Summary
Spx is a major regulator of stress responses in Firmicutes. In Streptococcus mutans, two Spx homologues, SpxA1 and SpxA2, were identified as mediators of oxidative stress responses but the regulatory circuits controlling their levels and activity are presently unknown. Comparison of SpxA1 and SpxA2 protein sequences revealed differences at the C-terminal end, with SpxA1 containing an unusual number of acidic residues. Here, we showed that a GFP reporter becomes unstable when fused to the last 10 amino acids of SpxA2 but remained stable when fused to the C-terminal acidic tail of SpxA1. Inactivation of clpP or simultaneous inactivation of clpC and clpE stabilized the GFP::SpxA2tail fusion protein. Addition of acidic amino acids to the GFP::SpxA2tail chimera stabilized GFP while deletion of the acidic residues destabilized GFP::SpxA1tail. Promoter reporter fusions revealed that spxA1 transcription is co-repressed by the metalloregulators PerR and SloR while spxA2 transcription is largely dependent on the envelope stress regulator LiaFSR. In agreement with spxA2 being part of the LiaR regulon, SpxA2 was found to be critical for the growth of S. mutans under envelope stress conditions. Finally, we showed that redox-sensing is essential for SpxA1-dependent activation of oxidative stress responses but dispensable for SpxA2-mediated envelope stress responses.
Keywords: Streptococcus mutans, Spx, ClpP, oxidative stress
Graphical Abstract
Abbreviated Summary
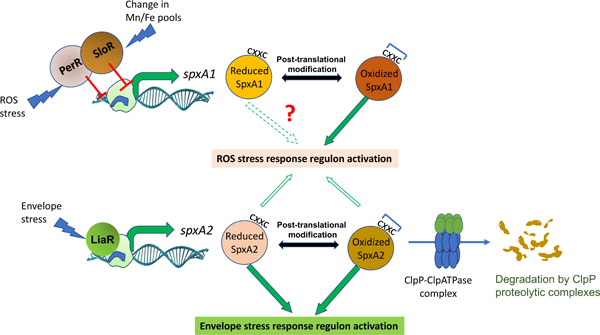
Streptococcus mutans encodes two Spx homologues. SpxA1 is resistant to ClpP proteolysis due to the presence of C-terminal acidic residues, transcriptionally regulated by the metalloregulators PerR and SloR, and relies on its redox sensing motif to activate an oxidative stress response. SpxA2 levels are controlled by the LiaFSR signal transduction system and posttranslationally by ClpP proteolysis. In addition, SpxA2 was critical for the growth of S. mutans under envelope stress conditions regardless of its redox status.
Introduction
An occasional inhabitant of the dental plaque, Streptococcus mutans is regarded as the major bacterial agent responsible for the initiation and progression of dental caries (Bowen, Burne, Wu, & Koo, 2018; K. Nakano, Nomura, Matsumoto, & Ooshima, 2010). Upon oral colonization, S. mutans metabolizes dietary carbohydrates to produce an acidic biofilm matrix that challenges the survival of less aciduric bacteria, which includes members of the mitis streptococci group such as S. sanguinis and S. gordonii (Lemos & Burne, 2008). Epidemiological studies have shown that high numbers of S. mutans in saliva and plaque are closely associated with active caries whereas high numbers of mitis streptococci are normally associated with oral health (Banas & Drake, 2018; Mira, Simon-Soro, & Curtis, 2017). It follows that S. sanguinis, S. gordonii and a few other streptococci associated with oral health are net producers of H2O2 that is inhibitory to the growth of S. mutans (Chen, Chakraborty, Zou, Burne, & Zeng, 2019; Zhu & Kreth, 2010). In addition to peroxigenic oral competitors, reactive oxygen species (ROS) in dental biofilms can derive from metabolic reduction of oxygen by the resident oral flora, including S. mutans’ own metabolism, and from the use of oral health and tooth bleaching products (Marquis, 1995).
In gram positive bacteria, activation of oxidative stress responses is largely dependent on the activity of the global transcription regulator Spx (Galvão et al., 2015; Kajfasz et al., 2012; Kajfasz et al., 2010; Whiteley, Ruhland, Edrozo, & Reniere, 2017; Zuber, 2004). The spx gene was first discovered in the context of reverting phenotypes associated with clpP and clpX mutants in the soil organism Bacillus subtilis, thus given the name Spx for suppressor of ClpP and ClpX phenotypes (M. M. Nakano, Hajarizadeh, Zhu, & Zuber, 2001). Subsequent studies revealed that B. subtilis Spx is subjected to ClpXP proteolytic control in vivo, and that toxic accumulation of Spx was responsible for many of the phenotypes associated with the clpP and clpX deletion strains (Garg, Kommineni, Henslee, Zhang, & Zuber, 2009; S. Nakano, Zheng, Nakano, & Zuber, 2002; Zhang & Zuber, 2007). The products of the clpP and clpX genes code for a serine peptidase and an ATPase, respectively. These subunits interact to form a barrel-shaped complex responsible for recognizing proteins that are captured and then subjected to ClpP degradation (T. A. Baker & Sauer, 2012). Spx proteolysis via ClpXP in B. subtilis was later shown to be greatly enhanced by the YjbH adapter protein, which interacts with the Spx C-terminal tail to facilitate ClpXP-mediated proteolysis (Engman & von Wachenfeldt, 2015; Frees, Savijoki, Varmanen, & Ingmer, 2007). More contemporary studies confirmed previous observations (S. Nakano et al., 2002) that the ClpCP protease also participates in Spx turnover, particularly under conditions that appear to destabilize ClpX (Rojas-Tapias & Helmann, 2019).
Previously, we identified two Spx homologues in S. mutans and showed that inactivation of either one of the Spx-encoding genes alleviated phenotypes of clpP and clpX mutants (Kajfasz et al., 2010). These genes were initially named spxA and spxB but later renamed spxA1 and spxA2, respectively, to avoid confusion with a pyruvate oxidase found in other streptococci that was also called spxB (Regev-Yochay, Trzcinski, Thompson, Malley, & Lipsitch, 2006; Xu, Itzek, & Kreth, 2014). Transcriptional and phenotypic characterizations of the S. mutans ΔspxA1 strain revealed that SpxA1 plays a major role in mediating oxidative stress survival by serving as the primary transcriptional activator of genes involved in cell detoxification (e.g., ahpCF, sodA, tpx), DNA repair (e.g., smn, smxA), and thiol homeostasis (e.g., trxA, trxB, gor) (Galvao et al., 2015; Kajfasz, Ganguly, Hardin, Abranches, & Lemos, 2017; Kajfasz et al., 2015). Not surprisingly, the ∆spxA1 strain was highly sensitive to oxidative stresses (Kajfasz et al., 2010). Though the ΔspxA2 strain did not show an oxidative stress tolerance phenotype, the ΔspxA1ΔspxA2 double mutant strain was hypersensitive to oxidative stresses when compared to its ΔspxA1 counterpart (Kajfasz et al., 2010; Kajfasz et al., 2015). In addition to what appears to be a secondary/ back-up role in the activation of oxidative stress responses, global transcriptional analysis suggested that SpxA2 may participate in the regulation of processes associated with cell envelope homeostasis (Kajfasz et al., 2010; Kajfasz et al., 2015). After the initial discovery of two Spx-like regulators in S. mutans, several studies revealed the presence of two Spx paralogs in other streptococci as well as in Listeria monocytogenes and Bacillus anthracis (Barendt, Birch, Mbengi, & Zuber, 2016; Barendt et al., 2013; Chen, Ge, Wang, Patel, & Xu, 2012; Port, Cusumano, Tumminello, & Caparon, 2017; Turlan, Prudhomme, Fichant, Martin, & Gutierrez, 2009; Zheng et al., 2014). In addition to the conserved role in activation of oxidative stress responses, Spx regulation has been more recently linked to activation of other types of stress, including antibiotic, osmotic and cell envelope stresses (Baek et al., 2015; Jousselin, Kelley, Barras, Lew, & Renzoni, 2013; Nilsson, Jakobsen, Givskov, Twetman, & Tolker-Nielsen, 2019; Renzoni et al., 2011; Rojas-Tapias & Helmann, 2018a; Villanueva et al., 2016). It is noteworthy that while the Spx protein of B. subtilis has been implicated in the regulation of multiple types of stress, including oxidative, heat and envelope stresses (Rojas-Tapias & Helmann, 2018a; Schafer et al., 2019; Zuber, 2004), the B. subtilis genome encodes a Spx paralogue, MgsR, that is part of the σB regulon and appears to participate in the ethanol stress response (Reder et al., 2008).
In the bacterial species studied to date, Spx levels were shown to be under ClpP proteolytic control (Chan, Garg, Lin, & Zuber, 2012; Chan, Hahn, & Zuber, 2014; Engman, Rogstam, Frees, Ingmer, & von Wachenfeldt, 2012; Garg et al., 2009; Kajfasz et al., 2009; S. Nakano et al., 2002; Pamp, Frees, Engelmann, Hecker, & Ingmer, 2006). The core genome of S. mutans encodes one copy of the clpP peptidase and five Clp ATPase-encoding genes named clpB, clpC, clpE, clpL and clpX. As the products of clpB and clpL lack the recognition tripeptide that mediates interaction with ClpP, only three of these S. mutans Clp ATPases interact with ClpP to form the ClpCP, ClpEP and ClpXP complexes (Frees et al., 2007). Using antibodies raised against the B. subtilis Spx, we showed that Spx accumulates in ΔclpP and ΔclpX strains of S. mutans but not in ΔclpC or ΔclpE strains (Kajfasz et al., 2009). However, due to the high degree of conservation at the protein level and nearly identical molecular weight of SpxA1 and SpxA2, it was not possible to discern if only one or both Spx proteins were accumulating in the ΔclpP and ΔclpX strains at that time. In addition to posttranscriptional control, the levels of Spx may also be regulated at the transcriptional level. In B. subtilis, spx is transcribed through five different promoters recognized by either the housekeeping sigma factor (σA) or by the alternative sigma factors σB, σM and σW. In addition, two transcriptional regulators, PerR and YodB, repress transcription from the major σA promoter such that spx promoters may be turned on or off in response to different environment inputs (Leelakriangsak, Kobayashi, & Zuber, 2007; Rojas-Tapias & Helmann, 2018a).
With the exception of B. subtilis, the transcriptional and posttranscriptional mechanisms controlling Spx levels have not been thoroughly investigated in other bacteria, particularly in organisms such as S. mutans that encodes two Spx homologs. In this study, we sought to identify the regulatory networks controlling the cellular levels and activity of the S. mutans SpxA1 and SpxA2 regulators. Using molecular genetics approaches, we showed that the cellular levels of SpxA1 and SpxA2 are controlled by distinct mechanisms, which includes the participation of different transcriptional regulators governing transcription of each gene and the unexpected observation that ClpP-mediated posttranslational control is restricted to SpxA2. In addition to the characterization of the regulatory circuits controlling Spx levels, we also demonstrate that the primary function of SpxA2 in S. mutans is to mediate an envelope stress response.
Results
Streptococcal Spx homologues differentiate mostly at their C-terminal end
Spx proteins belong to the ArsC_Spx sub-family (cd03032), which is divided into three sub-groups based on the presence of a redox-sensing CXXC motif in the amino terminus and a conserved glycine residue, crucial for the interaction of Spx with the RNAP α-CTD domain, located around the center of the protein sequence (Turlan et al., 2009). Two sub-groups in this sub-family are very closely associated consisting of 131 to 137 amino acid proteins with the conserved glycine residue always located at position 52 (Gly52) (M. M. Nakano et al., 2010). The other sub-group of the ArsC family contains relatively shorter proteins (ranging from 117 to 120 amino acids) with a glycine residue at the 50th amino acid position or absent in some cases. Previously, we showed that both S. mutans SpxA1 and SpxA2 have a Gly52 residue, and therefore belong in the first two sub-groups, whereas a third Spx paralog with a Gly50 residue failed to suppress phenotypes associated with clpP gene inactivation and therefore was not considered a true Spx protein (Kajfasz et al., 2010). Here, we focused our analysis on the two groups that contain the conserved Gly52 motif, often termed as SpxA1 and SpxA2 (Chen et al., 2012; Turlan et al., 2009). The length of SpxA1 proteins among streptococcal species range from 133 to 137 amino acids, whereas all streptococcal SpxA2 proteins are 132 amino acids long (Fig. 1). The difference in the length of streptococcal SpxA1 proteins is largely due to the presence of acidic amino acid residues at the protein C-terminal end in some species, including S. mutans and Streptococcus agalactiae (Fig. 1 and Fig. S1). On the other hand, the last four residues (RAAL) of all streptococcal SpxA2 proteins sequenced to date are 100% conserved (Fig. 1).
Figure 1:
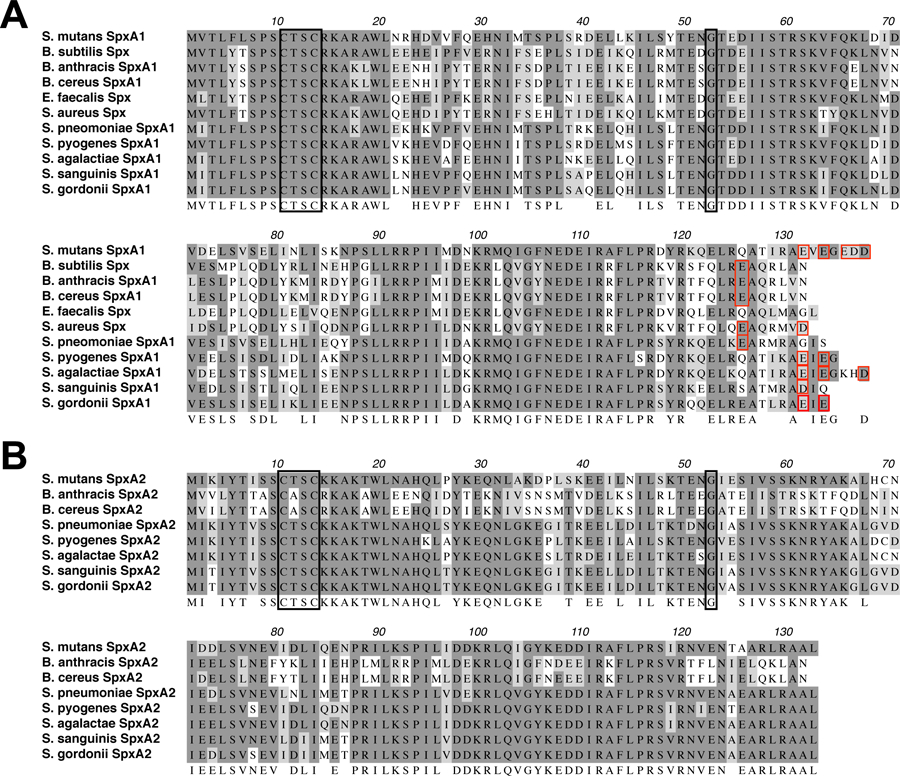
ClustalW alignment of SpxA1 (A) and SpxA2 (B) proteins from selected gram-positive bacteria. SpxA1 protein length varies from 131 to 137 amino acids whereas SpxA2 from all streptococcal species are 132 amino acids long. Identical residues are shown in dark shades and similar residues in light shades. A consensus is shown below the sequences. Black boxes indicate the conserved CXXC motif and glycine 52 residue. Red boxes indicate acidic residues present within the last 10 amino acids of each protein.
SpxA2 but not SpxA1 is subjected to ClpP proteolysis
In B. subtilis, Spx levels, herein SpxBsu, are kept in check by cooperative degradation by the ClpXP and ClpCP proteolytic systems (Chan et al., 2012; Engman et al., 2012; S. Nakano et al., 2002; Rojas-Tapias & Helmann, 2018b). Though both ClpCP and ClpXP can degrade SpxBsu, early evidence indicated that regulation of SpxBsu levels in vivo is primarily mediated by ClpXP (S. Nakano et al., 2002). Previously, we showed that Spx accumulated in both ΔclpP and ΔclpX strains of S. mutans, though at the time we were unable to determine if it was SpxA1, SpxA2, or both proteins that were accumulating in those mutant strains (Kajfasz et al., 2009). Attempts to monitor individual Spx protein stability in ΔclpPΔspxA1 or ΔclpPΔspxA2 strains by Western blotting were also unsuccessful, either because the detectable Spx band in the ΔclpP and ΔclpX cell extracts was a result of the accumulation of both proteins, or because inactivation of one spx gene affected transcriptional or posttranslational mechanisms controlling the levels of the other Spx protein (Kajfasz et al., 2009). A logical next step was to demonstrate degradation of recombinant S. mutans SpxA1 and SpxA2 proteins, herein SpxA1Smu and SpxA2Smu, by ClpXP in a reconstituted in vitro system (S. Nakano et al., 2002). However, under the conditions tested, both rSpxA1Smu and rSpxA2Smu remained fairly stable for 2 hours (Fig. S2). After that, both proteins started to degrade, though this degradation also occurred in the absence of purified ClpP (data not shown). In B. subtilis, the in vitro degradation of SpxBsu by ClpXP was very inefficient in the absence of the YjbH adaptor protein (Chan et al., 2014; Garg et al., 2009). Because streptococcal genomes do not encode YjbH homologs, we suspect that the S. mutans ClpXP system depends on the presence of a yet-to-be-identified adaptor protein for efficient degradation of Spx.
Previous studies from the Zuber lab showed that a motif comprised of the last 12 C-terminal amino acid residues of SpxBsu is recognized by the YjbH adaptor protein, facilitating Spx degradation by ClpXP (Chan et al., 2012). In addition, the addition of two aspartate residues to the C-terminal end was shown to render SpxBsu resistant to proteolysis. As mentioned above, SpxA1Smu and SpxA2Smu differ significantly at their C-terminal end, with SpxA1Smu displaying 5 acidic amino acid residues within the last 10 amino acids (Fig. 1). Considering that the addition of acidic residues to the protein C-terminus stabilized SpxBsu (Chan et al., 2012), we suspected that SpxA2Smu but not SpxA1Smu is naturally subjected to ClpP proteolysis. To circumvent the hurdles and shortcomings of in vivo detection and in vitro proteolysis reconstitution, we engineered recombinant proteins containing the last 10 amino acids of either SpxA1Smu or SpxA2Smu fused to an otherwise stable green florescence protein (GFP) (Jana, Tao, & Biswas, 2016), and then used these GFP::Spxtail chimera to investigate if the C-terminal of SpxA1Smu or SpxA2Smu served as recognition sites for ClpP-dependent proteolysis upon its expression in S. mutans (Fig. 2A). The stability of GFP over time was monitored in the S. mutans UA159 (parent) or ΔclpP strains by measuring fluorescence decay and Western blot before and after protein synthesis was halted with chloramphenicol. When expressed in the UA159 strain, the GFP protein alone remained stable for the duration of the experiment (Fig. 2B–C). Also in the UA159 strain, the GFP::SpxA1tail fusion protein remained stable for up to 12 hours whereas GFP::SpxA2tail was degraded by ∼ 80% within the initial 2 hours (Fig. 2B–C). Importantly, the GFP::SpxA2tail fusion protein was stabilized in the ΔclpP strain strongly indicating that SpxA2Smu is under ClpP proteolytic control (Fig. 2B–C).
Figure 2.
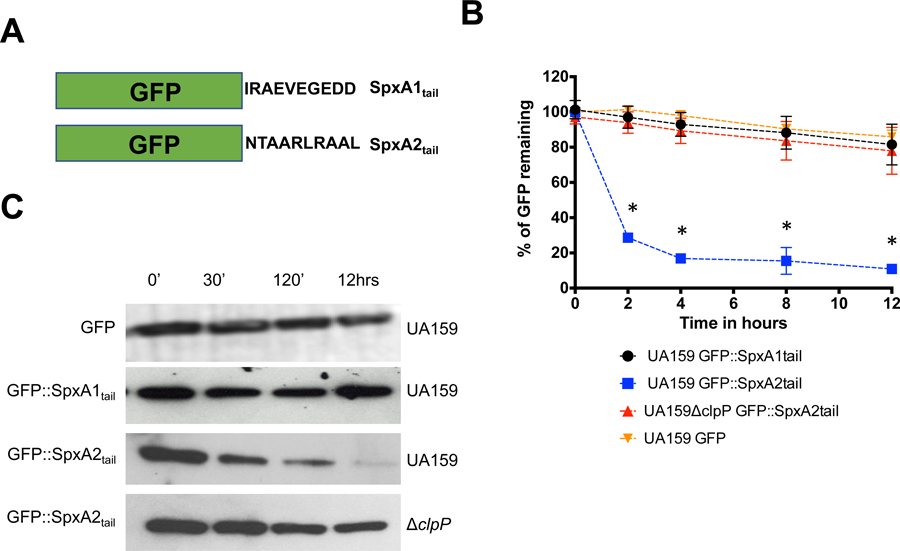
Stability of GFP::SpxA1tail and GFP::SpxA2tail fusion proteins in S. mutans. (A) Graphical representation of GFP fused to the last 10 C-terminal amino acids of either SpxA1Smu or SpxA2Smu. (B) Fluorescence decay of GFP::SpxA1tail and GFP::SpxA2tail expressed in UA159 (parent) or ΔclpP strains after addition of chloramphenicol. Asterisks indicate time points showing statistically significant differences (p ≤ 0.01, one-way ANOVA) in decay of GFP expression in the SpxA2tail construct when hosted in UA159 compared to the ΔclpP strain. (C) Western blot analysis of UA159 or ΔclpP expressing GFP::SpxA1tail and GFP::SpxA2tail probed with anti-GFP polyclonal antibody. The images shown are representative of 3 or more independent experiments.
To investigate whether C-terminal acidic residues can generally protect Spx against ClpP-mediated proteolysis, we next added 2 aspartate residues to the last 10 amino acids of the SpxA2Smu C-terminus, generating the GFP::SpxA2DDtail fusion protein. As anticipated, addition of aspartate residues stabilized the GFP:SpxA2DDtail fusion protein to levels that were almost identical to those described for the GFP:SpxA2tail in the ΔclpP strain (Fig. 3A and 3C). To further demonstrate the importance of the C-terminal residues for Spx stabilization, we engineered GFP::SpxA1tail fusion proteins with either 5 or 7 of the last C-terminal amino acids removed such that the acidic amino acids shown to stabilize SpxA1Smu were absent in these fusion proteins. In this case, the SpxA1Smu C-terminal residues lacking the last 5 (GFP::SpxA1-5tail) or 7 (GFP::SpxA1-7tail) acidic amino acids but still containing 10 C-terminal amino acids of SpxA1Smu was rapidly degraded (Fig. 3B–C), resembling the degradation kinetics observed for the SpxA2Smu C-terminal tail in the UA159 background that is shown in Figure 2. Collectively, these results reveal that the levels of SpxA2Smu are maintained by ClpP proteolysis, and that acidic residues at the SpxA1Smu C-terminus confers protection against proteolysis.
Figure 3.
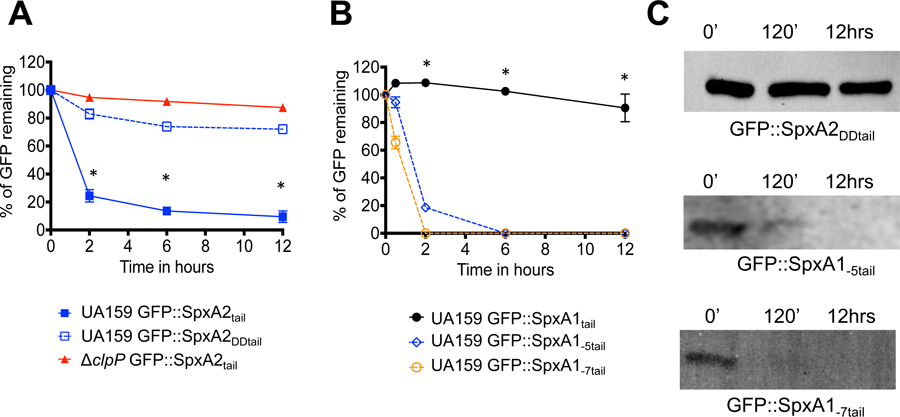
Stability of GFP::Spxtail fusion proteins with acidic residues added to the C-terminus of SpxA2 (SpxA2DDtail) or with acidic residues deleted from SpxA1 (SpxA1-5tail and SpxA1-7tail). (A) Fluorescence decay of GFP::SpxA2tail or GFP::SpxA2DDtail when expressed in UA159 or ΔclpP host strains. Asterisks indicate time points showing statistically significant differences (p ≤ 0.01, one-way ANOVA) in GFP::SpxA2tail decay compared to GFP::SpxA2DDtail when expressed in UA159. (B) Fluorescence decay of GFP::SpxA1tail, GFP::SpxA1-5tail or GFP::SpxA1-7tail expressed in UA159. Asterisks indicate time points showing statistically significant differences (p ≤ 0.01, one-way ANOVA) in decay of the GFP::SpxA1tail construct as compared to GFP::SpxA1-5tail or GFP::SpxA1-7tail. (C) Western blot analysis of S. mutans UA159 expressing GFP::SpxA2DDtail, GFP::SpxA1-5tail or GFP::SpxA1-7tail probed with anti-GFP polyclonal antibody. The images shown are representative of 3 or more independent experiments.
Both Spx proteins of S. pneumoniae are subjected to ClpP degradation
As mentioned above, the C-terminal acidic residues of SpxA1Smu are not widespread in other Spx proteins, seemingly limited to S. agalactiae and a small number of poorly characterized oral streptococcal species (Fig. S1). For a broader perspective of the posttranslational regulatory mechanisms controlling Spx levels in streptococci, we engineered GFP::Spxtail fusion proteins containing the last 10 C-terminal amino acids of the S. pneumoniae SpxA1 (SpxA1Spn) and SpxA2 (SpxA2Spn) proteins, neither of which possess acidic residues at their C-terminal end (Fig. 1). Here, both GFP::SPNSpxA1tail and GFP::SPNSpxA2tail fusion proteins were efficiently degraded in the S. mutans UA159 background and stabilized in the ΔclpP strain (Fig. 4). Thus, the differences in the posttranslational regulation of the S. mutans Spx proteins appears to be the exception and not the rule in the Streptococcus genus. We predict that SpxA1 homologs from S. agalactiae and the few oral streptococci that contain multiple acidic residues (Fig. S1) are resistant to ClpP proteolysis.
Figure 4.
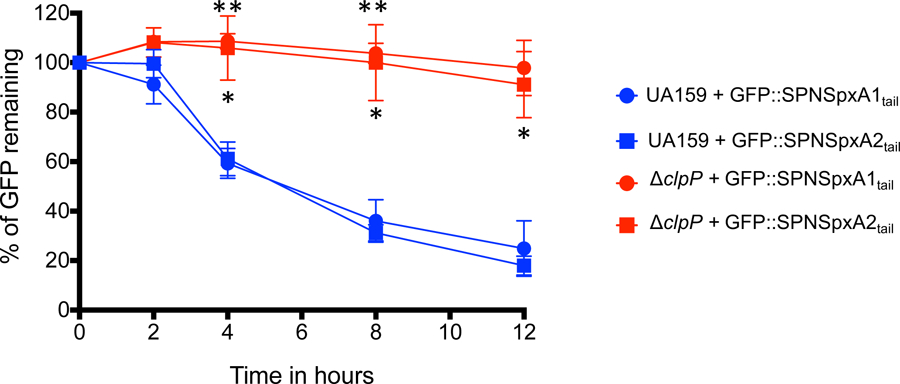
Stability of GFP::SPNSpx fusion proteins. Fluorescence decay of GFP::SPNSpxA1tail and GFP::SPNSpxA2tail expressed in UA159 or ΔclpP strains. Asterisks indicate time points showing statistically significant differences (p ≤ 0.01, one-way ANOVA) when the corresponding fusion protein is expressed in S. mutans UA159 or ΔclpP strains; a single asterisk indicates comparisons with the GFP::SPNSpxA1tail construct, while two asterisks indicate comparisons with the GFP::SPNSpxA2tail construct.
Simultaneous inactivation of clpC and clpE stabilized the GFP::SpxA2tail fusion protein
Next, we sought to identify the Clp ATPase(s) that interact with ClpP to degrade SpxA2Smu. First, we assessed the stability of the GFP::SpxA2tail fusion in single clp ATPase deletion strains (ΔclpC, ΔclpE and ΔclpX) generated in a previous study (Kajfasz et al., 2009). While our initial results indicate that SpxA2Smu is primarily under ClpXP proteolytic control (Kajfasz et al., 2009), single inactivation of clpX (or clpC) did not increase stability of the GFP::SpxA2tail (Fig. 5). Surprisingly, inactivation of clpE significantly increased GFP::SpxA2tail stability (Fig. 5). Due to the precedent of functional redundancy among different ClpP-Clp ATPase systems, including evidence that SpxBsu levels are controlled by both ClpCP and ClpXP (S. Nakano et al., 2002; Rojas-Tapias & Helmann, 2019), we next examined the stability of GFP::SpxA2tail in the 3 possible clp ATPase double mutant combinations. When compared to the partial stability observed in the ΔclpE strain, the GFP::SpxA2tail was further stabilized in the ΔclpXΔclpE strain, and almost completely stabilized in the ΔclpEΔclpC strain (Fig. 5). The simultaneous inactivation of clpC and clpX, the two primary Clp ATPases involved in degradation of SpxBsu (S. Nakano et al., 2002; Rojas-Tapias & Helmann, 2018b, 2019), did not increase GFP::SpxA2tail stability.
Figure 5.
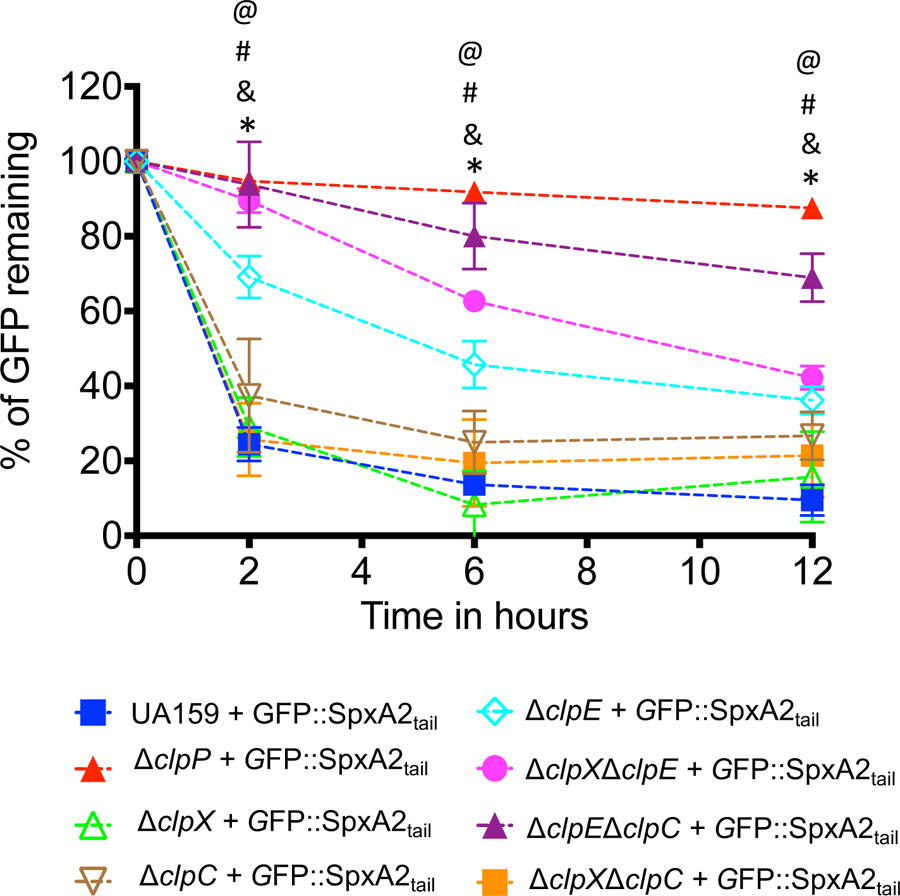
Fluorescence decay of GFP::SpxA2tail fusion proteins expressed in S. mutans UA159 and a panel of clp single and double mutant strains. Time points showing statistically significant differences (p ≤ 0.01, one-way ANOVA) when GFP::SpxA2tail expressed in S. mutans UA159 is compared to GFP::SpxA2tail expressed in ΔclpP (@), ΔclpE (*), ΔclpEΔclpC (#), or ΔclpXΔclpE (&) are indicated in the figure.
Transcriptional analyses of selected SpxA1- and SpxA2-regulated genes strengthen the GFP reporter studies
To obtain additional evidence that ClpP-mediated degration of SpxA2Smu but not of SpxA1Smu occurs in its native context, we used quantitative real-time PCR to compare the expression profile of SpxA1- and SpxA2-regulated genes in the UA159, ∆clpE and ∆clpP strains as a readout of SpxA1 and SpxA2 stability in vivo. Transcription of smu1412c, a gene coding for a hypothetical protein previously shown to be under SpxA2 positive regulation (Kajfasz et al., 2010), was significantly inceased (~ 3 fold) in the ∆clpP strain (Fig. 6A), a finding that supports the GFP::SpxA2tail protein chimera studies. This increase was not observed in the ∆clpE strain, possibly due to the redundant role of different ClpP systems (ClpEP, ClpCP and ClpXP) in SpxA2 degradation. On the other hand, transcription of sodA, a direct target of SpxA1 regulation (Kajfasz et al., 2017; Kajfasz et al., 2010; Kajfasz et al., 2015) was not altered in the ∆clpP and ∆clpE strains when compared to UA159 providing further evidence that SpxA1Smu is not a naturally subjected to ClpP proteolysis (Fig. 6B).
Figure 6.
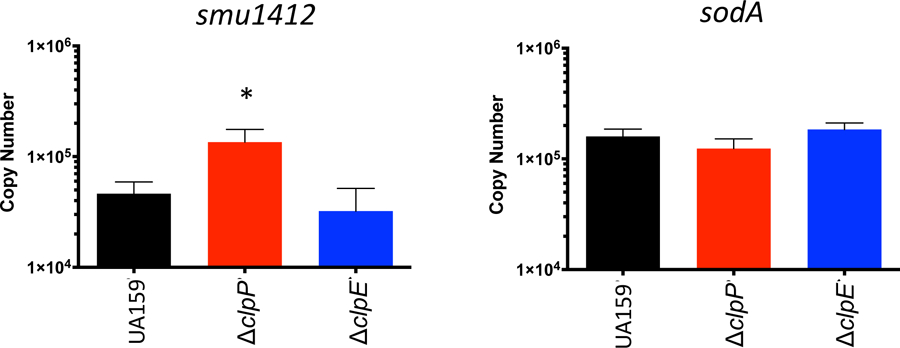
qRT PCR analysis of (A) SpxA1-regulated sodA or (B) SpxA2-regulated smu.11412c genes in different the ΔclpP and ΔclpE mutant background. (*) p< 0.05
Transcriptional regulation of spxA1 and spxA2
While Spx proteins were initially thought to be primarily regulated at the posttranslational level and functionally activated by means of a redox-sensing CXXX motif (Schafer & Turgay, 2019), previous studies have shown that spx genes may also be under transcriptional control (Pamp et al., 2006; Rojas-Tapias & Helmann, 2018a; Shankar, Mohapatra, Biswas, & Biswas, 2015). Using RACE-PCR, a single transcription initiation site was identified 27 nucleotides 5′ to the translational initiation site of spxA1smu that mapped to a putative σA-type promoter (TAGCCA-N17-TATAAT) (Fig. 7A). Similarly, a single transcriptional start also located 27 nucleotides from the start codon and a σA-type promoter (TCTTTA-N16-TAAGAT) were identified upstream of the spxA2 start codon (Fig. 7A). To determine the transcriptional profile of spxA1smu and spxA2smu, DNA fragments containing the promoter region of spxA1smu (PspxA1) and spxA2smu (PspxA2) were separately cloned in front of a promoterless chloramphenicol acetyl transferase (cat) reporter gene and integrated in the chromosome of UA159 using the one-step CAT integration vector pJL84 (Santiago, MacGilvray, Faustoferri, & Quivey, 2012). First, we measured CAT activity of PspxA1 and PspxA2 over the different phases of growth to find that CAT activity remained unaltered in both cases indicating that transcription from PspxA1 or PspxA2 is not growth phase dependent (Fig. S3). Based on this observation, the remaining experiments were performed with cells grown to an OD600 ∼ 0.4 (mid-log phase). Next, we determined the CAT-specific activity of PspxA1 or PspxA2 in cultures exposed to 0.5 mM H2O2, 0.5 mM diamide, or pH 6.0 for up to 30 minutes. CAT activity from either PspxA1 or PspxA2 was not altered after those treatments, indicating that neither spxA1 nor spxA2 levels are regulated by peroxide or acid stresses (Fig. 7B–C).
Figure 7.
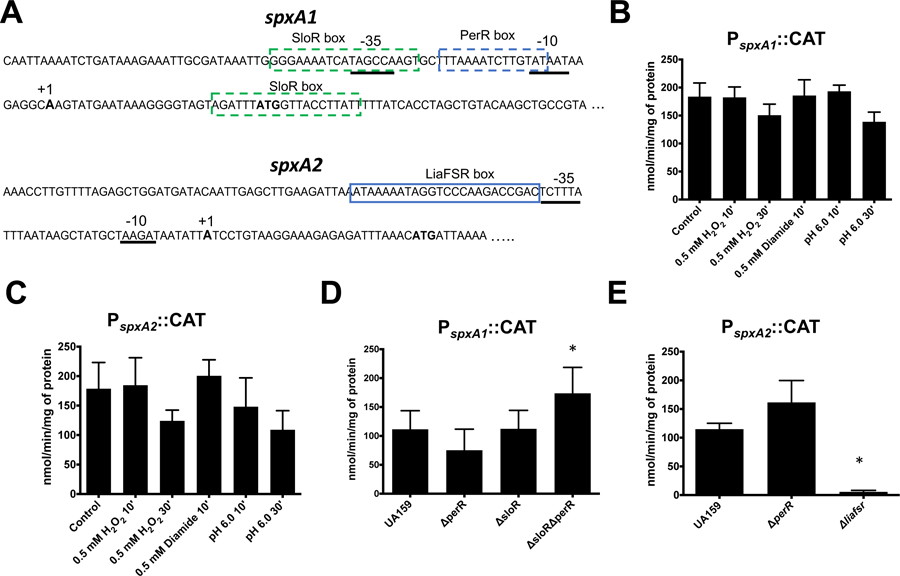
Transcriptional characterization of S. mutans spxA1 and spxA2. (A) Sequence of the PspxA1 and PspxA2 promoter regions. The σA-type −35 and −10 regions, mapped by 5’ RACE-PCR, are underlined. The transcriptional start site is shown in boldface and labeled with “+1”. The ATG start codon is also shown in boldface. Putative PerR- and SloR-binding sites in PspxA1 and LiaR-binding site in PspxA2 are shown inside boxes. (B-E) CAT activity driven from PspxA1 or PspxA2. (B-C) Cells were grown to mid-log phase and incubated in the presence of H2O2 (0.5 mM), diamide (0.5mM) or pH 6.0 for 10 or 30 minutes. (D-E) Mutant strains harboring the indicated CAT fusions were grown to mid-log phase. Asterisks indicates statistical significance (p value ≤ 0.01) when compared to control (B-C) or UA159 (D-E) by one-way ANOVA.
In addition to a previously identified SloR-binding motif (Crepps et al., 2016), in silico analysis revealed a potential regulatory site for PerR (NTANAANNATTNTAN) within the PspxA1 region (Fig. 7A). Both PerR and SloR are metalloregulators shown to function primarily as negative regulators of genes associated with peroxide stress and metal uptake, respectively (Crepps et al., 2016; Makthal et al., 2013). While the role of PerR in spxA1smu gene regulation has not been explored, the Spatafora lab recently provided evidence that SloR interacts, albeit weakly, with the spxA1 promoter region (Crepps et al., 2016). To further assess the participation of SloR and begin to explore the role of PerR in spxA1 gene regulation, we compared activity of the PspxA1-CAT reporter in the UA159 strain and its ΔsloR and ΔperR derivatives. Despite the high conservation (2 mismatches in 15 base consensus sequence) of the PerR-binding box identified in the spxA1 promoter region, loss of PerR resulted in a fairly small reduction in PspxA1 activity that had no statistical significance (Fig. 7D). Though contrary to the identification of the PerR box operator, this result is consistent with the finding that that PspxA1 was unresponsive to peroxide stress (Fig. 7B), the primary stress signal that alleviates PerR regulation. Despite previous evidence showing that SloR directly and specifically interacts with the spxA1 promoter region (Crepps et al., 2016), the CAT activity from PspxA1 also did not change in the ΔsloR strain (Fig. 7D), even in the presence of a high concentration of manganese (250 µM) that is known to increase SloR activity (data not shown). One possible explanation for the unaltered activity of PspxA1 in the ΔsloR or ΔperR strains is that repression of the spxA1 promoter by SloR and PerR is redundant such that one transcriptional repressor compensates for the absence of the other. It follows that the SloR-binding domain, known as SloR response element (SRE) (Spatafora et al., 2015) and PerR-binding domain are both AT-rich and one of the two putative SRE sites identified in the spxA1 promoter region is separated from the putative PerR-binding site by only two nucleotides (Fig. 7A). To test this possibility, we generated a double ΔsloRΔperR strain and found that PspxA1 activity in this double mutant strain displayed a statistically significant increase (∼ 65%) when compared to the parent strain UA159 (Fig. 7D).
When in silico analysis was performed to search the PspxA2 region for a conserved regulatory motif, only a previously identified LiaR-binding motif (Shankar et al., 2015; Suntharalingam, Senadheera, Mair, Levesque, & Cvitkovitch, 2009) was found. LiaR is the response regulator of the signal transduction system LiaFSR, which is responsible for orchestrating an envelope stress response against cell wall-targeting antibiotics and membrane damaging agents (Suntharalingam et al., 2009). Previously, Shankar and colleagues showed through a gel mobility shift assay that the S. mutans LiaR specifically binds to the PspxA2 region, but further studies to determine whether LiaR functions as a repressor or activator of spxA2 were not pursued at that time (Shankar et al., 2015). Here, we found that PspxA2 activity was strongly dependent on the LiaFSR system as PspxA2::CAT activity was nearly undetectable in a ΔliaFSR strain (Fig. 7E). This finding prompted us to examine PspxA2-CAT activity after exposure to a number of cell wall- or membrane-damaging agents that are sensed by LiaFSR in S. mutans or in closely-related organisms (Eldholm et al., 2010; Suntharalingam et al., 2009). In agreement with the strong dependence of LiaFSR for activation, CAT activity from PspxA2 was significantly higher in cells exposed to bacitracin, chlorhexidine or SDS while ampicillin or daptomycin failed to induce PspxA2 activity, at least under the conditions tested. (Fig. 8). As expected, the effect of bacitracin, chlorhexidine or SDS stresses on spxA2 transcription was sensed by the LiaFSR system as PspxA2 CAT activity remained close to the detection limit in the ΔliaFSR strain after exposure to those envelope stress agents (Fig. S4).
Figure 8.
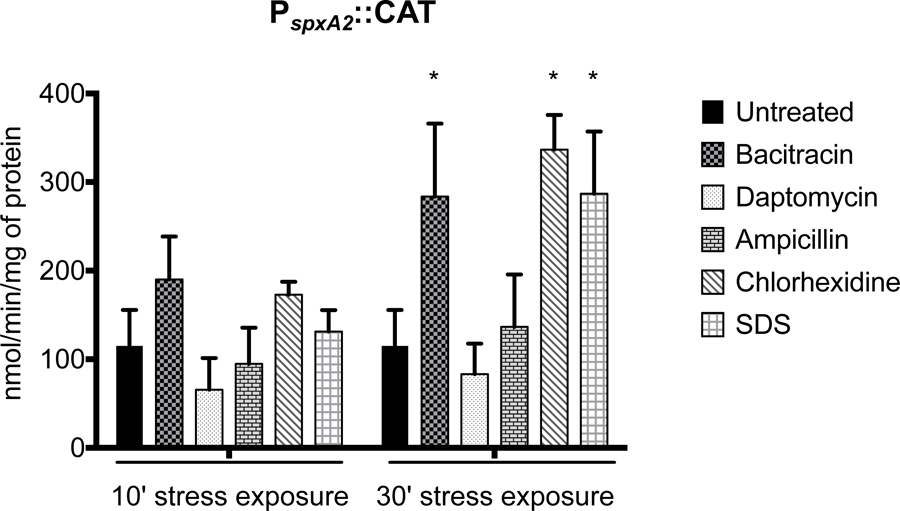
CAT activity driven from PspxA2 under selected cell envelope stress conditions. Cells were grown to mid-log phase and incubated in the presence of bacitracin (64 µg ml−1), daptomycin (2 µg ml−1), ampicillin (0.4 µg ml−1), chlorhexidine (0.5 µg ml−1) or SDS (0.00125% w/v) for either 10 or 30 minutes. Asterisks indicate statistical significance (p value ≤ 0.01) when compared to untreated control by one-way ANOVA.
Redox-sensing switch is essential for SpxA1Smu activity but not for SpxA2Smu
A common feature of Spx proteins is the presence of a conserved N-terminal CXXC disulfide switch (Fig. 1). In SpxBsu, oxidation of this motif results in a conformational change that promotes interaction of the Spx-RNAP complex with Spx-regulated promoters (M. M. Nakano et al., 2010). However, a recent study from the Helmann group revealed that redox-sensing is not essential for activation of SpxBsu-regulated genes during cell envelope stress (Rojas-Tapias & Helmann, 2018a). To determine the importance of the CXXC motif for SpxA1Smu and SpxA2Smu function, the first cysteine residue of the motif was replaced by a serine residue (SXXC) in each protein to create the SpxA1SXXC and SpxA2SXXC strains. In the absence of stress (i.e., plain BHI at 37°C), the ΔspxA2 strain displayed a slightly extended adaptation (lag) phase whereas the ΔspxA1 strain grew slower without reaching the same final growth yield of the parent and ΔspxA2 strains (Fig. 9A). The SpxA1SXXC and SpxA2SXXC strains grew almost as well as the parent strain in plain BHI indicating that redox sensing is largely dispensable for SpxA1Smu and SpxA2Smu activities in the absence of stress (Fig. 9A). As shown previously (Kajfasz et al., 2010), ΔspxA1 was hypersensitive to peroxide stress (Fig. 9B) and, in this environment, an intact CXXC motif proved essential for SpxA1 activity as the SpxA1SXXC strain phenocopied the ΔspxA1 strain (Fig. 9B). When compared to growth in the absence of stress, the ΔspxA2 strain displayed an even longer adaptation phase in the presence of H2O2, which was not observed in the SpxA2SXXC strain (Fig. 9B).
Figure 9.
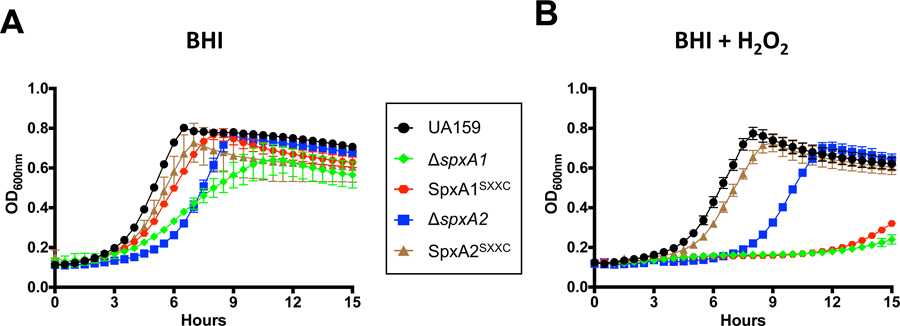
Growth curves of S. mutans UA159, Δspx and SpxSXXC strains in (A) BHI or (B) in the presence of sub-inhibitory concentration of H2O2 (0.4 mM).
Based on global transcriptional analysis, we have previously suggested that SpxA2Smu may have evolved to control cell envelope homeostasis while retaining a residual ability to regulate oxidative stress genes, particularly when in the absence of SpxA1Smu (Kajfasz et al., 2017; Kajfasz et al., 2010). The possible association of SpxA2Smu with envelope homeostasis is further supported by the discovery that spxA2 is under LiaFSR control (Fig. 7 and Fig. S4), and by previous studies that have implicated Spx regulation with envelope stress responses in other gram positive bacteria including, B. subtilis, E. faecalis and Streptococcus suis (Kajfasz et al., 2012; Rojas-Tapias & Helmann, 2018a; Zheng et al., 2014). To probe the role of SpxA2Smu and of its CXXC motif in cell envelope stress, we compared the ability of the parent, ΔspxA2 and SpxA2SXXC strains to grow in the presence of sub-inhibitory concentrations of cell wall (ampicillin or bacitracin) and membrane (chlorhexidine, daptomycin or SDS) stress agents using norfloxacin (DNA gyrase/replication inhibitor) and chloramphenicol (protein synthesis inhibitor) as antibiotic controls that do not target the cell envelope. When compared to the parent strain, the ΔspxA2 strain was dramatically more susceptible to all five cell envelope targeting antibiotics tested while it grew as well as the parent strain in the presence of norfloxacin or chloramphenicol (Fig. 10). When analyzed in conjuntion with the transcriptional studies shown in Figures 7 and 8, these results indicate that SpxA2Smu is an integral member of the LiaR regulon and a major player in envelope stress responses. Finally, the SpxA2SXXC strain phenocopied the parent strain (Fig. 10) indicating that redox-sensing is not necessary for activation of the SpxA2Smu regulon during envelope stress.
Figure 10.
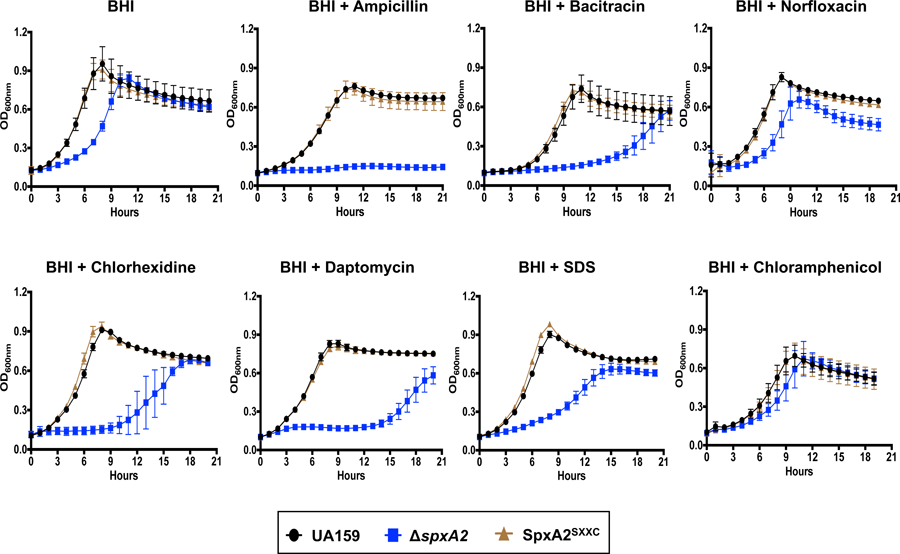
Growth curves of S. mutans UA159, ΔspxA2 and SpxA2SXXC strains in BHI or in the presence of sub-inhibitory concentrations of ampicillin (0.4 µg ml−1), bacitracin (32 µg ml−1), norfloxacin (1 µg ml−1), chloramphenicol (1 µg ml−1), chlorhexidine (125 ng ml−1), daptomycin (2 µg ml−1), or SDS (0.0005%).
Discussion
In several gram positive bacteria, activation of oxidative stress responses is primarily mediated by Spx, a transcription factor that physically interacts with the C-terminal domain of the RNAP to activate transcription of genes involved in thiol homeostasis and detoxification (Antelmann & Helmann, 2011; Barendt et al., 2013; S. Nakano, Erwin, Ralle, & Zuber, 2005; S. Nakano, Kuster-Schock, Grossman, & Zuber, 2003; Runde et al., 2014; Villanueva et al., 2016). In addition, Spx has been shown to participate in the regulation of other types of stress and in developmental processes such as competence and sporulation, and to repress transcription of ribosomal RNA and ribosomal protein genes (Galvao et al., 2017; M. M. Nakano et al., 2001; M. M. Nakano, Nakano, & Zuber, 2002; Schafer et al., 2019; Turlan et al., 2009). Previously, we identified two Spx homologs in S. mutans that, when individually deleted, suppressed phenotypes associated with clpP or clpX gene inactivations (Kajfasz et al., 2009). In follow-up studies, we found that most pathways linked to oxidative stress survival are regulated by SpxA1Smu and, to a much less extent, SpxA2Smu (Galvao et al., 2015; Kajfasz et al., 2017; Kajfasz et al., 2010). Subsequent work from other laboratories supported our initial findings by showing that the genomes of streptococcal species and other Firmicutes encode two Spx paralogues often exhibiting both overlapping and unique regulatory functions (Chen et al., 2012; Zheng et al., 2014).
The Spx regulator was first identified in B. subtilis and, to this day, most of the mechanistic understanding of how cellular Spx levels are controlled comes from investigations conducted with this gram positive soil organism (Leelakriangsak et al., 2007; S. Nakano et al., 2003; S. Nakano et al., 2002; Rojas-Tapias & Helmann, 2018a, 2019; Zhang & Zuber, 2007). At the posttranslational level, SpxBsu is controlled by both the ClpXP and ClpCP systems (S. Nakano et al., 2002; Rojas-Tapias & Helmann, 2018b, 2019). In our initial study, we showed that Spx accumulates in S. mutans ΔclpP and ΔclpX strains, but we were unable to determine if one or both proteins were specifically targeted by ClpXP for degradation (Kajfasz et al., 2009). The low expression levels typical of transcriptional regulators and the high degree of conservation between SpxA1Smu and SpxA2Smu proved to be major hurdles for studying the stability of these proteins in vivo. Upon stumbling on the inherent limitation of a reconstituted in vitro proteolysis assay and taking into account that the C-terminal residues of SpxBsu are determinant for ClpP-mediated proteolysis, we turned our focus onto the roles that the C-terminal amino acids may play in differentiating SpxA1Smu and SpxA2Smu. To accomplish this, we fused the last ten amino acids of either SpxA1Smu or SpxA2Smu to an otherwise stable GFP and expressed the corresponding GFP::SpxA1tail and GFP::SpxA2tail fusion proteins in S. mutans to monitor GFP stability over time. At the amino acid level, the greatest difference between SpxA1Smu and SpxA2Smu appeared exactly at the C-terminal domain, with SpxA1Smu displaying several acidic residues (4 of the last 5 are acidic residues) compared to a complete lack of acidic residues within the last 10 amino acids of SpxA2Smu (Fig. 1). Notably, substitution of the last two non-acidic amino acid residues of SpxBsu (alanine and asparagine) by two aspartic acid residues abolished ClpXP-mediated proteolysis without affecting Spx-YjbH interaction (Chan et al., 2014; S. Nakano et al., 2003). Because of the disproportionately high number of acidic residues present in the C-terminus of SpxA1Smu when compared to other Spx proteins, we predicted that SpxA1Smu is resistant to ClpP proteolysis, which we were able to confirm using the GFP::SpxA1tail and GFP::SpxA2tail reporters (Fig. 2). The significance of C-terminal acidic residues for Spx stabilization was further demonstrated by the addition of two acidic residues to the SpxA2 tail (GFP::SpxA2DDtail), which stabilized SpxA2Smu, or by the removal of the acidic residues of SpxA1Smu (GFP::SpxA1-5tail and GFP::SpxA1-7tail), which rendered SpxA1 susceptible to ClpP degradation (Fig. 3). Because the GFP reporter alone was stable for up to 12 hours in the parent UA159 strain, this later observation suggests that the amino acidic residues within the last 10 amino acids of SpxA1Smu could be masking a degron in the C-terminal tail of SpxA1 that may only be exposed under specific conditions.
As an attempt to show that ClpP-mediated degration of SpxA2 Smu but not of SpxA1Smu occurs in its native context, we compared the transcriptional profile of an SpxA1- (sodA) and of an SpxA2-regulated (smu1412c) gene (Kajfasz et al., 2010) in the UA159, ∆clpP and ∆clpE strains. While no changes in expression of sodA was observed between these strains, smu1412c was significantly upregulated in ∆clpP compared to parent strain UA159 (Fig. 6) suggesting accumulation of SpxA2 but not of SpxA1 in the absence of ClpP proteolysis. Unfortunately, attempts to validate these observations by reproducing the SpxA2Smu stabilization (SpxA2DD strain) and SpxA1Smu destabilization (SpxA1-7AA strain) in S. mutans were not conclusive. If in one hand, replacing the original spxA1 by a gene copy expressing a truncated SpxA1 lacking the last 7 amino acids (SpxA1-7AA strain) increased peroxide stress sensitivity, stabilization of SpxA2Smu (UA-SpxA2DD strain) did not increase S. mutans tolerance towards cell envelope stress agents (Fig. S5 and S6). These mixed results were unsurprising given that there could be other factors that ultimately control Spx levels and activity. Moreover, stabilization and the presumably accumulation of SpxA2Smu (UA-SpxA2DD strain) may not necessarily have to translate in the opposite phenotype of the ΔspxA2 strain.
Barring a handful of streptococcal species (Fig. S1), the C-terminal residues of SpxA1 proteins have none or very few acidic amino acids (one or two within the last 10 residues, none within the last 5 amino acids). In the lone Spx protein of S. aureus (SpxSau), the last C terminal amino acid is an aspartic acid; however, the SpxSau was shown to become stable upon clpP gene inactivation (Engman et al., 2012). To further demonstrate the importance of the C-terminal tail for Spx stability, we also showed that addition of the C-terminal tail of either SpxA1Spn or SpxA2Spn (the 2 pneumococcal Spx proteins), neither one containing acidic residues within their last 7–8 amino acids, destabilized GFP in a ClpP-dependent manner (Fig. 4). Thus, it appears that the scenario in which SpxA1 is stable, while SpxA2 is susceptible to ClpP degradation is limited to S. mutans and few additional streptococcal species. Because of the intimate relationship between streptococcal SpxA1 and SpxA2 proteins, seemingly ranging from cooperative to antagonistic (Chen et al., 2012; Port et al., 2017; Zheng et al., 2014), the different susceptibility of streptococcal SpxA1 proteins to ClpP degradation and how this may affect expression of the SpxA1 and SpxA2 regulons will demand analysis on a species-by-species basis.
In B. subtilis, Spx is degraded by both ClpCP and ClpXP complexes (S. Nakano et al., 2002; Rojas-Tapias & Helmann, 2019). It has been proposed that in the absence of disulfide stress, Spx levels are primarily controlled by ClpXP and the YjbH adaptor (Chan et al., 2012; Engman et al., 2012; Feng et al., 2013; Garg et al., 2009). When exposed to disulfide stress, the structural integrity of ClpX is severely compromised due to structural changes in its N-terminal Cys4 zinc-binding domain (Zhang & Zuber, 2007). It has been proposed that once ClpX becomes inactive, ClpCP degradation takes over to prevent accumulation of Spx (Rojas-Tapias & Helmann, 2019). Of note, the Spx paralog MgsRBsu protein was also shown to be subjected to both ClpCP and ClpXP posttranscriptional control (Reder, Pother, Gerth, & Hecker, 2012). In the study that led to the identification of spxA1Smu and spxA2Smu, we showed that Spx accumulates in ΔclpP and ΔclpX strains (Kajfasz et al., 2009), though at the time we were unable to make a distinction between SpxA1 and SpxA2. In this work, we confirmed the central role of ClpP in SpxA2Smu degradation using the GFP reporter. Further, we found that deletion of clpE partially increased the stability of the GFP::SpxA2tail fusion protein whereas single deletion of clpC or clpX did not. Stability of the GFP::SpxA2tail construct was further increased in the ΔclpEΔclpX double mutant and almost completely stabilized in the ΔclpCΔclpE strain (Fig. 5). We certainly did not expect ClpE, followed by ClpC, to be the major Clp ATPases involved in SpxA2Smu degradation and acknowledge that this observation may reflect the limitation of using chimeric GFP-Spxtail reporters. Nevertheless, our results indicate that all three Clp proteolytic systems, ClpCP, ClpEP and ClpXP, have the potential to degrade SpxA2Smu in vivo. We should also note that the Biswas group showed previously that SsrA-tagged proteins of S. mutans can be degraded by all three Clp proteolytic complexes, with ClpCP and ClpEP playing a more prominent role during heat stress (Tao & Biswas, 2015). In a separate study, they also characterized a tripeptide motif LPF as a ClpX degradation signal that functions only in selected S. mutans strains, including the UA159 strain used in our study (Jana et al., 2016). However, neither SpxA1Smu nor SpxA2Smu encode the LPF motif. Further studies are necessary for a complete understanding of how the different Clp ATPases regulate SpxA2Smu levels. Considering the absence of YjbH (ClpXP adaptor) and McsB (ClpCP adaptor) homologs in streptococci, subsequent studies should consider the use of genetic screens for the identification of adaptor proteins that may shed new light into the specific contributions of the different Clp ATPases to Spx degradation in vivo.
In addition to proteolytic control, B. subtilis spx transcription is coordinated through multiple promoters that are controlled by different sigma factors (σA, σB and σM), as well as two transcriptional regulators (Barendt et al., 2016; Eiamphungporn & Helmann, 2008; Leelakriangsak et al., 2007; Reyes & Zuber, 2008; Rojas-Tapias & Helmann, 2018a). While we cannot rule out that the spxA1Smu and spxA2Smu genes may also be transcribed by distal promoters, we focused our initial studies on the proximal σA-type promoters upstream the spxA1 and spxA2 coding regions. We found that the PerR and SloR metalloregulators have redundant roles as repressors of PspxA1 as only the simultaneous inactivation of perR and sloR increased PspxA1 activity (Fig. 7). Based on this evidence, we propose that full induction of spxA1 transcription may only occur in a low manganese/high H2O2 environment, when the DNA-binding capacity of SloR and PerR should be simultaneously impaired. This condition may be restricted to a few specific environments such as the initial stages of an infectious process when neutrophils and other professional phagocytes secrete large quantities of manganese-sequestering calprotectin and mediate an oxidative burst at the same time.
Recently, SpxBsu was shown to accumulate in response to cell wall stress and the B. subtilis Δspx strain displayed increased sensitivity to cell wall-targeting antibiotics (Rojas-Tapias & Helmann, 2018a). In support of the possible association of SpxA2Smu in cell envelope stress, a previous study revealed that spxA2Smu transcription is under LiaFSR regulation, a signal transduction system that activates a transcriptional response to cell envelope stress in Firmicutes (Shankar et al., 2015; Suntharalingam et al., 2009). In S. mutans, inactivation of liaFSR increased sensitivity to cell wall lipid II inhibitors and to cell membrane-disrupting agents (Suntharalingam et al., 2009). Here, we showed that transcription of spxA2Smu is strongly dependent on the LiaFSR signal transduction system (Fig. 8E), and that transcription of spxA2Smu is induced by some of the same cell wall and membrane-targeting agents that activate the LiaFSR system (Fig. 8). In addition, we provided conclusive evidence that SpxA2Smu plays a prominent role in envelope stress responses as the ΔspxA2 strain was hypersensitive to all five cell wall and membrane-targeting agents tested (Fig. 10). During revision of this manuscript, Baker and colleagues came to a similar conclusion by showing that transcription of S. mutans spxA2 was highly dependent on the LiaFSR system and that SpxA2Smu was required for growth under envelope stress conditions (J. L. Baker, Saputo, Faustoferri, & Quivey, 2020). Studies to identify new members of the SpxA2Smu regulon as a means to reveal the scope of SpxA2 regulation and to identify novel envelope stress genes are currently underway.
In addition to transcriptional and posttranslational mechanisms, Spx activity is also determined by reversible oxidation and disulfide bond formation of a N-terminal CXXC redox switch (S. Nakano et al., 2005). It has been shown that oxidation of the CXXC motif results in a conformational change that unfolds the helix α4 of the Spx protein, which facilitates interaction between the Spx-RNAP complex and target promoters (M. M. Nakano et al., 2010). Substitution of one or both cysteine residues of the motif by alanine significantly impaired competence development in B. subtilis and S. pneumoniae, as well as disulfide stress responses in B. subtilis and L. monocytogenes (Gaballa, Antelmann, Hamilton, & Helmann, 2013; S. Nakano et al., 2005; Rochat et al., 2012; Turlan et al., 2009; Whiteley et al., 2017). More recently new evidence indicated that the redox switch is not always essential for Spx activity, particularly during cell wall stress (Gaballa et al., 2013; Rojas-Tapias & Helmann, 2018a). More specifically, activation of the Spx regulon during cell wall stress occurred with SpxBsu found primarily in a reduced state (Rojas-Tapias & Helmann, 2018a). These authors discovered that Spx levels increased during envelope stress due to upregulation of the distal σM-regulated PM1 promoter, revealing that increased SpxBsu levels rather than its oxidation state was the determinant factor for activation of the Spx regulon during envelope stress (Rojas-Tapias & Helmann, 2018a). Here, we showed that redox-sensing via the CXXC motif is essential for SpxA1Smu-mediated activation of oxidative stress responses but dispensable for SpxA2Smu activity during envelope stress conditions (Fig. 9). Thus, it appears that the functions of a single Spx protein in B. subtilis is shared by two Spx proteins in S. mutans, with SpxA2Smu primarily functioning as a regulator of cell envelope stress and SpxA1Smu as an oxidative stress regulator.
In summary, we showed here that SpxA1Smu and SpxA2Smu levels are controlled by very distinct transcriptional and posttranslational mechanisms. Most notably, posttranslational control through targeted proteolysis appears to be restricted to SpxA2Smu as acidic amino acid residues at the extreme C-terminus of SpxA1Smu prevented ClpP-mediated degradation. On the other hand, SpxA1Smu activity was found to be largely dependent on its oxidation status whereas redox-sensing played a negligible role for SpxA2Smu activity. Finally, transcriptional and phenotypic characterizations provided unequivocal evidence that SpxA2Smu evolved to regulate responses associated with cell envelope stress while retaining the ability to partially compensate for the loss of SpxA1Smu during oxidative stress.
Experimental procedures
Bacterial strains and culture conditions
Strains and plasmids used in this study are listed in Table 1 and Table S1, respectively. All E. coli strains were routinely grown in Luria-Bertani (LB) media at 37°C. When required, kanamycin (100 μg mL−1) or ampicillin (100 μg mL−1) was added to LB broth or agar plates. Strains of S. mutans were routinely grown in brain heart infusion (BHI) at 37°C in a humidified 5% CO2 atmosphere. When required, kanamycin (1000 μg mL−1), erythromycin (10 μg mL−1), or spectinomycin (1500 μg mL−1) was added to the growth media. Due to the high background detected in BHI medium, the biofilm media (BM) supplemented with glucose (Loo, Corliss, & Ganeshkumar, 2000) was used to monitor GFP decay. For growth kinetics experiments, overnight cultures were diluted 1:20 into BHI and grown in a 5% CO2 atmosphere to an OD600 of 0.3, at which point 5 μl of the culture was used to inoculate wells of a 100-well plate containing 250 μL of the appropriate medium. The automated growth reader Bioscreen C (Oy Growth Curves Ab Ltd.) was used to monitor the ability of S. mutans UA159 and derivatives to grow in the presence of sub-inhibitory concentrations of H2O2, or the cell envelope stress agents ampicillin, bacitracin, chlorhexidine, daptomycin and SDS. To maintain an anaerobic environment, an overlay of 50 μL of sterile mineral oil was added to each well.
Table 1.
Streptococcus mutans strains used in this study.
| Strains | Relevant characteristics | Source |
|---|---|---|
| UA159 | wild-type | Lab stock |
| ∆clpP | clpP::Kan | (Kajfasz et al., 2009) |
| ∆clpC | clpC::Spec | (Kajfasz et al., 2009) |
| ∆clpX | clpX::Kan | (Kajfasz et al., 2009) |
| ∆clpE | clpE::Kan | (Kajfasz et al., 2009) |
| ∆clpC∆clpX | clpC::Spec; clpX::Kan | This study |
| ∆clpC∆clpE | clpC::Spec; clpE::Kan | This study |
| ∆clpE∆clpX | clpE::spec; clpX::Kan | This study |
| ∆spxA1 | spxA1::Spec | (Kajfasz et al., 2009) |
| ∆spxA2 | spxA2::Erm | (Kajfasz et al., 2009) |
| ∆perR | perR::Erm | This study |
| ∆liaFSR | liaFSR::spec | This study |
| GMS584 (∆sloR) | sloR::Erm | (Rolerson et al., 2006) |
| ∆perR∆sloR | sloR::Erm; perR::Spec | This study |
| UA159 SpxA1SXXC | SpxA1 with C-S substitution at position 10 | This study |
| UA159 SpxA2SXXC | SpxA2 with C-S substitution at position 10 | This study |
| SpxA1-7AA | SpxA1 with 7 amino acid C-terminal deletion | This study |
| UA-SpxA2DD | UA159 harboring pIB184::SpxA2DD, ErmR | This study |
Recombinant protein purification and in vitro proteolysis
To obtain N-terminal 6x-His-tagged rClpP, rClpX, rSpxA1 and rSpxA2 proteins, the full-length genes were individually ligated into the expression vector pET16b (Novagen), then introduced into the E. coli expression strain BL21 (DE3) via electroporation. Expression of recombinant proteins was achieved by growing cells in LB to an OD600 of 0.5 and inducing protein expression by adding 0.4 mM isopropyl-β-d-1-thiogalactopyrosinide (IPTG) to the exponentially-grown cultures for 16 h at 15°C. After cell lysis using the Avestin Emulsiflex C5 homogenizer (ATA Scientific), recombinant proteins were purified by affinity chromatography with Ni2+-nitrilotriacetic acid-agarose (Qiagen). Eluted recombinant proteins were dialyzed overnight in phosphate-buffered saline (PBS) at 4°C. Purity of recombinant proteins was analyzed by 12% SDS-PAGE followed by Coomassie blue staining. Concentration of purified proteins was determined using the bicinchoninic acid (BCA) assay (Thermo fisher). Aliquots of purified proteins were stored at −20°C in 10% glycerol for one-time use.
The in vitro proteolysis assay was carried out as previously described (S. Nakano et al., 2002) with slight modifications. Briefly, rClpP (4 μM) and rSpxA1 or rSpxA2 (4 μM) were incubated at 37°C in the presence of rClpX (2.5 μM) to a final volume of 50 μl of reaction buffer (25 mM MOPS [morpholinepropanesulfonic acid]-KOH [pH 7.0], 100 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 4 mM ATP, 2 mM phosphoenol pyruvate, 0.93 μM pyruvate kinase [Sigma]). Phosphoenol pyruvate and pyruvate kinase were used to regenerate ATP. At selected intervals, 10 μl aliquots were mixed with 5 μl of stop solution. The stability of SpxA1 or SpxA2 was analyzed by separating proteins on 15% SDS-PAGE followed by Coomassie blue staining.
Mutant construction and genetic manipulation
The ΔclpC, ΔclpE, ΔclpP and ΔclpX strains have been isolated previously (Kajfasz et al., 2009). To accommodate double deletions of the clp ATPases, we used a PCR ligation approach to create a new clpE deletion strain using a spectinomycin resistance cassette in place of the original kanamycin resistance cassette. The ΔclpCΔclpX and ΔclpEΔclpX strains were generated by transforming the ΔclpX strain with clpC::spec or clpE::spec ligation mixes. The ΔclpCΔclpE strain was generated by transforming the ΔclpC mutant with the clpE::kan ligation mix. To create the perR and liaFSR deletion mutants, the PCR ligation mutagenesis approach was again utilized (Lau, Sung, Lee, Morrison, & Cvitkovitch, 2002). Briefly, PCR fragments flanking the desired region to be deleted were ligated to a nonpolar spectinomycin (specr) or erythromycin (ermr) cassette and the ligation mix used to transform S. mutans UA159. To create the ΔperRΔsloR double mutant, perR::spec ligation mix was used to transform a sloR single mutant strain (Rolerson et al., 2006), a gift from Dr. Grace Spatafora (Middlebury College). The deletions were confirmed as correct by Sanger sequencing of the insertion site and flanking region.
To generate GFP-Spxtail chimeras, the gfp gene was PCR amplified with reverse primers that carried sequence of the respective Spx C-terminal 10 amino acids and a forward primer that recognizes GFP N-terminal end using the pUG vector (Hwang et al., 2016) as template. Following digestion with BamHI and XhoI, the amplified PCR products were ligated onto plasmid pIB184 (Biswas, Jha, & Fromm, 2008), which carries a strong P23 promoter (Tao & Biswas, 2015). The same approach was used to generate the S. pneumoniae GFP::SpxA1 and GFP::SpxA2 tails. To express SpxA2Smu with two additional acidic amino acids, the spxA2 gene was amplified using a reverse primer encoding two additional codons for aspartic acid before cloning into pIB184. To generate the SpxA1 C-terminal 5 or 7 amino acid deletion tail, primers were designed to match either SpxA1 amino acids corresponding to C-terminal 15th to 6th (SpxA1-5tail) or C-terminal 17th to 8th (SpxA1-7tail) amino acids along with a few C-terminal GFP amino acids and used with GFP N-terminal forward primer and cloned as described above. To generate promoter CAT fusions, the promoter region upstream spxA1 or spxA2 were PCR amplified with their own ribosome binding site and cloned into the one-step cat integration vector pJL84 (Santiago et al., 2012) and transformed into S. mutans UA159 and derivative strains such that a single copy of the PspxA1-cat or PspxA2-cat reporter was integrated at the mannitol utilization locus.
Overlap extension PCR (OE-PCR) was utilized to alter a single amino acid in the conserved CXXC motif of SpxA1 or SpxA2 to generate, respectively, the SpxA1SXXC and SpxA2SXXC strains. Primer sets 5’ C10SF/5’ C10SR and 3’ C10SF/3’ C10SR were used to amplify 2-kb fragments of the DNA flanking the site of mutation. The 2 kb PCR fragments carried an overlapping 26 base sequence, which was used to anneal the two PCR products in a ligase-free PCR reaction followed by overlap-extension (OE) amplification using the 5’ C10SF and 3’ C10SR primer sets. The overlap PCR product containing the single amino acid substitution was purified and used to transform the S. mutans ∆spxA1 and ∆spxA2 strains. Here, we took advantage of the antibiotic resistances of the ∆spxA1 and ∆spxA2 strains strictly as a screening tool. The mutant colonies were allowed to grow on BHI, then were patched onto both BHI and BHI containing the appropriate antibiotic for the original deletion mutation (spectinomycin for ∆spxA1 and kanamycin for ∆spxA2). Candidate transformants were sorted by loss of the ability to grow on BHI agar containing the antibiotic appropriate to the original mutation, then screened by PCR and confirmed by sequencing. A similar PCR-based approach was used to isolate the SpxA1-7AA strain. Primer sets 5’ C10SF/ SpxA1–7 5armrevERI and SpxA1–7 3armfwdERI/3’ C10SR were used to amplify 2-kb fragments of the DNA flanking the 7 amino acids deletion region. The PCR products were digested with EcoRI and ligated using T4 DNA ligase (NEB) before transforming into ∆spxA1 strain and colonies were selected based on loss of antibiotic resistance as described before. For generating stable SpxA2DD strain, the spxA2 gene was amplified using primer set pIB184SpxA2Fwd and SpxA2DDRev and cloned into pIB184. The plasmid expressing SpxA2DD was transformed into UA159 to generate strain UA-SpxA2DD. All primers used in the genetic manipulations are listed in Table S2.
Western blot analysis
Whole-cell protein lysates were obtained by homogenization in the presence of 0.1-mm glass beads using a bead beater (Biospec). Equal amounts of protein extracts (usually 50 μg per lane) were separated by 12% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore) using standard protocols. GFP detection was performed using rabbit anti-GFP polyclonal antibody (Thermo Fisher Scientific) diluted 1:2000 in PBS containing 0.01% Tween 20 and anti-rabbit horseradish peroxidase (HRP)-coupled antibody (Sigma-Aldrich). Protein concentration of the lysates was determined using the bicinchoninic acid (BCA) assay and equal loading was confirmed by staining the blots with Ponceau S (Figure S7).
CAT assay
Chloramphenicol acetyltransferase (CAT) activity was measured from cell cultures grown in BHI to different growth phases (early-, mid- and late-log), or grown to an OD600 of 0.4 and subjected to acid stress (pH 6.0), 0.5 mM H2O2, 0.5 mM diamide, or cell envelope stress (0.4 µg ml−1 ampicillin, 64 µg ml−1 bacitracin, 0.5 µg ml−1 chlorhexidine, 2 µg ml−1 daptomycin or 0.00125% SDS) for up to 30 minutes. Aliquots of the culture left untreated were used as controls. Cell-free lysates were prepared by homogenizing the cells in a bead-beater, and CAT activity determined by a spectrophotometric method (Chakraborty & Burne, 2017). Briefly, the CAT assay reaction mixtures were prepared fresh so that final concentrations of each component were as follows: 5,5-dithio-bis-nitrobenzoic acid (DTNB), 0.4 mg ml−1; Tris-HCl, 100 mM; and acetyl-CoA, 0.1 mM. Chloramphenicol (5 mM stock) and cell lysates were pre-warmed to 37°C just before the reactions were initiated. Changes in absorbance were recorded at OD412 in a 37°C spectrophotometer. Protein concentrations of cell lysates used in CAT assays were determined using the BCA assay. CAT activity was expressed as the amount of lysate needed to acetylate 1 nmol of chloramphenicol min−1 mg of protein−1.
qRT-PCR analysis
RNA was extracted from cultures grown to mid-exponential phase (OD600 ∼0.4) as previously described (Galvao et al., 2015). Briefly, cDNA from 1 μg of RNA was synthesized using a high-capacity cDNA reverse transcriptase kit containing random primers (Applied Biosystems). Gene-specific primers for the sodA and smu1412c (Table S2) were designed using Beacon Designer (version 2.0) software (Premier Biosoft International) to amplify a region of around 120 bp in length. Quantitative real-time PCR (qRT-PCR) were performed in an iCycler apparatus (Bio-Rad).
GFP decay assay
S. mutans UA159 and Δclp derivatives harboring different pIB184::GFP-Spx fusion tails were grown overnight in BHI supplemented with erythomycin and sub-cultured in BM-glucose media (Loo et al., 2000). Cultures were grown on 96-well plates at 37°C in a spectro-fluorimeter (Synergy H1, Biotek) to an OD600 of 0.25 when an inhibitory concentration of chloramphenicol (20 ug ml−1) was added to stop protein synthesis. GFP fluorescence was monitored in a timely manner and, to determine GFP decay rate, fluorescence was normalized to the fluorescence immediately before addition of chloramphenicol. The results are presented as percent fluorescence remaining over time of atleast 6 replicates.
5’ RACE-PCR
5’ Rapid Amplification of cDNA Ends (RACE)-PCR was used to determine the transcription start site of spxA1 and spxA2 according to the manufacturer’s protocol (Invitrogen). Briefly, 2 μg of RNA subjected to reverse transcription with gene-specific primer 1 (GSP1, Table S2) and SuperScript II Reverse Transcriptase, followed by RNase treatment and 3’poly dC tail addition with terminal deoxynucleotidyl transferase. The dC-tailed cDNA was then PCR amplified using a nested gene specific primer 2 (GSP2, Table S2) and Abridged Anchor Primer (AAP). The transcription start site was determined by sequencing the amplified cDNA.
Statistical Analysis
All data were analyzed using GraphPad Prism 6.0 software. For the GFP-decay assay, data were analyzed at each time point using ordinary one-way ANOVA with Dunnett’s multiple comparison post-test. To determine the statistical significance for the rate of degradation, geometric mean values of percentage of GFP-fluorescence remaining for each strain at a given time point were plotted with 99% confidence interval. Each respective figure legend indicates the control used for multiple comparisons. For CAT assays and qRT-PCR analyses, similar ordinary one-way ANOVA with Dunnett’s multiple comparison post-test were used. To determine the statistical significance under stress conditions, geometric mean values of CAT activities were plotted with 99% confidence interval from a single CAT construct and compared to a stress-free control strain. To determine the CAT activity in a different strain background, ordinary one-way ANOVA with Dunnett’s multiple comparison post-test were used as mentioned before, and significance were analyzed using parent strain CAT activity as control, plotting geometric mean values of CAT activities with 99% confidence interval.
Supplementary Material
Acknowledgements
This study was supported by NIH-NIDCR award R01 DE019783 to J.A.L. We thank Grace Spatafora (Middlebury College) and Indranil Biswas (University of Kanas) for the gift of the ΔsloR strain and pIB184 plasmid, respectively.
Footnotes
The authors have no conflict of interest.
References
- Antelmann H, & Helmann JD (2011). Thiol-based redox switches and gene regulation. Antioxid Redox Signal, 14(6), 1049–1063. 10.1089/ars.2010.3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek KT, Thogersen L, Mogenssen RG, Mellergaard M, Thomsen LE, Petersen A, … Frees D (2015). Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother, 59(11), 6983–6991. 10.1128/AAC.01303-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Saputo S, Faustoferri RC, & Quivey RG Jr. (2020). Streptococcus mutans SpxA2 relays the signal of cell envelope stress from LiaR to effectors that maintain cell wall and membrane homeostasis. Mol Oral Microbiol. 10.1111/omi.12282 [DOI] [PMC free article] [PubMed]
- Baker TA, & Sauer RT (2012). ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta, 1823(1), 15–28. 10.1016/j.bbamcr.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA, & Drake DR (2018). Are the mutans streptococci still considered relevant to understanding the microbial etiology of dental caries? BMC Oral Health, 18(1), 129 10.1186/s12903-018-0595-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendt S, Birch C, Mbengi L, & Zuber P (2016). Evidence that Oxidative Stress Induces spxA2 Transcription in Bacillus anthracis Sterne through a Mechanism Requiring SpxA1 and Positive Autoregulation. J Bacteriol, 198(21), 2902–2913. 10.1128/JB.00512-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendt S, Lee H, Birch C, Nakano MM, Jones M, & Zuber P (2013). Transcriptomic and phenotypic analysis of paralogous spx gene function in Bacillus anthracis Sterne. Microbiologyopen, 2(4), 695–714. 10.1002/mbo3.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Jha JK, & Fromm N (2008). Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology, 154(Pt 8), 2275–2282. 10.1099/mic.0.2008/019265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Burne RA, Wu H, & Koo H (2018). Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol, 26(3), 229–242. 10.1016/j.tim.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty B, & Burne RA (2017). Effects of Arginine on Growth, Virulence Gene Expression, and Stress Tolerance by Streptococcus mutans. Appl Environ Microbiol. 10.1128/AEM.00496-17 [DOI] [PMC free article] [PubMed]
- Chan CM, Garg S, Lin AA, & Zuber P (2012). Geobacillus thermodenitrificans YjbH recognizes the C-terminal end of Bacillus subtilis Spx to accelerate Spx proteolysis by ClpXP. Microbiology, 158(Pt 5), 1268–1278. 10.1099/mic.0.057661-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Hahn E, & Zuber P (2014). Adaptor bypass mutations of Bacillus subtilis spx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP. Mol Microbiol, 93(3), 426–438. 10.1111/mmi.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chakraborty B, Zou J, Burne RA, & Zeng L (2019). Amino Sugars Modify Antagonistic Interactions between Commensal Oral Streptococci and Streptococcus mutans. Appl Environ Microbiol, 85(10). 10.1128/AEM.00370-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Ge X, Wang X, Patel JR, & Xu P (2012). SpxA1 involved in hydrogen peroxide production, stress tolerance and endocarditis virulence in Streptococcus sanguinis. PLoS One, 7(6), e40034 10.1371/journal.pone.0040034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepps SC, Fields EE, Galan D, Corbett JP, Von Hasseln ER, & Spatafora GA (2016). The SloR metalloregulator is involved in the Streptococcus mutans oxidative stress response. Mol Oral Microbiol, 31(6), 526–539. 10.1111/omi.12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W, & Helmann JD (2008). The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol, 67(4), 830–848. 10.1111/j.1365-2958.2007.06090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldholm V, Gutt B, Johnsborg O, Bruckner R, Maurer P, Hakenbeck R, … Havarstein LS (2010). The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J Bacteriol, 192(7), 1761–1773. 10.1128/JB.01489-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engman J, Rogstam A, Frees D, Ingmer H, & von Wachenfeldt C (2012). The YjbH adaptor protein enhances proteolysis of the transcriptional regulator Spx in Staphylococcus aureus. J Bacteriol, 194(5), 1186–1194. 10.1128/JB.06414-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engman J, & von Wachenfeldt C (2015). Regulated protein aggregation: a mechanism to control the activity of the ClpXP adaptor protein YjbH. Mol Microbiol, 95(1), 51–63. 10.1111/mmi.12842 [DOI] [PubMed] [Google Scholar]
- Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, … Frees D (2013). Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res, 12(2), 547–558. 10.1021/pr300394r [DOI] [PubMed] [Google Scholar]
- Frees D, Savijoki K, Varmanen P, & Ingmer H (2007). Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol Microbiol, 63(5), 1285–1295. 10.1111/j.1365-2958.2007.05598.x [DOI] [PubMed] [Google Scholar]
- Gaballa A, Antelmann H, Hamilton CJ, & Helmann JD (2013). Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx. Microbiology, 159(Pt 10), 2025–2035. 10.1099/mic.0.070482-0 [DOI] [PubMed] [Google Scholar]
- Galvao LC, Miller JH, Kajfasz JK, Scott-Anne K, Freires IA, Franco GC, … Lemos JA (2015). Transcriptional and Phenotypic Characterization of Novel Spx-Regulated Genes in Streptococcus mutans. PLoS One, 10(4), e0124969 10.1371/journal.pone.0124969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão LC, Miller JH, Kajfasz JK, Scott-Anne K, Freires IA, Franco GC, … Lemos JA (2015). Transcriptional and Phenotypic Characterization of Novel Spx-Regulated Genes in Streptococcus mutans. PLoS One, 10(4), e0124969 10.1371/journal.pone.0124969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao LC, Rosalen PL, Rivera-Ramos I, Franco GC, Kajfasz JK, Abranches J, … Lemos JA (2017). Inactivation of the spxA1 or spxA2 gene of Streptococcus mutans decreases virulence in the rat caries model. Mol Oral Microbiol, 32(2), 142–153. 10.1111/omi.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SK, Kommineni S, Henslee L, Zhang Y, & Zuber P (2009). The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol, 191(4), 1268–1277. 10.1128/JB.01289-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Liu Y, Kim D, Sun V, Aviles-Reyes A, Kajfasz JK, … Koo H (2016). Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure. Sci Rep, 6, 32841 10.1038/srep32841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana B, Tao L, & Biswas I (2016). Strain-Dependent Recognition of a Unique Degradation Motif by ClpXP in Streptococcus mutans. mSphere, 1(6). 10.1128/mSphere.00287-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousselin A, Kelley WL, Barras C, Lew DP, & Renzoni A (2013). The Staphylococcus aureus thiol/oxidative stress global regulator Spx controls trfA, a gene implicated in cell wall antibiotic resistance. Antimicrob Agents Chemother, 57(7), 3283–3292. 10.1128/AAC.00220-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Ganguly T, Hardin EL, Abranches J, & Lemos JA (2017). Transcriptome responses of Streptococcus mutans to peroxide stress: identification of novel antioxidant pathways regulated by Spx. Sci Rep, 7(1), 16018 10.1038/s41598-017-16367-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Martinez AR, Rivera-Ramos I, Abranches J, Koo H, Quivey RG Jr., & Lemos JA (2009). Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J Bacteriol, 191(7), 2060–2068. 10.1128/JB.01609-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Mendoza JE, Gaca AO, Miller JH, Koselny KA, Giambiagi-Demarval M, … Lemos JA (2012). The Spx regulator modulates stress responses and virulence in Enterococcus faecalis. Infect Immun, 80(7), 2265–2275. 10.1128/IAI.00026-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, … Lemos JA (2010). Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol, 192(10), 2546–2556. 10.1128/JB.00028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Scott-Anne K, Gregoire S, Abranches J, & Lemos JA (2015). Transcription of Oxidative Stress Genes Is Directly Activated by SpxA1 and, to a Lesser Extent, by SpxA2 in Streptococcus mutans. J Bacteriol, 197(13), 2160–2170. 10.1128/JB.00118-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, & Cvitkovitch DG (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods, 49(2), 193–205. 10.1016/s0167-7012(01)00369-4 [DOI] [PubMed] [Google Scholar]
- Leelakriangsak M, Kobayashi K, & Zuber P (2007). Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis. J Bacteriol, 189(5), 1736–1744. 10.1128/JB.01520-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, & Burne RA (2008). A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology, 154(Pt 11), 3247–3255. 10.1099/mic.0.2008/023770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo CY, Corliss DA, & Ganeshkumar N (2000). Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol, 182(5), 1374–1382. 10.1128/jb.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makthal N, Rastegari S, Sanson M, Ma Z, Olsen RJ, Helmann JD, … Kumaraswami M (2013). Crystal structure of peroxide stress regulator from Streptococcus pyogenes provides functional insights into the mechanism of oxidative stress sensing. J Biol Chem, 288(25), 18311–18324. 10.1074/jbc.M113.456590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis RE (1995). Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol, 15(3), 198–207. [DOI] [PubMed] [Google Scholar]
- Mira A, Simon-Soro A, & Curtis MA (2017). Role of microbial communities in the pathogenesis of periodontal diseases and caries. J Clin Periodontol, 44 Suppl 18, S23–S38. 10.1111/jcpe.12671 [DOI] [PubMed] [Google Scholar]
- Nakano K, Nomura R, Matsumoto M, & Ooshima T (2010). Roles of oral bacteria in cardiovascular diseases--from molecular mechanisms to clinical cases: Cell-surface structures of novel serotype k Streptococcus mutans strains and their correlation to virulence. J Pharmacol Sci, 113(2), 120–125. 10.1254/jphs.09r24fm [DOI] [PubMed] [Google Scholar]
- Nakano MM, Hajarizadeh F, Zhu Y, & Zuber P (2001). Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol Microbiol, 42(2), 383–394. 10.1046/j.1365-2958.2001.02639.x [DOI] [PubMed] [Google Scholar]
- Nakano MM, Lin A, Zuber CS, Newberry KJ, Brennan RG, & Zuber P (2010). Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLoS One, 5(1), e8664 10.1371/journal.pone.0008664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Nakano S, & Zuber P (2002). Spx (YjbD), a negative effector of competence in Bacillus subtilis, enhances ClpC-MecA-ComK interaction. Mol Microbiol, 44(5), 1341–1349. 10.1046/j.1365-2958.2002.02963.x [DOI] [PubMed] [Google Scholar]
- Nakano S, Erwin KN, Ralle M, & Zuber P (2005). Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol, 55(2), 498–510. 10.1111/j.1365-2958.2004.04395.x [DOI] [PubMed] [Google Scholar]
- Nakano S, Kuster-Schock E, Grossman AD, & Zuber P (2003). Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A, 100(23), 13603–13608. 10.1073/pnas.2235180100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Zheng G, Nakano MM, & Zuber P (2002). Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J Bacteriol, 184(13), 3664–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Jakobsen TH, Givskov M, Twetman S, & Tolker-Nielsen T (2019). Oxidative stress response plays a role in antibiotic tolerance of Streptococcus mutans biofilms. Microbiology, 165(3), 334–342. 10.1099/mic.0.000773 [DOI] [PubMed] [Google Scholar]
- Pamp SJ, Frees D, Engelmann S, Hecker M, & Ingmer H (2006). Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol, 188(13), 4861–4870. 10.1128/JB.00194-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port GC, Cusumano ZT, Tumminello PR, & Caparon MG (2017). SpxA1 and SpxA2 Act Coordinately To Fine-Tune Stress Responses and Virulence in Streptococcus pyogenes. MBio, 8(2). 10.1128/mBio.00288-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reder A, Hoper D, Weinberg C, Gerth U, Fraunholz M, & Hecker M (2008). The Spx paralogue MgsR (YqgZ) controls a subregulon within the general stress response of Bacillus subtilis. Mol Microbiol, 69(5), 1104–1120. 10.1111/j.1365-2958.2008.06332.x [DOI] [PubMed] [Google Scholar]
- Reder A, Pother DC, Gerth U, & Hecker M (2012). The modulator of the general stress response, MgsR, of Bacillus subtilis is subject to multiple and complex control mechanisms. Environ Microbiol, 14(10), 2838–2850. 10.1111/j.1462-2920.2012.02829.x [DOI] [PubMed] [Google Scholar]
- Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, & Lipsitch M (2006). Interference between Streptococcus pneumoniae and Staphylococcus aureus: In vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol, 188(13), 4996–5001. 10.1128/JB.00317-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzoni A, Andrey DO, Jousselin A, Barras C, Monod A, Vaudaux P, … Kelley WL (2011). Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One, 6(6), e21577 10.1371/journal.pone.0021577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes DY, & Zuber P (2008). Activation of transcription initiation by Spx: formation of transcription complex and identification of a Cis-acting element required for transcriptional activation. Mol Microbiol, 69(3), 765–779. 10.1111/j.1365-2958.2008.06330.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T, Nicolas P, Delumeau O, Rabatinova A, Korelusova J, Leduc A, … Noirot P (2012). Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res, 40(19), 9571–9583. 10.1093/nar/gks755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Tapias DF, & Helmann JD (2018a). Induction of the Spx regulon by cell wall stress reveals novel regulatory mechanisms in Bacillus subtilis. Mol Microbiol, 107(5), 659–674. 10.1111/mmi.13906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Tapias DF, & Helmann JD (2018b). Stabilization of Bacillus subtilis Spx under cell wall stress requires the anti-adaptor protein YirB. PLoS Genet, 14(7), e1007531 10.1371/journal.pgen.1007531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Tapias DF, & Helmann JD (2019). Identification of Novel Spx Regulatory Pathways in Bacillus subtilis Uncovers a Close Relationship between the CtsR and Spx Regulons. J Bacteriol, 201(13). 10.1128/JB.00151-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolerson E, Swick A, Newlon L, Palmer C, Pan Y, Keeshan B, & Spatafora G (2006). The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J Bacteriol, 188(14), 5033–5044. 10.1128/JB.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runde S, Moliere N, Heinz A, Maisonneuve E, Janczikowski A, Elsholz AK, … Turgay K (2014). The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis. Mol Microbiol, 91(5), 1036–1052. 10.1111/mmi.12521 [DOI] [PubMed] [Google Scholar]
- Santiago B, MacGilvray M, Faustoferri RC, & Quivey RG Jr. (2012). The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J Bacteriol, 194(8), 2010–2019. 10.1128/JB.06737-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer H, Heinz A, Sudzinova P, Voss M, Hantke I, Krasny L, & Turgay K (2019). Spx, the central regulator of the heat and oxidative stress response in B. subtilis, can repress transcription of translation-related genes. Mol Microbiol, 111(2), 514–533. 10.1111/mmi.14171 [DOI] [PubMed] [Google Scholar]
- Schafer H, & Turgay K (2019). Spx, a versatile regulator of the Bacillus subtilis stress response. Curr Genet. 10.1007/s00294-019-00950-6 [DOI] [PubMed]
- Shankar M, Mohapatra SS, Biswas S, & Biswas I (2015). Gene Regulation by the LiaSR Two-Component System in Streptococcus mutans. PLoS One, 10(5), e0128083 10.1371/journal.pone.0128083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora G, Corbett J, Cornacchione L, Daly W, Galan D, Wysota M, … Glasfeld (2015). Interactions of the Metalloregulatory Protein SloR from Streptococcus mutans with Its Metal Ion Effectors and DNA Binding Site. J Bacteriol, 197(22), 3601–3615. 10.1128/JB.00612-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam P, Senadheera MD, Mair RW, Levesque CM, & Cvitkovitch DG (2009). The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol, 191(9), 2973–2984. 10.1128/JB.01563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, & Biswas I (2015). Degradation of SsrA-tagged proteins in streptococci. Microbiology, 161(Pt 4), 884–894. 10.1099/mic.0.000048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlan C, Prudhomme M, Fichant G, Martin B, & Gutierrez C (2009). SpxA1, a novel transcriptional regulator involved in X-state (competence) development in Streptococcus pneumoniae. Mol Microbiol, 73(3), 492–506. 10.1111/j.1365-2958.2009.06789.x [DOI] [PubMed] [Google Scholar]
- Villanueva M, Jousselin A, Baek KT, Prados J, Andrey DO, Renzoni A, … Kelley WL (2016). Rifampin Resistance rpoB Alleles or Multicopy Thioredoxin/Thioredoxin Reductase Suppresses the Lethality of Disruption of the Global Stress Regulator spx in Staphylococcus aureus. J Bacteriol, 198(19), 2719–2731. 10.1128/JB.00261-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Ruhland BR, Edrozo MB, & Reniere ML (2017). A Redox-Responsive Transcription Factor Is Critical for Pathogenesis and Aerobic Growth of Listeria monocytogenes. Infect Immun, 85(5). 10.1128/IAI.00978-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Itzek A, & Kreth J (2014). Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. Microbiology, 160(Pt 12), 2627–2638. 10.1099/mic.0.082156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, & Zuber P (2007). Requirement of the zinc-binding domain of ClpX for Spx proteolysis in Bacillus subtilis and effects of disulfide stress on ClpXP activity. J Bacteriol, 189(21), 7669–7680. 10.1128/JB.00745-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Xu J, Li J, Hu L, Xia J, Fan J, … Bei W (2014). Two Spx regulators modulate stress tolerance and virulence in Streptococcus suis serotype 2. PLoS One, 9(9), e108197 10.1371/journal.pone.0108197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, & Kreth J (2010). Role of Streptococcus mutans eukaryotic-type serine/threonine protein kinase in interspecies interactions with Streptococcus sanguinis. Arch Oral Biol, 55(5), 385–390. 10.1016/j.archoralbio.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P (2004). Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol, 186(7), 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


