Abstract
Single-cell RNA-sequencing (scRNA-seq) of the Caenorhabditis elegans nervous system offers the unique opportunity to obtain a partial expression profile for each neuron within a known connectome. Building on recent scRNA-seq data and on a molecular atlas describing the expression pattern of ∼800 genes at the single cell resolution, we designed an iterative clustering analysis aiming to match each cell-cluster to the ∼100 anatomically defined neuron classes of C. elegans. This heuristic approach successfully assigned 97 of the 118 neuron classes to a cluster. Sixty two clusters were assigned to a single neuron class and 15 clusters grouped neuron classes sharing close molecular signatures. Pseudotime analysis revealed a maturation process occurring in some neurons (e.g. PDA) during the L2 stage. Based on the molecular profiles of all identified neurons, we predicted cell fate regulators and experimentally validated unc-86 for the normal differentiation of RMG neurons. Furthermore, we observed that different classes of genes functionally diversify sensory neurons, interneurons and motorneurons. Finally, we designed 15 new neuron class-specific promoters validated in vivo. Amongst them, 10 represent the only specific promoter reported to this day, expanding the list of neurons amenable to genetic manipulations.
INTRODUCTION
The human brain comprises around 100 billion neurons. Early classification based on morphology already distinguished pyramidal, stellate, granule, bipolar, Purkinje or basket cells (1). Additional knowledge about their neurotransmitters, functional and electrophysiological properties further improved this classification. Recent progresses in molecular profiling using single-cell RNA-sequencing (scRNA-seq) allowed exploring neuronal diversity at the molecular level in human and mouse brain (2–4). In comparison, the nervous system of Caenorhabditis elegans adult hermaphrodite has a simple and well-described structure composed of only 302 neurons. Despite its anatomical simplicity, the neuronal diversity of C. elegans encompasses 118 neuron classes identified by the examination of their complete diagram of connectivity as revealed by serial sections and electron microscopy (5–7). Based on expression patterns at the single-cell resolution, the 118 anatomically defined neuron classes in the adult hermaphrodite would correspond to 118 or more unique molecular identities (8,9). However, the transcriptional profiles are only known for a few neuron classes purified by FACS-sorting (10–12).
The exquisitely described small circuits composed by few neurons in C. elegans allow exploring how cell-to-cell communication can shape behavior. The function of several C. elegans neuron classes has been defined using laser ablation, genetic and optogenetic methods (13,14). These later methods, however, rely on reliable promoters driving transgene expression in a single neuron class to be characterized. Currently, cell-specific promoters are described for only 29 neuron classes. This current lack of cell-specific promoters has limited the optogenetic and chemogenetic analysis of the neuronal functions to a fraction of the C. elegans nervous system.
Recently, scRNA-seq of the full C. elegans organism at L2 larval stage was delivered (15). Out of the 42,000 sequenced cells, the author yielded ∼7,000 single neuron transcriptomes. Based on gene co-expression patterns, a semi-supervised clustering analysis segregated these 7,000 neurons into ∼40 clusters and assigned 23 neuronal classes to 16 clusters; 10 clusters being assigned to single neurons classes. However, 112 out of the 118 adult neuron classes are theoretically present in the L2 larval stage (16).
Therefore, our aims were (i) to define molecular profiles for the remaining neuron classes and (ii) to determine new neuron-specific promoters. To these ends, a heuristic approach was used where we matched a molecular atlas made of ∼800 neuron class-specific marker genes (8) to the scRNA-seq dataset generated by Cao et al. (15). We performed a clustering analysis aiming to segregate ∼100 cell-clusters assigned to the expected ∼100 neuronal identities. Through several clustering iterations, we segregated 81 neuronal clusters and assigned 62 clusters to a single neuron class, from which we extracted the enriched genes. Based on these expression profiles, we designed 15 new neuron-specific promoters validated in vivo. We extracted enriched key transcription factors in three different lineages as potential terminal cell-fate selector. Finally, we explored whether and how differentially expressed genes drive neuronal diversity in C. elegans. We observed that sensory machinery genes generate diversity in sensory neurons, neuromodulatory genes in interneurons while genes involved in membrane potential tuning generate diversity in motor neurons.
MATERIALS AND METHODS
Dataset pre-processing
As starting dataset, we used the neuron clusters defined in (15) and containing 7,603 cells. This dataset is a sub-selection of clusters coming from a whole worm dataset using the neuroendocrine markers egl-21, egl-3, ida-1 and sbt-1. We added 1475 cells included in clusters expressing the neuron markers unc-104, unc-31, ric-4, snt-1 or unc-10 (17), further expanding the dataset to 9,078 cells. Most of these added cells end up in non-neuronal cluster far away from the neuronal clusters (Supplementary Figure S1C). The dataset clustering was performed using Seurat v3, and visualized using t-SNE or UMAP plots (see below). Louvain algorithm was used for all clustering settings after performing a PCA dimension reduction using Seurat v3 with default parameters, unless otherwise stated. All analysis described below are accessible on https://github.com/lorenzo-jr/ce-neuronclusters.
Clustering parameters
Two parameters strongly affect Louvain algorithm clustering output in Seurat: the number of PCs used and the resolution used. Each PC essentially represents a ‘metagene’, combining information across a set of correlated genes. The resolution parameter sets the ‘granularity’ of the clustering: increased resolution facilitates clusters segregation. The process of Selection for clustering parameter is available on GitHub, point 1. We first ranked the PCs according to the percentage of variance explained but did not observe any obvious elbow. To determine the optimal number of components and the appropriate resolution for the first iteration, we repeated the clustering analysis with 40–100 PCs and a resolution ranging from 1 to 8. To evaluate the quality of each clustering output, we designed an automatic assignment of neuronal identities for each cluster using specific genes signatures (see cluster assignment method). We finally selected the PCs and resolution parameter pair able to maximize the proportion of clusters with unique identities. The clustering parameters were set to 92 PCs and a resolution value of 4 (Supplementary Figure S1A and B). Using those parameters, the dataset was divided into 64 clusters including 20 directly assigned to only one neuron class (Figure 3). The first clustering iteration (PCs = 92, resolution = 4) was used as input for DoubletFinder, an algorithm using artificial nearest neighbors (pANN) to detect doublets (18). nExp was calculated assuming a 3.1% doublet formation rate (15), pN was set to 0.25; pK was set to 0.3 based on (18,19) (script details on GitHub, point 8). As a result, 274 cells doublet cells were removed from the gene expression analysis (Supplementary Figure S1C).
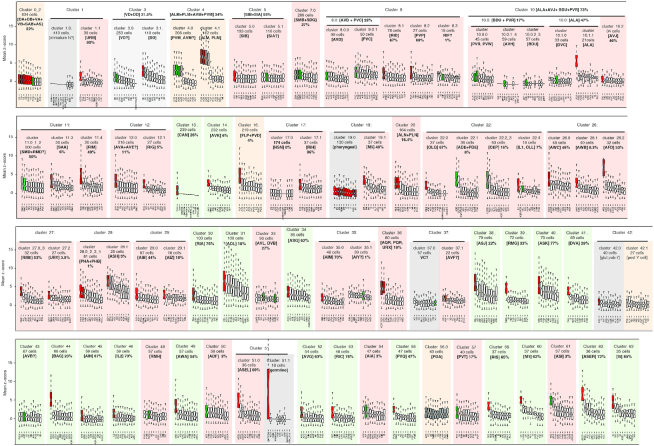
Figure 3.
Clusters assignment by combined z-score and multiple iterations. For each cluster, the 10 best-ranked combined z-scores are displayed as well as the quality of the assignment consensus (%) reached by the re-clustering iterations of the parent cluster. These two assessments helped us to assign each cluster to one or several neuron class. The automated assignment by ranked z-scores is displayed by the red colour within the notched box, gray if discarded for final assignment, green for manual assignment. The first iteration clusters automatically assigned to a single neuron class identity are shaded in light green. Clusters assigned through iterations are shaded in pink. Clusters detailed in pseudotime are shaded in orange. Clusters poorly assigned to any neuronal class are shaded in gray.
z-scores assignment method
More than 800 gene expression patterns at the single cell resolution were extracted from Wormbase.org and curated manually through literature screening. For each of the 112 neuron classes present in L2 stage, a combinatorial gene expression atlas was generated (Supplementary Table S1). A neuron class signature is defined by a set of 19–116 marker genes specifically expressed in this neuron class. The expression of marker genes in each cluster was used to assign clusters to the best matching neuronal class. For each cell in a given cluster, 112 combined z-scores were calculated by comparing the expression of marker genes to the neuron class signatures from Supplementary Table S1. For a given gene, each individual cell z-score was computed using standardized expression level of that gene (expression in the cell – mean expression/var expression). For a given cell and a given neuron class k, the individual z-scores were aggregated using Stouffer's method (20), according to the formula:
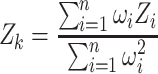 |
where Zk represents the combined z-score for a particular cell in a cluster for a specific neuron class signature k, Zi the individual z-score for a cell for a particular gene, n the genes composing the neuron class and ωi a gene specific weight calculated as 1 minus the fraction of the cells expressing that gene. The Zk scores were computed for all the cells within a cluster.
Hence, for each cluster a distribution of z-scores was calculated for the 112 neuron classes using all the cells present in the given cluster. For each cluster, the combined z-scores for all the 102 classes were ranked using their median values. A sequential t-test from the highest to lowest ranked neuron classes was applied iteratively to the successive pair of classes. The most specific neuron classes occurring before a significant break was detected (P < 0.05) and assigned to the cluster. A second independent filter was applied by computing a one sample upper-tailed t-test over the z-sores of each identity. Resulting P-values were adjusted by the false discovery rate method (21) and only statistically significant identities with adjusted P-values lower than 0.05 were kept. Finally, only the neuron classes selected by both methods were assigned to a cluster. Assignment of the first iteration clusters is available on GitHub, point 2. For each cluster, the corresponding boxplot with automatic assignment of neuronal identities is available (in red).
Clustering iterations
We performed a second and a third round of clustering on the 64 parent clusters obtained above. These additional iterations potentially segregate non-homogenous clusters, allowing the assignment of more clusters to their neuron class. Single-cell profiles coming from parent clusters were independently re-clustered with Seurat v3. Multiple re-clustering trials were generated for resolution values from 0.1 to 3 and for PCs values from 3 to 92. These conditions generated a total number of clustering trials over each parent cluster ranging from 151 to 330, depending on the cell number within the parent clusters. For each trial, the identities of the daughter clusters were assigned using the z-score assignment method (see above). Neuron class assignments were counted (in GitHub point 3 and 5). The neuron classes most often detected in the parent cluster represent a consensus on the identity(ies) present in the parent clusters (as shown in Figure 2). The consensus on the identities present in each parent cluster was used to detect the neuron class most likely present in each parent cluster. We chose the lowest PCs and resolution among the most representative clustering parameters to represent clustering results. Clustering iterations are available on GitHub, points 3, 4, 5 and 6.
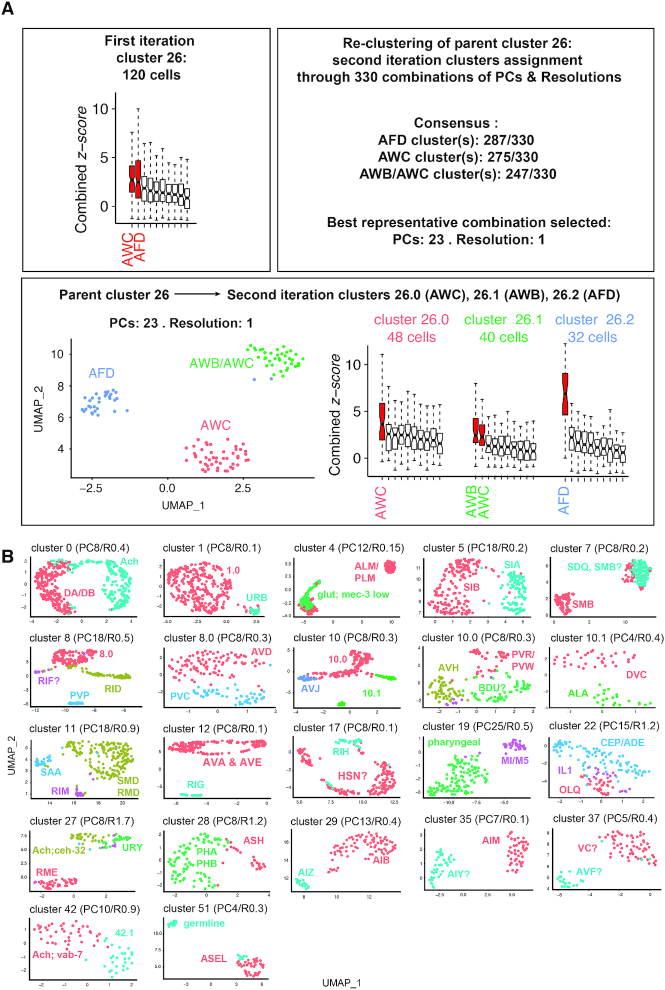
Figure 2.
Multiple iteration clustering approach and results. (A) Each parent cluster from the first iteration clustering was treated independently for re-clustering for all combinations of parameters ranging from 3 to 92 PCs and resolution from 0.1 to 3. The neuron classes assigned to every sub-clusters generated by this large number of re-clustering trials are analysed. A consensus is reached for parent cluster 26: it suggests the re-clustering of cluster 26 should generate 3 sub-clusters. These three sub-clusters should be assigned to AFD, AWC and AWB/AWC. This result is observed for PCs: 23/resolution: 1, used for representation and for gene expression lists. (B) Uniform Manifold Approximation and Projection (UMAP) projections are used to represent the second or third iteration clustering from parent clusters containing multiple neuron classes. Several of sub-clusters can be assigned to a single neuron classes using the combined z-scores.
Pseudotiming
Monocle 3 was used to infer trajectories on five sets of neuronal clusters (0_2, 16.0_16.1, 3.0_3.1, 4.0_4.1 and 42.2_56). Monocle 3 implements a principal graph-embedding procedure described in (22–24) using UMAP algorithm (25) to reduce data into low-dimensional space. First, in a pre-processing step the digital gene-count matrix was log-transformed after adding a pseudocount, and then scaled to unit variance and zero mean. The matrix was then projected into 100 top principal components. Selection of principal components was performed by visual inspection of the elbow on the explained variance plot. Before Graph-embedding takes place, two steps are required: UMAP dimensional reduction and clustering on UMAP space. Monocle 3 default parameters were used for the dimensional reduction step: number of neighbors to use during kNN graph construction set to 15, distance metric to be used when calculating UMAP was set to cosine and minimum distance set to 0.1. Clustering step was performed using Leiden algorithm (26), where k-nearest neighbor graph k value was set to 20 and resolution parameters were set to match the cluster partitions produced by our method. Finally, Monocle 3 principal graph-embedding procedure to learn trajectories was used with default parameters. Starting points for the trajectories were set manually. Genes selected for pseudotime analysis were selected based on the neuronal identities we wanted to differentiate. More details on markers and pseudotime on GitHub, point 9.
Differentially expressed TF
For each of the lineages, differentially expressed transcription factors (TF) were extracted from C. elegans embryogenesis visualizer (VisCello) from (27) (Supplementary Table S6). For each neurons, we extracted from our dataset the TF within the 20 most enriched genes against all neurons.
Comparison between gene lists
The read count for each gene was extracted for each cluster. We also computed lists of enriched genes compared to all other neuronal clusters of our dataset. The AVK bulk sequencing profile was generated by FACS-sequencing and compared to all neurons to compute the enriched genes (12). The comparison between the top 100 P-value ranked genes for each neuronal cluster versus AVK bulk sequencing was calculated by running a hypergeometric test for the overlap between lists considering a total of 5589 genes found in both datasets. The multiple comparisons between our enrichment lists and the enrichment lists from (27) and (28) were done similarly for the top 100 P-value ranked genes and considering a total of 13 988 and 5968 genes respectively, found in both datasets.
Reporter strain generation
The primers used to generate the promoters collection and the amplified regions size are described in Supplementary Figure S2. The promoter sequences were amplified from N2 genomic DNA, purified from gel extraction then fused with mKate or mEGFP sequence followed by the 3′UTR sequence of unc-54 or let-858, respectively. 30 ng/μl of the fusion PCR, 30 ng/μl of 1kb Plus DNA mass ladder (Invitrogen) and 30 ng/μl of the injection co-marker punc-122::mKate were co-injected in D1 animals to generate transgenic animals. Injections were carried out in N2, OH12312, OH13083, CZ13799, OH15262 and/or OH12734. After injection, three to five independent transgenic strains were maintained for at least three generations before imaging. To validate unc-86 involvement in RMG cell fate, we injected the transgene Ex[Pnlp-56::mKate::unc-54 3′UTR] in the OH12734 strain. Progeny of Ex[Pnlp-56::mKate/YFP] transgenic animals are either unc-86(n846) mutant or unc-86 rescued by a UNC-86 fosmid. We counted mKate expression in RMG neurons in unc-86(n846) mutant or based on YFP expression in unc-86 rescued. The quantification was divided as follow: YFP+/− & RMG WT, YFP+/− & RMG faint, YFP+/− & no RMG.
Microscopy
In order to identify the neurons expressing the reporter transgene, we performed confocal microscopy imaging (Zeiss LSM 510 and Zeiss LSM 780) with 20× or 40× objective. Animals were anesthetized with 25 mM sodium azide in M9 solution, transferred to 2% agarose mounting slides and imaged within 30 min of mounting. The images were analysed using Fiji imaging software. The expression patterns were analysed at L2, L4 and YA developmental stages for three to five independent transgenic strains. In order to identify the expression pattern, we used cell position, morphology and/or the NeuroPAL strain (OH15262), used as described (29). The images obtained are available in Supplementary Figure S2 together with the detailed evidences used to identify the neurons.
Gene ontology (GO) analysis
We generated gene ontology lists for the genes enriched in:
28 sensory neurons/24 clusters: 4.1: ALM, PLM, 16.0: FLP, 20.0: ALN, PLN, 22.0: OLQ, 22.1: ADE?, 22.2_3: CEP?, 22.4: IL1, 26.0: AWC, 26.1: AWB, 26.2: AFD, 27.2: URY?, 28.1: ASH, 31.0: ADL, 34.0: ASG, 36.0: AQR, PQR, URX, 38.0: ASJ, 40.0: ASK, 44.0: BAG, 46.0: IL2, 49.0: AWA, 50.0: ADF, 51.0: ASEL, 61.0: ASI, 62.0: ASER.
21 interneurons/20 clusters: 8.2: PVP, 10.0.1_4: AVH?, 10.2: AVJ, 12.0: AVA, 12.1: RIG, 14.0: AVK, 17.1: RIH, 29.0: AIB, 29.1: AIZ, 30.0: RIA, 33.0: AVL, DVB, 35.0: AIM, 35.1: AIY?, 41.0: DVA, 43.0: AVB, 45.0 AIN, 52.0: AVG, 53.0: RIC, 55.0: PVQ, 87.0: PVT?. We did not include 1.1 URB, 8.1: RID, 10.1.1: ALA, 39.0: RMG, 11.4: RIM, 37.1: AVF, 48.0: RMH that are mixed modalities.
15 motorneurons/8 clusters: 0_2: DA, DB, VA, VB, SAB, PDB, 3.1: VD, DD?, 5.0: SIB, 5.1: SIA, 7.0: SMB, 11.0_1_2: RMD, SMD, 11.3: SAA, 27.0_3: RME.
To get an equivalent number of genes per modality (∼200 genes per modality), we extracted the top 10 most enriched genes from each sensory neuron class, the top 10 most enriched genes from each interneuron class and the top 25 ones from each motor neuron class (8).
Nematode strains
The primers used to generate the reporter strains are described in Supplementary Figure S2.
OQ180: CZ13799; ulb180 Ex[psrsx-18::mKate; unc-122p::mKate]
OQ181: CZ13799; ulb181 Ex[pttr-39::mKate; unc-122p::mKate]
OQ182: N2; ulb182 Ex[pC18F10.2::mEGFP]
OQ183: N2; ulb183 Ex[pC41A3.1::mKate]
OQ184: N2; ulb184 Ex[pC50F7.5::mKate]
OQ185: N2; ulb185 Ex[pflp-33::mKate]
OQ186: N2; ulb186 Ex[pnlp-56::mKate; unc-122p::mKate]
OQ187: N2; ulb187 Ex[pocr-3::mKate; unc-122p::mKate]
OQ188: N2; ulb188 Ex[pY48G10A.6::mKate; unc-122p::mKate]
OQ189: OH12312; ulb189 Ex[pC18H7.6::mKate; unc-122p::mKate]
OQ190: OH12312; ulb190 Ex[pnlp-17::mKate; unc-122p::mKate]
OQ191: OH12312; ulb191 Ex[pT09B9.3::mKate; unc-122p::mKate]
OQ192: OH12312; ulb192 Ex[pY48G10A.6::mKate; unc-122p::mKate]
OQ193: OH13083; ulb193 Ex[ptwk-47::mKate; unc-122p::mKate]
OQ194: OH15262; ulb194 Ex[pC42D4.1::mEGFP]
OQ195: OH15262; ulb195 Ex[psem-2::mEGFP]
OQ196: OH15262; ulb196 Ex[pttll-9::mEGFP]
OQ197: OH15262; ulb197 Ex[pY58G8A.5::mEGFP]
CZ13799: juIs76 [unc-25p::GFP + lin-15(+)] II
N2 CGC
OH 12734: unc-86(n846) III; otEx5851 [unc-86(fosmid)::NLS::mChopti + lin-44::YFP].
OH15262: otIs669 V [NeuroPAL]
OH13083: otIs576 [unc-17(fosmid)::GFP + lin-44::YFP]; him-5(e1490) V
OH12312: pha-1(e2123); otIs388 [eat-4(fosmid)::SL2::YFP::H2B+pha-1(+)]III; him-5(e1490) V
RESULTS
Matching the clustering analysis to a molecular atlas
Each neuron class is theoretically detectable by a specific combinatorial gene expression signature. To assign each cell cluster to an anatomically defined neuron class in C. elegans, we designed an automated assignment strategy where the genes expressed in each cell cluster are compared to a molecular atlas made of 19–116 marker genes (Supplementary Table S1). While these expression patterns do not necessarily capture the accurate expression of each of the respective gene loci, this atlas provided us a unique proxy to assign objectively each cell-cluster to a neuron class.
From the original dataset generated by Cao et al. (15), we extracted 9,078 cells from all clusters expressing the neuron markers egl-21, egl-3, ida-1, sbt-1, unc-104, unc-31, ric-4, snt-1 and unc-10 (17). Our clustering was realized using the Louvain algorithm implemented in Seurat R package (30). Clustering parameters were optimized in order to identify a maximum of clusters automatically assigned to a single neuron class.
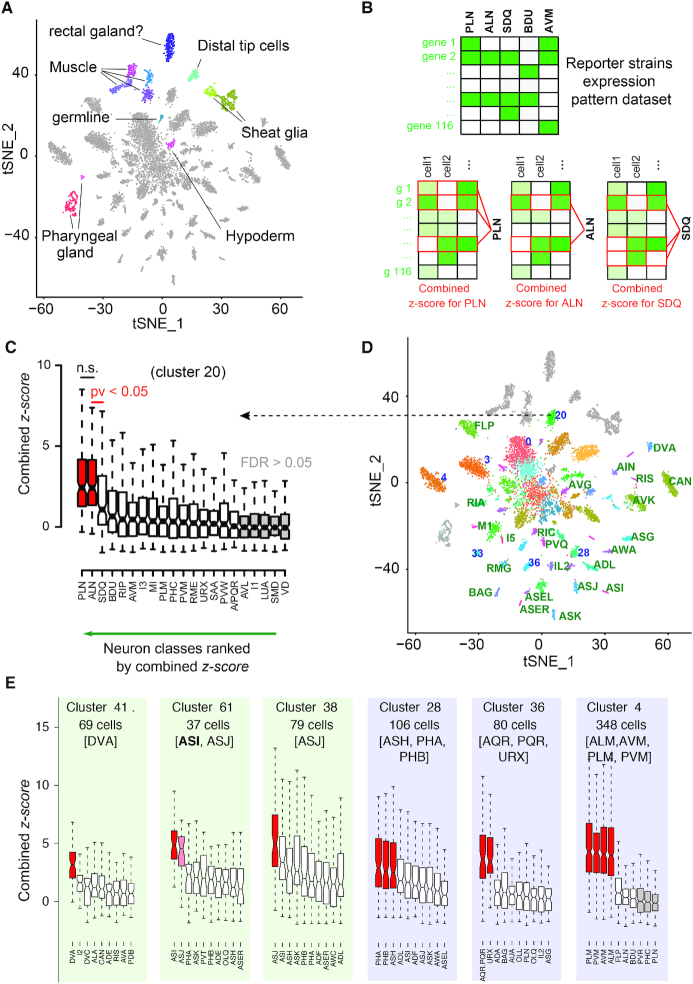
Using Louvain algorithm over 92 PCs and a resolution of 4, we identified 64 cell-clusters visualized with t-distributed Stochastic Neighbor Embedding (t-SNE) (Figure 1a). Each cluster was assigned to a cell identity –neuron class or tissue– based on a comparison with the 112 molecular signatures of L2 neuron classes (Supplementary Table S1). From genes expressed in all cells in a given cluster, we computed and ranked 112 combined z-score (Stouffer's method) corresponding to the comparison with the 112 molecular signatures. For example, cluster 20 is assigned to PLN and ALN, but not SDQ (Figure 1B and C). These 64 identified cell clusters included 53 clusters corresponding to neurons and 11 clusters corresponding to glial, hypodermal, pharyngeal and muscle cells (Figure 1A). The automatic assignment allowed us to automatically assign 20 clusters to a single neuron class (Figures 1D and 3). Among clusters not automatically assigned, some could easily be assigned manually: ASI or ASJ passed our threshold for assignment significance in clusters 38 and 61, but ASJ matched noticeably better with cluster 38 and ASI with cluster 61. We assigned ASI and ASJ to clusters 61 and 38, respectively (Figure 1E). We also observed that 7 first iteration clusters matched well with a subset of molecularly similar neuron classes (Figure 1E, purple shade). For example, cluster 28 is assigned to three nociceptive neuron classes (ASH, PHA and PHB) co-expressing many chemosensory GPCRs and responding to similar cues (31). Similarly, cluster 36 and cluster 4 contain a subset of sensory neurons sharing sensory machineries for oxygen or touch, respectively (32,33). Many other clusters were poorly or not assigned automatically by the first clustering iteration (Figures 1D and 3).
Figure 1.
t-SNE projection and neuron-class assignment principle. (A) Single cell profiles are projected in two dimensions using t-SNE. Cell-clusters corresponding to non-neuronal cells or to neurons are distinguished by color and label. (B) Neuron classes assignment to clusters using the z-score method. Gene's z-scores from the centred scaled dataset are used to assign neuronal classes to the clusters. A combined z-score (Stouffer's method) for all neuronal classes is obtained for each cell in the cluster, in this case PLN, ALN, SDQ and cluster 20. (C) Neuron classes are treated as independent groups and ranked by the median value of their combined z-scores. In the notched box plots, the notches display the 95th confidence interval around the median (black horizontal line); the box contains the interquartile and the whiskers extend to the most extreme data points which are no more than 1.5 times the interquartile range from the box. A sequential t-test from the highest to lowest ranked neuron class indicates where the assignment of neuron classes should stop (P<0.05). An additional false discovery rate (FDR) filter of <5% is applied. (D) Clusters assigned to a single neuron class are labelled in green (e.g., DVA); clusters assigned to a subset of molecularly similar neuron classes are labelled in blue (e.g. 36, corresponding to URX, AQR and PQR). (E) Based on their combined z-score, clusters 41 is assigned to the DVA neuron class; clusters 61 and 38 are assigned to ASI and ASJ, respectively; clusters 28, 36 and 4 are assigned to a set of molecularly similar classes of sensory neurons, which we could or could not further segregate with clustering iterations.
Altogether, 3 clustering iterations assign 62 clusters to a single neuron class
To validate the automated assignment of the first clustering iteration and to assess if a better cluster segregation could be obtained for the 24 poorly assigned clusters, each of the 64 parent clusters were isolated and treated independently for a second clustering iteration. Theoretically, this second iteration starting from a molecularly less complex collection of cells is expected to further divide each parent cluster. As we could not predict the best clustering parameters, we examined the results of all combinations of parameters from 3 to 92 PCs and resolution from 0.1 to 3. These ∼300 combinations often reached a consensus on the neuron classes’ content of the parent clusters: e.g. for cluster 40, 241 out of 310 combinations identified one to many clusters assigned to the ASK neuron class. The quality of this re-clustering consensus as well as the z-scores for the neuron classes assigned to the parent clusters offers two ways to measure the assignment confidence (Figures 2 and 3). Interestingly, 93 ± 19% (SD) of the re-clustering trials confirmed the neuron class of the 20 clusters automatically assigned by the first iteration. We applied the same logic for a third iteration of clusters 8 and 10.
In addition to cluster assignment validation, second and third re-clustering also succeeded to segregate poorly assigned parent clusters into better-assigned second iteration clusters. For example, the parent cluster 26 displayed a high z-score for AWC and AFD sensory neurons, suggesting it might contain several neuron classes. We observed that 285, 275 and 247 combinations of parameters segregated cluster 26 into 3 sub-clusters assigned to AFD, AWC and AWB/AWC, respectively (Figure 2A). This consensus is well represented by 23 PCs and a resolution of 1, splitting cluster 26 into AFD, AWC and a third sub-cluster assigned to both AWB and AWC (Figure 2A). Differential expression analysis shows AWB/AWC sub-cluster expresses AWB specific markers (e.g. srab-24 and srab-4), suggesting this sub-cluster should have been assigned to AWB. A lack of differentially expressed genes between AWB and AWC in Supplementary Table S1 potentially explains this inadequate assignment. Moreover, the re-clustering improved the z-score of AFD between the first and second iteration, suggesting the AFD sub-cluster is more homogeneously composed of AFD cells than the original parent cluster 26 (Figure 2A). This example above illustrates how re-clustering allowed us to split the poorly assigned clusters and/or to remove contaminating cells from the parent clusters. Altogether, the two clustering iterations coupled with the z-score assignment strategy automatically assigned 44 clusters, including 35 clusters assigned to a single neuron class and 2 clusters to ASER and ASEL. Manual intervention including removal of the previously assigned neuron classes and re-examining the genes enrichment lists for cell-specific markers allowed the assignment of another 25 clusters to a single identity. Fifteen clusters remained assigned to a set of two to six neuron classes molecularly too similar to be segregated (Figure 3 and Supplementary Table S2). Five clusters remained non-assigned: cluster 1.0 likely corresponds to immature neurons, 19.0 to many pharyngeal neuron classes, 27.1 to a cholinergic neuron expressing ceh-32, 42.0 to a glutamatergic neuron expressing vab-7, 10.0.5 contains only nine cells and was not assigned. We observed that cluster 42.1 and 56 could be assigned to PDA, a neuron that appears in L3 stage (see below, Figure 6). Based on markers, we also observed that cluster 17.0 potentially correspond to HSN an embryonically born neuron that acquire its final identity in the adult hermaphrodite (34) (Supplementary Figure S1E). Therefore, we took into account the 118 neurons present in the adult, although not all of them might be mature enough to be identified at the L2–L3 stage sequenced. Accordingly, 21 neuron classes are not confidently assigned to any cluster (e.g. AUA, URA). We assume the neurons of these 21 missing neuron classes populate the five non-assigned clusters or contaminate the assigned clusters (Supplementary Table S2).
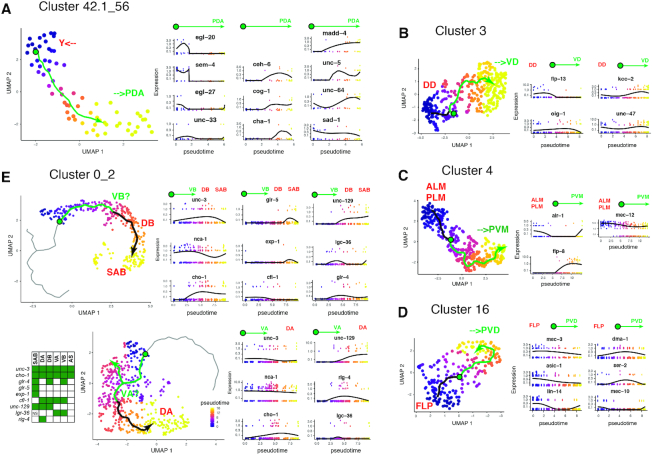
Figure 6.
Pseudotiming reveals the maturation of PDA neurons. (A) Pseudotiming connects cluster 42.1 (late Y cell) to cluster 56 (PDA neuron) by an inferred trajectory (angled arrow). The expression level of some markers along this abstract trajectory is reported in RNA count. We report expression of markers for the Y cell (egl-20, sem-4, unc-33 and egl-27) on one side and markers for the PDA neuron (ceh-6, cog-1 and cha-1) on the other side. Markers for synaptic genes (sad-1/SAD kinase, unc-64/Syntaxin) and guidance genes (unc-5/Netrin Receptor, madd-4/Punctin) are also reported. (B) For pseudotiming of cluster 3, we report expression levels for markers of DD (flp-13) and VD (oig-1) motorneurons as well as the gabaergic marker unc-47 and a potential maturation marker: kcc-2. (C) For pseudotiming of cluster 4, we report expression levels for markers of ALM_PLM (mec-12 high) and PVM (flp-8) neurons as well as a potential marker of maturation: alr-1. (D) For pseudotiming of cluster 16, we report expression levels for markers of FLP (asic-1, lin-14), FLP_PVD (dma-1, mec-10) or PVD (ser-2) neurons. (E) Pseudotiming connects clusters 0 to cluster 2 by an inferred trajectory along which we report expression levels for markers for the DA, VA, VB, DB, AS, SAB motorneurons. For all pseudotiming, the location of the putative immature precursors and their maturation is displayed on the trajectories by the green circle and arrow, respectively.
Expression profile validation for cluster 14 assigned to the AVK neuron class
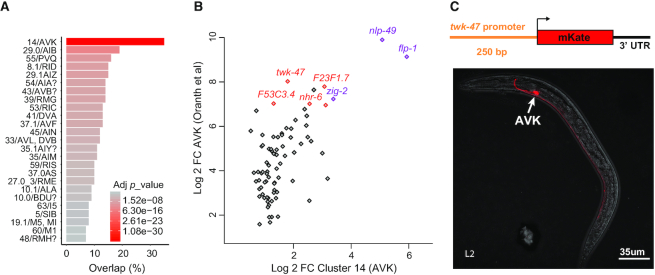
We next extracted the genes expressed in each assigned cluster and computed their most enriched genes (Supplementary Tables S3 and S4). To validate one of these lists, we compared each of our clusters to the bulk sequencing of the AVK neurons purified by FACS-sorting (12). Although both scRNA-seq and bulk sequencing have their own biases, we expect a strong match between cluster 14 we assigned to AVK and the AVK bulk sequencing. Indeed, the best match was observed between cluster 14 and the AVK bulk sequencing with 38 of the 100 most enriched genes present in both lists (p = 9.83e–40). This overlap included the known AVK marker flp-1 as well as new potential AVK markers such as twk-47 (Figure 4).
Figure 4.
AVK profiling overlap and reporter strain design strategy. (A) We observed an overlap of 35 genes between the 100 most enriched genes in cluster 14 assigned to AVK neuron class and the 100 most enriched genes from the AVK neuron bulk sequencing (adj. pv = 9.82e–40; P-value adjusted by Bonferroni correction). (B) The fold change comparison highlights the genes co-enriched in cluster 14 and in AVK bulk sequencing profile. The expression of nlp-49 and flp-1 mRNA in AVK were previously described (purple) (12,48). Several new potential AVK markers are highlighted in red, including twk-47. (C) Generation of a reporter strain expressing a 250 bp twk-47 promoter fused to the mKate sequence. The confocal image shows the specific expression of mKate in the AVK neurons.
In C. elegans, intergenic regions containing the promoters are short and in most case, a segment extending 1 kb upstream the ATG was shown to drive expression of a fluorescent protein reporter. We designed a reporter strain expressing mKate (a monomeric red fluorescent protein) under the control of a 250 bp twk-47 promoter. As expected, we observed expression of mKate only in two neurons that we could identify as the two AVK neurons, supporting the accuracy of our gene enrichment list for cluster 14 (Figure 4).
Predictions of cell-fate regulators
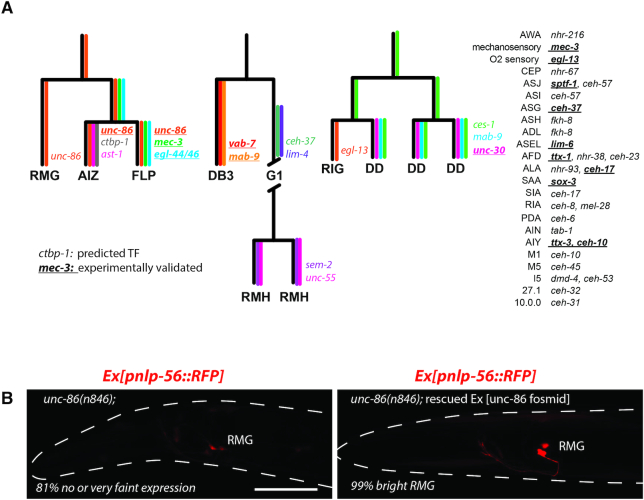
Neuronal diversification in C. elegans mostly occurs during embryogenesis. Most neurons derive from different lineages, showing neither clonal expansion nor correlation of lineage history with the terminal features of the neuron types ((35) and Supplementary Figure S1H). Virtually all neurons differentiate through asymmetric terminal cell division and asymmetric expression of transcription factors between sister cells. To identify potential regulators of cell fate decisions for neuron lineages not previously explored by (27), we identified transcription factors (TFs) enriched within these lineages and/or differentially expressed between mother, daughter and sister cells (Figure 5A and Supplementary Table S6). Based on these rules, one to three TFs are predicted for each explored lineage to play some role in cell fate decisions. Interestingly, 16 of these predicted TFs were previously proven experimentally to contribute to cell fate decisions in these neurons (36). To provide additional confidence in these predicted key transcription factors, we tested one of them. The POU transcription factor UNC-86 is enriched in the FLP, AIZ, and RMG lineage and is required for FLP and AIZ terminal differentiation (37). To evaluate cell differentiation of RMG in unc-86 mutants, we used the Pnlp-56::RFP marker identified by us and by (28) as specifically expressed in RMG. Expression of Pnlp-56::RFP in RMG required UNC-86, suggesting UNC-86 play a role in RMG differentiation (Figure 5B).
Figure 5.
Predictions of cell fate regulators. (A) We identified transcription factors enriched in neuron class and differentially expressed between sister cells. Based on these rules, predicted cell fate regulators for neuron classes’ differentiation are displayed. The expression pattern of 16 of these predicted cell fate regulators are indicated by color and gene name for three lineages. The 16 transcription factors in bold and underlined were also previously proven experimentally to contribute to neuron class differentiation. (B) Expression of an RMG molecular identity marker (Ex[Pnlp-56::RFP] transgenes) was analyzed in unc-86(n846) animals and in unc-86(n846) animals rescued by an UNC-86 fosmid. UNC-86 is required for expression of the RMG marker. 81% of the non-rescued unc-86(n846) had no or faint expression of Pnlp-56::RFP in RMG. Differential-interference contrast suggests neurons are present at the position of FLP and RMG in unc-86(n846). Scale bar: 30 μm.
Maturation of neurons classes during the L2 stage
The hermaphrodite L1 hatches with 222 differentiated neurons corresponding to 96 neuron classes. However, 80 postembryonic neurons corresponding to 22 neuron classes are later added to the nervous system (16). As previously observed for the DD neuron class (38,39), postembryonic neurons might follow a remodelling process during early larval stages and this maturation process might be captured in our dataset. Therefore our clusters assigned to postembryonic neurons classes were interesting to explore for the potential occurrence of a dynamic process. Rather than clustering cells into discrete clusters, pseudotiming algorithms arrange cells based on their progression along a continuous path that represents dynamic changes in gene expression along a process (neuronal maturation here). We attempted pseudotiming for each postembryonic cluster. Most interestingly, pseudotiming connected cluster 42.1 to cluster 56. Markers allowed us to assign this trajectory to the maturation process of the PDA neuron (Figure 6A). During the L2 stage, the Y cell (an epithelial cell initially part of the rectum) migrates away from the rectum and becomes a motor neuron named PDA at the L3 stage (40). The ceh-6, cog-1 and cha-1 markers suggested one side of the pseudotime trajectory would correspond to the matured cholinergic PDA neuron. The other side would correspond to a late stage of the Y cell, posterior to trans-differentiation: the cells still carried markers of Y cell such as egl-20, had lost epithelial markers such as che-14 and gained neuronal markers such as sad-1. The maturation process appeared to involve increased expression of the guidance proteins MADD-4/Punctin and UNC-5/Netrin receptor as well as the presynaptic protein UNC-64/Syntaxin. Pseudotiming placed DD and VD on each side of a trajectory within cluster 3 (Figure 6B), ALM/PLM and PVM on each side of a trajectory within cluster 4 (Figure 6C), and FLP and PVD on each side of a trajectory within cluster 16 (Figure 6D). The increased expression of markers of potential maturation such as kcc-2 in cluster 3 (41), alr-1 in cluster 4 (42), suggest—but do not prove—we capture the progression toward VD and toward PVM mature neurons, respectively. Despite its large number of cells, cluster 0_2 was not segregated through multiple iterations but assigned to DA, VA, DB, VB, SAB, AS and PDB. Among those classes VA, VB, and AS are generated at the end of the L1 stage. Pseudotiming organized motorneurons classes along a continuum, placing the embryonic born DA, SAB and DB at the end of 2 trajectories, while VA and VB might show some signs of maturation (e.g. increasing expression of cho-1) (Figure 6E).
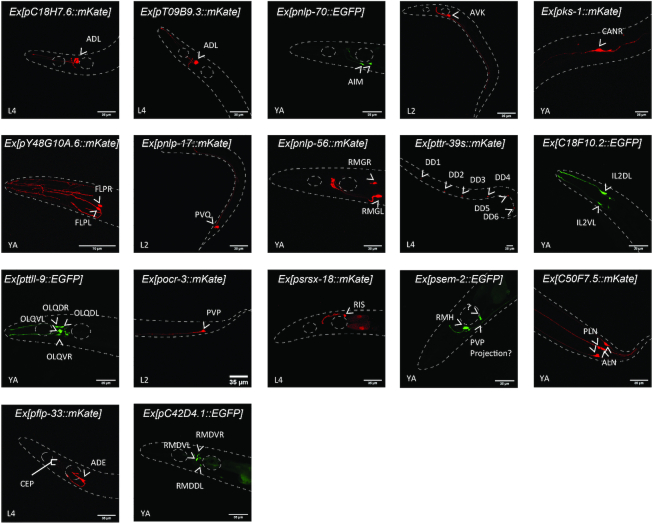
Gene enrichment lists provide 15 new neuron class-specific promoters
Identifying suitable promoters to specifically drive the expression of genetic tools in a given neuron class is an important step toward its functional characterization in vivo. Currently, only 29 neuron classes can be targeted using specific promoters (Table 1). Theoretically, each of the clusters assigned to a single neuron class could deliver potential specific markers within enriched genes. To validate the approach, we designed short (<1 kb) and medium (<2 kb) size promoters upstream the ATG of 17 genes highly enriched in clusters of interest (Figure 7 and Supplementary Figure S2). These promoters were fused to a monomeric EGFP (mEGFP) or mKate sequence in order to visualize their expression patterns in vivo (Figure 7 and Supplementary Figure S2). We confirmed the expression patterns of these promoters based on their cellular position, morphology, and/or with NeuroPAL, a polychromatic strain expressing four fluorescent proteins under overlapping drivers, designed to unambiguously identify individual neurons (29) (Supplementary Figure S2). First, we confirmed the validity of the clustering prediction by designing two promoters for cluster 31, poorly assigned to ADL. We designed promoter-mKate fusion for two enriched genes: T09B9.3 and C18H7.6. Both constructions showed mKate expression in the two ADL neurons, confirming the identity of cluster 31 (Figure 7 and Supplementary Figure S2). We next designed promoters for a set of parent clusters in order to identify new promoters for neuron classes lacking specific promoters: AIM, AVK, CAN, FLP, PVQ, IL2L/R and RMG neurons. Indeed, all the seven designed reporter strains showed a specific expression of the fluorescent protein in their corresponding neuron classes (Figure 7 and Supplementary Figure S2). Finally, we designed promoters for a set of clusters assigned after the second clustering iteration. Among those, we identified new specific promoters for DD, OLQ, PVP and RIS neurons (Figure 7 and Supplementary Figure S2). Interestingly, no specific promoters were described until now for FLP without PVD, or IL2 D/V without IL2 L/R (Table 1).
Table 1.
Neuron class-specific promoters. As a resource to drive expression selectively in neurons of interest, we gathered all known neuron class-specific promoters published by the C. elegans community as well as the ones we identified in this work
| Neuron | Genes | Ref | Neuron | Genes | Ref |
|---|---|---|---|---|---|
| ADE_CEP | flp-33 | us | BAG | gcy-9 | (68) |
| ADF | srh-142 | (50) | BAG | flp-17 | (69) |
| ADL | svh-1 | (51) | CAN | S pks-1 | us |
| ADL | T09B9.3 | us | DD | S ttr-39 | us |
| ADL | C18H7.6 | us | DVA | twk-16 | (70) |
| AFD | gcy-8 | (52) | DVC | ceh-63 | (71) |
| AIA | gcy-28 | (53) | FLP | Y48G10A.6 | us |
| AIM | nlp-70 | us | FLP+PVD | deg-3 | (72) |
| AIY | ttx-3 | (54) | FLP+PVD | des-2 | (73) |
| ALN_PLN | C50F7.5 | us | IL1 | aqp-6 | (74) |
| ASEL | gcy-6 | (55) | IL1 | Y111B2A.8 | (75) |
| ASEL | gcy-7 | (55) | IL1 | flp-3 | (76) |
| ASER | gcy-4 | (56) | IL2 | klp-6 | (77) |
| ASER | gcy-5 | (55) | IL2LR | C18F10.2 | us |
| ASG | gcy-15 | (56) | OLQ | ocr-4 | (78) |
| ASI | str-3 | (57) | OLQ | ttll-9 | us |
| ASJ | sptf-1 | (58) | PHA | srg-13 | (61) |
| ASJ | ssu-1 | (59) | PHA | gcy-17 | (56) |
| ASJ | trx-1 | (60) | PVD_FLP | F49H12.4 | (73) |
| ASK | sra-7 | (61) | PVD_FLP | dma-1 | (79) |
| ASK | sra-9 | (61) | PVP | ocr-3 | us |
| ASK | srg-2 | (61) | PVQ | nlp-17 | us |
| ASK | srg-8 | (61) | PVT | zig-2 | WB |
| ASK | srbc-64 | (62) | RIA | glr-3 | (80) |
| ASK | srbc-66 | (62) | RIA | glr-6 | (80) |
| AVJ | hlh-34 | (63) | RIC | tbh-1 | (81) |
| AVK | flp-1 | (64) | RIM | gcy-13 | (56) |
| AVK | twk-47 | us | RIM | cex-1 | (82) |
| AWA | odr-10 | (65) | RIS | srsx-18 | us |
| AWB | srab-16 | (66) | RIS | flp-11 | (83) |
| AWB | sru-38 | (66) | RMD | C42D4.1 | us |
| AWB | str-44 | (66) | RMG | nlp-56 | (28) and us |
| AWB | str-163 | (66) | RMH | sem-2 | (76) and us |
| AWC OFF | srsx-3 | (67) | URX_AQR_PQR | gcy-32 | (55) |
| AWC OFF | srt-26 | (67) | SAA | rig-5? | (76) |
| AWC OFF | srt-28 | (67) | |||
| AWC ON | str-2 | (67) | |||
| AWC ON | srt-45 | (67) | |||
| AWC ON | srt-47 | (67) |
WB: described in Wormbase, sequence not known.
Figure 7.
Confocal images of 17 designed reporter strains. The expression patterns of mKate (red) or mEGFP (green) driven by the corresponding gene promoter were analysed by confocal microscopy. White arrowheads indicate the neuron-classes expressing the fluorescent proteins. The animals were imaged between L2, L4 or young adult (YA) developmental stages. Details concerning neuron class identification, promoters’ size and primers’ sequences are described in Supplementary Figure S2.
This promoter screen also helped us to assign an identity to the poorly assigned cluster 48. A reporter strain for sem-2, enriched in cluster 48, showed a strong expression in RMH, suggesting cluster 48 can be assigned to RMH or does include RMH (Figure 7 and Supplementary Figure S2). Some of the selected promoters highlighted more than a single neuron class but confirmed our predictions. For example, C50F7.5 was enriched in cluster 20, assigned to the oxygen sensory neurons ALN and PLN. Accordingly to our prediction, the C50F7.5-mKate reporter strain showed a specific expression of mKate in both ALN and PLN neurons, allowing us to define a new marker for ALN and PLN (Figure 7 and Supplementary Figure S2). Similarly, flp-33 was expressed in the dopaminergic neurons ADE and CEP, but more importantly enriched in the ADE neurons cluster. In fact, the flp-33-mKate reporter strain strongly highlighted the ADE neurons and showed a weak expression of mKate in the CEP neurons (Figure 7 and Supplementary Figure S2). Nevertheless, some of the selected promoters failed to highlight a single neuron class: C42D4.1 was enriched in a cluster assigned to SMD and RMD. The reporter strain showed indeed an expression in RMD but also showed a weak expression in another non-identified neuron (Figure 7 and Supplementary Figure S2).
Gene Ontology (GO) analysis reveals functional diversification within neuronal modalities
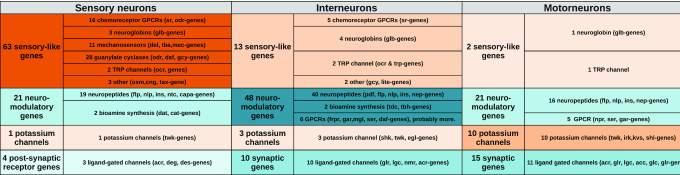
Similarly to the mouse brain (43), we observed that genes involved in sensory machineries, membrane potential, synaptic function and neuromodulation were over-represented within the differentially expressed genes of each neuron class (Supplementary Table S3). We reasoned that these specific genes contribute to the functional diversity observed between the 37 ‘sensory neurons’ classes, between the 44 ‘interneurons’ classes and between the 23 ‘motorneurons’ classes. To explore which classes of genes are involved, we selected 200 neuron class specific genes for these three neuronal modalities. The genes required for the sensory machineries (guanylate-cyclases, chemosensory receptors, Transient Receptor Potential channels etc.) mostly contributed to the diversity of the sensory neuron classes, while post-synaptic receptors are not highly diversified. The genes involved in neuromodulation (bioamine synthetic pathways, neuropeptides and their receptors) mostly contributed to the diversity of the interneuron classes. Finally, potassium channels contributed mostly to the diversity of the motor neuron classes (Table 2). These observations suggest the functional diversification of neurons involves different classes of genes, accordingly to neuronal modality.
Table 2.
Differential gene class enrichment between neuronal modalities. The molecular and biological functions of 200 neuron class specific genes were analysed independently for 28 sensory neurons clusters, 21 interneurons clusters and 15 motor neurons clusters. Genes were grouped accordingly to their putative or demonstrated functions in C. elegans
|
DISCUSSION
Although neuronal classification is a useful abstraction to neurophysiologists, the border between classes is difficult to resolve. Contrasting with complex brains, the full diversity of the C. elegans nervous system is already anatomically established at the single cell resolution. In order to generate the molecular profiles for each neuron class in C. elegans, we attempted to match the molecular information from scRNA-seq data to these anatomically defined neuron classes. Built on Cao et al. (15), our heuristic approach succeeded to assign 52 additional clusters to a single-neuron class. However, the simple one-to-one correspondence between neuron classes and cell-clusters did not apply to 15 of our clusters, assigned to a subset of neuron classes. This could be explained by several technical limitations in the depth of expression profiling, in molecular signatures and in the number of neurons sequenced, preventing us to identify genes that might segregate these classes. Alternatively, some neuronal classes might be molecularly too similar to each other to be robustly separated despite known differences in connectivity and functions (e.g. the motor neuron classes in cluster 0_2). In contrast, we easily segregated the chemosensory neuron class ASE in two sub-classes ASER and ASEL, known to display symmetric morphology but asymmetric gene expressions and functions (15). These two examples above illustrate the abstract border between neuron classes and sub-classes from the molecular point of view.
We anticipate our work will serve as a resource to facilitate future works on C. elegans nervous system. Indeed, only a few markers were available for each neuron class, and only a few neuron classes were profiled in depth (10,12,44,45). Our work provides a partial molecular profile for the 62 clusters we assigned to a single neuron class and 15 combined profiles for clusters assigned to a subset of neuron classes. Despite the shallow sequencing, zero-inflation effects and cluster size affecting the analysis (Supplementary Figure S1G) (46), we confirmed the quality of the expression profiles by the comparison with bulk sequencing and by the generation of neuron-specific promoters. Although the regulatory cis-elements of every gene are not known, the restricted expression patterns of the 17 promoters we tested here support the strength of this approach to deliver additional specific promoters. Partly because of the lack of specific promoters, little is known about the functions of some neuron classes (e.g. ALN, PLN, AIM and CAN). The 15 new markers we validated here already provide to the community a set of new specific promoters, of which 10 are for neuron classes without any known specific marker to this day. Some of these new promoters will be valuable tools to manipulate neurons hard to access currently. For example, optogenetic manipulation of RMG required to use specific illumination method or to restrict the expression using overlap between two promoters (47).
Recently, two other datasets corresponding to ∼85,000 embryonic cells and to ∼52 000 L4 larval stage neurons were sequenced by (27) and (28), respectively. In comparison to our approach, the authors used a small number of specific markers to assign each cell clusters to their corresponding neuron classes but had access to the lineage information. We observed an important overlap (80.5%) between our assigned clusters and the corresponding ones in the L4 stage animal (28) (Supplementary Table S5), supporting the results of both assignment efforts and suggesting additional markers for specific promoters (Supplementary Table S3). Similarly, we observed an important overlap (82.8%) between our assigned clusters and the neuron classes emerging at the end of the embryonic development (27) (Supplementary Table S5). Although, some neuron classes change their expression profiles only after circuit assembly (27) or mature through larval stage (38,39), these above observations suggest most of the neurons are committed to their transcriptomic identities at the end of the embryogenesis, and retain their identities throughout post-developmental stages. An interesting exception to such robust commitment at the end of embryogenesis is the trans-differentiation of the Y cell into PDA (40) that we partially observe in this dataset.
We extracted enriched transcription factors as potential cell fate regulators within three specific lineages. We show experimentally that unc-86 is necessary for the normal differentiation of RMG neurons. We also analysed differentially enriched gene ontologies between neuronal modalities (sensory-, inter- and motor neurons). While it is intuitively expected sensory machineries genes diversify the sensory neurons, we observed that other specific gene families contributed to the diversity in interneurons and motor neurons. On one hand, genes involved in neuromodulation were highly diverse between interneuron classes, suggesting a complex neuromodulation soup is generated by each interneuron and differentially controls these interneuron classes. On the other hand, genes involved in the tuning of membrane potential were highly diverse between motor neurons classes, suggesting their diversity involves a tight control of their membrane potential. Altogether, our work will contribute to the molecular characterisation of the neuronal diversity of C. elegans, a crucial step toward the understanding of its neuronal network.
DATA AVAILABILITY
All scripts and datasets used for the iterative clustering analysis plus the resulting outputs are available in the GitHub repository (https://github.com/lorenzo-jr/ce-neuronclusters). Representation of scatter plots using the log2(fold change) of marker genes for matching clusters between Lorenzo et al., Packer et al. and Taylor et al. are available on the resource of our website.
Supplementary Material
ACKNOWLEDGEMENTS
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We acknowledge the Shendure, Trapnell, and Waterston laboratories for the scRNA-seq dataset; Wormbase for the molecular atlas; the Hobert lab for the NeuroPAL strain, the Satija Lab for the Seurat v3 R package; Amandine Cornil for IT support, Sumeet Pal Singh for expertise and advises on scRNA analysis.
Contributor Information
Ramiro Lorenzo, Laboratory of Neurophysiology, ULB Neuroscience Institute (UNI), Université Libre de Bruxelles (ULB), Brussels, Belgium; Centro de Investigación Veterinaria de Tandil (CIVETAN), CONICET-CICPBA-UNCPBA, Facultad de Ciencias Veterinarias, Universidad Nacional del Centro (FCV-UNCPBA), Tandil, Argentina.
Michiho Onizuka, Laboratory of Neurophysiology, ULB Neuroscience Institute (UNI), Université Libre de Bruxelles (ULB), Brussels, Belgium.
Matthieu Defrance, Interuniversity Institute of Bioinformatics in Brussels, Université Libre de Bruxelles, Brussels, Belgium.
Patrick Laurent, Laboratory of Neurophysiology, ULB Neuroscience Institute (UNI), Université Libre de Bruxelles (ULB), Brussels, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds de la Recherche Scientifique (FNRS), Belgium; R.L. received support from the Fondation Wiener Anspach; Wallonie-Bruxelles International. Funding for open access charge: Fonds de la Recherche Scientifique (FNRS).
Conflict of interest statement. None declared.
REFERENCES
- 1. Cajal S. Sobre las fibras nerviosas de la capamolecular del cerebelo. Rev. Trimest. Histol. Norm. Patol. 1888; 2:33–49. [Google Scholar]
- 2. Zhu Y., Sousa A.M.M., Gao T., Skarica M., Li M., Santpere G., Esteller-Cucala P., Juan D., Ferrández-Peral L., Gulden F.O. et al.. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 2018; 362:eaat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tasic B., Menon V., Nguyen T.N., Kim T.K., Jarsky T., Yao Z., Levi B., Gray L.T., Sorensen S.A., Dolbeare T. et al.. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016; 19:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G. et al.. Molecular architecture of the mouse nervous system. Cell. 2018; 174:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. White J.G., Southgate E., Thomson J.N., Brenner S.. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986; 314:1–340. [DOI] [PubMed] [Google Scholar]
- 6. Albertson D.G., Thomson J.N.. The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976; 275:299–325. [DOI] [PubMed] [Google Scholar]
- 7. Cook S.J., Jarrell T.A., Brittin C.A., Wang Y., Bloniarz A.E., Yakovlev M.A., Nguyen K.C.Q., Tang L.T., Bayer E.A., Duerr J.S. et al.. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 2019; 571:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee R.Y.N., Howe K.L., Harris T.W., Arnaboldi V., Cain S., Chan J., Chen W.J., Davis P., Gao S., Grove C. et al.. WormBase 2017: molting into a new stage. Nucleic Acids Res. 2018; 46:D869–D874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hobert O., Glenwinkel L., White J.. Revisiting neuronal cell type classification in Caenorhabditis elegans. Curr. Biol. 2016; 26:R1197–R1203. [DOI] [PubMed] [Google Scholar]
- 10. Hallem E.A., Sternberg P.W.. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc. Natl Acad. Sci. U.S.A. 2008; 105:8038–8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lim M.A., Chitturi J., Laskova V., Meng J., Findeis D., Wiekenberg A., Mulcahy B., Luo L., Li Y., Lu Y. et al.. Neuroendocrine modulation sustains the C. elegans forward motor state. Elife. 2016; 5:e19887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oranth A., Schultheis C., Tolstenkov O., Erbguth K., Nagpal J., Hain D., Brauner M., Wabnig S., Steuer Costa W., McWhirter R.D. et al.. Food sensation modulates locomotion by dopamine and neuropeptide signaling in a distributed neuronal network. Neuron. 2018; 100:1414–1428. [DOI] [PubMed] [Google Scholar]
- 13. Gray J.M., Hill J.J., Bargmann C.I.. A circuit for navigation in Caenorhabditis elegans. Proc. Natl Acad. Sci. U.S.A. 2005; 102:3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagel G., Brauner M., Liewald J.F., Adeishvili N., Bamberg E., Gottschalk A.. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 2005; 15:2279–2284. [DOI] [PubMed] [Google Scholar]
- 15. Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J. et al.. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017; 357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sulston J.E., Horvitz H.R.. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977; 56:110–156. [DOI] [PubMed] [Google Scholar]
- 17. Stefanakis N., Carrera I., Hobert O.. Regulatory logic of Pan-neuronal gene expression in C. elegans. Neuron. 2015; 87:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McGinnis C.S., Murrow L.M., Gartner Z.J.. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019; 8:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfister R., Schwarz K.A., Janczyk M., Dale R., Freeman J.B.. Good things peak in pairs: a note on the bimodality coefficient. Front Psychol. 2013; 4:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whitlock M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 2005; 18:1368–1373. [DOI] [PubMed] [Google Scholar]
- 21. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc.: Ser. B (Methodological). 1995; 57:289–300. [Google Scholar]
- 22. Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J. et al.. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019; 566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C.. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017; 14:979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McInnes L., Healy J., Umap J.M.. Uniform manifold approximation and projection for dimension reduction. 2018; arXiv:06 December 2018, preprint: not peer reviewedhttps://arxiv.org/abs/1802.03426.
- 26. Traag V.A., Waltman L., van Eck N.J.. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019; 9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., Stefanik D., Tan K., Trapnell C., Kim J. et al.. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019; 365:eaax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor S.S., Santpere G., Reilly M., Glenwinkel L., Poff A., McWhirter R., Xu C., Weinreb A., Basavaraju M., Cook S.J. et al.. Expression profiling of the mature C. elegans nervous system by single-cell RNASequencing. 2019; bioRxiv doi:16 August 2019, preprint: not peer reviewed 10.1101/737577. [DOI]
- 29. Yemini E., Lin A., Nejatbakhsh A., Varol E., Sun R., Mena G.E., Samuel A.D.T., Paninski L., Venkatachalam V., Hobert O.. NeuroPAL: a neuronal polychromatic atlas of landmarks for whole-brain imaging in C. elegans. 2019; bioRxiv doi:20 June 2019, preprint: not peer reviewed 10.1101/676312. [DOI] [PMC free article] [PubMed]
- 30. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R.. Comprehensive integration of Single-Cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vidal B., Aghayeva U., Sun H., Wang C., Glenwinkel L., Bayer E.A., Hobert O.. An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol. 2018; 16:e2004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Busch K.E., Laurent P., Soltesz Z., Murphy R.J., Faivre O., Hedwig B., Thomas M., Smith H.L., de Bono M.. Tonic signaling from O2 sensors sets neural circuit activity and behavioral state. Nat. Neurosci. 2012; 15:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chalfie M., Sulston J.E., White J.G., Southgate E., Thomson J.N., Brenner S.. The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 1985; 5:956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lloret-Fernández C., Maicas M., Mora-Martínez C., Artacho A., Jimeno-Martín Á., Chirivella L., Weinberg P., Flames N.. A transcription factor collective defines the HSN serotonergic neuron regulatory landscape. Elife. 2018; 7:e32785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hobert O. Neurogenesis in the nematode Caenorhabditis elegans. WormBook 1-24. 2010; doi:10.1895/wormbook.1.12.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hobert O. A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscipl. Rev.: Dev. Biol. 2016; 5:474–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Serrano-Saiz E., Leyva-Díaz E., De La Cruz E., Hobert O.. BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr. Biol. 2018; 28:2813–2823. [DOI] [PubMed] [Google Scholar]
- 38. Petersen S.C., Watson J.D., Richmond J.E., Sarov M., Walthall W.W., Miller D.M.. A transcriptional program promotes remodeling of GABAergic synapses in Caenorhabditis elegans. J. Neurosci. 2011; 31:15362–15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson-Peer K.L., Bai J., Hu Z., Kaplan J.M.. HBL-1 patterns synaptic remodeling in C. elegans. Neuron. 2012; 73:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jarriault S., Schwab Y., Greenwald I.. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl Acad. Sci. U.S.A. 2008; 105:3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han B., Bellemer A., Koelle M.R.. An evolutionarily conserved switch in response to GABA affects development and behavior of the locomotor circuit of Caenorhabditis elegans. Genetics. 2015; 199:1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Topalidou I., van Oudenaarden A., Chalfie M.. Caenorhabditis elegans aristaless/Arx gene alr-1 restricts variable gene expression. Proc. Natl Acad. Sci. U.S.A. 2011; 108:4063–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C. et al.. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015; 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- 44. Spencer W.C., McWhirter R., Miller T., Strasbourger P., Thompson O., Hillier L.W., Waterston R.H., Miller D.M.. Isolation of specific neurons from C. elegans larvae for gene expression profiling. PLoS One. 2014; 9:e112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang J., Kaletsky R., Silva M., Williams A., Haas L.A., Androwski R.J., Landis J.N., Patrick C., Rashid A., Santiago-Martinez D. et al.. Cell-Specific Transcriptional Profiling of Ciliated Sensory Neurons Reveals Regulators of Behavior and Extracellular Vesicle Biogenesis. Curr. Biol. 2015; 25:3232–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vallejos C.A., Risso D., Scialdone A., Dudoit S., Marioni J.C.. Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat. Methods. 2017; 14:565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Macosko E.Z., Pokala N., Feinberg E.H., Chalasani S.H., Butcher R.A., Clardy J., Bargmann C.I.. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009; 458:1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chew Y.L., Grundy L.J., Brown A.E.X., Beets I., Schafer W.R.. Neuropeptides encoded by nlp-49 modulate locomotion, arousal and egg-laying behaviours in Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018; 373:20170368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kwon A.T., Arenillas D.J., Worsley Hunt R., Wasserman W.W.. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda). 2012; 2:987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bacaj T., Tevlin M., Lu Y., Shaham S.. Glia are essential for sensory organ function in C. elegans. Science. 2008; 322:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li C., Hisamoto N., Nix P., Kanao S., Mizuno T., Bastiani M., Matsumoto K.. The growth factor SVH-1 regulates axon regeneration in C. elegans via the JNK MAPK cascade. Nat. Neurosci. 2012; 15:551–557. [DOI] [PubMed] [Google Scholar]
- 52. Inada H., Ito H., Satterlee J., Sengupta P., Matsumoto K., Mori I.. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics. 2006; 172:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shinkai Y., Yamamoto Y., Fujiwara M., Tabata T., Murayama T., Hirotsu T., Ikeda D.D., Tsunozaki M., Iino Y., Bargmann C.I. et al.. Behavioral choice between conflicting alternatives is regulated by a receptor guanylyl cyclase, GCY-28, and a receptor tyrosine kinase, SCD-2, in AIA interneurons of Caenorhabditis elegans. J. Neurosci. 2011; 31:3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wenick A.S., Hobert O.. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell. 2004; 6:757–770. [DOI] [PubMed] [Google Scholar]
- 55. Yu S., Avery L., Baude E., Garbers D.L.. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl Acad. Sci. U.S.A. 1997; 94:3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ortiz C.O., Etchberger J.F., Posy S.L., Frøkjaer-Jensen C., Lockery S., Honig B., Hobert O.. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics. 2006; 173:131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peckol E.L., Troemel E.R., Bargmann C.I.. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc. Natl Acad. Sci. U.S.A. 2001; 98:11032–11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. González-Barrios M., Fierro-González J.C., Krpelanova E., Mora-Lorca J.A., Pedrajas J.R., Peñate X., Chavez S., Swoboda P., Jansen G., Miranda-Vizuete A.. Cis- and trans-regulatory mechanisms of gene expression in the ASJ sensory neuron of Caenorhabditis elegans. Genetics. 2015; 200:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carroll B.T., Dubyak G.R., Sedensky M.M., Morgan P.G.. Sulfated signal from ASJ sensory neurons modulates stomatin-dependent coordination in Caenorhabditis elegans. J. Biol. Chem. 2006; 281:35989–35996. [DOI] [PubMed] [Google Scholar]
- 60. Miranda-Vizuete A., Fierro González J.C., Gahmon G., Burghoorn J., Navas P., Swoboda P.. Lifespan decrease in a Caenorhabditis elegans mutant lacking TRX-1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 2006; 580:484–490. [DOI] [PubMed] [Google Scholar]
- 61. Troemel E.R., Chou J.H., Dwyer N.D., Colbert H.A., Bargmann C.I.. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995; 83:207–218. [DOI] [PubMed] [Google Scholar]
- 62. Kim K., Sato K., Shibuya M., Zeiger D.M., Butcher R.A., Ragains J.R., Clardy J., Touhara K., Sengupta P.. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science. 2009; 326:994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cunningham K.A., Hua Z., Srinivasan S., Liu J., Lee B.H., Edwards R.H., Ashrafi K.. AMP-activated kinase links serotonergic signaling to glutamate release for regulation of feeding behavior in C. elegans. Cell Metab. 2012; 16:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nelson L.S., Rosoff M.L., Li C.. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science. 1998; 281:1686–1690. [DOI] [PubMed] [Google Scholar]
- 65. Sengupta P., Chou J.H., Bargmann C.I.. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996; 84:899–909. [DOI] [PubMed] [Google Scholar]
- 66. Brear A.G., Yoon J., Wojtyniak M., Sengupta P.. Diverse cell type-specific mechanisms localize G protein-coupled receptors to Caenorhabditis elegans sensory cilia. Genetics. 2014; 197:667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lesch B.J., Bargmann C.I.. The homeodomain protein hmbx-1 maintains asymmetric gene expression in adult C. elegans olfactory neurons. Genes Dev. 2010; 24:1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guillermin M.L., Castelletto M.L., Hallem E.A.. Differentiation of carbon dioxide-sensing neurons in Caenorhabditis elegans requires the ETS-5 transcription factor. Genetics. 2011; 189:1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brandt J.P., Aziz-Zaman S., Juozaityte V., Martinez-Velazquez L.A., Petersen J.G., Pocock R., Ringstad N.. A single gene target of an ETS-family transcription factor determines neuronal CO2-chemosensitivity. PLoS One. 2012; 7:e34014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Puckett Robinson C., Schwarz E.M., Sternberg P.W.. Identification of DVA interneuron regulatory sequences in Caenorhabditis elegans. PLoS One. 2013; 8:e54971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Feng H., Reece-Hoyes J.S., Walhout A.J., Hope I.A.. A regulatory cascade of three transcription factors in a single specific neuron, DVC, in Caenorhabditis elegans. Gene. 2012; 494:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yassin L., Gillo B., Kahan T., Halevi S., Eshel M., Treinin M.. Characterization of the deg-3/des-2 receptor: a nicotinic acetylcholine receptor that mutates to cause neuronal degeneration. Mol. Cell. Neurosci. 2001; 17:589–599. [DOI] [PubMed] [Google Scholar]
- 73. Chatzigeorgiou M., Yoo S., Watson J.D., Lee W.H., Spencer W.C., Kindt K.S., Hwang S.W., Miller D.M., Treinin M., Driscoll M. et al.. Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nat. Neurosci. 2010; 13:861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang C.G., Lamitina T., Agre P., Strange K.. Functional analysis of the aquaporin gene family in Caenorhabditis elegans. Am. J. Physiol. Cell Physiol. 2007; 292:C1867–C1873. [DOI] [PubMed] [Google Scholar]
- 75. Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A. et al.. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007; 5:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vidal B., Santella A., Serrano-Saiz E., Bao Z., Chuang C.F., Hobert O.. C. elegans SoxB genes are dispensable for embryonic neurogenesis but required for terminal differentiation of specific neuron types. Development. 2015; 142:2464–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang F., Bhattacharya A., Nelson J.C., Abe N., Gordon P., Lloret-Fernandez C., Maicas M., Flames N., Mann R.S., Colón-Ramos D.A. et al.. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development. 2014; 141:422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tobin D.M., Madsen D.M., Kahn-Kirby A., Peckol E.L., Moulder G., Barstead R., Maricq A.V., Bargmann C.I.. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002; 35:307–318. [DOI] [PubMed] [Google Scholar]
- 79. Liu O.W., Shen K.. The transmembrane LRR protein DMA-1 promotes dendrite branching and growth in C. elegans. Nat. Neurosci. 2011; 15:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brockie P.J., Madsen D.M., Zheng Y., Mellem J., Maricq A.V.. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 2001; 21:1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Suo S., Kimura Y., Van Tol H.H.. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J. Neurosci. 2006; 26:10082–10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Piggott B.J., Liu J., Feng Z., Wescott S.A., Xu X.Z.. The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell. 2011; 147:922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Turek M., Besseling J., Spies J.P., König S., Bringmann H.. Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep. Elife. 2016; 5:e12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts and datasets used for the iterative clustering analysis plus the resulting outputs are available in the GitHub repository (https://github.com/lorenzo-jr/ce-neuronclusters). Representation of scatter plots using the log2(fold change) of marker genes for matching clusters between Lorenzo et al., Packer et al. and Taylor et al. are available on the resource of our website.