Abstract
The regulation of gene expression by small RNAs in Escherichia coli depends on RNA binding proteins Hfq and ProQ, which bind mostly distinct RNA pools. To understand how ProQ discriminates between RNA substrates, we compared its binding to six different RNA molecules. Full-length ProQ bound all six RNAs similarly, while the isolated N-terminal FinO domain (NTD) of ProQ specifically recognized RNAs with Rho-independent terminators. Analysis of malM 3′-UTR mutants showed that tight RNA binding by the ProQ NTD required a terminator hairpin of at least 2 bp preceding an 3′ oligoU tail of at least four uridine residues. Substitution of an A-rich sequence on the 5′ side of the terminator to uridines strengthened the binding of several ProQ-specific RNAs to the Hfq protein, but not to the ProQ NTD. Substitution of the motif in the malM-3′ and cspE-3′ RNAs also conferred the ability to bind Hfq in E. coli cells, as measured using a three-hybrid assay. In summary, these data suggest that the ProQ NTD specifically recognizes 3′ intrinsic terminators of RNA substrates, and that the discrimination between RNA ligands by E. coli ProQ and Hfq depends both on positive determinants for binding to ProQ and negative determinants against binding to Hfq.
INTRODUCTION
RNA-binding proteins are important contributors to major life processes, including the regulation of gene expression by RNAs (1). In many bacteria, the prominent role played by trans-encoded base-pairing small RNAs (sRNAs) in regulating gene expression requires a matchmaker protein called Hfq, which assists sRNA in pairing to complementary sequences in target mRNAs and affects sRNA stability (2–6). Global identification of new RNA/protein interactions has been enabled by deep-sequencing approaches (7), such as Grad-seq, which uses glycerol gradients to identify novel RNA/protein complexes (8), CLIP-seq, which uses crosslinking to define protein binding sites in the transcriptome (9) and RIL-seq which identifies protein-dependent RNA–RNA interactions (10). Recent studies using these approaches showed that another protein, named ProQ, is a global RNA binding protein in Escherichia coli and Salmonella enterica (11–13), and is involved in sRNA interactions with other RNA molecules (12,14–15). The pool of RNA ligands of ProQ is mostly distinct from that of Hfq and contains more mRNAs than sRNAs (11,12).
While ProQ was originally discovered in a search for genes that affect proline transport (16), this protein contributes to several physiological processes in E. coli and S. enterica (17)—including DNA metabolism (13,14), bacterial virulence (15) and adaptation to osmotic stress (12) and resource limitation (18). ProQ is active as a monomer (19), and it belongs to the FinO-domain family with representatives in numerous γ-proteobacteria (17,20). Other members of this family include the F-like plasmid FinO protein (21,22), Legionella pneumophila RocC protein (23) and S. enterica pCol1B9 plasmid-encoded FopA protein (J. Vogel, personal communication), which each interact with few RNAs to control expression of specific genes, and a minimal ProQ protein (NMB1681), which, like E. coli ProQ, is a global RNA-binding protein in Neisseria meningitidis (24,25).
The E. coli ProQ protein consists of the N-terminal FinO domain (NTD) (19,26–27), for which this protein family is named (28), and the C-terminal Tudor domain (CTD), connected by an extended linker enriched for positively charged residues (19). The surface of the FinO domain of ProQ displays patches of positively and negatively charged residues, which could contribute to RNA binding via electrostatic interactions or hydrogen bonding (17). It was shown that the isolated FinO domain binds a double-stranded model RNA as well as the full-length E. coli ProQ protein does, suggesting that the NTD is the main site of RNA binding in ProQ (29). In agreement with that suggestion, mapping of the ProQ surfaces affected by the binding of two natural ProQ ligands, the 3′-UTR of cspE mRNA and the sRNA SraB, using hydrogen-deuterium exchange (HDX) experiments, showed the strongest protection by each RNA on partly overlapping surfaces of the FinO domain (19). In further support, a study using a bacterial three-hybrid assay showed that mutations in the FinO domain of ProQ were detrimental for the binding of the cspE 3′-UTR and SibB sRNA in E. coli cells, and that this domain alone was sufficient for the binding of each RNA (30). Additionally, mutations in the homologous domain of L. pneumophila RocC negatively affected the RocR sRNA lifetime in cells (23), and the homologous domain of the FinO protein crosslinked to bound RNA (31). While the FinO domain of ProQ appears to be the main site of RNA binding, reported data suggest that other regions of ProQ also form interactions with RNA molecules. The HDX study suggested that the sRNA SraB contacts the linker and CTD of E. coli ProQ more extensively than the cspE 3′-UTR (19). In agreement with this observation, the linker and the CTD regions of ProQ contributed to the binding of SibB sRNA, but not the cspE 3′-UTR, when measured by the three-hybrid assay (30).
The ProQ protein binds structured RNA motifs in its ligands in E. coli and S. enterica (11,12). This preference for double-stranded RNA was initially observed in studies of E. coli ProQ binding to RNAs derived from FinP RNA (29). RNA ligands of S. enterica ProQ identified by the Grad-seq method were also observed to be highly structured (13). Further analysis of ProQ binding sites in the transcriptomes of E. coli and S. enterica using CLIP-seq method showed that ProQ bound to GC-rich double-stranded elements, including Rho-independent terminators (11). Similarly, RIL-seq analysis of transcripts crosslinked to E. coli ProQ showed that interacting regions of RNAs contained GC-rich motifs followed by uridine tracts (12). Other FinO-domain proteins—minimal N. meningitidis ProQ, L. pneumophila RocC, and F-like plasmid FinO—also preferably bind secondary structure motifs, including transcription terminators (23,25,32).
In contrast to the behavior of ProQ, the Hfq protein binds single-stranded RNA sequences, which include uridine- or adenosine-rich motifs (33,34). Hfq often binds single-stranded A-rich sequences, including ARN repeats, in 5′-UTRs of mRNA molecules and in ribosomal RNAs (35–37). On the other hand, the binding of Hfq to sRNAs is dependent on the 3′ terminal oligoU tails (38), and either single-stranded U-rich sequences (39), or single-stranded A-rich sequences (40–42) upstream of the terminator hairpin. Hence, although Hfq binds at Rho-independent transcription terminators in some of its RNA ligands, Hfq binding to these regions involves single-stranded sequences.
ProQ and Hfq bind to different, partly overlapping, pools of RNA ligands in E. coli and S. enterica (11–13). It was observed that the group of highly structured ProQ-binding RNAs identified using Grad-seq was distinct from that of Hfq-binding RNAs (13). Further studies using CLIP-seq revealed that ProQ recognizes its RNA ligands using a structural code, which is different from Hfq's recognition of RNA sequence motifs (11). Additionally, the RIL-seq analysis of RNA-RNA interactions on ProQ and Hfq showed that while the RNA sets most enriched on each protein were generally unique, there was some overlap (12). The distinctive features of ProQ RNA ligands include structured GC-rich RNA motifs and single-stranded oligoU tails of intrinsic terminators that are shorter than those in Hfq-specific RNAs (11,12). As it has been proposed that some of ProQ functions center on RNA 3′ ends (11), it is likely that properties of 3′ RNA regions could be important for the discrimination between RNA ligands by ProQ and Hfq proteins.
To better understand how E. coli ProQ recognizes its RNA ligands, we compared the binding of six natural RNA ligands to the full-length ProQ and the isolated NTD domain using an electrophoretic mobility shift assay. The data showed that the N-terminal FinO domain is the part of ProQ which is responsible for the recognition of Rho-independent terminator structures in RNA ligands of ProQ. Studying mutations in the Rho-independent terminator of malM 3′-UTR showed that binding of the NTD required a double-stranded region of at least 2 bp in length, and a 3′ oligoU tail of four or more uridine residues. Finally, results obtained using in vitro and in vivo binding assays showed that an A-rich sequence located on the 5′ side of the terminator hairpin in several ProQ-specific RNAs serves as a negative determinant of their binding to the Hfq protein. Overall, these data suggest that the sequences and structures of ProQ-specific RNAs have been optimized to ensure correct recognition by ProQ, while at the same time preventing their binding by Hfq.
MATERIALS AND METHODS
Preparation of RNAs
The DNA templates for the in vitro transcription were obtained by Taq polymerase extension of chemically synthesized overlapping oligodeoxyribonucleotides (Sigma-Aldrich) (Supplementary Table S1). RNAs were transcribed with T7 RNA polymerase and purified using denaturing gel electrophoresis, as described (43,44). RNA molecules were 5′-32P labeled using T4 polynucleotide kinase (Thermo Scientific), which was followed by phenol-chloroform extraction, denaturing gel purification and ethanol precipitation.
Protein overexpression and purification
The sequences of E. coli ProQ protein or its N-terminal domain (NTD) were cloned into pET15b vector (Novagen) using BamHI restriction site (Supplementary Table S2). In the constructs the coding sequence of the protein was preceded by His6-tag and TEV protease recognition sequence (ENLYFQ↓S). Constructs were overexpressed in BL21 Δhfq E. coli strain (a kind gift of Prof. Agnieszka Szalewska-Pałasz, University of Gdańsk). After harvesting, the cells were suspended in buffer consisting of 50 mM Tris, pH 7.5, 500 mM NaCl and 10% glycerol (buffer A), and frozen at −80°C. Thawed cells were mixed with protease inhibitor cocktail (Roche) and lysed by sonication, followed by clarification of the lysates by centrifugation. After lysis, the freezing and thawing of the samples was avoided during all steps in the purification procedure. Lysates were loaded onto HisTrap crude column (GE Healthcare) on FPLC system (AKTA pure), and the proteins were eluted with linear imidazole gradient (25–600 mM) in buffer A. To remove nucleic acids, the samples were loaded onto HiTrap heparin column (GE Healthcare), and eluted with linear NaCl gradient (0.1–2 M). Subsequently, the His6-tag was removed by TEV-His6 protease used at 100:1 OD ratio of total protein to TEV, at 4°C, overnight. The completion of the TEV cleavage was confirmed by sodium dodecyl sulphate-polyacrylamide gelelectrophoresis, and the samples were loaded onto HisTrap column and eluted in buffer A containing 25 mM imidazole. The preparation was further purified using gel filtration on HiLoad 16/60 Superdex 200 size exclusion column (GE Healthcare) and eluted in storage buffer containing 50 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol and 1 mM ethylenediaminetetraacetic acid. The samples were stored at −80 °C as 10 and 20 μl aliquots, and used without refreezing. The molecular weights of the purified proteins were determined by MALDI-TOF as 25964.9 Da for ProQ, and 14673.5 Da for NTD, which agrees well with the calculated mass of 25979.5 Da for ProQ, and 14669.5 Da for the NTD, each with an additional N-terminal serine residue remaining from TEV cleavage site. The protein concentration was determined by measuring the absorption at 280 nm using extinction coefficient of 9650 M-1 cm-1.
The E. coli Hfq protein was purified as previously described (40).
ProQ-binding assay
Prior to use, RNAs were denatured at 90°C for 2 min followed by refolding on ice for 5 min. The concentration series of ProQ or NTD were made by 2-fold dilutions from the highest concentration. A total of 1 nM 32P-labeled RNA was incubated with indicated concentrations of ProQ in the binding buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM MgCl2) for 30 min at RT. Incubations were performed in low-protein binding microplates, additionally pre-treated with a solution containing 0.0025% bovine serum albumin (BSA). After incubation, the samples were loaded onto a native 6% polyacrylamide gel (19:1), and run in 0.5×TBE at 4°C. Gels were vacuum-dried, exposed to phosphor screens and quantified using a phosphorimager with MultiGauge software (Fuji FLA-5000). In the data fitting the GraphPad Prism software was used. The equilibrium dissociation constant (Kd) values were calculated by fitting data to the quadratic equation (45),
 |
where Y is a fraction bound, Y0 is a bottom plateau, and ΔY is the difference between top and bottom plateaus.  is concentration of 32P-labeled RNA, and
is concentration of 32P-labeled RNA, and 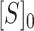 is the concentration of the ProQ protein or the NTD.
is the concentration of the ProQ protein or the NTD.
Hfq-binding assay
RNA binding to the E. coli Hfq protein was measured using double-filter retention assay, essentially as previously described (40,44). Prior to use 32P-RNA was denatured by heating at 90°C for 2 min followed by refolding on ice for 5 min. Then 20 μL of 200 pM 32P-RNA was mixed with 20 μL of the Hfq dilutions in binding buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM MgCl2) and incubated for 30 min at RT in low-protein binding microplates, additionally pre-treated with a solution containing 0.0025% BSA. Afterward, 35 μl aliquots were filtered and washed with 100 μl of 1× binding buffer. Membranes were dried and exposed to phosphorscreens, followed by quantification using a phosphorimager (Fujifilm FLA-5000) with ImageQuant software. The Kd values were determined by fitting data to the quadratic equation as described above for ProQ and the NTD.
Computational analysis of sequences of ProQ- and Hfq-specific RNAs
To analyze the nucleotide content of the sequence immediately 5′-adjacent to the terminator hairpin we used the databases of ProQ and Hfq binding sites in the transcriptomes of E. coli and S. enterica obtained using RIL-seq and CLIP-seq methods. The sequences of E. coli RNAs that bind to ProQ were obtained from published RIL-seq data (12) and CLIP-seq data (11). The sequences of E. coli sRNAs binding to Hfq were obtained from RIL-seq data (12). The sequences of S. enterica sRNAs binding to ProQ and those binding to Hfq were obtained from CLIP-seq data (9,11). For the analysis we selected the top 50 RNA ligands of each protein containing Rho-independent terminators (Supplementary Table S3). Specifically, from the database of individual RNAs from RIL-seq in LB (12), the top 50 RNA sequences annotated as mRNA 3′ UTRs or sRNAs were chosen for the analysis. From CLIP-seq data (9,11) 50 RNA sequences with the highest averaged read-counts were selected, in which the CLIP-seq peak covered a terminator loop or when it was located no further than 10 nt upstream from it. We note that, of the top 50 ProQ RNA ligands with intrinsic terminators, most were mRNA 3′ UTRs, while of the top 50 Hfq RNA ligands with terminators, most were sRNAs.
In these RNA ligands, a 10-nt sequence 5′-adjacent to the intrinsic terminator was analyzed. To define the sequences 5′-adjacent to the terminator hairpins the secondary structure of each RNA was analyzed using the RNAStructure software (46). The 3′ end of this sequence was defined as the nucleotide which was located opposite to the first uridine of the 3′ oligoU tail on the other side of the terminator hairpin. Afterward, the 10-nt sequences 5′-adjacent to the terminator hairpins were extracted and analyzed using WebLogo software (47) for the five datasets: RIL-seq data for ProQ and for Hfq in E. coli (12), CLIP-seq data for ProQ in E. coli (11) and CLIP-seq data for ProQ and for Hfq in S. enterica (9,11). Nucleotide frequency was calculated for each dataset using WebLogo software and compared with the whole-transcriptome nucleotide frequency in χ2 goodness-of-fit test. Subsequently, the statistical importance of nucleotide frequency at each position of the 10-nt sequence 5′-adjacent to the terminator hairpin in ProQ ligands was calculated in reference to each position of this sequence in Hfq ligands using the pLogo software (48). The pLogo analysis was performed for the E. coli ProQ dataset versus Hfq dataset obtained by RIL-seq (12) and for the S. enterica ProQ dataset versus Hfq dataset obtained by CLIP-seq (9,11).
Bacterial strains and plasmids used in the bacterial three hybrid assay
A complete list of plasmids, strains, and oligonucleotides used in this assay is provided in Supplementary Tables S4–7. NEB 5-alpha F’Iq cells (New England Biolabs) were used as the recipient strain for all plasmid constructions. Reporter strains KB483 (OL2–62-lacZ; Δhfq) and KB480 (OL2–62-lacZ; hfq+) were used as the reporter strains for bacterial three-hybrid (B3H) experiments (30) (Supplementary Table S4). Plasmids were constructed as specified in the Supplementary Tables S5 and 6. The E. coli (Ec) malM 3′-UTR (final 90 nts) was polymerase chain reaction (PCR) amplified using Ec genomic DNA and ligated into to pCDF-1XMS2hp (pCH1) between XmaI and HindIII to generate MS2hp-malM 3′-UTR (pKB1210). PCR mutagenesis to create site-directed mutants of malM 3′-UTR and cspE 3′-UTR was conducted with the Q5 site-directed mutagenesis kit (New England Biolabs) using end-to-end primers designed with NEBaseChanger. The full sequence of each hybrid RNA is provided in Supplementary Table S7.
Bacterial three-hybrid assay in E. coli
For B3H assays, reporter cells (KB480 or KB483) were freshly co-transformed with compatible pAC-, pBR- and pCDF-derived plasmids, as indicated. Transformed bacteria were inoculated into 1 ml LB broth supplemented with carbenicillin (100 μg/ml), chloramphenicol (25 μg/ml), tetracycline (10 μg/ml), spectinomycin (100 μg/ml) and 0.2% arabinose in a 2 ml 96-well deep well block (VWR), sealed with breathable film (VWR) and shaken at 900 rpm at 37°C. Overnight cultures were diluted 1:50 into 200 μl LB supplemented as above and cells were grown to mid-log (OD600 ≈ 0.6) in optically clear 200 μl flat bottom 96-well plates (Olympus) covered with plastic lids, as above. From here, 2 μl of cells were diluted 1:100 and plated on 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal)-indicator medium (containing arabinose and antibiotics as above along with 40 μg/ml X-Gal, 70 μM phenylethyl-β- D-thiogalactopyranoside (TPEG) and 1.5 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG)) and incubated overnight. Following overnight growth, plates were allowed to sit at 4°C for 1–2 days to allow color to develop and were photographed with a black-velvet background and oblique lighting. Brightness and levels were adjusted evenly across photographs. B3H interactions are interpreted as the β-gal activity in reporter cells containing all hybrid constructs (α-ProQ, λCI-MS2CP and MS2hp-bait hybrid RNA), relative to the highest activity from negative controls—cells containing plasmids where one of the hybrid constructs is replaced by an α empty, CI empty or MS2hp empty construct. In the context of qualitative plate-based assays, this is represented as the relative color of bacterial patches on X-gal containing media, where blue color represents higher β-gal activity, and therefore stronger RNA–protein interaction. Assays were conducted in duplicate on at least three separate days and a representative experiment is shown.
RESULTS
To study the recognition of RNA ligands by the E. coli ProQ protein, 6 functionally and structurally diverse RNA molecules were selected (Figure 1). In this set two were sRNAs expressed as independent transcripts—MicA and SibA, two were mRNA 3′-UTRs—malM-3′ and cspE-3′, and two were mRNA 5′-UTRs—lpp-5′ and hupA-5′. These RNAs have been identified in E. coli as ProQ ligands using CLIP-seq (11) and detected as single fragments or in chimeras on ProQ using RIL-seq (12). Among them, MicA is a trans-encoded base pairing sRNA, which is an important ligand of the Hfq protein (49), while SibA is a cis-encoded base pairing sRNA (50). malM-3′ is a processing product of 3′-UTR of the respective mRNA, and cspE-3′ is made as an independent transcription unit within 3′-UTR (12). The 5′-UTR of lpp mRNA was found in two of the most abundant chimeras identified on ProQ by RIL-seq (12). Additionally, the homologous region of the hupA 5′-UTR was identified in S. enterica as a target for regulation by the ProQ-dependent sRNA RaiZ (14). Hence, beyond their different sequences and structures, these RNAs also differ in their origins, making them representative of a diversity of ProQ RNA ligands.
Figure 1.
RNA molecules bound by ProQ protein in Escherichia coli, which were used in this study. The secondary structures of small RNAs (A) MicA (49), and (B) SibA (13) are presented according to the references. The secondary structures of (C) cspE-3′, (D) malM-3′, (E) lpp-5′ and (F) hupA-5′ were predicted using RNAstructure software (46). UAA stop codons and AUG start codons are shown in red and green, respectively. The lower case g denotes guanosine residue added on 5′ end of malM-3′ to enable T7 RNA polymerase transcription.
ProQ binds diverse RNA molecules with similar affinities
The binding affinity of the full-length E. coli ProQ protein to RNA molecules was determined using a gel shift assay (Figure 2). The electrophoretic mobility shift data showed that ProQ complexes with each of the RNAs were well resolved from unbound RNAs on the gel. In the range of ProQ concentrations used, five RNAs: MicA (72-nt), SibA (144-nt), cspE-3′ (52-nt), malM-3′ (91-nt) and lpp-5′ (86-nt), each formed one dominant complex with ProQ, while the 110-nt long hupA-5′ formed an additional higher order complex with ProQ (Figure 2A–F). The equilibrium binding constant (Kd) values ranged between 2.4 nM (for SibA) and 21 nM (for lpp-5′) (Figure 2G and Table 1). Aside from SibA, which bound ProQ more than 8-fold tighter than lpp-5′, the binding affinities of other RNAs with ProQ differed less than 3-fold among one another and were similar to the previously reported Kd of 31 nM for E. coli ProQ binding to a fragment of FinP RNA (29), and to low nanomolar Kd values estimated for SibA and RaiZ binding to S. enterica ProQ in a buffer with lower ionic strength (14). Overall, these results showed that the full-length E. coli ProQ binds these six natural RNA ligands with similar affinities.
Figure 2.
Equilibrium RNA binding to Escherichia coli ProQ protein and to the isolated FinO domain of ProQ (NTD). The binding of 32P-labeled RNAs MicA (A), SibA (B), cspE-3′ (C), malM-3′ (D), lpp-5′ (E) and hupA-5′ (F) to full-length ProQ protein and to the NTD was monitored using a gelshift assay. Free 32P-RNA is marked as R, RNA-ProQ complexes as RP, a higher order complex of hupA-5′ binding to ProQ as RP2 and RNA–NTD complexes as RN. Asterisk (*) indicates additional RP complexes that could not be quantified due to low signal-to-noise ratio. (G) The fitting of the ProQ binding data from A to F using the quadratic equation provided Kd values of 13 nM for MicA, 3.3 nM for SibA, 10 nM for cspE-3′, 14 nM for malM-3′, 26 nM for lpp-5′ and 10 nM for hupA-5′. Fractions bound in both complexes of hupA-5′ with ProQ were summed together for the hupA-5′ Kd calculations. (H) The fitting of the NTD binding data from A to F using the quadratic equation provided Kd values of 15 nM for MicA, 5 nM for SibA, 4.5 nM for cspE-3′ and 9.7 nM for malM-3′, while the affinities of lpp-5′ and hupA-5′ binding to the NTD were estimated as higher than 1 μM. The average equilibrium dissociation constant (Kd) values calculated from at least three independent experiments are shown in Table 1.
Table 1.
Equilibrium binding of different RNAs and their mutants with extended or mutated 3′-ends to the full-length ProQ and its N-terminal domain (NTD)
| K d [nM] | ||
|---|---|---|
| 32P-RNA | ProQ | NTD |
| MicA | 15 ± 5.1 | 21 ± 10 |
| SibA | 2.4 ± 0.85 | 4.7 ± 0.89 |
| cspE-3′ | 13 ± 4.5 | 4.5 ± 1.5a |
| malM-3′ | 12 ± 5.3a | 12 ± 4.8 |
| lpp-5′ | 21 ± 5.9a | >1000 |
| hupA-5′ | 9.3 ± 4.1b | >1000 |
| lpp-5′-mut | 3 ± 1.3 | 1.3 ± 0.83 |
| lpp-5′-term | 3.6 ± 1.3 | 5.1 ± 1 |
| lpp-5′-hp | 20 ± 2.8b | n. d. |
| hupA-5′-mut | 14 ± 8b | 16 ± 4.7 |
| hupA-5′-term | 10 ± 5 | 8.5 ± 0.96 |
| hupA-5′-hp | 18 ± 1.9b | n. d. |
| cspE-3′-ext+5 | 31 ± 14a | 17 ± 7.5 |
| cspE-3′-ext+22 | 17 ± 8.7 | n. d. |
| malM-3′-ext+5 | 33 ± 4.2 | n. d. |
| malM-3′-ext+17 | 8.4 ± 2.6b | n. d. |
The Kd values were obtained by fitting the data to the quadratic equation. The average Kd values with standard deviations were calculated from three to five independent experiments, except for malM-3′ binding to ProQ (nine replicates) and the NTD (eleven replicates), and cspE-3′ binding to ProQ (six replicates) and the NTD (seven replicates). All individual experiments of malM-3′ binding to the NTD are shown on Supplementary Figure S1.
aThe endpoint value of 100% was assumed in the data fitting.
bIn the Kd value calculations the fractions of both RNA–protein complexes were summed together.
n. d.: the binding was essentially undetectable up to 1 μM concentration of the NTD.
The isolated FinO domain of ProQ preferentially binds RNAs that contain Rho-independent terminators
Given that previous studies showed contributions of ProQ’s FinO domain to RNA binding (19,29–30), the binding of this isolated domain to the six RNAs was also examined (Figure 2, Table 1 and Supplementary Figure S1). The truncated protein construct (NTD) contained the N-terminal 130-aa of E. coli ProQ, which corresponds to the FinO domain, but was devoid of the linker and the C-terminal Tudor domain of ProQ (19,29). In the concentration range examined, all six RNAs formed single complexes with the NTD (Figure 2A–F). However, when the binding affinity of the isolated NTD to the six RNAs was compared, the data showed striking differences (Figure 2H and Table 1). The affinities of MicA, SibA and malM-3′ to the isolated NTD were similar to their affinities to the ProQ protein (Figure 2A, B, D and H; Table 1), and the affinity of cspE-3′ was 3-fold tighter than to the ProQ protein (Figure 2C and H; Table 1). In contrast, the binding of lpp-5′ and hupA-5′ did not achieve saturation up to 1 μM concentration of the NTD, and hence their affinities to the NTD were at least 50- and 100-fold, respectively, weaker than to the full-length ProQ (Figure 2E, F and H; Table 1). The fact that lpp-5′ and hupA-5′ mRNA fragments were the only RNAs from this set that did not possess Rho-independent transcription terminators (Figure 1) suggested that 3′-terminal hairpin structures could be required for tight RNA binding by the isolated FinO domain of ProQ.
Binding by the ProQ NTD can be restored by the addition of a Rho-independent terminator structure
To test if the presence of terminator-like motifs on the 3′ ends of lpp-5′ and hupA-5′ would improve their binding to ProQ’s FinO domain, several point mutations were introduced into their 3′ terminal sequences to create structures similar to Rho-independent terminators (lpp-5′-mut and hupA-5′-mut) (Figure 3A and B). Since both RNAs were predicted to have unstable secondary structures on the 3′ end, we designed a minimal set of mutations that would stabilize the double-stranded regions (mainly by changing G-U pairs into G-C pairs) and append short oligoU tails on the 3′ termini. The data showed that the presence of stable terminator-like structures in the lpp-5′-mut and hupA-5′-mut constructs strongly increased their affinity to the NTD. As a result, both mutants bound with similar affinities to the NTD and to ProQ (Figure 3C, Table 1 and Supplementary Figure S2). Overall, these data showed that adding a hairpin motif followed by a short single-stranded tail to the 3′ end of an RNA could confer on it the ability to be bound by the isolated FinO domain of ProQ.
Figure 3.
The Rho-independent terminators and similar structure motifs located on RNA 3′ ends are recognized by the FinO domain of ProQ. On top are presented the sequences of RNAs lpp-5′ mut (A), hupA-5′-mut (B), lpp-5′-term (D), hup-5′- term (E), lpp-5′-hp (G) and hupA-5′-hp (H) with point mutations and transplanted regions shown in red. The respective binding data with ProQ and the NTD are shown on graphs below each pair of mutants, in panels (C), (F) and (I), respectively. The fitting of the data using the quadratic equation provided Kd values of (C) 3.1 nM for lpp-5′-mut binding to ProQ and 7.6 nM to NTD, and 10 nM for hupA-5′-mut binding to ProQ and 18 nM to NTD; (F) 3 nM for lpp-5′-term binding to ProQ and 4.7 nM to NTD, and 15 nM for hupA-5′-term binding to ProQ and 8.2 nM to NTD; (I) 22 nM for lpp-5′-hp and 6.2 nM for hupA-5′-hp binding to ProQ. The binding of lpp-5′-hp and hupA-5′-hp to the NTD was essentially not detected up to 1 μM NTD concentration. Fractions bound in both complexes of hupA-5′-mut, lpp-5′-hp, and hupA-5′-hp with ProQ were summed together for the Kd calculations. Gels corresponding to the data in the plots are shown in Supplementary Figure S2. Average Kd values are shown in Table 1.
To test if a natural Rho-independent terminator could also restore the binding to the NTD, the intrinsic terminator structure from malM 3′-UTR was transplanted to the 3′ ends of lpp-5′ and hupA-5′ to create lpp-5′-term and hupA-5′-term constructs (Figure 3D and E). Binding data showed that lpp-5′-term and hupA-5′-term bound the NTD similarly to ProQ, as did lpp-5′-mut and hupA-5′-mut constructs (Figure 3C and F, Table 1 and Supplementary Figure S2). Additionally, 3′-terminal oligoU sequences were removed from the transplanted terminators to leave only double-stranded motifs on the 3′ ends, thus creating lpp-5′-hp and hupA-5′-hp constructs (Figure 3G and H). The binding of lpp-5′-hp and hupA-5′-hp to the NTD was essentially undetectable up to 1 μM concentration of the NTD (Figure 3I, Table 1 and Supplementary Figure S2), which suggests that the 3′ oligoU sequence contributes to the RNA binding of the NTD. In summary, these data showed that adding a natural intrinsic terminator hairpin with a 3′ terminal oligoU sequence to the 3′ ends of lpp-5′ and hupA-5′ RNAs enabled their binding by the ProQ’s FinO domain.
To test if the location of the Rho-independent terminator structures at the very 3′ ends is required for their recognition by the ProQ NTD, the sequences of cspE-3′ and malM-3′ RNAs, which bind tightly to the NTD (Table 1), were extended beyond their natural 3′ ends (Figure 4, Table 1 and Supplementary Figure S3). Each of these molecules was extended by either a shorter or longer length, resulting in four constructs named the cspE-3′-ext+5, cspE-3′-ext+22, malM-3′-ext+5 and malM-3′-ext+17 (Figure 4). All four constructs bound strongly to ProQ with <3-fold differences in binding affinities. Strikingly, however, these constructs differed markedly in binding to the isolated NTD (Figure 4). The cspE-3′-ext+5 construct, which was extended by 5 nt, bound the NTD only 3-fold weaker than cspE-3′ molecule. On the other hand, extension of malM-3′ by 5 nt showed a strong inhibitory effect, because the binding was not detected up to 1 μM concentration of the NTD. Similarly, the binding of both longer constructs, cspE-3′-ext+22 and malM-3′-ext+17, to the NTD was essentially undetectable up to 1 μM concentration of the NTD (Figure 4, Table 1 and Supplementary Figure S3). In summary, the shorter extension had a detrimental effect only on the NTD binding of malM-3′ RNA, while the longer extensions were detrimental for the binding of both RNAs. One possibility is that extended sequences could form new secondary structures, potentially restricting the flexibility of the 3′ oligoU tails or making them less available for binding. This suggests that it is not simply the presence of a stretch of uridines, and/or their single-stranded nature, but also their presence at the 3′ terminus of the RNA that is important for binding to the NTD.
Figure 4.
The extension of RNA sequence beyond the intrinsic terminator structure is detrimental for RNA binding by the FinO domain of ProQ. The sequences of cspE-3′ and malM-3′ were extended beyond the transcription terminators to obtain: (A) cspE-3′-ext+5 and cspE-3′-ext+22, and (B) malM-3′-ext+5 and malM-3′-ext+17 RNAs. The 3′ added sequences are shown in dark blue. The fitting of data using the quadratic equation provided Kd values of 13 nM for cspE-3′-ext+5 binding to ProQ and 24 nM for binding to NTD, 16 nM for cspE-3′-ext+22 binding to ProQ, 29 nM for malM-3′-ext+5 binding to ProQ and 10 nM for malM-3′-ext+17 binding to ProQ, while cspE-3′-ext+22, malM-3′-ext+5 and malM-3′-ext+17 binding to NTD was essentially undetectable up to 1 μM concentration of the protein. Fractions bound in both complexes of malM-3′-ext+17 with full-length ProQ were summed together for Kd calculations. The data in the plots for cspE-3′ and malM-3′ binding to ProQ or NTD are the same as in Figure 2G and H. Gels corresponding to the data in the plots are shown in Supplementary Figure S3. Average Kd values are shown in Table 1.
The double-stranded region in the lower part of the terminator hairpin is essential for tight RNA binding by the FinO domain
Several recent studies showed that ProQ binding sites in E. coli and S. enterica transcriptomes were enriched in double-stranded RNA structural elements (11–13). To better understand the contribution of the malM-3′ terminator hairpin to the FinO domain binding, three constructs with mutations that interrupt the continuous double-stranded region in the middle part of this hairpin were designed (Figure 5A). The results showed that a triple-adenosine bulge and double A-A and U-U mismatches, each inserted in the middle of the stem, did not have detrimental effects on the binding to the NTD (Figure 5B, Table 2 and Supplementary Figure S4). This could suggest that in each case the disturbed region of the helical structure was located outside of the region of the terminator stem necessary for the binding to the NTD.
Figure 5.
The lower part of the terminator hairpin and the 3′-terminal oligoU tail are involved in malM-3′ binding to the FinO domain. (A) The malM-3′ constructs with mutations introduced in the middle of the terminator stem. The nucleotides involved in the 4 bp at the bottom of the terminator stem are shown in light blue font, and the introduced mutations are shown in red font. (B) The fitting of the binding data in the plot using the quadratic equation provided Kd values of 8.9 nM for malM-3′-bulge, 8.2 nM for malM-3′-AA and 7.6 nM for malM-3′-UU. (C) The mutants with shorter terminator stems, which were constructed by gradual removal of the base-paired nucleotides from the top part of the stem. The nucleotides involved in the four base-pairs at the bottom of the terminator stem are shown in light blue font. (D) The fitting of the binding data in the plot using the quadratic equation provided Kd values of 9.1 nM for malM-3′-4bp, 10 nM for malM-3′-3bp and 7.2 nM for malM-3′-2bp, while the binding of malM-3′-1bp construct did not reach saturation up to 1μM concentration of the NTD. (E) The malM-3′ constructs with different lengths of 3′-terminal oligoU tails. Nucleotides added on the 3′ end of malM-3′-9U are marked with red font. (F) The fitting of data shown on the graph using the quadratic equation provided Kd values of 4.4 nM for malM-3′-9U, 7.1 nM for malM-3′-6U, 10 nM for malM-3′-5U and 25 nM for malM-3′-4U, while the binding of malM-3′-3U did not reach saturation up to 1 μM concentration of the NTD. The binding of malM-3′-2U and malM-3′-1U was essentially undetectable up to 1 μM concentration of the NTD. Data for wt malM-3′ are the same as in Figure 2H. Gels corresponding to the data in the plots are shown in Supplementary Figures S4 and 5. Average Kd values are shown in Tables 2 and 3.
Table 2.
Short double-stranded region of the terminator hairpin of malM-3′ RNA is sufficient for binding by the ProQ NTD
| K d [nM] | |
|---|---|
| 32P-RNA | NTD |
| malM-3′ | 12 ± 4.8a |
| malM-3′-bulge | 9.0 ± 2.5 |
| malM-3′-AA | 6.6 ± 1.4 |
| malM-3′-UU | 8.7 ± 2.1 |
| malM-3′-4bp | 8.8 ± 2.4 |
| malM-3′-3bp | 8.7 ± 2.4 |
| malM-3′-2bp | 6.4 ± 2 |
| malM-3′-1bp | >1000 |
The Kd values were obtained by fitting the data using the quadratic equation. The average Kd values with standard deviations were calculated from three or four independent experiments, except for malM-3′ (eleven replicates).
aData from Table 1.
To test how the length of continuous base-pairing in the terminator stem could affect the binding, additional constructs with shorter stems were designed (Figure 5C). These constructs were made in such a way that the base-paired nucleotides were gradually removed from the top part of the malM-3′ terminator stem, while the apical loop and base-paired nucleotides at the bottom of the stem remained. Binding data showed that those constructs, which were predicted to have 4, 3 or 2 bp of the stem remaining, bound the NTD with similar affinities as the wt malM-3′. However, the construct possessing only the nucleotides corresponding to the bottom base pair of the stem, did not bind to the NTD (Figure 5D, Table 2 and Supplementary Figure S4). As the nucleotide sequence corresponding to the stem of this construct would likely be unfolded, this is consistent with a stably base-paired structure being necessary for the NTD binding. In summary, the effects of deleting predicted base pairs and inserting disruptions into the double-stranded region of the RNA terminator hairpin together suggest that only two–three continuous base pairs preceding the U-stretch supports strong binding to the isolated NTD domain.
Shortening the 3′-terminal oligoU sequence to less than four uridine residues abolishes binding by the FinO domain of ProQ
It was recently observed that 3′ oligoU sequences were part of the sequence logo of ProQ binding sites detected by RIL-seq in E. coli (12), and that 3′ oligoU tails of RNA ligands of ProQ were shorter than those of RNA ligands of Hfq in S. enterica (11). To elucidate the role of the 3′ oligoU sequence in binding to the NTD of ProQ, versions of malM-3′ were designed with varying lengths of 3′ oligoU tails (Figure 5E). The natural terminator of malM-3′ contains seven 3′-terminal uridine residues (Figure 1). Extending the 3′-oligoU sequence of malM-3′ by two uridines in the malM-3′-9U construct had only a small effect on the binding affinity to the NTD (Figure 5F, Table 3 and Supplementary Figure S5). On the other hand, a series of constructs with truncations of the 3′ oligoU sequence resulted in a range of effects on the NTD binding. The constructs with five or six uridines remaining bound the ProQ NTD similarly to wt malM-3′. When one additional uridine residue was removed, the resulting malM-3′-4U construct bound the NTD ∼2-fold weaker than the wt malM-3′. Even more dramatic effects were observed when an additional uridine was removed: the binding of malM-3′-3U construct, which had only three uridine residues in the 3′ tail, did not reach saturation up to 1 μM concentration of the NTD, while the binding of the constructs with even shorter tails was essentially undetectable (Figure 5E-F, Table 3 and Supplementary Figure S5). Hence, at least four 3′-terminal uridine residues were necessary for the strong binding of malM-3′ to the isolated FinO domain of ProQ.
Table 3.
The length of single-stranded sequence following the Rho-independent terminators affects the binding of malM-3′ RNA to the N-terminal domain of ProQ
| K d [nM] | |
|---|---|
| 32P-RNA | NTD |
| malM-3′-9U | 5.1 ± 2.9 |
| malM-3′ (7U, wt) | 12 ± 4.8a |
| malM-3′-6U | 7.8 ± 1.2 |
| malM-3′-5U | 9.7 ± 1.5 |
| malM-3′-4U | 27 ± 11 |
| malM-3′-3U | >1000 |
| malM-3′-2U | n. d. |
| malM-3′-1U | n. d. |
The Kd values were obtained by fitting the data to the quadratic equation. The average Kd values with standard deviations were calculated from three to five independent experiments, except for malM-3′ (eleven replicates).
aData from Table 1.
n. d.: the binding was essentially undetectable up to 1 μM concentration of the NTD.
To corroborate the observation that the length of the 3′ oligoU sequence affects the RNA binding by the ProQ NTD, three constructs of cspE-3′ with truncated 3′ oligoU stretches were compared to the wt RNA (Supplementary Figure S6). The intrinsic terminator structure of cspE-3′ contains eight uridine residues in the 3′-oligoU sequence, with four of these residues predicted to be base-paired (Figure 1). When two 3′-terminal uridine residues were removed, the resulting construct cspE-3′-6U bound the NTD with the same affinity as the wt cspE-3′. However, when two more uridine residues were removed, the cspE-3′-4U construct, in which the remaining part of the 3′-tail was fully base-paired, bound the NTD about 10-fold weaker than the wt cspE-3′. Additionally, when the entire oligoU sequence was removed in the cspE-3′-0U construct, binding was not detectable (Supplementary Figure S6). These data support the conclusion that the minimum length of four uridine residues of the 3′-oligoU sequence is necessary for RNA binding by the NTD. The fact that the binding of cspE-3′-4U was weaker than the binding of malM-3′-4U could suggest that because the 3′-terminal uridines of cspE-3′-4U are base-paired with adenosine residues on the 5′ side of the terminator, they are less available for binding by the ProQ NTD.
RNAs bound by ProQ are enriched for A-rich sequences on the 5′ side of the intrinsic terminator
Binding of the ProQ NTD to intrinsic terminator structures (Figures 2–4) is reminiscent of the Hfq protein, which also binds the terminator hairpins with 3′ oligoU tails (39). This poses an interesting question of how these proteins discriminate between their RNA ligands, because the ProQ and Hfq-specific RNAs constitute mostly separate RNA pools in bacterial cells (11–13). One hint comes from the recent observation that RNA ligands of ProQ identified by CLIP-seq had significantly shorter single-stranded lengths of 3′ oligoU tails, which was caused by their partial involvement in base-pairing with nucleotide residues on the 5′ side of the hairpin stem (11). Another hint comes from the fact that an U-rich sequence 5′ adjacent to the Rho-independent terminator stem is a part of the Hfq-binding module in some of its RNA ligands, predominantly trans-encoded sRNAs (39). These observations led us to hypothesize that the sequences on the 5′ side of the terminator hairpin could contribute to discrimination between RNA molecules by the ProQ and Hfq proteins.
When we examined the sequences of ProQ-specific RNAs in previously reported datasets (11,12), we noticed that they contained an A-rich sequence on the 5′ side of the terminator (Figure 6, Supplementary Figure S7 and Table S3). We began by computing sequence logos for a set of the top 50 terminator-containing RNAs, either mRNA 3′ UTRs or sRNAs, which were most enriched in the RIL-seq data of either ProQ- or Hfq-binding RNAs in E. coli (12). We analyzed the nucleotide frequency in a 10-nt long sequence on the 5′ side of the terminator of these RNAs. In this region close to the terminator stem, adenosines were over-represented in the ProQ-specific RNAs (Figure 6A and Supplementary Table S3). This A-rich motif was also present in ProQ-specific RNAs identified in E. coli using CLIP-seq (Supplementary Figure S7A and Table S3) (11). In contrast, in Hfq-specific RNAs, uridines were most common in the corresponding sequence (Figure 6B and Supplementary Table S3). In further support, adenosines were significantly over-represented when ProQ-specific RNAs were directly compared to Hfq-specific RNAs (Figure 6C) (12,48). In the datasets obtained in S. enterica using CLIP-seq, adenosines were also over-represented in this region of ProQ-specific RNAs in comparison to Hfq-specific RNAs (Supplementary Figure S7B–D and Table S3) (11). In summary, the results of this analysis showed that an A-rich motif on the 5′ side of the terminator is a sequence feature present in ProQ-specific RNAs but not Hfq-specific RNAs.
Figure 6.

The sequence on the 5′ side of the intrinsic terminator hairpin in top RNA ligands of ProQ is enriched in adenosine residues. Nucleotide frequencies in the 10-nt long sequence 5′ adjacent to the terminator hairpin are shown for top 50 RNA ligands of ProQ and Hfq containing Rho-independent terminators, in the data obtained previously by RIL-seq in Escherichia coli (12). (A) Nucleotide frequencies obtained by WebLogo software for the top 50 E. coli ProQ ligand RNAs containing Rho-independent terminators in RIL-seq data, P < 2.2 × 10–16; (B) Nucleotide frequencies obtained by WebLogo for the top 50 E. coli Hfq ligand RNAs containing Rho-independent terminators in RIL-seq data, P = 6.5 × 10–12; (C) The statistically significant nucleotide frequencies at each nucleotide position for ProQ RNA ligands as compared to Hfq RNA ligands. Sequences were analyzed by pLogo software for top 50 E. coli ProQ RNA ligands as the foreground, and top 50 Hfq target RNAs as the background. Statistically significant P-values: A at position 7, P = 0.002; A at position 8, P = 1.08 × 10–5; A at position 9, P = 2.99 × 10–21; A at position 10, P = 9.78 × 10–9. The number of foreground sequences was marked on the figure as n(fg), and the number of background sequences as n(bg).
The A-rich motif, upstream of a stem-loop, disfavors Hfq binding
To determine the role of the A-rich motif in discriminating between ProQ and Hfq binding, this sequence was mutated to uridines in several RNAs and their binding to the ProQ NTD and to the Hfq protein were compared (Figures 7–9 and Supplementary Figures S8–10). For this analysis we selected such molecules that were at least 60-fold enriched in RIL-seq data for E. coli (12), that had well defined A-rich motifs, and for which the A-to-U substitutions were not predicted to have large-scale effects on secondary structure, as predicted by RNAStructure software. In addition to malM-3′ and cspE-3′, RNAs selected for the analysis were cspA-3′, hupB-3′, infA-3′, ihfA-3′ and gapA-3′ (Figure 7 and Supplementary Figure S9A-B). Because recent bacterial three-hybrid experiments used a longer sequence of cspE-3′ (30), we also analyzed a longer variant of this molecule, named cspE81-3′. We substituted a continuous stretch of nucleotides, which in two cases also included single guanosines predicted to base pair with uridines of the 3′ oligoU tail, for all of the RNAs except malM-3′ where the substitution was interrupted by one nucleotide (Figure 7).
Figure 7.
The mutations introduced into RNA molecules to test the role of A-rich motifs on the 5′ side of the terminator hairpins. The secondary structures of malM-3′, cspE-3′, cspE81-3′, cspA-3′, hupB-3′ and infA-3′ RNAs with mutations introduced into the A-rich regions marked with red font. The stop codons are marked in bold red font. The lower case g denotes guanosine residue added on 5′ end of malM-3′ and malM-3′-AtoU sequences to enable T7 RNA polymerase transcription.
Figure 8.
The binding of wt RNAs and their variants with mutations in the A-rich motifs to the FinO domain. The binding of 32P-labeled RNAs to the NTD was monitored using a gelshift assay. The fitting of the data using the quadratic equation provided Kd values of (A) 9.7 nM for malM-3′, 6.5 nM malM-3′-AtoU; (B) 4.5 nM for cspE-3′, 65 nM for cspE-3′-AtoU; (C) 12 nM for cspE81–3′, 13 nM for cspE81–3′-AtoU; (D) 32 nM for cspA-3′, 24 nM for cspA-3′-RtoU; (E) 19 nM for hupB-3′, 35 nM for hupB-3′-RtoU; (F) 27 nM for infA-3′, 47 nM for infA-3′-AtoU. Data for cspE-3′ and malM-3′ are the same as in Figure 2H. Gels corresponding to the data in the plots are shown in Supplementary Figure S8. The average equilibrium dissociation constant (Kd) values calculated from at least three independent experiments are shown in Table 4.
Figure 9.
The binding of wt RNAs and their variants with mutations in the A-rich motifs to the Escherichia coli Hfq protein. The binding of 32P-labeled RNAs to the Hfq protein was monitored using a double-filter retention assay. The fitting of the data using the quadratic equation provided Kd values of (A) 17 nM for malM-3′, 2.3 nM malM-3′-AtoU; (B) 4.7 nM for cspE-3′, 2.1 nM for cspE-3′-AtoU; (C) 40 nM for cspE81–3′, 2.8 nM for cspE81–3′-AtoU; (D) 51 nM for cspA-3′, 2.1 nM for cspA-3′-RtoU; (E) 22 nM for hupB-3′, 2 nM for hupB-3′-RtoU; (F) 31 nM for infA-3′, 1.7 nM for infA-3′-AtoU. Raw filter data are shown in Supplementary Figure S10. The average equilibrium dissociation constant (Kd) values calculated from at least three independent experiments are shown in Table 4.
The substitutions of the A-rich motif to uridines had either small effects or were detrimental for the binding of mutant RNAs to the ProQ NTD (Figure 8 and Table 4; Supplementary Figures S8 and 9). The mutations in malM-3′, cspA-3′, infA-3′, hupB-3′ and gapA-3′ had 2-fold or smaller effects on the binding affinity to the NTD. On the other hand, mutations in both variants of cspE-3′ and in ihfA-3′ had more than 10-fold detrimental effects on binding to the NTD (Table 4 and Supplementary Figure S9).
Table 4.
Equilibrium binding of wt RNAs and their variants with mutations in the A-rich motifs to the ProQ NTD and the Hfq protein
| K d [nM] | ||
|---|---|---|
| 32P-RNA | NTDa | Hfqb |
| malM-3′ | 12 ± 4.8c | 22 ± 7 |
| malM-3′-AtoU | 5.0 ± 1.8 | 2.2 ± 0.8 |
| cspE-3′ | 4.5 ± 1.5c | 5.9 ± 2.5 |
| cspE-3′-AtoU | 100 ± 51 | 1.7 ± 0.88 |
| cspE81-3′ | 7.6 ± 4.5 | 47 ± 11 |
| cspE81-3′-AtoU | 17 ± 4.6 | 2.6 ± 1.2 |
| cspA-3′ | 33 ± 10d | 49 ± 15d |
| cspA-3′-RtoU | 42 ± 18d | 1.3 ± 0.66 |
| hupB-3′ | 18 ± 1.7 | 30 ± 16d |
| hupB-3′-RtoU | 29 ± 7.2 | 1.9 ± 0.75 |
| infA-3′ | 19 ± 10 | 28 ± 12d |
| infA-3′-AtoU | 32 ± 13 | 1.2 ± 0.55 |
The Kd values were obtained by fitting the data to the quadratic equation. The average Kd values with standard deviations were calculated from three to five independent experiments, except for malM-3′ binding to the NTD (eleven replicates) and Hfq (six replicates) and cspE-3′ binding to the NTD (seven replicates) and Hfq (seven replicates).
aThe RNA binding to the NTD was measured using gelshift assay
bThe RNA binding to the Hfq protein was measured using double-filter retention assay
cData from Table 1.
dThe endpoint value of 100% was assumed in the data fitting.
In contrast to the neutral-to-negative effects on ProQ binding, substitutions of the A-rich motif to uridines strongly increased the affinities of these RNAs to the Hfq protein (Figure 9, Table 4, and Supplementary Figures S9-10). The binding of E. coli Hfq protein to the studied RNAs was measured using a double-filter retention assay, which was previously used to compare the binding of different sRNAs to the Hfq protein (40,44). The advantage of this assay is that the separation of the RNA–Hfq complex from free RNAs occurs rapidly, and it allows parallel comparison of up to eight different RNAs (44). For all of the mutants, except cspE-3′-AtoU, the binding to Hfq was at least 10-fold tighter than to their wt counterparts. The strongest effects were observed for cspA-3′-RtoU, ihfA-3′-AtoU and gapA-3′-AtoU, and smaller effects were seen for malM-3′-AtoU, hupB-3′-RtoU and infA-3′-AtoU (Figure 9, Table 4 and Supplementary Figure S9).
While the binding of the cspE-3′-AtoU mutant to Hfq was not affected by the substitutions, the longer construct cspE81-3′-AtoU bound Hfq 18-fold tighter than its wt version (Figure 9, Table 4, and Supplementary Figure S10). The wt cspE-3′ molecule has a long single-stranded sequence in its 5′ region, while the corresponding sequence is involved in secondary structure in the cspE81-3′ molecule (Figure 7B and C). Because this sequence could serve as an additional Hfq binding site in wt cspE-3′, it is likely that the effect of the A-to-U substitutions in cspE-3′ is masked by the Hfq binding to this additional sequence. Overall, these data showed that the substitution of adenosines to uridines on the 5′ side of the terminator hairpin had different effects on Hfq and ProQ NTD binding—increasing the affinity of the Hfq protein to these RNAs while at the same time not altering or decreasing the affinity of the ProQ NTD.
In vivo RNA-binding determinants for ProQ and Hfq
The above results indicate that, in a purified in vitro system, adenosines immediately upstream of intrinsic terminators serve as negative determinants for Hfq binding (Figures 7 and 9; Table 4). In contrast, these adenosines appear to be neutral or detrimental factors for the ProQ NTD binding (Figure 8). An important question is whether these adenosines play a similar role in controlling Hfq- and ProQ-interactions in vivo. To investigate this, we utilized a bacterial three-hybrid (B3H) system for RNA–protein interactions that has been demonstrated to detect both Hfq- and ProQ–RNA interactions in vivo (30,51). In this assay, expression of three hybrid components make reporter-gene (lacZ) transcription from a test promoter dependent on a productive interaction between an RNA ‘bait’ and a protein ‘prey’ (Figure 10A). Specifically, a ‘Bait’ RNA is expressed as an MS2hp-hybrid RNA, and is tethered to DNA upstream of the transcription start site by interaction between a CI-MS2CP ‘Adapter’ protein and the λ operator OL2 sequence. Prey protein is expressed as a fusion with the N-terminal domain of the alpha subunit (α) of RNA polymerase (RNAP).
Figure 10.
In vivo analysis of ProQ- and Hfq-binding determinants and cellular competition. (A) Design of a bacterial three-hybrid assay (B3H) system to detect interaction between a protein (P) of interest (here, ProQ or Hfq) and an RNA (here cspE 3′-UTR or malM 3′-UTR). Interaction between the protein moiety and RNA moiety fused, respectively, to the NTD of the alpha subunit of RNAP (α) and to one copy of the MS2 RNA hairpin (MS2hp) activates transcription from test promoter, which directs transcription of a lacZ reporter gene. Compatible plasmids direct the synthesis of the α-fusion protein (under the control of an IPTG-inducible promoter), the CI-MS2CP adapter protein (under the control of a constitutive promoter; pCW17) and the hybrid RNA (under the control of an arabinose-inducible promoter). (B) Colony-color phenotypes of B3H interaction between ProQ and cspE on X-gal-indicator medium. Δhfq reporter strain cells (KB483) were freshly transformed with three compatible plasmids: one that encoded λCI alone or the λCI-MS2CP fusion protein, another that encoded α alone or an α-ProQ fusion protein and a third that encoded a 1XMS2hp-cspE hybrid RNA or only the MS2hp moiety. The left-most column (cspE + ProQ) contains all three hybrid plasmids, and each additional column contains two hybrid plasmids and one negative control plasmid: α alone (-Prey), λCI alone (-Adapter) or MS2hp alone (-Bait). Transformed cells were spotted on LB-agar medium containing 0.2% arabinose, 1.5 μM IPTG, 40 μg/ml X-gal and 75 μM TPEG and grown overnight at 37°C. Duplicate spots are shown. (C andD) Top: Schematic of WT cspE 3′-UTR (C) or malM 3′-UTR (D) constructs and four adenosines that were mutated to uridines in the A-to-U mutants. Bottom: Results of a representative B3H plate-based assay detecting interactions between full-length ProQ, the N-terminal FinO domain of ProQ, or Hfq with cspE (C) or malM (D) RNA. β-gal assays were performed with Δhfq reporter strain cells containing three compatible plasmids: one (Adapter) that encoded λCI (−) or the λCIMS2CP(+) fusion protein, another (Prey) that encoded α alone (−), α-ProQ (ProQ), α-ProQ-NTD (NTD), or α-Hfq, and a third (Bait RNA) that encoded an MS2hp-cspE (C) or MS2hp-malM (D) hybrid RNA—either WT or AtoU mutant—or an RNA that contained only the MS2hp moiety (−). (E) Results of a representative B3H plate-based assay detecting interactions between ProQ and its NTD with malM RNA. β-gal assays were performed with Δhfq or hfq+ reporter-strain cells containing three compatible plasmids, as above.
We began our B3H analysis with a fragment of cspE that has been previously used in ProQ B3H assays, and with Δhfq reporter cells, which have yielded optimal signal-to-noise for B3H interactions with both ProQ and Hfq (30,51). Patches of Δhfq reporter cells grown on X-gal-indicator medium appeared much more blue when all three hybrid components were present than in each of the three negative-control conditions when half of each component was absent (-Adapter, -Prey or -Bait; Figure 10B). This increase in blueness indicates an increase in lacZ expression, which represents an interaction between cspE RNA and ProQ. Under these assay conditions, wt cspE 3′-UTR displays minimal interaction with Hfq (Figure 10C, column 1), but strong interaction with ProQ (full length ProQ or NTD only), as previously reported (30).
The cspE 3′-UTR contains four adenosines that are predicted to form base pairs with four of the eight uridines of the intrinsic terminator (Figure 10C, top). These adenosines were mutated to uridines and the interaction of wt and mutant RNA to both ProQ and Hfq was assessed using the B3H assay. A-to-U substitutions led to a substantial increase in RNA interaction with Hfq, without strongly affecting the degree of ProQ interaction (either full length ProQ or the NTD alone; Figure 10C, bottom, column 3). These results indicate that both wt and A-to-U cspE variants are expressed inside of cells and are capable of interacting in the B3H assay, but that Hfq interaction is substantially strengthened by the A-to-U substitution.
We next tested whether the B3H assay could detect interactions of malM 3′-UTR with Hfq and ProQ, and whether adenosines just upstream of its terminator would play an analogous role to those in cspE 3′-UTR. The same fragment of malM used in gel-shift experiments above was cloned into the MS2hp RNA-bait construct. Indeed, when all three hybrid components were present (MS2hp-malM, α-ProQ and CI-MS2CP adapter) in reporter cells, bacteria plated on X-gal-indicator medium appear much more blue than negative controls where half of each hybrid component is left out (Figure 10D, column 1), indicative of a ProQ-malM interaction. As with cspE, wt malM 3′-UTR interacts more strongly in this system with ProQ than with Hfq. When the four adenosines upstream of the malM terminator were mutated to uridines, in vivo binding with Hfq was strengthened without altering the apparent interaction with ProQ (Figure 10D, column 3). This in vivo result mirrors in vitro binding results (Figures 8 and 9).
The above in vivo experiments were conducted in the absence of endogenous Hfq, but in a native cellular context, ProQ may need to compete with Hfq for RNA binding, given their partially overlapping RNA ligands (11–13). We wondered whether competition with Hfq could affect how ProQ binds to RNAs lacking the A-rich motif. To test if such competition could be observed in the B3H system the binding of wt and mutant malM 3′-UTR to ProQ in Δhfq reporter cells was compared with the same interactions in reporter cells that express endogenous Hfq (hfq+; Figure 10E). Unlike in Δhfq cells where the ProQ-malM interaction appears unaffected by the A-to-U substitution, this substitution leads to an apparent loss of ProQ binding in hfq+ cells (Figure 10E). In other words, the presence of endogenous Hfq appears to have little effect on the interaction of ProQ with wt malM 3′-UTR, but strongly inhibits interaction of A-to-U malM 3′-UTR (Figure 10E). This competitive effect was similar with either the isolated NTD or ProQ suggesting that, even in the presence of the CTD and linker, competition with Hfq may drive RNA interactions with ProQ in vivo. Together these results are consistent with our model in which adenosines upstream of the terminator are a positive determinant for ProQ interaction by reducing competition with Hfq.
DISCUSSION
Recognition of intrinsic terminators by the ProQ NTD
The data presented here demonstrate that the FinO domain of E. coli ProQ specifically recognizes intrinsic transcription terminator structures within its RNA ligands (Figures 3 and 4). This observation is consistent with RIL-seq and CLIP-seq studies showing that enriched RNAs were dominated by mRNA 3′-UTRs and sRNAs (11,12). In further support, the homologous domain of L. pneumophila RocC, and the full-length F-like plasmid FinO bound the Rho-independent terminators of their specific RNA ligands tighter than their 5′ parts (23,52). On the other hand, the N. meningitidis minimal ProQ, which is composed only of the FinO domain, recognizes both the intrinsic terminators and the structured DUS regions (24,25). Hence, it is possible that the detailed structural properties of FinO-like domains from different proteins could determine their ability to specifically recognize Rho-independent transcription terminators.
Given that the N-terminal FinO domain of ProQ appears to specifically bind to terminator structures, an interesting and important question is the role of the linker and CTD. Our data show that binding of ProQ is much less dependent on the presence of the 3′ terminator structure than the isolated NTD alone, suggesting that either the linker or CTD may participate in non-terminator-specific RNA binding. Previous studies have also indicated that the linker and CTD of ProQ may have more non-specific contributions to the binding (19,30,53), and that these contributions may vary between RNAs. Studies using HDX RNA binding analysis and in vivo three-hybrid assay showed that these regions of ProQ make a larger contribution to the binding of RNAs SraB and SibB, which are longer than cspE 3′-UTR (19,30). The linker region, together with the NTD, was also required for mRNA-dependent ProQ binding to the ribosomes (53). Hence, it is possible that the linker and the CTD could form additional contacts with the same RNA molecules bound by the NTD or with other nearby RNAs. We hypothesize that such contacts could be involved in promoting RNA–RNA interactions, such as the RNA pairs reported for ProQ in E. coli and S. enterica (12,14–15).
Within a Rho-independent terminator, our study suggests that the lower part of the hairpin and 3′ oligoU tract are essential for the binding to the ProQ NTD (Figure 5). The observation that a four-uridine tail is sufficient for malM-3′ binding to the NTD (Figure 5E and F) is consistent with a recent observation that the single-stranded 3′ tails of ProQ-specific RNAs were shorter than those of Hfq-specific RNAs (11). However, the number of 3′-terminal uridines required for efficient ProQ binding may differ at the primary sequence level between RNAs, given that their secondary structures involve different numbers of uridines contributing to base pairs within their terminator hairpin. It is possible that the shorter length of 3′ oligoU tails of ProQ-specific RNAs could be one of the features that distinguish RNA ligands of ProQ from those of Hfq. The importance of the bottom part of the terminator stem and adjacent single-stranded sequences has also been observed in the interactions of the homologous FinO protein with FinP RNA (32,52). It was reported that both FinO and ProQ bound FinP terminator hairpins with single stranded extensions (29,52), and that the binding of FinO protected the lower part of the FinP RNA terminator hairpin from RNase degradation (32). Hence, these data suggest that the recognition of the junction including the lower part of the terminator hairpin and the 3′ attached oligoU tail could be a general property of RNA binding proteins from the FinO-domain family.
A study of RNA binding by ProQ mutants in E. coli suggested a model of RNA/ProQ interactions that could explain the roles of the lower part of the terminator stem and the 3′ U-tract in binding to the ProQ NTD (30). In this genetic screen for mutants affecting RNA binding by ProQ several conserved residues on the concave side of the NTD as well as in a β-hairpin at the intersection between concave and convex surfaces were found to be essential for the binding of cspE 3′-UTR and SibB sRNA (30). It has been proposed that the residues on the concave side of the NTD form an electrostatic scaffold that matches the diameter of an A-type double helix, and that it is possible for the 3′ tail of an intrinsic terminator bound on the concave side to wrap around the edge of the FinO domain toward the convex side (30). Such a model is consistent with our observation that both the bottom part of the stem and the 3′ tail are required for the RNA binding by the NTD (Figure 5), and with data showing the involvement of both sides of the FinO domain of the homologous FinO protein in binding to the FinP RNA (31).
A terminator adjacent A-rich motif serves as a negative determinant for Hfq binding
The strongest RNA ligands of ProQ both in E. coli and S. enterica datasets (9,11–12) have clear A-rich motifs on the 5′ side of the intrinsic terminator hairpin (Figure 6A and C; Supplementary Figure S7). This appears to be a distinctive feature of ProQ-specific RNAs, because in the strongest ligands of Hfq the same region is dominated by uridines (Figure 6B) (9,12). While Hfq can bind A-rich sequences, for example ARN motifs, in some of its RNA ligands, these sequences are located either in mRNA 5′-UTRs (35–37) or far upstream of intrinsic terminators in a subgroup of Hfq-specific sRNAs, called Class II sRNAs (40–42). However, in the dominant subgroup of Hfq-binding sRNAs, called Class I sRNAs, the Hfq-binding module consists of a U-rich motif just upstream of the intrinsic terminator, which is followed by a long 3′ oligoU-tail (39,54). Hence, it appears that the presence of the A-rich motif immediately upstream of the transcription terminator is a feature that is unique to ProQ-specific RNAs relative to Hfq-specific RNAs.
Our data presented here support a model in which the A-rich motif upstream of the terminator hairpin serves as a negative determinant of binding to the Hfq protein (Figures 7–10 and Supplementary Figure S9). One reason for the detrimental effect of this motif on Hfq binding may come from the fact that adenosines of the A-rich motif base-pair to uridines of the 3′ oligoU tail (Figure 7). As a result, the 3′ oligoU tail becomes involved in secondary structure, which makes it less available for binding to Hfq. This agrees with the observation that single-stranded oligoU tails of ProQ-binding RNAs are shorter than those of Hfq-binding RNAs (11). On the other hand, the absence of an U-rich motif, on the 5′ side of the terminator in ProQ-specific RNAs, likely prevents interactions with the rim of Hfq, which are known to contribute to Hfq-RNA interactions (39,54). Hence, regardless of which of these two mechanisms is more important, the presence of the A-rich sequence on the 5′ side of the terminator would weaken the RNA binding to Hfq, in agreement with our in vitro and in vivo data (Figures 9 and 10). This suggests that correct recognition of RNA ligands by ProQ is dependent in part on competition with Hfq. Indeed, our in vivo results with both cspE and malM 3′-UTRs (Figure 10) are consistent with adenosines immediately upstream of an intrinsic terminator being a positive determinant for ProQ binding—not through strengthening direct interactions with ProQ—but by serving as a negative determinant for Hfq binding, minimizing competition with this alternative RNA-binding protein.
In summary, we propose that recognition of RNAs by ProQ depends not only on the specific binding of the FinO-like NTD to select structures but also on competition with the Hfq protein. We show that the presence of the A-rich motif on the 5′ side of the intrinsic transcription terminators serves to increase their availability for binding to ProQ by limiting the access of Hfq. Further studies are needed to reveal how the functional differences between ProQ and Hfq correlate with which of them is preferred for binding at 3′ ends of individual RNAs. Interestingly, other proteins from this family, including FinO (21), FopA (J. Vogel, personal communication), meningococcal ProQ (25), and RocC (23), have different RNA binding specificities than ProQ (17). It remains to be seen if this RNA recognition specificity is determined by motifs in their RNA ligands, or rather by differences in the structure of FinO domains and their N- or C-terminal extensions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gisela Storz for sharing unpublished data, helpful discussions and critical comments on the manuscript. We thank Erik Holmqvist and Olke Uhlenbeck for helpful discussions. We thank Jakub Kosicki and Lechosław Kuczyński for advice on statistical analysis of RNA sequence comparisons, and Michał Szcześniak for advice on computational methods.
Author contributions’: E.M.S. analyzed RNA binding by ProQ and NTD, and cloned and purified both proteins; J.K. analyzed RNA binding by Hfq; E.M.S., J.K. and M.O. analyzed the binding data; M.M.B. analyzed the nucleotide frequency of A-rich sequences; C.M.G. and K.E.B. analyzed RNA binding in vivo using bacterial three-hybrid assay, E.M.S., J.K., K.E.B. and M.O. wrote the paper.
Contributor Information
Ewa M Stein, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, Uniwersytetu Poznańskiego 6, 61-614 Poznań, Poland.
Joanna Kwiatkowska, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, Uniwersytetu Poznańskiego 6, 61-614 Poznań, Poland.
Maciej M Basczok, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, Uniwersytetu Poznańskiego 6, 61-614 Poznań, Poland.
Chandra M Gravel, Program in Biochemistry, Mount Holyoke College, South Hadley, MA 01075, USA.
Katherine E Berry, Program in Biochemistry, Mount Holyoke College, South Hadley, MA 01075, USA; Department of Chemistry, Mount Holyoke College, South Hadley, MA 01075, USA.
Mikołaj Olejniczak, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University, Uniwersytetu Poznańskiego 6, 61-614 Poznań, Poland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre [2014/15/B/NZ1/03330, 2018/31/B/NZ1/02612 to M.O.]; Foundation for Polish Science - co-financed by the European Union within the European Regional Development Fund [TEAM/2011–8/5 to M.O. ]; Ministry of Science and Higher Education of the Republic of Poland, from the quality promoting subsidy, under the Leading National Research Centre (KNOW) program for the years 2014–2019 [01/KNOW2/2014]; Faculty of Biology, Adam Mickiewicz University [GDWB-03/2017 to J.K.]; National Institutes of Health [R15GM135878 to K.E.B.]; Henry R. Luce Foundation; Mount Holyoke College. Funding for open access charge: National Science Centre [2018/31/B/NZ1/02612] and Adam Mickiewicz University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gorski S.A., Vogel J., Doudna J.A.. RNA-based recognition and targeting: sowing the seeds of specificity. Nat. Rev. Mol. Cell Biol. 2017; 18:215–228. [DOI] [PubMed] [Google Scholar]
- 2. Wagner E.G.H., Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 3. Updegrove T.B., Zhang A., Storz G.. Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 2016; 30:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holmqvist E., Vogel J.. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018; 16:601–615. [DOI] [PubMed] [Google Scholar]
- 5. Kavita K., de Mets F., Gottesman S.. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018; 42:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vogel J., Luisi B.F.. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011; 9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hor J., Gorski S.A., Vogel J.. Bacterial RNA biology on a genome scale. Mol. Cell. 2018; 70:785–799. [DOI] [PubMed] [Google Scholar]
- 8. Smirnov A., Schneider C., Hor J., Vogel J.. Discovery of new RNA classes and global RNA-binding proteins. Curr. Opin. Microbiol. 2017; 39:152–160. [DOI] [PubMed] [Google Scholar]
- 9. Holmqvist E., Wright P.R., Li L., Bischler T., Barquist L., Reinhardt R., Backofen R., Vogel J.. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016; 35:991–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y.E., Reiss N., Bar A., Altuvia Y., Argaman L., Margalit H.. Global mapping of small RNA-target interactions in bacteria. Mol. Cell. 2016; 63:884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holmqvist E., Li L., Bischler T., Barquist L., Vogel J.. Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol. Cell. 2018; 70:971–982. [DOI] [PubMed] [Google Scholar]
- 12. Melamed S., Adams P.P., Zhang A., Zhang H., Storz G.. RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol. Cell. 2020; 77:411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smirnov A., Forstner K.U., Holmqvist E., Otto A., Gunster R., Becher D., Reinhardt R., Vogel J.. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smirnov A., Wang C., Drewry L.L., Vogel J.. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 2017; 36:1029–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Westermann A.J., Venturini E., Sellin M.E., Forstner K.U., Hardt W.D., Vogel J.. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica Serovar Typhimurium. mBio. 2019; 10:e02504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Milner J.L., Wood J.M.. Insertion proQ220::Tn5 alters regulation of proline porter II, a transporter of proline and glycine betaine in Escherichia coli. J. Bacteriol. 1989; 171:947–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olejniczak M., Storz G.. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers. Mol. Microbiol. 2017; 104:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Avrani S., Bolotin E., Katz S., Hershberg R.. Rapid genetic adaptation during the first four months of survival under resource exhaustion. Mol. Biol. Evol. 2017; 34:1758–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez G.M., Hardwick S.W., Maslen S.L., Skehel J.M., Holmqvist E., Vogel J., Bateman A., Luisi B.F., Broadhurst R.W.. Structure of the Escherichia coli ProQ RNA-binding protein. RNA. 2017; 23:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Attaiech L., Glover J.N.M., Charpentier X.. RNA chaperones step out of Hfq's shadow. Trends Microbiol. 2017; 25:247–249. [DOI] [PubMed] [Google Scholar]
- 21. Mark Glover J.N., Chaulk S.G., Edwards R.A., Arthur D., Lu J., Frost L.S.. The FinO family of bacterial RNA chaperones. Plasmid. 2015; 78:79–87. [DOI] [PubMed] [Google Scholar]
- 22. van Biesen T., Frost L.S.. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol. Microbiol. 1994; 14:427–436. [DOI] [PubMed] [Google Scholar]
- 23. Attaiech L., Boughammoura A., Brochier-Armanet C., Allatif O., Peillard-Fiorente F., Edwards R.A., Omar A.R., MacMillan A.M., Glover M., Charpentier X.. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8813–8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaulk S., Lu J., Tan K., Arthur D.C., Edwards R.A., Frost L.S., Joachimiak A., Glover J.N.. N. meningitidis 1681 is a member of the FinO family of RNA chaperones. RNA Biol. 2010; 7:812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bauriedl S., Gerovac M., Heidrich N., Bischler T., Barquist L., Vogel J., Schoen C.. The minimal menigococcal ProQ has an intrinsic capacity for structure-based global RNA recognition. Nat. Commun. 2020; 45:6147–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith M.N., Crane R.A., Keates R.A., Wood J.M.. Overexpression, purification, and characterization of ProQ, a posttranslational regulator for osmoregulatory transporter ProP of Escherichia coli. Biochemistry. 2004; 43:12979–12989. [DOI] [PubMed] [Google Scholar]
- 27. Smith M.N., Kwok S.C., Hodges R.S., Wood J.M.. Structural and functional analysis of ProQ: an osmoregulatory protein of Escherichia coli. Biochemistry. 2007; 46:3084–3095. [DOI] [PubMed] [Google Scholar]
- 28. Ghetu A.F., Gubbins M.J., Frost L.S., Glover J.N.. Crystal structure of the bacterial conjugation repressor finO. Nat. Struct. Biol. 2000; 7:565–569. [DOI] [PubMed] [Google Scholar]
- 29. Chaulk S.G., Smith Frieday M.N., Arthur D.C., Culham D.E., Edwards R.A., Soo P., Frost L.S., Keates R.A., Glover J.N., Wood J.M.. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry. 2011; 50:3095–3106. [DOI] [PubMed] [Google Scholar]
- 30. Pandey S., Gravel C.M., Stockert O.M., Wang C.D., Hegner C.L., LeBlanc H., Berry K.E.. Genetic identification of the functional surface for RNA binding by Escherichia coli ProQ. Nucleic Acids Res. 2020; 48:4507–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghetu A.F., Arthur D.C., Kerppola T.K., Glover J.N.. Probing FinO-FinP RNA interactions by site-directed protein-RNA crosslinking and gelFRET. RNA. 2002; 8:816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arthur D.C., Edwards R.A., Tsutakawa S., Tainer J.A., Frost L.S., Glover J.N.. Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucleic Acids Res. 2011; 39:4450–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Link T.M., Valentin-Hansen P., Brennan R.G.. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19292–19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sauer E., Weichenrieder O.. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13065–13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andrade J.M., Dos Santos R.F., Chelysheva I., Ignatova Z., Arraiano C.M.. The RNA-binding protein Hfq is important for ribosome biogenesis and affects translation fidelity. EMBO J. 2018; 37:e97631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soper T.J., Woodson S.A.. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008; 14:1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen J., Gottesman S.. Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev. 2017; 31:1382–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Otaka H., Ishikawa H., Morita T., Aiba H.. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13059–13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ishikawa H., Otaka H., Maki K., Morita T., Aiba H.. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012; 18:1062–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malecka E.M., Strozecka J., Sobanska D., Olejniczak M.. Structure of bacterial regulatory RNAs determines their performance in competition for the chaperone protein Hfq. Biochemistry. 2015; 54:1157–1170. [DOI] [PubMed] [Google Scholar]
- 41. Schu D.J., Zhang A., Gottesman S., Storz G.. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015; 34:2557–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kwiatkowska J., Wroblewska Z., Johnson K.A., Olejniczak M.. The binding of Class II sRNA MgrR to two different sites on matchmaker protein Hfq enables efficient competition for Hfq and annealing to regulated mRNAs. RNA. 2018; 24:1761–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011; 50:4427–4440. [DOI] [PubMed] [Google Scholar]
- 45. Johnson K.A. Kinetic Analysis for the New Enzymology. 2019; Austin: KinTek Corporation. [Google Scholar]
- 46. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Shea J.P., Chou M.F., Quader S.A., Ryan J.K., Church G.M., Schwartz D.. pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods. 2013; 10:1211–1212. [DOI] [PubMed] [Google Scholar]
- 49. Udekwu K.I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E.G.. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005; 19:2355–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fozo E.M., Kawano M., Fontaine F., Kaya Y., Mendieta K.S., Jones K.L., Ocampo A., Rudd K.E., Storz G.. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008; 70:1076–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Berry K.E., Hochschild A.. A bacterial three-hybrid assay detects Escherichia coli Hfq-sRNA interactions in vivo. Nucleic Acids Res. 2018; 46:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jerome L.J., Frost L.S.. In vitro analysis of the interaction between the FinO protein and FinP antisense RNA of F-like conjugative plasmids. J. Biol. Chem. 1999; 274:10356–10362. [DOI] [PubMed] [Google Scholar]
- 53. Sheidy D.T., Zielke R.A.. Analysis and expansion of the role of the Escherichia coli protein ProQ. PLoS One. 2013; 8:e79656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sauer E., Schmidt S., Weichenrieder O.. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:9396–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.











