Summary
Following agonist activation, G protein-coupled receptors (GPCRs) recruit β-arrestin, which desensitizes heterotrimeric G protein signaling and promotes receptor endocytosis1. Additionally, β-arrestin directly regulates many cell signaling pathways that can induce cellular responses distinct from that of G proteins2. Here we present a cryo-electron microscopy (cryoEM) structure of β-arrestin1 (βarr1) in complex with muscarinic acetylcholine-2-receptor (M2R) reconstituted in lipid nanodiscs. The M2R-βarr1 structure shows a multimodal network of flexible interactions, including binding of the βarr1 N-domain to phosphorylated receptor residues and βarr1 finger loop insertion into the M2R seven-transmembrane bundle, which adopts a conformation similar to that in the M2R-heterotrimeric Go protein structure3. Moreover, the cryoEM map reveals that the βarr1 C-domain edge engages the lipid bilayer. Through atomistic simulations, biophysical, biochemical, and cellular assays, we show that the C-edge is critical for stable complex formation, βarr1 recruitment, receptor internalization, and desensitization of G protein activation. Taken together, these data suggest the cooperative interactions of β-arrestin with both the receptor and phospholipid bilayer contribute to its functional versatility.
Activation of G protein-coupled receptors (GPCRs) leads to heterotrimeric G protein-mediated signaling that quickly returns to basal levels1. This remarkably conserved process of GPCR “desensitization” (Fig. 1a) is mainly orchestrated by two small families of proteins, GPCR kinases (GRKs) and arrestins (reviewed in 4). GRKs phosphorylate agonist-bound GPCRs on their carboxyl (C)-terminus or intracellular loops (ICLs), leading to arrestin recruitment (Fig. 1a). In humans, visual arrestin (arrestin1) and X-arrestin (arrestin4) are selectively expressed in the retina, while the ubiquitously expressed β-arrestins 1 and 2 (also known as arrestin2 and −3, respectively) regulate the hundreds of GPCRs found elsewhere. Arrestins are comprised of juxtaposed N- and C-terminal seven-stranded β-sandwich domains with a central crest of three loops (finger, middle, and C-loops)5,6. After the phosphorylated GPCR C-terminus engages arrestin’s N-domain7,8, conformational changes promote binding of central crest elements to the receptor 7-transmembrane (7TM) bundle, sterically blocking G protein coupling5. β-arrestins also act as adaptors for endocytic machinery, thereby increasing receptor internalization9. Besides modulating desensitization, β-arrestins potentiate many signaling pathways independently of G proteins2. Notably, certain “biased” GPCR agonists preferentially activate G protein or β-arrestin pathways, which could be exploited therapeutically to obtain more selective drugs2,10.
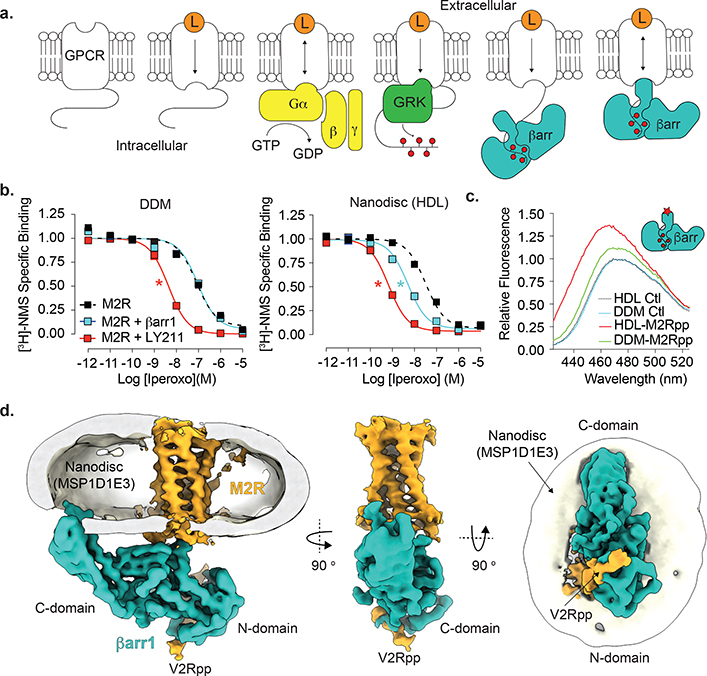
Fig. 1: βarr1 recruitment by M2R in a native lipid environment.
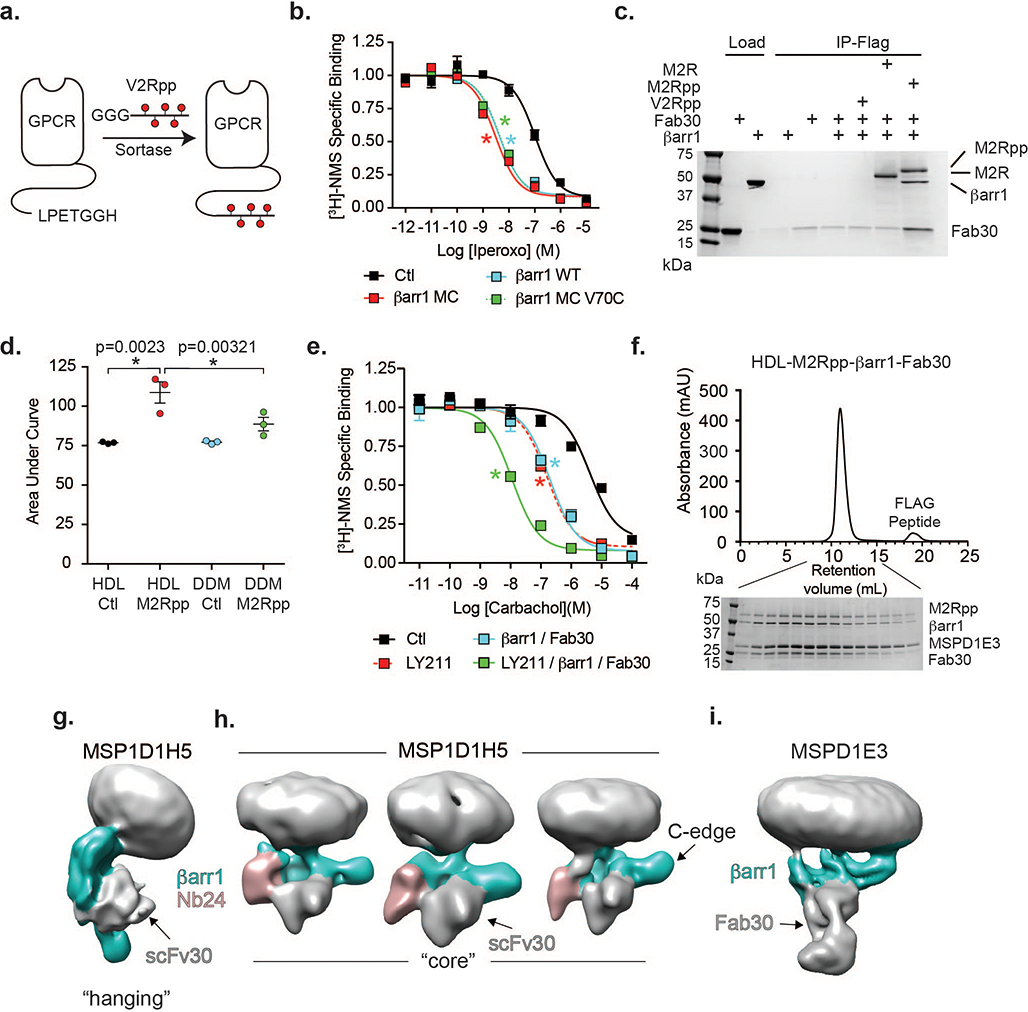
(a) Ligand (L)-induced conformational changes in GPCRs lead to heterotrimeric G protein activation (GTP hydrolysis) and subsequent GRK-mediated receptor phosphorylation. Initial binding of βarr to phosphorylated receptors leads to its coupling to the transmembrane (TM) bundle, sterically occluding further G protein binding. (b) βarr1 allosterically enhances iperoxo affinity to HDL-M2Rpp but not DDM-M2Rpp as determined by competition radioligand binding. The positive allosteric modulator LY211960 (LY211) enhanced iperoxo affinity regardless of reconstitution environment. Data are the mean of three independent experiments with error bars representing SE. (*) Indicates significance compared to control (one-way ANOVA). (LogIC50 values: DDM-Ctl, −7.10 ± 0.09; DDM-βarr1, −7.03 ± 0.06; DDM-LY211, −8.36 ± 0.04 (p<0.0001); HDL-Ctl, −7.49 ± 0.08; HDL-βarr1 −8.29 ± 0.08 (p<0.0007); HDL-LY211, −9.17 ± 0.05 (p<0.0001) (c) HDL-M2Rpp but not DDM-M2Rpp enhance βarr1 finger loop bimane (red star, inset) fluorescence. Curves represent difference in spectra obtained with antagonist (atropine) and agonist (iperoxo). Data represent means of three independent experiments. (d) Orthogonal views of cryoEM density map of the HDL-M2Rpp-βarr1 complex colored by subunit (orange, M2Rpp; teal, βarr1; gray/white, HDL particle). The orange-colored density on βarr1 corresponds to the phosphorylated C-terminal peptide (V2Rpp) ligated to the receptor. The nanodisc density, omitted in the middle panel for clarity, has been generated earlier in image processing before high-resolution refinement of the M2Rpp-βarr1 complex.
Notwithstanding numerous high-resolution GPCR-G protein structures obtained primarily by electron cryo-microscopy (cryoEM) (reviewed in 11), the only GPCR-arrestin structure to date is a crystal structure of rhodopsin fused to a constitutively active visual arrestin mutant8,12. Thus, knowledge of the molecular framework of GPCR-arrestin interactions remains limited, especially for the β-arrestins that modulate the vast majority of GPCRs. Here we report the cryoEM structure of β-arrestin1 (βarr1) in complex with human muscarinic acetylcholine-2 receptor (M2R) in high-density lipoprotein (HDL) particles (lipid nanodiscs) that mimic a native membrane. This structure provides new insights into βarr-mediated GPCR desensitization and signaling and highlights the importance of the membrane environment in these processes.
M2R-βarr1 cryoEM structure determination
Stabilizing GPCR-βarr complexes has been historically difficult because of the need for receptor phosphorylation and because of the low-affinity βarr-7TM interaction. To reconstitute a functional complex we chose M2R, a family A GPCR that regulates cardiac function13, because βarr1 has a relatively strong interaction with its 7TM core14. To ensure homogeneous phosphorylation, we used sortase to enzymatically ligate a synthetic phosphopeptide (pp) derived from the vasopressin-2-receptor (V2R) C-terminus onto the M2R C-terminus (M2Rpp)14 (Extended Data Fig. 1a). Although wild-type M2R lacks a C-terminus with phosphorylatable residues and is phosphorylated within ICL3, βarr1 enhances agonist affinity for M2Rpp comparably to natively phosphorylated M2R14,15. To enhance βarr1 stability, we generated a minimal cysteine variant truncated at residue 393 (βarr1-MC-393) with unaltered allosteric coupling to M2R (Extended Data Fig. 1b). This variant will be referred to as βarr1 unless specified.
Initially we characterized the ability of βarr1 to bind M2Rpp in n-dodecyl-β-D-maltoside (DDM-M2Rpp) detergent. βarr1 and the antibody fragment Fab30, which stabilizes the βarr1-V2Rpp interaction, co-immunoprecipitate with receptor only when it is phosphorylated (Extended Data Fig. 1c). Since βarr1 binding to the receptor 7TM core enhances agonist binding14,15, we monitored this interaction using radioligand competition binding to measure agonist affinity. The affinity of the agonist iperoxo for DDM-M2Rpp is increased by the positive allosteric modulator LY2119620 but not by βarr1 (Fig 1b). In contrast, both LY2119620 and βarr1 increase iperoxo affinity when M2Rpp is reconstituted into nanodiscs (HDL-M2Rpp), which provide a more physiological lipid bilayer (Fig. 1b). The importance of the lipid bilayer in βarr1-receptor binding was further tested by labeling the βarr1 finger loop, which inserts into the 7TM core, with monobromobimane (mBr), a probe whose fluorescence increases in hydrophobic environments. The V70C variant labeled for these experiments still couples to the M2Rpp 7TM core by radioligand binding (Extended Data Fig. 1b). We observed an agonist-dependent increase in bimane fluorescence with HDL-M2Rpp but not DDM-M2Rpp (Fig. 1c, Extended Data Fig. 1d) suggesting that βarr1 interacts with the M2Rpp 7TM core only in a lipid bilayer15.
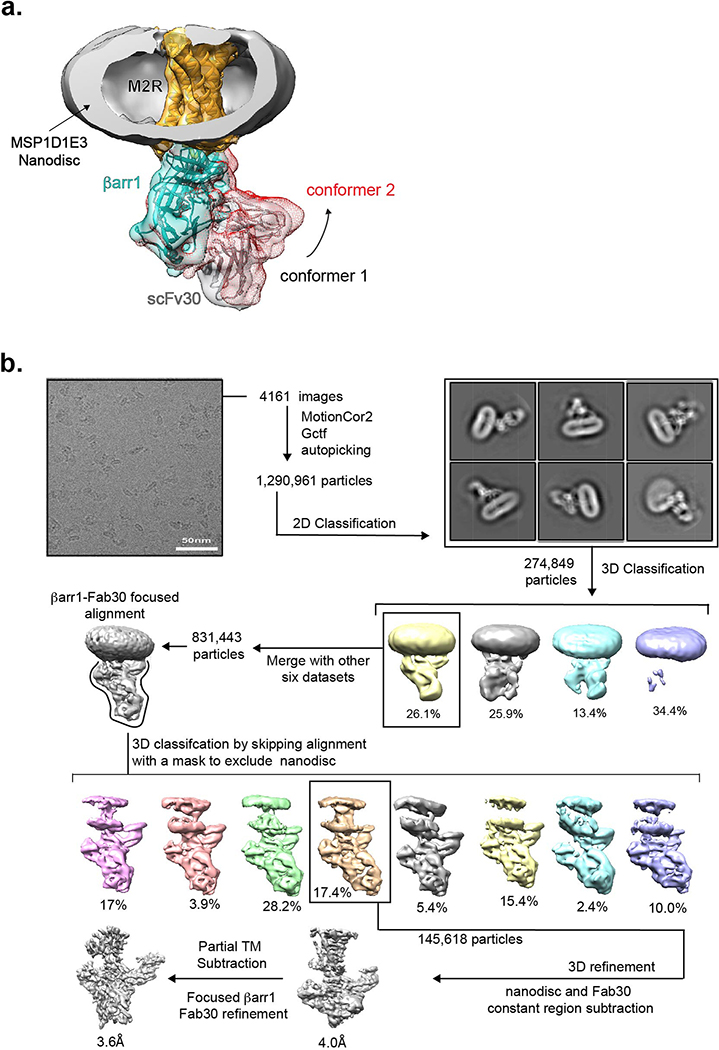
We optimized M2Rpp-βarr1 complexes for cryoEM using radioligand competition binding and negative stain EM16. The final complex—iperoxo/LY2119620-liganded M2Rpp in MSP1D1E3 lipid nanodiscs, bound to βarr1-MC-393 and the antibody fragment Fab30—was monodisperse and displayed agonist affinity 350-fold greater than receptor alone (Extended Data Fig. 1e,f). Fab30 was included to stabilize the βarr1-V2Rpp interaction and aid in cryoEM particle alignment7. We chose 12-nm diameter MSP1D1E3 nanodiscs since preliminary cryoEM analysis of an M2Rpp-βarr1 complex in smaller MSP1DH5 nanodiscs (~9-nm diameter) indicated a mixture of “hanging” conformations, with βarr1 interacting only with V2Rpp, and “core” conformations, with βarr1 further engaging the 7TM bundle (Extended Data Fig. 1g,h)17. We previously observed these conformations in our EM analysis of β2-adrenergic receptor-βarr1 complexes18. Low-resolution cryoEM maps of MSP1DH5 M2Rpp-βarr1 indicated conformational variability even among particles adopting the core interaction, with βarr1 “rocking” with respect to M2Rpp and the βarr1 C-domain edge occasionally approaching the lipid bilayer (Extended Data Fig. 1h). Given that hydrophobic residues in the visual arrestin C-domain interact with the membrane19, we hypothesized that the extended lipid surface of larger MSP1D1E3 nanodiscs would stabilize complexes for higher resolution studies.
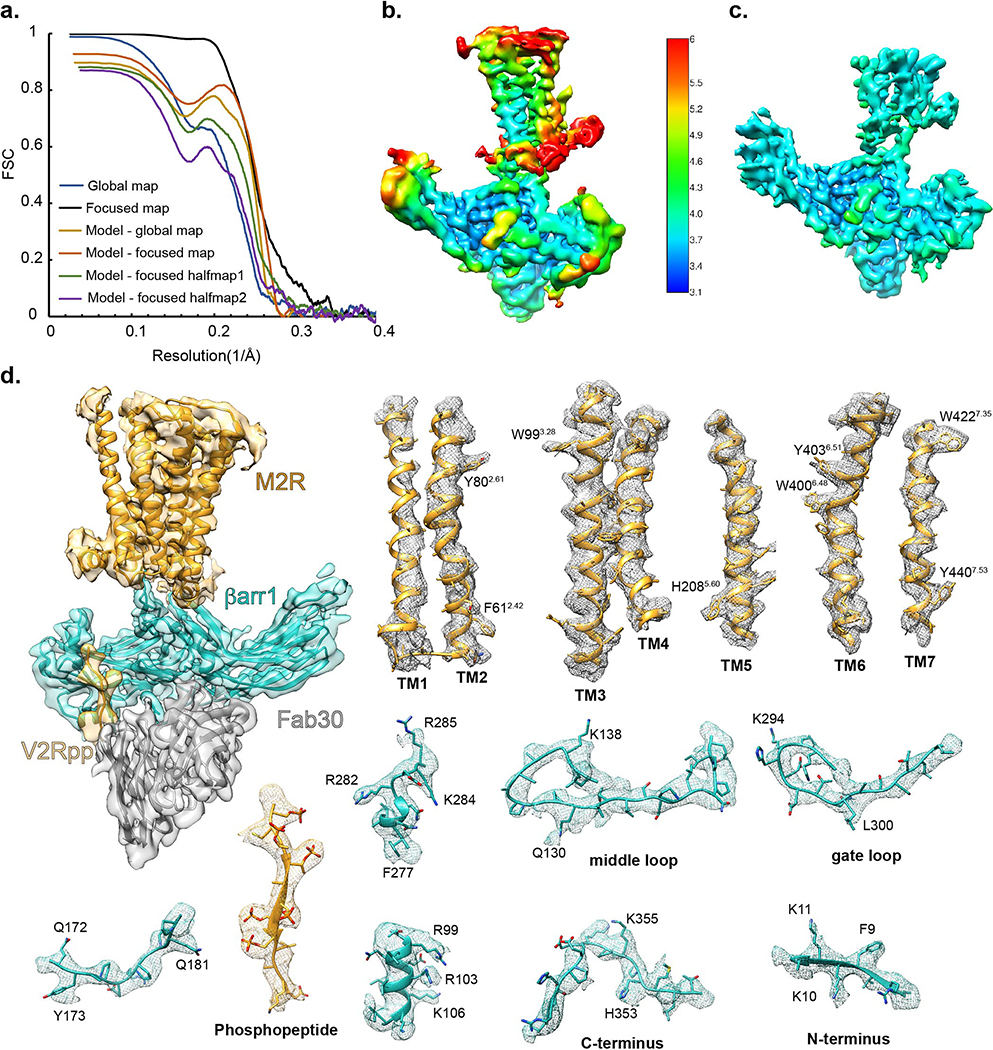
Gratifyingly, complexes in 12-nm nanodiscs showed more uniform interactions of βarr1 with the receptor 7TM core and the lipid surface (Extended Data Fig. 1i). Nevertheless, variability in the size and tilt of the lipid nanodisc relative to the βarr1-Fab30 density presented a challenge for structure determination. Low-resolution analysis confirmed small differences among particle classes in the angle of βarr1 relative to the receptor 7TM, indicating inherent flexibility (Extended Data Fig. 2a). Through 3D classification we identified a class of ~145,000 particle projections (17.4% of 831,443 particles with well-defined βarr1-Fab30 region) showing solid density for the 7TM portion in one position. Other 3D classes lacked defined receptor density, possibly due to a combination of specimen distortion at the air-water interface during cryo-sample preparation20 and potential projection misalignments due to the large nanodiscs. 3D refinement of the well-defined class enabled us to obtain a 3D map of the M2Rpp-βarr1 complex with a global indicated resolution of 4Å (Fig. 1d, Extended Data Fig. 2b). This map showed relatively lower resolution in the 7TM region, including poor density of the highly mobile TM1, but nevertheless allowed a confident comparison with M2R active-state structures. Subsequent map refinement focused on βarr1 and its interface with M2Rpp yielded a 3.6Å resolution map displaying well-resolved features employed for model building in this region (Extended Data Fig. 3).
Topography of the M2R-βarr1 complex
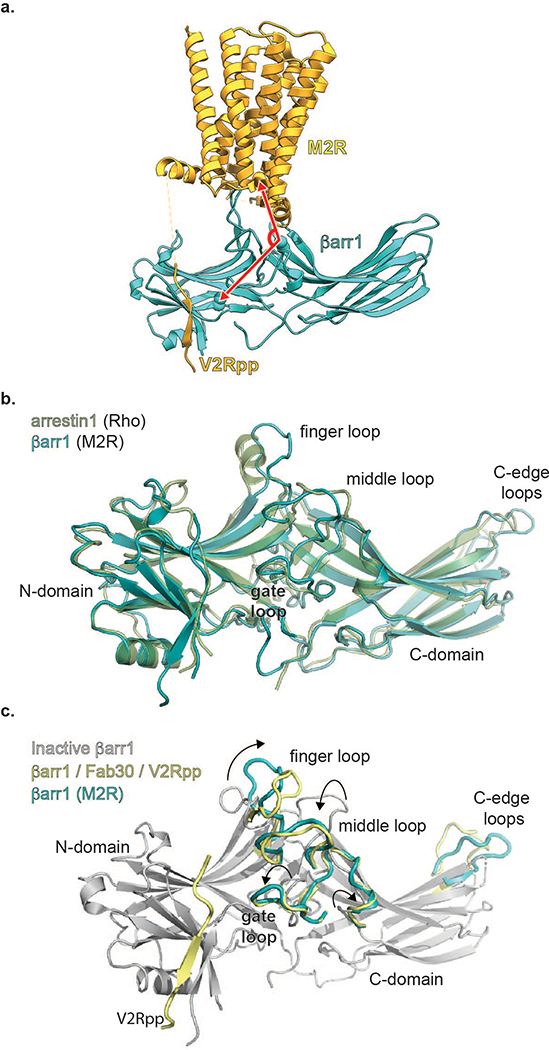
The cryoEM structure of M2Rpp-βarr1 in nanodiscs reveals a multimodal interaction network with: (1) the βarr1 N-domain engaging the phosphorylated receptor C-terminus, (2) βarr1 interdomain loops engaging the receptor 7TM core and ICL2, and (3) the βarr1 C-domain engaging the phospholipid bilayer (Fig. 2a-c). Topologically, the central crest region interacting with the 7TM bundle occupies a similar position as the Ras domain of G proteins. Comparison of the M2Rpp-βarr1 structure and our recent M2R-Go structure (PDB ID: 6OIK) reveals that the receptor 7TM bundle adopts similar active-state configurations characterized by the opening of the TM6 cytoplasmic portion3 (Fig. 2d). Relatively small shifts in the cytoplasmic ends of TM5, TM6 and TM7 are likely required to accommodate βarr1, although we cannot exclude effects of lipidic (M2Rpp-βarr1) versus detergent (M2R-Go) environments. The similarity of the M2R’s transducer-bound conformations is consistent with the similar allosteric enhancement of agonist binding to M2R by βarr1 and G protein14. In M2R-Go, opening of the cytoplasmic portion of TM6 accommodates the Go α5 helix. Similarly, TM6 displacement accommodates the βarr1 finger loop, which occupies a similar position as the Go Ras domain α5 helix, albeit to a slightly lower depth and involving a smaller interaction surface (Fig. 2d inset). This difference likely accounts for the lower affinity and inherent conformational variability of the βarr1–7TM interaction compared to M2R-Go.
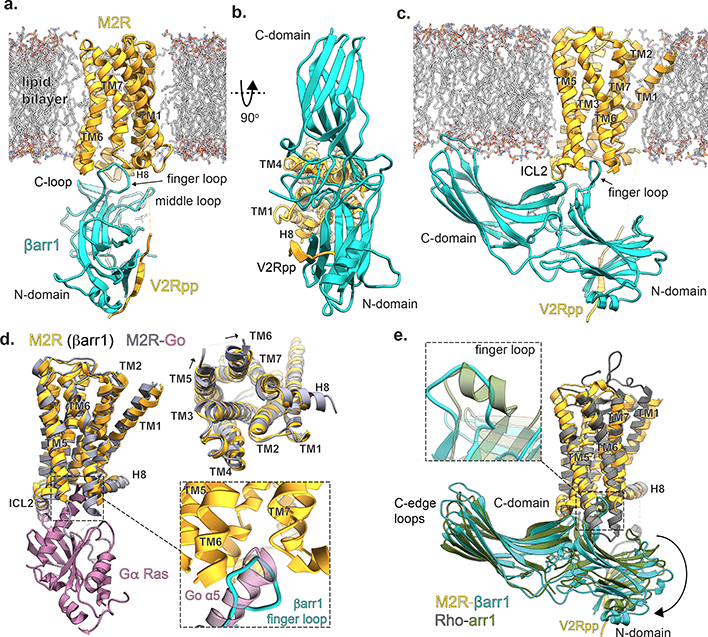
Fig. 2: Structure of the M2Rpp-βarr1 complex.
Orthogonal views (a,b,c) of the M2Rpp-βarr1 structure colored by subunit (orange, M2Rpp; teal, βarr1; gray sticks, model lipid bilayer). (d) Superposed structures of M2Rpp-βarr1 and M2R-Go complexes (M2R, gray; Gαo Ras domain, pink; PDBID: 6OIK) aligned by M2R. Go βγ subunits are omitted for clarity. Top right panel shows the cytoplasmic view of M2R transmembrane alignment in the absence of transducers. Bottom right inset shows similar insertion depth of βarr1 finger loop and Go α5 helix into the M2R TM bundle. (e) Superposition of M2Rpp-βarr1 and rhodopsin-arrestin1 structures (rhodopsin, gray; arrestin1, green; PDBID: 4ZWJ). Inset shows enlarged view βarr1 (M2Rpp) and visual arrestin (rhodopsin) finger loops.
In the M2Rpp-βarr1 cryoEM structure and rhodopsin-visual arrestin crystal structure (PDB ID: 5W0P)8, the arrestins show notable differences, despite their similar orientations relative to the 7TM bundles (Fig. 2e). βarr1 is further tilted ~7° towards the membrane (Extended Data Fig 4a), likely due to the interaction of the C-domain with the nanodisc phospholipids, although crystal packing interactions in the visual arrestin-rhodopsin structure might also contribute to these differences. In addition, the finger loops display differences in both positioning and structure. While the visual arrestin finger loop is modeled with an α-helical segment (residues E71-G77), the βarr1 finger loop adopts an extended loop configuration. The other arrestin interdomain loops adopt similar conformations in these two GPCR-arrestin structures (Extended Data Fig. 4b).
Interaction of βarr1 with M2R
βarr1 interacts with M2Rpp using three main interfaces: the phosphorylated C-terminus, the 7TM core, and ICL2 (Fig. 3). The phosphopeptide-βarr1 interaction is essentially identical to our crystal structure of V2Rpp-βarr1-Fab307, and six phosphorylated residues are well resolved in the 3.6Å map. The peptide binds to a positively charged crevice on the N-domain by displacing the βarr1 C-terminus, which destabilizes the arrestin polar core and allows the gate loop to flip toward the N-domain. A critical phosphorylated residue on the receptor C-terminus, T491 (T360 in V2Rpp), establishes interactions with βarr1 R25 (N-domain) and K294 (gate loop), stabilizing an activated conformation characterized by interdomain twisting21 (Fig. 3a). The fused V2Rpp is 36 residues long and provides ample flexibility so as not to limit the orientation of arrestin, also evident by the lack of observed density for the 23 residues that do not engage arrestin. Similarly, while we cannot rule out the possibility that natively phosphorylated ICL3 residues influence the relative orientation of βarr1, this seems unlikely given the length of this loop (152 residues) and its lack of order in our structure.
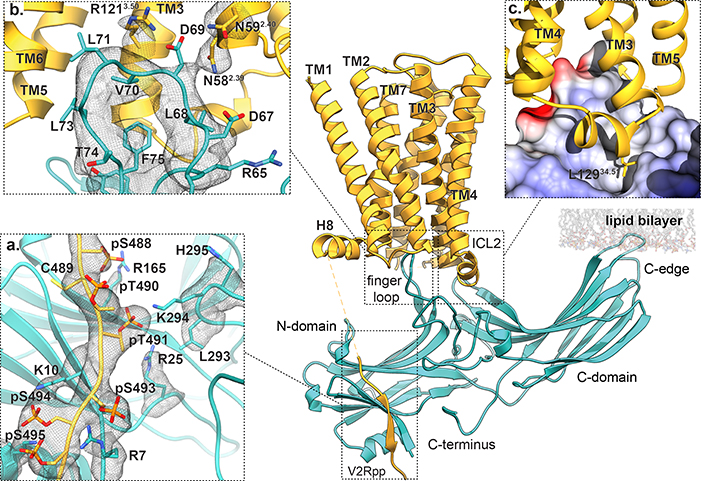
Fig. 3: Interaction regions between M2R and βarr1.
M2Rpp-βarr1 complex, with dashed boxes indicating main interaction sites between βarr1 (teal) and receptor (orange). (a) Enlarged view of the βarr1 finger loop inserting into the M2R TM bundle. The model includes side chain positions from Phenix refinement with OPLS3e electrostatics. (b) Enlarged view of βarr1 N-domain bound to the phosphorylated M2Rpp C-terminus. (c) Expanded view of interaction between M2R ICL2 (ribbon) and a hydrophobic cleft in βarr1 (rendered as electrostatic surface; red, blue and white graduations indicate negative, positive, and neutral surface potential, respectively). The mesh in all panels depict the 3.6Å cryoEM map contoured at σ=3.0 with a masked 2.0Å zone around the atoms depicted.
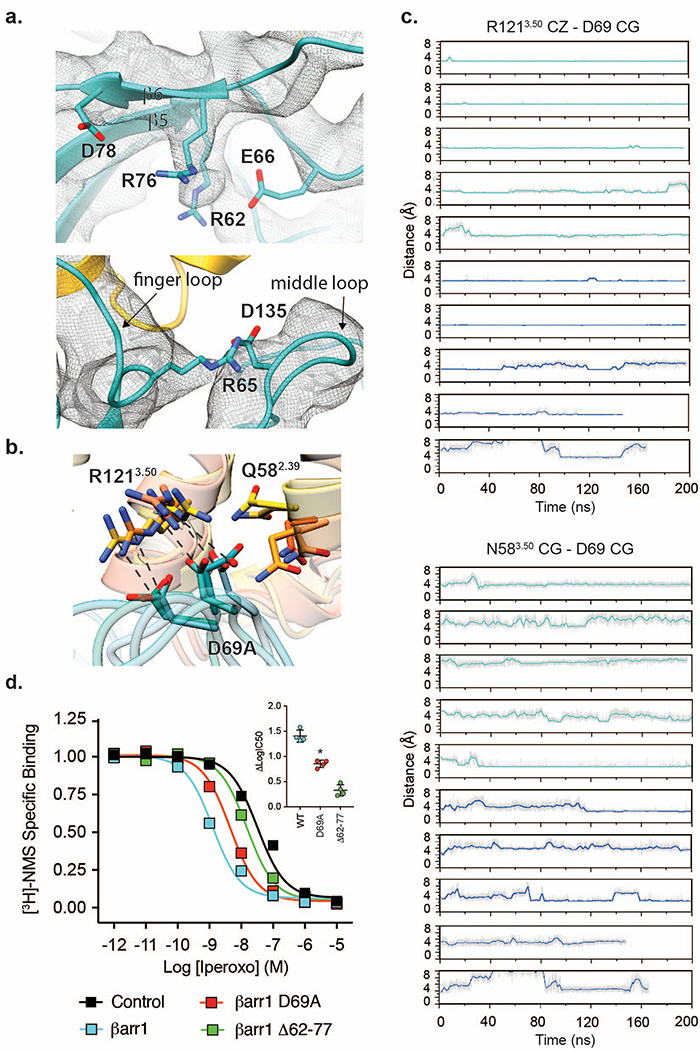
The second interface between the βarr1 finger loop and the 7TM bundle involves both hydrophobic and electrostatic interactions (Fig. 3b). The C-terminal finger loop region, including residues L68, V70, L71, and F75, is hydrophobically packed against the side of the receptor pocket formed by TMs 3, 5, 6 and ICL2. On the opposite side, the map of the N-terminal part of the finger loop displays partial side chain densities of ionic residues R65, E66, and D67, revealing both their position and relative mobility. A potential salt bridge between R65 and D135 (βarr1 middle loop) could stabilize the finger loop conformation, whereas E66, though only resolved through Cβ, points towards βarr1 R62 and K138 (middle loop) (Extended Data Fig. 5a). At the finger loop tip, D69 appears to be positioned to form hydrogen bonds or salt bridges with several receptor residues, including N582.39 and R1213.50 of the highly-conserved DRY motif (superscript numerals refer to Ballesteros-Weinstein numbering22). These interactions formed consistently across multiple, independent molecular dynamics (MD) simulations, suggesting that this interaction network stabilizes the M2Rpp–βarr1 complex (Extended Data Fig. 5b,c). Considering that R3.50-E6.30 salt bridges stabilize the inactive receptor state of M2R and other Class A GPCRs, βarr1 may stabilize an active receptor partly by engaging R3.50. βArr1 mutant D69A displays significantly reduced coupling to M2Rpp, supporting this hypothesis (Extended Data Fig. 5d).
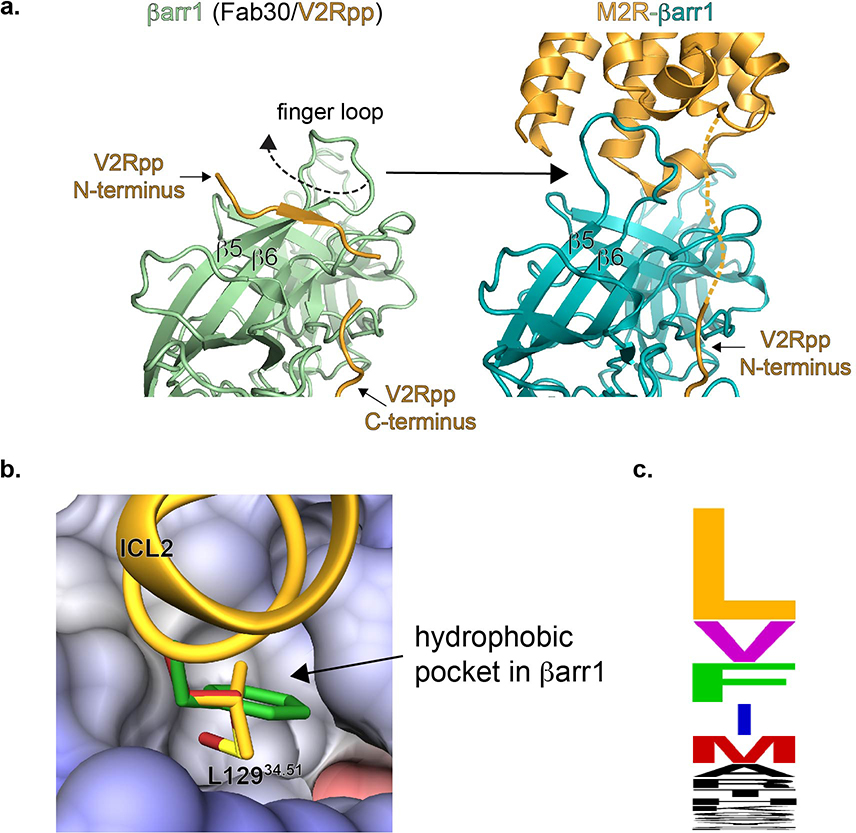
Overall, the βarr1 finger loop fold, the interdomain twist, and the V2Rpp C-terminus binding mode are very similar to the active βarr1-V2Rpp-Fab30 crystal structure7 (PDB:4JQI, Extended Data Fig. 4c). However, the crystal structure shows a large twist in the finger loop orientation, due to the N-terminal segment of the phosphopeptide inducing a twist in the flanking β-strands S5 and S6. This V2Rpp segment must disengage before finger loop insertion into M2Rpp and is not ordered in our map. The positioning of the N-terminal V2Rpp segment in the crystal structure could be due to lattice packing. Alternatively, these differences could reflect a multi-step binding mechanism, where initial βarr1 binding to the full length of the phosphopeptide is followed by release of the phosphopeptide N-terminus, βarr1 finger loop flipping, and burial of the finger loop’s hydrophobic residues in the receptor intracellular cavity (Extended Data Fig. 6a).
The M2Rpp ICL2 rests in a cleft between the βarr1 N- and C-domains that is comprised of portions of the finger, middle, gate, and C-loops. While this portion of the map lacks well-resolved sidechain features, the density suggests that ICL2 adopts the same helical conformation as in M2R-Go (Fig. 3c). This ICL2 orientation places L12934.51 facing a βarr1 hydrophobic cleft that is capable of accommodating even larger sidechains (Extended Data Figure 6b). While leucine is most prevalent at this position in other class A GPCRs that couple to βarr, larger hydrophobic amino acids (methionine, phenylalanine) are common (Extended Data Fig. 6c), suggesting one source of βarr’s promiscuity. Notably, the same hydrophobic ICL2 residue often engages in van der Waals interactions with G proteins3,23–25.
βarr1 C-domain interaction with membrane
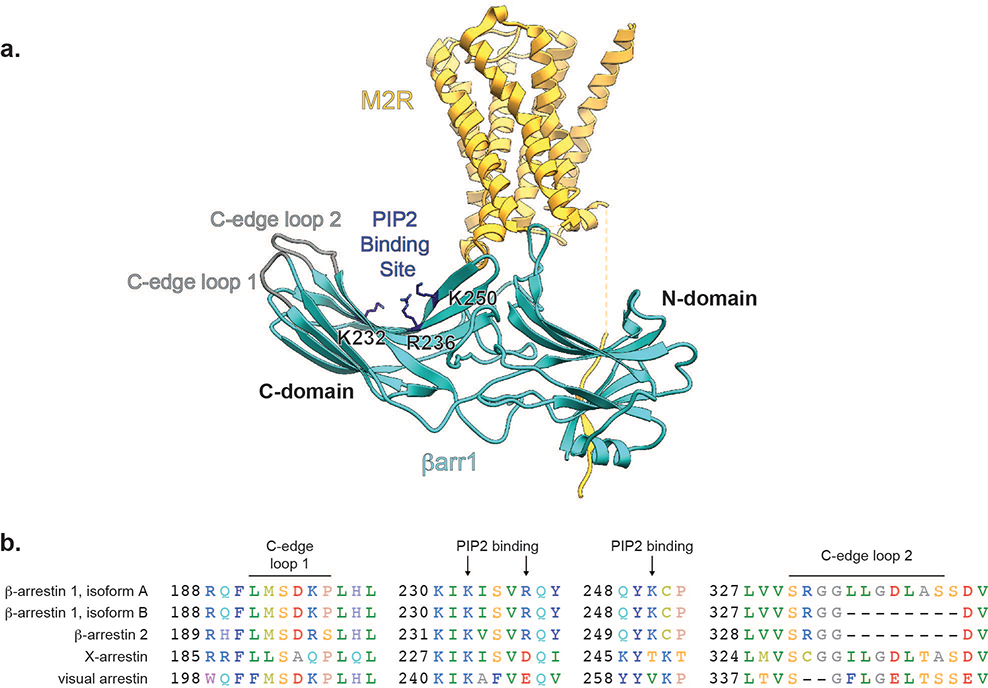
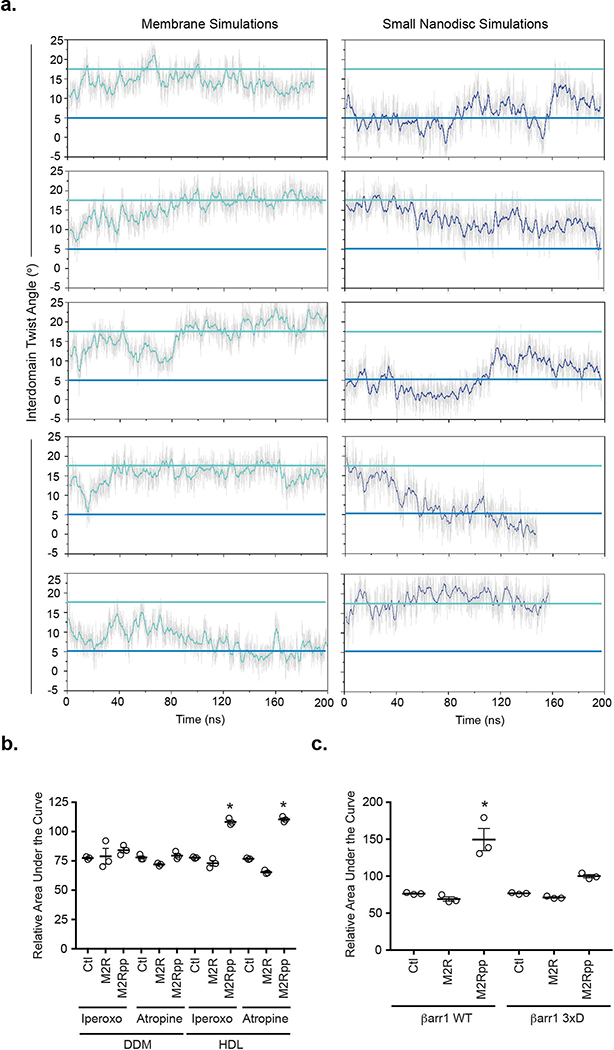
The cryoEM maps reveal that the βarr1 C-domain edge interacts with the nanodisc, with one C-edge loop contacting the membrane (loop 1, residues 191–196, LMSDKP) and a second loop burying itself in the lipid bilayer (loop 2, residues 330–340, SRGGLLGDLAS) (Extended Data Fig. 7). Interaction of arrestin’s C-edge with the membrane has been suggested by the rhodopsin-visual arrestin structure and by site-specifically labeling the visual arrestin C-edge with environmentally sensitive probes12,19. Given the higher affinity of βarr1 for the M2Rpp 7TM core in nanodiscs versus detergent (Fig. 1b,c), we hypothesized that the C-edge-lipid interaction has functional consequences. First, we performed multiple, independent MD simulations21 with M2Rpp-βarr1 initially embedded in either continuous membrane (allowing C-edge interaction) or a 9nm-diameter nanodisc (too small for C-edge interaction) (Fig. 4a, Extended Data Fig. 8a). In four of the five simulations employing the full membrane, βarr1 stayed in its active conformation, characterized by an average interdomain twist angle of ~17°, similar to the interdomain twist in the βarr1-V2Rpp-Fab30 crystal structure7. However, in small nanodiscs, βarr1 adopted a more heterogeneous conformational ensemble dominated by inactive conformations (interdomain twist angle 0–7°) (Fig. 4b), suggesting that the C-edge-membrane interaction stabilizes the active βarr1 conformation.
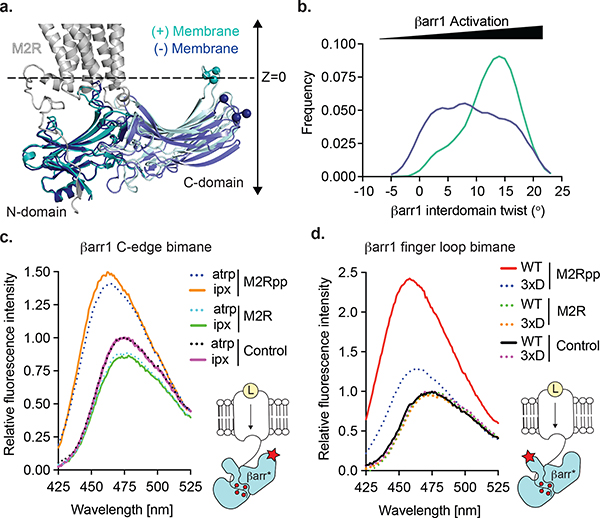
Fig 4: Lipid membrane anchoring of the βarr1 C-domain.
Atomistic simulations of βarr1 in complex with M2Rpp (in absence of Fab30) to investigate the (a) position of βarr1 C-edge and (b) βarr1 interdomain twist angle in the absence (dark blue) or presence (cyan) of a lipid membrane (dashed black line). (c) HDL-M2Rpp but not HDL-M2R enhances βarr1 C-edge bimane (inset, red star) fluorescence independent of antagonist (atropine, atrp) or agonist (iperoxo, ipx). (d) The ability of iperoxo-activated HDL-M2Rpp to increase βarr1 finger loop bimane (inset, red star) fluorescence requires receptor phosphorylation (compare to HDL-M2R) and is significantly reduced by the C-edge 3xD mutations (L335D, L338D, and S340D). Data representative of three independent experiments.
We site-specifically labeled C-edge loop 2 with mBr (L338mBr) to assess whether the βarr1-lipid interaction requires βarr1 binding to the M2Rpp phosphorylated tail or 7TM bundle19. An increase in βarr1 L338mBr fluorescence, reflecting C-edge insertion into a hydrophobic environment, requires M2Rpp phosphorylation and nanodisc reconstitution (Fig. 4c, Extended Data Fig. 8b). This is consistent with previous observations that visual arrestin engagement with membranes requires rhodopsin phosphorylation19. However, the change in βarr1 L338mBr fluorescence is not agonist (iperoxo)-dependent, indicating the C-edge-lipid interaction does not require βarr1–7TM interaction. Since the C-edge-membrane interaction is observed in nanodiscs but not detergent (Fig. 1b,c), we hypothesized that it might facilitate βarr1–7TM coupling. We sought to test this by attenuating the C-edge’s hydrophobicity with aspartic acid substitutions at βarr1 L335, L338, and S340 (βarr1 3xD) 19. Bimane fluorescence of finger loop-labeled βarr1 (Fig. 4d, Extended Data Fig. 8c) confirmed that the 3xD mutations significantly reduce βarr1 interaction with the HDL-M2Rpp 7TM bundle. Thus, maximal coupling of βarr1 to the M2Rpp 7TM bundle requires not only receptor phosphorylation but also βarr1 C-edge interaction with the phospholipid bilayer.
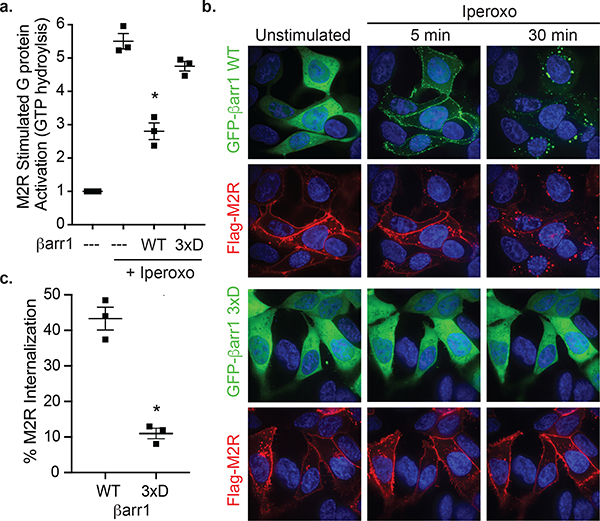
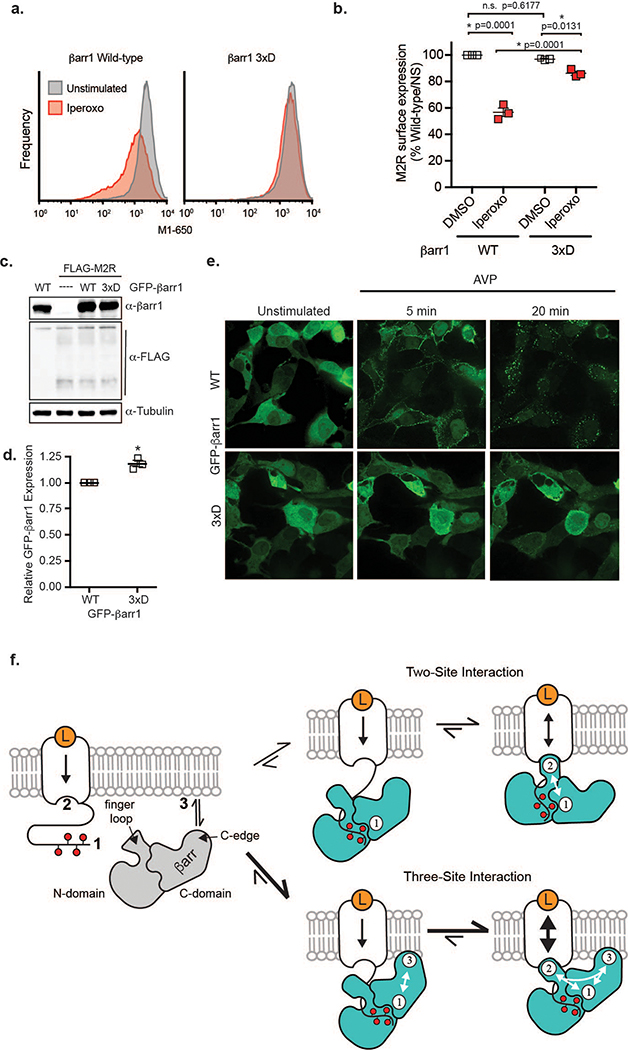
Furthermore, we asked whether the βarr1 C-edge-membrane interaction modulates βarr-mediated receptor desensitization and internalization. We measured the ability of purified WT and 3xD βarr1 to inhibit (desensitize) HDL-M2Rpp-mediated G protein activation in vitro. Iperoxo-induced activation of HDL-M2Rpp increases the GTPase activity of purified heterotrimeric Gi. While WT βarr1 blocks 50% of Gi activity, βarr1 3xD has almost no effect (Fig. 5a). We then compared the ability of GFP-βarr1 WT or 3xD to induce FLAG-M2R internalization in βarr1/βarr2-null HEK293 cells. After 5 minutes of iperoxo stimulation, GFP-βarr1 WT translocates from the cytoplasm to the plasma membrane. By 30 minutes, both GFP-βarr1 and FLAG-M2R (>40%) are largely internalized (Fig. 5b,c; Extended Data Fig. 9a,b). In contrast, GFP-βarr1 3xD, which expresses at similar levels as WT (Extended Data Fig. 9c,d), remains cytoplasmic even after 30 minutes, with little FLAG-M2R internalization (10%). Importantly, the impaired recruitment of GFP-βarr1 3xD to WT M2R and V2R (Extended data Fig. 9e) shows that membrane anchoring contributes to βarr1 recruitment to natively phosphorylated GPCRs, not only the engineered M2Rpp construct. Thus, the membrane interaction of βarr1 is critical for receptor desensitization and internalization.
Fig 5: βarr1 functionality depends on C-domain lipid interaction.
(a) Activation of purified heterotrimeric Gi protein by iperoxo-stimulated HDL-M2Rpp in vitro is reduced by βarr1 WT but not βarr1 3xD. Data are the mean of three independent experiments with error bars representing SE. (*) Denotes statistical significance (p<0.0001, one-way ANOVA) compared to WT βarr1 plus iperoxo. (b) Iperoxo stimulation of FLAG-M2R causes plasma membrane recruitment of GFP-βarr1 WT and subsequent receptor internalization, which is impaired by the 3xD mutations as assessed by confocal microscopy. βarr1, nuclei, and M2R colored green, blue, and red, respectively. Confocal images are representative of three independent experiments. (c) Quantification of FLAG-M2R internalization by flow cytometry in same cells as (b). Data are the mean of three independent experiments with error bars representing SE. (*) Denotes statistical significance (p=0.0008, unpaired two-sided T-test) compared to WT βarr1.
Three-site interaction model
Arrestin binding to GPCRs has been viewed as a two-part process: binding to receptor’s phosphorylated ICLs or C-terminus induces conformational changes in βarr, subsequently promoting its coupling to the receptor 7TM bundle. Our findings, supported by observations on visual arrestin-rhodopsin12,19 and the NTSR1-βarr1 complex (this issue), expand this model to include a critical βarr1-lipid interaction (Extended Data Fig. 9f). In this three-site interaction model, arrestin recruitment requires both GPCR phosphorylation and an interaction of βarr1 with the plasma membrane (Fig. 5b), which could increase the arrestin concentration at the cell membrane prior to receptor activation. The C-edge-membrane interaction subsequently enhances arrestin binding to the 7TM bundle (Fig. 4d) and desensitization of G protein activation (Fig. 5a).
While membrane anchoring may be a conserved property of arrestins, the mechanisms underlying membrane association could be distinct. The C-edge loop buried in the nanodisc is found in visual arrestin and the dominant form of βarr1, but not in βarr2 or an alternative βarr1 splice variant. The binding site of phosphatidylinositol 4,5-bisphosphate (PIP2), a phospholipid observed in the structure of the NTSR1-βarr1 complex (this issue), is conserved in βarrs but not retinal arrestins12,19,26,27 (Extended Data Fig. 7). Accordingly, the variability of membrane compositions could provide yet another level of regulation for GPCR desensitization, internalization, and signaling. Interestingly, βarr2 can stimulate MAP kinase signaling from clathrin-coated structures after dissociation from activated GPCRs28. These and other recent data suggest the lipid membrane stabilizes activation of β-arrestin when bound to and even after dissociation from the receptor21,28–30. Thus, evident from the structures of βarr1 bound to M2Rpp or NTSR1 (this issue), a complex cooperative network of low-affinity interactions involving both receptor and membrane phospholipids endow arrestins with remarkable plasticity to variably couple to hundreds of GPCRs.
Methods
Receptor expression and purification
Human muscarinic acetocholine-2-receptor (M2R) containing an N-terminal FLAG tag, C-terminal sortase ligation consensus sequence (LPETGGH), and 6xHIS-tag was cloned into pcDNA-zeo-teto and stably expressed in tetracycline-inducible Expi293F cells (Invitrogen)14. Cells were not authenticated or routinely tested for mycoplasma. Cells were grown to a density of 4–5 million cells per mL and then treated with Expi293F expression enhancers (Thermo Fisher), 5 μM atropine, and 5 μM kifunensine to restrict glycosylation. Expression was induced 18 hours thereafter by addition of 4 μg/mL doxycycline and 5 mM sodium butyrate. Cells were harvested 48 hours post-induction and stored at −80 °C until further processing. FLAG-M2R-LPETGG-His6 was purified as previously described14,31, with all purification steps conducted at 4 °C with protease inhibitors (benzamidine and leupeptin) unless stated otherwise. In brief, cells were lysed for 30 min by stirring in 10 mL/g wet cell mass of lysis buffer (10 mM Tris (pH 7.4), 2 mM EDTA, and 10 mM MgCl2, 5 units/mL benzonase, 5 μM atropine, and 2 mg/mL iodoacetamide). Membrane was pelleted at 30,000×g for 20 min and resuspended in 10 mL/g original cell pellet mass of solubilization buffer (20 mM HEPES (pH 7.4), 750 mM NaCl, 1% n-dodecyl-β-D-maltoside (DDM), 0.05% cholesterol hemisuccinate, 10% glycerol, 5 units/mL benzonase, 5 μM atropine, and 2 mg/mL iodoacetamide). After extensive dounce homogenization, solubilizing membrane was sequentially stirred at room temperature (RT) and 4 °C for 1 h each. Insoluble material was removed by centrifugation at 30,000×g for 30 min, and the supernatant was loaded onto M1-FLAG resin with 2 mM CaCl2 at 1–3 mL/min. M1-FLAG resin was washed with 5 column volumes each of wash buffer (20 mM HEPES (pH 7.4), 0.1% DDM, 0.01% CHS, 2 mM CaCl2, and 1 μM atropine) containing high (750 mM) and low (100 mM) NaCl at ratios of 4:0, 3:1, 2:2, 1:3, and 0:4, respectively. Receptor was eluted in elution buffer (20 mM HEPES (pH 7.4), 100mM NaCl, 0.1% DDM, 0.01% CHS, 0.2mg/mL FLAG-peptide, 1 μM atropine, and 5 mM EDTA), and glycosylation was removed by incubation with a 1:10 protein ratio of EndoH to M2R for 90 min at RT. The enzyme sortase was used to ligate the synthetic phosphopeptide GGG-V2Rpp (GGG-ARGRpTPPpSLGPQDEpSCpTpTApSpSpSLAKDTSS) on the C-terminus of M2R (M2Rpp) as previously described14. Monomeric receptor was collected by size exclusion chromatography on a Superdex200 Increase column (GE Healthcare Life Sciences).
HDL-reconstitution
DDM-M2R was pre-incubated with 2-fold molar excess atropine on ice for 30 min prior to reconstitution. Receptor (5μM) was mixed with 8 mM POPC/POPG and 150 μM MSP1D1E3 on ice for 1 hour in a final buffer composition of 20 mM HEPES (pH 7.4), 100 mM NaCl, and 0.5 mM EDTA. Detergent was removed using Biobeads (BioRad) (50 mg per 100 μL reaction volume) and rotated overnight at 4 °C. The supernatant was separated from the Biobeads using a 28 gauge needle and was diluted with 20 mM HEPES (pH 7.5) and 100 mM NaCl (HN buffer) to obtain a final concentration of 2 μM HDL-M2R. HDL-M2R was rotated with M1-FLAG resin for 30 min at RT and then an additional 15 minutes after adding 2 mM CaCl2. FLAG-M1 resin was quickly washed in column format with five resin volumes of HN buffer with 2 mM CaCl2. HDL-M2R was eluted with HN buffer containing 0.2 mg/mL FLAG peptide and 5 mM EDTA. Size exclusion chromatography was used to collect monomeric HDL-M2R and to remove FLAG-peptide.
βarr1 and Fab30 purification
To enhance expression and stability, a minimal cysteine (C59A, C125S, C140I, C150V, C242V, C251V, and C269S) and truncated (after amino acid 393) variant of rat β-arrestin1 (βarr1-MC-393) in pGEX4T was generated. βarr1-MC-393 was expressed and purified as previously described32. In brief, GST-βarr1-MC-393 was expressed in BL21(DE3) bacteria, lysed by sonication, and captured using glutathione sepharose. βarr1-MC-393 was cleaved from GST by thrombin digestion and further purified using HiTrap Q sepharose anion exchange. Fab30 was purified as described previously7.
HDL-M2Rpp βarr1 complex formation and purification
HDL-M2Rpp was pre-incubated for 30 min on ice with a 5-fold molar excess of iperoxo and LY211960 and then 90 min at RT with a 2-fold molar excess of βarr1-MC-393 and Fab30. The complex of HDL-M2Rpp-βarr1-Fab30 was separated from unbound βarr1-MC-393 and Fab30 by FLAG-M1 chromatography as described above. Following FLAG-M1 elution, the complex was subjected to size exclusion chromatography on a Superdex200 Increase column with HN containing 1 μM iperoxo and 2 μM LY211960. Peak fractions were concentrated to obtain a final concentration of 2 mg/mL for cryoEM analysis.
Co-immunoprecipitation
DDM-M2R or DDM-M2Rpp (4 μg) was pre-incubated with iperoxo (10 μM) for 30 min on ice before the addition of 2-fold molar excess purified βarr1-MC-393 and Fab30. Reactions were incubated at RT for 1 hour, rotated with FLAG-M1 resin for 20 min, washed with HN buffer containing 0.1% DDM, and eluted with HN buffer with 0.1% DDM, 0.4 mg/mL FLAG peptide, and 10 mM EDTA. Proteins were separated by SDS-PAGE and visualized by Instant Blue stain (Expedeon)
Radioligand Binding
Competition equilibrium binding experiments with DDM-M2R were conducted using a scintillation proximity assay (Perkin Elmer) in a final volume of 100 μL containing HN buffer with 0.1% DDM and 1 mg/mL BSA. DDM-M2Rpp was incubated for 1h at RT with 1 μM βarr1 or 10 μM LY2119620, 1 nM [3H]-N-methylscopolamine (NMS), and varying concentrations of iperoxo. Yttrium silicate (YSi) protein A beads (Perkin Elmer) coated with FLAG-M1 antibody were subsequently added for 30 min, and YSi emission was read on a Wallac 1450 Microbeta Plus. Radioligand binding with HDL-M2R was conducted in HN with 1 mg/mL BSA with the same final concentrations of components as above. Reactions proceeded for 90 min and were harvested onto glass fiber filters (GF-B) soaked with 0.3% polyethyleneimine using a Brandel harvester. Data from competition radioligand binding assays were analyzed using GraphPad software.
Bimane Fluorescence
Purified rat βarr1-MC-393 V70C, L337C, and V70C/L334D/L337D/S339D (3xD) were labeled overnight at 4 °C with a 3-fold molar excess of monobromobimane (mBr) (Sigma) and an additional 3-fold molar excess added for 1 h at RT the next day. Reactions were quenched with L-cysteine, and free mBr was removed by size exclusion chromatography. For bimane experiments, DDM-M2R or HDL-M2R was pre-incubated with 10 μM iperoxo or atropine and 2-fold molar excess Fab30 for 20 min at RT in HN buffer containing 1 mg/mL BSA. Bimane-labeled βarr1-MC-393 V70C (mBr-βarr1-V70C), βarr1-MC-393 L337C (mBr-βarr1-L337C), or βarr1-MC-393 V70C 3xD (mBr-βarr1-V70C/3xD) was added to a final concentration of 200 nM with a 1.5-fold molar excess of DDM- or HDL-M2R. For DDM-M2R, buffer additionally contained 0.1% DDM and 0.01% CHS. Reactions were equilibrated for 30 min in black, solid-bottom 96-well microplates (Corning) before fluorescence emission spectra were collected on a CLARIOstar plate reader (BMG Labtech) in top-read mode, with excitation at 370 nm (16 nm bandpass) and emission scanning from 400 nm to 600 nm (10 nm bandpass) in 1-nm increments. Wells for background subtraction contained all components except bimane-labeled βarr1-MC-393. Statistics were conducted by comparing the area under the curves (Graphpad prism) from at least three independent experiments.
CryoEM data acquisition and data processing
Prior to cryoEM preparation, all samples were screened for quality using conventional negative stain EM16. In preliminary cryoEM experiments, M2Rpp-βarr1-Nb24-scFv30 (Nb24: camelid nanobody) complex in the smaller (~9 nm diameter) MSP1D1H5 nanodiscs, at a concentration of 1 mg/mL, was applied to glow-discharged 200 mesh grids (Quantifoil R1.2/1.3), and vitrified using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4 °C and 100% humidity. Images were collected on a Titan Krios equipped with a Gatan K2 Summit direct electron camera, at a nominal magnification of 59000× with a pixel size of 0.86 Å. A total of 3589 movie stacks were acquired with dose-fractionation into 40 frames with an accumulated dose of 70 e/Å2. Dose-fractionated stacks were subjected to beam-induced motion correction using MotionCor233. Data processing was performed in RELION3.034. 487,582 particles projections were selected by template-based autopicking and subjected to 2D classification. A subset of ~70,000 particles showing density for both HDL-receptor and βarr1 were subjected to 3D classification. One out of six 3D classes showed a “hanging” arrestin conformation, and three classes showed a “core” interaction between receptor and arrestin but with “rocking” motion of arrestin relative to the receptor embedded nanodisc (Extended Data Fig. 1e,f).
M2Rpp-βarr1-Fab30 complex in the larger (~12nm diameter) MSP1D1E3 nanodiscs was prepared as described above, except that cryoEM images were collected on a Titan Krios equipped with the Gatan GIF Quantum LS Imaging energy filter using a Gatan K2 Summit direct electron camera in counted mode, corresponding to a pixel size of 1.06 Å. Each image was dose-fractionated into 40 frames with a dose rate of 7 e/pix/s and total exposure time of 8s, resulting in an accumulated dose of 50 electrons per Å2. Data were collected in seven independent sessions resulting in a total number of 30,454 movie stacks. Contrast transfer function parameters for each micrograph were determined by Gctf v1.0635. Auto-picking was performed with templates that were generated from manually picked particles. Particles were subsequently screened by 2D reference-free classification followed by 3D classification. Particle projections were pooled together after independent 3D classification of each dataset. A representative data processing workflow is shown in Extended Data Fig. 2b. In 2D and 3D classification, ~25% of particle projections showing well-defined density for complex components indicated an extra density connected to Fab30, which we suspect to be a non-specifically attached second Fab30. Since this domain was variable among complexes, a mask was applied to remove its density during 3D classification. Particles from each dataset contributing to 3D reconstructions with well-defined features were merged with the other six subsets obtained in a similar strategy, resulting a total number of 831,443 projections with well-defined particle components. Due to the variability in size and tilt of the large MSP1D1E3 nanodisc, which was dominating the projection alignment, the 831,443 particle projections were subjected to a focused alignment on βarr1-Fab30 density followed by 3D classification without alignment while masking out the nanodisc density. One 3D class accounting for 145,618 particles showed well defined transmembrane portion, and its particle projections were subjected to 3D refinement after subtracting from raw images the density of the nanodisc and the constant (CL/CH1) portion of Fab30 due to its relative flexibility, but leaving the scFv portion intact. This strategy resulted in a 3D reconstruction of the M2Rpp-βarr1-Fab30(scFv) with global indicated resolution of 4.0Å. In this map, the extracellular portion of M2Rpp showed the lowest relative resolution. To improve the densities for the βarr1 and M2Rpp interface we used the refinement parameters from the global map to subtract more than half of the extracellular receptor side together with nanodisc and the constant (CL/CH1) domains of Fab30 from raw particle images. The subtracted particles were imported to cisTEM36 for a refinement with local search, resulting in a focused map with global indicated resolution of 3.6 Å. Global resolution estimations were obtained with Phenix from two half maps at FSC cutoff of 0.143. Local resolution determination of the global M2Rpp-βarr1-Fab30(scFv) map and the focused Interface M2Rpp-βarr1-Fab30(scFv) map was calculated with blocres from the Bsoft package37 at FSC cutoff of 0.5 (Extended Data Fig. 3b,c and Extended Data Table 1).
Model building and refinement
The initial model was prepared by manually docking the cryoEM structure of active-state M2R (PDB:6OIK) and the crystal structure of activated V2Rpp-βarr1-Fab30 (PDB:4JQI) into the cryo-EM map. We employed the global M2Rpp-βarr1-Fab30(scFv) 4Å map for modeling the 7TM bundle and the focused Interface M2Rpp-βarr1-Fab30(scFv) 3.6Å map for modeling βarr1 and its interface with M2R. Iterative rounds of real-space refinement were performed with phenix.real_space_refine in Phenix38 and manual model building with Coot39. Rosetta was periodically used to assess if more optimal models existed. Additional refinement near the end of model building was performed with Phenix using the state-of-the-art OPLS3e force field with electrostatics. A final refinement was performed on the full complex with Phenix using the global M2Rpp-βarr1-Fab30(scFv) 4Å map (Extended Date Table 1). Independent FSC curves for model-map correlations were calculated between the resulting model and the two maps (Extended Data Fig. 3a). Model overfitting was evaluated through its refinement against the focused Interface M2Rpp-βarr1-Fab30 half maps after randomly displacing all atoms by 0.2 Å (Extended Data Fig. 3a).
Model Analysis
Electrostatic potential surfaces were calculated with APBS40 in Chimera. Charges were prepared with PDB2PQR41 using the PARSE force field42. Tilt angle of arrestin vs receptor was calculated between residues V37 (βarr1), I317 (βarr1), and R121 (M2) in the case of M2Rpp-βarr1 and V2042 (arr1), I2324 (arr1), and R135 (rhodopsin) for the rhodopsin-arrestin structure.
Molecular dynamics simulations
Starting from a model of the M2R-βarr1 complex derived from an earlier refinement (model available upon request), the OPM webserver43 was used to orient the structure with respect to the plane of the lipid bilayer. The aligned structure was then prepared further using CHARMM-GUI44 to place the structure in either a membrane bilayer or a MSP1D1–44 nanodisc. In both cases a 3:2 ratio of POPC to POPG was used for the lipid system. The systems were hydrated with TIP3P water and charge neutralized with 150 mM NaCl. Further system preparation was performed in VMD45, where palmitoylation was added to residue Cys457 of the receptor. Serine 488, 493, 494, 495 and threonine 490 and 491 were phosphorylated with dibasic phosphate. The systems were simulated in NAMD46 with the OPLS-AA/M force field47. OPLS-AA parameters for iperoxo and LY2119620 were obtained with the LigParGen server48. OPLS-AA parameters for POPC were taken from49, while POPG parameters were adapted from POPC and the OPLS-AA small molecule set. OPLS-AA/M parameters for phosphorylated serine and threonine were also developed for this work based on existing OPLS-AA parameters for phosphates50. Parameters for palmitoylated cysteine were developed for this work by combining OPLS-AA/M parameters for cysteine with the lipid parameters of Kulig et al. All simulations followed a 2-fs timestep in the NPT ensemble using a Langevin thermostat set at 300K with a dampening coefficient of 1 ps−1 and a Nosé-Hoover Langevin piston barostat set at 1 atm with a period of 50 fs and a decay of 25 fs. Non-bonded interactions were smoothed starting at 10 Å to 12 Å, where long-range interactions were treated with particle mesh Ewald. Systems were minimized for 2000 steps before being slowly heated from 0K to 300K in 20K increments with 0.4 ns of simulation at each increment. During the heating phase, harmonic restraints of 1 kcal/mol/Å2 were applied to all lipid, protein, and small molecule non-hydrogen atoms. The system was then equilibrated with harmonic restraints of 1 kcal/mol/Å2 applied to all non-hydrogen protein atoms for 2 ns, followed by 2 ns of simulation with restraints on only protein C alpha atoms. These restraints were then gradually reduced first to 0.3 kcal/mol/Å2 for 2 ns before being removed completely. This aggregate 12 ns of heating and equilibration and an additional 18 ns of production simulation were discarded from the final calculated quantities. Each system was then simulated for five replicates of up to 200 ns to produce the results used in this work. Trajectories were analyzed with the VMD software45. Interdomain rotation was calculated as described by Latorraca et. al21 using the inactive and active state crystal structures of arrestin (PDB:1CF151 for the inactive structure and PDB:4ZWJ12) to define the axis of rotation. These structures were chosen as references, as the rhodopsin-arrestin structure was thought to be the closest analogue to our structure; however results were qualitatively similar with other references.
Confocal microscopy
HEK293 (ATCC) cells lacking endogenous βarr1/2 as described elsewhere52,53 were transiently transfected with a 1:4 DNA ratio of pcDNA-teto-M2R and GFP-βarr1-WT or the −3xD variant, respectively. Cells were not authenticated or routinely tested for mycoplasma. Cells were split into 35 mm glass bottom microwell dishes (MatTek) and 24 h thereafter serum starved for 2 h in MEM media containing 20 mM HEPES (pH7.4) and 1 mg/mL BSA. Cells were incubated with Alexa-650-labeled FLAG-M1 antibody and NucBlue Live cell stain (Invitrogen) for 1 h at 37 °C, washed two times in starvation media, and imaged using confocal microscopy. In parallel, transfected cells split into 6-well plates were stimulated for 30 min at 37 °C +/− 1 μM iperoxo. Cells were immediately placed on ice and kept at 4 °C for the remainder of the experiment. Cells were washed twice with cold phosphate-buffered saline and then detached with 0.05% EDTA. Cells were resuspended in assay buffer (Hanks balanced salt solution, 20 mM HEPES pH 7.4, 3 mM CaCl2, 1 mg/mL BSA) and stained with Alexa-650-labeled FLAG-M1 antibody for 30 min at 4 °C. Cells were washed once with assay buffer before analysis by flow cytometry (Bio-Rad S3e Cell Sorter). Data were analyzed using FlowJo software, gating for GFP-positive singlet cells.
G-protein GTPase Assay
The GTPase activity of purified heterotrimeric Gαi was measured in vitro using the GTPase Glo Assay (Promega) with the following modifications. Final reaction consisted of 20 mM HEPES (pH7.4), 100 mM NaCl, 10 mM MgCl2, and 1 mg/mL BSA. HDL-M2Rpp (12.5 nM) was pre-incubated with iperoxo (10 μM), purified βarr1-MC-393 WT or βarr1-MC-393 3xD (1 μM), and Fab30 (1 μM) for 15 min at RT. G-protein (250 nM) and GTP (2.5 μM) were subsequently added, and reactions proceeded for 1 hour at RT before addition of GTPase Glo reagent and ADP, as described in manufacturer protocol. Luminescence was measured on a CLARIOstar plate reader (BMG Labtech).
Data and code availability statement
The atomic coordinates of the M2R-βarr1 structure have been deposited in the Protein Data Bank under accession number 6U1N. The EM maps of M2R-βarr1-Fab30(scFv) and InterfaceM2R-βarr1-Fab30(scFv) have been deposited in EMDB with the codes EMD-20612 and EMD-20948, respectively.
Extended Data
Extended Data Figure 1: M2R-βarr1 complex characterization.
(a) Schematic showing sortase-mediated ligation of GGG-V2Rpp onto GPCRs containing a C-terminal sortase consensus sequence (LPETGGH). (b) Competition radioligand binding experiments using [3H]-N-methylscopolamine (NMS) to measure the affinity of iperoxo for HDL-M2Rpp in the absence (control; Ctl) (logIC50 −6.98 ± 0.07) or presence of βarr1 (logIC50 −8.38 ± 0.07), βarr1-minimal cysteine (MC) (logIC50 −8.52 ± 0.07), and βarr1-V70C mutant (logIC50 −8.34 ± 0.06). (c) Co-immunoprecipitation (IP) of βarr1and Fab30 in the presence and absence of DDM-FLAG-M2Rpp and DDM-FLAG-M2R. Data representative of three independent experiments. d) Statistical analysis of bimane data from Fig. 1c. Data are the mean of three independent experiments with error bars representing SE. (*) Indicates significance (one-way ANOVA) (e) Competition radioligand binding experiments using [3H]-NMS to measure the affinity of the agonist carbachol for HDL-M2Rpp in the absence (Ctl; logIC50 −5.38 ± 0.12) and presence of LY2119620 (LY211) (logIC50 −6.7 ± 0.10), βarr1 and Fab30 (logIC50 −6.8 ± 0.07), or in combination (logIC50 −7.95 ± 0.06). (f) Size exclusion chromatography of final MSP1D1E3-M2Rpp-βarr1-Fab30 complex and SDS-PAGE analysis of peak fractions. (g,h) Low resolution cryoEM analysis of M2Rpp-βarr1-Nb24-scFv30 complex in MSP1D1H5 nanodiscs showing βarr in a “hanging” conformation (left map) or “core” conformations with “rocking” relative to the nanodisc density. (i) Low resolution cryoEM map of M2Rpp-βarr1-Fab30 complex in the larger MSP1D1E3 nanodiscs shows βarr1 in the “core” conformation involving an additional interaction of the C-domain with the lipid bilayer. All βarr1 variants are truncated at amino acid 393. Radioligand binding experiments are the means of three independent experiments with error bars representing SE. (*) Indicates significance compared to control (p<0.0001, one-way ANOVA).
Extended Data Figure 2: M2Rpp-βarr1 complex conformers and cryoEM reconstruction workflow.
(a) Conformational variability of the M2Rpp-βarr1 complex. Overlay of two low resolution reconstructions aligned on the 7TM portion reveal variability in the angle of the βarr1-Fab30 segment relative to the receptor. One map is shown as mesh and the other as solid surface. The constant domains of Fab30 have been masked out in the reconstructions. For clarity, only one receptor-nanodisc density is shown. (b) Flow chart of cryoEM data processing towards high-resolution reconstructions. All eight-particle classes show engagement of βarr1 with the lipid nanodisc but only one class (17.4% of particles) could be further processed to high resolution. A final focused refinement yielded a 3.6Å structure providing insights into the binding interface and orientation of βarr1 relative to M2Rpp.
Extended Data Figure 3: Map resolution and model validation.
(a) “Gold standard” Fourier Shell Correlation (FSC) plots of the reconstruction of M2Rpp-βarr1-Fab30(scFv) (global map, black) and focused refinement reconstruction for Interface M2Rpp-βarr1-Fab30(scFv) (focused map, blue). The red and brown curves represent the model-map correlation for global map and focused map, respectively. Refined model validation was also performed by calculating the correlation between model and focused half map 1 (green), and correlation between the randomly displaced model and focused half map 2 (purple). (b, c) Local resolution estimation map for M2R-βarr1-Fab30(scFv) and focused map of half-M2Rpp-βarr1-Fab30(scFv). (d) Overall model fit to the cryo-EM map. The 7TM portion (orange) comes from the full M2Rpp-βarr1-Fab30(scFv) map contoured at σ=7.5, whereas the βarr1-scFv portion comes from the focused refinement of Interface M2Rpp-βarr1-Fab30(scFv) contoured at σ=5.2.
Extended Data Figure 4: Structural comparisons of arrestins.
(a) The tilt of β-arrestin relative to the M2Rpp was measured by the angle (red arrow) between V37 (βarr1), I317 (βarr1), and R1213.50 (M2R). (b) Superposition of βarr1 in M2Rpp-βarr1 complex and visual arrestin in crystal structure of Rho-visual arrestin (PDBID:5W0P). (c) Superposition of βarr1 in inactive state (grey, PDB: 1G4M), in activated state by phosphopeptide (green, PDBID:4JQI) and in receptor-bound active state (gold, M2Rpp-βarr1). The arrows indicate rearrangements of central crest loops.
Extended Data Figure 5: Computational and experimental examination of finger loop/M2Rpp interactions.
(a) Depiction of the finger loop E61 and probable interaction partners R57 and R71 (above) and the R60 of the finger loop and D130 of the middle loop (below) from Phenix/OPLS3 refinement in the cryoEM map. The mesh depicts the 3.6Å cryoEM map contoured at σ=4.0 with a masked 3.0 Å zone around the atoms depicted (above) or contoured at σ=3.0 with a masked 3.0 Å zone around the atoms depicted (below). (b) An overlay of the last frame from 200 ns of simulation for 5 of the MD simulation trajectories. D69 of arrestin is depicted as bonds in addition to R3.50 and N2.39 of M2Rpp. Lines connect D69 and R3.50 from the same snapshot. (c) Plots of the R3.50 zeta carbon-D69 gamma carbon distance (left) and N2.39 gamma carbon-D69 gamma carbon distance (right) over the course of the MD simulations performed in this work. Grey lines correspond to raw data, while colored lines correspond to a 1-ns sliding average. Teal traces correspond to simulations with membrane, and blue traces correspond to simulations with small nanodisc. (d) Competition radioligand binding experiments using [3H]-N-methylscopolamine to measure the affinity of the agonist iperoxo for HDL-M2Rpp in the absence and presence of βarr1 WT, D69A and Δ62–77 mutant lacking the finger loop. Data are the mean of four independent experiments with error bars representing SE. Inset: difference in logIC50 between βarr1 variants and control (no βarr1), (*) denotes significance compared to WT βarr1 (p<0.0001, one-way ANOVA).
Extended Data Figure 6: Finger loop rearrangements and ICL2.
(a) Detail of crystal structure of activated βarr1with phosphopeptide (left panel, PDBID: 4JQI) showing the N-terminal portion of V2Rpp bound on β-strands 5 and 6, thereby twisting the finger loop fold. Unbinding of the N-terminal portion of V2Rpp is required for the finger loop to adopt the observed conformation in the M2Rpp-βarr1 cryoEM structure (right panel). The arrow shows the direction of finger loop untwisting. (b) Close-up of L129 (orange) in the cleft of βarr1 overlaid with a modeled phenylalanine (green) and methionine (red) at the same position. (c) Plot of the frequency of specific amino acids occurring in the second position of ICL2 for βarr-binding class A GPCRs. The size of the 1-letter code is correlated to the frequency with which that residue occurs at that position.
Extended Data Figure 7: Conservation of lipid binding regions in arrestin family.
(a) Ribbon model of the M2Rpp-βarr1 complex depicting the location of C-edge loops and phosphatidylinositol 4,5-bisphosphate (PIP2)-interacting residues (b) Sequence alignment of arrestin family to show differential conservation of C-edge loops and PIP2-binding motif.
Extended Data Figure 8: Effects of lipid membrane on βarr1 coupling to M2Rpp.
(a) Atomistic simulations of M2Rpp-βarr1 complex in membrane bilayer or a small nanodisc. Time courses are provided for the calculated interdomain twist angle for each replicate in membrane bilayer (left column) or a small nanodisc (right column). Raw data is provided in grey while a 1-ns rolling average is provided in teal for the membrane simulations and blue for the small nanodisc simulations. Horizontal teal and blue lines correspond to an active-like and inactive-like interdomain twist angle, respectively. (b) Statistical analysis of figure 4c data are the mean of three independent experiments with errors bars representing SE. (*) Indicates statistical significance from control (Ctl) within the same subset (p<0.0001, one-way ANOVA). (c) Statistical analysis of data from figure 4d; data represent the mean and standard error of three independent experiments and (*) denotes significance compared to Ctl (p<0.0001, one-way ANOVA).
Extended Data Figure 9: βarr1 C-edge-lipid interaction facilitates M2R internalization.
(a) Flow cytometry analysis of βarr1/2 null cells transiently transfected with FLAG-M2R and WT or 3xD GFP-βarr1. Cells were treated with vehicle or iperoxo for 30 minutes and subsequently stained with Alexa Fluor-650-labeled anti-FLAG M1 antibody. GFP+ singlet cells were gated for analysis. Figure is representative of data from three independent experiments. (b) Quantitation of FLAG-M2R surface staining by flow cytometry, as described above. Alexa Fluor-650-labeled anti-FLAG M1 staining was normalized to the mean fluorescence of unstimulated cells expressing WT β-arrestin1-GFP in each experiment. These data were used to calculate the percentage of receptor internalized in Fig. 5c. Data represent the mean and standard error from three independent experiments and asterisks (*) indicate statistical significance (one-way ANOVA). N.S. represents non-significance. (c) Expression of GFP-βarr1 wild-type or 3xD and FLAG-M2R in βarr1/2 null HEK293 cells as assessed by SDS-PAGE and western blot analysis. Tubulin used as loading control. Data representative of three independent experiments. (d) Quantification of GFP-βarr1 WT or 3xD expression by flow cytometry using βarr1/2 null HEK293 cells. Data represent the mean and standard error from three independent experiments with asterisk (*) representing statistical significance (p=0.0034, two-sided unpaired T-test). (e) Localization of GFP-βarr1 WT or 3xD in FLAG-Vasopressin-2-receptor overexpressing HEK293 cells treated with arginine vasopressin peptide (AVP) for the indicated time. Data representative of three independent experiments. (f) Three-site interaction network of GPCR-βarrestin binding. In the classic two-site interaction model, conformational changes in β-arrestin induced by binding to phosphorylated receptor (1) leads to transmembrane receptor core coupling (2) to sterically block G protein binding. Our findings suggest an expanded model including interaction of β-arrestin C-domain with the lipid bilayer (3) since it synergistically enhances the interaction of β-arrestin with the phosphorylated receptor tail/loops and transmembrane core. Vertical arrows in the receptor represent direction and strength of cooperativity between the extracellular orthosteric ligand-binding and intracellular transducer-binding sites.
Extended Data Table 1: CryoEM data table.
Information on CryoEM data collection, refinement, and validation statistics.
| M2R-βarr1-Fab30(scFv) (EMDB-20612) (PDB 6U1N) | InterfaceM2R-βarr1-Fab30(scFv) (EMDB-20948) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 47,169 | 47,169 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 50 |
| Defocus range (μm) | 1.2–2.2 | 1.2–2.2 |
| Pixel size (Å) | 1.06 | 1.06 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 11.7million | 11.7million |
| Final particle images (no.) | 145,618 | 145,618 |
| Map resolution (Å) | 4Å | 3.6Å |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 3.6–6 | 3.2–5 |
| Refinement | ||
| Initial model used (PDB code) | 6OIK 4JQI |
|
| Model resolution (Å) | 4.03 | |
| FSC threshold | 0.5 | |
| Map sharpening B factor (Å2) | 150 | |
| Model composition | ||
| Non-hydrogen atoms | 5863 | |
| Protein atoms | 5834 | |
| Ligands | 29 | |
| B factors (Å2) | ||
| Protein | 164.85 | |
| Ligand | 292.74 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.005 | |
| Bond angles (°) | 0.682 | |
| Validation | ||
| MolProbity score | 2.2 | |
| Clashscore | 18.0 | |
| Poor rotamers (%) | 1.0 | |
| Ramachandran plot | ||
| Favored (%) | 91.6 | |
| Allowed (%) | 8.4 | |
| Disallowed (%) | 0.0 | |
Acknowledgments
We are grateful to Georgina Hodgson, Jan Taylor, Quivetta Lennon, and Victoria Brennand for administrative assistance. Financial support was provided by the National Institutes of Health (Grants R01 HL16037 to R.J.L. and R01 NS092695 to G.S.) and the Mathers Foundation (G.S.). R.J.L is a Howard Hughes Medical Institute investigator. ALWK is a Howard Hughes Medical Institute medical research fellow. We thank Ali Masoudi for assistance in structural analysis of the M2R-βarr1 complex, Sanduo Zheng for assistance in initially screening M2R-βarr1 complexes by negative stain electron microscopy, and Asuka Inoue for βarr1/2-null HEK293 cells.
Footnotes
The authors declare no competing financial interests.
References
- 1.Rajagopal S & Shenoy SK GPCR desensitization: Acute and prolonged phases. Cell Signal 41, 9–16, doi: 10.1016/j.cellsig.2017.01.024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiter E, Ahn S, Shukla AK & Lefkowitz RJ Molecular mechanism of beta-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol 52, 179–197, doi: 10.1146/annurev.pharmtox.010909.105800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda S, Qu Q, Robertson MJ, Skiniotis G & Kobilka BK Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557, doi: 10.1126/science.aaw5188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurevich VV & Gurevich EV GPCR Signaling Regulation: The Role of GRKs and Arrestins. Front Pharmacol 10, 125, doi: 10.3389/fphar.2019.00125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q, Iverson TM & Gurevich VV Structural Basis of Arrestin-Dependent Signal Transduction. Trends Biochem Sci 43, 412–423, doi: 10.1016/j.tibs.2018.03.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheerer P & Sommer ME Structural mechanism of arrestin activation. Curr Opin Struct Biol 45, 160–169, doi: 10.1016/j.sbi.2017.05.001 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Shukla AK et al. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141, doi: 10.1038/nature12120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou XE et al. Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell 170, 457–469 e413, doi: 10.1016/j.cell.2017.07.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller WE & Lefkowitz RJ Expanding roles for beta-arrestins as scaffolds and adapters in GPCR signaling and trafficking. Curr Opin Cell Biol 13, 139–145 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Smith JS, Lefkowitz RJ & Rajagopal S Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov 17, 243–260, doi: 10.1038/nrd.2017.229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Nafria J & Tate CG Cryo-EM structures of GPCRs coupled to Gs, Gi and Go. Mol Cell Endocrinol 488, 1–13, doi: 10.1016/j.mce.2019.02.006 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Kang Y et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567, doi: 10.1038/nature14656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruse AC et al. Muscarinic acetylcholine receptors: novel opportunities for drug development. Nat Rev Drug Discov 13, 549–560, doi: 10.1038/nrd4295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staus DP et al. Sortase ligation enables homogeneous GPCR phosphorylation to reveal diversity in beta-arrestin coupling. Proc Natl Acad Sci U S A 115, 3834–3839, doi: 10.1073/pnas.1722336115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM & Onorato JJ Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J Biol Chem 272, 28849–28852, doi: 10.1074/jbc.272.46.28849 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Peisley A & Skiniotis G 2D Projection Analysis of GPCR Complexes by Negative Stain Electron Microscopy. Methods Mol Biol 1335, 29–38, doi: 10.1007/978-1-4939-2914-6_3 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Grinkova YV, Denisov IG & Sligar SG Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel 23, 843–848, doi: 10.1093/protein/gzq060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shukla AK et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222, doi: 10.1038/nature13430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lally CC, Bauer B, Selent J & Sommer ME C-edge loops of arrestin function as a membrane anchor. Nat Commun 8, 14258, doi: 10.1038/ncomms14258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noble AJ et al. Routine single particle CryoEM sample and grid characterization by tomography. Elife 7, doi: 10.7554/eLife.34257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latorraca NR et al. Molecular mechanism of GPCR-mediated arrestin activation. Nature 557, 452–456, doi: 10.1038/s41586-018-0077-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballesteros JA & Weinstein H in Methods in Neurosciences Vol. 25 (ed Sealfon Stuart C.) 366–428 (Academic Press, 1995). [Google Scholar]
- 23.Koehl A et al. Structure of the mu-opioid receptor-Gi protein complex. Nature 558, 547–552, doi: 10.1038/s41586-018-0219-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen SG et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555, doi: 10.1038/nature10361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishna Kumar K et al. Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell 176, 448–458 e412, doi: 10.1016/j.cell.2018.11.040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaidarov I, Krupnick JG, Falck JR, Benovic JL & Keen JH Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J 18, 871–881, doi: 10.1093/emboj/18.4.871 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parruti G et al. Molecular analysis of human beta-arrestin-1: cloning, tissue distribution, and regulation of expression. Identification of two isoforms generated by alternative splicing. J Biol Chem 268, 9753–9761 (1993). [PubMed] [Google Scholar]
- 28.Eichel K, Jullie D & von Zastrow M beta-Arrestin drives MAP kinase signalling from clathrin-coated structures after GPCR dissociation. Nat Cell Biol 18, 303–310, doi: 10.1038/ncb3307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichel K et al. Catalytic activation of beta-arrestin by GPCRs. Nature 557, 381–386, doi: 10.1038/s41586-018-0079-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuber S et al. beta-Arrestin biosensors reveal a rapid, receptor-dependent activation/deactivation cycle. Nature 531, 661–664, doi: 10.1038/nature17198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse AC et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106, doi: 10.1038/nature12735 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobles KN, Guan Z, Xiao K, Oas TG & Lefkowitz RJ The active conformation of beta-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of beta-arrestins1 and −2. J Biol Chem 282, 21370–21381, doi: 10.1074/jbc.M611483200 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi:ARTN e42166 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant T, Rohou A & Grigorieff N cisTEM, user friendly software for single-particle image processing. Elife 7, doi:ARTN e35383 10.7554/eLife.35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heymann JB & Belnap DM Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol 157, 3–18, doi: 10.1016/j.jsb.2006.06.006 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Adams PD et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106, doi: 10.1016/j.ymeth.2011.07.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132, doi: 10.1107/S0907444904019158 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041, doi: 10.1073/pnas.181342398 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolinsky TJ et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35, W522–525, doi: 10.1093/nar/gkm276 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang CL, Alexov E, Pyle AM & Honig B Calculation of pKas in RNA: on the structural origins and functional roles of protonated nucleotides. J Mol Biol 366, 1475–1496, doi: 10.1016/j.jmb.2006.12.001 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Lomize MA, Pogozheva ID, Joo H, Mosberg HI & Lomize AL OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40, D370–376, doi: 10.1093/nar/gkr703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jo S, Kim T, Iyer VG & Im W CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29, 1859–1865, doi: 10.1002/jcc.20945 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Phillips JC et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802, doi: 10.1002/jcc.20289 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson MJ, Tirado-Rives J, & Jorgensen WL Improved Peptide and Protein Torsional Energetics with the OPLS-AA Force Field. Journal of Chemical Theory and Computation, doi: 10.1021/acs.jctc.5b00356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodda LS, Cabeza de Vaca I, Tirado-Rives J & Jorgensen WL LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res 45, W331–W336, doi: 10.1093/nar/gkx312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulig W, Pasenkiewicz-Gierula M & Rog T Topologies, structures and parameter files for lipid simulations in GROMACS with the OPLS-aa force field: DPPC, POPC, DOPC, PEPC, and cholesterol. Data Brief 5, 333–336, doi: 10.1016/j.dib.2015.09.013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jorgensen WL, Maxwell DS, & Tirado-Rives J Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society, doi: 10.1021/ja9621760 (1996). [DOI] [Google Scholar]
- 51.Hirsch JA, Schubert C, Gurevich VV & Sigler PB The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell 97, 257–269, doi: 10.1016/s0092-8674(00)80735-7 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Luttrell LM et al. Manifold roles of beta-arrestins in GPCR signaling elucidated with siRNA and CRISPR/Cas9. Sci Signal 11, doi: 10.1126/scisignal.aat7650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Hayre M et al. Genetic evidence that beta-arrestins are dispensable for the initiation of beta2-adrenergic receptor signaling to ERK. Sci Signal 10, doi: 10.1126/scisignal.aal3395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The atomic coordinates of the M2R-βarr1 structure have been deposited in the Protein Data Bank under accession number 6U1N. The EM maps of M2R-βarr1-Fab30(scFv) and InterfaceM2R-βarr1-Fab30(scFv) have been deposited in EMDB with the codes EMD-20612 and EMD-20948, respectively.