Abstract
Base excision repair (BER) maintains genomic stability through the repair of DNA damage. Within BER, AP-endonuclease 1 (APE1) is a multifunctional enzyme that processes DNA intermediates through its backbone cleavage activity. To accomplish these repair activities, APE1 must recognize and accommodate several diverse DNA substrates. This is hypothesized to occur through a DNA sculpting mechanism where structural adjustments of the DNA substrate are imposed by the protein; however, how APE1 uniquely sculpts each substrate within a single rigid active site remains unclear. Here, we utilize structural and biochemical approaches to probe the DNA sculpting mechanism of APE1, specifically by characterizing a protein loop that intercalates the minor groove of the DNA (termed the intercalating loop). Pre-steady-state kinetics reveal a tyrosine residue within the intercalating loop (Y269) that is critical for AP-endonuclease activity. Using X-ray crystallography and molecular dynamics simulations, we determined the Y269 residue acts to anchor the intercalating loop on abasic DNA. Atomic force microscopy reveals the Y269 residue is required for proper DNA bending by APE1, providing evidence for the importance of this mechanism. We conclude that this previously unappreciated tyrosine residue is key to anchoring the intercalating loop and stabilizing the DNA in the APE1 active site.
INTRODUCTION
Base excision repair (BER) is the cell's primary defense against oxidative DNA damage. If left unrepaired, oxidative DNA lesions promote genomic instability, which can lead to a variety of consequences including cell death and human disease (1–4). Human apurinic/apyrimidinic (AP) endonuclease 1 (APE1) is an essential enzyme in the BER pathway. In addition to being embryonic lethal upon knockout (5), the biological significance of APE1 activity is demonstrated by enhanced sensitivity to DNA damaging agents upon expression of an inactive variant (6). APE1 is a multifunctional DNA nuclease having AP-endonuclease, 3′ phosphodiesterase, 3′ to 5′ exonuclease and even RNA cleavage activities (2,7–12). The APE1 AP-endonuclease activity functions to cleave at abasic (AP) sites during BER. Specifically, APE1 cleaves the phosphate backbone 5′ of an abasic site, generating 5′-phosphate and 3’-hydroxyl ends for further processing by DNA polymerase β. High-resolution crystal structures have been published detailing the APE1 AP-endonuclease reaction, revealing that an essential part of the APE1 cleavage mechanism is the rotation and flipping of the AP site out of the DNA duplex and into the active site (13,14). This rotation places the AP site in proper alignment with the catalytic residues for nucleophilic attack by an activated water molecule coordinated by a single metal ion. The term DNA sculpting is used to describe the bending and shaping of DNA substrates by repair enzymes, thus when a complex is formed, changes occur in the DNA structure as opposed to the protein active site (15,16). APE1 must sculpt the DNA into the rigid APE1 active site to facilitate catalysis and flipping of the AP site into the active site is one example of DNA sculpting (7,13–15,17).
The ability of APE1 to sculpt its DNA substrates is essential for obtaining the proper organization of the DNA in the active site to allow cleavage. The field has generally hypothesized that this is enabled by two loops that intercalate into the DNA double helix at the AP site and are thus poised to mediate DNA sculpting (Figure 1A). This is based largely on the structural location of M270 and R177 (13). The loop containing M270 intercalates the minor groove, while the loop containing R177 opposes M270 in the major groove (Figure 1B). This ‘double loop’ was hypothesized to cap the active site, stabilize the DNA conformation, and lock APE1 onto DNA for catalysis (13). Two tandem methionine residues (M270 and M271) have drawn interest, but when mutated to alanine, there was very little effect on APE1 activity (13). This previous work prompted many questions about the DNA sculpting mechanism of APE1 and the importance of the M270 intercalating loop (hereafter referred to as the intercalating loop, Figure 2A). To address this, we studied a previously unexplored residue in the DNA minor groove intercalating loop, Y269 (Figure 2B). Positioned directly next to M270, Y269 contacts the DNA backbone as well as D308, a residue responsible for coordinating the active site metal (Figure 2C). In this study, we mutated this tyrosine residue to an alanine as a means to understand its function in the active site and catalysis. Here, we report kinetic, binding, structural and single-molecule studies probing the APE1 Y269A variant and its effect on organization of the APE1 active site. This work demonstrates that this tyrosine is critical for APE1 DNA sculpting, thus providing molecular insight into how APE1 achieves substrate specificity and selection during BER.
Figure 1.

Key APE1 residues intercalate the DNA. (A) Overview of APE1:DNA complex with APE1 (green, cartoon representation) and the 21-mer DNA (gray, surface representation) highlighting the loops that intercalate the major and minor DNA grooves (shown in red). Dashed box indicates focused view shown in panel B. (B) A focused view of key APE1 residue side chains (green sticks) intercalating the DNA (transparent gray surface representation, excluding the THF, with white carbons in stick representation) (PDB ID: 5DFF).
Figure 2.

APE1 DNA intercalating loop. (A) Overview of high-resolution APE1:DNA complex with APE1 and 21-mer DNA showing the DNA bend (red dashed line) and DNA intercalating loop (black arrow). (B) A focused view of the APE1 AP-endonuclease product structure active site with intercalating loop (green, cartoon representation). Key protein side chains are indicated (green sticks). (C) Surface representation of APE1 (green) bound to DNA (gray) highlighting the position of the Y269 residue side chain (green sticks) (PDB ID: 5DFF).
MATERIALS AND METHODS
DNA sequences
To generate the 21-mer used for crystallization of the APE1 product complex, the following DNA sequences were used: nondamaged strand, 5′-GGA-TCC-GTC-GGG-CGC-ATC-AGC-3′; damaged strand, 5′-GCT-GAT-GCG-CXC-GAC-GGA-TCC-3′, where the underlined X represents abasic analog, tetrahydrofuran (THF). Substrate complex crystallization utilized a phosphorothioate-containing 21-mer (indicated by /) to prevent cleavage, as described previously (18), with the following sequences: nondamaged strand, 5′-GGA-TCC-GTC-GGA-CGC-ATC-AGC-3′; damaged strand, 5′-GCT-GAT-GCG-T/XC-GAC-GGA-TCC-3′. To generate the 30-mer used for kinetic and binding experiments, the following sequences were used: nondamaged strand, 5′-ATG-CGG-ATC-CGT-CGA-GCG-CAT-CAG-CGA-ACG-3′, damaged strand with a 5′ fluorescein label (indicated by asterisk), 5′-*CGT-TCG-CTG-ATG-CGC-XCG-ACG-GAT-CCG-CAT-3′. All sequences were purchased from IDT, resuspended, and their concentrations determined by absorbance at 260 nm. DNA substrates were prepared by annealing the correct pair of oligonucleotides (above). The annealing reactions were performed by incubating a solution of damaged and non-damaged complimentary oligonucleotides in 1:1 ratio using a PCR thermocycler set to heat to 95°C for 5 min and cool at 1°C min−1 to 4°C.
Protein expression and purification
Human APE1 Y269A was generated using full length or truncated (lacking N-terminal 42 amino acids) wild-type APE1 as a template by QuikChange II site-directed mutagenesis protocol and kit (Agilent). The Y269A mutation was confirmed via sequencing and then overexpressed in BL21-CodonPlus (DE3)-RP Escherichia coli and purified as previously described (17).
Crystallization and structure determination
For crystallization, annealed 21-mer abasic DNA was mixed with APE1 at a final concentration of 0.56 mM DNA and 10–12 mg/ml APE1, and subsequently incubated for 30 min. APE1 protein used for crystallography contains a truncation of the N-terminal 42 amino acids and the C138A mutation to aid in crystallization (19). APE1:DNA crystals were grown via sitting drop vapor diffusion using 2 μl protein/DNA mix combined with 2 μl reservoir solution (100 mM Bis Tris Propane, pH 6.5, 200 mM Sodium Fluoride, and 18–28% polyethylene glycol (PEG) 3350). Crystals were soaked in a cryosolution containing 20% ethylene glycol and 80% reservoir solution (with the addition of 50 mM MnCl2 for product complexes) before being flash frozen and subjected to X-ray diffraction. The APE1:DNA product structure was collected at 100 K on a Rigaku MicroMax-007 HF rotating anode diffractometer system at a wavelength of 1.54 Å. This system utilizes a Dectris Pilatus3R 200K-A detector and HKL3000R software was used for processing and scaling the data after collection (20). The APE1:DNA substrate structure was collected on the 4.2.2 synchrotron beamline at the Advanced Light Source of the Ernest Orlando Lawrence Berkeley National Laboratory, at a wavelength of 1.00 Å and then indexed and scaled using XDS (21,22). Initial models were determined using molecular replacement in PHENIX with a modified version of a previously determined APE1:DNA complex structure (Protein Data Bank accession number 5DFF) searching for 2 APE1:DNA complexes in the asymmetric unit. Refinement and model building were done with PHENIX and Coot, respectively, and figures were made using PyMol (Schrödinger LLC) (23,24).
Endonuclease activity assay
For kinetic characterization, pre-steady-state experiments were carried out using a rapid quench flow system (KinTek RQF-3). Reactions were initiated by rapidly mixing APE1 enzyme and annealed 30-mer DNA substrate solutions in reaction buffer (50 mM HEPES, pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.1 mg/ml bovine serum albumin (BSA)) at 37°C. DNA used for this assay contained a centrally located abasic site and 5′ fluorescein label (6-FAM) to monitor the conversion of substrate to product via fluorescence imaging. For multiple turnover experiments, final concentrations of 30 nM APE1 and 100 nM DNA were used. Single turnover conditions utilized final concentrations of 500 nM APE1 and 50 nM DNA. Reactions were subsequently quenched with 200 mM ethylenediaminetetraacetic acid (EDTA) at a range of time points from 0.002 to 1 s. Quenched reactions were mixed in 1:1 ratio with loading dye (100 mM EDTA, 80% deionized formamide, 0.25 mg/ml bromophenol blue and 0.25 mg/ml xylene cyanol), incubated at 95°C for 6 min and then separated on 15% denaturing polyacrylamide gel electrophoresis gels. Substrate and product bands were visualized using a GE Typhoon FLA 9500 imager in fluorescence mode and quantified using ImageJ software (25). Data were plotted and fit using Kaleidagraph (26). Multiple turnover kinetics generate biphasic time courses (Figure 3A) that give a rapid burst of product corresponding to the rate of cleavage (kobs), followed by a linear increase in the amount of product corresponding to the steady-state rate that describes the product release step (kss). To quantitatively determine these values, time courses were fit to equation: Product = A(1−e−kobst) + vsst, where A represents the amplitude of the rising exponential and kobs is the first order rate constant. The steady-state rate constant (kss) is the steady-state velocity (vss)/A, where A represents the fraction of actively bound enzyme. For single turnover experiments, only a burst phase is observed and, therefore, they are fit to equation: Product = A(1−e−kobst). Each time point in the curves represents an average of at least three independent experiments ± the error determined using standard deviation of the mean.
Figure 3.
Pre-steady-state kinetic and binding characterization of APE1 Y269A. (A) Multiple turnover and (B) single turnover kinetic time courses of product formation for the Y269A APE1 AP-endonuclease reaction with line of best fit (black line) and kinetic parameters. The time points are the mean and standard error of three independent experiments with error bars shown. Where error bars are not seen, they are present, but smaller than the data point. (C) Kinetic and binding parameters for wild-type and Y269A APE1, with fold change comparing the mutant and wild-type values. *represents previously published data (39)
Electrophoretic mobility shift assay (EMSA)
Samples were prepared to a volume of 10 μl by mixing 2 nM DNA (containing an abasic site analog, tetrahydrofuran, THF) with varying amount of full length wild-type or mutant APE1 (as indicated in figure legends) in a buffer containing 50 mM Tris (pH 8), 1 mM EDTA (pH 8), 0.2 mg/ml BSA, 1 mM DTT and 5% v/v sucrose for purposes of gel loading. Samples equilibrated for 30 min on ice before loading onto a 10% native 59:1 polyacrylamide gel. Native gels were run with electrophoresis buffer of 0.2× TBE and on ice for 1.5 h at a constant voltage of 120V and subsequently visualized using a GE Typhoon FLA 9500 imager in fluorescence mode. This allowed visualization of the DNA via the 5′ fluorescein label and the amount of DNA in each band to be quantified using ImageJ (25). Data were plotted and fit in Kaleidagraph using Equation (1):
 |
(1) |
where AT and BT represent the total concentration of APE1 and DNA, respectively, and AB is the concentration of APE1:DNA complex used to determine apparent affinity (KD,app). Each data point represents an average of at least three independent experiments ± the error determined using standard deviation of the mean.
Molecular dynamics simulations
Molecular dynamics (MD) simulations were performed for wild-type and the Y269A mutant in complex with DNA under explicit solvent (water) conditions. Model preparation and simulations were performed using the AMBER v16 suite of programs for biomolecular simulations (27). AMBER’s ff14SB (28) force-fields were used for the simulations. MD simulations were performed using NVIDIA graphical processing units (GPUs) and AMBER’s pmemd.cuda simulation engine using our lab protocols published previously (29,30).
A total of two separate simulations were performed (for wild-type APE1 and mutant Y269A) based on the X-ray crystal structures determined in this study. Hydrogen atoms were added by AMBER’s tleap program. After processing the coordinates of the protein and substrate, all systems were neutralized by addition of counter-ions and the resulting system was solvated in a rectangular box of SPC/E water, with a 10 Å minimum distance between the protein and the edge of the periodic box. The prepared systems were equilibrated using a protocol described previously (31). The equilibrated systems were then used to run 1.0 μs of production MD under constant energy conditions (NVE ensemble). The use of NVE ensemble is preferred as it offers better computational stability and performance (32). The production simulations were performed at a temperature of 300 K. As NVE ensemble was used for production runs, these values correspond to initial temperature at start of simulations. Temperature adjusting thermostat was not used in simulations; over the course of 1.0 μs simulations the temperature fluctuated around 300 K with RMS fluctuations between 2 and 4 K, which is typical for well equilibrated systems. A total of 1000 conformational snapshots (stored every 1000 ps) were collected for each system and were used for analysis.
RMSF10 calculations
Root mean square fluctuations (RMSF) were computed based on the conformational snapshots collected during the MD simulations. To identify global motions on slower time-scales from MD, for each of the three systems, the fluctuations associated with the first (slowest) 10 quasi-harmonic modes (RMSF10) were also computed and aggregated. It is well known that the slowest 10 modes contribute to the majority of the fluctuations in proteins (>80%) and the use of RMSF10, instead of all modes (RMSF), removes the faster stochastic motions of the protein, allowing focus on intrinsic dynamics of proteins (33). Both of these calculations were performed using AMBER’s ptraj analysis program. All trajectory conformations were first aligned to a common structure, to remove any translation and overall molecular rotation during the simulations.
Protein-substrate interactions
The energy for the APE1:DNA interactions (EApe1-DNA) were calculated as a sum of electrostatic (Eel) and van der Waals (Evdw) energy between atom pairs, based on an approach developed in previous publications (34,35). All APE1 and DNA atom pairs were included in the calculations and the resulting interaction energies were summed up per residue pair. The energies were calculated for 1000 snapshots, every 1000 ps, sampled during the full 1.0 μs simulation and were averaged over these 1000 snapshots.
Atomic force microscopy
The psTHF538 DNA substrate used for AFM, containing a single abasic site analog (THF) at position 375 (30% from end of the DNA) with a phosphorothioate linkage to residue 374 to prevent cleavage, was prepared as described previously (36,37). Briefly, a 37-mer oligonucleotide with the THF lesion modification was annealed/ligated into gapped pSCW01 plasmid. The 538-bp fragment was cut out of the plasmid by restriction digest and purified by gel extraction, followed by DNA clean up (Zymo Research) and phenol-chloroform extraction. Samples for AFM imaging were prepared as described previously (36). Purified full-length APE1 (WT or Y269A; 500 nM) was incubated with the 538-bp DNA substrate (100 nM) in binding buffer of 50 mM Hepes (pH 7.5), 150 mM NaCl, 10 mM MgCl2 and 1.4 mM DTT. Samples were incubated at room temperature for 25 min and then diluted by 1:25 in AFM deposition buffer of 25 mM Hepes (pH 7.5), 25 mM NaOAc and 10 mM Mg(OAc)2. Samples were then transferred to freshly cleaved mica, rinsed with filtered water, dried with nitrogen gas and imaged on a MultiModeV microscope (Bruker Corporation) in ScanAsyst PeakForce Tapping (Bruker) mode in air, at a scan rate of 0.977 Hz (38). Probes had a triangular tip with a nominal radius of 2 nm on a silicon nitride cantilever (SCANASYST-AIR, Bruker). Images were collected at a scan size of 1 μm × 1 μm and a resolution of 512 × 512 pixels. Data analysis, including binding position and bend angle measurement, was done using ImageJ and measurements were then binned and fit to a Gaussian distribution using Prism.
RESULTS
Kinetic and binding characterization of APE1 Y269A
To probe the role of the Y269 residue in the APE1 catalytic mechanism, we determined the activity of the APE1 Y269A mutant using pre-steady-state measurements of AP-site incision. In these kinetic experiments, the reactions contained an excess of substrate DNA to allow multiple turnovers of the cleavage reaction. As previously observed, the APE1 reaction resulted in a biphasic time course with an initial burst phase followed by a subsequent steady-state phase of product formation (Figure 3A) (14,17,39,40). For the Y269A mutant, the observed rate constant (kobs) of the burst phase is 14.9 ± 1.4 s−1. This represents a substantial decrease in AP-endonuclease activity compared to wild-type APE1, with at least a 13-fold decrease in the observed burst rate (Figure 3C) (39). This burst rate corresponds to the chemistry step, which for APE1 specifically defines the rate of phosphodiester backbone cleavage at the AP site. However, the Y269A mutant produced a steady-state rate similar to wild-type of 2.4 ± 0.2 s−1 (39). This indicates that while the Y269A mutant has a decreased rate of cleavage, the mutation does not affect the rate of enzyme turnover (i.e. product release). Because limiting the reaction to a single turnover can often provide a better estimation of kobs, which is the rate of cleavage, we also preformed experiments under single turnover conditions which contained an excess of APE1 enzyme (Figure 3B). Consistent with the multiple turnover data, the single turnover kobs for the Y269A mutant was determined to be 14.2 ± 1.4 s−1. Importantly, relative to wild-type APE1, the Y269A mutant has a reduced rate of cleavage by at least 60-fold (Figure 3C) (39).
One way that catalytic efficiency can be altered is through changes in binding affinity, therefore we further probed this mutant APE1 enzyme by determining its ability to bind the abasic DNA substrate. To this end, we utilized an electrophoretic mobility shift assay (EMSA) to identify complex formation between APE1 and abasic DNA, thus allowing for the determination of an apparent KD by varying the concentration of APE1 while maintaining a DNA concentration of 2 nM (41). For this assay, we utilized full-length wild-type or Y269A APE1 enzyme and 6-FAM labeled abasic DNA combined in the presence of EDTA to prevent catalysis. Our results indicate that both wild-type and Y269A APE1 exhibit a high affinity for the abasic substrate, with the Y269A mutant enzyme binding with less than a 2-fold weaker affinity compared to wild-type APE1 (Figure 3C and Supplementary Figure S1). While this 2-fold reduction is an approximation via EMSA analysis, these two affinity values are likely very similar. This suggests that the measured reduced catalytic activity cannot be solely accounted for by a simple reduction in DNA binding affinity.
Structural characterization of APE1 Y269A
To gain structural insight into the Y269A mutant protein, we collected X-ray crystallographic data of APE1 Y269A complexed with a 21-mer double-stranded DNA oligo containing a centrally located AP site analog (THF). We were able to obtain both product complex (bound to incised AP-DNA) and substrate complex (bound to intact AP-DNA) crystals that diffracted to a resolution of 2.20 and 2.77 Å, respectively (Table 1). The resulting structures each contained two APE1:DNA complexes in the asymmetric unit, and crystallized in the P212121 space group. To generate the product complex crystal, a substrate complex crystal was soaked in 50 mM MnCl2 prior to structure determination. Importantly, this approach relies on cleavage by APE1 to generate the product complex thus indicating that the substrate structure represents a catalytically competent complex.
Table 1.
Data collection and refinement statistics of APE1:DNA co-complexes
| Y269A product (AP-DNA) | Y269A substrate (AP-DNA) | |
|---|---|---|
| Data collection | ||
| Space group | P 21 21 21 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 45.38,150.23,156.45 | 45.66,148.32,153.24 |
| α, β, γ (°) | 90.00,90.00,90.00 | 90.00,90.00,90.00 |
| Resolution (Å) | 50.00–2.20 | 48.29–2.77 |
| R meas (%) | 0.073 (0.440) | 0.261 (2.670) |
| I/σI | 20.6 (2.3) | 7.4 (0.8) |
| cc1/2 | (0.792) | (0.395) |
| Completeness (%) | 99.5 (96.9) | 100.0 (100.0) |
| Redundancy | 5.9 (3.5) | 6.8 (6.8) |
| Refinement | ||
| Resolution (Å) | 43.58–2.20 | 48.29–2.77 |
| No. reflections | 91 210 | 27 446 |
| R work/ R free | 18.55/22.73 | 21.85/27.30 |
| No. atoms | ||
| Protein | 4379 | 4382 |
| DNA | 1669 | 1669 |
| Water | 363 | 39 |
| B-factors (Å2) | ||
| Protein | 28.73 | 46.81 |
| DNA | 62.14 | 75.36 |
| Water | 33.86 | 40.54 |
| R.m.s deviations | ||
| Bond length (Å) | 0.008 | 0.017 |
| Bond angles (°) | 1.018 | 1.432 |
| PDB ID | 6W4I | 6W4T |
Each structure was obtained from a single crystal. Values in parentheses are for highest-resolution shell.
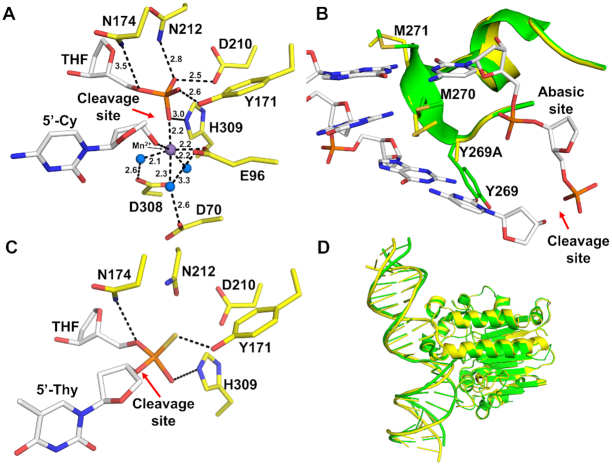
We obtained the APE1 Y269A product complex in the presence of MnCl2. This structure revealed an active site very similar to that of previously reported WT APE1 product structures (Figure 4A) (13,14,42). The abasic site is flipped out of the double helix and into the active site binding pocket where it is in position for cleavage of the phosphate backbone 5′ to the abasic site. A single Mn2+ is coordinated by E96 and three ordered water molecules, which additionally coordinate D70 and D308, as seen previously. The Mn2+ ion is also within hydrogen bonding distance of the 5′-phosphate and 3′-hydroxyl ends that are generated by the cleavage reaction. The 5′ phosphate is coordinated to the active site residues N212, D210, Y171, H309 and N174 (Figure 4A). Importantly, the intercalating loop containing the Y269A mutation, remains in the same intercalating position in the minor groove of DNA as observed for the wild-type enzyme (Figure 4B). The neighboring residue M270 is positioned into the minor groove opposite the intercalating R177 in the major groove.
Figure 4.
High-resolution structures of the Y269A APE1 mutant. (A) A focused view of the Y269A mutant product complex active site. The site of cleavage is indicated by the red arrow. The THF and 5’-Cy are shown in stick format (white carbons). Active site waters and Mn2+ are shown as blue and purple spheres, respectively. Key residues (yellow sticks) and distances (Å) are indicted. (B) Overlay of Y269A mutant (yellow) with previous structure for WT APE1 (green, PDB ID: 5DFF). DNA intercalating loops in their respective product complex structures are shown in cartoon representation. The DNA in white corresponds to wild-type APE1 (PDB ID: 5DFF). Key protein side chains are shown as sticks. (C) A focused view of the Y269A mutant substrate complex active site. Key protein side chains (yellow sticks) are indicted. The THF and 5’-Thy are shown in stick format (white carbons). (D) An overlay of the Y269A product complex (yellow) shown in cartoon representation with the previously determined WT APE1:DNA product complex (green, PDB ID: 5DFF) showing the variation in DNA bending.
To obtain substrate structures of APE1, active site mutations are often used to prevent catalysis (43). Here, in order to focus on the Y269A mutation and avoid making these additional mutations, a modified DNA oligo with a phosphorothioate backbone modification was used to prevent the cleavage reaction from occurring. Specifically, this modification places a sulfur substitution for a non-bridging oxygen 5′ to the abasic site which reduces the incision rate (14,18,44). The phosphorothioate substrate exists in two isomers, Sp and Rp. In the crystal structure, we observed both isomers in the active site with equal occupancy. The APE1 Y269A substrate structure revealed that the abasic site backbone oxygens are coordinated by N174, Y171 and H309 (Figure 4C). These contacts are seen in both substrate and product complexes. Residues N212 and D210 are in position to coordinate the nucleophilic water, which is not represented in the model due to resolution limitations (Figure 4C). Overall, both substrate and product Y269A structures are similar to wild-type APE1, although there is notable positional variation in the DNA including a slight change in bend angle (Figure 4D). This variation, high B-factors, as well as the difficulty in obtaining Y269A crystals, lead us to suspect that there are dynamics that are being overlooked by analyzing the crystal snapshot alone and we thus pursued computational MD simulations.
Molecular dynamics simulations
Microsecond (1 μs) time-scale MD simulations provided insights into the change in dynamics of the entire APE1 protein introduced by the single Y269A mutation. As depicted in Figure 5A, wild-type APE1 is mostly rigid with only slight dynamics. However, the introduction of the mutation Y269A significantly alters the dynamical behavior of APE1. Interestingly, wild-type and Y269A have very similar dynamics, except three regions (residues 175–200, 208–250 and 250–285) that show significant increase in dynamics. All three of these regions are loops involved in interacting with the DNA (Supplementary Figure S2). This includes the intercalating loop, as well as the R177 loop that opposes in the major groove. The increased dynamics in the region composed of residues 208–250 may be due to interactions with the DNA backbone. Overall, the MD simulations indicate that the Y269A mutation changes the dynamical behavior of APE1 including the intercalating loop (site of the Y269A mutation). This highlights the important role the intercalating loop, and the Y269 residue, play in stabilizing the APE1:DNA complex.
Figure 5.

Difference in microsecond time-scale dynamics for the wild-type and Y269A APE1. (A) The dynamics were characterized using MD simulations and aggregating top 10 quasi-harmonic modes (RMSF10). Quantitative comparison indicates that three regions show increase in dynamics (residues 175–200, 208–250 and 250–285) for the mutant Y269A. The areas of increased dynamics include R177 and Y269/Y269A (indicated by gray dashed lines). (B) Behavior of the intercalating loop was estimated based on the distance between M270 N (backbone nitrogen) and chain V base G10 atom N1 (WC face). A distance <11.0 Å is considered as ‘in’ helix conformation. For wild-type 930 out of 1000 (93.0%) conformations were below this cut-off, while for Y269A only 118 (11.8%) conformations met this criterion.
Computer simulations also reveal that the intercalating loop in the Y269A mutant quickly disengages from the duplex, while the wild-type appears to remain intrahelical (see Supplementary Movies S1 and S2). To quantify this movement, the fraction of conformations where the loop is intercalating into the helix was determined based on a distance cut-off. After structural considerations, a distance shorter than 11.0 Å between the backbone nitrogen of M270 and the Watson–Crick face (atom N1) of base G10 on chain V was considered as an ‘in’ helix conformation. As depicted in Figure 5B, for the wild-type, the intercalating loop remained intrahelical for most of the conformations during the simulation (93.0% conformations ‘in’ helix), while for the Y269A simulation the intercalating loop was outside the helix for the vast majority of conformations (11.8% ‘in’ helix). Note that in Figure 5B, the Y269A loop starts in an intrahelical position (as shown in crystal structure), but quickly disengaged from the helix.
Atomic force microscopy
X-ray crystallography structures of APE1 report the DNA bent at about 35° to be accommodated into the active site for catalysis (13,14). Although this is what is captured via crystallography, the biologically relevant and catalytically competent bend angle in solution remains unknown. To investigate this, we used atomic force microscopy (AFM) to obtain a single molecule view of APE1 interactions with abasic DNA (45). To this end, either wild-type or Y269A APE1 was incubated with a 538-bp DNA substrate containing a single abasic site analog (THF) positioned 30% from one end of the molecule. After imaging with PeakForce Tapping AFM, we first measured the position of each bound APE1 as a percentage of the total DNA contour length. Consistent with APE1 scanning ability, this analysis shows a broad distribution of APE1 binding position along the length of the abasic DNA substrate (Supplementary Figure S3). For both proteins, some binding to DNA ends and other non-specific sites was observed. However, the Gaussian distribution of internally bound wild-type and Y269A APE1 was centered at 30.1 and 32.3%, respectively (Supplementary Figure S3). In both cases, this closely corresponds to the location of the abasic site, suggesting that both wild-type and Y269A mutant APE1 have specificity for the abasic site lesion.
To gain further insight into the role of Y269 in DNA sculpting, we next used AFM to measure the DNA bend angles at sites of bound APE1, following a similar procedure to that used previously (46). As the MD simulations suggest, the interaction of APE1 with an abasic site causes a range of conformations with differing bend angles. Overall, there was a significant difference in the DNA bend angle distributions at sites of bound wild-type APE1 compared to Y269A APE1 (Figure 6A). With the Y269A mutant, there is an increase in the frequency of smaller bend angles (particularly 0–20°, Figure 6A red) and a decrease in sharper angles (80–120°, Figure 6A blue and purple). This suggests that without the Y269 residue anchoring the intercalating loop, APE1 is unable to bend the DNA as sharply causing a decrease in the observed catalysis (Figure 3C). DNA bending was observed at both specific (abasic) and non-specific sites. When wild-type APE1 was specifically bound to an abasic site, the DNA was bent 41.7 ± 30.8° (Figure 6B), while DNA with specifically bound Y269A APE1 was only bent 30.8 ± 26.1° (Figure 6C). In this case, the abasic site may facilitate some DNA bending even in the presence of the mutant. While the flexibility of the lesion may be contributing, the decrease in bend angle seen with the Y269A mutant is consistent with the general observation that the DNA is less bent without the Y269 anchor residue.
Figure 6.

Single molecule analysis of DNA bend angle at APE1 binding sites. (A) Analysis of DNA bend angle at all internal WT and Y269A binding sites (Chi-square test, P = 0.0018). (B) Histogram and Gaussian fitting of DNA bend angles at WT APE1 proteins specifically bound between 20 and 40% contour length (41.7 ± 30.8°, N = 47, red) and non-specifically bound (N = 50, white). (C) Histogram and Gaussian fitting of DNA bend angles at Y269A APE1 proteins specifically bound between 20 and 40% contour length (30.8 ± 26.1°, N = 45, red) and non-specifically bound (N = 43, white).
In addition to the observed reduction in bend angle at the abasic site, a similar trend was seen for non-specifically bound APE1. Interestingly, the DNA bend angle does not appear to be dependent on APE1 binding position for the wild-type protein, as a similar distribution of angles was observed for wild-type APE1 bound specifically at the abasic site and non-specifically along the rest of the DNA substrate (Figure 6B). The Y269A mutant, however, appears to almost completely lose its ability to bend the DNA at non-damaged sites (Figure 6C). Together, this AFM analysis highlights the importance of DNA sculpting at the abasic site in the APE1 mechanism and suggests that the Y269 residue anchors the intercalating loop to allow for proper orientation of the DNA in the APE1 active site.
DISCUSSION
This study utilized single turnover kinetics, crystallography, MD simulations and AFM to reveal that the aromatic residue Y269 in the DNA intercalating loop is essential for efficient DNA sculpting and subsequent catalysis. The APE1 DNA intercalating loop contains two tandem methionines (M270 and M271) that have previously drawn the attention of researchers in the field due to their proximity to the abasic site in APE1:AP–DNA complexes, as well as their intrahelical structural position (13,14,17,47) (Figure 1B). It was initially hypothesized that these methionine residues promote the flipping of the abasic site into the APE1 active site for catalysis. However, when this hypothesis was tested by mutagenesis, both mutants had similar steady-state catalytic rates as wild-type APE1 (13). This result suggests that the methionine residues are not playing a major role in DNA sculpting as previously hypothesized. Additionally, pre-steady-state kinetics of the APE1 exonuclease activity showed M270A had only a slight (<3-fold) effect on both the chemistry (kobs) and product release (kss) kinetic rates (17). This study included a comparison of the APE1 AP-endonuclease and exonuclease structures which highlighted the position of the DNA intercalating loop at the DNA bend site. In this position, the DNA intercalating loop can mediate the differential DNA sculpting required to accommodate both substrates. The data presented here suggests that the positioning and mechanistic function of the intercalating loop is conferred not through M270/M271, but through the Y269 residue. Despite interest in the intercalating loop, Y269 has largely gone uncharacterized due to its position adjacent to but not directly in the active site. We found that the single Y269A mutation decreased APE1 activity by >60-fold. In addition, this mutation results in increased loop dynamics and a loss of APE1-mediated DNA bending. Thus, we conclude that in the absence of this key tyrosine residue anchoring the intercalating loop in the active site, APE1 is less effective at stabilizing the DNA in the active site for catalysis.
BER involves the coordination of several enzymes that work together to protect the cell from unstable and cytotoxic repair intermediates. During BER, glycosylases are the first enzyme to identify and remove DNA base damage, generating the abasic substrate for APE1. One proposed model asserts that BER is coordinated around distortions in the DNA substrates (48). The idea that DNA intermediates are sequentially bent and passed by APE1 to polymerase β is supported by the fact that these enzymes bend the DNA to an increasingly sharper angle, as seen via crystallography (13,48,49). In other words, DNA bending by one enzyme may promote the binding of the next. Additionally, the concept of DNA intercalating loops involved in DNA sculpting has been reported in other BER enzymes. Like the AP-endonuclease activity of APE1, glycosylases require the DNA lesion to be flipped out of the helix and into the active site. Many DNA glycosylases use a ‘wedge residue’ on a DNA intercalating loop to scan for their target damaged bases (50,51). This wedge residue is required for DNA sculpting similar to the Y269 residue that was identified for APE1. APE1 also scans DNA for damage, displaying rapid rates of diffusion along DNA of ∼1 bp/2–5 μs (52,53). While the precise APE1 search mechanism remains unclear, the data presented here suggests that the Y269 residue could play a role in the DNA scanning of APE1 by allowing the intercalating loop to interrogate the flexibility of potential DNA substrates.
There are many types of nucleases in the cell, but DNA repair nucleases are unique because of their lack of sequence dependence. Unlike restriction nucleases, recognition must be accomplished using the DNA structure, not sequence specificity (54–57). These ‘structure-specific’ nucleases have been shown to sculpt the DNA to control their recognition and provide specificity to their damaged substrates (15). Similar to glycosylases, a theme among repair nucleases is the use of amino acid wedges to help sculpt the DNA substrate in the active site. These wedges are hydrophobic and often include an aromatic residue that stacks against the DNA, often against the strand complementary to the stand incision (15). Several nucleases have been reported to utilize these wedges such as UVDE (58), Vsr (59), Nfi (60,61), FEN1 (62), Exo1 (63) and Mre11 (64). For example, FEN1 uses a hydrophobic wedge to interrupt the linear structure of the DNA and force a 90° bend (15,62). In this study, we have identified a similar phenomenon in APE1, where it utilizes the intercalating loop with aromatic residue Y269 as an anchor for DNA sculpting. Importantly, sequence alignments have shown that this residue is conserved in Ape1-like proteins in different organisms (47,65). This highlights that the importance of the Y269 anchor residue is not limited to one enzyme, but is likely a mechanism present in many AP-endonucleases. Of note, the significance of this anchoring residue may be even more evident in the context of a complex cellular environment, including nucleosomes. To this point, APE1 activity is reduced in less-accessible regions of nucleosome substrates compared to accessible regions and naked DNA (66). Combined with our results, this supports a hypothesis in which APE1-mediated dynamic DNA bending is crucial for catalysis, particularly in the context of DNA wrapped around a nucleosome, as APE1 may be unable to appropriately bend the DNA into its active site for catalysis. Moreover, this also indicates that APE1 may use Y269, and the intercalating loop, as a mechanism of lesion recognition and/or substrate specificity.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 6W4I (Y269A product complex) and 6W4T (Y269A substrate complex).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jay Nix (Molecular Biology Consortium 4.2.2 beamline at Advanced Light Source) for aid in remote data collection and help with data analysis. This research used resources of the Advanced Light Source, which is a Department of Energy Office of Science user facility under Contract DE-AC02-05CH11231. We thank Dr Tyler Weaver for his assistance with the EMSA binding assay.
Contributor Information
Nicole M Hoitsma, Department of Biochemistry and Molecular Biology, University of Kansas Medical Center, Kansas City, KS 66160, USA.
Amy M Whitaker, Department of Biochemistry and Molecular Biology, University of Kansas Medical Center, Kansas City, KS 66160, USA.
Emily C Beckwitt, Program in Molecular Biophysics and Structural Biology, University of Pittsburgh, Pittsburgh, PA 15260, USA; UPMC Hillman Cancer Center, Pittsburgh, PA 15213, USA; Laboratory of DNA Replication, The Rockefeller University, New York, NY 10065, USA; Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10065, USA.
Sunbok Jang, UPMC Hillman Cancer Center, Pittsburgh, PA 15213, USA; Department of Pharmacology and Chemical Biology, University of Pittsburgh, Pittsburgh, PA 15261, USA.
Pratul K Agarwal, Department of Physiological Sciences and High-Performance Computing Center, Oklahoma State University, Stillwater, OK 74078, USA.
Bennett Van Houten, Program in Molecular Biophysics and Structural Biology, University of Pittsburgh, Pittsburgh, PA 15260, USA; UPMC Hillman Cancer Center, Pittsburgh, PA 15213, USA; Department of Pharmacology and Chemical Biology, University of Pittsburgh, Pittsburgh, PA 15261, USA.
Bret D Freudenthal, Department of Biochemistry and Molecular Biology, University of Kansas Medical Center, Kansas City, KS 66160, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01ES029203 to B.D.F., 5R01ES019566, 5R01ES028686 to B.V.H.]; National Institute of General Medical Sciences [GM105978 to P.K.A.]; Computing time provided by National Science Foundation funded XSEDE program [MCB180199, MCB190044]; American Heart Association [20PRE35210752 to N.M.H.]. Funding for open access charge: National Institutes of Health [R01ES029203 to B.D.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Whitaker A.M., Schaich M.A., Smith M.R., Flynn T.S., Freudenthal B.D.. Base excision repair of oxidative DNA damage: from mechanism to disease. Front. Biosci. (Landmark Ed.). 2017; 22:1493–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li M., Wilson D.M. III. Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal. 2014; 20:678–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schermerhorn K.M., Delaney S.. A chemical and kinetic perspective on base excision repair of DNA. Acc. Chem. Res. 2014; 47:1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caglayan M. Interplay between DNA polymerases and DNA ligases: influence on substrate channeling and the fidelity of DNA ligation. J. Mol. Biol. 2019; 431:2068–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xanthoudakis S., Smeyne R.J., Wallace J.D., Curran T.. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McNeill D.R., Lam W., DeWeese T.L., Cheng Y.C., Wilson D.M. III. Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol. Cancer Res. 2009; 7:897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whitaker A.M., Freudenthal B.D.. APE1: a skilled nucleic acid surgeon. DNA Repair (Amst.). 2018; 71:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tell G., Quadrifoglio F., Tiribelli C., Kelley M.R.. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid. Redox Signal. 2009; 11:601–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malfatti M.C., Balachander S., Antoniali G., Koh K.D., Saint-Pierre C., Gasparutto D., Chon H., Crouch R.J., Storici F., Tell G.. Abasic and oxidized ribonucleotides embedded in DNA are processed by human APE1 and not by RNase H2. Nucleic Acids Res. 2017; 45:11193–11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wong D., DeMott M.S., Demple B.. Modulation of the 3′→5′-exonuclease activity of human apurinic endonuclease (Ape1) by its 5′-incised Abasic DNA product. J. Biol. Chem. 2003; 278:36242–36249. [DOI] [PubMed] [Google Scholar]
- 11. Chen D.S., Herman T., Demple B.. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991; 19:5907–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dyrkheeva N.S., Lomzov A.A., Pyshnyi D.V., Khodyreva S.N., Lavrik O.I.. Efficiency of exonucleolytic action of apurinic/apyrimidinic endonuclease 1 towards matched and mismatched dNMP at the 3′ terminus of different oligomeric DNA structures correlates with thermal stability of DNA duplexes. Biochim. Biophys. Acta. 2006; 1764:699–706. [DOI] [PubMed] [Google Scholar]
- 13. Mol C.D., Izumi T., Mitra S., Tainer J.A.. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature. 2000; 403:451–456. [DOI] [PubMed] [Google Scholar]
- 14. Freudenthal B.D., Beard W.A., Cuneo M.J., Dyrkheeva N.S., Wilson S.H.. Capturing snapshots of APE1 processing DNA damage. Nat. Struct. Mol. Biol. 2015; 22:924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsutakawa S.E., Lafrance-Vanasse J., Tainer J.A.. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst.). 2014; 19:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dalhus B., Nilsen L., Korvald H., Huffman J., Forstrom R.J., McMurray C.T., Alseth I., Tainer J.A., Bjoras M.. Sculpting of DNA at abasic sites by DNA glycosylase homolog mag2. Structure. 2013; 21:154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whitaker A.M., Flynn T.S., Freudenthal B.D.. Molecular snapshots of APE1 proofreading mismatches and removing DNA damage. Nat. Commun. 2018; 9:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilson D.M. III, Takeshita M., Grollman A.P., Demple B.. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 1995; 270:16002–16007. [DOI] [PubMed] [Google Scholar]
- 19. He H., Chen Q., Georgiadis M.M.. High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I. Biochemistry. 2014; 53:6520–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 21. Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 25. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirsch P.D., Ekerdt J.G.. KaleidaGraph: graphing and data analysis. Version 3.5 for Windows Synergy Software. J. Am. Chem. Soc. 2000; 122:11755–11755. [Google Scholar]
- 27. Case D.A., Berryman J.T., Betz R.M., Cerutti D.S., Cheatham T.E. III, Darden T.A., Duke R.E., Giese T.J., Gohlke H., Götz A.W. et al.. AMBER 15. 2015; San Francisco: University of California. [Google Scholar]
- 28. Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C.. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory. Comput. 2015; 11:3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramanathan A., Agarwal P.K., Kurnikova M., Langmead C.J.. An online approach for mining collective behaviors from molecular dynamics simulations. J. Comput. Biol. 2010; 17:309–324. [DOI] [PubMed] [Google Scholar]
- 30. Ramanathan A., Savol A.J., Agarwal P.K., Chennubhotla C.S.. Event detection and sub-state discovery from biomolecular simulations using higher-order statistics: application to enzyme adenylate kinase. Proteins. 2012; 80:2536–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramanathan A., Agarwal P.K.. Evolutionarily conserved linkage between enzyme fold, flexibility, and catalysis. PLoS Biol. 2011; 9:e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beck D.A., Daggett V.. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004; 34:112–120. [DOI] [PubMed] [Google Scholar]
- 33. Ramanathan A., Agarwal P.K.. Computational identification of slow conformational fluctuations in proteins. J. Phys. Chem. B. 2009; 113:16669–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agarwal P.K. Cis/trans isomerization in HIV-1 capsid protein catalyzed by cyclophilin A: insights from computational and theoretical studies. Proteins. 2004; 56:449–463. [DOI] [PubMed] [Google Scholar]
- 35. Gagne D., Narayanan C., Nguyen-Thi N., Roux L.D., Bernard D.N., Brunzelle J.S., Couture J.F., Agarwal P.K., Doucet N.. Ligand binding enhances millisecond conformational exchange in Xylanase B2 from Streptomyces lividans. Biochemistry. 2016; 55:4184–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kong M., Beckwitt E.C., Springall L., Kad N.M., Van Houten B.. Single-molecule methods for nucleotide excision repair: building a system to watch repair in real time. Methods Enzymol. 2017; 592:213–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghodke H., Wang H., Hsieh C.L., Woldemeskel S., Watkins S.C., Rapic-Otrin V., Van Houten B.. Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E1862–E1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beckwitt E.C., Jang S., Carnaval Detweiler I., Kuper J., Sauer F., Simon N., Bretzler J., Watkins S.C., Carell T., Kisker C. et al.. Single molecule analysis reveals monomeric XPA bends DNA and undergoes episodic linear diffusion during damage search. Nat. Commun. 2020; 11:1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maher R.L., Bloom L.B.. Pre-steady-state kinetic characterization of the AP endonuclease activity of human AP endonuclease 1. J. Biol. Chem. 2007; 282:30577–30585. [DOI] [PubMed] [Google Scholar]
- 40. Schermerhorn K.M., Delaney S.. Transient-state kinetics of apurinic/apyrimidinic (AP) endonuclease 1 acting on an authentic AP site and commonly used substrate analogs: the effect of diverse metal ions and base mismatches. Biochemistry. 2013; 52:7669–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masuda Y., Bennett R.A., Demple B.. Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J. Biol. Chem. 1998; 273:30352–30359. [DOI] [PubMed] [Google Scholar]
- 42. Fairlamb M.S., Whitaker A.M., Freudenthal B.D.. Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5′ mismatches. Acta Crystallogr. D. Struct. Biol. 2018; 74:760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McNeill D.R., Wilson D.M. 3rd. A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol. Cancer Res. 2007; 5:61–70. [DOI] [PubMed] [Google Scholar]
- 44. Mundle S.T., Delaney J.C., Essigmann J.M., Strauss P.R.. Enzymatic mechanism of human apurinic/apyrimidinic endonuclease against a THF AP site model substrate. Biochemistry. 2009; 48:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beckwitt E.C., Kong M., Van Houten B.. Studying protein-DNA interactions using atomic force microscopy. Semin. Cell. Dev. Biol. 2018; 73:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kladova O.A., Bazlekowa-Karaban M., Baconnais S., Pietrement O., Ishchenko A.A., Matkarimov B.T., Iakovlev D.A., Vasenko A., Fedorova O.S., Le Cam E. et al.. The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation. DNA Repair (Amst.). 2018; 64:10–25. [DOI] [PubMed] [Google Scholar]
- 47. Redrejo-Rodriguez M., Vigouroux A., Mursalimov A., Grin I., Alili D., Koshenov Z., Akishev Z., Maksimenko A., Bissenbaev A.K., Matkarimov B.T. et al.. Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA. Biochimie. 2016; 128–129:20–33. [DOI] [PubMed] [Google Scholar]
- 48. Wilson S.H., Kunkel T.A.. Passing the baton in base excision repair. Nat. Struct. Biol. 2000; 7:176–178. [DOI] [PubMed] [Google Scholar]
- 49. Sawaya M.R., Prasad R., Wilson S.H., Kraut J., Pelletier H.. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997; 36:11205–11215. [DOI] [PubMed] [Google Scholar]
- 50. Nelson S.R., Dunn A.R., Kathe S.D., Warshaw D.M., Wallace S.S.. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2091–E2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dunn A.R., Kad N.M., Nelson S.R., Warshaw D.M., Wallace S.S.. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011; 39:7487–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu L., Kong M., Gassman N.R., Freudenthal B.D., Prasad R., Zhen S., Watkins S.C., Wilson S.H., Van Houten B.. PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1. Nucleic Acids Res. 2017; 45:12834–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jang S., Kumar N., Beckwitt E.C., Kong M., Fouquerel E., Rapic-Otrin V., Prasad R., Watkins S.C., Khuu C., Majumdar C. et al.. Damage sensor role of UV-DDB during base excision repair. Nat. Struct. Mol. Biol. 2019; 26:695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harrington J.J., Lieber M.R.. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994; 8:1344–1355. [DOI] [PubMed] [Google Scholar]
- 55. Harrington J.J., Lieber M.R.. DNA structural elements required for FEN-1 binding. J. Biol. Chem. 1995; 270:4503–4508. [DOI] [PubMed] [Google Scholar]
- 56. Huffman J.L., Sundheim O., Tainer J.A.. DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 2005; 577:55–76. [DOI] [PubMed] [Google Scholar]
- 57. Bennett R.J., Dunderdale H.J., West S.C.. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993; 74:1021–1031. [DOI] [PubMed] [Google Scholar]
- 58. Meulenbroek E.M., Peron Cane C., Jala I., Iwai S., Moolenaar G.F., Goosen N., Pannu N.S.. UV damage endonuclease employs a novel dual-dinucleotide flipping mechanism to recognize different DNA lesions. Nucleic Acids Res. 2013; 41:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsutakawa S.E., Jingami H., Morikawa K.. Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell. 1999; 99:615–623. [DOI] [PubMed] [Google Scholar]
- 60. Dalhus B., Arvai A.S., Rosnes I., Olsen O.E., Backe P.H., Alseth I., Gao H., Cao W., Tainer J.A., Bjoras M.. Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nat. Struct. Mol. Biol. 2009; 16:138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosnes I., Rowe A.D., Vik E.S., Forstrom R.J., Alseth I., Bjoras M., Dalhus B.. Structural basis of DNA loop recognition by endonuclease V. Structure. 2013; 21:257–265. [DOI] [PubMed] [Google Scholar]
- 62. Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G., Tomlinson C.G., Thompson P., Sarker A.H., Shen B. et al.. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011; 145:198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Orans J., McSweeney E.A., Iyer R.R., Hast M.A., Hellinga H.W., Modrich P., Beese L.S.. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011; 145:212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G. et al.. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008; 135:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hadi M.Z., Ginalski K., Nguyen L.H., Wilson D.M. 3rd. Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol. 2002; 316:853–866. [DOI] [PubMed] [Google Scholar]
- 66. Hinz J.M. Impact of abasic site orientation within nucleosomes on human APE1 endonuclease activity. Mutat. Res. 2014; 766–767:19–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 6W4I (Y269A product complex) and 6W4T (Y269A substrate complex).