Abstract
Methylation of miRNAs at the 2′-hydroxyl group on the ribose at 3′-end (2′-O-methylation, 2′Ome) is critical for miRNA function in plants and Drosophila. Whether this methylation phenomenon exists for mammalian miRNA remains unknown. Through LC–MS/MS analysis, we discover that majority of miR-21-5p isolated from human non-small cell lung cancer (NSCLC) tissue possesses 3′-terminal 2′Ome. Predominant 3′-terminal 2′Ome of miR-21-5p in cancer tissue is confirmed by qRT-PCR and northern blot after oxidation/β-elimination procedure. Cancerous and the paired non-cancerous lung tissue miRNAs display different pattern of 3′-terminal 2′Ome. We further identify HENMT1 as the methyltransferase responsible for 3′-terminal 2′Ome of mammalian miRNAs. Compared to non-methylated miR-21-5p, methylated miR-21-5p is more resistant to digestion by 3′→5′ exoribonuclease polyribonucleotide nucleotidyltransferase 1 (PNPT1) and has higher affinity to Argonaute-2, which may contribute to its higher stability and stronger inhibition on programmed cell death protein 4 (PDCD4) translation, respectively. Our findings reveal HENMT1-mediated 3′-terminal 2′Ome of mammalian miRNAs and highlight its role in enhancing miRNA’s stability and function.
INTRODUCTION
Accumulating evidence from us (1–4) and others (5–7) has demonstrated that extracellular miRNAs in human and animal serum, plasma and other body fluids display considerable stability and can be readily detected by Solexa sequencing, qRT-PCR and northern blot assays. The unique expression pattern of circulating miRNAs can serve as effective biomarker for various diseases including cancer (3,6,8). Our study of the mechanisms underlying the stability of extracellular miRNAs, particularly miRNAs in exosomes secreted by various cells, further showed that, in addition to the protection by miRNA-associated protein complexes such as Argonaute-2 (AGO2), modification of miRNAs might be also contribute to miRNA stability (9). However, the nature of such modification of miRNAs in mammalian cells remains largely unknown.
There are different kinds of modification of miRNAs, including 2′-O-methylation (10) and m7G (11), which all can affect miRNA biogenesis and functions thereby playing a role in miRNA-based gene regulation network (10,11). Methylation of miRNAs at the 2′-hydroxyl group on the ribose at the last nucleotide (3′-terminal 2′Ome) has been reported in plants (10) and Drosophila (12), which is crucial for enabling miRNAs to execute their function. Artificially introducing 3′-terminal 2′Ome in small interfering RNAs (siRNAs) further showed that such methylation can increase the stability of siRNAs in serum without affecting their RNA interference activities in mammalian cells (13). However, although piRNAs in animals and human are 2′-O-methylated at the terminal ribose of their 3′-end (14), whether such methylation occurs in mammalian miRNAs still remains elusive (15). In plants and Drosophila, 3′-terminal 2′Ome of miRNAs are mediated by a methyltransferase HEN1 (15). Although HENMT1, an HEN1 analogy, is identified in mammalian cells, it has been reported to be restrictively expressed in testis and only involved in 3′-terminal 2′Ome of animal and human piRNA (16). Whether HENMT1 is responsible for the possible methylation of mammalian miRNAs still remains unknown.
In the present study, we employed The Liquid Chromatograph Triple Quadrupole Mass Spectrometer (LS-MS/MS), as well as Solexa sequencing, qRT-PCR and northern blot assays following oxidation and β-elimination (Oxid/β-elim) procedure (10,17,18), to address whether 3′-terminal 2′Ome is found in mammalian miRNAs isolated from cancerous or non-cancerous tissues. Results derived from miRNAs directly isolated from non-small cell lung cancer (NSCLC) and the paired distal non-cancerous lung tissues clearly show that miRNAs in NSCLC and non-tumor lung tissues display different patterns of 3′-terminal 2′Ome. In NSCLC lung tissue, miR-21-5p is heavily methylated whereas it is not in non-tumor lung tissues. Our studies further identify HENMT1 as the methyltransferase responsible for 3′-terminal 2′Ome of mammalian miRNAs and highlight the role of 3′-terminal 2′Ome in enhancing miRNA stability and function.
MATERIALS AND METHODS
Tissue preparation
Cancerous and the distal non-cancerous lung tissue samples were obtained from NSCLC patients with the newly diagnosis without receiving any treatment at Nanjing Drum Tower Hospital of Nanjing University. This study has been approved by the Ethics Committee of the Nanjing Drum Tower Hospital of Nanjing University, and written informed consent has been obtained from all patients. HENMT1 knockout mice were purchased from Model Animal Research Center of Nanjing University (Nanjing, China). All experiments were performed in accordance with relevant guidelines and regulations. The other human tissues (heart, liver, lung, kidney, stomach, pancreas, spleen, colon and testis) from tissue bank (Nanjing University School of Life Sciences) were also included in this study.
RNA extraction, purification and detection
The tissues were frozen in liquid nitrogen and ground to a powder. Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer's instruction. To extract small RNA sized at 18–24nt, the large RNAs were removed from total RNA by selective precipitation with 5% polyethylene glycol 8000 in 0.5 M NaCl (final concentration). Small RNAs were then precipitated from the supernatants by the addition of 3 volumes of absolute ethanol and 0.1 volume of 3 M sodium acetate (pH5.2) with incubation overnight at −80°C. Then the small RNAs were separated by 15% PAGE containing 7 M urea (300 V, 35 min). The gel was stained with SYBR Green I solution and visualized by gel documentation system (Wealtec). Small RNAs sized at 18–24nt were excised from the gel and then extracted using 0.3 M NH4Cl and precipitated with glycogen and ethanol. The quality of the purified small RNAs was determined on an Agilent 2100 Bio-analyzer using a small RNA Kit (Agilent).
Northern blot analysis
Northern blot was performed as previously described (10). Briefly, small RNA (1.0 μg) and miRNA (0.5 μg) prepared from lung tissues or cultured cells were electrophoresed through a 7.5% polyacrylamide gel containing 7 M urea (300 V, 30 min). Then nucleic acids were transferred onto positively charged nylon membranes (Roche) in 1× Tris–borate–EDTA buffer at 100 mA for 1 h. Subsequently, the membranes were air-dried and UV crosslinked. The hybridization procedure was carried out with the indicated 3′-digoxigenin (DIG) labeled probes (Supplement Table S1) at 8 nM in DIG Easy Hyb buffer (Roche) at 42°C for 16 h. After hybridization, the blots were washed and detected using the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche). For oxidation/β-elimination, small RNAs or miRNAs were suspended in 1× borate/borax buffer with or without NaIO4 (final concentration 25 mM) (Sigma-Aldrich) and incubated for 30 min at room temperature in the dark. After adding 1/10 volume of 100% glycerol, the samples were incubated further for 10 min at room temperature in the dark. After purification, the RNA samples were re-suspended in 1× borate/borax buffer and adjusted with NaOH to a final concentration of 0.1 mM. The samples were incubated for 90 min at 45°C followed by purification and northern blots analysis.
AGO2 binding assay
The same amount of synthetic Cy3-labeled miR-21-5pCH3 and Cy5-labeled miR-21-5p (or synthetic Cy5-labeled miR-21-5p CH3 and Cy3-labeled miR-21-5p) were transfected into A549 cells. After 24 h, cells were lysed in lysis buffer and AGO2 was immunoprecipitated by anti-AGO2 antibody according to previous studies (9,19). Briefly, the cells were lysed with lysis buffer (20 mm Tris–HCl, 150 mm NaCl, 0.5% Nonidet P-40, 2 mm EDTA, 0.5 mm DTT, 1 mm NaF, 1 mm PMSF and 1% protease inhibitor cocktail from Sigma, pH 7.5) for 30 min on ice. The lysates were cleared by centrifugation (16 000 × g) for 10 min at 4°C and then immunoprecipitated with anti-AGO2 antibody (ab32381, Abcam) or normal IgG followed by protein G-Agarose beads. After extensive washing, the fluorescence intensities of Cy3 and Cy5 in the immunoprecipitated complex were detected, respectively.
Oxidation, qRT-PCR and Solexa sequencing
Small RNAs were suspended in 1× borate/borax buffer with or without NaIO4 (final concentration: 25 mM) (Sigma-Aldrich) and incubated for 30 min at room temperature in the dark. After adding 1/10 volume of 100% glycerol, samples were further incubated for 10 min at room temperature in the dark. After purification, RNA samples were precipitated using ethanol and used as input for Solexa sequencing by Illumina 3000 or qRT-PCR by all in one miRNA First Strand cDNA Synthesis Kit (GeneCopoeia, Guangzhou, China), containing poly(A) polymerase and RTase Mix, according to the manufacturer's instruction (20). After obtaining cDNAs, rTaq (Takara), dNTP (Takara), EvaGreen (Invitrogen), sense primer and Universal Adaptor PCR Primer (GeneCopoeia) (Supplemental Table S1) were added for qRT-PCR detection. The program was as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Each cDNA sample was tested in triplicate.
Western blotting
Western blot analysis was performed to assess the HENMT1 expression in tissue and cells. Proteins were extracted using RIPA buffer (Beyotime, Shanghai, China) with protease inhibitors (Roche cOmplete ULTRA tablets and Merck proteas inhibitor cocktail set III). The PVDF membranes (BioRad, USA) carrying the protein extract after blotting were blocked at room temperature for 1 h in blocking solution containing 5% dry milk in TBST. Membranes were incubated with primary antibodies in a solution containing 5% bovine serum albumin (BSA) in TBST: anti-HENMT1 primary antibody (HPA028464, Sigma) at 1 μg/ml and rabbit anti-GAPDH antibody diluted 1:1000 (Catalog No. ab9485-100, Abcam) for normalization. Membranes were then incubated over night at 4°C. After three washes with TBST for 10 min each, membranes were then incubated for 1 h at room temperature in HRP-conjugated secondary antibodies diluted 1:10 000 in a solution containing 5% dry milk in TBST. Membranes were illustrated with Clarity ECL substrate (BioRad).
HENMT1 immunofluorescence and immunohistochemistry
Immunofluorescence microscopy was used to identify the subcellular localization of HENMT1 in A549 cells. Cells were cultured on four-well chamber slides. At the time of harvest, cells were fixed with 4% paraformaldehyde and then permeabilized with 0.01% Triton X-100 for 10 min and subsequently probed with antibodies against HENMT1 followed by incubation with fluorescent-tagged secondary antibodies (488 nm). All samples were treated with DAPI dye for nuclear staining (358 nm). For confocal microscopy, the Nikon C2 Plus confocal microscope was used. HENMT1 immunohistochemistry was performed in cancerous and distal non-cancerous lung tissue sections as described previously with minor modification (21). Briefly, the tissues sections were fixed in 4% paraformaldehyde and washed in PBS containing 0.01% Triton X-100. The anti-HENMT1 primary antibody (HPA028464, Sigma) was diluted in blocking solution (25 μg/ml) and added to the lung tissue slides and incubated at 4°C overnight. An isotype-matched mAb (control IgG) with no specific affinity to HENMT1 was used as the negative control. The Image-Pro Plus software (version 5.0) was performed to calculate the average integrated optical density (IOD) of HENMT1 IHC staining in cancerous and non-cancerous lung tissue.
Cell transfection with plasmid
We designed three gRNAs for HENMT1 via the lentiviral transfection system (GenScript) and tested the effects of three gRNAs on gene silence by Western blotting prior to experiments. The gRNA with the best effect of gene silence for HENMT1 was selected for the following experiments. The gRNA was cloned into plentiCRISPRV2 plasmid and the sequence was as follows: gRNA#1: TGGTGTGCTGATGACAATCATGG; gRNA#2: GAGGATAAATTACGATGGAGAGG; gRNA#3: TACCGCTGTCTGTATAGTGGAGG. Cells were seeded in six-well plates or 10-mm dishes, and then were transfected with Lipofectamine 3000 (Invitrogen) the following day according to the manufacturer's instructions. Cells were treated with 2 μg puromycin for 7 days, and the remaining cells were then re-plated in a 96-well plate (single cell per well) for single-cell colony formation. The resulting colonies were sub-cultured and collected for western blotting to confirm the deletion or for the following assays.
In vitro miRNA stability assay
To monitor the effect of 3′-terminal 2′Ome on the stability of mammalian miRNAs, A549 and A549KO (HENMT1-KO) cells were treated by 5 μg/ml actinomycin D to block the transcription of new miR-21-5p, and then assessed miR-21-5p level at 0, 3, 6, 9, 12, 15, 18, 21 and 24 h post-treatment by qRT-PCR, respectively.
Luciferase reporter assay
A whole sequence of human PDCD4 and ULK1 3′-UTR was synthesized and inserted into the pMIR-report plasmid (Ambion) by GenScript Corporation (Nanjing, China). Efficient insertion was confirmed by sequencing. For the luciferase reporter assays, cells were cultured in 24-well plates, and each well was transfected with 0.5 μg firefly luciferase reporter plasmid, 0.5 μg β-galactosidase expression vector (Ambion), and equal amount of control plasmid or HENMT1 shRNA plasmid using Lipofectamine 3000 (Invitrogen). The β-galactosidase vector was used as a transfection control. Cells were assayed using the luciferase assay kits (Promega, Madison, WI, USA) 24 h after transfection. The reported data represented three independent experiments.
Microscale thermophoresis
A total of 10 nM of 5′-Cy5-miR-21-5p or 5′-Cy5-miR-21-5pCH3 were incubated with increasing concentrations (1–100 nM) of GST-AGO2 (Ag1032, ProteinTech) in a 10 μl reaction containing 5 mM DTT, 0.1 mg/ml BSA, 3% (w/v) Ficoll-400 and 5% (v/v) glycerol at 37°C for 30 min. RNA–protein complexes were then analyzed by microscale thermophoresis (MST) on a Monolith NT.115 system (NanoTemper Technologies). Results were analyzed using Prism software (GraphPad Prism for Windows, GraphPad Software, San Diego, CA, USA).
Purification of miR-21-5p and miR-26-5p
The total small RNAs from human cancer lung tissues and paired non-tumor lung tissues were extracted by miRcute miRNA Isolation Kit (Tiangen Biotech, Beijing) according to the manufacturer's instructions. The purification of miR-21-5p and miR-26-5p was carried out with a two-step strategy. Firstly, total small RNA was incubated with the biotinylated DNA oligonucleotide which is antisense to miR-21-5p or miR-26-5p with two extra A residues at its 5′-end to ensure that its molecular mass is larger than that of miR-21-5p and miR-26-5p in 0.5 × SSC at 50°C for 15 h (miR-21-5p probe: 5′-AATCAACATCAGTCTG ATAAGCTA-3′; miR-26-5p probe: 5′-AAAGCCTATCCTGGATTACTTGAA-3′). The biotinylated DNA was then captured by streptavidin magnetic particle (Roche) and washed with 0.5 × SSC. The miR-21-5p or miR-26-5p was eluted with DD water by incubating at 70°C for 5 min. Secondly, 1μl 21-bases ssDNA (10μM, complementary to human miR-21-5p:5′-TCAACATCAGTCTGATAAGCTA-3′ or miR-26-5p:5′ -AGCCTATCCTGGATTACTTGAA-3′) in buffer (20 nM Tris–HCl, 100 mM KCl, 5 mM MgCl2, pH 7.5) was added for every 5μg purified miR-21-5p or miR-26-5p. The mixture was incubated in 95°C for 2 min, followed by slowly decreasing to 25°C. To eliminate the unpaired RNAs, 1000 units Nuclease S1 (Thermo Fisher) was added to the solution. The reaction was allowed at room temperature for 30 min and terminated by 70°C incubation for 10 min. The 21-bases ssDNA was then degraded by adding 4 units DNaseI (New England Biolabs) and incubating at 37°C for 30 min. The remained RNAs (purified miR-21-5p or miR-26-5p) were then extracted by RNA Clean & Concentrator-5 kit (Zymo Research) according to the manufacturer's instructions. The quality of the purified miR-21-5p or miR-26-5p was determined by RNA sequencing.
Enzymatic digestion of small RNA (18-24nt) or miR-21-5p and LC-MS/MS
1 μg of small RNAs (18–24nt) or miR-21-5p was degraded with 0.2 unit nuclease P1 (Sigma-Aldrich) at 50°C for 3 h in 50 mM NH4OAc (pH5.3). For further digestion into mononucleosides, 0.04 unit phosphodiesterase I (Sigma-Aldrich) was added and incubated at 37°C for another 2 h (pH 8.8). Finally, small RNAs or miR-21-5p was incubated with 2 unit alkaline phosphatase (Takara) for 2 h at 37°C. The proteins in nucleoside digests were removed by Amicon® Ultra 3K device with centrifugation in 12 000 × g for 30 min. Subsequently, the metal ion in hydrolysates was removed by HyperSep™ Hypercarb™ SPE (ThermoFisher). Hydrolysates were-suspended with 80% acetonitrile (containing 0.1% formic acid) for LC-MS/MS analysis. These nucleosides were subjected to LC–MS/MS analysis using AB SCIEX TripleTOF® 4600 in positive ion mode.
Methylation assay
The methylation assay is carried out in a reaction mixture (total volume: 20 μl) containing 25 mM Tris–HCl (pH8.0), 50 mM KCl, 2.5 mM MgCl2, 0.05 mM EDTA, 2.5% glycerol, 5 mM DTT, 20 μM AdoMet, 10μM synthetic miR-21-5p and 5 μM recombinant HENMT1 proteins. The genes encoding full-length HENMT1 was cloned into the pET15b vector with an N-terminal His tag and transfected into BL21 (DE3) Escherichia coli cells. Recombinant HENMT1 protein was generated and purified as previously described (22). The mixtures were incubated at 37°C for 40 min. The RNAs were then extracted by RNA Clean & Concentrator-5 kit (Zymo Research) according to the manufacturer's instructions.
miRNA degradation Assay
For cleavage of miRNAs by PNPT1, 10 μg miRNA samples were incubated with 5 μl Recombinant Human PNPT1 protein (ab202628, abcam) at 37°C from 0.5 to 2 h. The RNA was re-purified using the RNA Clean & Concentrator-5 kit (Zymo Research) according to the manufacturer's instructions.
Statistical analysis
Data derived from at least three independent experiments were presented as the mean ± SEM. Statistical comparisons between two groups were performed by Student's t-test. P value < 0.05 was considered statistically significant.
RESULTS
Detection of 3′-terminal 2′Ome in human miRNAs
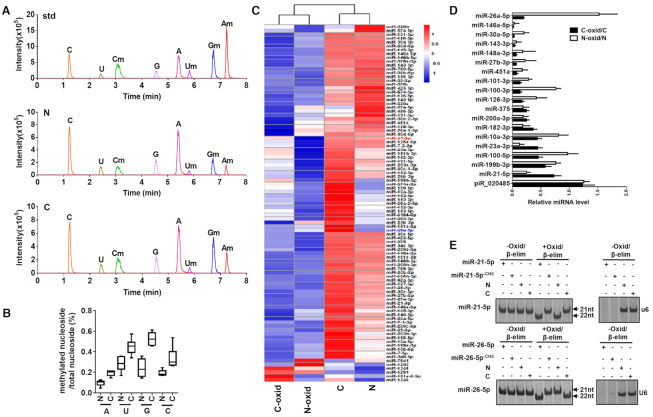
To characterize the modifications present in human miRNAs, we performed liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis of human miRNAs following enzymatic digestion (23). Small RNAs (18–24nt) were extracted from non-small cell lung cancer (NSCLC) and the paired distal non-cancerous lung tissues. RNAs were first digested by nuclease PI, phosphodiesterase I and alkaline phosphatase, and then subjected to LC–MS/MS analysis (Supplementary Figure S1). LC–MS/MS parameters for authentic nucleotides are listed in Supplementary Table S2. As shown in Figure 1A, LC–MS/MS chromatogram in MRM mode revealed the authentic nucleosides from the synthetic nucleosides (standard, std), small RNAs from NSCLC (C) and the paired non-cancerous lung tissues (N). Reference to synthetic standard nucleosides, small RNAs in both NSCLC and non-cancerous lung tissues displayed 2′Ome on nucleotides regardless of the base (A, G, T, C). Normalized to the total number of nucleotides, the percentages of 2′Ome on four nucleotides are generally higher in NSCLC lung tissue than non-cancerous lung tissue (Figure 1B).
Figure 1.
Detection of 2′Ome of human miRNAs at the 3′-end. (A) LC−MS/MS chromatogram in MRM mode of the synthetic nucleosides (standard, std) and authentic nucleosides isolated from non-cancerous lung tissue (N) and NSCLC lung tissue (C). (B) Comparison of nucleoside methylation in the indicated small RNAs (18–24nt) from non-cancerous lung tissue (N) and NSCLC lung tissue (C). (C) Heatmap of 2′Ome at the 3′-end of human miRNAs in non-cancerous lung tissue (N) and NSCLC lung tissue (C) after oxidation procedure. N-oxid: non-cancerous lung tissue with oxidation. C-oxid: NSCLC lung tissue with oxidation. (D) Detection of 2′Ome of human miRNAs at the 3′-end by qRT-PCR after oxidation procedure. (E) Detection of 2′Ome of human miRNAs at the 3′-end by northern blotting after Oxid/β-elim procedure.
To further test whether miRNAs in human tissue are methylated at 3′-end, we employed oxidation procedure (10). As previously reported (10), RNAs methylated at 3′-end are protected from oxidation, while the 3′-ends of non-methylated RNAs will be oxidized into dialdehyde, thus losing the capacity to be ligated in following ligation reactions (Supplementary Figure S2a). With a preserved 3′ methylated end, miRNAs can yield cDNA which can then be subjected to Solexa sequencing. Thus, only RNA that is methylated at the 2′OH group on the ribose of nucleotide at the 3′-end contributes to the cDNA library which is then Solexa sequenced. As all piRNAs are methylated at the 2′OH group on the ribose at the 3′-end (14,18), so the process of oxidation should not change piRNA contribution to the cDNA library. Indeed, our results confirmed that the contribution to the DNA library of piRNA was not affected by oxidation procedure (Supplementary Figure S2b and Table S3). In contrast, the counts of miRNAs in both NSCLC and non-cancerous lung tissues were significantly reduced after oxidation procedure (Supplementary Figure S2c). In line with this, we also observed that the sizes of lung tissue small RNAs were generally decreased, with a peak shift from 22nt to 20nt, following oxidation procedure (Supplementary Figure S2d). In agreement with previous reports (15), our results suggest that the majority of mammalian miRNAs may be not methylated at the 3′-end. However, several human miRNAs extracted from lung tissues were resistant to oxidation, and their levels detected by Solexa sequencing were not affected by oxidation treatment. As shown in heat-map of miRNA 2′Ome at the 3′-end (Figure 1C), considerable amount of miRNAs from both normal lung and NSCLC lung tissues were still detected by Solexa sequencing following oxidation procedure (SRA database with the accession numbers PRJNA575072), suggesting that these miRNAs in human lung tissues likely possess 3′-terminal 2′Ome. In fact, NSCLC and non-cancerous lung tissues displayed different 2′Ome patterns in miRNAs, which was validated by miRNA qRT-PCR assay after oxidation procedure (Figure 1D). As piRNA piR_020485 possesses 3′-terminal 2′Ome, we detected piR_020485 by qRT-PCR assay as a positive control in this experiment. As expected, level of piR_020485 was not affected by oxidation procedure (Supplementary Figure S3a). We also treated the synthetic unmethylated piRNA-020485 and methylated piRNA_020485CH3 with Oxid/β-elim procedure and assessed the piRNA by high-resolution denaturing PAGE (18). As shown in Supplementary Figure S3b, Oxid/β-elim procedure removed the 3′-terminal nucleoside of unmethylated piRNA_020485 but not methylated piRNA_020485CH3. In a similar fashion, synthetic unmethylated miR-21-5p and miR-26-5p (without any methyl groups) or methylated miR-21-5pCH3 and miR-26-5pCH3 (with a 2′-O-methyl on the last nucleotide at 3′-end) were also subjected to Oxid/β-elim treatment (Figure 1E). Whereas the unmethylated synthetic miRNAs (miR-21-5p, miR-26-5p) exhibited a mobility shift after Oxid/β-elim procedure, as anticipated the methylated miRNAs (miR-21-5pCH3, miR-26-5pCH3) did not exhibit a shift (Figure 1E). While our findings suggest that the majority of miRNAs in the lung are not methylated, we did observe two notable exceptions: miR-21-5p from NSCLC lung tissue and miR-26-5p from the paired non-cancerous lung tissue. Northern blot analysis also showed no shift of miR-21-5p in NSCLC lung tissue and miR-26-5p in non-cancerous lung tissue after Oxid/β-elim procedure, suggesting that both miR-21-5p in NSCLC tissue and miR-26-5p in non-cancerous lung tissue possess 3′-terminal 2′Ome (Figure 1E).
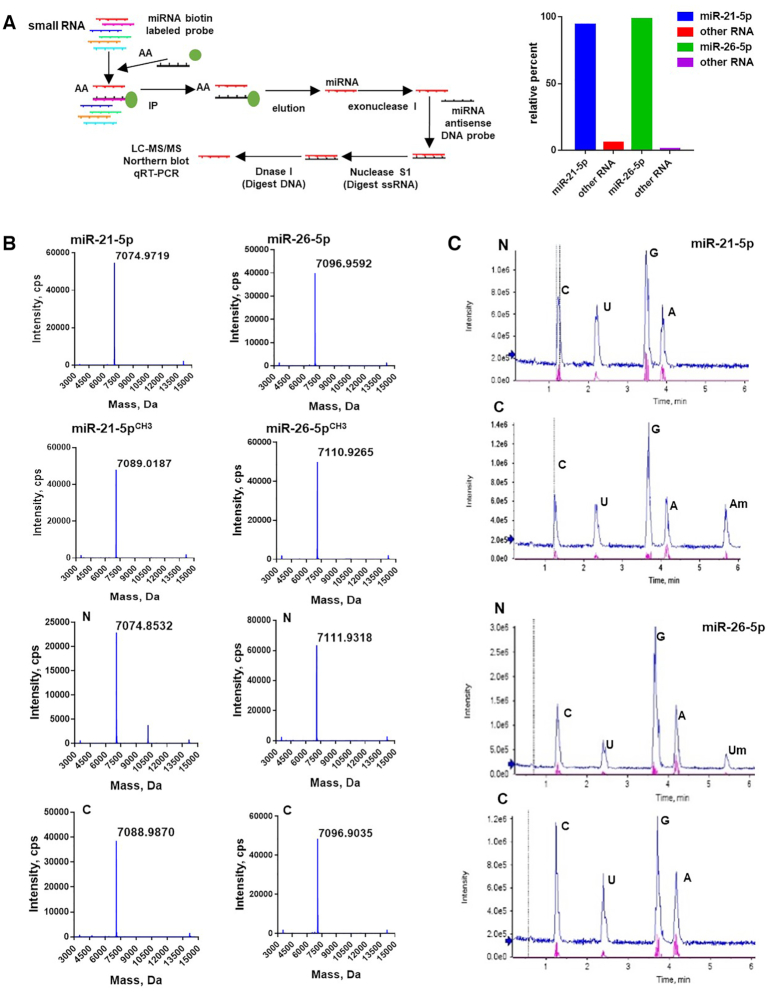
To further verify that the differential methylation of miR-21-5p in cancerous versus non-cancerous tissue, we employed a selective sequential enzymatic digestion strategy to isolate miR-21-5p directly from NSCLC and the paired distal non-cancerous lung tissues. As shown in Figure 2A, left, we extracted small RNA (<25nt) from both NSCLC and non-cancerous lung tissue. Firstly, the miR-21-5p was isolated from lung cancer tissues and non-cancerous lung tissue with a biotinylated antisense DNA oligonucleotide. The eluted miRNAs from the immunoprecipitates were then thoroughly mixed with single-strand oligonucleotides (ssDNA) that are completely complementary to miR-21-5p. The temperature was adjusted (heating to 95°C and then cooled to 25°C at a rate of 0.1°C/s) so that complementary stretches of nucleotides would anneal. We then eliminated unpaired nucleotides first by digestion with Nuclease S1 and followed by digestion with DNase1. Solexa sequencing showed that the final purified small RNA product contained more than 99% of miR-21-5p (Figure 2A, right). Finally, the purified miR-21-5p from both cancerous and non-cancerous lung tissue was then subjected to mass spectrometry analysis to determine the molecular mass and LC-MS/MS to determine nucleoside modification, respectively. As shown in mass spectrometry analysis (Figure 2B), purified miR-21-5p from non-cancerous lung tissue display only one major peak with a mass of 7074.8532 Da, which represents the unmethylated miR-21-5p. In contrast, in addition to a small peak of 7074.8532 Da, a significantly higher peak with a mass of 7088.9870 Da is detected in miR-21-5p purified from NSCLC lung tissue, which represents the miR-21-5p with 3′-terminal 2′Ome (Figure 2B). These results are consistent with the results from the LC–MS/MS analysis supporting that majority of miR-21-5p is methylated in cancerous tissue but is unmethylated in non-cancerous lung tissue (Figure 2C). As shown in Figure 2C, LC–MS/MS analysis confirmed the modification of miR-21-5p in NSCLC lung tissues as 3′-terminal 2′Ome. Methylation on adenosine (Am) was detected in the miR-21-5p from NSCLC but not paired non-cancerous lung tissue. The miR-26-5p was also analyzed by the same procedure. In agreement with previous result, we detected the modification of miR-26-5p in non-cancerous lung tissue (Figure 2B and C). Moreover, after oxidation procedure, we performed qRT-PCR assay to measure the methylation level of miR-21-5p in the serum derived from healthy volunteers or NSCLC patients before and after surgery. We found that the methylation levels of miR-21-5p were significantly higher in serum samples from NSCLC patients than in those from healthy volunteers (Figure 3a), while the methylation levels of miR-21-5p in NSCLC sera were markedly decreased after surgery to remove the cancer (Figure 3B). As miR-21-5p has been reported to act as an oncomiR in different cancers (24), we determined whether miR-21-5p is also methylated in gastric cancer and colon cancer. The level of miR-21-5p in cancer and non-cancerous tissues were detected by qRT-PCR after oxidation, and the results suggest that miR-21-5p in gastric cancer and colon cancer likely possesses the same methylation modification as in lung cancer (Supplementary Figure S4).
Figure 2.
Detection of 3′-terminal 2′Ome of miR-21-5p and miR-26-5p in NSCLC and non-cancerous lung tissues. (A) Left, depict of experimental strategy for isolating miR-21-5p and miR-26-5p from NSCLC and non-cancerous lung tissues. Right, purity of isolated miR-21-5p or miR-26-5p examined by Solexa sequencing. (B, C) Detection of 3′-terminal 2′Ome of miR-21-5p or miR-26-5p in NSCLC (C) and non-cancerous lung tissues (N) by Mass spectrometry (B) and LC–MS/MS (C).
Figure 3.
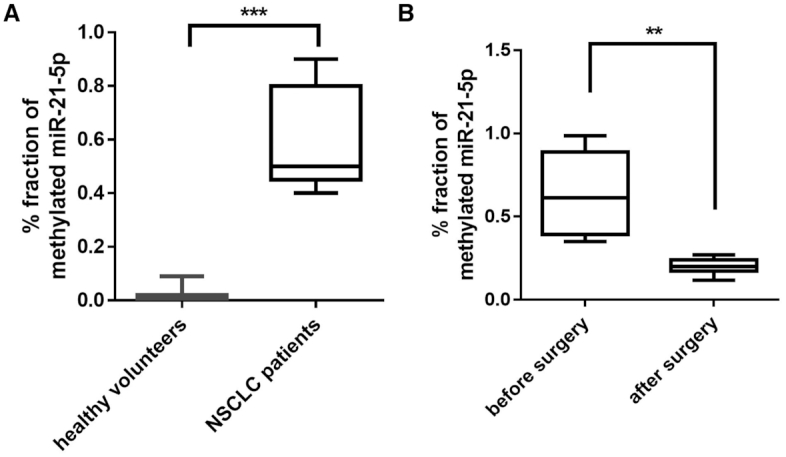
Level of methylated miR-21-5p in sera from NSCLC patients and healthy donors. (A) Elevation of methylated miR-21-5p level in serum samples from NSCLC patients (n = 21) than that from healthy donors (n = 11). (B) Decrease of methylated miR-21-5p level in serum samples from NSCLC patients (n = 20) before and after tumor removal by surgery. **P < 0.01, ***P < 0.001.
3′-Terminal 2′Ome of mammalian miRNAs is methylated by HENMT1
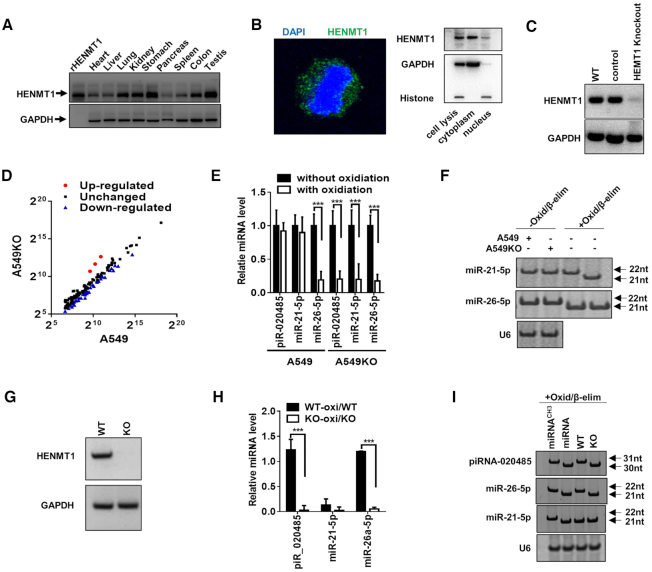
In plants, the 3′-terminal 2′Ome of miRNAs and siRNAs are mediated by the methyltransferase HEN1 (10). HEN1 homologs in mammalian cells, particularly HENMT1 (16,25), have been reported to methylate piRNAs in animals and AGO2-associated small RNAs in Drosophila (15). However, although in vitro biochemical assays using the purified human HENMT1 show that HENMT1 is able to carry out methylation on 20nt–30nt small single-stranded RNAs (22,26), it remains unknown whether HENMT1 can methylate mammalian miRNA at 3′-terminal. To explore the potential function of HENMT1 in mammalian miRNAs, we first determined the tissue distribution of human HENMT1. Although previous studies reported that HENMT1 expression was restricted to the testis (25), our results clearly showed expression of HENMT1 protein in almost all human tissues, including testis (Figure 4A). Moreover, both cell fraction assay (Figure 4B, left) and immunofluorescence labeling (Figure 4B, right) in alveolar A549 cell showed that HENMT1 was mainly located in the cytoplasm of A549 cell. To determine whether HENMT1 mediates mammalian miRNA methylation, we generated a stable HENMT1-knockout A549 cell strain (A549KO) via CRISPR/Cas9 technique (Figure 4C). The miRNA libraries generated from WT A549 and A549KO cells were then profiled by Solexa sequencing (SRA database with the accession numbers PRJNA576334). After normalization, the read counts of majority of miRNAs in A549KO cells were reduced more than two-fold compared to those in A549 cells after oxidation procedure (Figure 4D), suggesting that HENMT-1 mediates the methylation of many miRNAs in WT A549 cells. The reduction of miRNAs by HENMT1 knockout was further confirmed by qRT-PCR assay (Figure 4E) and high-resolution PAGE (Figure 4F).
Figure 4.
HEMT1-knockout impairs 3′-terminal 2′Ome of miRNAs in A549 cells and mice. (A) HENMT1 expression in human tissues. (B) HENMT1 expression and location in A549 cell. Left: localization of HENMT1 detected by immunofluorescence analysis; right: localization of HENMT1 detected by Western blot. (C) HENMT1 expression detected by Western blot in WT A549 cells (WT), scramble gDNA (control cells) and HEMT1-knockout A549 cells (HEMT1 Knockout) via Crisper/cas9 technique. (D–F) Levels of miRNAs in A549 or HENMT1-knockout A549 (A549KO) cells detected by high-through sequencing after oxidation (D), qRT-PCR after oxidation (E) and Northern blot (F) after Oxid/β-elim procedure. (G) HENMT1 expression detected by western blot in lung derived from wild type mice (WT) and HEMT1-knockout mice (KO). (H) Ratio of piR_020485, miR-21-5p and miR-26a-5p levels with oxidation versus piR_020485, miR-21-5p and miR-26a-5p levels without oxidation in lung tissues from WT and HENMT1-KO (KO) mice by qRT-PCR. (I) Northern blot analysis of piR_020485 and miR-26a-5p expression in lung tissues from WT and KO mice after Oxid/β-elim procedure.
We also generated a HENMT1-deficiency mouse strain using Crisper/Cas9 technique. As shown, HENMT1 is not detected in HENMT1-KO mouse lung by Western blot (Figure 4G). The qRT-PCR assay (Figure 4H) and high-resolution PAGE analysis (Figure 4I) of piR_020485, miR-21-5p and miR-26a-5p in WT and HENMT1-KO mice before and after Oxid/β-elim procedure showed that both piRNA and miRNAs were not protected against Oxid/β-elim procedure after HENMT1 knockout, suggesting that 3′-terminal 2′Ome of piRNAs and miR-26-5p in mouse lung tissues is likely mediated by HENMT1. Taken together, these results suggest that HENMT1 is a major methyltransferase responsible for 3′-terminal 2′Ome of not only piRNAs, but also mammalian miRNAs including miR-21-5p in NSCLC tissue and miR-26-5p in non-cancerous lung tissue.
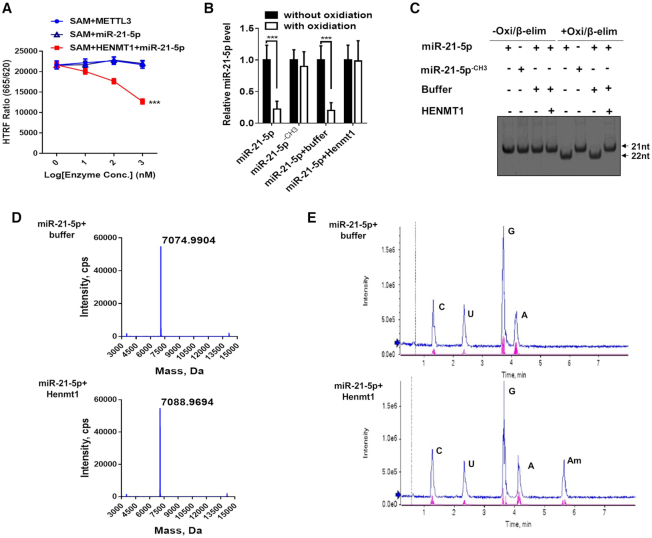
To validate the role of HENMT1 in mammalian miRNA methylation, we next performed an in vitro methyltransferase assay using synthetic miR-21-5p and purified HENMT1 recombinant protein. First, human HENMT1 was overexpressed in A549 cells and then purified via affinity column. Second, an orthogonal EPIgeneous Homogeneous Time-Resolved Fluorescence Methyltransferase Assay (27) was performed to examine the enzymatic activity of HENMT1 recombinant protein (Figure 5A). In this assay, synthetic miR-21-5p (without any methyl groups) serves as substrate. The assay measured the HENMT1 reaction product SAH, which competitively displaced d2-labeled SAH that was pre-bound to Lumi4-Tb-labeled anti-SAH, resulting in a loss of FRET signal (27). As shown in Figure 4A, synthetic miR-21-5p was successfully methylated by HENMT1 recombinant protein. To ascertain if the loss of FRET signal is associated with HENMT1 enzymatic activity, we performed qRT-PCR after oxidation (Figure 5b) and high-resolution PAGE assay (Figure 5C) after Oxid/β-elim to analyze the methylation statue of synthetic miR-21-5p. Both results confirmed that synthetic miR-21-5p was methylated by HENMT1 recombinant. Mass Spectrometry (Figure 5D) and LC–MS/MS (Figure 5E) were also performed to analyze the modification of synthetic miR-21-5p after the methyltransferase assay. Mass Spectrometry analysis indicated that the molecular weight of the synthetic miR-21-5p was increased from 7074.9904 Da to 7088.9694 Da after incubation with HENMT1 recombinant (Figure 5D). Moreover, LC–MS/MS analysis revealed a peak of the methylation of adenosine (Am) of miR-21-5p after incubation with HENMT1 recombinant (Figure 5E), confirming the 3′-terminal 2′Ome of miR-21-5p by HENMT1.
Figure 5.
In vitro assay of methylation of synthetic miR-21-5p by HEMT1. The methylation assay is carried out in 20 μl reaction buffer containing 10 μM synthetic miR-21-5p and 5μM HENMT1 recombinant proteins. After incubating at 37°C for 40 min, RNA was recovered and purified by ethanol precipitation. (A–E) Methylation of synthetic miR-21-5p at 3′-end by purified HENMT1 assessed by EPIgeneous™ methyltransferase assay (A), Northern blot (B), qRT-PCR (C), mass spectrometry (D) and LC–MS/MS (E), respectively.
3′-Terminal 2′Ome enhances miR-21-5p's stability and association with AGO2
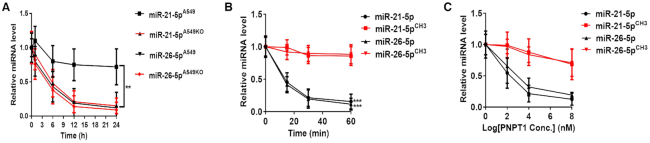
Previous studies have demonstrated that 2′Ome of plant miRNAs at the 3′-terminal can serve as a protective mechanism against miRNA degradation (15). To monitor the effect of 3′-terminal 2′Ome on the stability of mammalian miRNAs, the decay of miR-21-5p in WT A549 and HENMT1-knockout A549KO cells was assessed by qRT-PCR after blocking the transcription of new miR-21-5p via treatment with actinomycin D (ActD). As shown in Figure 6A, the relative level of miR-21-5p in WT A549 cells was much higher than in A549KO cells, suggesting that 3′-terminal 2′Ome of miRNA may contribute to miR-21-5p stability. In contrast, the level of miR-26-5p, which is unmethylated in lung cancer A549 cells, displayed no difference between WT and HENMT1-KO cells (Figure 6A). We also transfected 293T cells, which contain very low endogenous miR-21-5p level, with synthetic miR-21-5p (without methyl groups) and miR-21-5pCH3 (miR-21-5p with a 2′-O-methyl on its 3′-end nucleotide). As shown in Figure 6B, miR-21-5pCH3 was degraded much slower than miR-21-5p. The similar results were obtained in miR-26-5p and miR-26-5pCH3 (Figure 6B). These observations collectively suggest that 2′-O-methyl group on the 3′-terminal nucleotide protects mammalian miRNAs against the cleavage by 3′→5′ exoribonucleases.
Figure 6.
3′-terminal 2′Ome of human miR-21-5p increases its stability. (A) The half-life of miR-21-5p and miR-26-5p in A549 cells and A549KO (HENMT1-knockout) cells. (B, C) Direct degradation of synthetic miR-21-5p/miR-21-5pCH3 and miR-26-5p/miR-26-5pCH3 by PNPT1 recombination protein in a time-dependent (B) and concentration-dependent (C) manner.
As a 3′→5′ exoribonuclease, polyribonucleotide nucleotidyltransferase 1 (PNPT1) plays a critical role in decay of mammalian RNAs including miRNAs (28). To further examine the protective role of 3′-terminal 2′Ome of miR-21-5p against the degradation by 3′→5′ exoribonucleases, we compared the cleavage of synthetic miR-21-5p/miR-21-5pCH3 and miR-26-5p/miR-26-5pCH3 by purified PNPT1. As shown in Figure 6C, when synthetic miRNAs with or without 3′-terminal 2′Ome were incubated with various concentrations of PNPT1, cleavage of non-methylated miR-21-5p and miR-26-5p by PNPT1 was significantly faster than that of methylated miR-21-5pCH3 and miR-26-5pCH3. This result supports the notion that 3′-terminal 2′Ome of mammalian miRNAs can stabilize miRNAs through protecting them against the cleavage by 3′→5′ exoribonucleases.
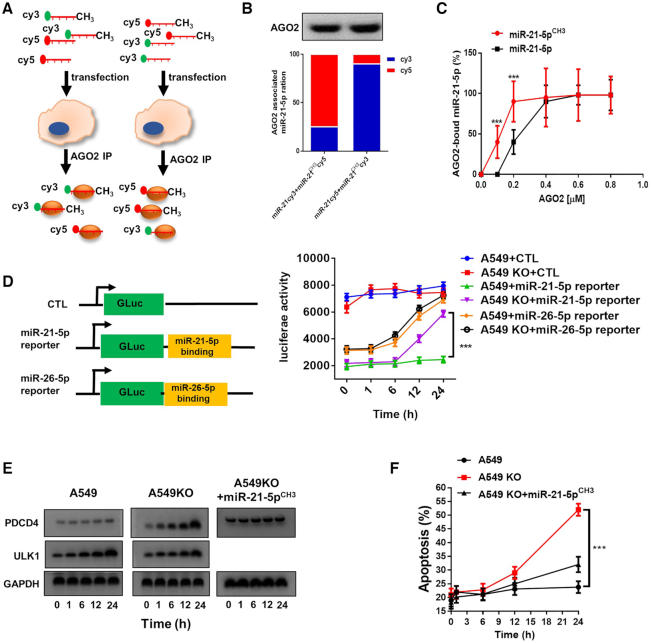
Previous studies have also suggested that modification by 3′-terminal 2′Ome can enhance the binding affinity of siRNAs to AGO2, which in turn, promotes the cellular activity of siRNAs (29). To explore whether 3′-terminal 2′Ome of mammalian miRNAs can increase the binding affinity of miRNAs to AGO2, we performed an AGO2-binding assay in A549 cells following transfection with the same amount of fluorescently labeled synthetic miR-21-5p and miR-21-5pCH3 (Supplementary Figure S5a and b). To avoid of the influence by fluorescence labeling, we switch the labeling of miR-21-5p or miR-21-5pCH3 from Cy3 to Cy5 or verse versa. As depicted in Figure 7A, at 24 h post-transfection with synthetic Cy3-labeled miR-21-5pCH3 and Cy5-labeled miR-21-5p or Cy5-labeled miR-21-5pCH3 and Cy3-labeled miR-21-5p, AGO2 in A549 cell was immunoprecipitated using anti-AGO2 antibody and the fluorescence of Cy3 and Cy5 were detected, respectively. The results showed that, regardless of Cy3 or Cy5 labeling, the level of miR-21-5pCH3 was significantly higher than miR-21-5p in the anti-AGO2 immunoprecipitated complex (Figure 7B), suggesting that miR-21-5pCH3 possesses a higher affinity to AGO2 complex. Direct protein-miRNA binding analysis (30) using purified AGO2 recombinant and synthetic miR-21-5p and miR-21-5pCH3 further confirmed higher affinity of miR-21-5pCH3 to AGO2 than that of miR-21-5p (Figure 7C).
Figure 7.
3′-Terminal 2′Ome of human miR-21-5p increases its association with AGO2 complex and inhibition on PDCD4 expression. (A) Depict of experimental strategy of assaying binding affinity of miR-21-5p or miR-21-5pCH3 with AGO2 complex using Cy3- or Cy5-labeled miRNAs. (B) Binding affinity of miR-21-5p and miR-21-5pCH3 with AGO2 complex detected by fluorescence intensity measurement. (C) Kinetics of binding of miR-21-5p and miR-21-5pCH3 with AGO2 detected by microscale thermophoresis. (D) Binding of miR-21-5p or miR-26-5p with AGO2 detected by the luciferase activity in A549 or A549KO cells transfected with miR-21-5p or miR-26-5p reporter plasmid. (E) PDCD4 and ULK1 protein level in A549 and A549KO cells at different time points after serum depletion. (F) Apoptosis of A549 and A549KO cells induced by serum depletion.
Given that 3′-terminal 2′Ome of miR-21-5p can enhance its stability and AGO2 binding, we further monitored the influence of such methylation on the cellular function of miR-21-5p. Firstly, a luciferase report plasmid consisting of a miR-21-5p binding sequence was constructed and transfected into WT A549 and A549KO cells. As expected, since both the stability and AGO2-binding affinity of miR-21-5p were significantly decreased after HENMT1 knockout, the level of miR-21-5p was rapidly reduced in A549KO cells, resulting in an increase of luciferase activity (Figure 7D). In contrast, without rapid reduction of miR-21-5p in WT A549 cells, the luciferase activity remained unchanged (Figure 7D). For non-methylated miR-26-5p in A549 cells, its level was not affected by HENMT1 knockout. As shown in Figure 7D, decrease of miR-26-5p level in both HENMT1-KO and WT cells resulted in an increase of luciferase activity. This result provides another piece of evidence that 3′-terminal 2′Ome of mammalian miRNA can increase its stability and AGO2-binding. Programmed cell death protein 4 (PDCD4), which promotes cancer cell apoptosis, has been proven as a typical target gene of miR-21-5p (31). We thus determined the effect of miR-21-5p on protein level and cellular function of PDCD4 in WT A549 or A549KO cells. In line with that both the stability and AGO2-binding of miR-21-5p in A549KO cells were significantly lower than those in WT A549 cells, the protein level of PDCD4 was significantly higher in A549KO cells compared to WT A549 cells (Figure 7E). In contrast, the level of ULK1, a target gene of non-methylated miR-26-5p (32), was significantly increased in both A549KO and WT cells (Figure 7E). In agreement with this, due to upregulation of PDCD4 expression, the apoptosis of A549KO cells induced by serum depletion was significantly higher than that of A549 cells (Figure 7F).
DISCUSSION
Employing various technical platforms including LC–MS/MS, Solexa sequencing, qRT-PCR and northern blot assays following Oxid/β-elim procedure, we demonstrated for the first time the existence of 3′-terminal 2′Ome in mammalian cells in cancerous and non-cancerous tissue. In fact, NSCLC lung tissue and the paired distal non-cancerous lung tissue displayed different 3′-terminal 2′Ome pattern, evidenced by significant 3′-terminal 2′Ome of miR-21-5p in NSCLC while 3′-terminal 2′Ome of miR-26-5p in non-cancerous lung tissues. Moreover, we showed that HEMT1 served as the methyltransferase responsible for 3′-terminal 2′Ome of mammalian miRNAs.
First, LC–MS/MS result provided the direct evidence that miR-21-5p in NSCLC lung tissue but not the paired non-cancerous lung tissue possesses 3′-terminal 2′Ome (Figure 2). In the past, one major barrier in analyzing the specific modification of tissue miRNAs by LC–MS/MS is the purity of isolated miRNAs. To overcome this, we employed a selective sequential enzymatic digestion strategy to directly isolate miR-21-5p from NSCLC and the paired distal non-cancerous lung tissues (Figure 2A). Examined by Solexa sequencing, the purity of the isolated miR-21-5p was higher than 99%, which is suitable for LC–MS/MS analysis. As shown in Figure 2C, LC–MS/MS analysis clearly showed 3′-terminal 2′Ome of miR-21-5p in NSCLC but not non-cancerous lung tissues. The finding of 3′-terminal 2′Ome in NSCLC miR-21-5p was further validated by other technical platforms currently available for examination of miRNA modification, including qRT-PCR and high-resolution PAGE following oxidation or Oxid/β-elim treatment. Second, we identified the HENMT1 as the methyltransferase responsible for 3′-terminal 2′Ome of mammalian miRNAs. As a HEN1 homolog expressed in mammalian cells (25), HENMT1 has been reported to methylate piRNAs in animals and the AGO2-associated small RNAs in Drosophila (15). However, although purified human HENMT1 is able to methylate small single-stranded RNAs (22,26), whether it can methylate mammalian miRNAs at the 3′-end remains unknown. Different from the previous report that HENMT1 was restrictively expressed in testis (25), we found that HENMT1 protein was expressed in almost all human tissues with major distribution in the cell cytoplasm (Figure 4A and B) and significantly increased in lung cancer tissues compared to non-cancerous lung tissues (Supplementary Figure S6). HENMT1 knockout in mouse and culture cells both markedly reduced 3′-terminal 2′Ome of mammalian miRNAs. In addition, an in vitro assay using purified HENMT1 recombinant protein and synthetic miR-21-5p clearly showed that HENMT1 mediated 3′-terminal 2′Ome of miR-21-5p.
We have also explored the potential function of 3′-terminal 2′Ome in mammalian miRNAs. The miRNA decaying assay indicated that miR-21-5p with 3′-terminal 2′Ome had a significantly longer half-life than unmethylated miR-21-5p (Figure 6A), suggesting that 3′-terminal 2′Ome can stabilize miR-21-5p. This finding is in agreement with previous report in plant (15). Given that 3′→5′ exoribonucleases serve as a critical part of miRNA decaying machinery in mammalian cells (33), we further tested whether 3′-terminal 2′Ome can protect miRNAs against the cleavage by PNPT1. In vitro cleavage assay using purified PNPT1 recombinant protein and synthetic miR-21-5p and miR-21-5pCH3 clearly indicated that 3′-terminal 2′Ome protected miRNA 3′→5′ degradation by PNPT1 (Figure 6B and C). Previous study in flies by Ameres et al. (34) showed that AGO2-bound small RNAs, but not those bound to AGO1, exhibited 3′-terminal 2′Ome, suggesting that the modification of 3′-terminal 2′Ome may facilitate the binding of small RNAs to AGO2 instead of AGO1. Combining the experimental strategies of reverse fluorescence labeling of methylated or unmethylated miRNAs, AGO2 immunoprecipitation, and direct miRNA-AGO2 binding assay, we found that methylated miR-21-5pCH3 exhibited a higher affinity to AGO2 than unmethylated miR-21-5p did (Figure 7A–C). Higher affinity of methylated miR-21-5pCH3 to AGO2 compared to unmethylated miR-21-5p was further confirmed by Luciferase report assay (Figure 7D). In line with the notion that 3′-terminal 2′Ome of miR-21-5p enhances the stability and the AGO2-binding of miR-21-5p, functional studies demonstrated that defect of 3′-terminal 2′Ome in miR-21-5p, induced by HENMT1 knockout in A549KO cells, resulted in an impaired miR-21-5p function, leading to an enhanced expression of its target gene PDCD4 in A549KO cells (Figure 7E), which in turn, promoted cell apoptosis at the absence of serum (Figure 7F).
Although our results show the existence of 3′-terminal 2′Ome in mammalian miRNAs and the involvement of HENMT1 in miRNA methylation, the data also clearly demonstrate the differential methylation of miRNAs in lung cancer and non-cancerous tissues. In addition, oxidation in combination of qRT-PCR assay and sequencing indicates that majority miRNAs are not influenced by methylation (Figure 1). The mechanisms that control the selective methylation of mammalian miRNAs at 3′-terminal under various pathophysiological conditions and its associated function still remain largely unknown. Unlike plant miRNAs, which need methylation because extensive base pairing between miRNAs and their targets (35,36), mammalian miRNAs generally lack of completely base pairing with their target genes. The future study is thus necessary to explore how mammalian miRNAs are selectively methylated and whether the methylation of mammalian miRNAs in also involved in the recognition of miRNA targets. The selective methylation of miRNA may play a role in fine-tuning the function of different groups of miRNAs under different condition. For instance, miR-21-5p generally serves as an oncomiR in promoting cancer progression (24), methylation of miR-21-5p in lung cancer tissue may enhance its oncomiR function through increasing its stability or binding with AGO2.
In summary, the present study is the first to identify the 3′-terminal 2′Ome of mammalian miRNAs and uncover its potential role in enhancing miRNAs’ function through protecting them against cleavage by 3′→5′ exoribonucleases and increasing their binding to RNA effector protein AGO2 complex.
DATA AVAILABILITY
The data of RNA-seq in this study can be viewed in SRA database with the accession numbers PRJNA575072 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA575072) and PRJNA576334 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA576334).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Qihan Chen (Nanjing University) and Ms Yao Tang (Nanjing University) for their help in the purification of miRNAs. The authors also thank Dr Jill Leslie Littrell (Georgia State University, Atlanta, GA) for critical reading and constructive discussion of the manuscript.
Author contributions: K.Z., X.C., T. W. and C.Z. designed research; H.L, Z.J, W.R. and Z.L performed research and analyzed data; J.W. collected the patients’ samples; Y.L., X.S., S.Q., Y.W., Q.Z. contributed new reagents/analytic tools; K.Z. and H.L wrote the paper.
Contributor Information
Hongwei Liang, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Zichen Jiao, Department of Cardiothoracic Surgery, Nanjing Drum Tower Hospital, Medical School of Nanjing University, Nanjing, China.
Weiwei Rong, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Shuang Qu, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Zhicong Liao, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Xinlei Sun, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Yao Wei, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Quan Zhao, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Jun Wang, Department of Emergency Medicine, Nanjing Drum Tower Hospital, Medical School, Nanjing University, 210008 Nanjing, China.
Yuan Liu, Center for Inflammation, Immunity and Infectious Diseases, Georgia State University, Atlanta, GA 30032, USA.
Xi Chen, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Tao Wang, Department of Cardiothoracic Surgery, Nanjing Drum Tower Hospital, Medical School of Nanjing University, Nanjing, China.
Chen-Yu Zhang, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
Ke Zen, State Key Laboratory of Pharmaceutical Biotechnology, Department of Gastroenterology, Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu 210093, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2018YFA0507100]; National Natural Science Foundation of China [31801088]; Natural Science Foundation of Jiangsu Province [BK20170076]; Postdoctoral Science Foundation of China [2018M642210]; Jiangsu Province Postdoctoral Fund [2018K050A]. Funding for open access charge: Ministry of Science and Technology of China [2018YFA0507100]; National Natural Science Foundation of China [31801088]; Natural Science Foundation of Jiangsu Province [BK20170076]; Postdoctoral Science Foundation of China [2018M642210]; Jiangsu Province Postdoctoral Fund [2018K050A].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X. et al.. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008; 18:997–1006. [DOI] [PubMed] [Google Scholar]
- 2. Chen X., Liang H., Zhang J., Zen K., Zhang C.Y.. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012; 22:125–132. [DOI] [PubMed] [Google Scholar]
- 3. Zen K., Zhang C.Y.. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012; 32:326–348. [DOI] [PubMed] [Google Scholar]
- 4. Chen X., Liang H., Zhang J., Zen K., Zhang C.Y.. Horizontal transfer of microRNAs: molecular mechanisms and clinical applications. Protein Cell. 2012; 3:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cortez M.A., Bueso-Ramos C., Ferdin J., Lopez-Berestein G., Sood A.K., Calin G.A.. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011; 8:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mori M.A., Ludwig R.G., Garcia-Martin R., Brandao B.B., Kahn C.R.. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019; 30:656–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turchinovich A., Tonevitsky A.G., Burwinkel B.. Extracellular miRNA: a collision of two paradigms. Trends Biochem. Sci. 2016; 41:883–892. [DOI] [PubMed] [Google Scholar]
- 8. Quinn J.F., Patel T., Wong D., Das S., Freedman J.E., Laurent L.C., Carter B.S., Hochberg F., Van Keuren-Jensen K., Huentelman M. et al.. Extracellular RNAs: development as biomarkers of human disease. J. Extracell. Vesicles. 2015; 4:doi:10.3402/jev.v4.27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li L., Zhu D., Huang L., Zhang J., Bian Z., Chen X., Liu Y., Zhang C.Y., Zen K.. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012; 7:e46957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu B., Yang Z.Y., Li J.J., Minakhina S., Yang M.C., Padgett R.W., Steward R., Chen X.M.. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005; 307:932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pandolfini L., Barbieri I., Bannister A.J., Hendrick A., Andrews B., Webster N., Murat P., Mach P., Brandi R., Robson S.C. et al.. METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol. Cell. 2019; 74:1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abe M., Naqvi A., Hendriks G.J., Feltzin V., Zhu Y., Grigoriev A., Bonini N.M.. Impact of age-associated increase in 2′-O-methylation of miRNAs on aging and neurodegeneration in Drosophila. Genes Dev. 2014; 28:44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoerter J.A., Walter N.G.. Chemical modification resolves the asymmetry of siRNA strand degradation in human blood serum. RNA. 2007; 13:1887–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kirino Y., Mourelatos Z.. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007; 14:347–348. [DOI] [PubMed] [Google Scholar]
- 15. Ji L.J., Chen X.M.. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012; 22:624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim S.L., Qu Z.P., Kortschak R.D., Lawrence D.M., Geoghegan J., Hempfling A.L., Bergmann M., Goodnow C.C., Ormandy C.J., Wong L. et al.. HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLos Genet. 2015; 11:e1005782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alberti C., Manzenreither R.A., Sowemimo I., Burkard T.R., Wang J., Mahofsky K., Ameres S.L., Cochella L.. Cell-type specific sequencing of microRNAs from complex animal tissues. Nat. Methods. 2018; 15:283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T.. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007; 14:349–350. [DOI] [PubMed] [Google Scholar]
- 19. Wei Y., Li L.M., Wang D., Zhang C.Y., Zen K.. Importin 8 regulates the transport of mature MicroRNAs into the cell nucleus. J. Biol. Chem. 2014; 289:10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang N., Qu S., Sun W., Zeng Z., Liang H., Zhang C.Y., Chen X., Zen K.. Direct quantification of 3′ terminal 2′-O-methylation of small RNAs by RT-qPCR. RNA. 2018; 24:1520–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng X., Zhang X., Su J., Zhang Y., Zhou W., Zhou J., Wang C., Liang H., Chen X., Shi R. et al.. miR-19b downregulates intestinal SOCS3 to reduce intestinal inflammation in Crohn's disease. Sci. Rep. 2015; 5:10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan C.M., Zhou C., Brunzelle J.S., Huang R.H.. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc. Natl Acad. Sci. U.S.A. 2009; 106:17699–17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan M., Wang Y., Hu Y., Feng Y., Dai C., Wu J., Wu D., Zhang F., Zhai Q.. A high-throughput quantitative approach reveals more small RNA modifications in mouse liver and their correlation with diabetes. Anal. Chem. 2013; 85:12173–12181. [DOI] [PubMed] [Google Scholar]
- 24. Kumarswamy R., Volkmann I., Thum T.. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011; 8:706–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kirino Y., Mourelatos Z.. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007; 13:1397–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mickute M., Nainyte M., Vasiliauskaite L., Plotnikova A., Masevicius V., Klimasauskas S., Vilkaitis G.. Animal Hen1 2′-O-methyltransferases as tools for 3′-terminal functionalization and labelling of single-stranded RNAs. Nucleic Acids Res. 2018; 46:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimos M., Burton M., Urbain D., Caudron D., Martini M., Famelart M., Gillard M., Barrow J., Wood M.. Development of an HTRF assay for the detection and characterization of inhibitors of catechol-O-methyltransferase. J. Biomol. Screen. 2016; 21:490–495. [DOI] [PubMed] [Google Scholar]
- 28. Ruegger S., Grosshans H.. MicroRNA turnover: when, how, and why. Trends Biochem. Sci. 2012; 37:436–446. [DOI] [PubMed] [Google Scholar]
- 29. Kuhn C.D., Joshua-Tor L.. Eukaryotic Argonautes come into focus. Trends Biochem. Sci. 2013; 38:263–271. [DOI] [PubMed] [Google Scholar]
- 30. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frankel L.B., Christoffersen N.R., Jacobsen A., Lindow M., Krogh A., Lund A.H.. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008; 283:1026–1033. [DOI] [PubMed] [Google Scholar]
- 32. Jin F., Wang Y., Li M., Zhu Y., Liang H., Wang C., Wang F., Zhang C.Y., Zen K., Li L.. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death. Dis. 2017; 8:e2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Z., Qin Y.W., Brewer G., Jing Q.. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip. Rev. RNA. 2012; 3:593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D.. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010; 328:1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ren G., Xie M., Zhang S., Vinovskis C., Chen X., Yu B.. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl Acad. Sci. U.S.A. 2014; 111:6365–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ren G., Chen X., Yu B.. Small RNAs meet their targets: when methylation defends miRNAs from uridylation. RNA Biol. 2014; 11:1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of RNA-seq in this study can be viewed in SRA database with the accession numbers PRJNA575072 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA575072) and PRJNA576334 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA576334).