Abstract
Adenylate/uridylate-rich elements (AREs) are the most common cis-regulatory elements in the 3′-untranslated region (UTR) of mRNAs, where they fine-tune turnover by mediating mRNA decay. They increase plasticity and efficacy of mRNA regulation and are recognized by several ARE-specific RNA-binding proteins (RBPs). Typically, AREs are short linear motifs with a high content of complementary A and U nucleotides and often occur in multiple copies. Although thermodynamically rather unstable, the high AU-content might enable transient secondary structure formation and modify mRNA regulation by RBPs. We have recently suggested that the immunoregulatory RBP Roquin recognizes folded AREs as constitutive decay elements (CDEs), resulting in shape-specific ARE-mediated mRNA degradation. However, the structural evidence for a CDE-like recognition of AREs by Roquin is still lacking. We here present structures of CDE-like folded AREs, both in their free and protein-bound form. Moreover, the AREs in the UCP3 3′-UTR are additionally bound by the canonical ARE-binding protein AUF1 in their linear form, adopting an alternative binding-interface compared to the recognition of their CDE structure by Roquin. Strikingly, our findings thus suggest that AREs can be recognized in multiple ways, allowing control over mRNA regulation by adapting distinct conformational states, thus providing differential accessibility to regulatory RBPs.
INTRODUCTION
Precise control of gene expression is essential for every organism, in every cell type and tissue, but appears to be particularly crucial during development and in immune responses of higher eukaryotes. Consequently, dysregulated levels of gene products are a major cause of uncontrolled immune responses, autoimmune diseases and various types of cancer. A key point for regulation of gene expression is at the post-transcriptional level. Steady-state mRNA levels result from their balanced synthesis and decay. mRNA half-lives are primarily controlled by cis-regulatory elements within the 3′-UTR of the mRNA, which are recognized by trans-acting factors such as microRNAs, but even more often by RNA-binding proteins (RBPs). RBP interactions with target mRNAs trigger downstream effects including mRNA decay, translational inhibition or long-term protection from decay factors leading to an accumulation of certain mRNAs. Such post-transcriptional regulation is crucial to balance the mRNA levels of pro-inflammatory cytokine-encoding mRNAs. These are inherently unstable to promote the resolution of inflammation, preventing unintended tissue damage and autoimmune reactions. cis-regulatory elements mediating this instability are either miRNA binding sites, stem-loop (SL) elements or adenylate/uridylate (AU)-rich elements (AREs) (1,2).
AREs are the currently best-studied cis-regulatory elements that control mRNA fate, owing to their early discovery and broad investigation over the past decades (3). AREs were initially identified in mRNAs encoding early response genes (4,5), but soon after that also identified in mRNAs of growth factors, pro-inflammatory cytokines and proto-oncogenes, where they likewise induce transcript degradation (6–9). In line with that, aberrantly expressed transcripts of tumor-promoting transcription factors like c-fos are characteristic for their lack of AREs (9). However, AREs are now considered a widespread and heterogeneous group of elements that occur in numerous transcripts. They are listed in databases such as AREsite2, which provide a bioinformatic starting point for the characterization of AU-rich elements in a given target gene (10).
AREs had first been subdivided in three major classes with different sequences, where the currently best-studied class comprises AREs with a core sequence of AUUUA, primarily embedded in an (A)U-rich context (3,11). However, various, less-defined but primarily U-rich sequences are able to mediate mRNA decay (9,11). Notably, AREs are most effective in their function when organized in repeats of their core motifs and a total sequence length of up to 150 nucleotides. In addition, they specially act as cis-element, as they appear to be both miRNA target elements and targets of regulatory RBPs in inducing mRNA decay, occasionally in a directly overlapping manner (12–14).
Prominent example proteins that promote target mRNA decay are Tristetraprolin (TTP or ZFP36), Butyrate response factor 1 (BRF1 or ZFP36L1) and K-homology splicing regulatory protein (KSRP), all of which recognize one or more linear ARE core motifs (3,15). Of note, such linear AREs are also targeted by mRNA-protecting RBPs, e.g. by the ELAV (embryonic lethal abnormal visual system)-like proteins, such as human antigen R (HuR or ELAVL1) ((16–18). Interestingly, the first protein found to recognize AREs (19), the ARE/poly(U)-binding factor 1 (AUF1 or hnRNP D) is capable of both stabilization and destabilization of its mRNA targets. While it is still enigmatic how AUF1 exerts its dual function, its (de-)stabilizing activity seems to be dependent on cell type, the specific AUF1 protein isoform as well as the target itself [reviewed in (20)]. AUF1 shows a relatively promiscuous target sequence specificity, recognizing not only canonical AREs, but more generally U-rich targets (21,22).
In general, these ARE-BPs harbor multiple RNA-binding domains (RBDs) and recognize clustered regions of short single-stranded RNA cis-elements, e.g. multiple copies of AREs organized in hubs. Here, affinity and specificity are increased through multivalent interactions using multiple copies of the same or sets of different RBDs, which is a unifying principle in driving affinity and specificity for RNA-recognition by multi-domain RBPs (18,23–27). While by default, AREs are regarded as linear single-stranded RNA motifs, earlier studies have suggested that the transient presence of expanded structures in ARE regions interferes with the recognition by ssRNA-specific RBPs (23,28), in particular for the canonical AUF1 RRM (RNA recognition motif) domains (29–31). This will in vivo lead to an intrinsic dynamic regulation of mRNA stability through the formation of transient RNA secondary structure (Figure 1A), and at the same time influence the potential presence of miRNA target sites.
Figure 1.
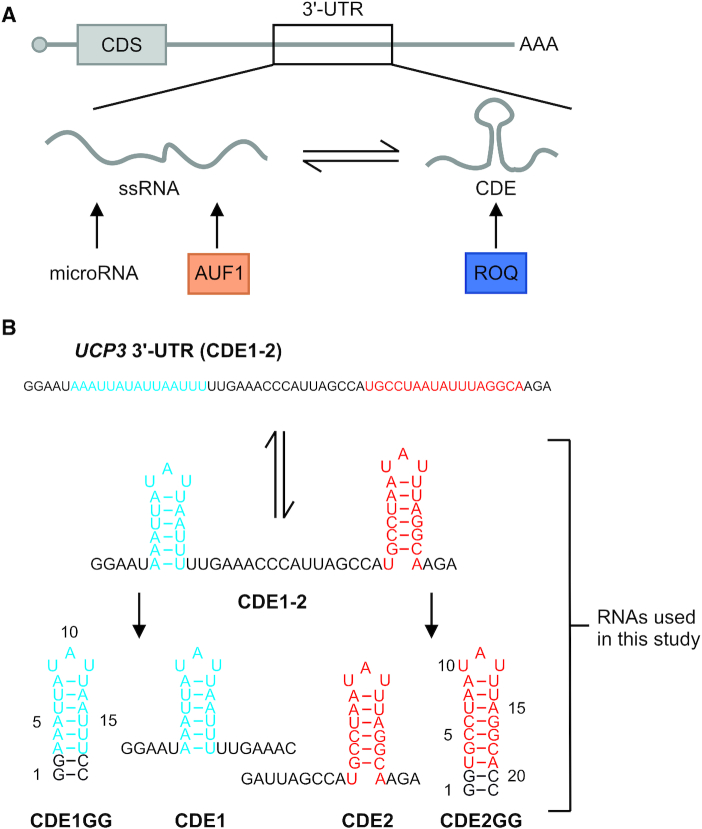
Introduction to AREs, their cognate RBPs and the RNA constructs used in the present study. (A) Scheme of AREs as integral part of an mRNA’s 3′-UTR (upper panel). Lower panel, a potential ARE can exist in an equilibrium between its single-stranded form (ssRNA, e.g. recognized by the protein AUF1) and a structured form, the latter exemplified by a CDE-like shaped element bound by the highly specific Roquin ROQ domain. The target ARE conformers are mutually exclusive and can lead to redundant or competitive regulation of the ARE-containing mRNA through the respective cognate RBP. (B) The previously described regulatory ARE region in the UCP3 mRNA 3′-UTR shown as linear sequence and as its structured conformer with the two CDEs bound by Roquin (44). Below that, the two CDEs are shown as isolated wild type species and as stabilized SLs with the underlying numbering of bases as used in this study. See also Supplementary Figure S1.
Recently, small SL elements (constitutive/alternative decay elements, CDE/ADE) have been identified as an alternative way to promote mRNA decay through highly specific recognition by novel types of domains, most prominently shown for Roquin and Regnase. Both proteins are essential regulators of immune responses, preventing excessive cytokine production and autoimmune reactions by regulating a shared set of target mRNAs through a common SL element (32–37). Herein, Roquin uses its unique ROQ domain for specific binding to CDE and ADE SLs (38–42).The involvement of the ROQ domain in mRNA repression has long been assumed independent from the simultaneous presence of single-stranded AREs.
The strictness of the CDE sequences in functional targets for Roquin has been a matter of debate for years now, including contradictive findings (36,38,43,44). Certainly, a major requirement for a functional SL represents its accessibility in vivo, i.e. not being masked by other RBPs or unfolded because of low thermodynamic stability. An ongoing redefinition and expansion of the CDE ‘consensus’ has recently led to the identification of AREs that were suggested to fold into CDEs (44). Interestingly, in the UCP3 mRNA 3′-UTR, such CDE-AREs appear as a tandem recognized by the Roquin ROQ domain (Figure 1B). Despite strong in vitro and cellular data underlining the regulation, this earlier study could not unambiguously prove these RNA moieties to be bound by Roquin in a CDE-like manner (i.e. comparable to the Tnf CDE (38)). The latter, however, needs to be taken into account considering the recently identified capabilities of the Roquin ROQ domain to bind single-stranded AU-containing sequences (45). Further, the high AU-content of CDE-AREs suggests that these elements exhibit a bi-functional nature, switching between a structured and linear form. As such, CDE-AREs would be hotspots of RBPs competing for these AREs (e.g. AUF1), independent of their preference in recognizing ssRNA or structured RNA. However, we to date lack any atom-resolved evidence of stem-looped AREs, a feature that had been suggested to affect the function of AUF1 before (30,31).
We here provide structural evidence for the existence of CDE-like folded AREs, both in an unbound as well as ROQ domain-bound form, by NMR and X-ray crystallography. By investigating the UCP3 CDEs 1 and 2, we show that, despite a low intrinsic thermostability, both CDEs are locked in stable complexes with the ROQ domain and engage with affinities comparable to the most canonical CDE found in the 3′-UTR of Tnf mRNA (46). Besides, we show that the UCP3 CDE1 is also a target of the AUF1 RRM1-2 domains in its unfolded form and prove a direct competition of the two proteins for the same CDE-ARE, underlining a central role for the equilibrium between the linear and structured forms of AREs.
MATERIALS AND METHODS
Plasmids
The Roquin ROQ domain used in this study was expressed from the pETTrx1a vector, obtained from Gunther Stier (EMBL); its cloning has been described before (45). For protein purification of the RNA recognition motifs of AUF1 (AUF1_RRM2 and AUF1_RRM1-2), coding sequences were generated by PCR amplification using pCMV-hnRNP D37 as a template (47) and introduced into XbaI and XhoI sites of an His6- and thioredoxin-tag encoding plasmid based on pETTrx1a, i.e. including a TEV (tobacco etch virus) site for proteolytic removal of all tags.
For RNA synthesis, CDE1, CDE2, CDE1GG, CDE2GG and CDE1-2 (tandem) sequences together with the T7 promoter were generated by hybridization of complementary oligonucleotides and introduced into the NcoI and HindIII sites of an HDV ribozyme encoding plasmid based on the pSP64 vector (Promega). RNAs were transcribed as HDV ribozyme fusions to obtain uniform 3′ ends.
For the luciferase reporter system, 3′-UTR variants were generated by hybridization of complementary oligonucleotides and introduced downstream of the firefly luciferase open reading frame into the multiple cloning site of pDLP (48) using NotI and HindIII restriction sites.
All protein and RNA sequences are summarized in Supplementary Tables S1 and S2.
Protein production
The proteins were produced as His6-Trx-fusion proteins containing a TEV cleavage site from a single freshly transformed clone grown in an overnight culture and used to inoculate an expression culture in either LB or M9 minimal media containing 100 μg/l kanamycin. Cells were grown to an OD600 of 0.7–0.9, protein production was induced with 0.5 mM isopropyl-β-d-thiogalactopyranosid, and cultures were grown overnight at 37°C. Cell pellets were lysed by sonification in 150 mM sodium chloride, 50 mM Tris pH 8.3, 4 mM β-mercaptoethanol. For affinity purification via immobilized metal ion affinity chromatography (IMAC), manual Ni2+-NTA (nickel-nitrilotriacetic acid agarose) columns were used with the lysis buffer complemented with 50, 100 and 300 mM imidazole. The eluted protein was TEV-cleaved overnight during dialysis against 1 M sodium chloride, 50 mM Tris pH 8.0, 4 mM β-mercaptoethanol at 4°C. For the reverse IMAC the cleaved protein was loaded onto Ni2+-NTA columns and washed with the same buffer containing 50, 100 and 300 mM Imidazole. The flow-through containing the target protein was concentrated prior to size exclusion chromatography. For that, a Superdex S75 16/60 column was run in 1 M sodium chloride, 20 mM Tris pH 7.0 and 2 mM Tris-(2-carboxyethyl)-phosphin (TCEP). The protein was frozen in liquid nitrogen, stored at −80°C and buffer exchanged to 150 mM sodium chloride, 20 mM Tris pH 7.0, 2 mM TCEP using Amicon® centrifugal filter units prior to use. For NMR titration experiments the proteins were concentrated to 40–50 μM or 1–1.5 mM stock concentrations when used for competition experiments.
In vitro transcription of RNAs
pHDV constructs were linearized with HindIII and purified by phenol extraction. 2 mg of linearized DNA were transcribed overnight at 37°C using the following conditions: 200 mM Tris–HCl pH 8.0, 20 mM magnesium acetate, 50 mM dithiothreitol (DTT), 2 mM spermidine, 4 mM nucleoside triphosphates (NTP) (each) and 100 μg of in-house-made T7 polymerase. After transcription, precipitated pyrophosphate was pelleted by centrifugation and 20% (v/v) EDTA (0.5 M, pH 8.0) was added to the supernatant. After ethanol precipitation, the RNA was purified using denaturing polyacrylamide gel electrophoresis. The RNA was detected via ultraviolet (UV) shadowing, excised and eluted from the gel in 0.3 M sodium acetate pH 6.5 at 4°C overnight. To remove the remaining gel slices, the supernatant was filtered and the RNA was again precipitated using ethanol. Finally, the RNA was resolved in double-distilled water and stored at –20°C. The purified RNA was folded by heating to 95°C for 5 min prior to injection into five equivalents of ice-cold water. For buffer exchange (NMR buffer: 25 mM potassium phosphate pH 6.2, 50 mM potassium chloride) and concentration, we used Vivaspin® concentrators (molecular weight cut-off 3.000 Da).
NMR spectroscopy
NMR experiments for RNA structure determination and RNA-protein interactions were performed at the BMRZ at the Goethe-University Frankfurt. Spectra were acquired on Bruker AV600, AV700, AV800 and AV950 spectrometers equipped with triple-resonance cryoprobes.
1H–15N-HSQCs of ROQ alone and with UCP3 CDE RNA variants were measured at 40 μM in 20 mM Tris, 150 mM sodium chloride, 2 mM TCEP, 0.02% NaN3, pH 7.0 at 298 K. Spectra for AUF1 RRM2 were recorded at 100 μM and for tandem RRM1-2 at 500 μM in the same buffer. For titration and competition experiments AUF1 RRM1-2, concentrations were adjusted to 40 μM. 1D-1H spectra for the imino proton region of the individual CDE versions were recorded at concentrations of 20 μM with 2k scans at 900 MHz proton frequency. Both CDE1 and CDE2 were titrated in molar ratios of 0.25, 0.5, 1.0, 1.5 and 2.0 to 15N-labeled ROQ. Tandem CDE1-2 was titrated in steps of 0.125, 0.25, 0.5, 1.0 and 1.5 molar ratios. To test competition for RNA binding, the complex of 15N-labeled ROQ and CDE1 was titrated with unlabeled AUF1 RRM1-2 in steps of 0.5, 1.0, 2.0 and 3.0 molar ratio. The reverse experiment was done with 15N-labeled AUF1 RRM1-2/CDE1 complex and unlabelled ROQ (1.0, 2.0, 4.0 molar ratio). To saturate the first binding protein (ROQ or AUF1, respectively) CDE1 was added in small excess (1:1.125). For each titration and competition step, separate imino proton spectra were recorded. To assess the stabilizing effect of Roquin binding, 1D imino spectra were recorded of 20 μM RNA samples titrated with 0.5 and 1.0 (all RNAs), additionally 2.0 (CDE1, CDE2, tandem CDE1-2), and additionally 4.0 (CDE1 and CDE2) molar ratios of ROQ. All protein and RNP spectra were recorded at 298 K.
All samples for the CDE1/2GG structure calculations contained 100 μM sodium tri-methyl-silyl-propane-sulfonate as an internal reference. CDE1GG samples contained 220 μM RNA ([H2O]/[D2O] = 9:1), 350 μM RNA (100% D2O) and 300 μM RNA (100% D2O) for unlabelled and uniformly 13C15N-labeled samples, respectively. CDE2GG samples contained 900 μM RNA ([H2O]/[D2O] = 9:1) and 500 μM RNA (100% D2O) or 450 μM RNA ([H2O]/[D2O] = 9:1) and 1.3 mM RNA (100% D2O) for unlabeled and uniformly 13C,15N-labeled samples, respectively. Distance restraints were obtained from homonuclear 2D-1H,1H-NOESY spectroscopy on unlabeled RNA samples. NOESY spectra were acquired at 600 MHz (CDE1GG) / 950 MHz (CDE2GG) with 2048 × 656 (CDE1GG) or 4096 × 712 (CDE2GG) points. Angular restraints for angles β, ϵ and χ were obtained from J-modulated HCP (49), PFIDS (50) and Γ-HCN (51) experiments (see Supplementary Figures S3 and S4). T1 and hetNOE rates were determined from 13C-correlated 2D sensitivity enhanced experiments. We used relaxation delays of 10, 50, 100, 200, 400, 700, 1000 and 1500 ms for T1 determination. Temperature compensation for spin-lock heating was achieved by implementation of spin-lock pulses distributing identical heating during the relaxation delay of the experiments. HetNOE experiments were acquired in an interleaved manner. Fitting of relaxation curves was conducted using the software implemented in the Bruker Dynamic Center (Bruker Biospin).
Structure calculation of free CDEs
Structure calculation was performed with a modified version of CNS 1.2 (ARIA 1.3) (52). We included all distance restraints obtained from NOESY data (Table 1). Angular restraints for angles β, ϵ and χ were experimentally determined and incorporated with error margins of ±15°. Angles α, γ and ξ were taken from the work of Nozinovic et al. (53) and were incorporated as constraints only for stem residues with error margins of ±20°. Error margins of angular restraints of the loop-closing base pair were set to ±50° to allow backbone adaptation to loop geometry. No violations of angular restraints were observed. Data were corrected for spin diffusion with a rotational correlation time of 4.3 ns for both structures [calculated by HydroNMR (54)]. Refinement of structures was carried out with Xplor-NIH (55). All restraints applied in the original structure calculation were employed in addition to the statistical torsion potential torsionDBRNA provided with the Xplor-NIH software package.
Table 1.
Statistics of CDE1GG and CDE2GG NMR structure calculations. Statistics and accession numbers derived from structures obtained with ARIA/CNS and when additionally refined with Xplor-NIH
| NMR statistics | ||
| PDB ID (CNS) | 6XXB | 6XXA |
| PDB ID (Xplor-NIH) | 6XWW | 6XWJ |
| RNA name | CDE1GG | CDE2GG |
| NOE restraints | ||
| unambiguous: | ||
| intraresidual | 144 | 298 |
| sequential | 59 | 134 |
| medium range | 5 | 6 |
| long range | 14 | 28 |
| ambiguous: | ||
| intraresidual | 0 | 6 |
| sequential | 0 | 9 |
| medium range | 0 | 1 |
| long range | 0 | 2 |
| Base pair restraints | ||
| Planarity | 7 | 9 |
| H-bond | 32 | 46 |
| Dihedral restraints | ||
| sugar pucker | 95 | 105 |
| backbone | 117 | 94 |
| glycosidic bond | 16 | 21 |
| Violations (Xplor-NIH) | ||
| Distance > 0.5 Å | 1 | 2 |
| Dihedral > 10° | 0 | 1 |
| RMSDs/Å (CNS) | ||
| Stem (without terminal) | 1.1 | 1.6 |
| Loop | 2.2 | 2.3 |
| All atom | 1.7 | 1.7 |
| RMSDs/Å (Xplor-NIH) | ||
| Stem (without terminal) | 0.17 | 0.27 |
| Loop | 0.39 | 0.21 |
| All atom | 0.33 | 0.32 |
Circular dichroism (CD) spectroscopy
CD melting curves were acquired on a JASCO J-810 CD-spectrometer in a 0.2 μm quartz cuvette. Samples contained 10 μM RNA, 25 mM potassium phosphate and 50 mM potassium chloride. The wavelength showing maximal circular dichroism was determined from prior CD-spectra and was in the range from 260 to 265 nm. CD-melting curves were measured at a temperature-sampling rate of 1°C/min in a range from 5 to 95°C with a sampling rate of 10/min. Data was smoothed with a Savitzky-Golay filter (10 points). Melting curves were fit to a sigmoidal curve in the respective sigmoidal region with errors <2%. The melting temperature was obtained as the point with the highest slope.
Isothermal titration calorimetry (ITC)
ITC measurements were performed with either MicroCal PEAQ- or VP-ITC devices (Malvern, United Kingdom) in NMR buffer. In all experiments, ROQ domain protein was titrated from a stock of 10–20-fold excess to 20–40 μM of UCP3 CDE1/2GG or Tnf CDE RNAs provided in the reaction cell. In individual runs, we used 19–25 injections of protein with 150–180 s of spacing at 283 K and a stirring speed of 750 rpm. Raw data were analyzed with the NITPIC software tool (56) and heat production was fitted to a one-site binding model. For all runs, we performed a buffer subtraction using NMR buffer instead of RNA.
Crystallization, diffraction data collection and processing
The crystallization experiments for ROQ-RNA complexes were performed at the X-ray Crystallography Platform at Helmholtz Zentrum München. The crystals of Roquin-1 ROQ (171–326) with UCP3 CDE2GG were grown in 250 mM ammonium sulphate and 30% (v/w) PEG 4000. The crystals of Roquin-1 ROQ (171–326) with UCP3 CDE1GG were grown in 90 mM Bis-tris propane pH 6.5, 290 mM sodium tartrate and 19% (v/w) PEG 3350. Crystallization was performed using the hanging-drop vapor-diffusion method at 292 K in 24-well plates and a protein concentration of 11.5 mg/ml. The crystals appeared after two days. For the X-ray diffraction experiments, the crystals were mounted in a nylon fiber loop and flash cooled to 100 K in liquid nitrogen. The cryoprotection was performed for five seconds in reservoir solution complemented with 25% (v/v) 2-methyl-2,4-pentanediol for the structure with CDE2GG, or 25% (v/v) ethane-1,2-diol for the structure with CDE1GG. X-ray diffraction data for the ROQ-RNA complexes were collected using a Pilatus 2M-F detector at 1.0 Å wavelength on the PXIII X06DA beamline at SLS (Villigen, Switzerland). The best data set was indexed and integrated using XDS (57) and scaled using SCALA (58). Intensities were converted to structure-factor amplitudes using the program TRUNCATE (59). Table 2 summarizes data collection and processing statistics.
Table 2.
Data collection and refinement statistics for the ROQ domain in complex with UCP3 CDEs
| Data collection | ||
| PDB ID | 6TQB | 6TQA |
| RNA name | CDE1GG | CDE2GG |
| Beamline | SLS PXIII X06DA | SLS PXIII X06DA |
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P42212 | P21 |
| Cell dimensions a, b, c (Å) | 86.99, 86.99, 72.99 | 44.53, 160.66, 68.01 |
| α, β, γ (°) | 90, 90, 90 | 90, 107.96, 90 |
| No. of molecules per asymmetric unit | 2 (1 protein + 1 RNA) | 8 (4 protein + 4 RNA) |
| Resolution (Å) | 47–1.6 | 50–2.4 |
| (1.64–1.60)* | (2.46–2.40)* | |
| R merge | 4.7 (73.6) | 10.0 (83.7) |
| I / σI | 23.16 (2.46) | 14.05 (2.15) |
| CC (1/2) | 100 (79.7) | 100 (63.1) |
| Completeness (%) | 96.9 (75.5) | 99.6 (99.8) |
| Redundancy | 8.4 (6.3) | 5.2 (5.1) |
| Refinement | ||
| Resolution (Å) | 47-1.6 | 50-2.4 |
| (1.642–1.600) | (2.462–2.400) | |
| No. reflections | 36 355 (2052) | 33 655 (2618) |
| R work/Rfree | 15.63/19.72 | 19.68/25.90 |
| (23.80/24.10) | (29.40/34.60) | |
| No. atoms | 2195 | 6,942 |
| Protein | 1224 | 4847 |
| RNA | 400 | 1796 |
| Water | 241 | 295 |
| Ions | 7 | 4 |
| B-factor overall | 28.25 | 55.20 |
| RMSDs | ||
| Bond lengths (Å) | 0.013 | 0.01 |
| Bond angles (°) | 1.836 | 1.796 |
| Ramachandran plot | ||
| Most favored (%) | 100 | 96 |
| Additional allowed (%) | 0 | 4 |
*Values in parentheses are for highest-resolution shell.
Structure determination and refinement
The crystal structures of ROQ-RNA were solved by molecular replacement using PHASER (60). The native Roquin-1 ROQ structure served as a search model (PDB: 4QI0 (38)) for solving the structure of Roquin-1 ROQ (aa 171–326) with CDE2GG and the Roquin-1 ROQ structure in complex with Tnf CDE served as a search model (PDB: 4QI2 (38)) for solving the structure of Roquin-1 ROQ (aa 171–326) with CDE1GG. Model building was performed in COOT (61). The RNA molecules were modeled manually. The refinement was performed in REFMAC5 (62) using the maximum-likelihood target function including translation, liberation and screw-rotation displacements of a pseudo-rigid body (63). The final model of the structure with CDE2GG is characterized by R and Rfree factors of 19.7 and 25.9%, the final model of the structure with CDE1GG is characterized by R and Rfree factors of 15.8 and 19.7%, respectively (Table 2). The stereochemical analysis of the final model was done in PROCHECK (64) and MolProbity (65).
Cell culture
HEK293 cells (Leibniz-Institute DSMZ, Germany, ACC 305) were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS Superior, Biochrom), 1 mM sodium pyruvate (Thermo Fischer Scientific) and Pen Strep (Thermo Fisher Scientific) at 37°C in a 5% CO2 humidified incubator.
Transient transfection and luciferase assay
For transfection of pDLP reporter plasmids, 100 000 HEK293 cells were seeded in 24-well plates. 24 h after seeding, cells were transfected with 100 ng reporter plasmid using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's protocol. Firefly and Renilla luciferase activity were measured 24 h post-transfection using the Dual Luciferase Reporter Assay System (Promega).
RNA affinity purification and Western blot
Purified RNA was dephosphorylated using calf intestine phosphatase (Roche). For 5′ end biotinylation, 600 pmol RNA were incubated with 0.2 mM γ-S-ATP (Biomol) and T4 polynucleotide kinase (New England Biolabs) for 30 min at 37°C. Biotin-long-arm maleimide (Vector Laboratories) was added and incubated for 30 min at 65°C. Unincorporated label was depleted by lithium chloride precipitation. Biotinylated RNA (200 pmol) was conjugated to Dynabeads M-280 (Invitrogen) in incubation buffer (10 mM Tris–HCl pH 7.4, 150 mM potassium chloride, 0.5 mM DTT, 0.05% NP40, 100 U/ml RNasin) for 2 h at 4°C with continuous rotation. Whole cell protein lysate (1 mg) of HEK293 cells together with 200 μg yeast tRNA (Sigma-Aldrich) and 5 mg Heparin (Sigma-Aldrich) was added to the beads and incubated 1 h at 4°C followed by 15 min at room temperature with continuous rotation. Beads were washed five times with incubation buffer, resuspended in 30 μl protein loading dye and boiled at 95°C for 10 min. Eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Bio-Rad). The primary antibody was anti-AUF1 (1:10 000; rabbit; Millipore; 07-260) and the secondary antibody was Horseradish-conjugated anti-rabbit (1:7000; goat; Jackson ImmunoResearch). Blots were developed using ECL Select (Life Technologies). Imaging was performed on a ChemiDoc Imaging system (Bio-Rad).
RESULTS
Recognition of UCP3 AU-rich elements is CDE-like
Recognition of AREs by regulatory RBPs has mainly been described for binding of single-stranded (ss) RNAs to RBDs including ZnF and RRM domains, while the formation of ARE secondary structure had been suggested to interfere with ARE-mediated regulation, e.g. in the protein AUF1 (30) (Figure 1A). This general hypothesis has recently been challenged by CDE-like AREs bound by the protein Roquin (44). The 3′-UTR of UCP3 harbors two AU-rich elements with the potential to fold into tri-nucleotide hairpin loops, reminiscent of canonical CDEs (36). CDE1 contains only AU nucleotides and CDE2 harbors the canonical ARE motif 5′-AUUUA-3′ preceded by four AU nucleotides (Figure 1B and Supplementary Figure S1). While regulation of the UCP3 mRNA was shown to function through these sequences, we still lack an unambiguous evidence for whether the unstructured, single-stranded ARE RNA species or only the CDE-folded AREs are the responsible target of the ROQ domain within the functional UCP3 mRNP.
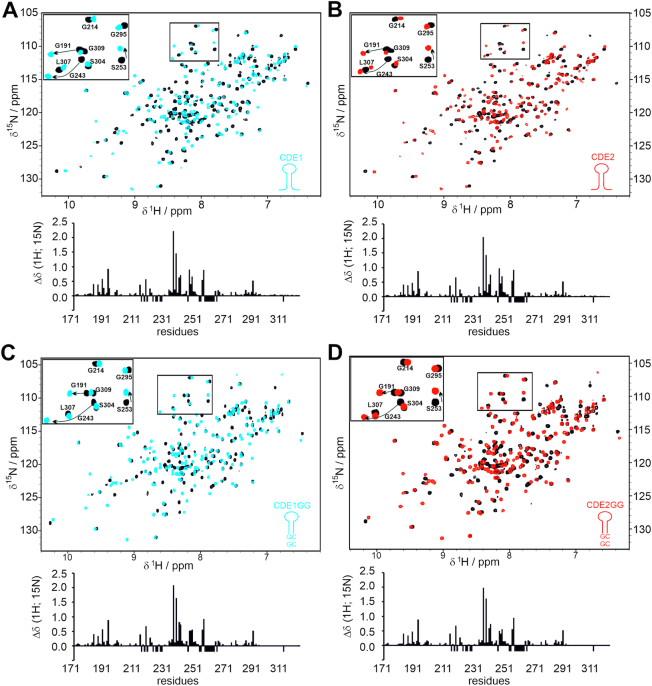
Thus, we sought to investigate the recognition mode of those CDE-AREs by Roquin. Taking into account the high intrinsic lability of AU-rich sequences, we decided to use solution NMR spectroscopy, which is unique in monitoring also weak molecular interactions under near physiological environment and at atomic resolution. We recorded 1H-15N-HSQC spectra comparing CDE1 and CDE2 individually in their free and ROQ domain-bound forms. As shown in Figure 2A and B, native individual CDE RNAs provoke strong chemical shift perturbations (CSPs) in the ROQ domain and the subsequent plot patterns are reminiscent of the CSPs that had been observed with the bona fide Tnf CDE before (38) (Supplementary Figure S2A). In contrast, titration of ROQ with ssRNAs was recently found to cause significantly smaller CSPs, in line with a 10–100-fold lower affinity (45), e.g. for the Nfkbiz 15-mer LBE RNA (Supplementary Figure S2A). This finding supports the assumption that UCP3 AREs are bound by the ROQ domain in a CDE-like manner. Notably, HSQC spectra of ROQ with either CDE are well superimposable despite obvious differences in CDE sequences (Supplementary Figure S2B). This underlines a highly similar, CDE-shape recognition rather than recognition of a linear sequence. The presence of RNA structure in CDE1 and 2 upon binding to ROQ is also evidenced by the observation of imino proton signals indicating the formation of H-bonds between complementary bases in a stem (Supplementary Figure 2E). In contrast, we only detected imino signals for the free GC-containing CDE2, while the pure-AU CDE1 did not reveal the presence of long-lived, i.e. NMR-detectable base pairs in its free form (Supplementary Figures S1 and S2) underlining the low intrinsic stability of AU-rich base pairs, while the underlying sequences suggest to be present in an unfolded, linear form to certain extent of their life time.
Figure 2.
Roquin recognizes wild type and stabilized UCP3 AREs in a CDE-like manner. (A and B) Overlay of 1H-15N-HSQCs showing ROQ alone and in complex with UCP3 wild type CDE1 (A) and CDE2 (B). The bar plots below the spectral overlays show the combined CSPs of amide resonances along with the primary ROQ domain sequence. (C and D) The same as in panels (A) and (B), but with CDE1GG (C) and CDE2GG (D) SLs from the UCP3 3′-UTR. The insets consistently show an equivalent zoom-in from all overlays highlighting a part of the glycine region. Selective amide assignments are given for orientation. Negative bars indicate residues with missing assignments in at least one of the two spectra or prolines. See also Supplementary Figure S2.
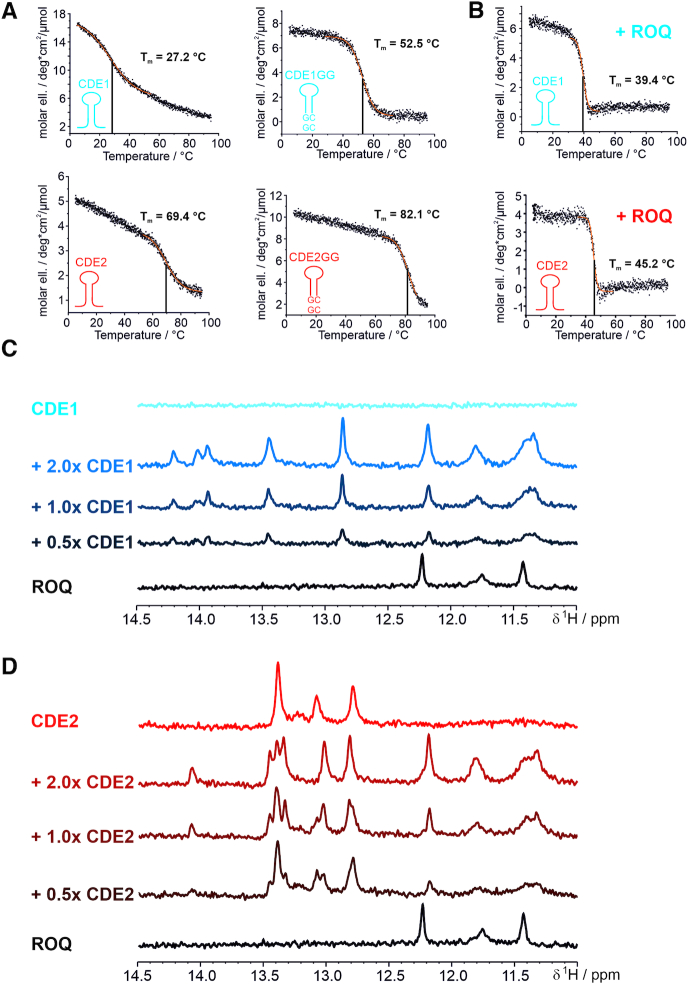
To exclude that only a small fraction of RNA is bound as SL, we next constructed stabilized CDEs with an additional two GC base pairs at the lower stem (Figure 1B). 1D-1H NMR spectra highlighting the respective imino proton regions show that these CDE-ARE versions are thermodynamically more stable in their free forms (Supplementary Figure S1), thus increasing the overall likelihood to be engaged by the ROQ domain in a CDE-preformed shape. HSQC titrations show that the stabilized CDE-AREs provoke an identical pattern in CSP plots; both in significantly shifting residues and absolute intensities (Figure 2C and D). Thus, we conclude that both wild type (WT, non-stabilized) and stabilized UCP3 CDE-AREs are equally bound as structured, CDE-like RNAs.
Structures of folded AU-rich elements
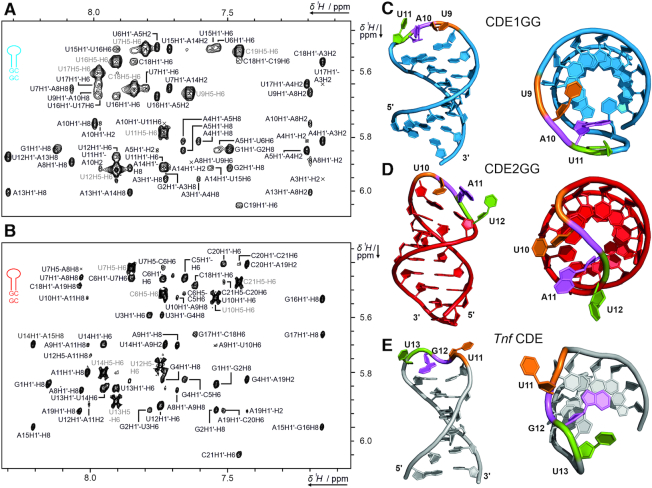
We next investigated whether the UCP3 CDE-AREs form the expected CDE-like SLs in the absence of Roquin, to test for their potency in acting as a pre-structured, unique cis-regulatory element, despite their AU-rich content. Given the apparent high intrinsic dynamics of the WT CDEs, yet the identical binding behavior towards ROQ, we determined the structures and dynamics of apo CDE1GG and CDE2GG by solution NMR (Figure 3 and Supplementary Figures S3 and S4). Spectral regions to demonstrate NOE data quality are displayed in Figure 3A and B for CDE1GG and CDE2GG, respectively, and in Supplementary Figures S3 and S4.
Figure 3.
Solution structures of free CDE1GG and CDE2GG RNAs. (A and B) Zoom-in of the assigned aromatic H1′ region of the 1H,1H-NOESYs of CDE1GG (A) and CDE2GG (B). The intraresidual H5/H6 cross peak assignment is shown in light grey. (C and D) The three dimensional structures of CDE1GG (blue) and CDE2GG (red) after refinement with the force-field RNA-ff1 (55) (see also Table 1). Two orientations for each CDE show the side-on view with the loops pointing into the direction of the viewer, and a top down view displaying the loop-orientation with respect to the helical main axis. Tri-loop residues are labeled in sequential order (orange, magenta and green respectively) to highlight differences in loop geometry. (E) NMR structure of the Tnf CDE (43). See also Supplementary Figures S3 and S4.
For CDE1GG, the ten lowest energy structures converged to an all-residue-all-atom RMSD of 1.7 Å (stem RMSD without terminal residues: 1.1 Å, loop RMSD: 2.2 Å) after structure calculation with ARIA/CNS (Table 1 and Supplementary Figure S3). The CDE1GG stem is composed of eight Watson-Crick base pairs spanning residues 1–8 and 12–19. While residues G1 and C19 show increased RMSD values, common for the termini in RNA SL structures, residues in the core of the stem converge well within the 10 best structures. In contrast, the loop residues are considerably less converged. For example, the loop residue U9 mostly stacks with the A8 residue in direction of the major groove of the stem, while in two structures of the bundle, U9 points to the solvent (Supplementary Figure S3G).
For CDE2GG, the ten lowest energy structures converged to an all-residue-all-atom RMSD of 1.7 Å (stem RMSD without terminal residues: 1.6 Å, loop RMSD: 2.3 Å) (Table 1 and Supplementary Figure 4). The stem consists of nine well-converged Watson–Crick base pairs. In contrast to the structure of CDE1GG, the central purine of the tri-loop (A11) does not point in the direction of the major groove, but in the opposite direction (Supplementary Figure S4G). In nine of the ten structures, the flanking loop residues U10 and U12 point towards the same direction as A11. In a single, deviating structure, we observed a flip of the first loop residue (U10). Notably, the analogous residue in the structure of CDE1GG (U9) showed a similar degree of structural heterogeneity as CDE2GG U10. Increased RMSD may partially be the result of lack of NOE data, but also a consequence of loop dynamics faster than NMR time-scale. As both the CDE1GG and the CDE2GG structures show increased RMSD values for loop nucleotides compared to the base-pairing stem (Table 1), we suggest their increased dynamics to facilitate protein recognition.
The increased dynamics of the two RNA SLs are also evident from heteronuclear relaxation data (Supplementary Figures S3F and S4F). We found around 40% increase of sugar C1′ proton R1 rates and over 15% increase in hetNOE intensity on aromatic protons for residues 8–12 (CDE1GG) and 9–13 (CDE2GG), respectively, compared to other residues. Thus, loops and loop flanking residues show increased dynamics compared to stem residues. In the context of molecular recognition by ROQ (Figure 2), the different loop geometries between the two CDEs suggest that the large dynamics facilitate folding of CDEs into a conformation, susceptible for engagement with the ROQ domain.
We additionally refined the best resulting structures with the RNA-specific force field RNA-ff1 (55), to accommodate to the backbone angles found in X-ray structures of RNAs deposited in the PDB (Table 1 and Figure 3C and D). This procedure removes most of the angular deviations in the loop and therefore does not account for the specifics of loop flexibility. The resulting structure represents the lowest energy and thus most populated structure free in solution. The all-atom RMSD of the refined structures is 0.33 Å for CDE1GG and 0.32 Å for CDE2GG, respectively (Table 1). We attribute the strong tightening of the bundle to parametrization introduced with the torsion potential of the RNA-ff1 force field. In line with that, the additional refinement led to improved backbone conformations reflected by an increased backbone suiteness from 0.19 to 0.69 for CDE1GG and 0.14 to 0.75 for CDE2GG, a value measuring the overall agreement of backbone parameters with a test set of RNA structures (66).
In comparison, the solution structure of the apo Tnf CDE (43) shows a structural arrangement of the loop residues oriented towards the major groove form, i.e. comparable to CDE1GG (Figure 3E). Additionally, a widening of the major groove is observed in the Tnf CDE, attributed to a purine stack in the 3′ side of the stem. As the UCP3 CDE-AREs do not feature this type of purine stack, this effect is not visible in our RNA SL structures. It was further shown that inversion of the two closing base pairs below the tri-loop in the Tnf CDE, and thus distortion of the purine stack, induced large differences in the loop structure, accompanied by loss of Roquin binding capability (43). Interestingly, UCP3 CDE1GG and CDE2GG differ from each other by an inversion of the penultimate loop-closing base pair (Figure 1B), which potentially accounts for the structural alteration observed in the loop, although their binding competence for the ROQ domain appears unaffected (Figure 2 and Supplementary Figure S2). Altogether, we assume that the less rigid AU-rich loop-closing sequence in CDE1 and CDE2 allows for loop dynamics, beneficial for adoption of a ROQ-binding-competent structure.
Structures of folded AU-rich elements in complex with the ROQ domain
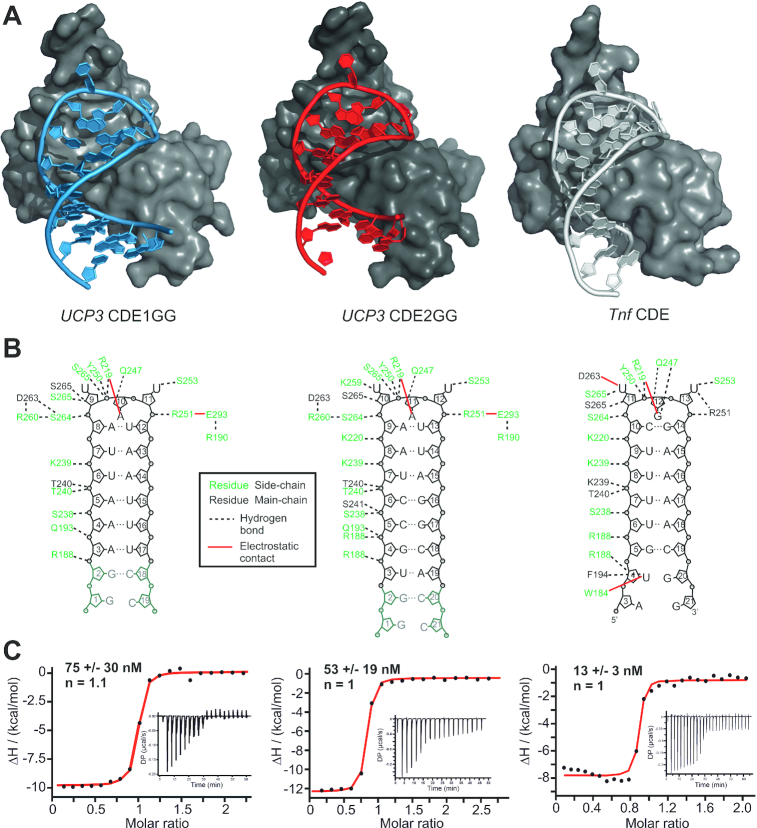
To finally prove that the UCP3 CDE-AREs are tightly complexed by Roquin in the characteristic arrangement, we crystallized the ROQ domain with the two stabilized SLs. We obtained well-diffracting crystals with both SLs and found ROQ-CDE1GG present with one complex molecule per asymmetric unit at a resolution of 1.6 Å (Table 2, Figure 4A and Supplementary Figure S5). ROQ-CDE2GG crystallized with four complex molecules per unit (2.4 Å resolution) that show an overall RMSD of 0.35 Å, while the complex appears as monomer according to the observed line width in NMR spectra. This is in line with all previously published crystal structures of SL RNAs in complex with the core ROQ domain.
Figure 4.
Crystal structures of CDE1GG and CDE2GG in complex with the ROQ domain. (A) Overall structures of ROQ (grey surface) in complex with CDE1GG (blue cartoon) and CDE2GG (red cartoon). For comparison, the same view of ROQ in complex with the Tnf CDE is included (grey cartoon) (38). (B) Internal contact map of ROQ domain residues with UCP3 CDE1GG and CDE2GG in panel (A). For comparison, the same is shown for ROQ in complex with the Tnf CDE (38). (C) Representative ITC curves of ROQ titrated with the RNAs as shown in (B) reveal binding affinities comparable to the Tnf CDE. KD values are mean +/− standard deviation from three independent experiments. See also Supplementary Figure S5.
Importantly, in neither of the two complexes the additional, stabilizing GC base pairs participate in direct interactions with the ROQ domain, indicating that the wild type CDE SLs are sufficient for a canonical CDE-like recognition by Roquin. In case of CDE1GG, this also reflects the first CDE structure with a pure-AU interface with the ROQ domain, adding functional impact and relevance to the so far unlikely concept of structured AREs. In addition, the high resolution for CDE1GG allowed for the observation of two distinct RNA conformations at the 5′ end of the stem, i.e. the first five nucleotides, while the complement strand appears as a single conformer (Supplementary Figure S5B). Although, one of the two conformations appears to be more compacted over the other, we find comparable and complete H-bonding patterns in respective Watson-Crick base pairs. In addition, both conformers share a highly conserved interaction of the Arginine 188 side chain with the backbone of the adenine nucleotide at position 3 (Figure 4B and Supplementary Figure S5C). It indicates that the two forms do not reflect two independent RNP folds, but are likely to be interchanging with each other in solution. This characteristic underlines the special feature of relatively labile CDE SL folds, which are likely to be stabilized by the ROQ domain.
For both complexes, we found a conserved network of interactions between protein and RNA, reflecting the previously described contacts between ROQ and the Tnf CDE (Figure 4A and B). Consequently, the overall fold of all three CDEs in the bound form is highly comparable underlining the earlier suggestion that ROQ recognizes its target RNAs through shape- rather than sequence-specific interactions and locks them in a canonical arrangement that is unique for CDEs (Supplementary Figure S5D). Most remarkably, all three CDEs shown in the overlay resemble an identical orientation of loop nucleotides, which is not reflected by the respective free forms (Supplementary Figure S5E and F). Previous work on the Tnf CDE has shown the role of CDE-intrinsic sequence features for the recognition by the ROQ domain (38,43,44). Comparison of the free and ROQ-bound Tnf CDE also suggested that major rearrangements of the loop nucleotides are necessary for complex formation (Supplementary Figure S5G). In line with that, comparing all individual unbound RNAs of our study with their respective conformations when bound to ROQ suggests, that the ROQ domain either provokes an induced-fit mechanism during engagement with the three CDEs, or that all of them are able to sample the final fold in solution (conformational selection).
Previous data showed a mutual stabilization of both the ROQ domain and CDE RNA upon complex formation (38), which was suggested to be a driving force for the high-affinity interaction between them and underlines the central role of CDE elements for Roquin-mediated post-transcriptional regulation (36,38,40–43,46). Using EMSA experiments, we previously estimated 100 nM affinity between the ROQ domain and the wild type UCP3 tandem CDE-AREs (44) (Figure 1B). Mutational analysis suggested similar contributions from either UCP3 CDE, with affinities in the low micromolar range. This affinity is in line with a CDE found in the Ox40 3′-UTR (39), but considerably higher than measured for the Tnf CDE (38,43). However, the measurements of the single CDEs were performed in the context of the UCP3 tandem RNA, with possible interference from the surrounding sequence context, due to transient interactions and thus weakening of the CDE fold. In order to quantify the individual binding affinity of the UCP3 CDEs, we used ITC and determined KD values for ROQ binding to CDE1GG and CDE2GG. The resulting KDs show tight, low-nanomolar binding, comparable to that of the Tnf CDE (Figure 4C). This reveals two remarkable features: First, these affinities are in full agreement with the ROQ-UCP3 ARE complex arrangement in a CDE-like manner as proven by the NMR data and crystal structures. Importantly, they do not support binding of the CDE-AREs as linear sequence, as ssRNA recognition by the ROQ domain occurs with 2–3 orders of magnitude lower affinity (45). Previous studies on CDEs and ADEs suggested high on-rates during complex formation as one cause for the high affinity (39). From the affinities observed, we here assume a similar kinetic basis, based on preformed SL RNAs. Second, the low KD values suggest a functionally relevant and active recognition of structured ARE elements by a single high-affine RBD, the ROQ domain. The latter is remarkable as the high-affine recognition of AREs had so far only been described for single-stranded ARE species together with the concerted action of multiple RBDs (17,67). In the case of AUF1 for example, both RRMs are necessary for and contribute equally to high affinity binding (68).
Biophysical investigation of UCP3 CDE-ARE-ROQ complexes
Next, we analyzed a potential CDE-stabilizing effect of the Roquin ROQ domain. Such stabilization had been suggested for the Tnf CDE before (38), while no clear experimental proof was provided. Thus, for the UCP3 CDEs, we first assessed differences in stability between the free UCP3 CDEs and their complexed form with ROQ in CD-melting experiments (Figure 5A). All SLs showed comparable and characteristic CD spectra with a maximum ellipticity in the range of wavelengths between 262 and 265 nm and a minimum between 235 and 245 nm (Supplementary Figure S6). As expected, melting points (Tm) largely differed between stabilized and non-stabilized CDEs, owing to the two stabilizing GC base pairs and rationalizing them for structure determination of free RNAs. In line with that, we also found a remarkable difference in melting temperatures of CDE1 (27°C) versus CDE2 (69°C) and CDE1GG (52°C) versus CDE2GG (82°C) (Figure 5A), where in the latter the effect of stabilization is less pronounced due to the intrinsic two GCs base pairs in CDE2 variants (compare RNA sequences in Figure 1B). This finding underlines the drastic lability of pure ARE sequences, but suggests at the same time an inherent potency to be also recognized in their linear form.
Figure 5.
The ROQ domain locks the UCP3 CDE-AREs in the CDE-fold, independent of their thermodynamic instability. (A and B) CD melting curves of CDE1 and CDE2, their respective stabilized constructs (GG) (A) and after addition of equimolar amounts of ROQ domain (B). Respective sigmoidal fitting regions are indicated in orange. Melting points and their errors are indicated as obtained from the fitting procedure. (C and D) Imino proton spectra of the non-stabilized CDE1 (C) and CDE2 (D) at room temperature alone and in complex with different ratios of ROQ domain. As a control, the same region is shown for the ROQ domain alone in order to indicate protein-derived signals. See also Supplementary Figure S6.
The Tm of ROQ alone is 42°C (Supplementary Figure S6) and thus below the melting points of CDE2 and the stabilized CDE variants. We thus used CDE1 to examine the role of the ROQ domain in CDE stabilization. We observed a strong stabilizing effect of ROQ on CDE1 by CD spectroscopy (Figure 5B) evidenced by an increase of Tm from 27°C to 39°C. We thus reason, Roquin captures and stabilizes the transient SL of the pure-AU CDE1 at physiological conditions, while the cis-element alone will be mainly unfolded.
To further verify the stabilizing role of ROQ towards the UCP3 CDEs at atomic level, we tested the ability of the ROQ domain to shift the equilibrium of unfolded CDE1 and CDE2 to the folded species in dependence of protein-RNA ratios (Figure 5C and D). To this end, we used the detectability of imino protons in SL RNAs, which indicate H-bonding of base-paired nucleotides, i.e. structured RNA (Supplementary Figure S1). Indeed, both wild type CDE1 and CDE2 revealed a significant increase in imino proton resonances in response to the presence of the ROQ domain. This suggests formation of stable ROQ-CDE1 and -CDE2 complexes with RNA-structure lifetimes observable during the NMR experiment (here a few hundred milliseconds) that clearly exceed the lifetime of imino protons in the free RNAs. For CDE2, we also observed small changes in imino proton chemical shifts between the apo RNA and the 1:1 complex with the ROQ domain (Figure 5D). We thus conclude that the ROQ domain, upon binding, induces the formation and promotes the stabilization of the CDE-fold in the wild type UCP3 3′-UTR. Surprisingly, this feature appears to be independent from the obvious differences in intrinsic stabilities between the two UCP3 SLs as determined by CD spectroscopy.
This observation suggests that tight binding to the two CDEs is additionally controlled by a low off-rate. Such a binding mechanism mainly requires the apical AUAUU pentamer, while the actual stem sequence is not of apparent relevance, presuming a general stem formation. This is in line with the completely sequence independent consensus for active Roquin binding sites suggested by mutational analyses of the UCP3 CDE-AREs (44).
Independent recognition of the two UCP3 CDEs by Roquin
The accumulated data suggest simultaneous and highly similar binding of ROQ to the two UCP3 CDEs in their endogenous context. To verify that, we made use of a tandem UCP3 RNA construct (CDE1-2, Supplementary Table S1) that harbors both CDEs in close sequential proximity (Figure 1B). To prevent dimerization observed at high RNA concentrations with the wild type sequence (Supplementary Figure S7A), a point mutation had to be introduced in the linker region between the two CDEs. Mutations in the linker region were shown not to affect mRNA repression in vivo (44). In order to ensure that the tandem construct used for NMR studies retains full repressive capacity, it was tested in a luciferase reporter assay. As expected the point mutation had no significant effect on the regulation in vivo (Supplementary Figure S7B). Thus, the construct is suitable for further analyses.
Earlier data had given good evidence for a 1:2 complex between the UCP3 RNA and ROQ domain (44). Consequently, to directly monitor the formation of comparable complexes (compare also Figure 2) at atomic resolution we titrated tandem UCP3 to the ROQ domain and recorded HSQC spectra (Supplementary Figure S2). Excess RNA (>0.5× with respect to the number of available CDEs) allows the interpretation of detectable complex resonances. The overall occupancy of CDEs showed a 1:1 distribution, which indicates that ROQ does not discriminate between the two CDEs for the formation of tight complexes. Consequently, in the tandem complex, both CDE-complexes appear with a comparable lifetime, indicating similar complex stability. However, we find a number of resonances that suggest a slight preference for CDE2, which is in line with its relatively higher intrinsic stability measured by CD. Strikingly, the two SLs behave fully independent as evidenced by the unaffected chemical shifts of individual imino protons (Supplementary Figure S1).
Competition of RBPs for AU-rich elements
The NMR and CD data clearly demonstrate that ROQ binding can rigidify the tandem-CDE regulatory hub within the 3′-UTR of the UCP3 mRNA. However, in the absence of Roquin binding, this region will be largely unstructured and thus accessible for RBPs that specifically recognize linear AREs. Thus, Roquin will be able to compete with these RBPs by removing their ssRNA target sites in the 3′-UTR, while conversely these RBPs will be able to compete with Roquin by masking the AREs and preventing them from adopting a CDE-fold.
To investigate competition for identical ARE-foldamers by two RBPs we included the well-studied protein AUF1 (hnRNP D) into the Roquin-UCP3 interactome (Figures 1A and 6A). The high affinity of AUF1 to AU-rich sequences has already been described (69), and such sequences are present in the studied UCP3 tandem CDE (Figure 6A). Additionally, previous work has shown that structure formation in AREs abolished binding and mRNA regulation by AUF1 (30,31). AUF1 thus represents a promising study case for the regulatory capacity that centers on dynamic CDE-AREs. The AUF1 protein comprises two canonical RRM domains, arranged in tandem that account for RNA-binding (69). With NMR, we found that the two AUF1 RRM domains are independent from each other, as shown by a 15N-HSQC overlay comparing tandem RRM1-2 and RRM2. Here, the single RRM2 domain provides resonances that are in full overlay with the respective resonances in the tandem RRM1-2 indicating that the two RRMs are structurally independent from each other with respect to their two-site RNA-binding capacity (Supplementary Figure S8A). As a prerequisite for the following experiments, we also ruled out an interaction of AUF1 RRMs with the Roquin ROQ domain (Supplementary Figure S8B).
Figure 6.
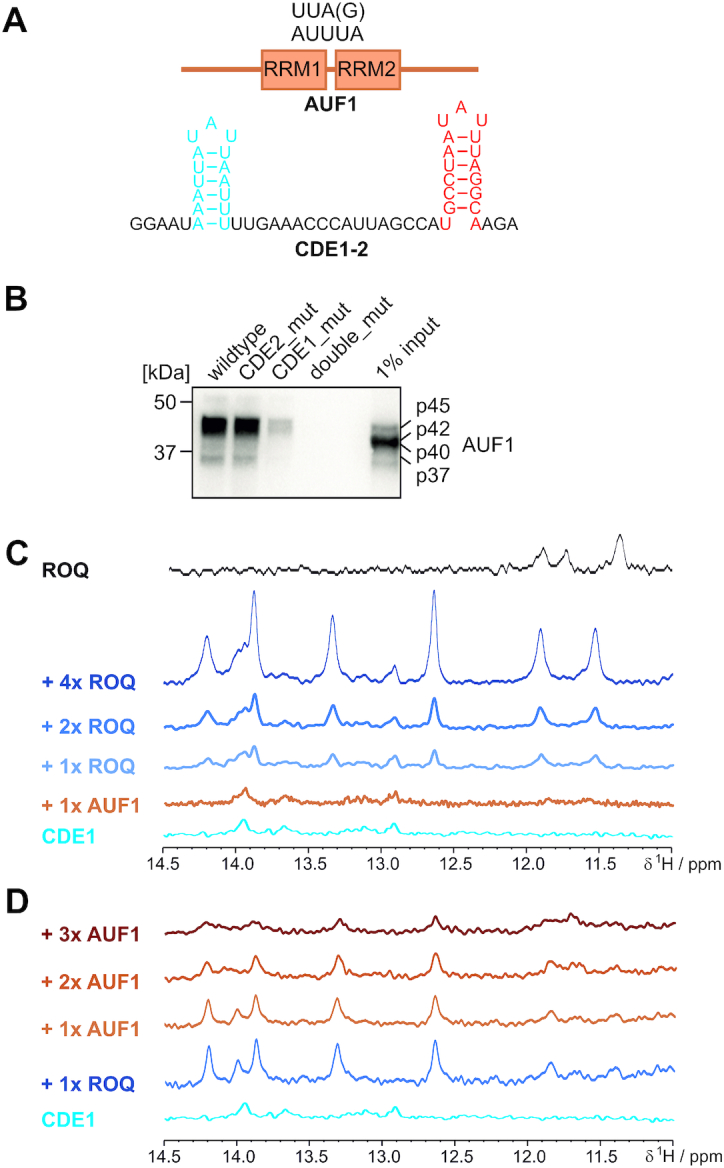
Competition of Roquin with AUF1 for the UCP3 ARE hub. (A) Top, schematic presentation of AUF1 domain arrangement with approximate position of the tandem RRM1-2 domains. Below that, the wild type UCP3 ARE hub is shown as used in this study and for the pulldown experiment in panel (B). (B) Analysis of Roquin binding to the UCP3 ARE hub. For RNA affinity purification, HEK293 whole cell lysates were incubated with the indicated UCP3 RNAs. AUF1 was visualized by Western blot using anti-AUF1 antibody. (C and D) NMR-based in vitro competition experiments between the Roquin ROQ domain and the AUF1 tandem RRM1-2. (C) Imino proton spectra of WT UCP3 CDE1 alone, in a 1:1 complex with AUF1 and after addition of increasing amounts of ROQ. (D) The same as in (C), but starting from a 1:1 complex of CDE1 with ROQ and subsequent titration of increasing amounts of AUF1 RRM1-2. For panels (C) and (D), the same spectral region is shown for ROQ alone for clarity towards protein-derived peaks. See also Supplementary Table S1 and Supplementary Figure S8.
We next tested whether AUF1 is capable of binding to the tandem UCP3 RNA using RNA immunoprecipitation. Notably, AUF1 isoforms can readily be isolated from cell lysates using the wild type UCP3 tandem CDE (Figure 6B). Further, overlays of 15N-HSQC spectra confirm the interaction of the two AUF1 RRMs with both individual UCP3 CDEs and the tandem construct. The spectra show large chemical shift perturbations accompanied by line broadening which is in line with the nanomolar affinities that had been reported for AUF1-ARE interactions earlier (70). As expected for canonical RRM domains, binding is primarily mediated by the intrinsic RNP motifs (Supplementary Figure S8C–E).
Interestingly, mutants of the RNA within CDE1 or 2 of the tandem UCP3 RNA revealed a significantly more pronounced loss in binding for CDE1, which is solely composed of AU residues (Figure 6A and B). This is in line with AUF1’s primary preference for extended AU-rich regions (71,72) and suggests a slight preference for the CDE1 region within the tandem RNA construct.
We thus decided to monitor the presence of imino protons in CDE1 as a measure of the RBP-competition for different RNA-foldamers. The wild type CDE1 did not show a significant proportion of structure on the NMR time scale (Figure 5C). Addition of AUF1 to the wild type CDE1 yielded no visible increase in RNA structure (Figure 6C), in line with the previously described target specificity of AUF1 for linear AREs (70). We next added increasing stoichiometric amounts of Roquin ROQ domain to the mixture. As expected, we observed a stepwise increase in imino signals indicating the stabilization of the CDE-fold in a ROQ-RNA complex (compare to Figure 5). Taking into account the high affinity interaction of AUF1 RRMs with CDE1 (Supplementary Figure S8), it suggests that AUF1 has to be occluded from the RNA-interface in favor of the ROQ domain. ROQ-binding sequesters the former linear ARE and thereby excludes AUF1 from its potential regulatory function on dynamic AREs. To exclude the potential scenario of both RBPs being bound to CDE1, we next started from a pre-formed ROQ-CDE1 complex and added increasing stoichiometric amounts of AUF1 RRM1-2 (Figure 6D). As expected, the increase in AUF1 led to a stepwise loss of imino resonances in CDE1 indicating that AUF1 is able to replace Roquin, and thus unfolds the CDE1.
To estimate the occurrence and folding of further CDE-AREs in vivo, we analyzed whether AU-pure CDEs overlap with binding sites of AUF1 derived from PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) data in HEK293 cells (22). A total of 146 AU-pure CDEs with a UAU tri-loop, i.e. analogous to CDE1, were predicted in the 3′-UTRs of 128 human genes (44). Of these, 48 genes (38%), containing 55 AU-pure CDEs (38%) in their 3′-UTRs, are recognized by AUF1 (Supplementary Table S3). Although these 48 genes together harbor 119 binding sites, only 4 binding sites (3%) overlap with CDE-AREs. Importantly, most of the genes targeted by AUF1 are also bound by Roquin-1 in HEK293 cells (37 genes, 77%), as shown by PAR-CLIP (42). Together, the data suggest that CDE-AREs are widespread and most often excluded from AUF1 binding, likely due to their structured nature and/or subsequent occupation by competing trans-acting factors. Interestingly, the four CDE-AREs bound by AUF1 were also recognized by the mRNA stabilizing factor HuR (22), thus further extending the landscape of possible protein interactors.
In summary, the data suggest competitive binding between Roquin ROQ and the AUF1 RRMs to the same cis-regulatory RNA sequence, while targeting different folding states and actively interfering with the equilibrium between the two forms.
DISCUSSION
The ROQ domain recognizes UCP3 cis-regulatory AREs in a pre-formed CDE-like manner
While AREs and CDEs had long been conceived as separate entities, functioning as parallel, redundant or eventually cooperative cis-elements (29,34,46), we have recently shown that AREs can be functioning as CDEs (44) and consequently to be regulated by the protein Roquin. Their low intrinsic half-lives as SLs suggested that they could fulfil additional roles as linear cis-elements. However, an unambiguous, i.e. structural proof, for the existence of CDE-shaped AREs and their recognition by Roquin in a CDE-like manner had remained open.
We show here that the two adjacent Roquin-regulatory cis-elements in the 3′-UTR of the UCP3 mRNA indeed form CDE-like folds. This holds true for the CDEs alone (Figure 3) and in complex with the protein (Figure 4). We also show that in solution, Roquin engages with these CDEs in a manner identical to the binding of the canonical Tnf CDE (38) and with similar, low nanomolar affinities. Thus, our data proof the previously described functional role for the UCP3 SLs as CDEs. Moreover, they generally suggest CDE-AREs as a subclass of CDEs that allow for unique regulatory features. Such features are particularly obvious for the pure-AU CDE1 of UCP3, which did not reveal base pairing in its free form, suggesting it to be unfolded in average on the NMR-observed time scale. Notably, we found CDE1 to be stabilized in its folded form by the Roquin protein, with a melting temperature of 27°C in its free form, while ROQ domain binding increases CDE1 stability to 39°C. We speculate that this narrow, physiologically relevant temperature window for stabilization by an RBP expresses the functional dynamics of interactions between Roquin and transiently folded AREs. Under physiological conditions, those CDE-AREs are in constant exchange with the unfolded form, and Roquin is replaced by competing RBPs through the adjustment of relative protein levels.
For Roquin, it has been discussed that RNA binding requires a certain RNA flexibility, while the stable ROQ-CDE complex is of a highly conserved geometry. Interestingly, this geometry is also present in ROQ-ADE complexes (39). It clearly highlights the shape-specificity of Roquin for RNA, rather than a particular sequence requirement. We found that both UCP3 CDEs in complexes with the ROQ domain reflect an identical RNP arrangement, which is furthermore identical to all other ROQ-CDE structures (38,40,73–75). Interestingly, other than for the Tnf CDE, the two stabilized UCP3 CDEs adopt in their free forms a fold, which is reminiscent of the ROQ-bound form. It remains open, whether this pre-arranged element supports their interaction with ROQ and if it is a particular feature of the UCP3 CDEs.
Taking into account the labile nature of free UCP3 CDEs (Supplementary Figure S1), we assume that complex stability is mainly driven by a low off-rate, where the ROQ domain locks CDEs in a compact structure. Ever since the discovery of CDEs, their intrinsic instability has been suggested to account for their regulatory potency. Indeed, former work has shown that GCs are by default underrepresented in CDEs (36) and a higher GC content does not per se increase affinity for ROQ, as e.g. seen for the CDE-like SL in the Ox40 3′-UTR (38,39). This is also supported by the U-rich SLs found in a PAR-CLIP study that are recognized by Roquin and show an almost exclusive content of AUs in their stems (42). In fact, the role of intrinsic lability for enabling an induced-fit with the ROQ domain was correlated with a preference for polyA-stretches along the 3′-side of the stem, i.e. this unique geometry was suggested to be a driving force of high-affine binding to Roquin as shown for the Tnf CDE (38). However, we recently showed that these stretches are no requirement for active CDEs (44).
As contradictive findings from mutational data on CDEs have been reported (36,38,43,44), our data essentially suggest binding to be facilitated by the pure possibility that a potential CDE can be formed in vivo. In principle, a plethora of short (∼20 nt) sequences will be capable of adopting CDE-like folds Accordingly, hundreds of low-populated, potential CDEs have been predicted in a genome-wide bioinformatic screen (44). Despite their potential to form a stable complex with Roquin, they were thought to be mainly unfolded at physiological conditions or to be hidden in a larger RNA secondary structure context. Nevertheless, such sequences might also be functional Roquin targets, when they transiently adopt a CDE-like fold.
In sum, we present the first pure-AU CDE that resembles a bona-fide CDE-like fold and thus extend the possible cis-element target group of Roquin for mRNA regulation on a structural basis. Indeed, while all previous studies have missed to provide clear structural evidence for a CDE-like fold of AREs, we here unambiguously prove the existence of such CDE-AREs in the free and ROQ-bound form with high resolution. However, while we assume a gain in complex stability for both RNA and ROQ, the question remains whether the rate-limiting step in vivo will be the prior formation of CDE-folded SLs; assuming a constant pressure against RNA unfolding (76) and the competition with miRNAs and proteins that favor linear AREs (77).
Opposing specificity for UCP3 ARE conformers by the Roquin ROQ domain vs. AUF1 RRMs
Our experiments show such a competition between Roquin and AUF1 for the same ARE in vitro, targeting either the folded or the linear form, respectively. As such, we suggest a mutually exclusive, i.e. functionally competitive or redundant action of RBPs on AREs with transient structure like the herein described CDE-AREs. Notably, the AUF1 isoform p45 is capable of dissolving viral SL structures, thus promoting genome replication of flaviviruses (78,79), and we found this isoform to be most effectively enriched after pulldown with the UCP3 CDEs (Figure 6). At the same time, an increase in RNA structure at its target cis-element environment clearly interferes with regulation of mRNA fate by the AUF1 isoform p37 (30). Strikingly, ARE structure formation was shown to differentially affect trans-factor binding (31), indicating a complex interplay of cis-trans interactions for mRNA regulation depending on mRNA structure and the exact preferences of regulatory RBPs.
Two scenarios are thus to be considered for post-transcriptional regulation: First, the relative proportions of RNA forms will decide about the accessibility for the respective RBPs. Herein, the interconversion between folded and unfolded RNA is a decisive parameter for the possible RNPs to be formed. Second, relative affinities and amounts of available protein will decide about the occupancy of AREs and their tight stabilization in the linear or folded forms, respectively. The individual AUF1 RRMs have an affinity of 300–800 nM for AREs, depending on the exact RNA sequence and protein isoform (69). Those numbers are in the identical order of affinity with Roquin binding to CDEs and ADEs (Figure 4C and (38,39)). Consequently, mRNA fate will depend on the relative RBP levels and their local availability. As shown in Figure 6, replacement of ROQ requires an excess of AUF1 on the investigated time scale. This is consistent with the finding that potentially folded CDE-AREs are underrepresented as binding sites of AUF1. In the case of counter-acting RBPs that compete for (CDE-)AREs, folding probability, affinity and availability of the trans-acting factors represent critical contributions and regulatory levels for individual fates of target mRNAs.
In immune-regulatory proteins like Regnase and Roquin, high-affine and specific binding of structured decay elements is accompanied by the recognition of adjacent AREs through the co-existing zinc finger domains. Despite the discussable gain in specificity and affinity (42,45,80), the role of ARE recognition by the Roquin ZnF has appeared to be clearly subordinated to the function of the ROQ domain after the discovery of CDE and ADE elements (36,39,46), with the major role for the ROQ domain in mRNA regulation (38,40). Our findings now provide an additional option, in that the Roquin ROQ domain would also account for mRNA regulation through pure AU-rich sites, which can fold into CDE-shaped SLs. This would increase the likelihood of Roquin to engage with its mRNA target regions when either using its ZnF or ROQ domains for identical AU-rich cis-elements.
Interestingly, genome-wide bioinformatic analyses predicted >100 CDEs consisting exclusively of AUs (see Supplementary Table S3). Moreover, several of these show evolutionary conservation, highly suggestive of further instances of CDE-AREs (44). This might explain possible target overlap with proteins that do not apparently recognize CDEs. In this context, recent work has also presented the novel RBP Arid5a that appears to counteract Roquin and Regnase binding, possibly by destabilization or refolding of SL elements (81,82). Further, Roquin recognition of an SL element in the Pten 3′-UTR competes with miRNA binding (83). Since miRNAs recognize ssRNA sequences by direct base pairing, at least partial melting of the Pten SL would be necessary.
Our findings pose the question whether also other cis-elements recognized by sequence-specific proteins in their linear form, might be structured more often and thus could be bound by RBPs with shape-specific domains. Strikingly, considering cis-element accessibility significantly increases the prediction of in vivo binding sites of RBPs, compared to predictions based only on sequence preferences (84). This suggests that linear motifs are often masked within RNA folds, limiting recognition by trans-acting factors that require single strand regions. There are >1000 proteins encoded in the human genome that are capable of interacting with mRNA, while binding preferences for the vast majority of them are unknown (85,86). Likely, the herein presented findings of protein competition for differently folded forms of the same ARE can be extended to other cis-elements that have so far been regarded as purely linear acting motifs. In essence, our data imply a role for transiently folded cis-elements in mRNA regulation. Our work highlights how cis-elements are used as signal-integrative hubs, and mRNA regulation is a consequence of cis-element availability/accessibility and the relative abundance of competitive RBPs.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures were deposited in the Protein Data Bank under the accession numbers 6TQB and 6TQA for the ROQ domain with UCP3 CDE1GG and CDE2GG, respectively. UCP3 CDE1GG and CDE2GG NMR structural ensembles were deposited under the accession numbers 6XXW and 6XJW in the Protein Data Bank and under IDs 34483 and 34482 in the BMRB, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Frankfurt BMRZ NMR facility for excellent support. We are grateful to Michael Sattler (Technical University and Helmholtz Centre Munich) and the Bavarian NMR centre for the possibility to perform some of the NMR and ITC experiments.
Notes
Present address: Johannes Braun, Center for Thrombosis and Homeostasis, Johannes Gutenberg University Medical Center, Mainz 55131, Germany.
Contributor Information
Oliver Binas, Institute for Organic Chemistry and Chemical Biology, Goethe University Frankfurt and Center for Biomolecular Magnetic Resonance (BMRZ), 60438 Frankfurt, Germany.
Jan-Niklas Tants, Institute for Molecular Biosciences, Goethe University Frankfurt and Center for Biomolecular Magnetic Resonance (BMRZ), 60438 Frankfurt, Germany.
Stephen A Peter, Department of Biology, Technical University of Darmstadt, Darmstadt 64287, Germany.
Robert Janowski, Institute of Structural Biology, Helmholtz-Zentrum München, 85764 Neuherberg, Germany.
Elena Davydova, Institute of Structural Biology, Helmholtz-Zentrum München, 85764 Neuherberg, Germany.
Johannes Braun, Department of Biology, Technical University of Darmstadt, Darmstadt 64287, Germany.
Dierk Niessing, Institute of Structural Biology, Helmholtz-Zentrum München, 85764 Neuherberg, Germany; Institute of Pharmaceutical Biotechnology, Ulm University, 89081 Ulm, Germany.
Harald Schwalbe, Institute for Organic Chemistry and Chemical Biology, Goethe University Frankfurt and Center for Biomolecular Magnetic Resonance (BMRZ), 60438 Frankfurt, Germany.
Julia E Weigand, Department of Biology, Technical University of Darmstadt, Darmstadt 64287, Germany.
Andreas Schlundt, Institute for Molecular Biosciences, Goethe University Frankfurt and Center for Biomolecular Magnetic Resonance (BMRZ), 60438 Frankfurt, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB902/B14, WE5819/3-1 to J.E.W., SFB902/B16, SCHL2062/2-1 to A.S., SFB902/A1 to H.S.]; Johanna Quandt Young Academy at Goethe [2019/AS01 to A.S.]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1. Baumjohann D., Heissmeyer V.. Posttranscriptional gene regulation of T follicular helper cells by RNA-Binding proteins and microRNAs. Front. Immunol. 2018; 9:1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yoshinaga M., Takeuchi O.. Post-transcriptional control of immune responses and its potential application. Clin Transl Immunol. 2019; 8:e1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beisang D., Bohjanen P.R.. Perspectives on the ARE as it turns 25 years old. Wiley Interdiscip. Rev RNA. 2012; 3:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A.. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen C.Y., Shyu A.B.. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 1994; 14:8471–8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kruys V.I., Wathelet M.G., Huez G.A.. Identification of a translation inhibitory element (TIE) in the 3′ untranslated region of the human interferon-beta mRNA. Gene. 1988; 72:191–200. [DOI] [PubMed] [Google Scholar]
- 7. Jones T.R., Cole M.D.. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol. Cell. Biol. 1987; 7:4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaw G., Kamen R.. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986; 46:659–667. [DOI] [PubMed] [Google Scholar]
- 9. Wilson T., Treisman R.. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988; 336:396–399. [DOI] [PubMed] [Google Scholar]
- 10. Fallmann J., Sedlyarov V., Tanzer A., Kovarik P., Hofacker I.L.. AREsite2: an enhanced database for the comprehensive investigation of AU/GU/U-rich elements. Nucleic Acids Res. 2016; 44:D90–D95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khabar K.S. Hallmarks of cancer and AU-rich elements. Wiley Interdiscip. Rev. RNA. 2017; 8:e1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bolognani F., Perrone-Bizzozero N.I.. RNA-protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 2008; 86:481–489. [DOI] [PubMed] [Google Scholar]
- 13. Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J.. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005; 120:623–634. [DOI] [PubMed] [Google Scholar]
- 14. Ma F., Liu X., Li D., Wang P., Li N., Lu L., Cao X.. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J. Immunol. 2010; 184:6053–6059. [DOI] [PubMed] [Google Scholar]
- 15. von Roretz C., Di Marco S., Mazroui R., Gallouzi I.E.. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip. Rev. RNA. 2011; 2:336–347. [DOI] [PubMed] [Google Scholar]
- 16. Tran H., Maurer F., Nagamine Y.. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell. Biol. 2003; 23:7177–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X., Tanaka Hall T.M.. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 2001; 8:141–145. [DOI] [PubMed] [Google Scholar]
- 18. Ma W.J., Chung S., Furneaux H.. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997; 25:3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 1991; 11:2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. White E.J., Matsangos A.E., Wilson G.M.. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip. Rev. RNA. 2017; 8:doi:10.1002/wrna.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson G.M., Sun Y., Lu H., Brewer G.. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 1999; 274:33374–33381. [DOI] [PubMed] [Google Scholar]
- 22. Yoon J.H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K.M., Noh J.H., White E.J., Martindale J.L. et al.. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014; 5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barreau C., Paillard L., Osborne H.B.. AU-rich elements and associated factors: are there unifying principles. Nucleic Acids Res. 2005; 33:7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gronland G.R., Ramos A.. The devil is in the domain: understanding protein recognition of multiple RNA targets. Biochem. Soc. Trans. 2017; 45:1305–1311. [DOI] [PubMed] [Google Scholar]
- 25. Schneider T., Hung L.H., Aziz M., Wilmen A., Thaum S., Wagner J., Janowski R., Müller S., Hüttelmaier S., Niessing D. et al.. Combinatorial recognition of clustered RNA elements by a multidomain RNA-binding protein, IMP3. Nat. Commun. 2019; 10:2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schlundt A., Tants J.N., Sattler M.. Integrated structural biology to unravel molecular mechanisms of protein-RNA recognition. Methods. 2017; 118–119:119–136. [DOI] [PubMed] [Google Scholar]
- 27. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bohjanen P.R., Petryniak B., June C.H., Thompson C.B., Lindsten T.. AU RNA-binding factors differ in their binding specificities and affinities. J. Biol. Chem. 1992; 267:6302–6309. [PubMed] [Google Scholar]
- 29. Paschoud S., Dogar A.M., Kuntz C., Grisoni-Neupert B., Richman L., Kuhn L.C.. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol. Cell. Biol. 2006; 26:8228–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson G.M., Sutphen K., Chuang K., Brewer G.. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J. Biol. Chem. 2001; 276:8695–8704. [DOI] [PubMed] [Google Scholar]
- 31. Fialcowitz E.J., Brewer B.Y., Keenan B.P., Wilson G.M.. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J. Biol. Chem. 2005; 280:22406–22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vinuesa C.G., Cook M.C., Angelucci C., Athanasopoulos V., Rui L., Hill K.M., Yu D., Domaschenz H., Whittle B., Lambe T. et al.. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005; 435:452–458. [DOI] [PubMed] [Google Scholar]
- 33. Matsushita K., Takeuchi O., Standley D.M., Kumagai Y., Kawagoe T., Miyake T., Satoh T., Kato H., Tsujimura T., Nakamura H. et al.. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009; 458:1185–1190. [DOI] [PubMed] [Google Scholar]
- 34. Fu M., Blackshear P.J.. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 2017; 17:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H.H., Ori D., Uehata T., Tartey S., Akira S., Suzuki Y. et al.. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 2015; 161:1058–1073. [DOI] [PubMed] [Google Scholar]
- 36. Leppek K., Schott J., Reitter S., Poetz F., Hammond M.C., Stoecklin G.. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013; 153:869–881. [DOI] [PubMed] [Google Scholar]
- 37. Wilamowski M., Gorecki A., Dziedzicka-Wasylewska M., Jura J.. Substrate specificity of human MCPIP1 endoribonuclease. Sci. Rep. 2018; 8:7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schlundt A., Heinz G.A., Janowski R., Geerlof A., Stehle R., Heissmeyer V., Niessing D., Sattler M.. Structural basis for RNA recognition in Roquin-mediated post-transcriptional gene regulation. Nat. Struct. Mol. Biol. 2014; 21:671–678. [DOI] [PubMed] [Google Scholar]
- 39. Janowski R., Heinz G.A., Schlundt A., Wommelsdorf N., Brenner S., Gruber A.R., Blank M., Buch T., Buhmann R., Zavolan M. et al.. Roquin recognizes a non-canonical hexaloop structure in the 3′-UTR of Ox40. Nat. Commun. 2016; 7:11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tan D., Zhou M., Kiledjian M., Tong L.. The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat. Struct. Mol. Biol. 2014; 21:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schuetz A., Murakawa Y., Rosenbaum E., Landthaler M., Heinemann U.. Roquin binding to target mRNAs involves a winged helix-turn-helix motif. Nat. Commun. 2014; 5:5701. [DOI] [PubMed] [Google Scholar]
- 42. Murakawa Y., Hinz M., Mothes J., Schuetz A., Uhl M., Wyler E., Yasuda T., Mastrobuoni G., Friedel C.C., Dolken L. et al.. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-kappaB pathway. Nat. Commun. 2015; 6:7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Codutti L., Leppek K., Zalesak J., Windeisen V., Masiewicz P., Stoecklin G., Carlomagno T.. A distinct, Sequence-Induced conformation is required for recognition of the constitutive decay element RNA by Roquin. Structure. 2015; 23:1437–1447. [DOI] [PubMed] [Google Scholar]
- 44. Braun J., Fischer S., Xu Z.Z., Sun H., Ghoneim D.H., Gimbel A.T., Plessmann U., Urlaub H., Mathews D.H., Weigand J.E.. Identification of new high affinity targets for Roquin based on structural conservation. Nucleic Acids Res. 2018; 46:12109–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Essig K., Kronbeck N., Guimaraes J.C., Lohs C., Schlundt A., Hoffmann A., Behrens G., Brenner S., Kowalska J., Lopez-Rodriguez C. et al.. Roquin targets mRNAs in a 3′-UTR-specific manner by different modes of regulation. Nat. Commun. 2018; 9:3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stoecklin G., Lu M., Rattenbacher B., Moroni C.. A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway. Mol. Cell. Biol. 2003; 23:3506–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kemmerer K., Fischer S., Weigand J.E.. Auto- and cross-regulation of the hnRNPs D and DL. RNA. 2018; 24:324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kemmerer K., Weigand J.E.. Hypoxia reduces MAX expression in endothelial cells by unproductive splicing. FEBS Lett. 2014; 588:4784–4790. [DOI] [PubMed] [Google Scholar]
- 49. Richter C., Reif B., Worner K., Quant S., Marino J.P., Engels J.W., Griesinger C., Schwalbe H.. A new experiment for the measurement of nJ(C,P) coupling constants including 3J(C4′i,Pi) and 3J(C4′i,Pi+1) in oligonucleotides. J. Biomol. NMR. 1998; 12:223–230. [DOI] [PubMed] [Google Scholar]
- 50. Schwalbe H., Samstag W., Engels J.W., Bermel W., Griesinger C.. Determination of 3J(C,P) and 3J(H,P) coupling constants in nucleotide oligomers with FIDS-HSQC. J. Biomol. NMR. 1993; 3:479–486. [Google Scholar]
- 51. Rinnenthal J., Richter C., Ferner J., Duchardt E., Schwalbe H.. Quantitative gamma-HCNCH: determination of the glycosidic torsion angle chi in RNA oligonucleotides from the analysis of CH dipolar cross-correlated relaxation by solution NMR spectroscopy. J. Biomol. NMR. 2007; 39:17–29. [DOI] [PubMed] [Google Scholar]
- 52. Linge J.P., Habeck M., Rieping W., Nilges M.. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003; 19:315–316. [DOI] [PubMed] [Google Scholar]
- 53. Nozinovic S., Furtig B., Jonker H.R., Richter C., Schwalbe H.. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010; 38:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garcia de la Torre J., Huertas M.L., Carrasco B.. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000; 147:138–146. [DOI] [PubMed] [Google Scholar]
- 55. Schwieters C.D., Kuszewski J.J., Clore G.M.. Using Xplor–NIH for NMR molecular structure determination. Progr. NMR Spectrosc. 2006; 48:47–62. [Google Scholar]
- 56. Keller S., Vargas C., Zhao H., Piszczek G., Brautigam C.A., Schuck P.. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 2012; 84:5066–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 59. Collaborative Computational Project, N The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 60. Read R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D. Biol. Crystallogr. 2001; 57:1373–1382. [DOI] [PubMed] [Google Scholar]
- 61. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 63. Winn M.D., Murshudov G.N., Papiz M.Z.. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003; 374:300–321. [DOI] [PubMed] [Google Scholar]
- 64. Laskowski R., MacArthur M.W., Moss D.S., Thornton J.M.. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993; 26:283–291. [Google Scholar]
- 65. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Richardson J.S., Schneider B., Murray L.W., Kapral G.J., Immormino R.M., Headd J.J., Richardson D.C., Ham D., Hershkovits E., Williams L.D. et al.. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution). RNA. 2008; 14:465–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hudson B.P., Martinez-Yamout M.A., Dyson H.J., Wright P.E.. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004; 11:257–264. [DOI] [PubMed] [Google Scholar]
- 68. Meyer A., Golbik R.P., Sanger L., Schmidt T., Behrens S.E., Friedrich S.. The RGG/RG motif of AUF1 isoform p45 is a key modulator of the protein's RNA chaperone and RNA annealing activities. RNA Biol. 2019; 16:960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kajita Y., Nakayama J., Aizawa M., Ishikawa F.. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J. Biol. Chem. 1995; 270:22167–22175. [DOI] [PubMed] [Google Scholar]
- 70. Zucconi B.E., Ballin J.D., Brewer B.Y., Ross C.R., Huang J., Toth E.A., Wilson G.M.. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J. Biol. Chem. 2010; 285:39127–39139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Katahira M., Miyanoiri Y., Enokizono Y., Matsuda G., Nagata T., Ishikawa F., Uesugi S.. Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA. J. Mol. Biol. 2001; 311:973–988. [DOI] [PubMed] [Google Scholar]
- 72. Nagata T., Kurihara Y., Matsuda G., Saeki J., Kohno T., Yanagida Y., Ishikawa F., Uesugi S., Katahira M.. Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0. J. Mol. Biol. 1999; 287:221–237. [DOI] [PubMed] [Google Scholar]
- 73. Schlundt A., Niessing D., Heissmeyer V., Sattler M.. RNA recognition by Roquin in posttranscriptional gene regulation. Wiley Interdiscip. Rev. RNA. 2016; 7:455–469. [DOI] [PubMed] [Google Scholar]
- 74. Sakurai S., Ohto U., Shimizu T.. Structure of human Roquin-2 and its complex with constitutive-decay element RNA. Acta Crystallogr. F Struct. Biol. Commun. 2015; 71:1048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Q., Fan L., Hou F., Dong A., Wang Y.X., Tong Y.. New insights into the RNA-Binding and E3 Ubiquitin ligase activities of Roquins. Sci. Rep. 2015; 5:15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S.. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014; 505:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Plass M., Rasmussen S.H., Krogh A.. Highly accessible AU-rich regions in 3′ untranslated regions are hotspots for binding of regulatory factors. PLoS Comput. Biol. 2017; 13:e1005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Friedrich S., Schmidt T., Geissler R., Lilie H., Chabierski S., Ulbert S., Liebert U.G., Golbik R.P., Behrens S.E.. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J. Virol. 2014; 88:11586–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Friedrich S., Engelmann S., Schmidt T., Szczepankiewicz G., Bergs S., Liebert U.G., Kummerer B.M., Golbik R.P., Behrens S.E.. The host factor AUF1 p45 supports flavivirus propagation by triggering the RNA switch required for viral genome cyclization. J. Virol. 2018; 92:e01647-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Glasmacher E., Hoefig K.P., Vogel K.U., Rath N., Du L., Wolf C., Kremmer E., Wang X., Heissmeyer V.. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat. Immunol. 2010; 11:725–733. [DOI] [PubMed] [Google Scholar]
- 81. Hanieh H., Masuda K., Metwally H., Chalise J.P., Mohamed M., Nyati K.K., Standley D.M., Li S., Higa M., Zaman M.M. et al.. Arid5a stabilizes OX40 mRNA in murine CD4(+) T cells by recognizing a stem-loop structure in its 3′UTR. Eur. J. Immunol. 2018; 48:593–604. [DOI] [PubMed] [Google Scholar]
- 82. Masuda K., Ripley B., Nishimura R., Mino T., Takeuchi O., Shioi G., Kiyonari H., Kishimoto T.. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl Acad. Sci. U.S.A. 2013; 110:9409–9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Essig K., Hu D., Guimaraes J.C., Alterauge D., Edelmann S., Raj T., Kranich J., Behrens G., Heiseke A., Floess S. et al.. Roquin suppresses the PI3K-mTOR signaling pathway to inhibit T helper cell differentiation and conversion of treg to Tfr cells. Immunity. 2017; 47:1067–1082. [DOI] [PubMed] [Google Scholar]
- 84. Li X., Quon G., Lipshitz H.D., Morris Q.. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010; 16:1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 86. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures were deposited in the Protein Data Bank under the accession numbers 6TQB and 6TQA for the ROQ domain with UCP3 CDE1GG and CDE2GG, respectively. UCP3 CDE1GG and CDE2GG NMR structural ensembles were deposited under the accession numbers 6XXW and 6XJW in the Protein Data Bank and under IDs 34483 and 34482 in the BMRB, respectively.