Abstract
Escherichia coli ItaT toxin reportedly acetylates the α-amino group of the aminoacyl-moiety of Ile-tRNAIle specifically, using acetyl-CoA as an acetyl donor, thereby inhibiting protein synthesis. The mechanism of the substrate specificity of ItaT had remained elusive. Here, we present functional and structural analyses of E. coli ItaT, which revealed the mechanism of ItaT recognition of specific aminoacyl-tRNAs for acetylation. In addition to Ile-tRNAIle, aminoacyl-tRNAs charged with hydrophobic residues, such as Val-tRNAVal and Met-tRNAMet, were acetylated by ItaT in vivo. Ile-tRNAIle, Val-tRNAVal and Met-tRNAMet were acetylated by ItaT in vitro, while aminoacyl-tRNAs charged with other hydrophobic residues, such as Ala-tRNAAla, Leu-tRNALeu and Phe-tRNAPhe, were less efficiently acetylated. A comparison of the structures of E. coli ItaT and the protein N-terminal acetyltransferase identified the hydrophobic residues in ItaT that possibly interact with the aminoacyl moiety of aminoacyl-tRNAs. Mutations of the hydrophobic residues of ItaT reduced the acetylation activity of ItaT toward Ile-tRNAIlein vitro, as well as the ItaT toxicity in vivo. Altogether, the size and shape of the hydrophobic pocket of ItaT are suitable for the accommodation of the specific aminoacyl-moieties of aminoacyl-tRNAs, and ItaT has broader specificity toward aminoacyl-tRNAs charged with certain hydrophobic amino acids.
INTRODUCTION
The toxin-antitoxin (TA) system in bacteria is an acquired strategy required for growth and survival in various environments. The bacterial TA module is a gene pair of a toxin and an antitoxin encoded within an operon (1–3). Proteinaceous toxins inhibit various pivotal cellular processes, such as DNA synthesis, protein synthesis and cell wall synthesis, and thereby repress and/or regulate bacterial growth. Antitoxins are RNAs or proteins that neutralize the toxin activities, by repressing the toxin expression or inhibiting the toxin activity under normal physiological conditions. The TA system is divided into six classes (types I–VI), based on the properties of the antitoxin (4). In the type II TA system, proteinaceous antitoxins repress the activity of the toxins through protein-protein interactions. Under normal environmental conditions, the antitoxin forms a tight complex with the cognate toxin, thus masking the toxin activity. When bacteria encounter environmental stresses, such as nutrient starvation or antibiotic exposure, the antitoxins are degraded by proteinases, such as Lon and ClpP (5). As a result, the masking of the toxin activities by the antitoxin is released and the bacteria repress their own growth (3,6–8).
Recently, a new type II toxin that belongs to the GNAT (Gcn5-related N-acetyltransferase) family was identified (9–13). These GNAT family toxins catalyze the acetylation of the α-amino group of the aminoacyl-moiety of aminoacyl-tRNAs, using acetyl-CoA as an acetyl group donor. TacT, TacT2 and TacT3 from Salmonella enterica Typhimurium acetylate various aminoacyl-tRNAs and inhibit overall protein synthesis, and their activities are associated with intracellular persistence in macrophages (9,14). A recent analysis showed that TacT, TacT2 and TacT3 acetylate different sets of aminoacyl-tRNA species in vitro (14). AtaT from the enterohemorrhagic Escherichia coli O157:H7 strain specifically acetylates initiator methionyl-tRNAMet (Met-tRNAfMet) in vitro, and it may also inhibit the initiation step of protein synthesis by blocking the interaction with the initiator factor (10,15,16). GmvT from the Shigella sonnei pINV plasmid (12) and KacT from Klebsiella pneumoniae (13,17,18) are also GNAT family toxins, and may acetylate aminoacyl-tRNAs and inhibit protein synthesis, although their target aminoacyl-tRNAs have not been clarified. ItaT, recently identified from the E. coli HS strain, specifically acetylates isoaccepting isoleucyl-tRNAIles (Ile-tRNAIle) and inhibits protein synthesis in vitro (11). While a number of GNAT family toxins targeting aminoacyl-tRNAs have been identified from various organisms over the last few years, as described above, the molecular mechanism of the aminoacyl-tRNA recognition and the specificities of the GNAT family toxins have remained elusive.
Here, we present functional and structural analyses of ItaT, Ile-tRNAIle acetyltransferase toxin, from E. coli HS (11). We show that, in addition to Ile-tRNAIle, Val-tRNAVal and Met-tRNAMet are acetylated by ItaT in vivo and in vitro. Neither Leu-tRNALeu, Ala-tRNAAla nor Phe-tRNAPhe is significantly acetylated by ItaT. Thus, the ItaT toxin has broader specificity toward aminoacyl-tRNAs charged with certain hydrophobic amino acids. We also identified a hydrophobic pocket in the ItaT structure for aminoacyl moiety recognition. Comparisons of the side chain structures of the aminoacyl moieties of substrate aminoacyl-tRNAs suggest that the size, shape, and hydrophobicity of the pocket in the vicinity of the catalytic site of ItaT select a specific group of aminoacyl-tRNAs for acetylation.
MATERIALS AND METHODS
Plasmid constructions
The DNA fragment encoding the E. coli HS itaRT module (11), the itaR-itaT operon, was purchased from Eurofins, Japan. The synthesized nucleotide sequence is shown in Supplementary Table S1. For ItaR-ItaT complex overexpression in E. coli, the itaR-itaT operon sequence was cloned between the NdeI and XhoI sites of the pET22b vector (Merck Millipore, Japan), yielding the plasmid pET22b-ItaRT. The ItaT in the complex has a hexahistidine tag at the C-terminus. To obtain the plasmid for the expression of the inactive ItaT (G115D) mutant protein, the G115D mutation was introduced into pET22b-ItaRT by the overlap PCR method, and then the itaT coding DNA bearing the G115D mutation was PCR amplified and cloned into the NdeI and XhoI sites of pET-22b, yielding pET22b-ItaT(G115D). For the evaluation of the toxicity of ItaT in vivo, the DNA fragment containing the itaT sequence was PCR amplified from pET22-ItaRT and cloned between the NdeI and HindIII sites of the pBAD33 vector, purchased from ATCC (ATCC® 87402™), yielding the plasmid pBAD33-ItaT. To obtain the plasmids containing itaT variants, the mutations were introduced by the overlap PCR method using pBAD33-ItaT as the template. The oligonucleotide sequences used for the plasmid constructions are listed in Supplementary Table S2.
Expression and purification of ItaT and its variants
Escherichia coli BL21(DE3) (Novagen-Merck Millipore) was transformed with pET-22b-ItaT(G115D), pET22b-ItaRT or its variants and grown in LB medium containing 50 μg/ml ampicillin at 37°C until the OD660 reached 0.5. The expression of ItaT(G115D), the ItaR-ItaT complex and its variants was induced by adding IPTG (isopropyl-β-d thiogalactopyranoside) at a final concentration of 0.1 mM and continuing the culture for 20 h at 20°C. The harvested cells were sonicated in buffer, containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 10 mM imidazole, 50 μg/ml lysozyme and 0.1 mM PMSF (phenylmethylsulfonyl fluoride), and the lysate was centrifuged. The supernatant was first applied to a Ni-NTA agarose column (Qiagen, Japan). The column was washed with buffer, containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, and 10 mM imidazole, and the protein was eluted from the column with buffer containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol and 250 mM imidazole. The proteins were further purified using a HiTrap Heparin column (GE Healthcare, Japan), and finally purified on a HiLoad 16/60 Superdex 200 column (GE Healthcare, Japan) equilibrated with buffer containing 20 mM Tris–HCl, pH 7.0, 500 mM NaCl and 10 mM β-mercaptoethanol.
For purification of the wild-type ItaT (or its variants) and the ItaR–ItaT complex (or its variant complexes) we first used a Ni-NTA agarose column and a HiTrap Heparin column, as described above. The ItaR-ItaT complex (or its variant complexes) was loaded onto a Ni-NTA column and denatured by adding denaturing buffer (8 M urea, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl and 5 mM β-mercaptoethanol). ItaT (or its variants) with a C-terminal histidine tag was retained on the column, and ItaR was washed out from the column. The denatured ItaT (or its variants) was refolded with a stepwise concentration gradient of urea (6, 4, 2, 1, 0.5 and 0 M) on the column. Finally, ItaT was eluted from the column with buffer containing 50 mM Tris–Cl, pH 7.0, 500 mM NaCl, 10 mM β-mercaptoethanol, 10% (v/v) glycerol and 400 mM imidazole, and further purified on a Heparin Sepharose 6 Fast Flow column (GE Healthcare, Japan).
Preparation of aminoacyl-tRNA synthetases
The DNA fragment encoding the E. coli isoleucyl-tRNA synthetase (IleRS) gene was PCR amplified from the genomic DNA and cloned between the NdeI and XhoI sites of the pET-22b plasmid. The primers used for the PCR are listed in Supplementary Table S2. The IleRS protein was expressed in E. coli BL21(DE3), and the overexpressed IleRS was purified by chromatography on Ni-NTA and HiTrap Heparin columns, as described above. Finally, the IleRS protein was purified on a HiLoad 16/60 Superdex 200 column, in buffer containing 20 mM Tris–HCl, pH 7.0, 200 mM NaCl and 10 mM β-mercaptoethanol. Methionyl-tRNA synthetase (MetRS) was prepared as described previously (16). The plasmids for the overexpression of other aminoacyl-tRNA synthetases (ARSs: AlaRS, ValRS, LeuRS and PheRS) were kind gift from Dr Shimizu (RIKEN, Japan), and the proteins were prepared in the same manner as described above.
Preparation of tRNAs
The synthetic DNA fragments of the E. coli tRNALeu, tRNAAla and tRNAPhe genes were inserted between the SacI and PstI sites of the pBSTNAV3 plasmid (19). The pBSTNAV3 plasmids encoding tRNAfMet, tRNAmMet, tRNAVal and tRNAIle were described previously (20,21). The nucleotide sequences of these tRNA genes encoding tRNAPhe, tRNAAla and tRNALeu are listed in Supplementary Table S1. The E. coli JM101Tr (22) strain was transformed by the pBSTNAV3 plasmid encoding the respective gene and cultured in 2× YT medium containing 50 μg/ml ampicillin, at 37°C for 24 h. The total tRNA fraction was prepared as described, with modifications (20,21). After the deacylation of the amino acyl-tRNAs, total RNAs were dissolved in buffer containing 20 mM Tris–Cl, pH 7.4, 0.1 mM EDTA, and 8 mM Mg(OAc)2, loaded on a HiLoad 16/10 Q-Sepharose HP column (GE Healthcare, Japan) and separated by a linear NaCl gradient (0.2–1.0 M) in the buffer. The tRNAIle, tRNAfMet, tRNAmMet, tRNAVal, tRNAAla, tRNALeu or tRNAPhe enriched fractions were detected by the aminoacylation activity, using the respective cognate aminoacyl-tRNA synthase and radiolabeled amino acid, pooled and ethanol-precipitated.
The tRNA fractions prepared as described above were aminoacylated by their cognate aminoacyl-tRNA synthetases, and the amounts of the enriched isoacceptor tRNAs in the respective tRNA fractions were measured. For estimations of the tRNAIle isoacceptors in the tRNA preparation, the reaction mixture (10 μl volume), containing 20 mM Tris–HCl, pH 7.4, 150 mM KCl, 7 mM MgCl2, 10 mM ATP, 10 mM β-mercaptoethanol, 9.4 μM tRNA preparation, 180 μM l-[methyl-14C]-isoleucine (50 Ci/mol; American Radiolabeled Chemicals, Inc.), and 1 μM isoleucyl-tRNA synthetase (IleRS), was incubated at 37°C for 30 min. An aliquot (9 μl) was spotted onto a Whatman 3MM filter (GE Healthcare, Japan) and the radioactivities on the filters were quantified with a liquid scintillation counter (Hitachi-Aloka Medical), as described (23). For the estimation of other tRNA isoacceptors in each tRNA preparation, the cognate purified aminoacyl-tRNA synthetase and amino acid were used for the reaction.
In vitro acetylation assay
First, a reaction mixture (15 μl volume), containing 20 mM Tris–HCl, pH 7.4, 150 mM KCl, 7 mM MgCl2, 10 mM β-mercaptoethanol, 2.7 mM ATP, 8.8 μM each tRNA isoacceptor in the respective preparation, 250 μM cognate amino acid and 1 μM cognate aminoacyl-tRNA synthetase, was incubated at 37°C for 1 h. Afterwards, a 15 μl portion of the next reaction mixture, containing 100 μM [acetyl-1-14C]-acetyl Coenzyme A (acetyl-CoA, 60 Ci/mol, Perkin Elmer, Japan) and 0.2 μM ItaT (or its variants), was added and incubated at 37°C. An aliquot (9 μl) was spotted onto a Whatman 3MM filter at the indicated time, and the radioactivities on the filters were measured as described above.
In vivo toxicity assay for itaT and its variants
Escherichia coli strain MG1655, purchased from NBRB E. coli strain (NIG, Japan) was transformed with pBAD33-ItaT or its variants, inoculated in LB containing 50 μg/ml chloramphenicol in the presence of 0.5% (w/v) glucose, and cultured at 37°C. To evaluate the toxicities of ItaT in liquid medium, the overnight cultures were diluted to an OD660 of 0.03 in fresh liquid LB containing 50 μg/ml chloramphenicol, supplemented with either 0.1% (w/v) glucose or 0.1% (w/v) arabinose. The cultures were continued at 37°C, and the OD660 values of the cultures were measured. To evaluate the toxicities of ItaT and its variants by the spot assay, the overnight LB cultures were serially diluted, and 3 μl portions were spotted on LB agar plates containing 50 μg/ml chloramphenicol, supplemented with either 0.1% (w/v) glucose or 0.1% (w/v) arabinose, and then the plates were incubated overnight at 37°C.
Crystallization and structural determination of ItaT
For crystallization of the ItaT(G115D) protein, 0.2 μl of the protein solution (2 mg/ml) was mixed with 0.2 μl of reservoir solution, containing 100 mM Tris–HCl, pH 7.6, 30 mM sodium citrate, and 26% (v/v) PEG3350, and the crystals were generated by the hanging drop vapor diffusion method at 20°C. Data sets were collected on beamline 17A at the Photon Factory at KEK, Japan. The crystals were flash cooled in 1.1× concentrated reservoir solution, supplemented with 30% (v/v) ethylene glycol as a cryoprotectant. The data were indexed, integrated, and scaled with XDS (24). The homology model structure of ItaT was built by the SWISS-MODEL server (25) using the K. pneumoniae KacT structure (18), and the initial phase was determined by the molecular replacement method, using the homology model as the search model, by Phaser. The structure was refined with phenix.refine (26), and manually modified with Coot (27).
Preparation of acetylated aminoacyl-tRNAs from E. coli upon induction of ItaT
Escherichia coli strain MG1655 was transformed with either pBAD33-ItaT or empty pBAD33 and cultured overnight. The overnight cultures were diluted to an OD660 of 0.03 into fresh liquid LB (3 ml) containing 50 μg/ml chloramphenicol, and the cultures were continued at 37°C until the A660 reached 0.2. At this point, 0.02% (w/v) arabinose was added. At 15 min after inoculation, the cells were harvested, and suspended in buffer containing 50 mM NaOAc, pH 5.0, 0.5 mM EDTA and 0.2 M NaCl. The RNA was extracted by phenol saturated with 300 mM NaOAc, pH 5.2, followed by isopropyl alcohol precipitation. The RNA was dissolved in 250 μl of cold 200 mM NaOAc, pH 5.0, and acetylated by adding acetic anhydride-D6 (Sigma Aldrich, Japan) as described (28). Afterwards, the RNA was ethanol precipitated and rinsed with 70% cold ethanol. The RNA was dissolved in cold buffer containing 50 mM NaOAc, pH 5.0, 0.5 mM EDTA and 0.2 M NaCl, and loaded onto 100 μl of Q-Sepharose FF (GE Healthcare, Japan). The resin was washed with buffer containing 50 mM NaOAc, pH 5.0, 0.5 mM EDTA and 0.2 M NaCl. The tRNA was eluted with buffer containing 50 mM NaOAc, pH 5.0, 0.5 mM EDTA and 0.6 M NaCl, ethanol precipitated and rinsed with 70% ethanol. The pellet was dissolved in 2 mM NaOAc, pH 5.0.
LC/MS spectrometry
The purified acetylated aminoacyl-tRNAs described above were digested with RNase One Ribonuclease (Promega, Japan), in a reaction mixture (25 μl volume) containing 25 mM NH4OAc and 2.5 units enzyme, at 37°C for 60 min. The digests were subjected to an LC/MS analysis using a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific), equipped with a Dionex UltiMate 3000 LC System (Thermo Fisher Scientific) and an SunShell C18 column (2.6 μm, 2.1 × 150 mm, ChromaNik Technologies Inc.). Elution was carried out at flow rate of 0.075 ml/min using multi-linear gradient with 5 mM ammonium acetate (pH 5.3) (solvent A) and 100% acetonitrile (Fuji Film) (solvent B). The following gradient was used:0-30% B from 0–40 min; 30–80% B from 40–41 min; 80% B from 41–46 min; 80-0% B from 46–47 min; and 0% B from 47–60 min. Ions were scanned by the use of the positive polarity mode over an m/z range of 110–950 (29).
RESULTS
Induction of ItaT in vivo is toxic
Recently, E. coli ItaT, isoleucyl-tRNA acetyltransferase toxin, was reported to acetylate the α-amino group of the aminoacyl moiety of isoleucyl-tRNAIlein vitro, in good accordance with its name (11). To verify the aminoacyl-tRNA substrates of ItaT in vivo, the ItaT toxin was expressed in E. coli using the pBAD expression system, in which the expression of ItaT can be tightly controlled by adding arabinose, and the aminoacyl-tRNAs in the cells were analyzed by LC/MS spectrometry. The induction of ItaT toxin in E. coli suppresses cell growth on an agar plate (Figure 1A), and in the liquid medium, after the induction of ItaT toxin, the growth of cells bearing the ItaT expression plasmid was suppressed due to the toxicity of ItaT (Figure 1B).
Figure 1.
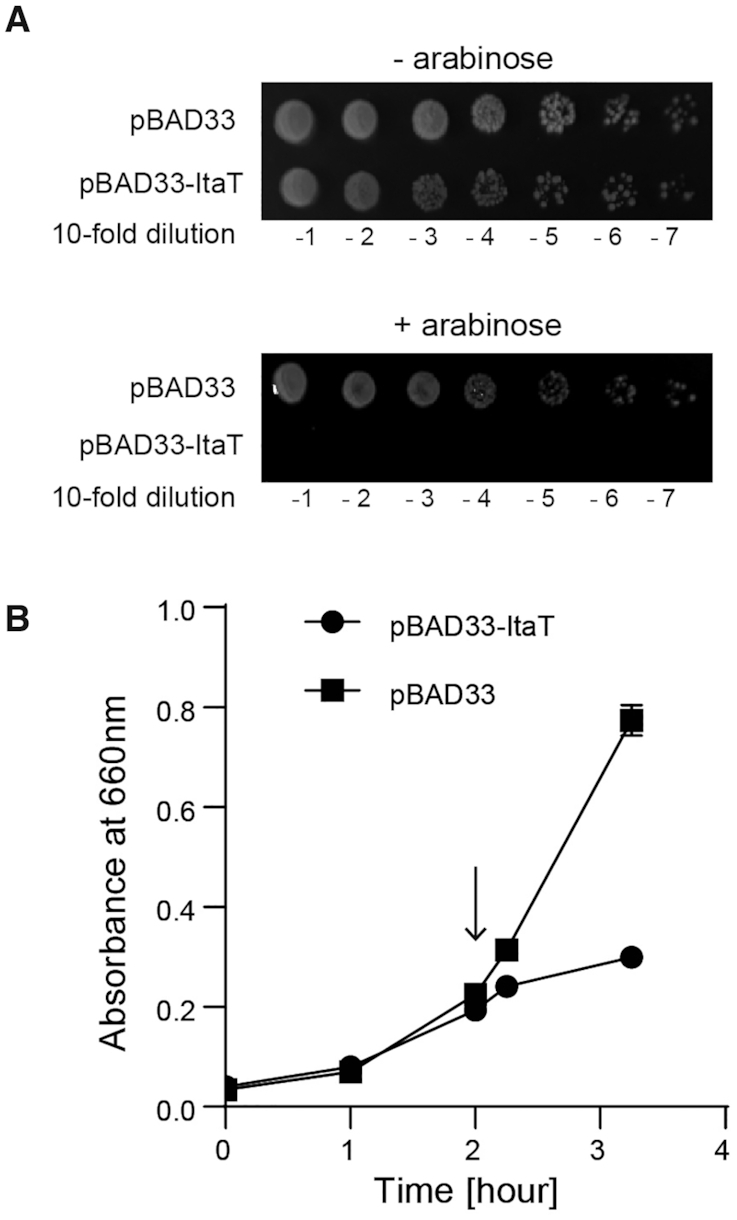
Induction of ItaTin vivo is toxic. (A) The killing activity of ItaT in vivo. Overnight cultures of E. coli MG1655 transformed with pBAD33 and pBAD33-ItaT were serially diluted, and the dilutions were spotted on LB agar plates containing 50 μg/ml chloramphenicol supplemented with 0.1% (w/v) arabinose (lower panel) or without arabinose (upper panel). The plates were incubated at 37°C. (B) Growth curves of E. coli MG1655 transformed with pBAD33 and pBAD33-ItaT. Induction of ItaT by the addition of 0.1% (w/v) arabinose suppresses the growth of E. coli harboring pBAD33-ItaT in LB containing 50 μg/ml chloramphenicol. When the OD660 reached 0.2, arabinose was added to the medium and the culture was continued at 37°C. The arrow in the graph indicates the induction of ItaT by arabinose addition.
Acetylation of aminoacyl-tRNAs by ItaT in vivo
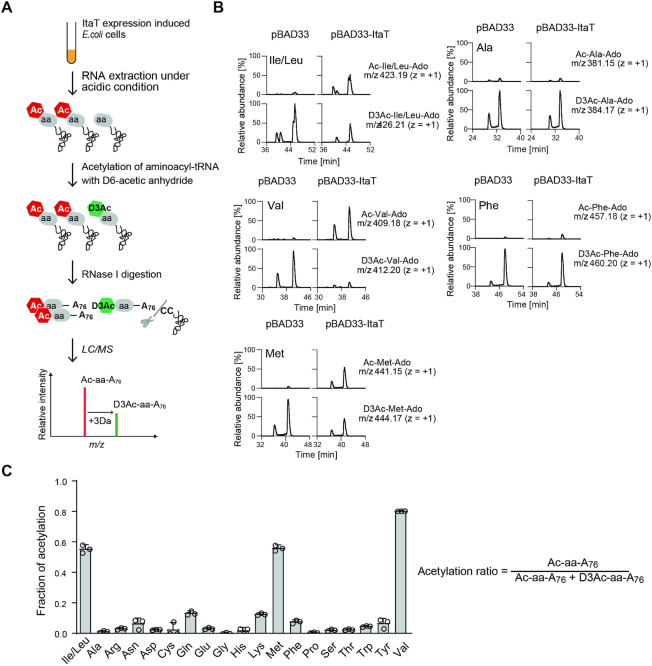
To analyze the aminoacyl-tRNA species acetylated by ItaT in vivo, after ItaT induction in E. coli, the RNA fraction was immediately prepared from the cells, under acidic and cold conditions to avoid the hydrolysis of aminoacyl-tRNAs (aa-tRNAs) and acetylated aminoacyl-tRNAs (Ac-aa-tRNAs) produced by ItaT in vivo. The purified RNA fraction was subsequently treated with stable isotopic acetic anhydride-D6 [(CD3CO)2O; D is deuterium], to chemically convert the remaining aa-tRNAs (i.e. aa-tRNAs not acetylated by ItaT in vivo) to D3-acetyl aminoacyl-tRNAs (D3Ac-aa-tRNAs) in vitro. The RNAs were then digested with RNase I, and the amounts of Ac-aa-A76 (A76 is the 3′-terminal adenosine of the tRNA molecule) and D3Ac-aa-A76 in the digests were quantified by LC/MS spectrometry. The scheme of the analysis is depicted in Figure 2A.
Figure 2.
Acetylation of Ile-tRNAIle, Val-tRNAVal and Met-tRNAMet isoacceptors in vivo. (A) Schematic diagram of detection and quantification of acetyl-aminoacyl-tRNAs, produced by the action of ItaT in vivo (See details in Materials and Methods). (B) LC/MS analysis of RNase I-digested fragments of acetyl-aminoacyl-tRNAs. Identification of the molecular masses corresponding to Ac-Ile/Leu-A76 (acetyl-isoleucyl/leucyl-adenosine, m/z 423.19), Ac-Val-A76 (acetyl-valyl-adenosine, m/z 409.18) and Ac-Met-A76(acetyl-methionyl-adenosine, m/z 441.15) derived from Ac-Ile/Leu-tRNAIle/Leu, Ac-Val-tRNAVal and Ac-Met-tRNAMet, respectively, produced by the action of ItaT in vivo. Ac-Ala-A76 (acetyl-alanyl-adenosine) and Ac-Phe-A76 (acetyl-phenylalanyl-adenosine) were not detected. Identification of the molecular masses corresponding to D3Ac-Ile/Leu-A76 (m/z 426.21), D3Ac-Val-A76 (m/z 412.20), D3Ac-Met-A76 (m/z 444.17), D3Ac-Ala-A76 (m/z 384.17) and D3Ac-Phe-A76 (m/z 460.20), derived from the in vitro aminoacyl-tRNAs chemically acetylated by acetic anhydride-D6. The two observed peaks in each Ac-aa-A76 represent structural isomers of 3′-acetyl-aminoacyl-A76 and 2′-acetyl-aminoacyl-A76, as observed for the separation of 3′-O-methyl and 2′-O-methyl nucleosides (38). (C) Quantification of the acetylation of aminoacyl-tRNAs by the action of ItaT in vivo. The fractions of individual acetylated aminoacyl-tRNAs by ItaT in vivo were estimated as the ratio of the amount of Ac-aa-A76 to the sum of the amounts of Ac-aa-A76 and D3Ac-aa-A76. The bars in the graphs are SD of more than three independent experiments.
The LC/MS analyses of RNase I-digested RNAs prepared from cells with ItaT induction revealed the molecular masses corresponding to acetyl isoleucyl (or leucyl)-adenosine (Ac-Ile/Leu-A76, m/z = 423.19), acetyl valyl-adenosine (Ac-Val-A76, m/z = 409.18), and acetyl methionyl- adenosine (Ac-Met-A76, m/z = 441.15) (Figure 2B). In the RNA preparation from control E. coli harboring the empty pBAD33 vector, the molecular masses corresponding to these Ac-aa-A76 molecules (aa: Ile/Leu, Val or Met) were not observed. These results suggested that Ile/Leu-tRNAIle/Leu (Ile-tRNAIle or Leu-tRNALeu), Val-tRNAVal and Met-tRNAMet are acetylated by the action of ItaT in vivo. The molecular masses corresponding to acetyl alanyl-adenosine (Ac-Ala-A76, m/z = 381.15) and acetyl phenylalanyl-adenosine (Ac-Phe-A76, m/z = 457.18) were barely detectable, suggesting that Ala-tRNAAla and Phe-tRNAPhe are not acetylated by ItaT in vivo (Figure 2B).
The molecular masses corresponding to D3Ac-Ile/Leu-A76 (m/z = 426.21), D3Ac-Val-A76 (m/z = 412.2), and D3Ac-Met-A76 (m/z = 444.17), which are derived from their respective aa-tRNAs acetylated by acetic anhydride-D6in vitro, were quantified (Figure 2B). The fractions of individual aminoacyl-tRNAs acetylated by ItaT in vivo were estimated as the ratio of the amount of Ac-aa-A76 to the sum of the amounts of Ac-aa-A76 and D3Ac-aa-A76 in the ItaT induced cells. Among the twenty kinds of aminoacyl-tRNAs (Supplementary Figure S1), significant fractions of Ile/Leu-tRNAIle/Leu (Ile-tRNAIle or Leu-tRNALeu), Val-tRNAVal and Met-tRNAMet were acetylated in vivo by the action of ItaT (Figure 2C). For Ile/Leu-tRNAIle/Leu, we could not definitely determine whether Ile-tRNAIle or Leu-tRNALeu or both are acetylated by ItaT in vivo, since isoleucine and leucine are structural isomers. However, about 60% of the Ile/Leu-tRNAIle/Leu isoacceptors in the cells are estimated to be acetylated by the action of ItaT in vivo (Figure 2C). As described below, since Ile-tRNAIle is acetylated by ItaT more efficiently than Leu-tRNALeuin vitro, over 60% of the Ile-tRNAIle isoacceptors in the cells would be acetylated by the action of ItaT in vivo. For Val-tRNAVal, about 80% of the Val-tRNAVal isoacceptors in the cells are acetylated by the action of ItaT in vivo. For Met-tRNAMet, in addition to the molecular mass corresponding to Ac-Met-A76, the molecular mass corresponding to N-formyl methionyl adenosine (fMet-A76, m/z = 427.13) was observed in the RNA preparation from both ItaT-induced and control E. coli (Supplementary Figure S2A).When the intensity of fMet-A76 in each sample is expressed relative to the intensity of D3Ac-Phe-A76 or D3Ac-Gly-A76 in the sample, the intensities of fMet-A76 are not significantly changed (Figure 2B). This observation suggests that the amounts of fMet-tRNAfMet in the cells are not significantly altered by the action of ItaT induction in vivo. Thus, about 60% of the remaining Met-tRNAMet isoacceptors in the cells are estimated to be acetylated by the action of ItaT in vivo.
Acetylation of aminoacyl-tRNAs by ItaT in vitro
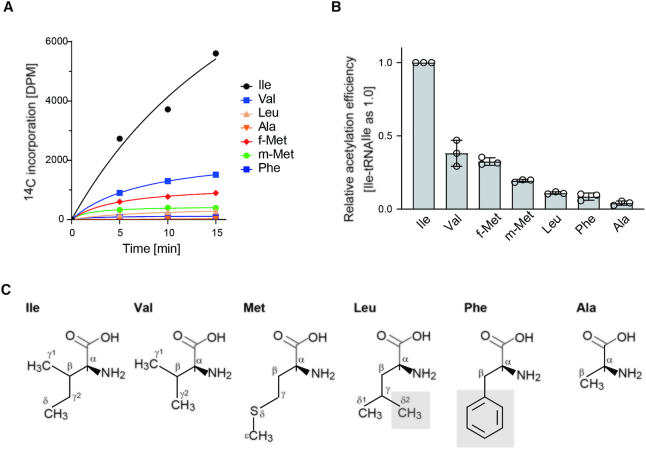
The analyses of the aminoacyl-tRNA species acetylated by the action of ItaT in vivo (Figure 2) suggest that ItaT acetylates Val-tRNAVal and Met-tRNAMet in addition to Ile-tRNAIle isoacceptors. To confirm the results obtained by the in vivo analyses, the acetylation of various aminoacyl-tRNAs by ItaT was analyzed in vitro (Figure 3). In the analyses, prior to the acetylation by ItaT, the concentration of each aminoacyl-tRNA was adjusted so the same amounts of aminoacyl-tRNAs were used for the assays. The results showed that in addition to Ile-tRNAIle isoacceptors, Val-tRNAVal and Met-tRNAMets (methionyl initiator tRNAMet; Met-tRNAfMet and methionyl elongator tRNAMet: Met-tRNAmMet) isoacceptors were significantly acetylated (Figure 3A). The acetylation efficiencies of the aminoacyl-tRNAs, calculated from the initial velocities of acetylation, revealed that Val-tRNAVal isoacceptors and Met-tRNAfMet are acetylated efficiently to extents of 30–40% of Ile-tRNAIle, and Met-tRNAmMet is acetylated to an extent of about 20% of the Ile-tRNAIle isoacceptors in vitro (Figure 3B). On the other hand, Leu-tRNALeu isoacceptors are acetylated by ItaT to an extent of less than 10% of Ile-tRNAIle isoacceptors. Thus, as described above, the molecular mass corresponding to the Ac-Ile/Leu-A76 observed by the mass spectrometric analysis of the RNA digests prepared from Ita-T-induced E. coli (Figure 2A) would be derived mostly from the Ac-Ile-tRNAIle isoacceptors, rather than the Ac-tRNALeu isoacceptors.
Figure 3.
Acetylation of Ile-tRNAIle, Val-tRNAVal and Met-tRNAMet isoacceptors in vitro. (A) Time courses of the acetylations of Ile-tRNAIle (Ile), Val-tRNAVal (Val), Met-tRNAfMet (f-Met: methionyl-initiator tRNAMet), Met-tRNAmMet (m-Met: methionyl-elongator tRNAMet), Leu-tRNALeu (Leu), Ala-tRNAAla (Ala) and Phe-tRNAPhe (Phe) by ItaT in vitro. (B) Quantification of the acetylation efficiencies of various aminoacyl-tRNAs in (A) by ItaT in vitro. The initial velocities of the acetylation of aminoacyl-tRNAs were calculated. The graph shows the relative acetylation efficiencies of the tested aminoacyl-tRNAs. The acetylation efficiency of Ile-tRNAIle isoacceptors was taken as 1.0. The bars in the graphs are SD of more than three independent experiments. (C) Chemical structures of amino acids, Ile, Val, Met, Leu, Phe and Ala. The shaded groups on the side chains of Leu and Phe would sterically clash with the amino acid residues in the aminoacyl moiety binding pocket of ItaT (see Discussion).
Since the side chains of isoleucine, valine and methionine are hydrophobic, we further tested the acetylation of other aminoacyl-tRNAs charged with hydrophobic amino acids, such as alanine and phenylalanine, by ItaT in vitro (Figure 3C). The results showed that the Ala-tRNAAla and Phe-tRNAPhe isoacceptors are acetylated to extents less than 10% of that of Ile-tRNAIle (Figure 3A, B). These in vitro analyses are consistent with the in vivo observations, in which the fractions of acetylated Ala-tRNAAla and Phe-tRNAPhe isoacceptors were less than 5% of the total Ala-tRNAAla and Phe-tRNAPhe, respectively, in vivo when ItaT is induced (Figure 2B, C). Altogether, the size and shape of the hydrophobic side chain of the aminoacyl-moiety of aminoacyl-tRNAs would be recognized by the catalytic pocket of ItaT. The detailed mechanism of the specificities of ItaT is discussed below.
Overall structure of ItaT
The amino acid sequence alignment of ItaT with the closely related GNAT family toxins revealed that G115 is located in the putative acetyl-CoA (Ac-CoA) binding sites (Supplementary Figure S3), and the mutations of the corresponding glycine to the aspartic acid in other GNAT toxins, such as AtaT, abolished the toxic activity in vivo (16). Thus, to understand the mechanism for the acetylation of a specific group of aminoacyl-tRNAs by ItaT, the ItaT(G115D) mutant protein was overexpressed in E. coli and crystallized, and the structure was determined.
The crystal belongs to the space group P21 and contains two dimer forms of ItaT in the asymmetric unit. The initial phase was determined by molecular replacement, using a homology model of ItaT constructed from the structure of K. pneumoniae KacT (18) as the search model. The structure was model-built and refined to an R factor of 23.7% (Rfree = 29.6%) at 2.8 Å resolution. The details of the crystallographic data collection and refinement statistics are provided in Table 1.
Table 1.
Data collection and refinement statistics
| ItaT (G115D) | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 70.18, 45.22, 120.38 |
| β (°) | 92.77 |
| Wavelength (Å) | 0.98000 |
| Resolution (Å)* | 50–2.8 (2.899–2.799) |
| No. of measured reflections | 260534 |
| No. of unique reflections | 19048 |
| R sym* | 0.193 (1.866) |
| I / σI * | 13.9 (1.5) |
| CC 1/2* | 99.7 (60.7) |
| Completeness (%)* | 99.9 (99.2) |
| Redundancy* | 13.7 (14.0) |
| Refinement | |
| Resolution (Å) | 20–2.8 |
| No. reflections | 18977 |
| R work/Rfree (%) | 0.2373/0.2958 |
| No. atoms | |
| Protein | 5518 |
| Water | 31 |
| B-factors (Å2) | |
| Protein | 73.00 |
| Water | 44.35 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.00 |
*Values in parentheses are for the highest-resolution shell.
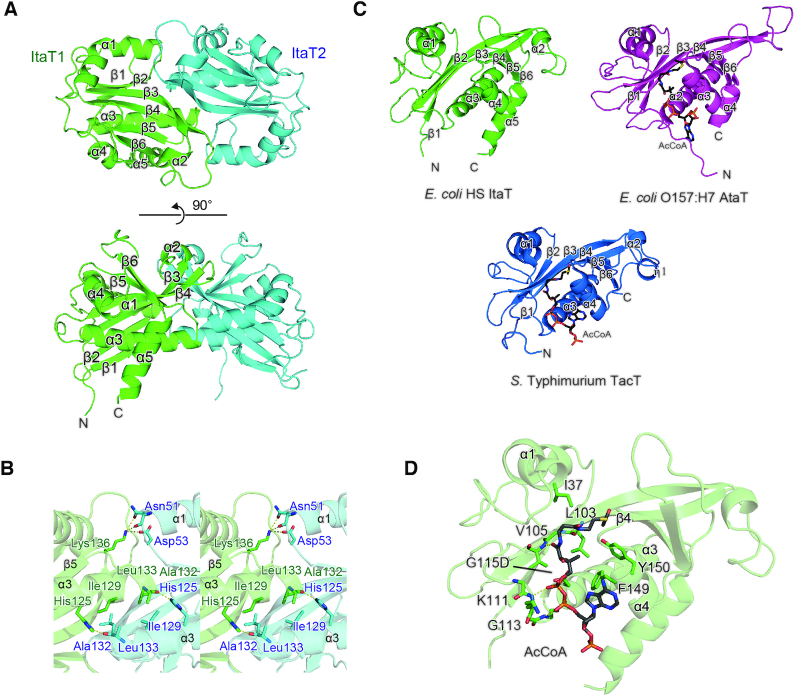
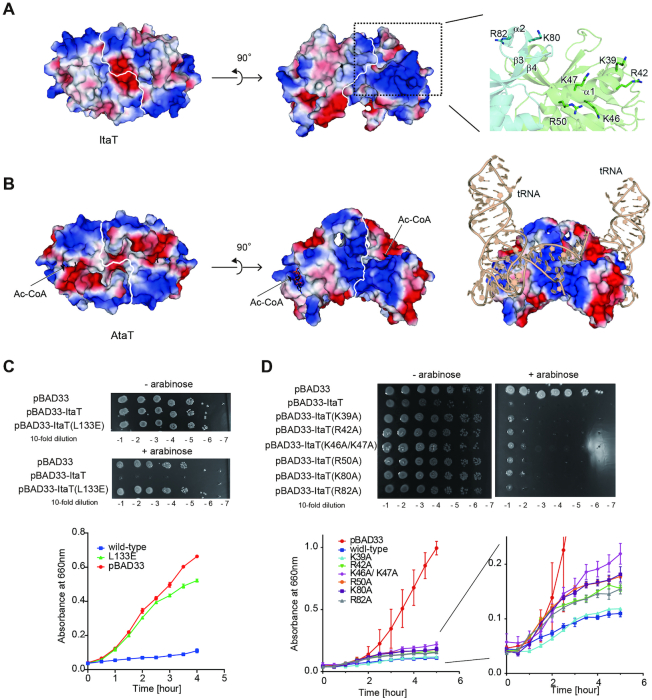
ItaT forms a dimer in the crystal, similar to other closely related GNAT toxins such as S. enterica Typhimurium TacT, K. pneumoniae KacT and E. coli AtaT (9,15,18) (Figure 4A). The two subunits of the ItaT dimer interact with each other through hydrogen-bond and hydrophobic interactions (Figure 4B). H125 in α3 of one subunit forms a hydrogen bond with the main chain carbonyl oxygen of A132 in α3 of the other subunit. I129, A132 and L133 in α3 of one subunit interact with I129, A132 and L133 in the other subunit through hydrophobic interactions. N51 and D53 in α1 also form hydrogen-bonds with K136 in the loop between α3 and β5 in the other subunit. The topology of the ItaT molecule adopts a GNAT fold (30), consisting of a mixed α/β fold with five α-helices and six β-strands, as also observed in the structures of closely related toxins (30–32) (Figure 4C, Supplementary Figure S3). The Ac-CoA binding site of ItaT is quite homologous to those of other GNAT toxins. Ac-CoA was modeled in this site in the ItaT(G115D) structure (Figure 4D). The conserved Ac-CoA binding motif, (Q/RxxGxG/A), resides in the loop between β4 and α3. G115, which was mutated to D115 for crystallization, resides in the loop. Thus, the G115D mutation in ItaT prevents Ac-CoA binding to the pocket, and thereby inhibits the toxicity of ItaT.
Figure 4.
Overall structure of ItaT. (A) Ribbon model of the ItaT dimer. ItaT1 and ItaT2 are colored green and cyan, respectively. Amino acid residues 1–8 and 1–9 were not modeled in the structures of ItaT1 and ItaT2, respectively. (B) Interface between two ItaT molecules in the ItaT dimer. The subunits are colored as in (A). (C) Comparison of the structure of ItaT with those of E. coli AtaT and S. enterica Typhimurium TacT bound with AcCoA. The AcCoA molecule is depicted by a stick model (9,15). (D) Acetyl-CoA (Ac-CoA) binding site in ItaT. The Ac-CoA molecule was modeled in the structure and is depicted by a stick model.
Possible aminoacyl moiety binding site in ItaT catalytic pocket
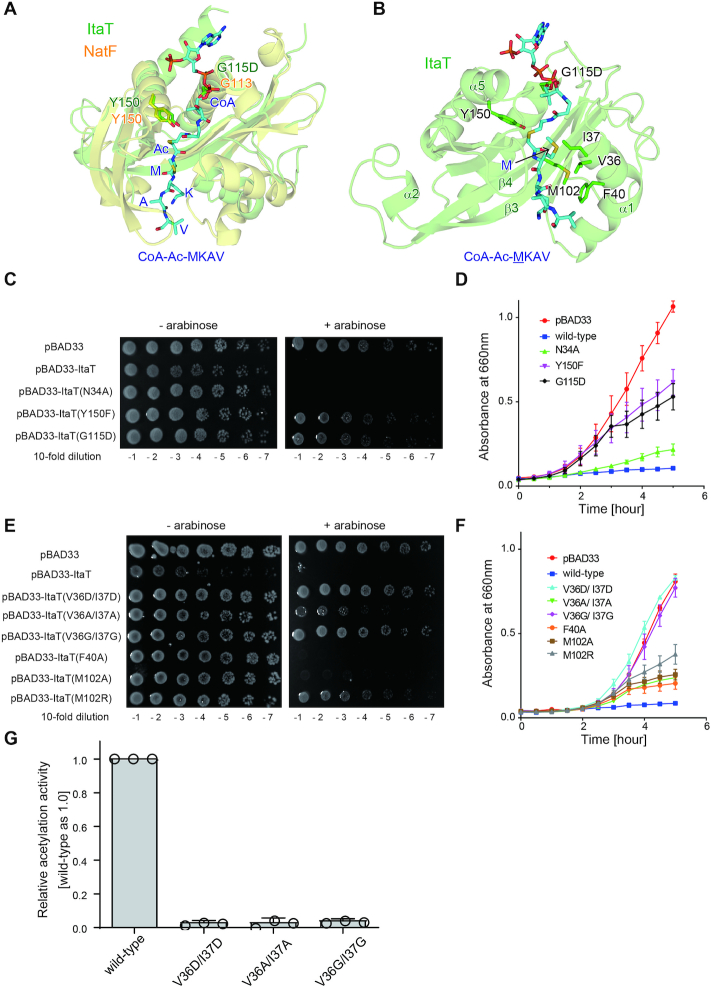
To identify the binding site of the aminoacyl moiety of aminoacyl-tRNAs in the catalytic pocket of ItaT, the ItaT structure was superimposed onto the structure of the protein N-terminal acetyltransferase, NatF (also named Naa60), complexed with a bisubstrate analog, CoA-Ac-MKAV (33).
NatF belongs to the GNAT protein family and catalyzes the N-terminal acetylation of transmembrane proteins (34). CoA-Ac-MKAV represents the transition analog of the acetyl reaction of the N-terminal amino group of the MKAV peptide, using Ac-CoA as an acetyl donor (33). In the acetylation of the N-terminal amino group of protein by NatF, the N-terminal amino group of the protein nucleophilically attacks the acyl-carbon of the acetyl group of Ac-CoA. Y150 in NatF acts as a general acid that donates a proton to the sulfur of the tetrahedral intermediate of the reaction, and promotes the release of the acetylated product (33,35). The amino acid sequence alignment of NatF and ItaT revealed the highly homologous catalytic sites, including the Ac-CoA binding sites (Figure 5A). G115, the Ac-CoA interacting residue, and Y150 in ItaT superimposed well onto the corresponding G113 and Y150 in NatF, respectively (Figure 5B). Thus, Y150 in ItaT also acts as a general acid for the acetylation of the α-amino group of aminoacyl-tRNAs. Consistent with this observation, in vivo toxicity assays of ItaT and its mutants demonstrated that ItaT with Y150F or G115D had reduced toxic activities (Figure 5C, D), and the hydroxyl group of Tyr150 is important for catalysis.
Figure 5.
Possible aminoacyl moiety binding site in ItaT. (A) Superimposition of the structure of ItaT (green) onto that of NatF complexed with the bisubstrate analog, CoA-Ac-MKAV (33), depicted by a stick model. (B) Detailed view of the structure of the active pocket of ItaT complexed with CoA-Ac-MKAV, from the structure of NatF complexed with the analog in (A). For clarity, the structure of NatF was omitted. (C) The killing activities of ItaT variants with mutations in Ac-CoA binding sites in vivo, as in Figure 1A. (D) Growth curves of E. coli MG1655 transformed pBAD33-ItaT and its variants in (C), in LB containing 50 μg/ml chloramphenicol and 0.1% (w/v) arabinose at 37°C. (E) The killing activities of ItaT variants with mutations in the putative aminoacyl moiety binding site in vivo. (F) Growth curves of E. coli MG1655 transformed with pBAD33-ItaT and its variants in (E). (G) The relative acetylation activities of ItaT variants (wild-type ItaT was taken as 1.0) under standard conditions. The reaction mixtures were incubated at 37°C for 1 h. The bars in the graphs are SD of more than three independent experiments.
The superimposition also identified the possible aminoacyl-moiety binding site in the ItaT catalytic pocket (Figure 5B). In the superimposition, the side chain of the methionine of CoA-Ac-MKAV in the NatF complex structure resides in the proximity of the hydrophobic residues V36, I37, F40 and M102 of ItaT. As described, ItaT acetylates Ile-tRNAIle, Val-tRNAVal and Met-tRNAMet, aminoacyl-tRNAs charged with hydrophobic amino acids, in vivo and in vitro (Figures 2 and 3). Thus, these hydrophobic residues would interact with the side chain of the aminoacyl moiety of aminoacyl-tRNAs. The V36D/I37D and V36G/I37G ItaT mutants lacked toxicities in vivo (Figure 5E, F). The V36A/V37A mutations in ItaT also reduced the toxicity in vivo, although the effects were smaller than the V36D/I37D or V36G/I37G mutation. Consistent with these results, the V36D/I37D, V36G/I37G and V36A/I37A mutations of ItaT all reduced the acetylation efficiency toward Ile-tRNAIle isoacceptors in vitro, to extents of <5% of wild type ItaT (Figure 5G). The F40A and M102A mutations in ItaT also reduced the toxicity of ItaT in vivo, and the M102R mutation had greater effects on the reduction of ItaT toxicity in vivo (Figure 5E, F). Altogether, these results suggest that the hydrophobic side chains of the aminoacyl moieties of aminoacyl-tRNAs (Ile-tRNAIle, Val-tRNAVal and Met-tRNAMet) would be recognized by these hydrophobic residues in the active pocket of ItaT.
Possible tRNA binding residues in ItaT
The electrostatic potential surface of the ItaT dimer revealed the highly biased distribution of charged residues. The positively charged area is distributed on α1 (K39, R42, K46, K47 and R50) in one subunit and α2 (K80 and R82) in another subunit (Figure 6A) In the closely related AtaT, the dimer formation is required for acetylation activity and toxicity of AtaT, and it was suggested that the dimer formation is required for tRNA binding (16) (Figure 6B). When L133 residues located at the dimer interface of dimeric ItaT (Figure 4B) was mutated (L133E), the ItaT toxicity was reduced (Figure 6C). Thus, ItaT dimer formation would be also required for tRNA binding and the positively charged residues in α1 in one subunit and α2 in another subunit would interact with tRNA. In consistent with this notion, the K46A/K47A and K80A mutations in ItaT slightly reduced the toxicities of ItaT in liquid medium, and the K46A/K47A mutations in ItaT slightly reduced the toxicity and on the plate (Figure 6D).
Figure 6.
Possible RNA binding region in ItaT. (A) Electrostatic potential of the dimer surface of ItaT. The clustered positively charged residues on α1 in one subunit, enclosed in the dotted box, are highlighted on the right. Positively and negatively charged areas are colored blue and red, respectively. (B) Electrostatic potential of the AtaT dimer. The positively charged region toward the Ac-CoA binding pocket spans across the dimer interface. tRNA binding model onto the AtaT dimer. (C) The killing activity of ItaT variant with L133E mutation in the dimer interface in vivo, as in Figure 1A (upper panel). Growth curves of E. coli MG1655 transformed pBAD33-ItaT and L133E variant, in LB containing 50 μg/ml chloramphenicol and 0.2% (w/v) arabinose at 37°C (lower graph). (D) The killing activities of ItaT variants with mutations in the positively charged area in vivo, as in (C) (upper panel). Growth curves of E. coli MG1655 transformed pBAD33-ItaT and its variants in upper panel, as in (C) (lower left graph), and the magnified view (lower right graph). The bars in the graphs are SD of more than three independent experiments.
DISCUSSION
ItaT, recently identified in the Escherichia coli HS strain, reportedly acetylates the α-amino group of the aminoacyl-moiety of Ile-tRNAIle isoacceptors specifically, and inhibits protein synthesis in vitro (11). In this study, to clarify the molecular mechanism by which ItaT specifically targets Ile-tRNAIle isoacceptors for acetylation, we analyzed the structure and functions of ItaT.
The LC/MS analysis of the in vivo target aminoacyl-tRNAs of ItaT for acetylation showed that, in addition to the Ile-tRNAIle isoacceptors, significant fractions of the Val-tRNAVal and Met-tRNAMet isoacceptors are acetylated by the action of ItaT in vivo (Figure 2B, C). The acetylation of these aminoacyl-tRNAs in vivo is quite rapid. By fifteen minutes after ItaT induction, 60–80% of these aminoacyl-tRNAs are acetylated in vivo. Consistent with the in vivo observations, the in vitro acetylation reactions of various aminoacyl-tRNAs showed that, in addition to the Ile-tRNAIle isoacceptors, Val-tRNAVal and Met-tRNAMet isoacceptors were acetylated significantly in vitro, although the acetylation efficiency of Ile-tRNAIle was greater than those of the Val-tRNAVal or Met-tRNAMet isoacceptors (Figure 3A, B). Further, the Ala-tRNAAla and Phe-tRNAPhe isoacceptors were minimally acetylated in vivo and in vitro (Figures 2C and 3A, B), and the Leu-tRNALeu isoacceptor was barely acetylated in vitro (Figure 3A, B). Thus, the specificity of ItaT for the substrate aminoacyl-tRNAs is broader than initially reported.
The claim in the recent study reporting the specific acetylation of Ile-tRNAIle isoacceptors by ItaT was based on the observation that ItaT caused ribosome stalling at the isoleucine codons in model mRNAs during translation in vitro, while the translation of a specific mRNA without an isoleucine codon was not affected by ItaT in vitro (11). Our in vitro analysis demonstrated that Ile-tRNAIle isoacceptors are more efficiently acetylated than Val-tRNAVal or Met-tRNAMet isoacceptors in vitro (Figure 3A, B). Thus, it is likely that only the effects of the acetylation of Ile-tRNAIle were observed under the conditions used for the reported assays, and the effects of the acetylation of Val-tRNAVal or Met-tRNAMet isoacceptors could not have been detected.
The structural analysis of ItaT (Figure 4) and the comparison of the ItaT structure with the structure of NatF complexed with the bisubstrate analog (Figure 5A) identified the hydrophobic residues (V36, I37, F40 and M102) in ItaT that possibly interact with the side chain of the aminoacyl-moiety of aminoacyl-tRNAs (Figure 5B). In particular, V36 and I37 would be important for the toxicity of ItaT in vivo and the acetylation of Ile-tRNAIlein vitro (Figure 5E, F, G). Thus, these hydrophobic residues would constitute the aminoacyl moiety binding pocket.
The substrate specificities of ItaT in vivo and in vitro, as described above (Figures 2 and 3), collectively suggest that the specificity of ItaT could be explained by the side chain structure of the aminoacyl moiety of the substrate aminoacyl-tRNA (Figure 3C). The shape and size of the aminoacyl moiety binding pocket in ItaT, in which V36, I37, F40 and M102 compose the wall, would be most suitable for the accommodation of the side chain of isoleucine. The branched methyl group (-Cγ1H3) from the Cβ position and the ethyl group (-Cγ2H2-CδH3) in isoleucine (Figure 3C) would snugly fit in the pocket. Valine also has a methyl group (-Cγ2H3) at the Cβ-position, and the ethyl group (-CγH2-CδH3) in isoleucine is replaced with a methyl group (-Cγ2H3). The side chain of valine can be accommodated in the pocket, but the hydrophobic interaction between the side chain of valine and the pocket in ItaT would be weaker than that between isoleucine and the pocket. Methionine lacks the branched methyl group at the Cβ-position, and the methyl group (-CδH3) in isoleucine is replaced with -Sδ-CϵH3. Thus, the hydrophobic interactions between the methionine side chain and the pocket would become even weaker. These observations can explain the order of the acetylation efficiencies of Ile-tRNAIle, Val-tRNAVal and Met-tRNAMet by ItaT in vitro (Figure 3A, B). Leucine lacks the branched methyl group (-CH3) at the Cβ position, and instead has two methyl (-CδH3) groups at the Cγ position. One of the methyl groups branching from the Cγ position would not be accommodated in the pocket of ItaT and would sterically clash with it; thereby, Leu-tRNALeu would not be a good substrate for acetylation by ItaT. Further, alanine is too small to snugly fit into the hydrophobic pocket of ItaT, and phenylalanine is too big and sterically clashes with the pocket. Thus, Ala-tRNAAla and Phe-tRNAPhe are also poor substrates of ItaT. The molecular mechanism observed in the selection of substrate aminoacyl-tRNAs by ItaT is similar to those observed in the aminoacyl-tRNA protein transferases, such as leucyl/phenylalanyl-tRNA-protein transferase (36,37), in that the size and shape of the hydrophobic pocket of the enzyme select the specific group of aminoacyl tRNAs as substrates.
As compared with the electrostatic potentials of other closely related toxins, such as AtaT, the distributions of the positively charged areas in ItaT are distinct from those of AtaT (Figure 6B). The previous tRNA docking model onto AtaT (16) showed that the positively charged areas between the two subunits interact with the acceptor stem (Figure 6B), and the mutations of the basic residues reduced the toxicity of AtaT in vivo (15). AtaT reportedly acetylated the initiator Met-tRNAMet (Met-tRNAfMet) specifically, but not the elongator Met-tRNAMet (Met-tRNAmMet), in vitro (10), implying that AtaT would recognize some sequential or structural features of the tRNAMet acceptor helix, rather than the aminoacyl-moiety. On the other hand, as revealed in this study, ItaT recognizes the Ile-tRNAIle, Val-tRNAVal Met-tRNAfMet and Met-tRNAmMet isoacceptors. There are no apparent shared sequences in their acceptor stem regions, except for the A73C74C75A76 sequence (Supplementary Figure S4). The discriminator bases at position 73 of tRNAAla, tRNAPhe and tRNALeu, are also A73 (Supplementary Figure S4). Thus, the A73 of tRNAs would not be positive determinant of the selection of aminoacyl-tRNA for acetylation by ItaT.
The suppression effects on the toxicity of ItaT by the mutations in the positively charged area (Figure 6D) are generally smaller than those of the mutations in the putative aminoacyl moiety binding pocket (Figure 5E, F). Therefore, the mechanism of the acetylation of specific aminoacyl-tRNAs by ItaT would be mainly governed by the aminoacyl moiety, rather than the tRNA body, and only the top of the acceptor helix or the 3′-part of the tRNA would interact with the basic residues proximal to the catalytic site of ItaT without any sequence specificity. It cannot be excluded that, in addition to the acceptor helix, other region of tRNA might interact with the dimer form of ItaT to enhance the acetylation of specific aminoacyl tRNAs. The elucidation of the exact mechanism of substrate aminoacyl-tRNA recognition by ItaT awaits further structural analyses of ItaT complexed with aminoacyl-tRNAs.
The recently identified Type II GNAT family toxins, which target aminoacyl-tRNAs, have various substrate specificities. However, the molecular mechanism underlying the different aminoacyl-tRNA recognition manners and specificities of these GNAT family toxins is still obscure. Further detailed comparative structural and biochemical analyses of these GNAT toxins would clarify the overall view of the molecular mechanisms of these toxins.
DATA AVAILABILITY
Coordinates and structure factors for the crystal structure of ItaT(G115D) have been deposited in the Protein Data Bank, under the accession code 7BYY.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the beamline staff of BL-17A (KEK, Tsukuba) for technical assistance during data collection. We thank Kaili Xu for assistance with the preparation of aminoacyl-tRNA synthetases.
Contributor Information
Chuqiao Zhang, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
Yuka Yashiro, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
Yuriko Sakaguchi, Department of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, Bunkyo-ku, Tokyo 113-8656, Japan.
Tsutomu Suzuki, Department of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, Bunkyo-ku, Tokyo 113-8656, Japan.
Kozo Tomita, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research (A) [18H03980 to K.T., in part]; Grant-in-Aid for Early-Career Scientists [19K16069 to Y.Y.] from JSPS; Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [26113002 to K.T.]; Takeda Science Foundation (to K.T.); Uehara Memorial Foundation; Tojuro Iijima Foundation for Food Science and Technology; Takahashi Industrial and Economic Research Foundation. Funding for open access charge: Grant-in-Aid for Scientific Research (A) from JSPS (to K.T.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Harms A., Brodersen D.E., Mitarai N., Gerdes K.. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell. 2018; 70:768–784. [DOI] [PubMed] [Google Scholar]
- 2. Hayes F., Van Melderen L.. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011; 46:386–408. [DOI] [PubMed] [Google Scholar]
- 3. Gerdes K., Bech F.W., Jorgensen S.T., Lobnerolesen A., Rasmussen P.B., Atlung T., Boe L., Karlstrom O., Molin S., Vonmeyenburg K.. Mechanism of postsegregational killing by the hok gene-product of the parb system of plasmid r1 and its homology with the relf gene-product of the escherichia-coli relb operon. EMBO J. 1986; 5:2023–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Page R., Peti W.. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016; 12:208–214. [DOI] [PubMed] [Google Scholar]
- 5. Muthuramalingam M., White J.C., Bourne C.R.. Toxin-antitoxin modules are pliable switches activated by multiple protease pathways. Toxins. 2016; 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harms A., Maisonneuve E., Gerdes K.. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016; 354:9. [DOI] [PubMed] [Google Scholar]
- 7. Maisonneuve E., Gerdes K.. Molecular mechanisms underlying bacterial persisters. Cell. 2014; 157:539–548. [DOI] [PubMed] [Google Scholar]
- 8. Yamaguchi Y., Inouye M.. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011; 9:779–790. [DOI] [PubMed] [Google Scholar]
- 9. Cheverton A.M., Gollan B., Przydacz M., Wong C.T., Mylona A., Hare S.A., Helaine S.. A salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell. 2016; 63:86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jurenas D., Chatterjee S., Konijnenberg A., Sobott F., Droogmans L., Garcia-Pino A., Van Melderen L.. AtaT blocks translation initiation by N-acetylation of the initiator tRNA(fMet). Nat. Chem. Biol. 2017; 13:640–646. [DOI] [PubMed] [Google Scholar]
- 11. Wilcox B., Osterman I., Serebryakova M., Lukyanov D., Komarova E., Gollan B., Morozova N., Wolf Y.I., Makarova K.S., Helaine S. et al.. Escherichia coli ItaT is a type II toxin that inhibits translation by acetylating isoleucyl-tRNAIle. Nucleic Acids Res. 2018; 46:7873–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McVicker G., Tang C.M.. Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nat. Microbiol. 2016; 2:16204–16211. [DOI] [PubMed] [Google Scholar]
- 13. Qian H.L., Yao Q.Q., Tai C., Deng Z.X., Gan J.H., Ou H.Y.. Identification and characterization of acetyltransferase-type toxin-antitoxin locus in Klebsiella pneumoniae. Mol. Microbiol. 2018; 108:336–349. [DOI] [PubMed] [Google Scholar]
- 14. Rycroft J.A., Gollan B., Grabe G.J., Hall A., Cheverton A.M., Larrouy-Maumus G., Hare S.A., Helaine S.. Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat. Commun. 2018; 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jurenas D., Van Melderen L., Garcia-Pino A.. Mechanism of regulation and neutralization of the AtaR-AtaT toxin-antitoxin system. Nat. Chem. Biol. 2019; 15:285–294. [DOI] [PubMed] [Google Scholar]
- 16. Yashiro Y., Yamashita S., Tomita K.. Crystal structure of the enterohemorrhagic Escherichia coli AtaT-AtaR toxin-antitoxin complex. Structure (London, England: 1993). 2019; 27:476–484. [DOI] [PubMed] [Google Scholar]
- 17. Yeo C.C. GNAT toxins of bacterial toxin-antitoxin systems: acetylation of charged tRNAs to inhibit translation. Mol. Microbiol. 2018; 108:331–335. [DOI] [PubMed] [Google Scholar]
- 18. Qian H., Yu H., Li P., Zhu E., Yao Q., Tai C., Deng Z., Gerdes K., He X., Gan J. et al.. Toxin-antitoxin operon kacAT of Klebsiella pneumoniae is regulated by conditional cooperativity via a W-shaped KacA-KacT complex. Nucleic Acids Res. 2019; 47:7690–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meinnel T., Blanquet S.. Maturation of pre-tRNA(fMet) by Escherichia coli RNase P is specified by a guanosine of the 5′-flanking sequence. J. Biol. Chem. 1995; 270:15908–15914. [DOI] [PubMed] [Google Scholar]
- 20. Guillon J.M., Meinnel T., Mechulam Y., Lazennec C., Blanquet S., Fayat G.. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNA(fMet) formyltransferase. J. Mol. Biol. 1992; 224:359–367. [DOI] [PubMed] [Google Scholar]
- 21. Meinnel T., Blanquet S.. Maturation of pre-tRNA(fMet) by Escherichia-coli RNase-P is specified by a guanosine of the 5′-flanking sequence. J. Biol. Chem. 1995; 270:15908–15914. [DOI] [PubMed] [Google Scholar]
- 22. Hirel P.H., Lévêque F., Mellot P., Dardel F., Panvert M., Mechulam Y., Fayat G.. Genetic engineering of methionyl-tRNA synthetase: in vitro regeneration of an active synthetase by proteolytic cleavage of a methionyl-tRNA synthetase–beta-galactosidase chimeric protein. Biochimie. 1988; 70:773–782. [DOI] [PubMed] [Google Scholar]
- 23. Martinez A., Yamashita S., Nagaike T., Sakaguchi Y., Suzuki T., Tomita K.. Human BCDIN3D monomethylates cytoplasmic histidine transfer RNA. Nucleic Acids Res. 2017; 45:5423–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. XDS. Acta Crystallogr. D-Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D-Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D-Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker S.E., Fredrick K.. Preparation and evaluation of acylated tRNAs. Methods. 2008; 44:81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suzuki T., Ikeuchi Y., Noma A., Suzuki T., Sakaguchi Y.. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007; 425:211–229. [DOI] [PubMed] [Google Scholar]
- 30. Dyda F., Klein D.C., Hickman A.B.. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000; 29:81–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ud-Din A., Tikhomirova A., Roujeinikova A.. Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT). Int. J. Mol. Sci. 2016; 17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vetting M.W., de Carvalho L.P.S., Yu M., Hegde S.S., Magnet S., Roderick S.L., Blanchard J.S.. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005; 433:212–226. [DOI] [PubMed] [Google Scholar]
- 33. Støve S.I., Magin R.S., Foyn H., Haug B.E., Marmorstein R., Arnesen T.. Crystal structure of the Golgi-associated human Nα-Acetyltransferase 60 reveals the molecular determinants for substrate-specific acetylation. Structure (London, England: 1993). 2016; 24:1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Damme P., Hole K., Pimenta-Marques A., Helsens K., Vandekerckhove J., Martinho R.G., Gevaert K., Arnesen T.. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet. 2011; 7:e1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen J.Y., Liu L., Cao C.L., Li M.J., Tan K., Yang X., Yun C.H.. Structure and function of human Naa60 (NatF), a Golgi-localized bi-functional acetyltransferase. Sci. Rep. 2016; 6:31425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suto K., Shimizu Y., Watanabe K., Ueda T., Fukai S., Nureki O., Tomita K.. Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 2006; 25:5942–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe K., Toh Y., Suto K., Shimizu Y., Oka N., Wada T., Tomita K.. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature. 2007; 449:867–871. [DOI] [PubMed] [Google Scholar]
- 38. Yang Z., Ebright Y.W., Yu B., Chen X.. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006; 34:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the crystal structure of ItaT(G115D) have been deposited in the Protein Data Bank, under the accession code 7BYY.