Abstract
Cobalamin riboswitches encompass a structurally diverse group of cis-acting, gene regulatory elements found mostly in bacterial messenger RNA and are classified into subtypes based on secondary and tertiary characteristics. An unusual variant of the cobalamin riboswitch with predicted structural features was identified in Bacillus subtilis over a decade ago, but its structure and mechanisms of cobalamin selectivity and translational control have remained unsolved. We present the crystal structure of the aptamer domain of this atypical cobalamin riboswitch and a model for the complete riboswitch, including its expression platform domain. We demonstrate that this riboswitch binds to multiple cobalamin derivatives and correlate its promiscuous behavior to its structure and unique arrangement of peripheral elements. Comparative structural analyses between conventional cobalamin riboswitches and the B. subtilis cobalamin riboswitch reveal that the likely basis for this promiscuous ligand binding is intrinsic structural adaptability encoded in the RNA structure. It suggests that cobalamin selectivity might ultimately be viewed as existing on a spectrum of affinity for each derivative rather than as belonging to distinct types based on ligand specificities. Our work provides an interesting and notable example of functional coupling of ligand-sensing and adaptive folding by a structured RNA molecule.
INTRODUCTION
Riboswitches are ligand-specific, cis-acting regulatory structural elements found in untranslated regions (UTRs) of messenger RNA (mRNA) transcripts (1–6). Although a wide variety of riboswitches have been identified to date, ranging from those that coordinate metal ions to others that discern the aminoacylation state of transfer RNAs (tRNAs) (7–21), a feature deemed universal to all riboswitches is that they are very specific to binding their cognate ligand (1,22). The vast majority of known riboswitches were discovered in the 5′ UTR of bacterial mRNA transcripts; however, 3′ UTR riboswitches exist as well, such as the thiamin pyrophosphate-sensing riboswitch of the thiamin biosynthetic gene THIC in plant seeds (23). In general, 5′ UTR riboswitches consist of an aptamer domain for ligand binding and an expression platform domain for regulating translation of the gene encoded by the mRNA in response to structural changes in the aptamer domain. Most 5′ UTR riboswitches employ premature transcriptional termination or translational activation/initiation as a mechanism by which they affect gene expression through negative feedback control (24). Therefore, there is a direct relationship between the gene downstream of the riboswitch and its corresponding ligand, whereby the gene encodes for an enzyme needed for either synthesizing or importing the ligand. As a result, riboswitches provide prokaryotic organisms a relatively rapid and direct means to regulate gene expression in cis at the mRNA level (1).
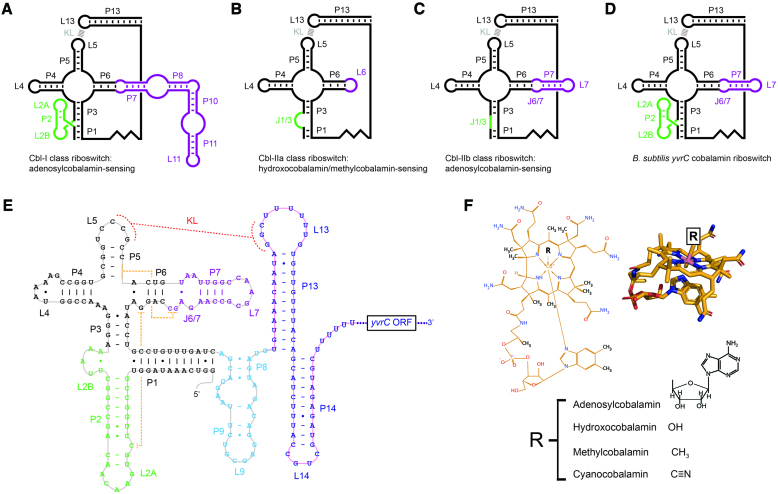
Although the initially discovered group of riboswitches included cobalamin riboswitches (14,15,25), the first crystal structures of them were not solved until a decade later and well after many other riboswitch crystal structures became available. Based on these crystal structures (26,27) and prior bioinformatic analyses conducted on a large set of cobalamin riboswitch homologues (25), it was proposed that cobalamin riboswitches can be classified into two classes (26,28), Cbl-I and Cbl-II, according to the peripheral secondary structural elements found surrounding a conserved, ligand binding core. Cobalamin riboswitches from different classes and sub-classes are largely specific for particular derivatives of cobalamin, which differ by the axial group that coordinates the cobalt in the corrin ring (Figure 1, Panel F). Thus, it was proposed that ligand specificity correlates directly with specific combinations of peripheral elements surrounding the structural core of the riboswitch (26,28). In particular, Cbl-I and Cbl-IIb riboswitches bind adenosylcobalamin (coenzyme B12), whereas Cbl-IIa riboswitches bind hydroxocobalamin or methylcobalamin (26).
Figure 1.
Architectural classification of cobalamin riboswitches. (A) Cbl-I class riboswitches bind specifically to adenosylcobalamin and consist of a loop–stem–loop P2 module (L2A–P2–L2B, green) and a large extension of its P6 stem (purple). (B) Cbl-IIa class riboswitches bind specifically to hydroxocobalamin or methylcobalamin and consist of a bulged segment (J1/3, green) and a small extension of its P6 stem (typically a loop, purple). (C) Cbl-IIb class riboswitches bind specifically to adenosylcobalamin and consist of a short mismatched or paired segment (J1/3, green) and a small extension of its P6 stem (typically a short hairpin, purple). (D) The B. subtilis yvrC cobalamin riboswitch consists of a loop–stem–loop P2 module (L2A–P2–L2B, green) and a small hairpin extension of its P6 stem (purple) and appears to be a structural hybrid of Cbl-I and Cbl-IIa/b cobalamin riboswitches. The kissing-loop (KL, grey) interaction between loops L5 (aptamer domain) and L13 (expression platform domain) is common to all Cbl-I and Cbl-IIa/b classes and the B. subtilis yvrC cobalamin riboswitch. (E) The primary sequence of the B. subtilis yvrC cobalamin riboswitch is superimposed on its conventional secondary structure schematic. The crystallized fragment corresponds to the P1-P9 region only (grey path). The P13–P14 region (pink path) was not part of the studied fragment. As in (D), the P2 module and the extension of the P6 stem are colored in green and purple, respectively. The stem-loops that comprise the majority of the expression platform domain are colored in blue. The region between the aptamer domain and the P13/L13 and P14/L14 hairpins (both in dark blue) actually forms a stem-loop (P8–P9/L9, light blue), even though it had been assumed previously to be a single-stranded linker. The KL interaction between loops L5 and L13 is depicted by orange dotted lines. Important long-range base-pairing interactions revealed by the crystal structure are depicted by yellow dotted lines. ORF denotes open reading frame. (F) The chemical and three-dimensional structures of cobalamin (gold). A particular cobalamin derivative is defined by the R-group moiety as shown in the figure.
A clear exception to the current classification scheme of cobalamin riboswitches is a cobalamin-sensing element located directly upstream of the open reading frame of the yvrC gene-encoding transcript in Bacillus subtilis, which exhibits secondary structural features that are characteristic of both cobalamin riboswitch classes. Although the ability of the B. subtilis cobalamin riboswitch to bind and respond to adenosylcobalamin was verified as part of its initial identification (25), its precise structural and functional characteristics have remained difficult to predict and thus poorly understood. In this study, we provide biochemical data demonstrating that the B. subtilis cobalamin riboswitch binds not only to adenosylcobalamin, but also to hydroxocobalamin and methylcobalamin. Additionally, we determined the crystal structure of a large region of the B. subtilis cobalamin riboswitch, comprising the complete aptamer domain and a part of the expression platform, bound to adenosylcobalamin at 3.28 Å and show that it utilizes a different peripheral element, as compared to Cbl-I cobalamin riboswitches, to buttress the adenosylcobalamin-binding core. Comparative structural analyses with the few available Cbl-I and Cbl-II cobalamin riboswitch structural models provide a simple explanation for how the atypical architecture of the B. subtilis cobalamin riboswitch can undergo a structural rearrangement to bind to hydroxocobalamin or methylcobalamin in addition to adenosylcobalamin. To the best of our knowledge, this may be the first example of an RNA molecule exhibiting intrinsic structural adaptability as the basis for promiscuous ligand derivative binding. Having determined the structure of the complete aptamer domain together with the poorly-predicted region constituting the expression platform, we also provide a model for the complete riboswitch consisting of both aptamer and expression platform domains.
MATERIALS AND METHODS
Cloning, in vitro RNA transcription, and RNA purification
The B. subtilis cobalamin riboswitch occurs naturally in the 5′ UTR region of the mRNA transcript encoding for an uncharacterized ABC transporter substrate-binding lipoprotein, yvrC. The aptamer domain of this riboswitch consists of 120 nucleotides and is followed by a putative 29-nucleotide single-stranded region that is a part of the expression platform. The gene encoding this 149-nucleotide RNA construct was pieced together by DNA oligonucleotides (Integrated DNA Technologies, Coralville, IA) and sub-cloned into pUC19 for in vitro RNA transcription. In brief, pairs of partly complementary sense and antisense DNA oligonucleotides were annealed at an equimolar ratio to generate four overlapping gene fragments (annotated sequences in Supplementary Data). The fragments were designed to have long overhangs such that they could be readily ligated and subcloned upon mixing with gel-purified pUC19 that was linearized by digestion at the compatible restriction sites (i.e. SacI and SphI). The ligation mixture was transformed into Escherichia coli DH5α (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) for replication and subsequent plasmid extraction. All clones were sequence-confirmed by Sanger sequencing. For transcription, the DNA template encoding the B. subtilis cobalamin riboswitch was cloned into the multiple cloning site of pUC19 with an upstream T7 RNA polymerase promoter element and a FokI restriction site immediately 3′ to the coding region. The plasmid was transformed into and thus replicated in cultures of E. coli DH5α grown in Terrific Broth media (Research Products International, Mount Prospect, IL) and purified by conventional alkaline lysis methods (29), followed by phenol-chloroform extraction and selective PEG 6000 precipitation, to remove contaminating host cellular proteins and RNAs, respectively. The purified plasmid DNA was subsequently digested with FokI (New England BioLabs, Ipswich, MA) and purified further by phenol-chloroform extraction and ethanol precipitation. The B. subtilis cobalamin riboswitch was co-transcribed with ligand (either adenosylcobalamin, hydroxocobalamin, methylcobalamin or cyanocobalamin at 0.5 mM) and produced in vitro on the milligram scale by run-off transcription using recombinant T7 RNA polymerase and standard reaction conditions (30). Following in vitro transcription, the RNA product was separated from other reaction components by electrophoresis in a native 10% acrylamide (37.5:1 acrylamide:bisacrylamide) gel ran in 89 mM Tris–HCl, pH 7.6, 89 mM boric acid (1× TB buffer), after which the principal RNA band was located by UV shadowing, extracted into 50 mM potassium acetate, pH 7.0, 200 mM potassium chloride by passive diffusion at 4°C, and stored at –20°C following ethanol precipitation. The riboswitch RNA samples produced in this manner were sufficiently homogenous that no further purification steps were necessary.
Preparation, handling, and storage of cobalamin ligand solutions
The cobalamin derivatives (adenosylcobalamin, hydroxocobalamin, methylcobalamin, and cyanocobalamin) were purchased in powder form (Sigma-Aldrich, St. Louis, MO, USA) and stored at the temperature (−20°C or 2–8°C) specified by the supplier before resuspension and use. For ligand binding studies, fresh ligand stock solutions were prepared prior to use at concentrations of either 1 or 10 mM. For crystallization trials, ligand stock solutions that were either newly prepared or stored in the dark at 4°C were used. There were no observed differences in diffraction quality between crystals grown with newly resuspended adenosylcobalamin versus crystals grown with adenosylcobalamin that had been stored aqueously in the dark at 4°C for up to several days. Care was taken when performing all experiments to minimize ambient lighting and thus to maximize sample quality against light-mediated degradation; however, no experiments were conducted in the dark. All binding and crystallographic studies were conducted in conditions consisting of a stoichiometric excess of ligand to RNA.
Analytical electrophoretic mobility shift assay (EMSA)
The conformation of the apo riboswitch and the same riboswitch co-transcribed with ligand (adenosylcobalamin, hydroxocobalamin, methylcobalamin, or cyanocobalamin) were assessed by EMSA. All samples were electrophoresed in a native 10% acrylamide gel in 1× TB buffer at 4°C or at room temperature (the temperature had no apparent effects on the results) and visualized subsequently by gel staining with Toluidine Blue O (Sigma-Aldrich, St. Louis, MO) resuspended in 1X TB buffer (0.02% w/v). Up to 10 μg of concentrated RNA was loaded per sample well in equal amounts and with the addition of glycerol (10% v/v). Purified RNA samples were in 25 mM Tris–HCl, pH 8.0, 50 mM potassium chloride, 10 mM magnesium chloride, 0.5 mM cobalamin derivative. The observed gel mobility differences between samples were consistent irrespective of whether this assay was performed with crude transcription products or with purified RNA samples, affirming that ligands incorporated by co-transcription remain bound to the riboswitch even through gel extraction and ethanol precipitation. Since only the ligand was varied across the samples, any observed differences in gel migration was attributed to conformational changes caused by the incorporation of a particular ligand.
Analytical size-exclusion chromatography and multi-angle light scattering (SEC-MALS)
The same RNA sample preparation procedure used for EMSA, as described above, was used for SEC-MALS experiments. Approximately 200 μg of apo riboswitch or riboswitch co-transcribed with ligand were sequentially analyzed by size exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare, Chicago, IL, USA) in 25 mM Tris–HCl, pH 8.0, 50 mM potassium chloride, 10 mM magnesium chloride at room temperature. The elution profile of each RNA sample was detected and examined by UV absorbance, multi-angle light scattering, quasi-elastic (dynamic) light scattering, and refractive index changes using an Agilent 1260 series HPLC (Agilent Technologies, Santa Clara, CA, USA) in series with a DAWN HELEOS II detector, a Wyatt QELS detector, and a Wyatt T-rEx detector (Wyatt Technology Corporation, Santa Barbara, CA), respectively. All data were analyzed using the ASTRA V software (version 5.3.4.19; Wyatt Technology Corporation, Santa Barbara, CA, USA) and a dn/dc (refractive index increment) value of 0.17 ml/g, which was confirmed using in-house RNA standards and is consistent with literature values (31).
Crystallization of the Bacillus subtilis cobalamin riboswitch bound to adenosylcobalamin
Crystallization of natively-purified B. subtilis cobalamin riboswitch co-transcribed with adenosylcobalamin was achieved in several conditions in which PEG 400 acted as the primary precipitating agent and in a wide range of temperatures (4–30°C). In brief, natively-purified RNA was precipitated in ethanol, pelleted by centrifugation, dried in a Savant DNA 110 SpeedVac, and resuspended in 50 mM Tris–HCl, pH 8.0, 100 mM potassium chloride, 20 mM magnesium chloride. The RNA was then adjusted to a concentration of 10 mg/ml, incubated at 37°C for 10 min, and an equal volume of 1 mM adenosylcobalamin was added. The resulting RNA was incubated at room temperature for 30 min, at a final concentration of 5 mg/ml, in 25 mM Tris–HCl, pH 8.0, 50 mM potassium chloride, 10 mM magnesium chloride, 0.5 mM adenosylcobalamin. To optimize crystal growth and size, classic crystallization parameters such as pH, temperature, ionic strength, precipitant concentration and additives were varied and tested. Crystallization trials were performed with many replicates to obtain the largest crystals, which grew infrequently. Crystals of adenosylcobalamin-bound B. subtilis cobalamin riboswitch grew best in 50 mM sodium cacodylate, 15% (v/v) PEG 400, 0.2 mM cobalt hexaammine, 80 mM magnesium acetate, 1 mM spermine between pH 6.0 to pH 6.5 at 14°C by vapor diffusion in a hanging drop format. The RNA crystals were rose red in color due to the presence of adenosylcobalamin and the crystals that yielded the best diffraction following dehydration (as described below) had a hexagonal prism morphology, sharp edges, and a diameter between 200 and 300 μm.
Post-crystallization improvement of the X-ray diffraction limit of crystals of the Bacillus subtilis cobalamin riboswitch bound to adenosylcobalamin
Crystals of the adenosylcobalamin-bound B. subtilis cobalamin riboswitch were reliably reproducible, typically growing to be ∼50–100 μm in diameter. Without cryo-protection, these crystals diffracted poorly and disorderedly when directly flash frozen out of the mother liquor with liquid nitrogen; however, when soaked in the crystallization solution supplemented with either 25% (v/v) glycerol or sucrose, the crystals diffracted anisotropically but consistently better to between 12 and 15 Å resolution in the best direction. In an attempt to improve the diffraction limit of these crystals, we found that by introducing either glycerol or sucrose into the crystals in a stepwise fashion helped to consistently improve the diffraction limit to 6–8 Å. Specifically, we soaked the crystals in their corresponding crystallization solution supplemented with 5% serial increments of glycerol or sucrose to a final concentration of 25% (v/v) prior to flash freezing with liquid nitrogen. Interestingly, the same stepwise addition of 25% (v/v) PEG 200, 25% (v/v) PEG 400 (the crystallization precipitant for these crystals), and 25% (v/v) xylitol had detrimental effects on diffraction.
In parallel to our approach of serially increasing the concentration of cryo-protectant within the crystals, which yielded diffraction that was at best to 6 Å resolution, we attempted to dehydrate the crystals as an alternate way to improve diffraction. Specifically, we systematically tested the effects of dehydrating the crystals for 12–48 h at the same temperature in which they grew by equilibrating them against crystallization solutions containing increasing concentrations of PEG 400 (i.e. from 15% (v/v) to 40% (v/v)) and found that when comparing similarly-sized crystals obtained from the same mother liquor, dehydration against crystallization solutions containing 25–30% (v/v) PEG 400 consistently yielded better than 2 Å improvement in diffraction.
Multiple rounds of optimization of crystal growth occasionally yielded crystals that were between 200 and 300 μm in diameter. By dehydrating these larger crystals against the corresponding crystallization solution with PEG 400 increased to a concentration between 25 and 30% (v/v), we were ultimately able to obtain 3.0 to 3.5 Å diffraction data for the adenosylcobalamin-bound B. subtilis cobalamin riboswitch.
Structure determination
X-ray diffraction data were collected at 100 K at the Life Sciences - Collaborative Access Team (LS-CAT) beamlines at the Advanced Photon Source, Argonne National Laboratory, using either Rayonix MX225 or MX300 CCD detectors. For data collection, crystals of the adenosylcobalamin-bound B. subtilis cobalamin riboswitch were cryo-protected with PEG 400, increased by dehydration to an effective concentration of 25–30% (v/v), and flash frozen with liquid nitrogen. Processing of diffraction data was done using MOSFLM (32) or XDS (33) for indexing and AIMLESS (34) for scaling.
Molecular replacement using a homology model of the core generated manually from the crystal structure of a Cbl-I class cobalamin riboswitch (PDB IDs: 4GXY) (27) produced a clear solution, but it was not possible to trace the molecule outside the core. To overcome this, we collected anomalous diffraction data by soaking the crystals at 287 K after dehydration with a tantalum tetradecabromide cluster, iridium (III) hexaammine, barium chloride or cobalt (III) hexaammine at concentrations between 1 and 10 mM for 0.1–12 h. An initial single isomorphous replacement with anomalous scattering (SIRAS) map to ∼6 Å resolution calculated with SHARP (35) using the tantalum data combined with the molecular replacement solution produced a more complete molecule, albeit at low resolution. To increase the resolution of the electron density map, a map was calculated by multiple isomorphous replacement with anomalous scattering (MIRAS) with SHARP (35) using the molecular replacement model and the iridium, cobalt, and barium data. The experimental map, to 4.1 Å resolution, was continuous and clearly showed many ridges corresponding to the phosphates of the RNA backbone (Supplementary Figure S1). After noticing significant diffraction anisotropy, the data were re-processed using the Staraniso server (36) to produce an anisotropic data set suitable for refinement. All iterative rounds of manual model building and refinement were performed with Coot (37) and both Refmac5 (38) and Buster (39,40), respectively, and all structural figures were depicted using PyMOL (41). The cobalt and iridium anomalous difference maps were used to help identify cobalt (III) hexaammine and magnesium hexahydrate molecules. All cobalt (III) hexaammine molecules had a corresponding peak in the cobalt anomalous map, but only five of eight putative magnesium hexahydrate molecules had a corresponding iridium anomalous peak. The final model consists of 148 nucleotides, 4 magnesium ions, 8 magnesium hexahydrate molecules, 7 cobalt (III) hexaammine molecules and 1 adenosylcobalamin molecule with an R-factor of 26.4%, an R-free of 27.9%, and good stereochemistry (root mean square deviation (rmsd) of 0.003 Å and 0.807° for bonds and angles, respectively). The statistics for data collection and refinement are shown in Tables 1 and 2. The electron density maps for the final structure are shown in Supplementary Figures S1 and S2.
Table 1.
Data collection and phasing statistics
| SIRAS phasing | MIRAS Phasing | ||||
|---|---|---|---|---|---|
| Data collection | Native | Ta | Ir | Co | Co/Ba |
| Detector type/source | RayonixCCD/APS | RayonixCCD/APS | RayonixCCD/APS | RayonixCCD/APS | RayonixCCD/APS |
| Wavelength (Å) | 0.97872 | 1.25237 | 0.92249 | 1.60395 | 1.54985 |
| Resolution rangea (Å) | 40.15–4.14 (4.25–4.14) | 56.82–5.90 (6.22–5.90) | 58.54–4.60 (4.85–4.60) | 80.46–4.90 (5.17–4.90) | 80.73–4.50 (4.74–4.50) |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 |
| a = b, c (Å) | 117.71, 260.72 | 116.39, 262.84 | 117.08, 261.53 | 118.0, 261.01 | 118.38, 261.97 |
| Measured reflectionsa | 69 490 (5154) | 59 907 (8612) | 125 511 (17 512) | 108 777 (15 795) | 139 261 (20 041) |
| Unique reflectionsa | 8710 (621) | 3135 (429) | 6431 (899) | 5430 (754) | 6994 (989) |
| Completenessa (%) | 99.7 (100.0) | 99.7 (100.0) | 99.9 (100.0) | 99.8 (100.0) | 99.8 (100.0) |
| Anomalous completenessa | – | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 99.9 (100.0) |
| Mean [I/σ(I)]a | 14.8 (2.8) | 23.7 (6.3) | 24.8 (7.9) | 22.0 (20.0) | 19.7 (8.1) |
| Multiplicitya | 8.0 (8.3) | 19.1 (20.1) | 19.5 (19.5) | 20.0 (20.9) | 19.9 (20.3) |
| Anomalous multiplicitya | 23.5 (23.5) | 11.2 (11.0) | 11.0 (10.5) | 11.4 (11.4) | 11.2 (10.8) |
| Rmeasurea | 0.101 (0.789) | 0.077 (0.557) | 0.089 (0.425) | 0.091 (0.310) | 0.101 (0.437) |
| CC(1/2) | 0.998 (0.834) | 0.999 (0.947) | 0.999 (0.983) | 0.999 (0.992) | 0.998 (0.990) |
| MFIDb | – | 0.158 | 0.107 | 0.179 | |
| Phasing | |||||
| Number of sites | – | 2 | 3 | 2 | 3 |
| Phasing Power | |||||
| Dispersive (centric/acentric) | – | 0.631/0.460 | 0.742/0.647 | 0.796/0.601 | 0.784/0.621 |
| Anomalous (acentric) | – | 1.135 | 0.532 | 0.668 | 0.974 |
| FOM (centric/acentric) | 0.576/0.453 | 0.445/0.452 | |||
aAll numbers in parenthesis are for highest resolution shell.
bMean fractional isomorphous difference against Native.
Table 2.
Data collection and refinement statistics
| Data collection | |
| Detector type/source | Rayonix CCD/APS LS-CAT |
| Wavelength (Å) | 0.97872 |
| Space group | P6522 |
| Cell parameters | a = b = 118.28 Å, c = 261.55 Å |
| Number of data sets | 5 |
| Resolution range (Å)a | 39.18–3.28 (3.46–3.28) |
| Total number of observations | 720 160 (41 046) |
| Unique reflections | 15 332 (760) |
| Mean (I)/σ(I) | 18.2 (1.8) |
| Completeness (spherical) (%)b | 88.3 (30.7) |
| Completeness (ellipsoidal) (%) | 92.8 (43.8) |
| Multiplicity | 47 (54) |
| R merge (all I+ & I−) | 0.160 (4.129) |
| R merge (within I+/I−) | 0.176 (4.153) |
| R meas (all I+ & I−) | 0.162 (4.168) |
| R meas (within I+/I−) | 0.179 (4.227) |
| R pim (all I+ & I−) | 0.025 (0.561) |
| R pim (within I+/I−) | 0.36 (0.784) |
| CC(1/2) | 0.996 (0.598) |
| Refinement | |
| Number of reflections working/test | 14,553/785 |
| R (working set; %) | 26.37 (37.6) |
| R free (test set; %) | 27.93 (61.2) |
| Structure quality | |
| RNA atoms | 3174 |
| Other atoms | 227 |
| RMS deviations in bond lengths (Å) | 0.003 |
| RMS deviations in bond angles (°) | 0.807 |
| Average B factor (Å2) (RNA) | 137.4 |
| Average B factor (Å2) (all atoms) | 136.8 |
aNumbers in parenthesis correspond to the highest resolution shell.
bAnisotropic data were processed used the Staraniso server (36).
RESULTS
An atypical cobalamin riboswitch in Bacillus subtilis shows promiscuous ligand derivative binding
Cobalamin riboswitches are classified into two classes, Cbl-I or Cbl-II, based on observations made in the few presently available cobalamin riboswitch crystal structures with the ensuing conjecture that specific combinations of peripheral structural elements confer exclusive binding specificity for either adenosylcobalamin (Cbl-I and Cbl-IIb) or hydroxocobalamin/methylcobalamin (Cbl-IIa) (26,27). Further biochemical and bioinformatics studies expanded upon this observation (28), but the premise of two major classes with distinct specificities remains. In general, cobalamin riboswitches consist of a four-way junction formed by stems P3 - P6 (Figure 1). A kissing loop capping stem P5 and interacting with a distal element in the expression platform domain is also common to all cobalamin riboswitches. The major differences between the distinct types are centered on the P1/P3 stems and the P6 stem. Cbl-I cobalamin riboswitches include a loop–stem–loop between the P1 and P3 stems and an elaborate extension of the P6 stem (P7–P11), which folds back toward the riboswitch core to engage the ligand binding region and thereby stabilize the core. Conserved residues in the P7–P11 extension act to stabilize a tertiary configuration in which there is space between two nucleobases to accommodate the adenosyl moiety of adenosylcobalamin. In contrast, Cbl-II class cobalamin riboswitches lack the loop–stem–loop between the P1 and P3 stems and contain a simple stem-loop addition to the P6 stem that projects away from the riboswitch core. Due to the absence of a cavity to accommodate the adenosyl moiety of adenosylcobalamin, Cbl-IIa cobalamin riboswitches cannot bind efficiently to adenosylcobalamin (26). Therefore, the difference in substrate discrimination between Cbl-I and Cbl-IIa riboswitches was explained based on the ability to accommodate the adenosyl moiety in adenosylcobalamin by Cbl-I cobalamin riboswitches; there is no available structural information on Cbl-IIb class riboswitches that helps to explain their specificity mechanism.
In retrospect, the concept that cobalamin riboswitches can be classified according to secondary structural characteristics is, at the very least, consistent with if not suggested by prior bioinformatics studies in which multiple sequences of purportedly representative cobalamin riboswitches were aligned to demonstrate that there are indeed recurrent patterns of variation in certain peripheral elements surrounding the conserved four-way junctional core (25,42). Although consensus models derived in this manner might not reveal unique or hidden features of specific homologues, they often provide a useful first step toward examining structure-function relationships in riboswitches. As a result of these early efforts, an atypical RNA aptamer variant was identified in B. subtilis within the 5′ UTR of the mRNA transcript of a four-gene (yvrC-yvrB-yvrA-yvqK) operon and immediately upstream of the leading yvrC gene, which encodes for an uncharacterized ATP-Binding Cassette cobalamin transporter. Based on sequence-based secondary structure predictions, the B. subtilis cobalamin riboswitch was noted to be unusual as it consists of a relatively large L2A/P2/L2B loop-stem-loop in combination with a modest extension of the P6 stem (J6/7, P7 and L7) (Figure 1). According to the current classification scheme, cobalamin riboswitches generally possess either small or large peripheral elements at the P2 and P7 stem-equivalent regions (25,42), which would leave the B. subtilis cobalamin riboswitch as an outlier and not a clear member of either class.
Although the B. subtilis cobalamin riboswitch has been shown to bind adenosylcobalamin and thus affect the expression of an adjoined reporter gene in vitro (25), in the absence of structural data, it remained unclear mechanistically how this riboswitch acts to regulate gene expression of its native operon. In order to investigate the ligand binding and structural properties of the B. subtilis cobalamin riboswitch, we generated an in vitro expression construct spanning the first 147 nucleotides of the riboswitch (gray path in Figure 1, Panel E) and optimized conventional in vitro transcription and gel electrophoresis-based purification methods for high-purity and milligram-scale production of the riboswitch (see Materials and Methods). Our initial attempts to incorporate adenosylcobalamin into the B. subtilis cobalamin riboswitch post-transcriptionally were unsuccessful, as assessed by EMSA and the underlying assumption that ligand binding would induce a sufficient conformational change in the RNA molecule to alter its gel migratory behavior. In these attempts, we varied multiple binding parameters such as incubation time, temperature, and solvent conditions, including different combinations of monovalent and divalent ions and their concentrations. Ultimately, we determined that relatively homogenous samples of ligand-bound riboswitch can be produced on a scalable fashion up to milligram-quantities simply by transcribing the RNA with T7 RNA polymerase with the ligand present at millimolar concentrations. This suggests that adenosylcobalamin incorporation into the B. subtilis cobalamin riboswitch may occur during co-transcriptional folding of the RNA, much like the FMN and other fuse-like riboswitches that require the presence of ligand during transcription to properly achieve the effector state (43).
Given the unusual combination of peripheral elements discerned by secondary structure predictions of the B. subtilis cobalamin riboswitch, we considered the possibility that its aptamer domain might exhibit different ligand binding properties as compared to conventional cobalamin riboswitches in a way that is reminiscent of variants of other riboswitch classes, such as purine riboswitches (44,45) and ykkC RNAs (46,47), which utilize different specificity mechanisms but share a common structural scaffold to exhibit altered ligand specificities. Having found an efficient way to incorporate ligand into the riboswitch, we carried out in vitro transcriptions of the same RNA construct with several cobalamin derivatives: adenosylcobalamin, hydroxocobalamin, methylcobalamin, and cyanocobalamin. Both the crude products of transcription and purified RNA samples were examined using EMSA, and ligand binding was assessed by changes in the electrophoretic migratory behavior of the RNA (Figure 2). Whereas the apo riboswitch ran as two closely-migrating conformers, the adenosylcobalamin-bound riboswitch ran as a single and relatively more compact species, as reflected by a subtle downward shift of its corresponding gel band. Similarly, both hydroxocobalamin-bound and methylcobalamin-bound riboswitch samples also ran as a single conformer, albeit with an even slighter downward shift than the adenosylcobalamin-bound form. These migratory patterns for the riboswitch when bound to adenosylcobalamin, hydroxocobalamin, and methylcobalamin were preserved irrespective of whether the samples were purified by gel extraction followed by precipitation with ethanol. As expected, there were no observed differences between the apo riboswitch and the riboswitch transcribed in the presence of cyanocobalamin, suggesting that the B. subtilis cobalamin riboswitch does not bind to cyanocobalamin. Of note, cyanocobalamin is the synthetic form of vitamin B12 and thereby serves as a negative control for non-specific ligand binding. Finally, for further comparison, we examined the E. coli cobalamin riboswitch in the same manner and confirmed with EMSA that, as expected for its classification as a Cbl-I cobalamin riboswitch, it is indeed specific for adenosylcobalamin (Figure 2).
Figure 2.
Promiscuous ligand binding by the B. subtilis yvrC cobalamin riboswitch. Co-transcriptional ligand loading was an effective approach in generating relatively homogenous samples of cobalamin-bound riboswitches, as demonstrated by both electrophoretic mobility shifts and size-exclusion chromatography (SEC) with multi-angle light scattering (MALS). (A) Left: The E. coli cobalamin riboswitch is specific for adenosylcobalamin (A, red), whereas the B. subtilis cobalamin riboswitch binds to adenosylcobalamin, hydroxocobalamin (H, green), and methylcobalamin (M, purple). Subtle gel migration differences were observed between the adenosylcobalamin versus hydroxocobalamin/methylcobalamin-bound B. subtilis riboswitch samples (green line). As expected, neither riboswitch associates with cyanocobalamin, a synthetic cobalamin derivative (C, orange, and C*, yellow, correspond to two different preparations). The electrophoretic mobility shift patterns of the RNAs were the same when there is no ligand (ᴓ, black) and when co-transcribed with cyanocobalamin. Right: Denaturing gel in 8 M urea shows that differences in native gel migration were not due to RNA sample degradation. (B) The B. subtilis cobalamin riboswitch exhibits conformational variation when bound to different cobalamin derivatives. SEC-MALS analysis shows that the B. subtilis cobalamin riboswitch binds to adenosylcobalamin, hydroxocobalamin, and methylcobalamin primarily as an RNA dimer. The differences observed in the various elution peak positions (vertical arrows) probably reflect conformational variation in the adenosylcobalamin versus the hydroxocobalamin/methylcobalamin-bound forms of the riboswitch. No difference in elution time was observed between the B. subtilis cobalamin riboswitch without ligand and with cyanocobalamin, consistent with the native gel findings.
To further characterize the apparent differences uncovered by EMSA between the ligand-bound forms of the B. subtilis cobalamin riboswitch, we conducted SEC-MALS experiments. As with EMSA, the addition of cyanocobalamin had no effect on the elution profile of the RNA compared to the apo form, corroborating our finding that the B. subtilis cobalamin riboswitch does not bind to cyanocobalamin. Furthermore, the apo RNA eluted as two separate peaks which, based on the MALS data, correspond to monomeric and dimeric species. In contrast, the adenosylcobalamin-bound form elutes purely as a dimer. Both the hydroxocobalamin-bound and methylcobalamin-bound forms eluted with similar profiles, consisting mostly of a dimer peak. Interestingly, the peak elution times of both hydroxocobalamin-bound and methylcobalamin-bound forms were shifted relative to that of the adenosylcobalamin-bound form, suggesting that there is a difference in the shape of these ligand-bound forms since the mass remains constant, as shown by the MALS data. Taken together, we concluded that whereas the B. subtilis cobalamin riboswitch binds to all cobalamin derivatives tested except for cyanocobalamin, its overall conformation when bound to adenosylcobalamin differs from when it is bound to either hydroxocobalamin or methylcobalamin. Furthermore, this difference in conformation is substantial enough to manifest as a discernable change in the gel filtration elution profile and EMSA.
Overall structure of the cobalamin riboswitch variant in Bacillus subtilis
The overall structure of the B. subtilis cobalamin riboswitch is reminiscent of that of other folded riboswitches of comparable size, exhibiting a pseudo-planar architecture, a ligand binding pocket assembled within a proximal junction, and multiple peripheral elements consisting of stems and stem-loops that act to stabilize the core of the aptamer domain through tertiary interactions (Figure 3). Unlike other cobalamin riboswitch structures, the crystallized B. subtilis cobalamin riboswitch construct includes a sizable extension that links the aptamer domain to the expression platform. This long extension (P8–P9/L9), which was predicted to be unstructured, forms an unusual hairpin consisting of several non-Watson-Crick base-pairs in its stem and a long distal loop (Figures 1, 3, and Supplementary Figure S3).
Figure 3.
Overall structure of the B. subtilis yvrC cobalamin riboswitch. (A) Schematic diagram showing the tertiary arrangement of secondary structural elements (color coded; grey elements were not observed directly in the crystal structure) with the primary sequence threaded through. Adenosylcobalamin (light orange) illustrates the location of the ligand binding pocket, which is within a cleft formed by the P3/P6 and P4/P5/P13 helical stacks. The ligand is in part bordered by the P13/L13 hairpin (dark grey) of the expression platform domain whose placement is mediated by a conserved kissing-loop (KL) interaction between 5′-CCG-3′ in L5 and 5′-GGC-3′ in L13. (B) Cartoon representation of the crystal structure of the B. subtilis cobalamin riboswitch shown in two orthogonal views. The diagram shows the P9/L9 hairpin of a symmetry related molecule (P9*/L9*), which is found in a position that mimics the KL interaction between L5 and P13. The view on the left shows clearly the adenosyl moiety of adenosylcobalamin (light orange) stacked between the P3 and P6 stems of the aptamer domain, which is the key mechanism for adenosylcobalamin specificity.
A common feature observed in both the B. subtilis cobalamin riboswitch and previously solved Cbl-I (PDB IDs: 4GMA, 4GXY) (26,27) and Cbl-IIa (PDB IDs: 4FRN, 4FRG) (26) cobalamin riboswitches is the near co-planar arrangement of the P3, P4, P5 and proximal P6 stems, which make up the core of the aptamer domain, and the adjoining single-stranded linker regions that come together to form the ligand binding pocket (26,27). The docking of the L6 loop into the L4 loop, observed in both Cbl-I and Cbl-IIa cobalamin riboswitches, is also present in the B. subtilis cobalamin riboswitch. Although the construct used does not contain the P13 stem (for consistency and ease of comparison with other cobalamin riboswitches, we named the stem-loop containing the kissing loop as P13 and L13, respectively, even though it would correspond to P10 and L10 if named sequentially), which contains a kissing loop sequence that binds to the P5 stem–loop, a loop from the P9 stem from a symmetry related molecule interacts with the P5 loop by mimicking the kissing loop that has been observed in the structures of both Cbl-I and Cbl-IIa cobalamin riboswitches. Thus, the spatial arrangement of one monomer and a symmetry-related molecule helped us build a model of a complete riboswitch, including the kissing loop interaction between L5 and L13 that spans one side of the ligand binding site. Overall, the structures of the core of the aptamer domain of all known Cbl-I and Cbl-IIa cobalamin riboswitches, and the B. subtilis cobalamin riboswitch, are very similar (Figure 4). The rmsd between the core nucleotides of the B. subtilis cobalamin riboswitch and those of the Thermoanaerobacter tengcongensis, Symbiobacterium thermophilum and the env8 environmental marine metagenome cobalamin riboswitches are 2.4, 2.5 and 2.3 Å, respectively.
Figure 4.
Structural conservation of the ligand binding core of cobalamin riboswitches. The superposition of crystal structures of representative cobalamin riboswitch from T. tengcongensis (Cbl-I class; core in dark cyan and peripheral elements in light cyan) (PDB ID: 4GMA) (26), the env8 environmental marine metagenome (Cbl-IIa class; core in dark purple and peripheral elements in light purple) (PDB ID: 4FRN) (26), and B. subtilis (core in dark blue and peripheral elements in light blue) are shown in two orthogonal views. (A) A side view illustrates the structural similarity of the core regions (in dark colors) surrounding the ligand (adenosylcobalamin from the B. subtilis structure is shown here and colored in orange). The kissing-loop interaction between the L5 (from the aptamer domain; in dark colors) and L13 (from the expression platform domain; in light colors) loops is conserved across all known cobalamin riboswitches. The P2 module in both the T. tengcongensis and the B. subtilis structures are positioned and oriented similarly, even though the distal loop of the T. tengcongensis P2 module is disordered in the crystal structure. (B) Top view shows the near co-planarity of the aptamer domain core structural elements, as well as the P2 module present in both the B. subtilis and the Cbl-I class structures. The L4 loop in both representative Cbl-I and Cbl-IIa structures forms a T-loop, whereas the L4 loop in the B. subtilis crystal structure forms a T-loop/tetraloop hybrid consisting of the residues U52, A53, A54 and A55.
Unlike Cbl-IIa cobalamin riboswitches, which consists only of a short loop between the P1 and P3 stems, the B. subtilis cobalamin riboswitch contains a relatively large L2A/P2/L2B loop–stem–loop, or P2 arm, analogous to that of the Cbl-I riboswitches (Figure 1). In both known structures of a Cbl-I riboswitch (26,27), the distal loop (L2A) is disordered but appears to extend away from the core of the aptamer domain. In contrast, this loop is well-ordered in the B. subtilis cobalamin riboswitch and, with the extrusion of C25, forms a GAAA tetraloop. Interestingly, both the proximal loop (L2B) of the P2 arm of Cbl-I riboswitches and the short loop between the P1 and P3 stems in Cbl-IIa riboswitches form T-loop motifs (48), but the former points downward and away from the adenosylcobalamin binding pocket, whereas the latter points upward and forms tertiary contacts with residues adjacent to the hydroxocobalamin binding pocket (Figures 4 and 5). A T-loop is also present in the L2B loop of the B. subtilis riboswitch structure but in an orientation resembling that observed in Cbl-I cobalamin riboswitches. Although the P2 arm of the B. subtilis cobalamin riboswitch is overall structured and oriented much like that of Cbl-I riboswitches, it consists of a larger L2A loop, which contains two internal base-pair interactions, G20–C28 and U21–A27 (Figure 3), that were not previously predicted based on sequence analysis. More importantly, a base-pair (C19–G103) is formed between a nucleotide in the P2 stem with one of two extruded bases (G103 and U105) in the P6 stem that appears to anchor the two stems together and help create the adenosylcobalamin binding pocket. This connection between the P2 arm and the P6 stem is further stabilized through base-stacking of G103 between G20 and A29, which embeds G103 within the P2 stem. A cobalt hexaammine molecule, which is likely to be substituting for a hexahydrated magnesium molecule, was found at this position in an anomalous Fourier difference map (Supplementary Figure S2).
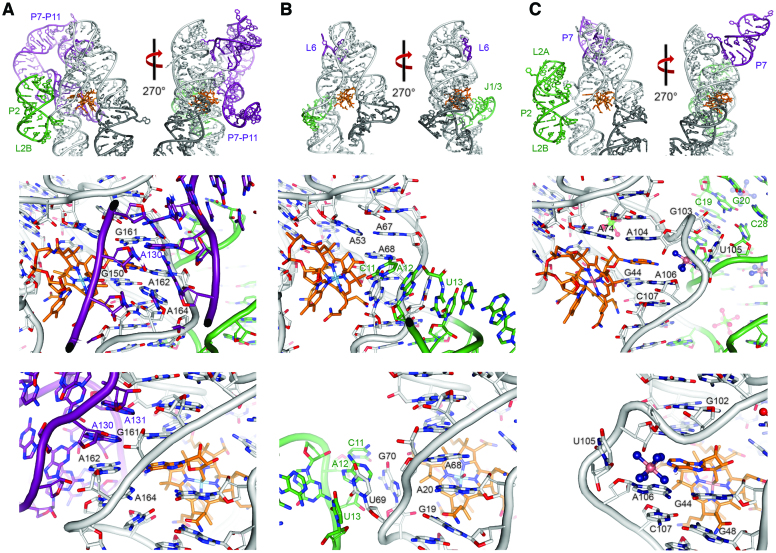
Figure 5.
Structural mechanism of cobalamin derivative selectivity and specificity. For each cobalamin riboswitch crystal structure, the P2 module and P6 extensions of the aptamer domain are colored in green and purple, respectively, and the P13/L13 stem-loop of its expression platform domain is colored in dark grey. (A) Top: Crystal structure of a Cbl-I class cobalamin riboswitch from T. tengcongensis (PDB ID: 4GMA) (26) shown in two orthogonal views. Middle: The adenosylcobalamin binding site. Recognition of the adenosyl moiety involves intercalation between two bases. The gap is formed via interactions with peripheral elements in the long P7-P11 extension (purple), whereas the smaller, P2 peripheral element (green) does not play a major role. Bottom: The P2 stem (green) does not interact significantly with the binding site region. (B) Top: Crystal structure of a Cbl-IIa class cobalamin riboswitch from the env8 environmental marine metagenome (PDB ID: 4FRN) (26) shown in two orthogonal views. Middle: Hydroxocobalamin recognition does not require the presence of a gap in the stacked bases and, unlike Cbl-I ribowitches, there is no participation of the J6 region. However, the J1/3 (green) peripheral element interacts with the binding site. Bottom: The J1/3 element (green) helps form the binding site. (C) Top: Crystal structure of the B. subtilis yvrC cobalamin riboswitch shown in two orthogonal views. Middle: Similarly to the Cbl-I riboswitches, the B. subtilis riboswitch forms a gap where the adenosyl moiety is intercalated. Unlike Cbl-I riboswitches, the P7 peripheral element does not participate in the formation of the binding site, but in common with both Cbl-I and Cbl-IIa riboswitches, the P2 module (green) helps form the binding site. Bottom: The cobalamin binding site does not involve extensive direct interactions with the major peripheral elements.
Although the B. subtilis cobalamin riboswitch L4 loop contains a sequence that conforms to the T-loop consensus (5′-UAAAA-3′) (48,49), it does not form a T-loop, unlike the L4 loops observed in other known cobalamin riboswitch structures. Instead, it forms a four-nucleotide loop consisting of residues U52-A53-A54-A55 while the subsequent A56 forms a separate base-stack with the adjacent G57 directly facing the J6/7 region. Interestingly, this UAAA loop forms neither a T-loop, since it consists of only four residues, nor a UAAA tetraloop, but rather a T-loop/tetraloop hybrid; when including the inserted base from the L6 loop, the overall tertiary conformation of the loop resembles that of a T-loop (48).
In addition to sharing overall similarities, the B. subtilis cobalamin riboswitch also exhibits structural features that are specific to either Cbl-I or Cbl-IIa cobalamin riboswitches. Cbl-II cobalamin riboswitches contain a short P6 extension (Figures 1 and 5), and in one particular example, its distal P6 loop docks directly into the L4 loop (PDB IDs: 4FRN, 4FRG) (26). In contrast, Cbl-I riboswitches have significantly longer extensions (Figures 1 and 5), which fold back toward the core of the aptamer domain such that conserved adenosines (A130 and A131 in the T. tengcongensis cobalamin riboswitch, PDB ID: 4GMA) in a distal (J11/10) region can engage and modify the ligand binding pocket to allow for binding of adenosylcobalamin. The P6 extension the B. subtilis cobalamin riboswitch consists of a simple P7/L7 stem-loop extension connected through a short J6/7 region (Figures 1 and 3), and similar to Cbl-I cobalamin riboswitches, an internal J6/7 junction docks into the L4 loop to allow for the P7/L7 stem-loop to extend distally and away from the aptamer core.
Although the linker region between the aptamer and expression platform domains of the B. subtilis cobalamin riboswitch was previously predicted to be single-stranded and unstructured (25), the crystal structure shows clearly that the linker region actually forms a long and stacked stem with two short helices connected by a junctional region (Figure 3). Interestingly, this long stem likely evaded detection by sequence-based secondary structure prediction algorithms due to the unusual amount of unpaired and non-Watson-Crick base-pairings. In the crystal, the loop at the tip of this linker region (L9) makes contacts with the L5 loop of a symmetry-related molecule to facilitate crystallization and to mimic the kissing loop interaction observed in other cobalamin riboswitch structures (26,27). This kissing loop interaction likely mediates dimer formation of the riboswitch fragment in solution (Supplementary Figure S3).
Structural basis for binding of adenosylcobalamin
When bound to adenosylcobalamin, the ligand binding pocket of the B. subtilis cobalamin riboswitch is similar in architecture to that of Cbl-I cobalamin riboswitches, involving mostly the P3 stem, the P5/L5 stem-loop, and the J3/4 and J6/3 linker regions between the P3/P4 and P6/P3 stems, respectively (Figure 5). In all three now known adenosylcobalamin binding riboswitch structures, an open space within the continuous base-stack that is formed by many of the J3/4 and J6/3 residues allows for insertion of the adenosyl moiety of adenosylcobalamin, thereby providing a mechanism for the specificity and selectivity of this ligand. In both Cbl-I cobalamin riboswitch structures (26,27), conserved residues within the long P6 extension (A130 and A131 in PDB ID: 4GMA, corresponding to A124 and A125 in PDB ID: 4GXY, respectively) (26,27) base-stack with two J6/3 adenosines (A162 and A164 in PDB ID: 4GMA, corresponding to A157 and A159 in PDB ID: 4GXY, respectively) to help create this opening (Figure 5). Furthermore, in both Cbl-I structures, A162 directly faces and is co-planar to the adenosyl group of the bound adenosylcobalamin, forming a trans A·A Watson–Crick–Hoogsteen pairing. Within the aptamer core of the B. subtilis cobalamin riboswitch, a similar space is created to allow for the insertion of the adenosyl moiety of adenosylcobalamin. However, in the B. subtilis structure, the buttressing is mediated by the P2 stem. In particular, the P2 arm accepts G103, which is flipped out of the P6 stem to form a Watson–Crick base-pair with C19 while the J6/3 nucleotide U105 faces outward and toward P2 to create a gap that accommodates the adenosyl group of the adenosylcobalamin. Unlike the Cbl-I riboswitches, there is no nucleobase facing the adenosyl group. Instead, a cobalt hexaammine molecule, most likely substituting for a hydrated magnesium, is present between the adenosyl moiety and the main phosphate backbone of the P2 stem. The adenosyl moiety of adenosylcobalamin stacks between the G44/A106 and A74/A104 non-Watson-Crick base-pairs (Figure 5). As expected, Cbl-IIa riboswitches do not have a pocket as they do not bind adenosylcobalamin. Instead, the nucleobases in the corresponding region form a continuous stack, whereas the gap found in Cbl-I and the B. subtilis cobalamin riboswitch is occupied by a base (Figure 5). Despite the apparent conservation of the general architecture of the adenosylcobalamin-binding pocket between the B. subtilis cobalamin riboswitch and Cbl-I cobalamin riboswitches, the key role of the long P6 extension in Cbl-I cobalamin riboswitches appear to be uniquely replaced by the P2 arm in the B. subtilis riboswitch.
Structural model of the complete B. subtilis cobalamin riboswitch consisting of both aptamer and regulatory domains
The linker region between the aptamer and expression platform domains of the B. subtilis cobalamin riboswitch forms an unusual hairpin structure that includes several non-Watson-Crick base-pairs as part of its long stem. Although the region of the expression platform domain immediately distal to this hairpin is absent in the crystal structure, it is predicted to consist of two simple stacked stem-loops (25) (Figures 1 and 3), and so we were able to model confidently the remaining portion of the riboswitch to construct a complete structural model of the B. subtilis cobalamin riboswitch (Figure 6). For the modeling, we took advantage of the fact that the L9 loop of a symmetry related molecule forms a kissing loop interaction with the L5 loop, in a similar configuration as was observed in two previous cobalamin riboswitch structures that included parts of their regulatory domain (26). This allowed us to orient the modeled P13 stem with respect to the aptamer domain as well as part of the L13 loop forming the kissing loop interaction with L5. Furthermore, the two modeled stems are arranged to form a long stack, placing the 3′ end of the full-length model near the 5′ end of the riboswitch. The 3′ end of our experimental structure and the 5′ end of the modeled stems are close in space, a proximity that was imposed by the length and orientation of the stems due to the L5/L13 interaction, further supporting the arrangement of the modeled stems with respect to the experimental structure. In addition, the model also shows precisely how this complete assembly, when bound to adenosylcobalamin, leads to the formation of the third stem-loop (P14) in the expression platform, which is immediately followed by a poly-U segment. In the absence of the interaction with adenosylcobalamin, a conformational rearrangement has to occur that leads to the dissolution of the P14 stem, thus providing access upstream of the poly-U sequence at the 3′ end. For this to happen, the proposed structure has to respond to the absence of adenosylcobalamin by breaking some stable interactions, but the exact mechanism for this conformational transition is not apparent from the structure. Although the full B. subtilis cobalamin riboswitch model does not offer extensive clues on how the structure changes in the absence of ligand to allow transcription to proceed, the model does provide unique insight into the structural mechanism of translational control by Rho-independent termination upon binding to adenosylcobalamin.
Figure 6.
Model of the complete B. subtilis yvrC cobalamin riboswitch suggests structural basis of functional coupling between aptamer and expression platform domains for Rho-independent transcription termination. (A) Model of the full B. subtilis cobalamin riboswitch based on the crystal structure. The P13/L13 and P14/L14 stem–loops of the expression platform domain were modeled as stacked stem-loops and their position based on a symmetry related P8–P9/L9 hairpin, which mimics the KL interaction in the crystal. In the model, the position of the P13/P14 stem was not optimized. The adenosylcobalamin is shown as a space-filling model (orange). Based on the primary sequence, regions that have the potential to form complementary base-paring interactions are colored in either red or cyan. (B) The schematic diagram on the left was drawn based on the B. subtilis cobalamin riboswitch structural model and is color-coded as in (A). The KL interaction is denoted by dashed lines (gray). Based on sequence complementary, two potential models for the unbound or ligand-free state can be drawn. In both cases, dissolution of the P14 stem–loop directly upstream of the poly-U region needs to occur to avert premature transcription termination. In the top right model, melting of P13 and P14 leads to the formation of a new stem with elements from the old P13 and P14 and the disappearance of the anti-terminator stem just upstream of the poly-U region. In the bottom right model, base-pairing between elements of P2 and P13 lead to the melting of P2, P13 and P14. The mechanism for these potential structural rearrangements remains unclear.
DISCUSSION
Revisiting the classification of cobalamin riboswitches
Cobalamin riboswitches are currently grouped into two major classes based on their configuration of peripheral structural elements, which is thought to dictate their preference for associating with either adenosylcobalamin (Cbl-I) or methylcobalamin/hydroxocobalamin (Cbl-IIa) (26,27). However, a recent study has shown that the ligand selectivity of cobalamin riboswitches can be altered dramatically by even small changes in the peripheral elements, implying that it is possible some cobalamin riboswitches might not act strictly as a sensor for one natural derivative of cobalamin over others (28). These recent data suggest that there is likely a complex interplay between the peripheral structural elements and the ligand binding pocket situated in the conserved core domain (28). This also raises the possibility that ligand selectivity might fall on an affinity spectrum for each cobalamin derivative and thus ultimately be related to the tendency of formation of a ligand binding pocket that favors one derivative over another. Although promiscuous ligand binding has been previously observed in other riboswitch classes, such as the yjdF riboswitch (50), which binds to a large number of azaaromatic compounds, and the TPP riboswitch (51), which binds to thiamine pyrophosphate or pyrithiamine pyrophosphate, it is unclear if the B. subtilis cobalamin riboswitch shares a similar mechanism for ligand selectivity.
In contrast to most cobalamin riboswitches that classify clearly as either Cbl-I or Cbl-II based on their predicted secondary structures, a cobalamin riboswitch found in B. subtilis consists of peripheral structural elements that are a blend of both Cbl-I and Cbl-II class elements. This makes the classification of this riboswitch not as straightforward as for other cases. In the B. subtilis cobalamin riboswitch structure, the ligand binding pocket is buttressed by the peripheral P2 arm instead of by a long P6 extension (Figure 5), a characteristic of adenosylcobalamin-binding Cbl-I riboswitches. Furthermore, the observed ligand binding promiscuity precludes ligand specificity as an alternate means to group the riboswitch in one or another class. The B. subtilis cobalamin riboswitch binds adenosylcobalamin and responds to it in vivo (25), but in vitro it can also bind methylcobalamin and hydroxocobalamin, although the difference in affinity among the three ligands is not known as it was not possible to incorporate the ligand to the apo riboswitch. It is possible that the unusual structural and functional properties of the B. subtilis cobalamin riboswitch are sufficiently unique to justify classifying it as the first and only known member of a novel cobalamin riboswitch subclass or that the existing classification scheme needs to be expanded to include the unusual B. subtilis cobalamin riboswitch.
RNA structural adaptability as basis of promiscuous ligand derivative binding
Based on EMSA and SEC-MALS data, we determined that the B. subtilis cobalamin riboswitch binds to adenosylcobalamin, hydroxocobalamin, and methylcobalamin, but not to cyanocobalamin. We also concluded that the riboswitch likely binds to adenosylcobalamin in a different conformation than when bound to hydroxocobalamin or methylcobalamin. To explain this apparent structural adaptation, we compared its structure to known Cbl-I and Cbl-IIa cobalamin riboswitches. Selectivity of the ligand is easy to explain for adenosylcobalamin, where the adenosyl moiety is recognized by entering a space created by the movement of key nucleotides and buttressed in different manners by peripheral elements. Cbl-IIa cobalamin riboswitches, which do not recognize adenosylcobalamin, do not have this opening, which is occupied by a riboswitch nucleobase instead (26). The mechanism of discrimination of hydroxocobalamin and methylcobalamin is not as clear and likely involves more subtle interactions between the riboswitch and the ligand. In Cbl-IIa cobalamin riboswitches, there are interactions between the J1/3 loop and the ligand (Figure 5, Panel B); these interactions are not present in Cbl-I or the B. subtilis cobalamin riboswitch as J1/3 is replaced by the P2 stem (Figure 5, Panels A and C). One possible explanation for the reduced selectivity in the B. subtilis cobalamin riboswitch is that the gap needed to accommodate the adenosyl group collapses by a rearrangement of the structure. For example, U105 could swing backward, thereby closing the gap and creating a continuous strand of stacked bases, as observed in Cbl-IIa molecules (Figure 5, Panels B and C). A simple rotation of the P2 arm in the B. subtilis riboswitch structure would disengage it from the ligand binding core, altering its structure and correspondingly orienting the T-loop formed by the proximal L2B loop into a position much like in Cbl-IIa cobalamin riboswitches, and thereby allow for hydroxocobalamin and methylcobalamin binding (52). As the P2 stem is necessary to maintain the structure around the binding site, it is possible that the P2 arm of the B. subtilis cobalamin riboswitch acts as an adaptive sensor that undergoes structural reconfiguration depending on which ligand the riboswitch accommodates into the ligand binding pocket. Given the likely association between the unique structural and functional properties of the B. subtilis cobalamin riboswitch and its relatively large P2 arm in combination with a diminutive P6 extension, it may be possible to use these structural features as restraints for potentially identifying a new cobalamin riboswitch subclass by secondary structure-based sequence analysis, as has been done before (53). With a simple structural rearrangement around the binding core, the B. subtilis cobalamin riboswitch could accommodate other related ligands. This rearrangement would alter the structure in a global manner, consistent with the biochemical observations. In contrast, Cbl-I cobalamin riboswitches may not exhibit the same degree of plasticity as the ligand binding site is formed with the aid of the much more complex P6 extension and disengagement of this region may not lead to a change in the adenosyl moiety binding pocket.
Structural model for a complete riboswitch utilizing Rho-independent transcription termination as mechanism for translational control
The structural model we propose for the entire B. subtilis cobalamin riboswitch is the first atomic model of a complete riboswitch that utilizes Rho-independent transcription termination as a mechanism for transcriptional control, a mechanism that is still not fully understood (54). Our model provides key insights into how this atypical cobalamin riboswitch in B. subtilis responds to adenosylcobalamin to repress gene expression of the entire operon through transcription termination. Furthermore, our model also highlights how there need not be extensive physical crosstalk between the aptamer and expression platform domains to affect gene regulation; in our model, the tertiary interaction between the L5 and L13 loops is sufficient to stabilize specific hairpin formation in the expression platform. As the L5/L13 interaction may depend on the presence of the ligand, suggested by its close proximity with the sugar-phosphate backbone of residues G161 and G162 in L13, it is also possible that in its absence the stabilization provided by the kissing loop interaction disappears, leading to a less structurally stable expression platform. Conversely, the formation of the L5/L13 interaction may also be necessary to anchor the P4/P5 coaxial stack in place relative to P6/P7, to position P13/P14 parallel to P1/P8/P9, and to enable the entire riboswitch to effectively encircle and clamp down on the ligand as a means to stabilize the ligand-bound state of the aptamer domain.
For transcription termination, it is unclear from the sequence and structure what changes need to occur to switch from the termination to the anti-termination conformation to allow access to the poly-U segment at the 3′ end. The stacked P13/P14 stems form what appears to be a stable region as there are 19 canonical Watson–Crick base-pairs and two non-canonical U·G wobble base-pairs. It has been assumed that in the presence of ligand this region would form a terminator, but in the absence of ligand it would rearrange to form an anti-terminator stem-loop that exposes the region 5′ of the poly-U segment (42) via alternative base-pairings. Sequence analysis of the region forming the P13 and P14 stems does show some possible alternative base-pairings, but none of them would lead to stems that could potentially be of comparable length and stability as the P13/P14 stem and that would expose the region upstream of the poly-U segment. Clearly, another set of interactions with the aptamer domain have to occur to offset the absence of base-pairing interactions. Interestingly, a region comprising the P2 and L2A regions (Figure 6) shows very high sequence complementarity to 10 nucleotides in the 5′ end of the P13 stem, suggesting that these two regions could interact to disrupt both the P2 and the P13/P14 stems. As the P2 stem is involved in stabilizing the cobalamin binding site, it is possible that this interaction could only occur in the absence of cobalamin binding, which would suggest a mechanism of regulation based on the presence or absence of cobalamin and a large conformational change with many alternative pairings and a different overall structure. A major drawback of this model is that the P2/L2A interaction would disrupt the P13 stem, but it may leave the P14 stem largely untouched, which could act as a transcription terminator. Overall, it is not clear from the structure and the sequence what alternative structure would form in the absence of cobalamin that would lead to the exposure of the poly-U segment. Additional experiments to probe the structure of the apo-riboswitch are needed to be able to discern the mechanism of control.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under the accession number 6VMY.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the beamline scientists at LS-CAT/Sector 21 at the Advanced Photon Source, Argonne National Laboratory, for their assistance and the Structural Biology Facility, Keck Biophysics Facility, and High Throughput Analysis Laboratory at Northwestern University for their support. We thank Bryan Dorsey and Lilien Voong for their suggestions and assistance.
Notes
Present address: Clarence W. Chan, Department of Pathology, The University of Chicago, 5841 S. Maryland Avenue, Chicago, IL 60637-1470, USA.
Contributor Information
Clarence W Chan, Department of Molecular Biosciences, Northwestern University, 2205 Tech Drive, Evanston, IL 60208-3500, USA.
Alfonso Mondragón, Department of Molecular Biosciences, Northwestern University, 2205 Tech Drive, Evanston, IL 60208-3500, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01 GM058443, R35 GM118108 to A.M.]. C.W. C. was supported by the Medical Scientist Training Grant [4T32 GM008152], the Molecular Biophysics Training Grant [5T32 GM008382], and the Achievement Rewards for College Scientists Foundation Fellowship. LS-CAT/Sector 21 at the Advanced Photon Source, Argonne National Laboratory was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility and the Keck Biophysics Facility is acknowledged. Funding for open access charge: NIH [R35 GM118108].
Conflict of interest statement. None declared.
REFERENCES
- 1. Garst A.D., Batey R.T.. A switch in time: detailing the life of a riboswitch. Biochim. Biophys. Acta. 2009; 1789:584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Serganov A., Nudler E.. A decade of riboswitches. Cell. 2013; 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breaker R.R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012; 4:a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Serganov A., Patel D.J.. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu. Rev. Biophys. 2012; 41:343–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Serganov A., Patel D.J.. Molecular recognition and function of riboswitches. Curr. Opin. Struct. Biol. 2012; 22:279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Serganov A., Patel D.J.. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007; 8:776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dann C.E. 3rd, Wakeman C.A., Sieling C.L., Baker S.C., Irnov I., Winkler W.C.. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007; 130:878–892. [DOI] [PubMed] [Google Scholar]
- 8. Grundy F.J., Henkin T.M.. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993; 74:475–482. [DOI] [PubMed] [Google Scholar]
- 9. Mandal M., Boese B., Barrick J.E., Winkler W.C., Breaker R.R.. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003; 113:577–586. [DOI] [PubMed] [Google Scholar]
- 10. Mandal M., Breaker R.R.. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004; 11:29–35. [DOI] [PubMed] [Google Scholar]
- 11. Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R.. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004; 306:275–279. [DOI] [PubMed] [Google Scholar]
- 12. Miranda-Rios J., Navarro M., Soberon M.. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl Acad. Sci. U.S.A. 2001; 98:9736–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A., Nudler E.. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002; 111:747–756. [DOI] [PubMed] [Google Scholar]
- 14. Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R.. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002; 9:1043. [DOI] [PubMed] [Google Scholar]
- 15. Nou X., Kadner R.J.. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J. Bacteriol. 1998; 180:6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sudarsan N., Lee E.R., Weinberg Z., Moy R.H., Kim J.N., Link K.H., Breaker R.R.. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008; 321:411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sudarsan N., Wickiser J.K., Nakamura S., Ebert M.S., Breaker R.R.. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003; 17:2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Winkler W., Nahvi A., Breaker R.R.. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002; 419:952–956. [DOI] [PubMed] [Google Scholar]
- 19. Winkler W.C., Cohen-Chalamish S., Breaker R.R.. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. U.S.A. 2002; 99:15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R.. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004; 428:281–286. [DOI] [PubMed] [Google Scholar]
- 21. Winkler W.C., Nahvi A., Sudarsan N., Barrick J.E., Breaker R.R.. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003; 10:701–707. [DOI] [PubMed] [Google Scholar]
- 22. Soukup J.K., Soukup G.A.. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004; 14:344–349. [DOI] [PubMed] [Google Scholar]
- 23. Wachter A., Tunc-Ozdemir M., Grove B.C., Green P.J., Shintani D.K., Breaker R.R.. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007; 19:3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lai E.C. RNA sensors and riboswitches: self-regulating messages. Curr. Biol. 2003; 13:R285–291. [DOI] [PubMed] [Google Scholar]
- 25. Nahvi A., Barrick J.E., Breaker R.R.. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004; 32:143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson J.E. Jr, Reyes F.E., Polaski J.T., Batey R.T.. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012; 492:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peselis A., Serganov A.. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat. Struct. Mol. Biol. 2012; 19:1182–1184. [DOI] [PubMed] [Google Scholar]
- 28. Polaski J.T., Webster S.M., Johnson J.E. Jr, Batey R.T.. Cobalamin riboswitches exhibit a broad range of ability to discriminate between methylcobalamin and adenosylcobalamin. J. Biol. Chem. 2017; 292:11650–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sambrook J., Russell D.W.. Molecular Cloning: a Laboratory Manual. 2001; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 30. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patel T.R., Chojnowski G., Astha, Koul A., McKenna S.A., Bujnicki J.M.. Structural studies of RNA-protein complexes: A hybrid approach involving hydrodynamics, scattering, and computational methods. Methods. 2017; 118:146–162. [DOI] [PubMed] [Google Scholar]
- 32. Powell H.R. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 1999; 55:1690–1695. [DOI] [PubMed] [Google Scholar]
- 33. Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr. D. Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W.. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D. Biol. Crystallogr. 2003; 59:2023–2030. [DOI] [PubMed] [Google Scholar]
- 36. Tickle I.J., Flensburg C., Keller P., Paciorek W., Sharff A., Vornhein C., Bricogne G.. STARANISO. 2018; Cambridge: Global Phasing Ltd. [Google Scholar]
- 37. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 39. Roversi P., Irwin J., Bricogne G.. A Bayesian approach to high-resolution X-ray crystallography: accurate density studies with program buster. Acta Crystallogr. A. 1996; 52:C343–C343. [Google Scholar]
- 40. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schrodinger, L.L.C. 2015; Version 1.8 edn.
- 42. Vitreschak A.G., Rodionov D.A., Mironov A.A., Gelfand M.S.. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003; 9:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wickiser J.K., Winkler W.C., Breaker R.R., Crothers D.M.. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005; 18:49–60. [DOI] [PubMed] [Google Scholar]
- 44. Gilbert S.D., Reyes F.E., Edwards A.L., Batey R.T.. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009; 17:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matyjasik M.M., Batey R.T.. Structural basis for 2′-deoxyguanosine recognition by the 2′-dG-II class of riboswitches. Nucleic Acids Res. 2019; 47:10931–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peselis A., Serganov A.. ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands. Nat. Chem. Biol. 2018; 14:887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sherlock M.E., Sadeeshkumar H., Breaker R.R.. Variant bacterial riboswitches associated with nucleotide hydrolase genes sense nucleoside diphosphates. Biochemistry-US. 2019; 58:401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chan C.W., Chetnani B., Mondragon A.. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip. Rev. RNA. 2013; 4:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krasilnikov A.S., Mondragon A.. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003; 9:640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li S.S., Hwang X.Y., Stav S., Breaker R.R.. The yjdF riboswitch candidate regulates gene expression by binding diverse azaaromatic compounds. RNA. 2016; 22:530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Serganov A., Polonskaia A., Phan A.T., Breaker R.R., Patel D.J.. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006; 441:1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Polaski J.T., Kletzien O.A., Drogalis L.K., Batey R.T.. A functional genetic screen reveals sequence preferences within a key tertiary interaction in cobalamin riboswitches required for ligand selectivity. Nucleic Acids Res. 2018; 46:9094–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weinberg Z., Nelson J.W., Lunse C.E., Sherlock M.E., Breaker R.R.. Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc. Natl Acad. Sci. U.S.A. 2018; 115:Eb326–Eb326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones C.P., Ferre-D’Amare A.R.. Long-range interactions in riboswitch control of gene expression. Annu. Rev. Biophys. 2017; 46:455–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under the accession number 6VMY.