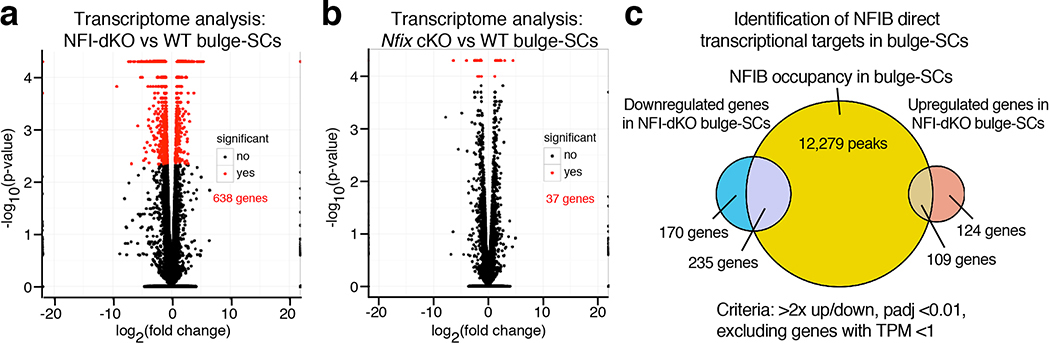
To probe deeper into NFI-TF effects on SC homeostasis, we transcriptionally profiled Nfib/x null and WT bulge-SCs, purified by fluorescence activated cell sorting (FACS) (CD34+INTEGRIN α6+YFP+SCA1neg) at 2 weeks post-ablation. Heatmaps of independent replicates revealed 638 differentially expressed genes (>2x cutoff, padj<0.01, TPM>1), while alone, Nfib23 and Nfix loss elicited few changes (Fig. 3a, Extended Data Fig. 6a,b and Supplementary Table 3).
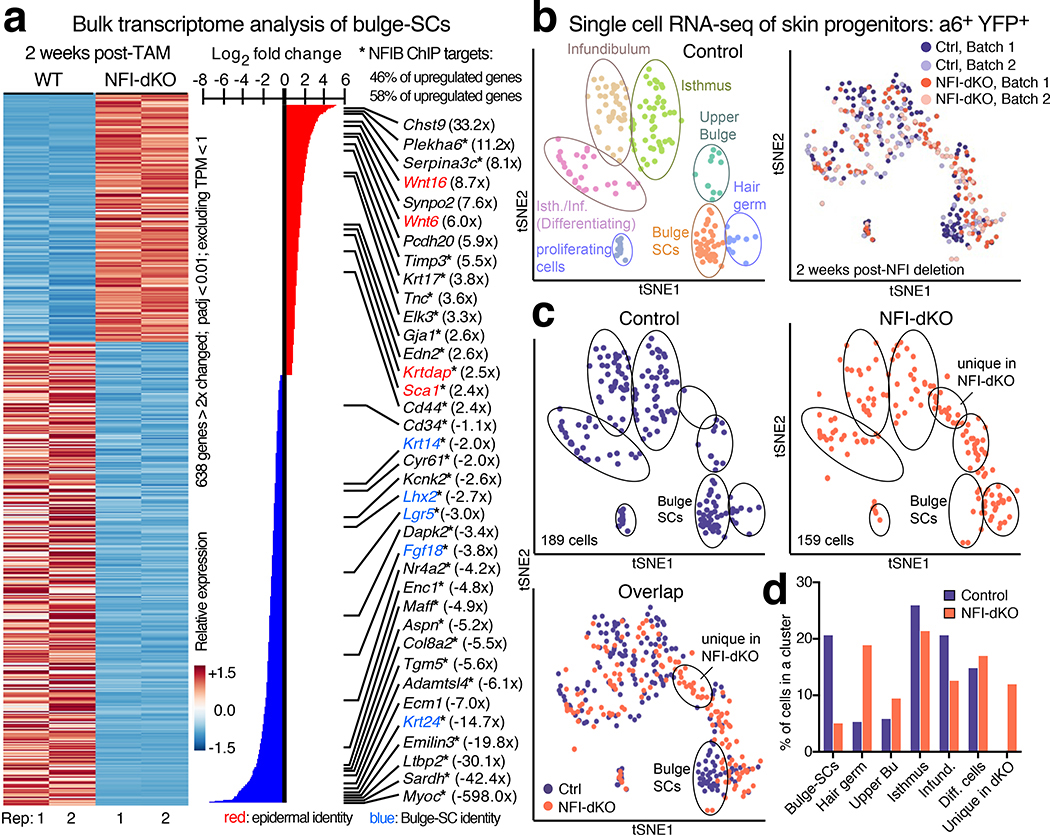
Fig. 3 |. Loss of Nfib and Nfix Alters Bulge-SC Identity.
a, Heatmap of mRNAs differentially expressed between FACS-purified CD34+/INTEGRIN α6+/ YFP+/SCA1neg bulge-SCs from WT and NFI-dKO mice, 2 weeks post-TAM. Duplicate datasets from 2 mice are shown. padj<0.01, p values were calculated from unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method. n=23491 genes were analyzed/genotype. Fold-changes in parentheses. Asterisks denote direct NFIB transcriptional targets. Note that Cd34 levels (green) were comparable in the two cohorts purified and analyzed. b, (left) Single cell RNA-seq analyses of INTEGRIN α6+/YFP+ skin progenitors. Unbiased clustering of transcriptomes of individual basal progenitors from control (Sox9-CreER;Nfixfl/fl;R26-YFP) telogen skin. Each cell is represented as a dot, colored by a clustering algorithm and plotted on the tSNE graph. (right) tSNE analysis of 2 batches of single-cell RNA-seq libraries shows minimal batch to batch variation (2 mice). c, Overlap of transcriptomes of control and Nfib/Nfix-deficient HF progenitors (2 mice). Note the emergence of a new cluster, not seen in control HF progenitors. d, Quantifications of progenitors per cluster in control vs. Nfib/Nfix-deficient HFs. Note that among the progenitor pools of Nfib/Nfix-dKO HFs, in addition to the unique cluster, there is also a relative loss of bulge-SCs with more cells exhibiting features of upper bulge cells and more committed, lineage-primed hair germ cells.
To distinguish primary consequences of Nfib/x loss from bystander effects, we compared transcriptomes with our prior NFIB ChIP-seq analyses23 (Extended Data Fig. 6c). Among the most highly downregulated genes in NFI-deficient bulge-SCs were major stemness genes Lhx2, Lgr5 and Krt24, all of which were direct NFIB-ChIP targets (Fig. 3a, blue)13,17,51. Upregulated were Edn2, consistent with NFIB’s role in suppressing melanocyte differentiation23, and also epidermal lineage differentiation genes such as Krtdap and Sca1, which exhibited NFIB-ChIP occupancy (Fig. 3a, red).
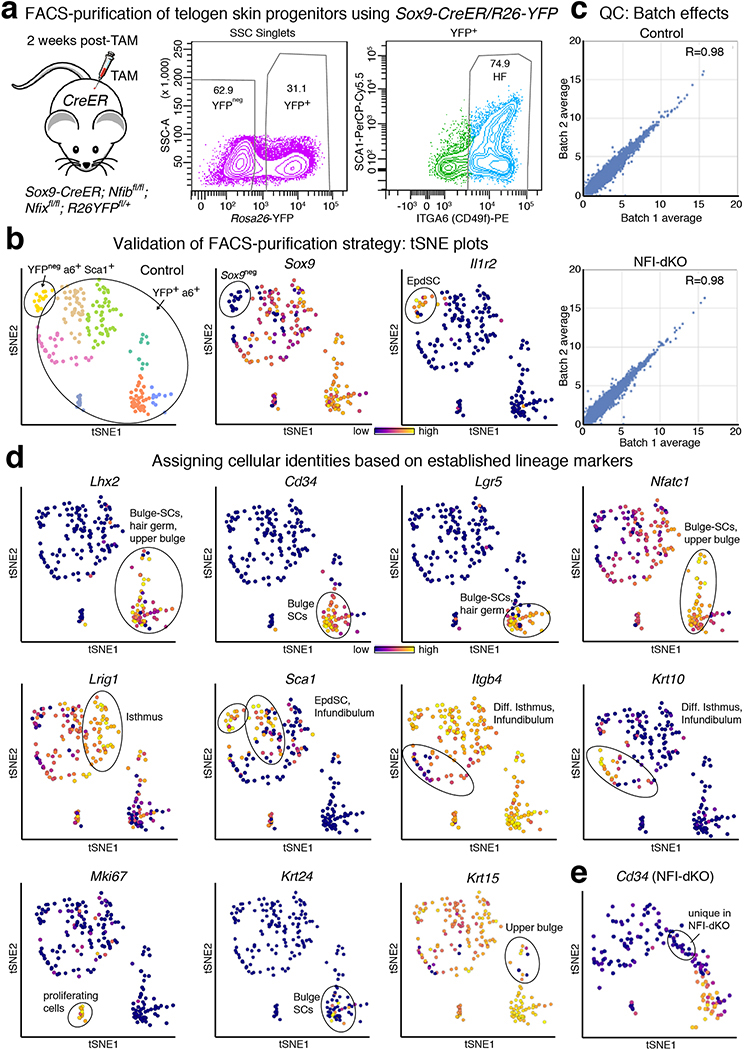
Turning to single cell RNA-sequencing, we used Smart-seq252 to transcriptionally profile 378 FACS-purified INTEGRIN α6+YFP+ progenitors from telogen-phase HFs at 2 weeks post-ablation. Non-targeted epidermal SCs (INTEGRIN α6+SCA1+YFPneg) were included as reference, and external RNA spike-ins were used to estimate technical noise53 (Extended Data Fig. 7a,b). ~5,000–6,000 genes were detected per cell, with minimal batch-to-batch variation (Fig. 3b, Extended Data Fig. 7c). Dimensional reduction by principal component (PC) analysis, t-distributed stochastic neighbor embedding (tSNE), and unsupervised hierarchical clustering revealed eight distinct expression patterns comprising cells of all telogen K14+ (Krt14+) skin progenitors: bulge-SCs (Cd34hiLgr5+Krt24+), upper bulge (Cd34negKrt15lo), hair germ (Cd34loLgr5+), isthmus (Lrig1hiSca1neg), infundibulum (Lrig1loSca1+), and spiked in epidermal SCs (Sca1hiYFPneg) (Fig. 3b, Extended Data Fig. 7d and Supplementary Table 4). Additionally, one subset of cells appeared to undergo differentiation (Itgb4lowKrt10+), and another consisted of proliferative cells (Mki67+), both typically found in small numbers in the isthmus/infundibulum.
By overlaying NFI-dKO and control transcriptomes, we evaluated how loss of NFI-TFs impacted different hair progenitor populations (Fig. 3c,d and Extended Data Fig. 7e). Most notable was a marked decline in bulge-SC representation, with a concomitant elevation in neighboring upper bulge54 and hair germ55. Additionally, a Cd34neg cluster between upper bulge and isthmus was present only in NFI-dKO and not control HF progenitors (Fig. 3c, Extended Data Fig. 7e). Overall, these major early transcriptome changes paralleled the accompanying decline in bulge-SC identity following NFIB/X loss, and further substantiated the reliance of bulge-SCs on NFIB/X.
NFIB Promotes Stemness by Establishing Chromatin Landscapes Accessible to Bulge-SC TFs
To elucidate the chromatin landscape changes that elicit the observed transcriptomic changes, we employed ATAC-seq38 on bulge-SCs at 2 weeks post-TAM (Extended Data Fig. 8a, Supplementary Table 5). Comparing the ATAC-seq read densities, we discovered that ~11% of all peaks (~8000 peaks) were uniquely accessible in WT but lost upon Nfib/x ablation (Fig. 4a). 3048 (37.2%) of these peaks were bound by NFIB, suggesting direct effects of NFI-loss at these loci.
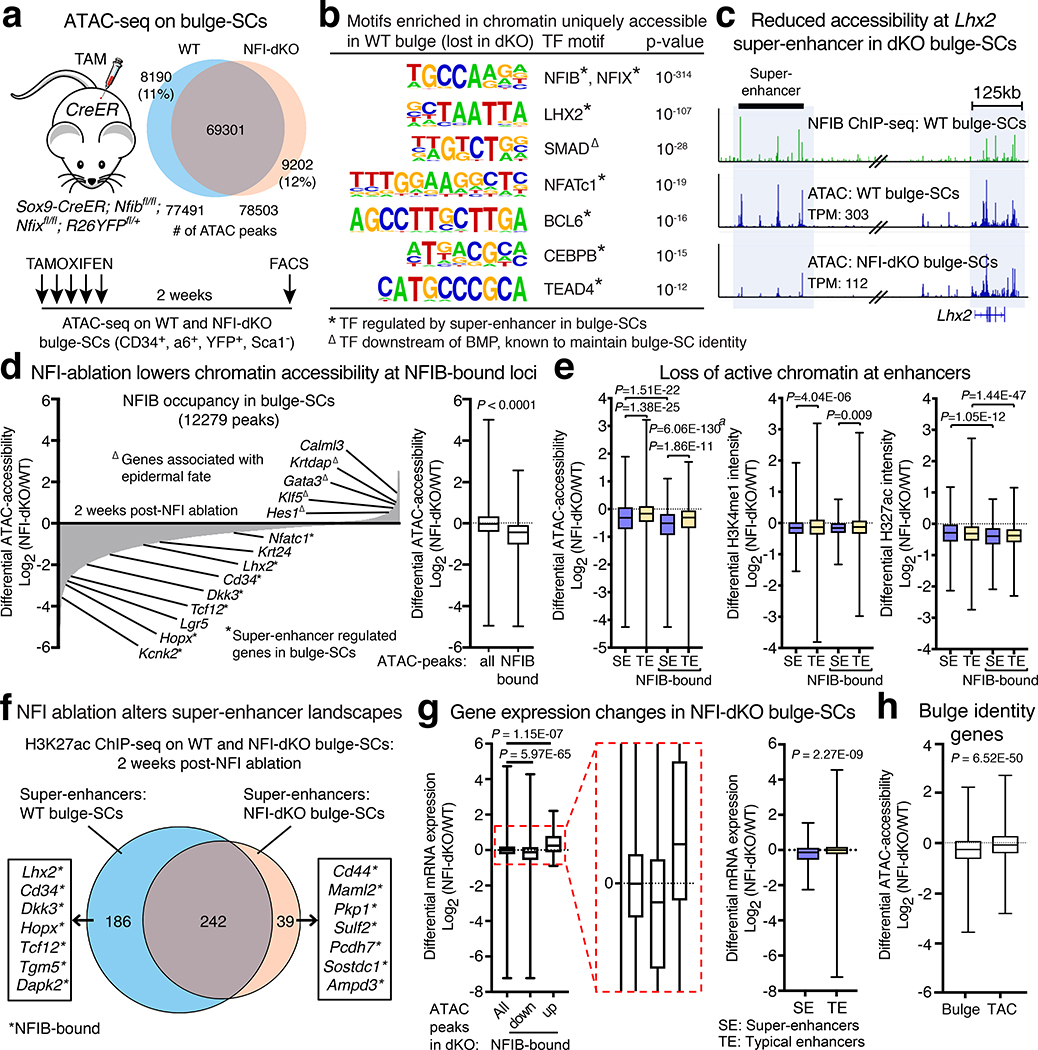
Fig. 4 |. NFI-TFs Maintain Bulge-SC Chromatin Landscapes.
a, ATAC-seq of chromatin isolated from FACS-purified WT or NFI-dKO bulge-SCs, analyzed at 2 weeks post-TAM. 2 independent experiments. b, TF-motif analysis of chromatin uniquely accessible in WT bulge-SCs (lost in NFI-dKO) reveals enrichment of motifs for bulge-SC identity TFs. P-values calculated by analyzing n=8190 WT-unique ATAC peaks (hypergeometric distribution analysis). c, NFIB ChIP-seq and ATAC-seq tracks of bulge-SC TF gene Lhx2 and its super-enhancer (black bar) in NFI-dKO and control bulge-SCs. d, (left), Differential chromatin accessibility at NFIB ChIP-occupied loci in WT vs. NFI-dKO bulge-SCs. (right), Upon NFI-ablation, NFIB-bound loci exhibit a greater reduction in chromatin accessibility than the genome as a whole. The median (line), first and third quartiles (box), and whiskers (highest and lowest values) are shown. N=86681 (all) and n=12279 NFIB-bound ATAC peaks were analyzed. e, Differential chromatin accessibility (ATAC-seq) and levels of histone modifications (ChIP-seq) at total and NFIB ChIP-occupied enhancers in WT vs. NFI-dKO bulge-SCs. Super-enhancers (SE) and typical enhancers (TE) for bulge-SCs were previously defined13. The median (line), first and third quartiles (box), and whiskers (highest and lowest values) are shown. Number of peaks analyzed: ATAC: n=2145 (SE), n=17244 (TE), n=779 (NFIB-bound-SE), n=4940 (NFIB-bound-TE); H3K4me1: n=3238 (SE), n=20857 (TE), n=961 (NFIB-bound-SE), n=5181 (NFIB-bound-TE); H3K27ac: n=3237 (SE), n=20884 (TE), n=961 (NFIB-bound-SE), and n=5191 (NFIB-bound-TE). f, H3K27ac super-enhancer landscapes in WT and NFI-dKO bulge-SCs. g, (left) NFI-dependent gene expression changes (RNA-seq) depending upon accessible chromatin and NFIB ChIP-occupancy. N=22860 (all), n=2480 (NFIB-bound ATAC-peaks decreased in NFI-dKO), n=51 genes (NFIB-bound ATAC-peaks increased in NFI-dKO). (right) Boxplots comparing the relative impact of NFI-status on gene expression (regulated by SEs or TEs) in bulge-SCs. N=300 SE genes and n=10515 TE genes. In boxplots (left and right), the median (line), first and third quartiles (box), and whiskers (highest and lowest values) are shown. h, Differential chromatin accessibility at identity genes in WT vs. NFI-dKO bulge-SCs or transit-amplifying cells (TACs). The median (line), first and third quartiles (box), and whiskers (highest and lowest values) are shown. N=2987 Bulge-SE ATAC peaks and n=2954 TAC-SE ATAC peaks. Unpaired two-tailed t-test used for all analyses.
Analysis of silenced ATAC peaks revealed sequence motifs for known, super-enhancer regulated bulge-SC master regulators15 (Fig. 4b). Notably, bulge-SC TF genes were among those whose peaks were rendered inaccessible early after Nfib/x ablation, indicating that NFIB/NFIX binding is required to maintain chromatin accessible at these loci (Fig. 4c,d; Extended Data Fig. 8b,c). Indeed without NFI-TFs, 40% of super-enhancers displayed >2x reduced accessibility (FDR<0.1, Figure 4e, Extended Data Fig. 8d). This decommissioning was reflected at both mRNA and protein levels for bulge-SC TFs (Fig. 3a, Extended Data Fig. 8e).
Given NFI’s impact on chromatin accessibility, we performed ChIP-seq on bulge-SCs to profile active histone marks, H3K4me1 and H3K27ac, and polycomb-repressive histone mark H3K27me3. Centering on NFIB ChIP-peaks, we queried the status of histone modifications at these loci in NFI-deficient versus control bulge-SCs. Whereas H3K27me3 was unchanged upon loss of NFIB-occupancy, H3K4me1 and H3K27ac were markedly reduced in chromatin encompassing regions (± 5kb) that had previously been occupied by NFI (Extended Data Fig. 8f,g).
Remarkably, at only 2 weeks post NFI-ablation and while HFs were in telogen, 26% of super-enhancers displayed >2x reduced H3K27ac levels (Fig. 4e). Without Nfib/Nfix, bulge-SCs exhibited 35% fewer super-enhancers, many of which were associated with bulge-SC identity genes (Fig. 4f, Supplementary Table 6). The prominent effect of NFI-loss on stemness genes was consistent with the relative enrichment of NFIB occupancy in super-enhancers compared to typical enhancers (Extended Data Fig. 1a), and provided compelling evidence for NFIB’s role in promoting open and active chromatin environments at these sites to drive stemness (Fig. 4g).
NFI Roles in Lineage Fate Choices
In human lung cancer, NFIB over-expression led primarily to the enhancement of open chromatin domains32. In NFI-deficient bulge-SCs, ~12% of ATAC peaks (~9000 peaks) showed increased accessibility, which was nearly comparable to peaks diminished in accessibility (Fig. 4a). However, our NFIB ChIP-seq data revealed that ~80% of NFIB-bound chromatin domains exhibited diminished accessibility upon NFI loss, whereas a smaller fraction displayed enhanced accessibility, suggesting that other factors are involved in activating new loci in NFI-dKO bulge-SCs (Fig. 4d). Irrespective of whether increased or decreased in accessibility, chromatin peaks that bound NFI factors correlated with the transcriptional status of their genes (Fig. 4g). Most intriguingly, among ATAC peaks that both bound NFI and gained accessibility upon NFI loss were epidermal TF genes, rather than the transit-amplifying cell (TAC) genes that become activated during normal HF regeneration when bulge-SC master regulators are suppressed (Fig. 4d,h)55. In this regard, NFIB/X loss was distinct from TCF3/4 loss, which promotes hair lineage progression18.
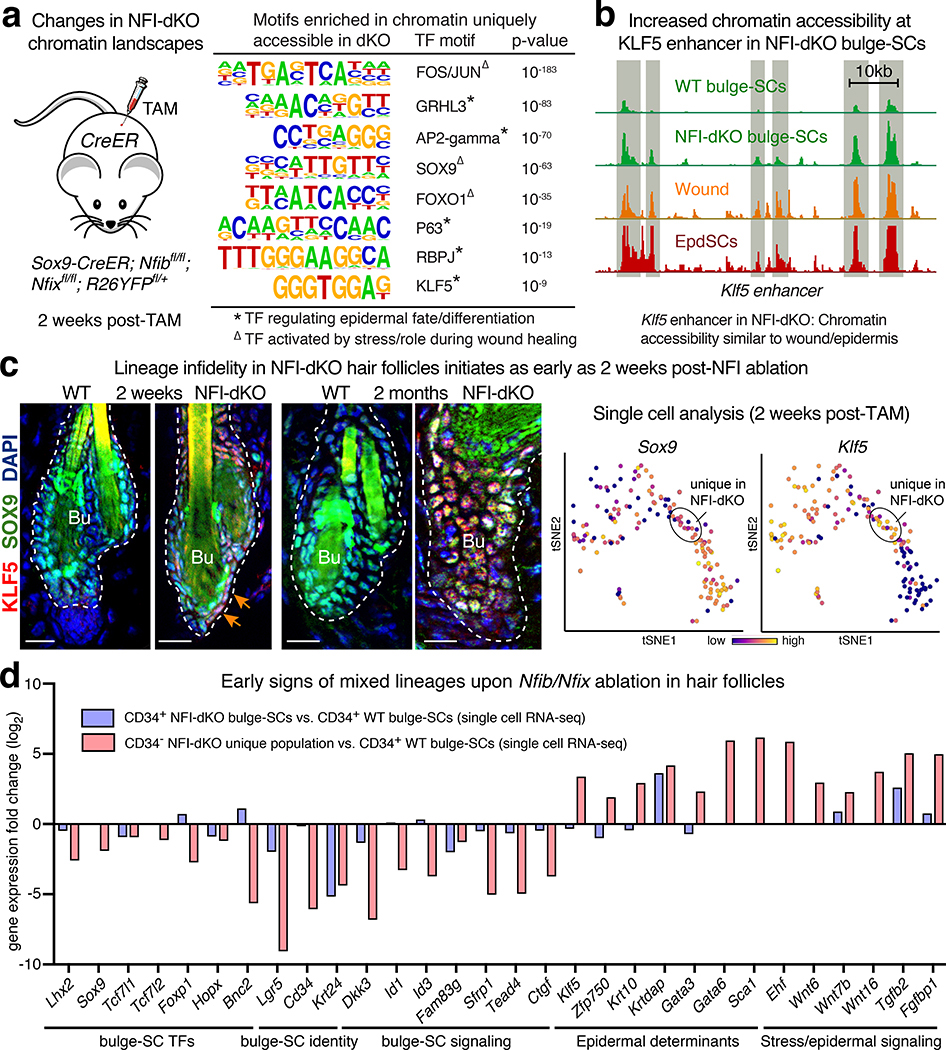
TF-motif scanning of newly accessible peaks revealed binding sites for NOTCH/RBPJ, KLF5 and other epidermal lineage TFs (Fig. 5a). Moreover, the Klf5 locus contained NFIB-occupied sites in WT bulge-SCs, and this locus became more accessible upon NFIB/X loss (Fig. 5b and 4d). Curiously, SOX9 motifs were also enriched in chromatin sites that opened upon NFI loss (Fig. 5a). Of the bulge-SC TFs, SOX9 is unusual in that its expression often persists even when the niche microenvironment has changed10,13. Immunofluorescence confirmed dual SOX9 and KLF5 expression in Nfib/x-null bulge-SCs (Fig. 5c). Known as ‘lineage infidelity’10, this condition is characteristic of wounded, stress-induced states. The presence of additional wound/stress-induced TF motifs, e.g. AP1 (FOS/JUN) and FOXO1, in newly accessible Nfib/x null chromatin further pointed to this state (Fig. 5a). Collectively, the activation of these TFs upon NFI loss suggested a mechanism by which the ATAC peaks that don’t bind NFI may be opened.
Fig. 5 |. NFI-Deficiency Leads to Lineage Infidelity in Bulge-SCs.
a, Motif analysis identifies enriched TF motifs associated with bulge-SC ATAC-peaks that were gained within two weeks of Nfib/Nfix ablation. Note that these de novo peaks harbor motifs for TFs that are activated in normal EpdSCs and by bulge-SCs exposed to wound/stress situations. P-values were calculated by comparing n=9202 NFI-dKO unique ATAC peaks to random background sequences (hypergeometric distribution analysis). b, ATAC tracks reveal enhanced chromatin accessibility within Klf5 regulatory regions in Nfib/Nfix-dKO compared to control bulge-SCs. Note similarities between Nfib/Nfix-dKO bulge-SC Klf5 ATAC profiles and those of wounded WT bulge and WT EpdSCs. c, Immunofluorescence (left) reveals induced co-expression of SOX9 and KLF5 (arrows) within 2 weeks of Nfib/Nfix ablation is even more prominent at 2 months, suggestive of chronic lineage infidelity. (right) tSNE plots showing Sox9/Klf5 co-expression in the unique population of NFI-dKO progenitors that began to emerge early after Nfib/x targeting. Single cell analysis from 2 biologically independent mice per group. d, Expression levels of bulge-SC, EpdSC and stress-related genes among the two weeks CD34+ bulge-SC populations of NFI-dKO and WT skins, as well as the CD34neg de novo NFI-dKO progenitor population. Note similar but more pronounced changes in the CD34neg de novo NFI-dKO progenitors, which our combined analyses indicate is a more advanced state of bulge-SCs following NFI loss. All scale bars = 20μm. Bu, bulge. Dashed lines, HF-dermal border.
Lineage infidelity cells accounted for the unique population of CD34neg HF progenitors that emerged de novo shortly after Nfib/x ablation (Fig. 3c, 5c and Extended Data Fig. 7e). Moreover, compared to WT CD34+ bulge-SCs, the lineage infidelity Nfib/x-null population displayed accentuated transcriptional differences over those seen for CD34+ Nfib/x-null bulge-SCs. While all Nfib/x-null bulge-SCs eventually lost CD34 (Fig. 1j and Extended Data Fig. 7e), even by 2 weeks post-NFI ablation, this CD34neg population of bulge-SCs had diverged more considerably than the lingering CD34+ bulge-SCs (Fig. 5d and Supplementary Table 7). Thus, the rapid reduction of bulge-SCs appeared to be due to loss of bulge-SC identity, rather than apoptosis, which was minor at 2 weeks post-TAM (Extended Data Fig. 2e).
Relation Between NFIB and NFIX and Restoration of Tissue Homeostasis after Wounding
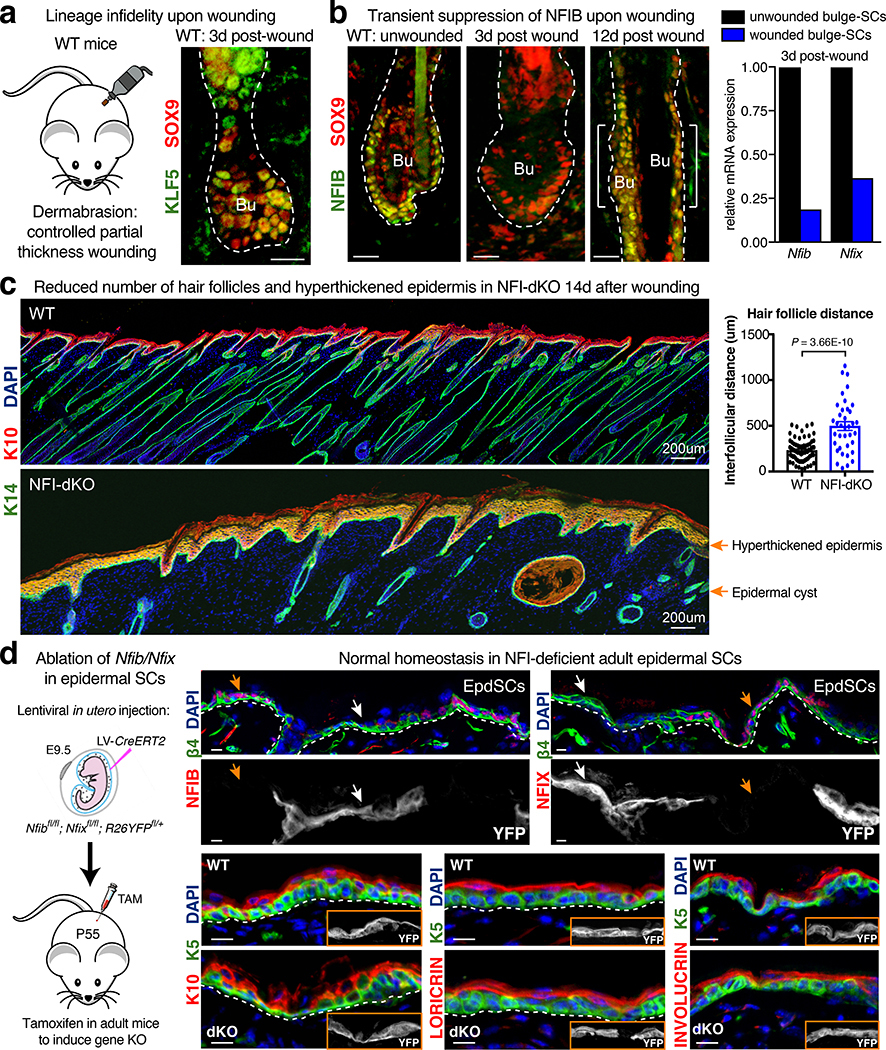
Since transient lineage infidelity is essential for SCs to achieve normal wound-healing, we wondered whether sustained lineage infidelity caused by NFI loss might be deleterious to the repair process. To test this possibility, we first examined NFI status in telogen-phase WT skin subjected to a shallow wound (dermabrasion) that removes overlying epidermis and uppermost dermis/HFs, and stimulates bulge-SCs to activate lineage infidelity and repair the damage. Interestingly, bulge-SCs markedly downregulated both Nfib and Nfix during the response, and then restored expression following healing (Fig. 6a,b).
Fig. 6 |. NFI-TF Dynamics Play an Essential Role During Wound-Repair.
a, Dermabrasion selectively removes EpdSCs while leaving behind bulge bulge-SCs. This mobilizes bulge-SCs to re-epithelialize the epidermis10. Mobilized bulge-SCs co-express epidermal (KLF5) and follicular (SOX9) TFs throughout the repair process. b, Transient suppression of Nfib and Nfix in mobilized bulge-SCs during wound-repair. c, Following dermabrasion wounding, stimulated WT bulge-SCs that do not participate in re-epithelialization of the epidermis will launch a new hair cycle. Without NFI TFs, this does not happen, and instead the wounded skin shows epidermal hyperthickening. (right) Quantifications of interfollicular distances, which also widen, reflective of global HF degeneration in NFI-dKO skin. Mean and standard deviation are shown. P-value was calculated from unpaired, two-tailed t-test based on n=74 WT HFs and n=37 NFI-dKO HFs. d, In utero delivery of LV-CreER for inducible ablation of Nfib/Nfix in the adult epidermis does not impair EpdSCs or their ability to maintain homeostasis. K5 marks basal epithelial cells. K10 labels differentiating cells. LORICRIN and INVOLUCRIN are markers of terminal epidermal differentiation. Representative images of three biological replicates are shown in a, b, c, d. Scale bars = 20μm, unless otherwise specified. Bu, bulge. Dashed lines, HF-dermal border. See also Source Data.
We next performed this assay on our mutant mice. At 14d post-wounding, repaired epidermis was even thicker than normal and was YFP+, indicating its derivation from Sox9-CreER targeted bulge-SCs (Fig. 6c). Moreover, when we conditionally ablated Nfib/x in the epidermis, epidermal growth and differentiation appeared to be normal (Fig. 6d).
DISCUSSION
NFI Factors Have a Unique Importance in Bulge-SCs
Many of the key factors that promote spatio-temporally choreographed differentiation programs in HFs have been uncovered55. Loss-of-function studies have revealed roles for these TFs in regulating BMP-mediated quiescence, WNT-mediated hair cycling and/or suppression of sebaceous or epidermal differentiation. In contrast, how this TF cohort maintains hair regenerative capacity of bulge-SCs and how dysregulation results in human disease states is not clear. Another key issue is how, upon injury, bulge-SCs enter ‘lineage infidelity’ to survive and become epidermal SCs following wound-repair10.
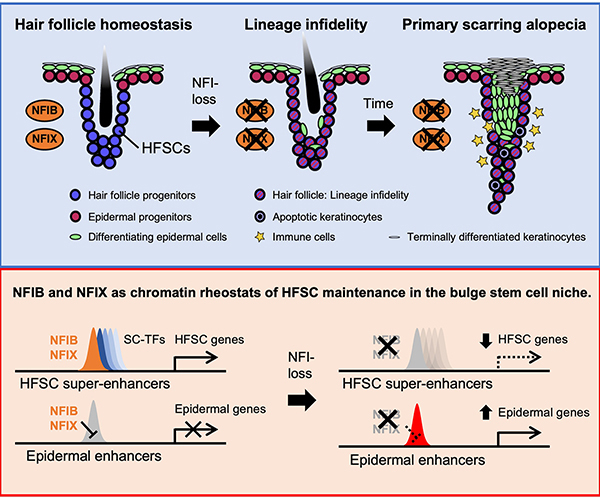
Here, we unearthed a critical role for NFI-TFs in maintaining HFs but not epidermis. Thus, upon NFI loss, the ability of SCs to sustain epidermal homeostasis appeared unscathed, while their HF regenerating capacity was progressively lost (Extended Data Fig. 9). NFI protein function appeared to make the difference in these two SC lineages, as the bulge began acquiring epidermal fate features within 2 weeks after Nfib/x ablation. Moreover, upon injury, where bulge-SCs are challenged to re-epithelialize epidermis10, WT bulge-SCs downregulated NFIs in transiently adopting their lineage infidelity state, while Nfib/x-null bulge-SCs produced an epidermis that was thicker than normal. NFI-TFs formed a barrier to plasticity and were continuously required to maintain bulge-SC identity. In this regard, NFIB/X’s function appeared distinct from even SOX9, which despite suppressing epidermal fate, is essential to maintain the lineage infidelity state10,16. Indeed, the phenotypic consequences of NFIB/X loss stood apart from all other bulge SC-TFs characterized to date.
Relevance of HFSC Maintenance to Human Scarring Alopecia
Unexpectedly, the histopathological features of NFI-dKO mice bore a striking resemblance to human scarring alopecia, a group of human inflammatory hair disorders typified by permanent destruction of HFs and loss of sebaceous glands, without affecting the epidermis. Although the etiology of this disease remains unknown and is likely to be complex, the prevailing view is that this disorder is rooted in immune dysfunction, and researchers and clinicians have proposed that loss of ‘immune privilege’ in the HF may spark PCA pathogenesis43,50. Curiously, however, PCA is irreversible in humans, and at least in our Nfib/x mutant mice exhibiting PCA, immune suppression rescued hair retention but not other HF phenotypes.
Overall, our findings suggest that the underlying sequence of events resulting in HF degeneration and a PCA-like phenotype (including defective lipid biogenesis56,57) can be recapitulated by a defect in HFSCs, in this case hardwired within the chromatin remodeling that occurs when NFIB/X are missing. While we do not discount the notion that there could be (and likely are) species to species differences, the absence of anti-NFI immunostaining in a number of human PCA cases further suggests that SC defects contribute to disease progression. Importantly, our data in mice suggest a possible reason why immunosuppression, despite ameliorating symptoms in the clinic, ultimately does not provide relief to many patients49.
Finally, although we do not present human genetic data, if NFI mutations are involved in some cases of human PCA, dominant negative rather than loss of function mutations would be predicted, given NFI’s genetic complexity in skin.
Lessons Learned Regarding NFI Functions in HFSCs
Our findings revealed that NFIB/NFIX provide chromatin access for the H3K4me1 and H3K27ac modified nucleosomes known to be particularly enriched at super-enhancers. Although histone acetylation on typical enhancers was also reduced in NFI-dKO bulge-SCs, the clustered binding sites and cooperative binding of super-enhancer regulated TFs within super-enhancers made these large open chromatin domains particularly sensitive to NFI-loss, as evident from transcriptomic analyses. NFIB/X loss in bulge-SCs was also required to de-repress enhancers governing epidermal and stress-coping TFs. This inverse correlation appeared to require additional TFs secondarily induced by NFIB/X loss. In this regard, our findings favor a role for NFIB/X as regulators of fate control, rather than general governors of chromatin.
NFI’s dual role in keeping some chromatin domains open while others silent also appeared to differ from the metastatic cancer state, where NFIB locus amplification results nearly unilaterally in expanding active chromatin regions32. We speculate that these distinctions may be analogous to those which distinguish the chromatin states of squamous cell carcinoma versus wound-healing in skin10. There, oncogenic RAS amplifies MAPK-driven super-activation of ETS2, which then opens new chromatin domains over those that occur in a wound state where ETS2 is induced at lower levels10,58. For the natural physiological context of SCs, NFI’s dual ability not only to maintain SC identity but also to prevent plasticity arising from activating the wrong SC fates becomes critical to tissue homeostasis.
ONLINE CONTENT
Any methods, additional references, Nature Research reporting summaries, data availability and associated accession codes are available online.
METHODS
Mice and Procedures
Rosa26-Flox-Stop-Flox-YFP (R26-YFPfl/+, C57BL6/J background), Sox9-CreER (C57BL6/J background41), Nfibfl/fl (C57BL6/J background23), and Nfixfl/fl (C57BL6/J background30) mice were used in this study. Mice were maintained in the Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility (AAALAC) of The Rockefeller University, and procedures were performed with Institutional Animal Care and Use Committee (IACUC)-approved protocols. The study is compliant with all relevant ethical regulations regarding animal research.
Five mice were housed per cage, with a 12hr light/dark cycle, and were provided food and water ad libitum. Mouse experiments were performed on age-matched and strain-matched pairs (usually littermates) randomly assigned to experimental groups. Male and female mice were used. To generate Nfib and Nfix single or double conditional knock out mice, Sox9-CreER, R26-YFPfl/+, Nfibfl/fl and/or Nfixfl/fl mice were crossed. At second telogen (Postnatal days 50–54), 50μg/g of tamoxifen was administrated to the mice. Mouse backskin was analyzed at 2 weeks or 2 months post-Tamoxifen treatment, hence mice were typically ~2–4 months old at time of analyses. Immunosuppression was achieved by daily i.p. injections of dexamethasone at 1mg/kg body weight.
Human patient samples
De-identified formalin-fixed paraffin-embedded tissue sections with clinical information were archived from the tissue bank in the department of pathology, Hanyang University Medical Center, Seoul, Korea. Hanyang University Institutional Review Board (IRB) approved the use of de-identified tissue sections for the study. Additional tissue sections of non-lesional hairy skin biopsy were obtained from NYU Langone Medical Center with approval of NYU IRB. We have complied with all relevant ethical regulations regarding research involving human participants, and Hanyang University IRB and NYU IRB had obtained informed consent from patients. De-identified patient information can be found in Supplementary Table 2.
Fluorescence Activated Cell Sorting and Analysis
To isolate quiescent bulge-SCs at the second telogen and at 2 weeks post-Tamoxifen treatment (Postnatal day ~68), mice backskins were harvested, and subcutaneous fat and blood vessels were removed from skin with a scalpel. Skins were placed dermis side down in 0.25% trypsin-EDTA (Gibco) for 45min. After digestion, single cell suspensions were obtained by scraping the dermal side gently using a blunt scalpel and by shaking vigorously. Cell suspensions were washed with FACS buffer (5% calcium-free FBS in calcium-free PBS) and filtered through 70μm and 45μm strainers. Cells were pelleted and washed once with FACS buffer. Cell suspensions were incubated with appropriate antibodies in FACS buffer for 30min on ice and washed with FACS buffer.
The following antibodies were used for staining (see Supplementary Table 8 for complete antibody information): CD34-eFluor660 (1:50), SCA1-PerCP-Cy5.5 (1:1000), CD49f-PE (1:500). Doublets and dead cells were excluded based on forward scatter, side scatter and DAPI (0.2 μg/ml) fluorescence. Cell-intrinsic YFP (Rosa26-YFP) was used as positive selection marker. For ATAC-seq and ChIP-seq, cells were collected in FACS buffer. For RNA-seq, cells were directly sorted into 750μl TRIzol LS (Invitrogen) and stored at −80oC. Sorting was performed using the 85μm nozzle.
Single-cells were sorted using a BD FACSAria II (BD Biosciences) into 96-well PCR plates (Bio-Rad) containing 2μl of lysis buffer (0.2% Triton X-100, 2U/μl RNaseOUT (Thermo Scientific), 0.25μM oligo-dT30VN primer and 1:8,000,000 diluted ERCC spike-in RNAs (Ambion). Doublets/dead cells were excluded based on forward scatter, side scatter and DAPI fluorescence. Sorting was done using the 100μm nozzle with the sort mode set to ‘single cell’. After sorting, plates were briefly centrifuged, snap-frozen in liquid nitrogen and stored at −80oC. FACS analyses were performed by FACSDiva software (BD Biosciences).
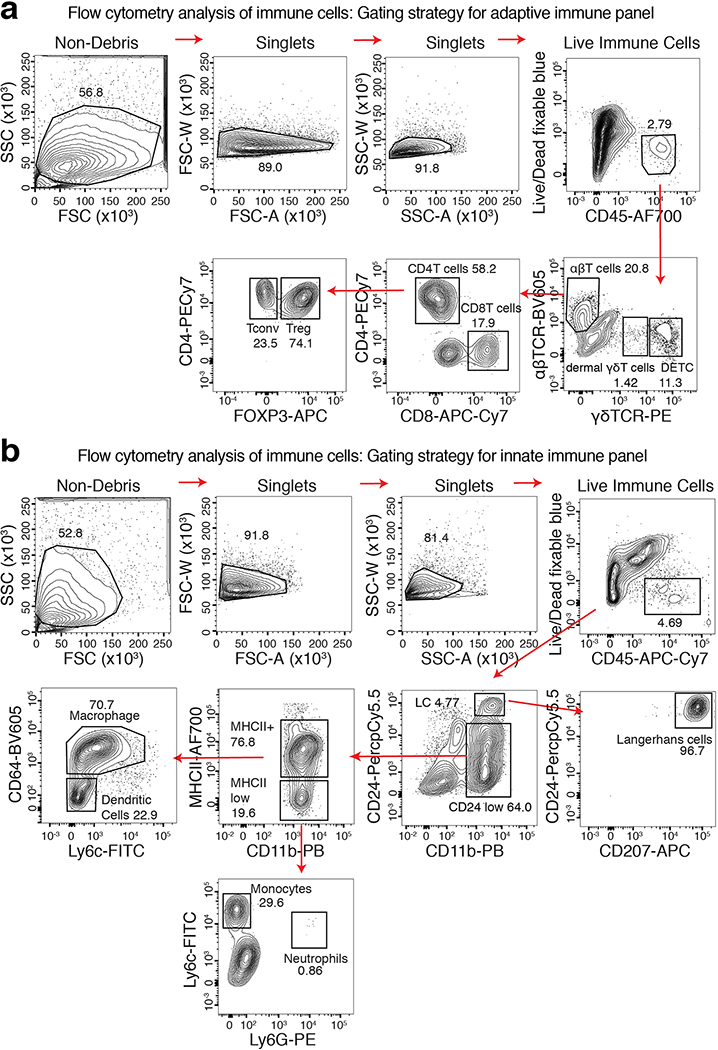
To analyze immune cells, backskin was minced in RPMI1640 media with L-glutamine (Gibco), 1mM sodium pyruvate (Lonza), 10mM acid-free HEPES (Gibco), 100U/ml penicillin and streptomycin (Gibco). Liberase TL (Roche) was added to the media (25 g/ml) and backskin was digested for 120min at 37 °C. Digestion was stopped by addition of 20ml of 0.5M EDTA (Life Technologies) and 1ml of 10% DNase (Sigma) solution. Dissociated cells were filtered with 70um strainer and stained with the following antibodies (see Supplementary Table 8 for complete information): Ly6c-FITC (1:500), Ly6g-PE (1:500), CD11b-PacBlue (1:1000), MHCII-AF700 (1:1000), CD64-BV605 (1:100), CD45-APC-Cy7 (1:200), CD24-Percp-Cy5.5 (1:500), CD207-APC (1:100), CD45-AF700 (1:200), TCRb-BV605 (1:200), gdTCR-PE (1:400), FOXP3-APC (1:100), CD4-PE-Cy7 (1:200), and CD8b-APC-Cy7 (1:200). Dead cells were excluded using LIVE/DEAD Fixable Blue Dead Cell Stain Kit (Molecular Probes) for UV excitation. FACS analyses were performed using LSRII FACS Analyzers and results were analyzed using FlowJo software.
For adaptive immune cell panels, we first gated on single cells. Among the CD45+ live cell population, we identified TCRγδHi T cells as dendritic epidermal T cells (DETCs), and TCRγδmid as γδT cells. In addition, in the TCRαβ+ population, we identified CD8+ or CD4+ T cells. Within the CD4+ T cell population, we further identified FOXP3+ Treg cells or the FOXP3− Tconv cells.
Innate immune cell panel: We first gated on single cells. In the CD45+ population, we first identified CD24Hi CD11b+ CD207+ cells as Langerhans cells. In the CD11b+ population, we further used class II MHC levels to identify MHCII positive and negative populations. In the CD24low MHCII+ CD11b+ cells, we identified CD64− Ly6c− cells as dendritic cells, and the rest of the MHCII+ CD11b+ cells were identified as macrophages. In the CD24low MHCII− CD11b+ population, the Ly6gHi Ly6clow cells were identified as neutrophils, and Ly6cHi Ly6glow cells were identified as monocytes.
RNA-seq and Quantitative RT-PCR
Total RNA was purified using the Direct-zol RNA MicroPrep kit (Zymo Research). After adding 750μl of 100% ethanol to samples, the lysate was loaded to RNA binding columns. To remove genomic DNA, DNase I was treated on the column for 15min at room temperature. After several washing steps, the RNA was eluted in DNase/RNase-free water. Quality of RNA samples was determined using an Agilent 2100 Bioanalyzer, and all samples for sequencing had RNA integrity (RIN) numbers >9. cDNA library construction using the Illumina TrueSeq mRNA sample preparation kit was performed by the Weill Cornell Medical College Genomic Core facility (New York, NY), and cDNA libraries were sequenced on an Illumina HiSeq 4000 instrument using a 50bp single-end-reads setting.
Single cell cDNA synthesis and sequencing library generation
Single cell RNA-seq libraries were prepared using the Smart-seq2 protocol52 with few modifications. Briefly, plates were thawed on ice and incubated for 3min at 72oC to lyse cells. Revers transcription (4U/μl Maxima H- transcriptase), template switching reaction and PCR pre-amplification (15 cycles) were performed according to the protocol. PCR product was cleaned up using 0.8X AMPure XP beads (Beckman Coulter) and the quality and quantity of cDNA libraries were measured by Agilent 2200 TapeStation and Qubit Florometer (Thermal Fisher Scientific). To exclude empty wells and low-quality libraries, cDNA libraries were assessed by qPCR with a primer pair of GAPDH, a housekeeping gene (forward: 5’-GTCGTGGAGTCTACTGGTGTCTTCAC-3’, reverse: 5’-GTTGTCATATTTCTCGTGGTTCACACCC-3’). Samples showing high Ct value (>35 cycles) or no PCR amplification were discarded. 50–100pg of each cDNA library was used for generating Illumina sequencing library using a Nextera XT DNA library preparation kits (Illumina). After the final PCR amplification, samples were pooled and cleaned by 0.9X AMPure XP beads. Pooled sequencing libraries were sequenced on an Illumina Nextseq 500 instrument using a 38/37bp paired-end-reads setting.
ATAC-seq
ATAC-seq libraries were made from freshly FACS-sorted cells. Library preparation and analyses were performed as described38 with minor modifications. Briefly, FACS-sorted cells were pelleted and incubated with cold lysis buffer (10mM Tris-HCl, pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% IGEPAL CA-630). After removing lysis buffer by centrifugation, samples were immediately subjected to a transposition reaction at 37°C for 30min with 10μL transposase enzyme (Illumina Nextera DNA Preparation Kit). Transposed DNA was purified using QIAGEN MiniElute PCR purification kit and PCR amplified with 10–15 cycles. Library concentration and quality was assayed by D1000 Tape Station (Agilent) prior to sequencing. Samples were sequenced on an Illumina HiSeq 2000 instrument using a 50bp paired-end-reads setting.
ChIP-seq
ChIP-seq was performed using the ‘ChIPmentation’ approach59. Briefly, H3K4me1, H3K27ac and H3K27me3 ChIP-seq experiments were performed on 200,000 FACS-sorted bulge-SCs from mice. Cells were cross-linked in 1% (wt/vol) formaldehyde fixation buffer, resuspended and lysed. To solubilize and shear cross-linked DNAs, lysates were subjected to a Covaris S220 focused ultrasonicator (140 PIP, 5% duty factor, 200 CPB, and 4°C). The resulting whole-cell extract was incubated overnight at 4oC with 4μL of Dynabeads Protein G magnetic beads (Life Technologies), which had been pre-incubated with 2ug of anti-H3K4me1 (Abcam Cat#ab8895), anti-H3K27ac (Abcam, Cat#ab4729) or anti-H3K27me3 (Millipore, Cat#07–449) antibodies. After chromatin immunoprecipitation, samples were washed, and tagmentation reaction was performed on beads using Nextera DNA sample preparation kit (Illumina). After several washes, bound complexes were eluted and reverse–cross-linked. ChIP DNA was prepared for sequencing by PCR using Illumina barcoded primer sets. After amplification, a range of fragment sizes between 300–700bp was selected using AmpureXP beads purification. Sequencing was performed on the Illumina Nextseq500 Sequencer using a 38/37bp paired-end high-output mode setting.
Immunofluorescence and Microscopy
Mouse backskins were dissected and either embedded directly in OCT (VWR Frozen Section Compound Premium CLR), or fixed with 2% paraformaldehyde (PFA) in PBS for 1hr at 4°C to preserve the cytoplasmic fluorescent YFP signal. Then, tissues were incubated with 30% sucrose for overnight followed by embedding in OCT. Frozen tissue blocks were sectioned at 12μm on a cryostat (Leica), and mounted on SuperFrost Plus slides (Fisher). Non-prefixed tissue sections were incubated with 2% PFA for 5min and rinsed 5 times with PBS. Tissue sections were blocked for 1hour at room temperature in blocking solution (5% normal donkey serum, 0.5% bovine serum albumin, 2.5% fish gelatin and 0.3% Triton X-100 in PBS). Sections were incubated with primary antibodies diluted in blocking solution at 4°C overnight. Sections were then washed three times with PBS and incubated with secondary antibodies in blocking solution at room temperature for 1hr. Finally, sections were washed three times with PBS and mounted with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific).
The following antibodies and dilutions were used (see complete antibody info in Supplementary Table 8): NFIB (1:1000), NFIX (1:1000), GFP (1:1000), SOX9 (1:1000), LHX2 (1:2000), K5 (1:200), K10 (1:1000), K6 (1:1000), K24 (1:2000), K14 (1:1000), CD104/β4-INTEGRIN (1:100), INVOLUCRIN (1:1000), LORICRIN (1:1000), KLF5 (1:50), CD45 (1:100), ADIPOPHILIN (1:1000), active Caspase 3 (1:500), FOXP3 (1:100), pNFkB (phospho-p65 (Ser536), 1:50), SCD1 (1:200), and PPARgamma (1:100). RRX, Alexa Fluor-488 or Alexa Fluor-647-conjugated secondary antibodies (1:500, Life Technologies) were used. Nuclei were stained using 4’6’-diamidino-2-phenylindole (DAPI). EdU click-it reaction was performed according to manufacturer’s directions (Thermo Fisher). Images were acquired with an Axio Observer.Z1 epifluorescence microscope equipped with a Hamamatsu ORCA-ER camera (Hamamatsu Photonics), and with an ApoTome.2 (Carl Zeiss) slider that reduces the light scatter in the fluorescent samples, using 20X objective, controlled by Zen software (Carl Zeiss). Images were processed using ImageJ and Adobe Photoshop CS5.
Immunohistochemistry
For immunohistochemistry analysis of human skin tissue samples, formalin-fixed paraffin-embedded sections were deparaffinized with CitriSolv Hybrid (Decon Labs) and rehydrated by sequentially incubating in 100%, 95%, 70%, and 50% ethanol in water. To expose antigenic epitopes, heat-induced antigen retrieval was performed using Antigen Retrieval Reagent-Acidic buffer (pH 6.0) (R&D) for 20min at 95°C. After quenching endogenous peroxidase activity by 3% hydrogen peroxide, sections were incubated with the following primary antibodies at 4 °C overnight (see Supplementary Table 8): NFIB (1:100), NFIX (1:100), K14 (1:2000). After several washes with PBS, primary antibody staining was visualized using peroxidase-conjugated anti-rabbit IgG (1:1000) followed by the DAB substrate kit for peroxidase visualization of secondary antibodies (Vector Laboratories).
In situ hybridization to probe for microRNAs
Frozen sections were fixed with 4% PFA in PBS for 20min and subjected to 10min acetylation followed by 10min proteinase K (5μg/mL) treatment. At 50ºC, sections were pre-incubated with hybridization buffer for 4hr before incubation with miRNA probes (Exiqon miRCURY LNA™ Detection probe, 3’end DIG-labeled with Roche labeling kit) at 0.1μM O/N, and then washed with 5X SSC followed by 3 times of 0.2X SSC for 30min each. Sections were then equilibrated at RT with B1 buffer, blocked for 1hr, incubated with anti-DIG antibody (Roche) at 4ºC O/N, washed with B1 buffer followed by B3 buffer, and developed with BM Purple (Roche). Bright field images were acquired on a Leica Axioskop2 using a 10×/0.8 air objective.
16S fluorescence in situ hybridization (FISH) and tissue clearing for whole-mount visualization
To prepare samples for 16S-FISH, backskins were fixed in fresh ice cold 4% PFA overnight at 4°C, then washed with cold PBS before being permeabilized in 0.1% PBS-Tween (PBST) overnight at 4°C. Backskins were additionally fixed and permeabilized through a methanol series: 25% 1×40 min, 50% 1×40 min, 75% 1×40 min and 100% 2×40 min in PBST. Samples were rehydrated with reverse series of graded methanol/PBST, followed by 3×10 min of 0.1% PBST washes. To probe for bacterial 16S, FISH with hybridization chain reactions (HCR) was employed60. Samples were pre-hybridized with pre-warmed probe hybridization buffer for 30min at 45°C before incubating with 1pmol of each probe, mixed together, overnight at 45°C. The sequences of the probes (probeBase) were: EUB338: GCUGCCUCCCGUAGGAGU; Univ1390: GACGGGCGGUGUGUACAA; Scrambled (non-EUB, negative control): ACUCCUACGGGAGGCAGC. Following hybridization, samples were washed with pre-warmed probe wash buffer for 2×5 min and 2×30 min at 45°C, then washed with 5X SSCT (containing 0.1% Tween-20) for 3×5 min at RT. Samples were pre-amplified with amplification buffer for 30min at RT before incubating with 30pmol of fluorescently labeled hairpins overnight at RT. Samples were washed with 5X SSCT for 2×5 min, 2×30 min and 1×5 min at RT before fixation in 4% PFA for 5–15min at RT and incubation with DAPI/PBST (2μg/ml) overnight at 4°C. Samples were then washed with 0.1% PBST before being cleared for imaging. Probe hybridization buffer, probe wash buffer and probes were purchased from Molecular Technologies.
For tissue clearing, backskins were transferred through increasing concentrations of ethanol diluted in distilled water and adjusted to pH9.0: 30% for 40min, 50% for 40min, and 70% for 40min. Backskins were then incubated in 100% ethanol for 1hr before transferring into ethyl cinnamate (Sigma) for clearing.
Images were acquired with an inverted spinning-disk confocal system driven by iQ Live Cell Imaging software (Andor) using a 20x N.A. 0.75 air objective and 561nm, 642nm and UV laser lines, and analyzed using Imaris software. For spot analysis and quantification, sphere sizes were set to 2.5–3, and the estimated x-y diameter for spots was 0.6–1.5μm.
Partial thickness wound (dermabrasion)
Animals were anaesthetized with ketamine/xylazine and administered buprenorphine analgesia. Skin was shaved and remaining hair cleared with hair removal cream. Skin was gently stretched between two fingers and epidermis removed using a small rotary drill (Dremel) with a polishing wheel attachment (model 520) to create a partial-thickness wound.
High Titer Lentivirus Production
293FT cells (Thermo Fisher, Cat#R70007) were used. These cells were not found in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample, and were not authenticated. Production of VSV-G pseudotyped lentivirus was performed by calcium phosphate transfection of 293FT cells with pLKO-PGK-CreERT2 plasmids and helper plasmids pMD2.G and pPAX2 (Addgene plasmids #12259/#12260). Viral supernatant was collected 46h after transfection and filtered through a 0.45-μm filter. For in utero lentiviral transduction, viral supernatant was concentrated by ultracentrifugation. Final viral particles were resuspended in viral resuspension buffer (20mM Tris pH 8.0, 250mM NaCl, 10mM MgCl2, 5% sorbitol) and 1μl of viral suspension was injected in utero into E9.5 embryos61. CreERT2 activity was induced in adult mice, at second telogen using tamoxifen.
QUANTIFICATION AND STATISTICAL ANALYSIS
Bulk RNA-Seq data analysis
Sequencing reads from RNA-seq were aligned to the mouse reference genome (Version mm10 from UCSC) using Bowtie262 (version 2.2.9) with default parameters. Expression values of each gene were quantified as transcript per million (TPM) using RSEM63 (v1.2.30). Differential gene expression analyses were performed on normalized raw counts using DEseq264 in R software. Genes with fold change >2 and false discovery rate (FDR) <0.01 were considered differentially expressed, and genes with TPM<1 in both WT and NFI-dKO were excluded. Differentially expressed genes were presented by a heatmap with z-scores normalized expression value.
Single cell RNA-Seq data analysis
1). Reads alignment and gene quantification
Sequencing reads from single cell RNA-seq libraries were aligned to the mouse reference genome (Version mm10 from UCSC) combined with sequences for ERCC spike-ins as artificial chromosomes using Bowtie262 (version 2.2.9) with default parameters for paired-end reads. Expression values of each gene were quantified as transcript per million (TPM) using RSEM63 (v1.2.30). TPMs were transformed to log2(TPM+1). For downstream analyses, cells with <2000 genes detected and genes expressed in <5 cells were removed.
2). Identification of highly variable genes, cell clustering and t-SNE visualization
All analyses and visualization of data were conducted in a Python environment built on the Numpy, SciPy, matplotlib, scikit-learn package and pandas libraries65. To distinguish true biological variability from technical noise in single cell experiments, we used the statistical model for identifying highly variable genes compared to ERCC spike-ins as described by53. Briefly, genes with higher level of variation (above the technical variation) and false discovery rate (FDR) value <0.1 were considered as highly variable.
To identify cell clusters, principal component analysis (PCA) was performed on the list of highly variable genes, and the first 10 principal components were used to perform t-distributed stochastic neighbor embedding (t-SNE)66. By unsupervised hierarchical clustering using Euclidean distance and ward’s method, cell clusters were identified. Expression levels (log-transformed) of individual genes were represented by colors indicated in the t-SNE plots.
3). Differential expression test and cluster identity annotation
To identify differentially expressed genes between clusters, normalized raw counts were applied to DEseq2 package64 in R software, and then genes with fold change >2 and FDR <0.1 were considered differentially expressed. Corrected p-values (FDR) were calculated using the Benjamini and Hochberg method. Signature genes of each cluster were defined as genes >2x differentially expressed, and unique for each cluster compared to all other clusters. Cluster identification was performed using all cells (combining control and NFI-dKO cells) to have enough numbers of cells per cluster. By comparing the list of signature gene of each clusters and lineage markers of HF basal progenitor cells, all HF progenitor populations were successfully annotated. To compare the changes of cell composition between control and NFI-dKO HF, the number of cells in each cluster were counted.
ATAC-seq data analysis
1). ATAC-seq Alignment, Peak Calling, and Visualization
Sequencing reads from ATAC-seq libraries were aligned to the mouse reference genome (Version mm10 from UCSC) using Bowtie67 (version 1.1.2) with parameters “-X 2000” and “-m 1” for 50bp paired-end reads. Duplicates were removed using Picard (https://github.com/broadinstitute/picard/releases/tag/2.7.1). Peaks were called using MACS2 (version 2.1.1.20160309)68 with the parameter “-keep-dup all”. Independent biological replicates showed a high degree of similarity (Pearson correlation coefficient, r = 0.94 ~ 0.96), so the replicate data were subsequently combined and processed. ATAC-seq signal tracks were presented by Integrative Genomics Viewer (IGV) software69.
2). Differential analysis of ATAC-seq peaks
To identify differential accessible chromatin regions, a set of non-redundant ATAC peaks in duplicates of control and NFI-dKO bulge-SCs were identified, and ATAC-seq signals of the peaks counted using bedtools. After normalization with total aligned non-redundant reads of each sample, differential accessible peaks (fold changes >2 and FDR <0.1) were calculated using DEseq264 R package.
3). Motif analysis of ATAC-seq peaks
De novo sequence motifs discovery was performed using HOMER with default parameters70. ATAC-seq peaks unique to control and NFI-dKO bulge-SCs were used to identify sequence motifs that matched known transcription factor binding sites.
4). Comparison of ATAC-seq peaks and expression changes of associated genes
ATAC-seq peaks of control and NFI-dKO bulge-SCs were annotated with associated genes using HOMER70. Log2 fold gene expression changes of control/NFI-dKO were presented as box plots.
ChIP-seq reads alignment and visualization
H3K4me1, H3K27ac, and H3K27me3 ChIP-seq reads were aligned to the mouse reference genome (Version mm10 from UCSC) using Bowtie2 with default parameters. To generate a heatmap showing the histone modification changes around NFIB peaks, 25-bp bins of genomic regions within ±5kb of the NFIB ChIP-seq peaks23 were identified, and the density matrix was calculated with normalization by reads per genomic content (RPGC) and then used to generate a heatmap using DeepTools271. Super-enhancers were identified using Rank Ordering of Super-Enhancers (ROSE) algorithm11. Super-enhancers were identified using new H3K27ac data (generated for this study using ‘ChIP-mentation’, hence the number of bioinformatically defined SEs differs slightly from our prior study13). Typical enhancers were defined as H3K27ac peaks ± 2.5 kb outside of transcription start sites.
Statistics and reproducibility
Statistical and graphical data analyses were performed using Microsoft Excel and Prism 8 (Graphpad) software. For measurements, three biological replicates and two or more technical replicates were used, where applicable. To determine the significance between two groups, comparisons were made using unpaired two-tailed Student’s t-test or ANOVA, as appropriate. Multiple testing correction was done by the Benjamini and Hochberg method. In box and whisker plots, the middle line is plotted at the median, the upper and lower hinges correspond to the first and third quartiles, and the upper and lower whiskers display the full range of variation (min. to max.). Experiments were repeated on ≥3 pairs of sample and control sets, and representative images shown in Figures were reproducible in both male and female mice.
DATA AVAILABILITY
ChIP-seq, ATAC-seq, RNA–seq and scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE135142, GSE135143, GSE135144, GSE135145, and GSE135146 (super-series). Previously published sequencing data on bulge-SC super-enhancers that were re-analyzed here are available under accession code GSE61316. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
CODE AVAILABILITY
Codes for analysis of the figures related to single cell RNA-seq data are available at https://github.com/hanseulyang01/NFI-single-cell-analysis.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Elaine Fuchs (fuchslb@rockefeller.edu).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. NFI-TFs Maintain Bulge-SC Identity and Prevent Ectopic Differentiation.
a, Enrichment of NFIB within chromatin of super-enhancers, compared to typical enhancers. Comparisons were made with 377 randomly selected typical enhancers and their flanking sequence extended 5’ and 3’ to match the average length of super-enhancers (average of 3 analyses is shown). b, Table showing examples of bulge-SC super-enhancer regulated genes with NFIB ChIP-occupancy. c, mRNA expression levels (TPM) of NFI family members in bulge-SCs. Mean TPM from 2 independent replicates are shown. d, Validation of Nfib and Nfix gene knockout and antibody specificity. INTEGRIN β4 (β4) marks basal epithelial cells. Inner bulge cells are β4neg, adjacent to the hair shaft and are critical to anchor the hair. e, Images of WT and Nfix cKO mice at 2 months post-TAM. Nfix-ablation on its own has no impact on the hair coat. f, Immunofluorescence using K24 antibody to label bulge-SCs. Ablation of Nfib or Nfix alone does not affect bulge-SCs, whereas combined ablation results in a loss of K24+ bulge-SCs. g, Images of WT or NFI-dKO mice at 4 weeks post-TAM. h, Immunofluorescence comparing telogen HFs of WT, Nfib cKO, Nfix cKO and Nfib/Nfix-dKO mice. K5 marks basal epithelial cells. Note aberrant K10+ (differentiating) cells only in double Nfib/Nfix targeted (dKO) mice. i, In situ hybridization using scramble or mir-203 probes on mouse skin. Note expansion of signal for the epidermal differentiation microRNA, mir-203, into bulge of NFI-dKO skin. j, Tape strip assay to evaluate hair anchoring. Tape stripping applies a mild tug to the hairs, which will be released from the coat if anchorage is weak. All scale bars = 20μm. Bu, bulge. Dashed lines, HF-dermal border. For d-j, at least three biological replicates were used; representative images are shown. See also Source Data.
Extended Data Fig. 2. NFI-dKO Mice Exhibit Features of Primary Cicatricial Alopecia.
a, Immunofluorescence showing HF degeneration in NFI-dKO mice at 2 months post-TAM. YFP labels Sox9-CreER-targeted HFs. K6 labels inner bulge cells anchoring the hair. b, Immunofluorescence showing hyperkeratosis (K10) and follicular plugging of infundibulum in NFI-dKO HFs. K5 marks basal epithelial cells. c, Loss of PPARG, SCD1 and ADIPOPHILIN, lipid-related markers of mature sebocytes, in NFI-dKO follicles. Mean and standard deviation are shown. 30 HFs per genotype, pooled from n=3 mice. P value is from unpaired, two-tailed t-test. d, Hematoxylin & Eosin image of NFI-dKO skin at 2 months post-TAM. Note follicular remnants and fibrous tracts (arrows). e, Immunofluorescence and quantifications of active CASPASE3+ (apoptotic) cells in NFI-dKO HFs. Mean and standard deviation are shown. For 2 weeks data, n=80 HFs per genotype (total, pooled from 4 mice). For 2 months data, n=59 HFs (WT) or n=74 HFs (NFI-dKO), (total, pooled from 3 mice). P values are from unpaired, two-tailed t-test. f, (left) Flow cytometry analysis of immune cells at 2 months post-NFI deletion. Mean and standard deviation are shown. n=4 mice/genotype. P values are from unpaired, two-tailed t-test. (right) Immunofluorescence analysis of FOXP3+ regulatory T-cells (Tregs) around the HF bulge niche. g, Fluorescence in situ hybridization (FISH) of pan-bacterial 16S rRNA (rainbow colors) in cleared skin whole-mounts, co-labeled for DAPI (gray) at 2 months post-TAM. Spot analysis of 16S-FISH signal was used to quantify bacterial load per μm3 of skin. Mean and standard deviation are shown. n=10 (WT) and 16 (NFI-dKO) HFs from 2 biologically independent mice/genotype. Scale bars = 20μm, unless otherwise specified. Bu, bulge. Inf, Infundibulum. Dashed lines, HF-dermal border. For a-d and f, at least three biological replicates were used; representative images are shown. See also Source Data.
Extended Data Fig. 3. Skin Immune Cell Profiling by Flow Cytometry.
a, Flow cytometry gating strategy for adaptive immune cell profiling. b, Flow cytometry gating strategy for innate immune cell profiling. Plots are shown for a representative WT mouse analyzed at 2 months post-NFI ablation. See Methods for details on immune cell identification.
Extended Data Fig. 4. Absence of Skin Immune Infiltration at 2 weeks post-NFI loss.
a, Fluorescence in situ hybridization (FISH) of pan-bacterial 16S rRNA (rainbow colors) in cleared skin whole-mounts, co-labeled for DAPI (gray) at 2 weeks post gene knockout. Spot analysis of 16S-FISH signal was used to quantify cutaneous bacterial load. Representative images of two biological replicates. b, Immunofluorescence showing skin immune cells (CD45+) are not changed at 2 weeks following Nfib/Nfix knockout (mean and standard deviation are shown). K5 marks basal epithelial cells. n=3 mice. c, Flow cytometry analysis of immune cell composition at 2 weeks post-NFI deletion. Mean and standard deviation are shown. n=4 mice/genotype. d, mRNA expression (from RNA-seq) of immune-related genes in bulge-SCs at 2 weeks post-TAM. All scale bars = 20μm. Bu, bulge. Inf, Infundibulum. Dashed lines, HF-dermal border. See also Source Data.
Extended Data Fig. 5. Immunosuppression does not prevent bulge-SC loss in NFI-dKO mice.
All analyses in mice were done at 2 months post-NFI deletion. Dexamethasone (DEX) was administered continuously since gene deletion to evaluate the long-term effect of immunosuppression on bulge phenotypes. a, Immunofluorescence analysis of FOXP3+ regulatory T-cells (Tregs) around the HF bulge niche. K14 marks basal epithelial cells. b, Flow cytometry analysis of immune cell composition at 2 months post-NFI deletion. Mean and standard deviation are shown. n=3 WT mice in PBS and DEX groups, n=5 NFI-dKO mice in PBS group, n=4 NFI-dKO mice in DEX group. P-values are from unpaired, two-tailed t-test. c, Immunofluorescence analysis of phosphorylated (active) NF-kB in the HF bulge. INTEGRIN β4 (β4) marks basal epithelial cells. d, Images of WT and NFI-dKO mice with or without DEX. Note DEX led to hair coat retention in NFI-dKO mice. e, Immunosuppression fails to rescue ectopic epidermal differentiation (K10) in the NFI-dKO bulge. f, Analysis of human scalp biopsies. Immunohistochemistry shows reduced expression of NFIB and NFIX in scarring alopecia patients compared to normal human scalp skin. g, Immunohistochemical analysis of human scalp biopsies using anti-K14 antibody, a marker of outer root sheath (ORS, progenitor) cells, where NFIB and NFIX are normally expressed. All scale bars = 20μm, unless otherwise indicated. Bu, bulge. Inf, Infundibulum. Dashed lines, HF-dermal border. For a, c- g, at least three biological replicates were used; representative images are shown. See also Source Data.
Extended Data Fig. 6. Bulk Transcriptome Analysis of NFI-dKO bulge-SCs.
a, b, Volcano plots showing differential gene expression of WT vs. NFI-dKO (a) or WT vs. Nfix cKO (b) bulge-SCs. Note that Nfix ablation on its own has little effect on bulge-SC transcriptomes. n=23491 genes were analyzed/genotype. c, Overlap of WT vs. NFI-dKO gene expression changes and NFIB ChIP-occupancy to identify transcriptional targets sensitive to NFIB levels. All transcriptome analyses were performed on 2 mice/genotype. Statistical analysis was performed using unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method.
Extended Data Fig. 7. Single Cell Transcriptome Analysis of Telogen Skin Epidermis.
a, FACS-purification of 2nd telogen skin progenitors from Sox9-CreER/Nfixfl/fl/R26-YFP (non-phenotypic) vs. NFI-dKO mice. Hair follicles were YFP+ and INTEGRIN α6+, while epidermal SCs (EpdSCs) were YFPneg, INTEGRIN α6+ and SCA1+. Representative plots for three biological replicates. b, Validation of FACS-purification strategy for single cell RNA-seq analysis. tSNE plot showing low Sox9 expression in the YFPneg cluster (EpdSC), whereas all YFP+ populations are Sox9+ (HF). n=2 mice per group. c, Correlation plots of single-cell RNA-seq libraries shows minimal batch to batch variation. n=2 mice per group. Correlation coefficients were calculated by Pearson’s method. d, tSNE plots showing expression of known epidermal lineage markers to determine the identity of individual clusters. n=2 mice per group. e, tSNE plot showing the unique cluster in NFI-dKO mice is Cd34neg. n=2 mice per group.
Extended Data Fig. 8. NFI-TFs are Required to Maintain Bulge-SC Identity.
a, Replicate analysis of ATAC-seq experiments show correlation coefficients of >0.94, indicating good reproducibility. Correlation coefficients were calculated by Pearson’s method. b, Reduction of chromatin accessibility at bulge-SC TF-bound loci upon loss of NFI-TFs. Note loci co-occupied by NFIB and bulge-SC TFs show a greater decrease of chromatin accessibility. In boxplots, the median (line), first and third quartiles (box), and whiskers (highest and lowest values) are shown. TF bindings sites are based on prior in vivo ChIP-seq on bulge-SCs13,16–18,20,72. ATAC-seq (this study) was used to determine differential accessibility at bulge-SC TF ChIP-bound loci. Statistics was analyzed using unpaired, two-tailed t-test. Number of peaks analyzed: SOX9: n=1813 (NFIB-bound), n=1175 (no NFIB binding); LHX2: n=1363 (NFIB-bound), n=1453 (no NFIB binding); NFATc1: n=1503 (NFIB-bound), n=4199 (no NFIB binding); pSMAD1: n=1547 (NFIB-bound), n=1778 (no NFIB binding); TCF3: n=7840 (NFIB-bound), n=7668 (no NFIB binding); TCF4: n=5346 (NFIB-bound), n=4371 (no NFIB binding); TLE: n=2901 (NFIB-bound), n=2840 (no NFIB-binding); bulge SE H3K27ac: n=970 (NFIB-bound), and n=2017 (no NFIB binding). c, Comparison of bulge-SC-TF ChIP-peaks reveals high co-occupancy with NFIB. d, Differential chromatin accessibility at NFIB ChIP-occupied super-enhancers in WT vs. Nfib/Nfix-dKO bulge-SCs (measured by ATAC-seq). e, Immunofluorescence analysis shows gradual reduction in bulge-SC marker LHX2 over time. Scale bars = 20μm. Bu, bulge. Dashed lines, HF-dermal border. Representative images for three biological replicates. f, Replicate analysis of 2 independent ChIP-seq experiments show correlation coefficients (r) of >0.94, indicating good reproducibility. Number of peaks analyzed: H3K4me1: n=128538 (WT), n=119965 (NFI-dKO); H3K27ac: n=65493 (WT), and n=82522 (NFI-dKO). Correlation coefficients were calculated by Pearson’s method. g, Heatmap of H3K4me1, H3K27ac and H3K27me3 ChIP-seq read densities centered on NFIB-bound peaks, depicting how they change with NFI status in bulge-SC chromatin. Note that Nfib/Nfix ablation associates with reduced H3K4me1 and H2K27ac but not H3K27me3 at NFIB-bound loci. See also Source Data.
Extended Data Fig. 9. Model of NFIB and NFIX Function in the HF SC Niche.
Although our studies focused on using Sox9-CreER mice to ablate NFI proteins in the HF, we also show that LV-CreER ablation of NFI proteins in the epidermis does not affect its SCs or its differentiation program. Rather, NFIB and NFIX act on the bulge-SCs and without them, a primary scarring alopecia phenotype is generated. At the chromatin level, NFI proteins act in the bulge-SC niche to maintain chromatin accessibility of bulge-SC super-enhancers while repressing epidermal enhancers. When NFI proteins are absent, many bulge-SC super-enhancers are silenced while some epidermal enhancers become ectopically activated, leading to a lineage infidelity state.
Supplementary Material
Supplementary Table 1. Histopathological assessment of NFI-dKO skin and comparison to human primary cicatricial alopecia.
Supplementary Table 2. Human patient information and clinical diagnosis of primary scarring alopecia patients.
Supplementary Table 3. The list of differentially expressed genes (>2-fold change, FDR < 0.01) in NFI-dKO vs WT bulge-SCs from bulk transcriptome analysis together with NFIB ChIP-seq occupancy. n = 2 mice per group were analyzed. P values were calculated from unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method.
Supplementary Table 4. List of signature genes of cell clusters (fold >2, FDR <0.1, VS other cells) in single cell transcriptome analysis of skin progenitors. n = 2 mice per each group were analyzed. P values were calculated from unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method.
Supplementary Table 5. The list of ATAC peaks that are differentially accessible in WT and NFI-dKO bulge-SCs.
Supplementary Table 6. The list of super-enhancers in WT and NFI-dKO bulge-SCs.
Supplementary Table 7. The list of differentially expressed genes (>2-fold change, FDR < 0.1) of the unique cell population in NFI-dKO vs WT bulge-SCs, from single cell transcriptome analysis. n = 2 mice per each group were analyzed. P values were calculated from unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method.
Supplementary Table 8. List of antibodies used in this study.
Acknowledgments
We thank E. Wong, M. Nikolova, J. Racelis and P. Nasseir for technical assistance; and L. Polak, J. Levorse and L. Hidalgo for assistance with mouse handling and experiments. We thank RU FACS facility for cell sorting, Rockefeller Genomics Resource Center and Weill Cornell Genomics Resource Center for high-throughput sequencing and Comparative Bioscience Center (AAALAC accredited) for care of mice in accordance with National Institutes of Health (NIH) guidelines. E.F. is an Investigator of the Howard Hughes Medical Institute. R.C.A. was the recipient of an Anderson Cancer Center Graduate Fellowship. H.Y. was the recipient of a Kwanjeong Educational Foundation Graduate Fellowship. Y.G was a postdoctoral fellow of the American Federation of Aging Research. N.I. is the recipient of an NIH Predoctoral National Research Service Award (NRSA) F31 Fellowship from NIAMS. The work was supported by grants from the National Institutes of Health to E.F. (R01-AR31737).
Footnotes
COMPETING INTERESTS
The authors declare the following competing interests: R.C.A. is currently employed at Regeneron Pharmaceuticals. E.F. is on the Scientific Advisory Board of L’Oreal and Arsenal Biosciences.
ADDITIONAL INFORMATION
Supplemental information is available for this paper.
REFERENCES
- 1.Avgustinova A, and Benitah SA Epigenetic control of adult stem cell function. Nat Rev Mol Cell Biol. 17, 643–658 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Scadden DT Nice neighborhood: emerging concepts of the stem cell niche. Cell 157, 41–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzales KAU, and Fuchs E Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev. Cell 43, 387–401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paksa A, and Rajagopal J The epigenetic basis of cellular plasticity. Curr Opin Cell Biol. 49, 116–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arwert EN, Hoste E, and Watt FM Epithelial stem cells, wound healing and cancer. Nat. Rev. Cancer 12, 170–180 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Dvorak HF Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med 315, 1650–1659 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Hsu Y-C, Pasolli HA, and Fuchs E Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu YC, Li L, and Fuchs E Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 157, 935–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page ME, Lombard P, Ng F, Göttgens B, and Jensen KB The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 13, 471–482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge Y, et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hnisz D, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 58, 362–370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adam RC, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trompouki E et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adam RC, et al. Temporal layering of signaling effectors drives chromatin remodeling during hair follicle stem cell lineage progression. Cell Stem Cell 22, 398–413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadaja M, et al. SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev. 28, 328–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folgueras AR, et al. Architectural niche organization by LHX2 is linked to hair follicle stem cell function. Cell Stem Cell 13, 314–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lien WH, et al. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat Cell Biol. 16,179–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horsley V, Aliprantis AO, Polak L, Glimcher LH, and Fuchs E NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132, 299–310 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genander M, et al. BMP signaling and its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell 15, 619–633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lay K, Kume T, and Fuchs E FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proc Natl Acad Sci U S A. 113, E1506–1515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Siegenthaler JA, Dowell RD, and Yi R Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science 351, 613–617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CY, et al. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature 495, 98–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Driller K, et al. Nuclear factor I X deficiency causes brain malformation and severe skeletal defects. Mol Cell Biol. 27, 3855–3867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matuzelski E, et al. Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord. Dev Biol. 432, 286–297 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Steele-Perkins G, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Molecular and Cellular Biology 25, 685–698 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gründer A, et al. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mechanisms of Development 112, 69–77 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Das Neves L, et al. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci U S A. 96, 11946–11951 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deneen B, et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Campbell CE, et al. The transcription factor Nfix is essential for normal brain development. BMC Dev. Biol. 13, 8–52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen KS, Lim JWC, Richards LJ, and Bunt J The convergent roles of the nuclear factor I transcription factors in development and cancer. Cancer Lett. 410, 124–138 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Denny SK, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 166, 328–342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chávez S, and Beato M Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci U S A. 94, 2885–2890 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu R, et al. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106, 309–318 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Ferrari S, et al. Chromatin domain boundaries delimited by a histone-binding protein in yeast. J Biol Chem. 279, 55520–55530 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Pjanic M, et al. Nuclear Factor I genomic binding associates with chromatin boundaries. BMC Genomics 14, 99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waki H, et al. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet. 7, e1002311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meisterernst M, Gander I, Rogge L, and Winnacker EL A quantitative analysis of nuclear factor I/DNA interactions. Nucleic Acids Res 16, 4419–4435 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruse U, and Sippel AE Transcription factor nuclear factor I proteins form stable homo- and heterodimers. FEBS Letters 348, 46–50 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Soeda T, et al. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48, 635–644 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pratt CH, King LE Jr., Messenger AG, Christiano AM, and Sundberg JP Alopecia areata. Nat Rev Dis Primers. 3, 17011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harries MJ, and Paus R The pathogenesis of primary cicatricial alopecias. Am. J. Pathol 177, 2152–2162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su Y, et al. Cicatricial alopecia. IntechOpen 10.5772/intechopen.78971 (2018). [DOI] [Google Scholar]
- 45.Harries M, Hardman J, Chaudhry I, Poblet E, and Paus R Profiling the Human Hair Follicle Immune System in Lichen Planopilaris and Frontal Fibrosing Alopecia: Can Macrophage Polarization Differentiate These Two Conditions Microscopically? Br. J. Dermatol DOI: 10.1111/bjd.18854 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Volc-Platzer B, et al. Accumulation of gamma delta T cells in chronic cutaneous lupus erythematosus. J. Invest. Dermatol 100, 84S–91S (1993). [DOI] [PubMed] [Google Scholar]
- 47.Lay K, et al. Stem cells repurpose proliferation to contain a breach in their niche barrier. Elife pii: e41661. doi: 10.7554/eLife.41661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenblum MD, et al. Expression of CD200 on epithelial cells of the murine hair follicle: a role in tissue-specific immune tolerance? J Invest Dermatol 123, 880–887 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Rigopoulos D, Stamatios G, and Ioannides D Primary scarring alopecias. Curr. Probl. Dermatol 47, 76–86 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Anzai A, Wang EHC, Lee EY, Aoki V, and Christiano AM Pathomechanisms of immune-mediated alopecia. Int. Immunol pii: dxz039. doi: 10.1093/intimm/dxz039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 40, 1291–1299 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nature Methods 10, 1093–1095 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Yang H, Adam RC, Ge Y, Hua ZL, and Fuchs E Epithelial-mesenchymal micro-niches govern stem cell lineage choices. Cell 169, 483–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karnik P, et al. Hair follicle stem cell-specific PPARgamma deletion causes scarring alopecia. J Invest Dermatol 129, 1243–1257 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Y, et al. Scd1 is expressed in sebaceous glands and is disrupted in the asebia mouse. Nat. Genet 23, 268–270 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Yang H, et al. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. Elife 4, e10870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidl C, Rendeiro AF, Sheffield NC, and Bock C ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi HM, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol 28,1208–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beronja S, Livshits G, Williams S, and Fuchs E Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat Med 16, 821–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, and Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B, and Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, and Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pedregosa F et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 12, 2825–2830 (2011) [Google Scholar]
- 66.Van der Maaten L, and Hinton G Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605 (2008). [Google Scholar]
- 67.Langmead B, Trapnell C, Pop M, and Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson JT, et al. Integrative genomics viewer. Nature Biotechnology 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keyes BE, et al. Nfatc1 orchestrates aging in hair follicle stem cells. Proc Natl Acad Sci U S A 110, E4950–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Histopathological assessment of NFI-dKO skin and comparison to human primary cicatricial alopecia.
Supplementary Table 2. Human patient information and clinical diagnosis of primary scarring alopecia patients.
Supplementary Table 3. The list of differentially expressed genes (>2-fold change, FDR < 0.01) in NFI-dKO vs WT bulge-SCs from bulk transcriptome analysis together with NFIB ChIP-seq occupancy. n = 2 mice per group were analyzed. P values were calculated from unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method.
Supplementary Table 4. List of signature genes of cell clusters (fold >2, FDR <0.1, VS other cells) in single cell transcriptome analysis of skin progenitors. n = 2 mice per each group were analyzed. P values were calculated from unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method.
Supplementary Table 5. The list of ATAC peaks that are differentially accessible in WT and NFI-dKO bulge-SCs.
Supplementary Table 6. The list of super-enhancers in WT and NFI-dKO bulge-SCs.
Supplementary Table 7. The list of differentially expressed genes (>2-fold change, FDR < 0.1) of the unique cell population in NFI-dKO vs WT bulge-SCs, from single cell transcriptome analysis. n = 2 mice per each group were analyzed. P values were calculated from unpaired, two-tailed t-test and corrected using the Benjamini and Hochberg method.
Supplementary Table 8. List of antibodies used in this study.
Data Availability Statement
ChIP-seq, ATAC-seq, RNA–seq and scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE135142, GSE135143, GSE135144, GSE135145, and GSE135146 (super-series). Previously published sequencing data on bulge-SC super-enhancers that were re-analyzed here are available under accession code GSE61316. All other data supporting the findings of this study are available from the corresponding author on reasonable request.