Abstract
The aim of this study was to compare the predictive performance of the Genomic Best Linear Unbiased Predictor (GBLUP) and machine learning methods (Random Forest, RF; Support Vector Machine, SVM; Artificial Neural Network, ANN) in simulated populations presenting different levels of dominance effects. Simulated genome comprised 50k SNP and 300 QTL, both biallelic and randomly distributed across 29 autosomes. A total of six traits were simulated considering different values for the narrow and broad-sense heritability. In the purely additive scenario with low heritability (h2 = 0.10), the predictive ability obtained using GBLUP was slightly higher than the other methods whereas ANN provided the highest accuracies for scenarios with moderate heritability (h2 = 0.30). The accuracies of dominance deviations predictions varied from 0.180 to 0.350 in GBLUP extended for dominance effects (GBLUP-D), from 0.06 to 0.185 in RF and they were null using the ANN and SVM methods. Although RF has presented higher accuracies for total genetic effect predictions, the mean-squared error values in such a model were worse than those observed for GBLUP-D in scenarios with large additive and dominance variances. When applied to prescreen important regions, the RF approach detected QTL with high additive and/or dominance effects. Among machine learning methods, only the RF was capable to cover implicitly dominance effects without increasing the number of covariates in the model, resulting in higher accuracies for the total genetic and phenotypic values as the dominance ratio increases. Nevertheless, whether the interest is to infer directly on dominance effects, GBLUP-D could be a more suitable method.
Keywords: Artificial Neural Network, genomic selection, nonadditive effects, Random Forest, Support Vector Machine
Introduction
Genome-wide dense marker availability on a commercial scale has brought a new paradigm to animal breeding. The use of such information for genome-enabled prediction of breeding values has allowed accelerating the genetic gains by providing early and more accurate prediction than pedigree-based approaches (Meuwissen et al., 2013). Nevertheless, whole-genome prediction models have been typically formulated considering only additive effects, ignoring possible nonadditive relationships, for instance, dominance effects caused by allele interactions at the same locus. Including this effect in genomic evaluation has theoretical advantages such as exploring the specific combining ability for enhancing progeny performance and may increase the accuracy of breeding values prediction, avoiding an overestimation, especially if dominance variance ratio is large (Toro and Varona, 2010; Aliloo et al., 2016).
Variance component estimates for dominance effects using either pedigree or genomic-based analyses have been ranging from null to a substantial contribution to the total genetic variance of different complex traits (Fuerst and Sölkner, 1994; Rodriguez-Almeida et al., 1995, Van Tassell et al., 2000; Gallardo et al., 2010; Su et al., 2012; Nagy et al., 2013; Aliloo et al., 2016). Nonetheless, accounting for nonadditive effects may increase the model parameterization with the construction and inversion of large and dense matrices, leading to an intensive computational cost (Su et al., 2012). Additionally, although parametric models as Genomic Best Linear Predictor (GBLUP) and Bayesian regressions have proven to be robust for genomic prediction, such models rely on strong assumptions that may not hold in practice, regarding mainly with the statistical distribution of marker effects (genetic architecture), presence of Hardy–Weinberg equilibrium (HWE), independence among loci and, model orthogonality (Okut et al., 2013; Vitezica et al., 2013; Hill and Mäki-Tanila, 2015).
Recently, machine learning theory has been expanded to a genomic prediction scope, mainly due to its theoretical flexibility to cope with complex relationships between markers and phenotypes. Such approaches can deal with the dimensionality problem adaptively, without imposing any specific relationship between phenotypes and genotypes, providing appealing attributes that make them well suited for genomic data analysis (Gonzalez-Récio et al., 2014).
Previous studies using simulated data reported similar or better prediction ability for machine learning methods such as Support Vector Regression, Random Forest (RF) and Artificial Neural Networks (ANNs) when compared with GBLUP or Bayesian regression models (González-Recio and Forni, 2011; Ogutu et al., 2011; Howard et al., 2014; Ghafouri-Kesbi et al., 2016). Nevertheless, comparisons have been performed mainly for scenarios under purely additive effects which may not represent real data conditions. Thus, this study aimed to compare the predictive performance of GBLUP and machine learning methods in simulated populations presenting different levels of dominance.
Material and Methods
The procedures performed in this research do not require any formal consent of the Institutional Animal Care and Use Committee.
Simulated data
The simulation procedures were performed according to previous simulation studies considering dominance genetic effects (Toro et al., 2010; Wittenburg et al., 2011; Nishio et al., 2014; Martini et al., 2017). Genotype data including markers and QTL spread across the genome were simulated using QMSim software (Sargozalei and Schenkel, 2009). First, a historical population with 1,000 animals was simulated with random mating and constant population size during 1,000 generations and then gradually reduced to 100 individuals in additional 1,020 generations. This step aimed to create the linkage disequilibrium and to allow mutation-drift equilibrium establishment. Recurrent mutation process was assumed for both marker and QTL, with a mutation rate of 5 × 10−4. For expanding the resultant population, the remained animals (50 males and 50 females) were randomly mated by an additional five generations assuming five offspring per dam and exponential growth of the number of dams.
Finally, 100 males and 1,500 females from the last generation of the expanded population were assumed to be the base population (G0) and additional five generations were simulated as recent population (G1 to G5), assuming one offspring per dam with equal probability of being male or female, resulting in a total of 9,100 animals (G0 to G5). The replacement rates of sires and dams were kept constant at 60% and 20%, respectively. Minor allele frequencies of markers and QTL on the G0 population were set to be ≥0.05. The simulated genome comprised 50k SNP markers and 300 QTL, both biallelic and randomly distributed across 29 autosomes, with a total length of 2,320 cM.
The resultant simulated populations were used to model complex traits affected by purely additive effects or presenting different degrees of dominance. Simulations of genotypic values were performed in terms of breeding values and dominance deviations, with the assumption of HWE (Falconer and Mackay, 1996), as described by Vitezica et al (2013):
where z and w are coded as
and
where , and are the true alleles frequencies for and at the jth QTL and is the allele substitution effect. The additive effect for each QTL was sampled from a gamma distribution with shape and scale parameters of 0.42 and 1.66, respectively, with positive and negative signs drawn with an equal chance (Hayes and Goddard, 2001; Meuwissen et al., 2001). Following previous studies (Wittenburg et al., 2011; Nishio and Satoh, 2014; de Almeida Filho et al., 2016), the dominance deviations were determined as , where is the degree of dominance, which was initially drawn from a normal distribution N(0, 1). Finally, the additive and dominance effects for each QTL were scaled to achieve the desirable variances for each scenario.
Final additive () and dominance () variances were computed as (Falconer and Mackay, 1996)
and
Consequently, the total genetic variance () was partitioned as
Residual effects were sampled from a normal distribution, with , and added to the total genetic effects to achieve a phenotypic variance of 1 for all scenarios. Therefore, the observed phenotypes were computed as
Six traits were simulated considering different narrow-sense (h2 = 0.10 and 0.30) and broad-sense (H2 = 0.10, 0.15, 0.20, 0.30, 0.45, and 0.60) heritability values. After stochastic simulation procedure, 1,500 animals were randomly selected from generation G1 to G4 to compose the reference population and, 500 animals from G5 were randomly selected as testing population, for which only genotypes were assumed to be known. We repeated this procedure for 10 replicates in each scenario.
Prediction models
Genomic best-unbiased prediction
The general model for the GBLUP can be written as
where y is the vector of phenotypes, 1n is a vector of 1’s, u is an overall mean, Z is an incidence matrix relating the animals to the additive effects, g is a vector of direct genomic breeding values, and e is a vector of residuals. The GBLUP model extended for dominance effects (GBLUP-D) can be represented as follows:
in which W is an incidence matrix relating the animals to dominance deviations and d is a vector of genomic dominance deviations. Both g and d were assumed to be normally distributed with g~ N(0, G) and d~ N(0, D), where G and D are genomic relationship matrices for additive and dominance effects, respectively, with and representing the respective variances of such effects. The G matrix was constructed as described by VanRaden (2008):
where is an n × matrix (n is the number the individuals and is the number of markers), in which the elements for the ith individual and jth marker are equal to , , and for the aa, Aa, and AA genotypes, respectively, with representing the expected frequencies of the allele A at the jth marker in the population. Similarly, the D relationship matrix was constructed as follows Vitezica et al. (2013):
in which is an n × matrix with the elements coded as , , and for aa, Aa, and AA genotypes, respectively. The GBLUP and GBLUP-D models were fitted using the BGLR package (Pérez and De Los Campos, 2014).
Random Forest
The RF algorithm (Breiman, 2001) uses an ensemble of unpruned decorrelated decision trees, built from B bootstrap samples of the training data set and randomly selecting a subset of the original predictor variables as candidates for splitting tree nodes. This ensemble of weak learners can be used for prediction of an unobserved data by averaging all B predictors as
where represents an individual b tree architecture in terms of split variables, cut point at each node and terminal node values and X is the raw SNP matrix, coded as 0, 1, and 2 for the aa, Aa, and AA genotypes, respectively. In the RF model, samples that are not selected (roughly one-third of the total observations) on bootstrapping, termed out-of-bag (OOB) samples, are used as internal validation from which the OOB error is computed. A common error measure adopted in regression problems is the mean square error of the OOB data:
in which N is the number of observations in the OOB samples, is the average of the predictions for the ith animal from trees in which it was OOB, and is the realized value of the animal.
In the RF approach, variable predictors can be ranked by the importance of their contributions to predictive accuracy. One of the possible ways for computing the variable importance measure (VIM) is by accounting how much OOB error increases when a given variable predictor (e.g., an SNP) is randomly permutated on the OOB data while all other predictor variables left unchanged. The relative variable importance can be calculated as the difference between the original predictive measure (without permuting the variable on the OOB sample) and that of the OOB with the permuted variable. This step is repeated for each covariate (SNP) and the decrease of accuracy is averaged over all trees in the RF. Important variables for the outcome prediction are expected to have higher VIM since the permutation of such variables on the validation data will increase the prediction error. We implemented the RF model in the randomForest R package (Liaw and Wiener, 2002). The model parameters used in the present study were fixed as ntree = 1,000 (number of trees to grow) and mtry = (number of SNP selected at each tree node) and nodesize as default.
Support Vector Machines
Support Vector Machines (SVMs) utilize linear models to implement nonlinear regressions, by mapping the predictors in a feature space of different dimensions using kernels inner products, followed by linear regression on the resulting observed space. The general model can be viewed as (Hastie et al., 2009):
where x stands for the markers genotypes (coded as 0, 1, and 2, as mentioned before), represents a linear or nonlinear transformation of the original input space featured by a given kernel function (h), here, the radial basis function, is a constant and are the weights for each variable in the feature space. For the risk minimization, we adopted the ε-insensitive loss function:
in which
is a function which sets an insensitive tube around the residuals, ignoring the errors within the tube (less than ε), C is a regularization parameter that controls the trade-off between the complexity of the loss function and the training error, and denotes the norm under a Hilbert Space. According to Hastie et al. (2009), if and are the constants that minimize H, the solution function has the following form:
where , are positive weights given to each observation and the inner product is an N × N symmetric and positive definite kernel matrix.
The parameters ε and C were defined as and , respectively, while the kernel bandwidth was defined by the grid-search procedure on the training data. The kernlab R package (Karatzoglou et al., 2004) was used on the model construction.
Artificial neural network
A multilayer perceptron neural network, with a single hidden layer and two neurons, was used in this study to predict the total genetic values. For reducing computational costs, only the top 1% of the most relevant SNP ranked according to the importance scores in the RF algorithm were used as input variables in the ANN model. The 1% threshold was based on the study by Li et al. (2018), which reported that different subsets of the most relevant SNP identified by the RF method (varying from 500 to 50,000 or from 0.08% to 7.7%, considering as percentage values) maintained or improved prediction accuracy of total genetic values when compared with predictions using the whole high-density panel (651,253 SNP markers) in Brahman cattle.
The model can be described as
where is the marker genotype for the animal i and loci j, s is the number of neurons, is the weight for the kth neuron, is the bias for the kth neuron, is the weight of the jth input to the net and is a given activation function, with
The Gauss–Newton algorithm, implemented in the brnn R package (Pérez-Rodrigues and Gianola, 2013), was used to perform the weights updates. To allow better generalization for the ANN architecture, Bayesian regularization was used on the learning process. The objective function to be minimized is
where D denotes a given dataset used on the network, λ is the network weights, and M is a given network architecture, in terms of the number of neurons and activation functions. Bayesian regularization produces shrinkage of the parameters estimates in order to reduce its variance and, the objective function becomes:
in which) is the sum of squares of the network weights and α and β are positive regularization parameters.
Assessing prediction accuracy
In GBLUP and GBLUP-D models, the prediction accuracy of the additive (a), dominance (d), and total genetic effects (g) were measured as the Pearson correlation between predicted and true values (, , , respectively). For the GBLUP-D model, the prediction of the total genotypic effect was calculated by summing up and whereas in GBLUP, was equal to In the machine learning methods (ML), no additive or dominance structures were imposed in the genotype matrix, so that it is not possible to compute directly the predicted values for a, d, or g. However, to assess which effects (additive and/or dominance) the ML methods are capturing, the prediction accuracy was assessed by the correlation between the predicted responses and the true additive, dominance, and total genetic values (, , , respectively). Mean-squared error (MSE) of the total genetic values predictions was also computed to compare the prediction ability of methods.
Since the true additive and dominance effects are unknown in real populations, there were also evaluated in this study, the correlations between the observed simulated phenotypes and the predicted responses, i.e., the estimated genomic breeding values in the GBLUP, the estimated total genetic values in the GBLUP-D and the predicted outcome for RF, SVM, or ANN. All predictive ability metrics were averaged over the 10 replicates in each scenario.
Results and Discussion
Extent of linkage disequilibrium
Genomic prediction accuracy is highly dependent on the extent of linkage disequilibrium (LD) between markers and QTL on the population (Meuwissen et. al., 2001). The LD between marker and QTL (generally unobserved) can be viewed as the proportion of the variation caused by the alleles at a QTL, explained by the marker (Hayes, 2009). Thus, high LD between markers in a specific genomic region is expected to capture the effects of alleles for each QTL in that region (Meuwissen et. al., 2001; Goddard, 2009).
In the present study, the simulations were performed to mimic the extent of LD in real beef cattle populations. In general, the LD measured by the r2 statistic, considering adjacent markers, has been reported in the literature ranging from 0.17 to 0.31, for different breeds and panel densities (Lu et al., 2012, Espigolan et al., 2013; Pérez O’Brien et al., 2014; Fernandes Junior et al., 2016). In our study, the average r2 between adjacent markers across all replicates was equal to 0.24 ± 0.004, thus within the interval reported by those authors.
Genomic additive and dominance relationship matrices
Summary statistics of the off-diagonal elements of additive and dominance genomic relationship matrices (G and D, respectively) were computed for a specific replicate. The G matrix off-diagonal elements presented an average of −0.0038 ± 0.04, with values ranging from −0.1715 to 0.759, and, for the D matrix, these presented mean equal to 0.0000197 ± 0.0212 with values ranging from −0.136 to 0.307. The G and D matrices are covariance structures estimated from the SNP markers data, with the genomic relationship coefficient being interpreted as a conditional correlation between homologous alleles in different gametes. In this regard, the negative off-diagonal elements imply that some individuals present less molecular resemblance than the population average, provided that this population is in HWE (Toro et al., 2002; Powell et al. 2010). The standard deviation of the off-diagonal elements of G is approximate twice the observed for D, which is expected since the additive relationship matrix is supposed to be more informative than the dominance relationship matrix. Further, the means of the off-diagonal on both G and D matrices are very close to zero, indicative of HWE on the simulated population (Vitezica et al., 2013). When the population is not in HWE, and contrasts are not necessarily orthogonal, which violates the model assumptions and may cause bias on genetic parameter estimates (Bolormaa et al., 2015; Vitezica et al., 2013). However, a recent study proposes an approach to build nonadditive genomic relationship matrices on populations deviating from HWE (Vitezica et al., 2017).
Prediction accuracy
In the purely additive scenario with low heritability (h2 = 0.10), GBLUP presented slightly better prediction accuracy of breeding values than machine learning methods, with accuracies ranging from 0.359 to 0.376 depending on the model (Table 1). The ANN model provided higher accuracies in the scenarios with moderate heritability (h2 = 0.30), for which accuracy ranged from 0.563 to 0.635 (Table 1). Possibly, the ANN model has been benefited from the predictive variables selection performed a priori by the RF model, such a result needs further investigation. As expected, higher correlations between predicted and true breeding values across models were observed as the narrow-sense heritability increased (from 0.10 to 0.30), since the contribution of additive genetic effects to the phenotypic variation has increased.
Table 1.
Prediction accuracies1 and standard deviations obtained by GBLUP and different machine learning methods for simulated traits with different proportions of additive variance (h2) and dominance deviations variance (d2)
| Methods | ||||||
|---|---|---|---|---|---|---|
| Scenario | Effect2 | GBLUP | GBLUP-D | RF | SVM | ANN |
| h 2 = 0.10 d2 = 0.00 | Add | 0.376 (0.06) | 0.373 (0.06) | 0.359 (0.09) | 0.360 (0.06) | 0.357 (0.09) |
| Dom | — | — | — | — | — | |
| Gen | 0.376 (0.06) | 0.328 (0.06) | 0.359 (0.09) | 0.360 (0.06) | 0.357 (0.09) | |
| h 2 = 0.10 d2 = 0.05 | Add | 0.411 (0.08) | 0.413 (0.08) | 0.399 (0.09) | 0.377 (0.07) | 0.378 (0.09) |
| Dom | — | 0.180 (0.06) | 0.062 (0.04) | 0.004 (0.03) | 0.007 (0.03) | |
| Gen | 0.340 (0.08) | 0.345 (0.08) | 0.364 (0.09) | 0.313 (0.08) | 0.306 (0.09) | |
| h 2 = 0.10 d2 = 0.10 | Add | 0.393 (0.04) | 0.396 (0.04) | 0.419 (0.05) | 0.369 (0.03) | 0.383 (0.05) |
| Dom | — | 0.237 (0.09) | 0.134 (0.10) | 0.018 (0.03) | 0.001 (0.07) | |
| Gen | 0.261 (0.05) | 0.306 (0.05) | 0.395 (0.07) | 0.249 (0.05) | 0.273 (0.05) | |
| h 2 = 0.30 d2 = 0.00 | Add | 0.595 (0.03) | 0.592 (0.03) | 0.585 (0.05) | 0.579 (0.03) | 0.632 (0.04) |
| Dom | — | — | — | — | — | |
| Gen | 0.595 (0.03) | 0.579 (0.03) | 0.585 (0.05) | 0.579 (0.03) | 0.632 (0.04) | |
| h 2 = 0.30 d2 = 0.15 | Add | 0.575 (0.05) | 0.575 (0.05) | 0.589 (0.07) | 0.566 (0.04) | 0.619 (0.04) |
| Dom | — | 0.286 (0.07) | 0.163 (0.06) | 0.016 (0.05) | 0.041 (0.07) | |
| Gen | 0.460 (0.05) | 0.485 (0.05) | 0.575 (0.06) | 0.454 (0.04) | 0.527 (0.05) | |
| h 2 = 0.30 d2 = 0.30 | Add | 0.575 (0.06) | 0.582 (0.06) | 0.611 (0.04) | 0.563 (0.06) | 0.635 (0.04) |
| Dom | — | 0.350 (0.05) | 0.185 (0.06) | 0.021 (0.04) | 0.053 (0.06) | |
| Gen | 0.408 (0.05) | 0.488 (0.05) | 0.555 (0.04) | 0.406 (0.05) | 0.478 (0.04) |
1Prediction accuracies for the breeding values (a), dominance deviations (d), and total genetic effects (g) were assessed as the Pearson correlation between predicted and true effects (), , and , respectively) in GBLUP and GBLUP-D models and by the correlation between predicted responses and the true effects (a, d, or g) for machine learning methods. Prediction accuracies are presented as the average of 10 replicates.
2Add, additive effects; Dom, dominance effects; Gen, total genetic effects.
In the absence of dominance effects, the prediction accuracies of breeding values were similar for both GBLUP and GBLUP-D models. Consistent with Nishio and Satoh (2014), at increasing levels of dominance, using the GBLUP-D model led to only a small improvement in the prediction accuracy for the breeding values (Table 1). These results indicate that the expected response to genetic selection would be similar in both models, especially in the scenarios with moderate heritability in the narrow sense.
The differences between the GBLUP and GBLUP-D models in the purely additive scenarios were more pronounced for the total genetic values prediction, as the inclusion of dominance in the absence of such effect is a confounding factor. On the other hand, as the dominance variance increased, the opposite was observed and total genetic values predictions become more accurate when considering both additive and dominance effects (Table 1).
Regardless of the used method, there was an overall decrease in the accuracies of the total genetic values predictions when dominance effects were present. These results are an indication that dominance effects may not be effectively accounted for in the prediction models as the additive effects are (de Almeida Filho et al., 2016), suggesting that a considerably larger data set is required to accurately predict the dominance deviations in comparison to the additive effects.
The accuracies of dominance deviation predictions ranged from 0.180 to 0.350 in the GBLUP-D and from 0.060 to 0.185 in the RF models, throughout the different scenarios. For the ANN and SVM models, those accuracies were close to zero (Table 1). Similarly, Nishio and Satoh (2014) have reported accuracy values for dominance deviations between 0.148 and 0.348, using the GBLUP-D model in a simulated population with five chromosomes.
As highlighted before, it is known that the genomic selection accuracy depends directly on the magnitude of LD, as a consequence, the proportion of additive variance explained by an observed marker decreases linearly as the r2 between such marker and the causal variant decreases. For the dominance variance, such a relationship reduces by a factor of r4, which implicates that a much larger LD extent is necessary to detect dominance effects (Wei et al., 2014). In the present study, the average LD measured by r2 statistic reflects the pattern found in commercial beef cattle populations, undoubtedly, other aspects such as the trait architecture in terms of number and distribution of QTL effects, presence of high order nonlinear marker relationships (e.g., epistasis, genotype by environmental interactions, and imprinting) and number of animals in the training set are expected to impact on the observed accuracies as well (Goddard et al., 2011).
Among machine learning methods, only the RF algorithm was capable to capture implicitly (i.e., without partition of additive and dominance effects) the dominance signals (Table 1), probably due to the broader notion of interactions in the regressions trees, meaning that the RF ability to model different subgroups of interaction effects is based on how the partition of the data space is performed at each node of the regression trees (Chen and Ishwaran, 2012). In the SVM, the input information is transformed into some N × N symmetric and positive definite kernel matrix for which nonlinear relationships can be accommodated depending on the used kernel function. Thus, building dominance-specific kernels for models based on Reproducing Kernel Hilbert Spaces regressions (Long et al., 2011) could be an alternative for directly handling dominance effects in the SVM model. Conversely, in the ANN, the nonlinearity is covered by the activation function in the hidden neurons, here the used activation function was the hyperbolic tangent, well known for its great flexibility (Ehret et al., 2015). A straightforward way to model both additive and dominance effects in ANN models would be using directly the G and D matrices as input variables in the net architecture (Pérez-Rodrigues and Gianola, 2013). However, this approach would highly increase computational requirements (compared with models using only G), which could be unfeasible in practical applications. Further, there are no clear advantages of such ANN architecture (using G matrix) for genome-enabled prediction over a benchmark approach such as GBLUP (Howard et al., 2014; Ehret et al., 2015). It is worth mentioning that ANN and SVM are powerful methods for covering other nonadditive effects such as epistasis (Beam et al., 2014; Howard et al., 2014), not considered here.
The higher accuracies for dominance deviations predictions using the GBLUP-D model could be explained by the fact that this method handles such effect directly. In contrast, it can be noted that as the dominance variance increases, compared with the parametric models, the RF method predictions tended to present higher correlations with the breeding values and, notably, with the total genotypic values (Table 1). Note that the RF method does not provide interpretable inferences about the additive or dominance effects since it combines all sources of genetic effects (additive and dominance in the present study) in a unique overall prediction. Nonetheless, the RF predictions would be useful to identify most productive animals or with susceptibility to diseases by capturing additive and nonadditive signals.
Overall, MSEs for total genetic values predictions obtained with GBLUP-D were lower than those for GBLUP, especially when the dominance variance contribution was sizable (Figure 1). Although the RF model has presented the highest accuracies for the total genetic predictions, in some cases this method provided higher MSE values when compared with those observed in GBLUP-D, particularly in scenarios with moderate variance for the additive and/or dominance effects (Figure 1d–f). In turn, the ANN model presented the poorest predictive ability in the low heritability scenarios, with the MSE values approximately twice those obtained in the other methods (Figure 1a–c). Since the ANN model was built considering only the top 1% SNP ranked with the RF algorithm, one could attribute this result to the fact that, at low heritability levels, the power for detecting relevant regions deeply decreases (van den Berg et al., 2013). Moreover, it is known that ANN models are prone to over-fitting which affects its predictive ability and generalization capability (Lawrence et al., 1997). One must highlight that the ANN implementation in the present study is rather simplistic, possibly, applying deeper and more complex architectures (e.g., Multiple Hidden Layer and Convolutional Neural Networks) could improve the predictive ability for total genetic effects. Anyhow, although deep learning (DL) has gained much attention in quantitative genomics over recent years, hitherto, evidence of the better performance of DL for genomic prediction of complex traits over parametric models and other machine learning methods is controversial, and sometimes, discouraging (Bellot et al., 2018; Ma et al., 2018; Abdollahi‑Arpanah et al., 2020).
Figure 1.
MSEs (reported as the average of 10 replicates) and standard deviations for total genetic values predictions obtained with genomic best linear unbiased predictor, ignoring or including dominance effects (GBLUP and GBLUP-D, respectively) and different machine learning methods (RF, SVM, ANN) in simulated traits considering different levels of broad-sense heritability (H2): (a) H2 = 0.10 (h2 = 0.10 and d2 = 0.00); (b) H2 = 0.15 (h2 = 0.10 and d2 = 0.05); (c) H2 = 0.20 (h2 = 0.10 and d2 = 0.10); (d) H2 = 0.30 (h2 = 0.30 and d2 = 0.00); (e) H2 = 0.45 (h2 = 0.30 and d2 = 0.15); (f) H2 = 0.60 (h2 = 0.30 and d2 = 0.30).
Figure 2 shows the average accuracy for breeding values, dominance deviations, and total genotypic values obtained with the RF and GBLUP methods, considering the scenario with h2 = 0.30 and d2 = 0.15 and increasing the reference sample size. The gains in accuracy for breeding values prediction were equivalent in GBLUP and GBLUP-D models and higher than the observed for RF (Figure 2a). The accuracy of dominance effects improved from 0.181 to 0.418 for the GBLUP-D model when the number of animals in the reference population increased from 500 to 3,500 (Figure 2b). The increase in the number of animals is probably related to an increase in the number of animals with heterozygous genotypes at each locus, improving the prediction accuracy of dominance deviations. Conversely, in the RF method, there was observed only a little improvement in the correlation between predicted values and dominance deviations. Consequently, despite the superiority of the RF over parametric methods to predict total genetic values, the observed differences in the prediction accuracies between GBLUP-D and RF quickly decreased with the training population increasing. With a training set of 3,500 animals, GBLUP-D and RF methods presented similar prediction accuracies for the total genetic effects, both performing better than the GBLUP model (Figure 2c).
Figure 2.
Average accuracies and standard errors (computed with 10 replicates) for prediction of breeding values (a), dominance deviations (b) and total genetic effects (c), according to the number of animals in the training population and used method in a simulated trait with broad-sense heritability (H2) equal to 0.45 (h2 = 0.30 and d2=0.15). Methods assessed were the genomic best linear unbiased predictor, ignoring or including dominance effects (GBLUP and GBLUP-D, respectively) and RF.
In practice, the true additive genetic and dominance effects are unknown on real populations, so the validation of genomic prediction models including dominance effects has been based on the correlation between predicted total genetic effects and adjusted phenotypes (Su et al., 2012; Ertl et al., 2014; Aliloo et al., 2016). Increasing the dominance variance, the accuracy derived by the correlation between phenotypes and the estimated total genetic effects also increased for GBLUP-D and RF models, considering a reference population with 1,500 animals (Figure 3).
Figure 3.
Average phenotype prediction accuracy (standard errors) obtained with genomic best linear unbiased predictor, ignoring or including dominance effects (GBLUP and GBLUP-D, respectively) and different machine learning methods (RF, SVM, ANN) for simulated complex traits presenting low (h2 = 0.10; top) or moderate (h2 = 0.30; bottom) narrow-sense heritability values and different proportions of phenotypic variance attributed to dominance effects (d2).
In the low narrow-sense heritability scenarios, as the proportion of phenotypic variance attributed to dominance effects (d2) increased from 0 to 0.10, the prediction accuracies improved from 0.097 to 0.141 in GBLUP-D and from 0.105 to 0.145 in RF, respectively. Similarly, in the moderate narrow-sense heritability scenarios, as d2 increased from 0 to 0.30, the phenotype prediction accuracies improved from 0.322 to 0.375 in GBLUP-D and from 0.325 to 0.434 in RF, respectively (Figure 3). There was no evident improvement in the phenotype prediction using GBLUP-D when broad-sense heritability was equal to 0.45 (h2 = 0.30 and d2 = 0.15) compared with the results obtained with GBLUP. This result is probably because the residual noise masks the total genetic predictions when accuracy improvement of such an effect is not substantial. However, as depicted in Figure 2, the accuracy of the total genetic values prediction tends to improve by increasing the training sample size, thus, gains on the phenotype prediction accuracies are also expected.
The phenotype of an animal can be viewed as a combination of its total genetic merit and environmental deviations. Once the total genetic merit is a function of both additive (breeding values) and nonadditive (e.g., dominance and epistasis) genetic effects (Falconer and Mackay, 1996), the assessment of future performance based on the total genetic merit instead of the breeding values is expected to identify more accurately the most productive animals. Such a strategy can be used to support culling decisions and to improve the overall herd production. In dairy cattle, Aliloo et al. (2016) reported better predictions of phenotypes including dominance effects on the genomic analysis, albeit the observed differences were not significant, except for fat yield in the Holstein cows (P < 0.01).
Another practical use of dominance information in a breeding program would be to explore mating allocation for a specific combining ability of the parents to maximize the offspring’s productive performance. Previous studies have shown that an extra response is expected by the appropriate design of future mating pairs (Toro and Varona, 2010; Su et al., 2012). Mating strategies for obtaining economic advantages of dominance effects are possible even for closed populations (Toro, 1993, 1998). However, predicting an animal performance with the RF method requires the knowledge of its realized genotype, thus, exploring mate allocation techniques may not possible with such an approach.
In practice, there are some limitations on using models considering both additive and nonadditive genetic effects for genomic predictions. A common issue to be considered is the high computational cost, since accounting for every possible nonadditive effect rapidly increases the model parameterization. Although models based on genomic relationship matrices have relaxed those constraints, construction and inversion of such matrices are still challenging since both additive and nonadditive genomic relationship matrices are dense.
Machine learning methods offer a general framework to cope with nonlinear effects. In the present study, we provide insights into the behavior of ML methods when complex traits are affected by both additive and dominance genetic effects. In general, our results have pointed out the RF algorithm as an adequate approach to predict total genetic values and phenotypic performance for complex traits under the presence of dominance effects, without imposing any specific genetic structure on markers data. The RF method presented superior results in comparison to those obtained with the GBLUP approach and competitive with the model expanded to account directly for the dominance deviations (GBLUP-D).
Association mapping with RF algorithm
In the RF approach, VIMs can be used to identify relevant regions affecting the traits of interest. In regression problems, importance scores are generally based on the average percentage increase in MSE when generating a prediction of the OOB data randomly permuting the ith variable of interest, whereas all others remain unchanged (Breiman, 2001). Since the tree structures generated by the RF algorithm are informative to explore the different types of relationships between the explanatory variables, the importance scores for SNP can potentially reflect both additive and nonadditive contribution to the phenotype prediction (Yao et al., 2013). However, posterior analyses are necessary for assessing the nature of identified effects.
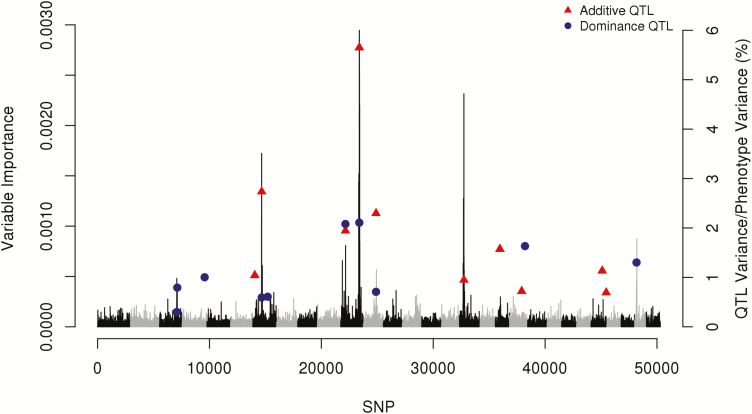
The RF model provided reasonable importance scores for the markers, generally with stronger peaks near to regions presenting the most relevant QTL effects (Figure 4). This is indicative that RF is a promising alternative tool for prescreening candidate genes, mainly with major effects. High importance scores were also assigned for regions showing a strong contribution to the dominance variance, although additive effects have contributed more effectively to the QTL detection (Figure 4). This is partially due to the genetic architecture of the trait considered, which presents higher variance for the additive than dominance deviations (h2 = 0.30 and d2 = 0.15), being more probable in real situations. Another reason is that dominance effects are more difficult to detect (Bolormaa et al., 2015).
Figure 4.
Manhattan plot for SNP importance scores (percent increase in mean squared error) in the RF analysis and, real positions and percentage of phenotypic variance related to QTL with the highest additive and/or dominance effects over 29 autosomes.
Our results are in agreement with those from Waldmann (2016), using a simulated dataset, this author reported that RF detected all non-additive effects (both dominance and epistasis effects), although they were not well separated from adjacent noise.
RF also has been successfully applied to the genome-wide association on real livestock data. Examining the structures of individual trees within the RF, Yao et al. (2013) identified SNPs potentially presenting additive and epistatic effects associated with residual feed intake in dairy cattle. In a Canchim beef cattle population, the RF approach identified rather plausible genomic regions associated with backfat thickness, providing an SNP subset explaining ~50% of the deregressed estimated breeding values variance (Mokry et al., 2013).
Conclusions
We have investigated the predictive ability of GBLUP and different machine learning methods in the presence of dominance effects. According to the found results, among machine learning methods, only the RF method was capable to cover implicitly dominance effects, i.e., without increasing the number of covariates in the model, providing higher accuracies for the total genetic values as the dominance ratio increases. Nevertheless, whether the interest is to infer directly on dominance effects, GBLUP-D could be a more suitable method.
Glossary
Abbreviations
- ANN
artificial neural network
- GBLUP
genomic best linear unbiased predictor
- GBLUP-D
GBLUP extended for dominance effects
- LD
linkage disequilibrium
- MSE
mean squared error
- OOB
out-of-bag
- QTL
quantitative trait loci
- RF
random forest
- RKHS
reproducing kernel Hilbert space
- SNP
single nucleotide polymorphisms
- SVM
support vector machine
- VIM
variable importance measure
Funding
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES (Finance code 001). The valuable suggestions of reviewers are gratefully acknowledged by the authors.
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Abdollahi‑Arpanahi R., Gianola D., and Peñagaricano F.. . 2020. Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet. Sel. Evol. 52:1–15. doi: 10.1186/s12711-020-00531-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliloo H., Pryce J. E., González-Recio O., Cocks B. G., and Hayes B. J.. . 2016. Accounting for dominance to improve genomic evaluations of dairy cows for fertility and milk production traits. Genet. Sel. Evol. 48:8. doi: 10.1186/s12711-016-0186-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Filho J. E., Guimarães J. F., E Silva F. F., de Resende M. D., Muñoz P., Kirst M., and Resende M. F. Jr. 2016. The contribution of dominance to phenotype prediction in a pine breeding and simulated population. Heredity (Edinb.). 117:33–41. doi: 10.1038/hdy.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam A. L., Motsinger-Reif A., and Doyle J.. . 2014. Bayesian neural networks for detecting epistasis in genetic association studies. BMC Bioinformatics 15:1–12. doi: 10.1186/s12859-014-0368-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot P., de Los Campos G., and Pérez-Enciso M.. . 2018. Can deep learning improve genomic prediction of complex human traits? Genetics. 210:809–819. doi: 10.1534/genetics.118.301298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg I., Fritz S., and Boichard D.. . 2013. QTL fine mapping with Bayes C(π): a simulation study. Genet. Sel. Evol. 45:19. doi: 10.1186/1297-9686-45-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolormaa S., Pryce J. E., Zhang Y., Reverter A., Barendse W., Hayes B. J., and Goddard M. E.. . 2015. Non-additive genetic variation in growth, carcass and fertility traits of beef cattle. Genet. Sel. Evol. 47:26. doi: 10.1186/s12711-015-0114-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. 2001 Random Forests. Machine Learning. 45:5–32. doi: 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Chen X., and Ishwaran H.. . 2012. Random forests for genomic data analysis. Genomics. 99:323–329. doi: 10.1016/j.ygeno.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret A., Hochstuhl D., Gianola D., and Thaller G.. . 2015. Application of neural networks with back-propagation to genome-enabled prediction of complex traits in Holstein-Friesian and German Fleckvieh cattle. Genet. Sel. Evol. 47:22. doi: 10.1186/s12711-015-0097-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl J., Legarra A., Vitezica Z. G., Varona L., Edel C., Emmerling R., and Götz K. U.. . 2014. Genomic analysis of dominance effects on milk production and conformation traits in Fleckvieh cattle. Genet. Sel. Evol. 46:40. doi: 10.1186/1297-9686-46-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espigolan R., Baldi F., Boligon A. A., Souza F. R., Gordo D. G., Tonussi R. L., Cardoso D. F., Oliveira H. N., Tonhati H., Sargolzaei M., . et al. 2013. Study of whole genome linkage disequilibrium in Nellore cattle. BMC Genomics. 14:305. doi: 10.1186/1471-2164-14-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., and Mackay T. F. C.. . 1996. Introduction to quantitative genetics. et al. Essex, UK: Longman. [Google Scholar]
- Fernandes Júnior G. A., Rosa G. J., Valente B. D., Carvalheiro R., Baldi F., Garcia D. A., Gordo D. G., Espigolan R., Takada L., Tonussi R. L., . et al. 2016. Genomic prediction of breeding values for carcass traits in Nellore cattle. Genet. Sel. Evol. 48:7. doi: 10.1186/s12711-016-0188-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst C., and Sölkner J.. . 1994. Additive and nonadditive genetic variances for milk yield, fertility, and lifetime performance traits of dairy cattle. J. Dairy Sci. 77:1114–1125. doi: 10.3168/jds.S0022-0302(94)77047-8 [DOI] [PubMed] [Google Scholar]
- Gallardo J. A., Lhorente J. P., and Neira R.. . 2010. The consequences of including non-additive effects on the genetic evaluation of harvest body weight in Coho salmon (Oncorhynchus kisutch). Genet. Sel. Evol. 42:19. doi: 10.1186/1297-9686-42-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafouri-Kesbi F., Rahimi-Mianji G., Honarvar M., and Nejati-Javaremi A.. . 2016. Predictive ability of random forests, boosting, support vector machines and genomic best linear unbiased prediction in different scenarios of genomic evaluation. Anim. Prod. Sci. 57:229–236. doi: 10.1071/AN15538 [DOI] [Google Scholar]
- Goddard M. 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica. 136:245–257. doi: 10.1007/s10709-008-9308-0 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., and Meuwissen T. H.. . 2011. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. 128:409–421. doi: 10.1111/j.1439-0388.2011.00964.x [DOI] [PubMed] [Google Scholar]
- González-Recio O., and Forni S.. . 2011. Genome-wide prediction of discrete traits using Bayesian regressions and machine learning. Genet. Sel. Evol. 43:1–12. doi: 10.1186/1297-9686-43-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Recio O., Rosa G. J. M., and Gianola D.. . 2014. Machine learning methods and predictive ability metrics for genome-wide prediction of complex traits. Livest. Sci. 116:217–231. doi: 10.1016/j.livsci.2014.05.036 [DOI] [Google Scholar]
- Hastie T. J., Tibshirani R., Friedman J.. . 2009. The elements of statistical learning. New York: Springer. doi: 10.1007/978-0-387-84858-7 [DOI] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., and Goddard M. E.. . 2009. Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92:433–443. doi: 10.3168/jds.2008-1646 [DOI] [PubMed] [Google Scholar]
- Hayes B. J., and Goddard M. E.. . 2001. The distribution of effects of genes affecting quantitative traits in livestock. Genet. Sel. Evol. 33(3): 209–229. doi: 10.1186/1297-9686-33-3-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., and Mäki-Tanila A.. . 2015. Expected influence of linkage disequilibrium on genetic variance caused by dominance and epistasis on quantitative traits. J. Anim. Breed. Genet. 132:176–186. doi: 10.1111/jbg.12140 [DOI] [PubMed] [Google Scholar]
- Howard R., Carriquiry A. L., and Beavis W. D.. . 2014. Parametric and nonparametric statistical methods for genomic selection of traits with additive and epistatic genetic architectures. G3 (Bethesda). 4:1027–1046. doi: 10.1534/g3.114.010298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatzoglou A., Smola A., Hornik K., and Zeileis A.. . 2004. kernlab – an S4 package for kernel methods. J. Stat. Softw. 11:1–20. doi: 10.18637/jss.v011.i09 [DOI] [Google Scholar]
- Lawrence S., Giles C. L., and Tsoi A. C.. . 1997. Lessons in neural network training: overfitting may be harder than expected. In Proceedings of the Fourteenth National Conference on Artificial Intelligence, AAAI-97 Menlo Park, California: AAAI Press; p. 540–545. [Google Scholar]
- Li Y., Raidan F. S. S., Li B., Vitezica Z. G., and Reverter A.. . 2018. Using Random Forests as a prescreening tool for genomic prediction: impact of subsets of SNPs on prediction accuracy of total genetic values. In: Proceedings of the World Congress on Genetics Applied to Livestock Production, Auckland, 2018 http://www.wcgalp.org/system/files/proceedings/2018/using-random-forests-prescreening-tool-genomic-prediction-impact-subsets-snps-prediction-accuracy.pdf – [accessed December 20, 2019]. [Google Scholar]
- Liaw A., and Wiener M.. . 2002. Classification and regression by randomForest. R News. 2:18–22. Available from https://www.r-project.org/doc/Rnews/Rnews_2002-3.pdf. [Google Scholar]
- Long N., Gianola D., Rosa G. J., and Weigel K. A.. . 2011. Marker-assisted prediction of non-additive genetic values. Genetica. 139:843–854. doi: 10.1007/s10709-011-9588-7 [DOI] [PubMed] [Google Scholar]
- Lu D., Sargolzaei M., Kelly M., Li C., Voort G. V., Wang Z., Plastow G., Moore S., and Miller S. P.. . 2012. Linkage disequilibrium in angus, charolais and crossed beef cattle. Front. Genet. 152(3):1–10. doi: 10.3389/fgene.2012.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Qiu Z., Song J., Li J., Cheng Q., Zhai J., and Ma C.. . 2018. A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta. 248:1307–1318. doi: 10.1007/s00425-018-2976-9 [DOI] [PubMed] [Google Scholar]
- Martini J. W., Gao N., Cardoso D. F., Wimmer V., Erbe M., Cantet R. J., and Simianer H.. . 2017. Genomic prediction with epistasis models: on the marker-coding-dependent performance of the extended GBLUP and properties of the categorical epistasis model (CE). BMC Bioinformatics. 18:3. doi: 10.1186/s12859-016-1439-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., and Goddard M. E.. . 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 157:1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., and Goddard M. E.. . 2013. Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 1:221–37. doi: 10.1146/annurev-animal-031412-103705 [DOI] [PubMed] [Google Scholar]
- Mokry F. B., Higa R. H., de Alvarenga Mudadu M., Oliveira de Lima A., Meirelles S. L., Barbosa da Silva M. V., Cardoso F. F., Morgado de Oliveira M., Urbinati I., Méo Niciura S. C., . et al. 2013. Genome-wide association study for backfat thickness in Canchim beef cattle using Random Forest approach. BMC Genet. 14:47. doi: 10.1186/1471-2156-14-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I., Gorjanc G., Curik I., Farkas J., Kiszlinger H., and Szendrő Z.. . 2013. The contribution of dominance and inbreeding depression in estimating variance components for litter size in Pannon White rabbits. J. Anim. Breed. Genet. 130:303–311. doi: 10.1111/jbg.12022 [DOI] [PubMed] [Google Scholar]
- Nishio M., and Satoh M.. . 2014. Including dominance effects in the genomic BLUP method for genomic evaluation. PLoS One. 9:e85792. doi: 10.1371/journal.pone.0085792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogutu J. O., Piepho H. P., and Schulz-Streeck T.. . 2011. A comparison of random forests, boosting and support vector machines for genomic selection. BMC Proc. 5 Suppl. 3:S11. doi: 10.1186/1753-6561-5-S3-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okut H., Wu X. L., Rosa G. J., Bauck S., Woodward B. W., Schnabel R. D., Taylor J. F., and Gianola D.. . 2013. Predicting expected progeny difference for marbling score in Angus cattle using artificial neural networks and Bayesian regression models. Genet. Sel. Evol. 45:34. doi: 10.1186/1297-9686-45-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez P., and de los Campos G.. . 2014. Genome-wide regression and prediction with the BGLR statistical package. Genetics. 198:483–95. doi: 10.1534/genetics.114.164442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez O’Brien A. M., Mészáros G., Utsunomiya Y. T., Sonstegard T. S., Garcia J. F., Tassell C. P. V., Carvalheiro R., Silva M. V. B., and Sölkner J.. . 2014. Linkage disequilibrium levels in Bos indicus and Bos taurus cattle using medium and high-density SNP chip data and different minor allele frequency distributions. Livest. Sci. 166:121–132. doi: 10.1016/j.livsci.2014.05.007 [DOI] [Google Scholar]
- Pérez-Rodriguez P., and Gianola D.. . 2013. Brnn: brnn (Bayesian regularization for feed-forward neural networks) http://CRAN.R-project.org/package=brnn – [accessed May 12, 2019].
- Powell J. E., Visscher P. M., and Goddard M. E.. . 2010. Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 11:800–805. doi: 10.1038/nrg2865 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Almeida F. A., Van Vleck L. D., Willham R. L., and Northcutt S. L.. . 1995. Estimation of non-additive genetic variances in three synthetic lines of beef cattle using an animal model. J. Anim. Sci. 73:1002–1011. doi: 10.2527/1995.7341002x [DOI] [PubMed] [Google Scholar]
- Sargolzaei M., and Schenkel F. S.. . 2009. QMSim: a large-scale genome simulator for livestock. Bioinformatics. 25:680–681. doi: 10.1093/bioinformatics/btp045 [DOI] [PubMed] [Google Scholar]
- Su G., Christensen O. F., Ostersen T., Henryon M., and Lund M. S.. . 2012. Estimating additive and non-additive genetic variances and predicting genetic merits using genome-wide dense single nucleotide polymorphism markers. PLoS One. 7(9):1–7. doi: 10.1371/journal.pone.0045293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro M. A. 1993. A new method aimed at using the dominance variance in closed breeding populations. Genet. Sel. Evol. 25:63–74. doi: 10.1186/1297-9686-25-1-63 [DOI] [Google Scholar]
- Toro M. A. 1998. Selection of grandparental combinations as a procedure designed to make use of dominance genetic effects. Genet. Sel. Evol. 30:339–349. doi: 10.1186/1297-9686-30-4-339 [DOI] [Google Scholar]
- Toro M., Barragán C., Óvilo C., Rodrigañez J., Rodriguez C., and Silió L.. . 2002. Estimation of coancestry in Iberian pigs using molecular markers. Conserv. Genet. 3:309–320. doi: 10.1023/A:1019921131171 [DOI] [Google Scholar]
- Toro M. A., and Varona L.. . 2010. A note on mate allocation for dominance handling in genomic selection. Genet. Sel. Evol. 42:33. doi: 10.1186/1297-9686-42-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Van Tassell C. P., Misztal I., and Varona L.. . 2000. Method R estimates of additive genetic, dominance genetic, and permanent environmental fraction of variance for yield and health traits of Holsteins. J. Dairy Sci. 83:1873–1877. doi: 10.3168/jds.S0022-0302(00)75059-4 [DOI] [PubMed] [Google Scholar]
- Vitezica Z. G., Legarra A., Toro M. A., and Varona L.. . 2017. Orthogonal estimates of variances for additive, dominance, and epistatic effects in populations. Genetics. 206:1297–1307. doi: 10.1534/genetics.116.199406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitezica Z. G., Varona L., and Legarra A.. . 2013. On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics. 195:1223–1230. doi: 10.1534/genetics.113.155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann P. 2016. Genome-wide prediction using Bayesian additive regression trees. Genet. Sel. Evol. 48:42. doi: 10.1186/s12711-016-0219-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W. H., Hemani G., and Haley C. S.. . 2014. Detecting epistasis in human complex traits. Nat. Rev. Genet. 15:722–733. doi: 10.1038/nrg3747 [DOI] [PubMed] [Google Scholar]
- Wittenburg D., Melzer N., and Reinsch N.. . 2011. Including non-additive genetic effects in Bayesian methods for the prediction of genetic values based on genome-wide markers. BMC Genet. 12:74. doi: 10.1186/1471-2156-12-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Spurlock D. M., Armentano L. E., Page C. D. Jr, VandeHaar M. J., Bickhart D. M., and Weigel K. A.. . 2013. Random Forests approach for identifying additive and epistatic single nucleotide polymorphisms associated with residual feed intake in dairy cattle. J. Dairy Sci. 96:6716–6729. doi: 10.3168/jds.2012-6237 [DOI] [PubMed] [Google Scholar]