Abstract
PURPOSE
Smoldering multiple myeloma (SMM) is a precursor condition of multiple myeloma (MM) with a 10% annual risk of progression. Various prognostic models exist for risk stratification; however, those are based on solely clinical metrics. The discovery of genomic alterations that underlie disease progression to MM could improve current risk models.
METHODS
We used next-generation sequencing to study 214 patients with SMM. We performed whole-exome sequencing on 166 tumors, including 5 with serial samples, and deep targeted sequencing on 48 tumors.
RESULTS
We observed that most of the genetic alterations necessary for progression have already been acquired by the diagnosis of SMM. Particularly, we found that alterations of the mitogen-activated protein kinase pathway (KRAS and NRAS single nucleotide variants [SNVs]), the DNA repair pathway (deletion 17p, TP53, and ATM SNVs), and MYC (translocations or copy number variations) were all independent risk factors of progression after accounting for clinical risk staging. We validated these findings in an external SMM cohort by showing that patients who have any of these three features have a higher risk of progressing to MM. Moreover, APOBEC associated mutations were enriched in patients who progressed and were associated with a shorter time to progression in our cohort.
CONCLUSION
SMM is a genetically mature entity whereby most driver genetic alterations have already occurred, which suggests the existence of a right-skewed model of genetic evolution from monoclonal gammopathy of undetermined significance to MM. We identified and externally validated genomic predictors of progression that could distinguish patients at high risk of progression to MM and, thus, improve on the precision of current clinical models.
INTRODUCTION
Multiple myeloma (MM) is an incurable plasma cell malignancy with significant inter- and intrapatient heterogeneity. It is almost always preceded by asymptomatic precursor stages, namely monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM).1,2 Patients with SMM have a higher risk of progression to MM (10%/year) compared with MGUS (1%/year).3 Although some patients progress rapidly, others remain in an MGUS-like state for years.
CONTEXT
Key Objective
The identification of the patients with smoldering multiple myeloma (SMM) who will eventually progress to overt malignancy can allow for early intervention to prevent end-organ damage and potentially achieve long-term remission. Current risk models are based on solely clinical markers and often lack precision and accuracy to predict progression. We hypothesized that genomics can improve the prediction of progression from SMM to overt multiple myeloma (MM).
Knowledge Generated
Most genetic alterations have already occurred by the time of SMM diagnosis, which suggests that next-generation sequencing (NGS) can be used at that stage for reliable prognostication. Alterations in the mitogen-activated protein kinase and DNA repair pathways or MYC are independent risk factors of progression, whose addition to current clinical risk models significantly improves prediction of progression.
Relevance
Clinical-grade NGS at the SMM stage can be used to identify patients at high risk of progression to MM who might benefit from early intervention approaches for better outcomes.
Current prognostic models do not fully capture SMM progression risk because patients who are considered to be intermediate or low risk by those criteria can still progress. This might be because these models are based mainly on tumor burden markers and may not adequately reflect the underlying biology that could be critical for disease progression. Thus, there is an urgent need for novel prognostic markers that can accurately identify patients with SMM who are at risk for progression and could benefit from early treatment. Herein, we studied 214 samples from patients with SMM to comprehensively characterize the genomic landscape of SMM and identify biomarkers of progression to MM.
METHODS
We used next-generation sequencing (NGS) technologies to study 214 samples from patients with SMM at the time of diagnosis, including 5 serial samples. We performed whole-exome sequencing (WES) on 72 matched tumor-normal samples and 94 tumor-only samples and targeted deep sequencing on 48 samples. Patients who presented at diagnosis with MM symptoms, including hypercalcemia, renal impairment, anemia, or bone lytic lesions or who had any myeloma-defining event were excluded from the analysis.4 Patients with light-chain and nonsecretory SMM were included because they are understudied subtypes and their biology needs to be better understood. All samples were obtained after written informed consent in accordance with the Declaration of Helsinki. Time-to-event end points were estimated using the Kaplan-Meier method. Differences in survival curves were assessed using log-rank tests. Median follow-up was calculated using the reverse Kaplan-Meier method. Time to progression (TTP) was measured from the date of diagnosis to the date of documented progression to MM. Cox proportional hazards modeling was performed to assess the impact of genetic alterations on risk of disease progression. Statistical analysis is described in detail in the Data Supplement (online only).
RESULTS
The Genomic Landscape of SMM
The median age in our cohort was 62 years (range, 34-85 years). Patients were stratified into low-, intermediate-, and high-risk groups on the basis of the Mayo 2008 criteria5 and the revised Mayo 2018 criteria6 (Data Supplement).
The genomic landscape of the entire cohort is illustrated in Figure 1, with single nucleotide variants (SNVs), somatic copy number alterations (SCNAs) and translocations displayed in separate sections, and patients sorted according to their clinical risk stage. Immunoglobulin heavy chain translocations commonly seen in MM were present in 76 patients (36%) as identified by fluorescence in situ hybridization, while SCNAs were the most common genomic alterations and were present in 189 patients (88%). Hyperdiploidy (HRD; ie, with ≥ 48 chromosomes in the genome) was found in 55% of patients; hypodiploidy (defined as < 45 chromosomes) was found in only 10 patients (4.6%) and whole-genome doubling (ploidy > 2.5) in 6 (2.8%). The median mutation density in patients with SMM was 1.4 mutations/Mb, and SNVs in genes significantly mutated in MM were present in 118 patient samples (55%). Forty-six percent of those had alterations in the mitogen-activated protein kinase (MAPK) pathway (KRAS, NRAS, BRAF, and PTPN11). DNA repair pathway alterations (TP53 and ATM SNVs and deletion [del] 17p) were found in 21 (10%). SNVs in genes of nuclear factor-κB, protein processing, and cell cycle pathways were found in 22%, 21%, and 6.7% of patients, respectively. Biallelic inactivation events that affected TP53, RB1, CDKN2C, ZNF292, DIS3, or FAM46C were present in only 6% of patients. Copy number neutral loss of heterozygosity (LOH) was observed in 44 patients (27%), with chromosome 16 most frequently affected (20%), followed by chromosomes 1 (16%) and 6 (11%). Of note, 36 LOH events were clonal, and 31 events were subclonal. No significant focal amplifications were discovered in our data set (n = 166) by GISTIC2. However, 4 focal deletions were considered significant (del 1p22.1, del 6q27, del 14q24.3, and del 14q32.31), all of which contained putative tumor suppressors (Data Supplement). In fact, patients with del 1p22.1 and del 14q24.3 had a shorter TTP (P = .009 and .02, respectively; Data Supplement). Of note, the most common MM whole-chromosome and arm-level copy number alterations (CNAs), including gains of 1q and chromosomes 3, 5, 7, 9, 11, 15, 19, and 21 as well as del 13q and del 16q, were significant events (Data supplement). In terms of alteration co-occurrence, we observed that HRD often significantly co-occurred with mutations in KRAS and NRAS, while t(11;14) was significantly mutually exclusive with HRD and gain of 1q (Fig 2A). On the other hand, t(4;14) co-occurred with mutations in DIS3, BRAF, del 13q, and gain 1q. Del 13q was shown to co-occur infrequently with HRD and rarely with NRAS; however, it co-occurred frequently with del 16q and gain 1q. Our analysis also showed that del 1p frequently co-occurred with del 8p and 17p.
FIG 1.

The genomic profile of the 214 patients with smoldering multiple myeloma divided into single nucleotide variants (top 6 panels), somatic copy number alterations (CNAs; yellow panel), and translocations (bottom panel). The risk stratification according to the Mayo 2018 risk model (low, intermediate, and high risk colored as red, green, and blue, respectively). The sex of the patient is identified by the colors pink (female) and blue (male). amp, amplification; del, deletion; HRD, hyperdiploidy; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor-κB.
FIG 2.

(A) Correlation matrix of the 214 patients who demonstrated the significant associations and co-occurrence of different multiple myeloma (MM) drivers (adjusted P < .05). The size of the bubble corresponds to the odds ratio. The blue color indicates negative correlations, while the red color depicts positive ones. (B) The clonal proportions of recurrent somatic copy number alterations and (C) single nucleotide variants across the samples. amp, amplification; del, deletion; HRD, hyperdiploidy.
The Clonal Architecture and Phylogeny of SMM
We observed heterogeneity in the clonality of mutations that affected recurrently mutated genes in MM. Certain genes (TP53, KLHL6, DIS3, MAX, NFKBIA, and CCND1) carried clonal alterations more frequently, while other genes were more frequently subclonal (FAM46C, NFKB2, LTB, and TRAF3), which likely represent later events in the disease course (Fig 2C). Of note, mutations in KRAS, NRAS, and BRAF were subclonal in 64%, 70%, and 86% of patients, respectively. In contrast, our analysis showed that SCNAs were clonal in most of the samples, especially trisomies of odd-numbered chromosomes that were clonal in 98%, with the exception of trisomies 9, 19, and 21, which were found to be subclonal in a few samples (Fig 2B). Moreover, del 13q, 14q, and 16q and gain 1q were also clonal in 78%, 76%, and 72%, and 68%, respectively. These data suggest that SCNAs are largely founder events, while SNVs in the MAPK pathway are later events that contribute to tumor progression.
We next analyzed serial samples from 5 patients sampled at 2 time points of at least 1 year apart (range, 1-8 years). We observed evidence of clonal heterogeneity in all 5, which indicated that clonal branching had already happened at the smoldering stage. While copy number abnormalities were mostly clonal in all 5 samples, we observed evidence of subclonal copy number events in 3 patients (SMM_060, del 13q/del 1p; SMM_002, del 17p; SMM_093, del 14q, del 20p; Figs 3A-3C); all these events were associated with significant clonal expansion at the late time points, which suggests that copy number events have strong driving potential and eventually become clonal, even when acquired during progression. The majority of alterations present at progression were already present at the SMM stage; however, in 1 of 5 patients (SMM_060) sampled with a 5-year interval, there was evidence of a newly acquired subclone carrying a KRAS mutation as well as del 13q/del 1p, during progression. In another patient (SMM_064), a subclone carrying mutations in KRAS, TP53, CDKN2C, and DIS3 seemed to have occurred during progression; however, both time points had low tumor purity that precluded such conclusion. In 1 patient who has not progressed (SMM_077) and was sampled twice at the SMM stage with an 8-year interval, no new alterations or increase in tumor burden was observed (Fig 3D), which suggests that the presence of clonal branching and heterogeneity is not enough to lead to disease progression but, instead, either the functional impact of genomic alterations or tumor cell-extrinsic factors could be the determining factor. In all 5 patients, we observed changes in the cancer cell fraction of subclones.
FIG 3.

Fish plots of four samples of smoldering multiple myeloma (SMM) with two serial samples at different time points. (A-C) Time points correspond to time of SMM diagnosis and time of progression to multiple myeloma (MM). (D) Both time points are at the SMM stage because the patient has not progressed to date. amp, amplification; del, deletion.
Identifying Genomic Predictors of Progression From SMM to MM
To define biomarkers of progression, we used a subset of patients (n = 85) who did not receive any treatment in clinical trial setting before progression to MM. Their baseline characteristics are reported in the Data Supplement. Median follow-up time for all patients was 6.2 years. Median TTP was 3.9 years. In this cohort, 53 patients (62%) have progressed, while 32 (38%) have remained asymptomatic. The genomic landscape is illustrated according to progression status in the Data Supplement.
Patients who harbored MYC aberrations (translocations or amplifications) had the shortest median TTP (8.4 v 51.6 months; P < .001) followed by those with MAPK pathway mutations (14.4 v 60 months; P < .001) and DNA repair pathway alterations (15.6 v 50.4 months; P = .004; Figs 4A-4C). Moreover, t(4;14), as well as del 1p, del 8p, and biallelic deletion events (including TP53, RB1, DIS3, MAX, and CDKN2A), were associated with shorter TTP (Data Supplement).
FIG 4.

Kaplan-Meier curves for analysis of time to progression in patients with (A) mitogen-activated protein kinase (MAPK) pathway mutations (KRAS and NRAS); (B) MYC alterations, including translocation and amplifications; and (C) DNA repair pathway alterations (deletion 17p, TP53, and ATM single nucleotide variants [red] compared to the absence of these alterations [blue]). (D) Forest plots of multivariable Cox regression of the genomic alterations and the clinical risk model with genetic features selected after bootstrap forward/backward variable selection with Mayo 2018 criteria.
MAPK pathway mutations and MYC alterations were associated with higher bone marrow infiltration at the time of diagnosis (P = .001 and < .001, respectively). MAPK pathway mutations were the only alteration that significantly correlated with M-protein levels (P < .001), while none of these high-risk genomic alterations correlated with free light chain ratio (Data Supplement). Of note, although not a high-risk feature, t(11;14) was associated with the light chain SMM subtype (P = .002) and lower M-protein levels compared to other primary events (Data Supplement).
Developing a Genomic Model for Prediction of Progression to MM
We searched for independent risk factors of progression from SMM to MM to develop a genomic model for prediction of progression (Data Supplement). Four genomic features were independent predictors of progression: MYC aberrations, alterations in the DNA repair and MAPK (KRAS, NRAS SNVs) pathways, and t(4;14) translocation (Data Supplement). Of note, in a multivariable model that accounted for the Mayo 2018 clinical risk stratification, all but t(4;14) were independent risk factors of progression (Fig 4D). Thus, our model is predicated upon these three genomic alterations.
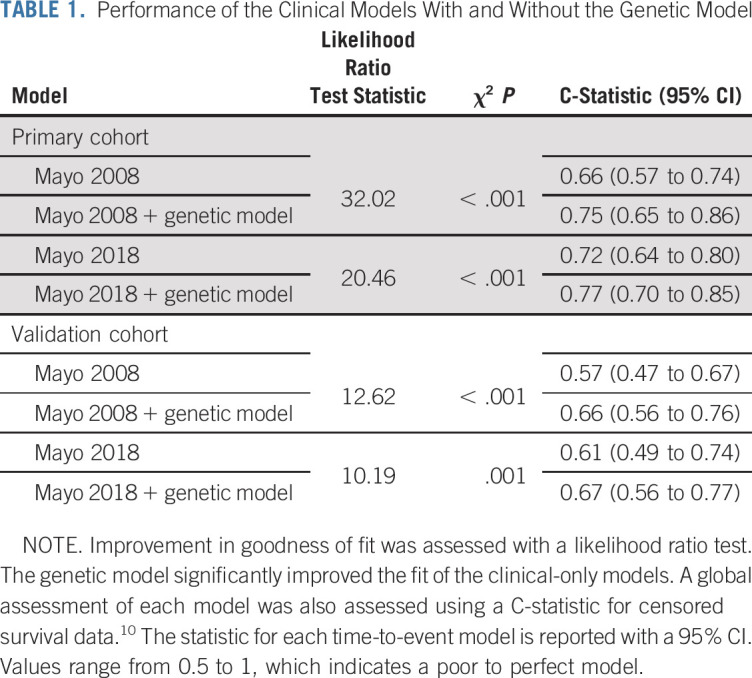
Patients with ≥ 1 high-risk genomic alterations had significantly shorter TTP (1.2 v 7.2 years, respectively; P < .001; Fig 5C). Moreover, patients with 0, 1, or ≥ 2 high-risk factors, had a significantly different TTP (Data Supplement), while patients with any of these high-risk alterations by the Mayo 2018 model (1.3 v 3.4 years; P = .006). Furthermore, clinically intermediate- and high-risk patients with these genomic alterations had significantly shorter TTP (2.6 v 9.3 years [P = .004] and 1.2 v 3.5 years [P = .001], respectively; Figs 5A and 5B), and our results were independent of the clinical model used (ie, Mayo 2008 or 2018; Data Supplement). Of note, high-risk genomic alterations were found in patients described as low risk by both models, in whom they conferred a significantly increased risk of progression (Data Supplement). Importantly, our genomic model improved the prediction of progression when added to the Mayo 2008 or 2018 models (C-statistic, 0.66 v 0.75 and 0.72 v 0.77, respectively; likelihood ratio test P < .001; Table 1; Data Supplement).
FIG 5.
Kaplan-Meier curves for analysis of time to progression of (A) clinically high-risk patients with (red) or without (blue) the high-risk genomic alterations, (B) clinically intermediate-risk patients with or without the high-risk genomic alterations, (C) patients with or without the high-risk genomic alterations in the Dana-Farber multicenter cohort, and (D) patients with or without the high-risk genomic alterations in the Mayo Clinic validation cohort.
TABLE 1.
Performance of the Clinical Models With and Without the Genetic Model
External Validation of the Genomic Prediction Model
To test the robustness and generalizability of our model, we validated it in an external cohort of 72 patients with SMM whose tumor DNA had been previously sequenced.7 Forty-seven patients in this cohort progressed to MM with a median TTP of 5 years. The cohort’s characteristics are listed in the Data Supplement. Again, we found that patients with any of the high-risk genomic alterations (n = 47) had a higher risk of progression (2.5 v 10 years; P = .001; Fig 5D). Similarly, patients with 0, 1, or ≥ 2 high-risk factors had different risks of progression (Data Supplement). Moreover, low-, intermediate-, and high-risk patients, as defined by the Mayo 2018 model, with high-risk genomic alterations had shorter TTP (9.7 v not reached [P = .028] and 2.3 v 6.9 years [P = .01], respectively; Data Supplement). These findings held true when we used the Mayo 2008 model as well (Data Supplement). Importantly, in a multivariable analysis that accounted for clinical risk group in this cohort, the genomic model was an independent risk factor of progression; when combined with the 2008 or 2018 clinical models, the genomic model performed better than the clinical model alone (C-statistic, 0.57 v 0.66 and 0.61 v 0.67, respectively; likelihood ratio test P < .001 and .001; Table 1; Data Supplement).
Mutational Signatures in SMM and Their Impact on Progression
We analyzed our WES data for specific mutational signatures that play an important role in myeloma pathogenesis and development: aging (COSMIC SBS1 and SBS5), adenosine-induced deaminase (AID), and apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) signatures. As expected, the majority of mutations were associated with aging signatures, while there was approximately equal contribution of AID (n = 217) and APOBEC (n = 250) signatures. Among myeloma recurrently mutated genes, the majority of events were attributable to aging signatures. However, we observed APOBEC-associated mutations in 14 MM driver genes, and AID-associated mutations in 11 (Data Supplement). Of note, all 3 mutations in ZNF292 were attributed to APOBEC, and t(14;16) had a significantly higher number of APOBEC mutations compared with the rest of the cohort (P = .005).
AID-associated mutations were observed in most of the patients, with no significant difference between progressors and nonprogressors (P > 0.99; Data Supplement). However, we found APOBEC to be significantly enriched in patients who progressed, after accounting for total mutation burden (P = .029; Data Supplement). Patients with versus those without APOBEC-associated mutations and those with more than the median number of APOBEC mutations (2.5) had shorter TTP (Data Supplement). Of note, although approximately 40% of APOBEC-associated mutations were clonal, patients with > 50% subclonal APOBEC-associated mutations were enriched for progressors (P = .046). Although APOBEC can be an early event in certain patients, these data indicate that continuous activity may underlie disease progression.
DISCUSSION
In this study, we leveraged NGS technology to analyze diagnostic tumor samples of 214 patients with SMM. We observed that SMM has similar genetic makeup to newly diagnosed MM, with similar mutation density and clonal heterogeneity. Although most of the SCNAs were clonal, del 1p and del 17p were mainly subclonal, which suggests that not all SCNAs are acquired early in a single catastrophic event and that in some cases, the clonal CNA profile could reflect the serial acquisition of certain SCNAs that eventually grew into clonal status. Indeed, in 3 of our patients with serial samples available, some clonal SCNAs at progression were subclonal at SMM diagnosis. On the other hand, SNVs that affect the most common MM drivers were mainly subclonal, which suggests that they are acquired later during disease progression. Nevertheless, mutations in KLHL6, TP53, DIS3, and MAX were mostly clonal, which indicates that in certain instances, driver mutations can perhaps be acquired early and drive clonal expansion.
Taken together, these findings indicate that MM disease progression, from tumorigenesis all the way to SMM and overt MM, is likely governed by the serial acquisition of genetic alterations, most of which will have been acquired by the time of SMM diagnosis. It should, however, be noted that in all 5 patients with serial samples, we found evidence of clonal evolution, with subclonal cancer cell fractions changing over time, which argues against the existence of a static model of progression in which the exact same clonal composition is observed in both the SMM and the MM stage.8 On the basis of our findings, we can expect some new driver alterations to be acquired between SMM diagnosis and progression to MM in a few patients; however, for the majority of patients, the tumor’s genetic makeup will have been largely shaped by the time of SMM diagnosis. This right-skewed model of MM genetic evolution, whereby the bulk of genetic alterations have already occurred by the time of SMM diagnosis, allows for reliable prognostication on the basis of genetic biomarkers discovered at SMM diagnosis.
Given these observations, we asked whether we could leverage diagnostic SMM samples to identify genomic predictors of progression that reflect an aggressive underlying biology and act independently of tumor burden. Specifically, we analyzed 85 untreated patients, excluding those who had been treated on SMM clinical trials. We found that mutations affecting genes in the MAPK and DNA repair pathway, as well as MYC translocations or amplifications, were all associated with a higher risk of progression to MM, even after accounting for the two currently used clinical risk stratification models. Namely, patients with SMM who carried any of the aforementioned alterations were at higher risk of progression compared with those in the same clinical risk group without them. Furthermore, we successfully validated our model in an external cohort of patients with SMM. In both cohorts, the genetic model improved the prediction of progression when added to the current clinical models, as assessed by both a likelihood ratio test and the C-statistic, which suggests that these genetic biomarkers are robust and can be reliably used for improved prognostication.
In an era where clinical-grade NGS is available in many centers, we could envision the stratification of patients with SMM being based on both clinical and genomic biomarkers. One of the limitations of the current clinical models is that they do not accurately predict progression risk in SMM patients. Our model helped identify genetic alterations that define a biologically aggressive subtype and are as important a predictor as increased tumor burden. And while MAPK/DNA repair pathway alterations and MYC aberrations should certainly be part of the prognostic genomic features, on the basis of the results from this and other studies,7 there are more features that seem to be significant predictors of progression, which suggests that larger cohorts are necessary to assess their impact. Such candidates include gene expression signatures in SMM,9 del 1p, del 8p, and APOBEC activity.
In conclusion, sequencing a large cohort of patients with SMM has allowed us to understand that SMM is a genetically mature tumor with slight differences from overt myeloma, which suggests a right-skewed model of genetic evolution from MGUS to MM whereby most driver genetic alterations have already occurred by the SMM stage. Therefore, we believe that genomic profiling of patients’ tumors at the time of SMM diagnosis represents an improved strategy for identifying patients at high risk of progression who could benefit from early intervention.
ACKNOWLEDGMENT
We thank all the patients and their families for their contributions to this study.
PRIOR PRESENTATION
Presented at the American Society of Hematology 59th Annual Meeting and Exposition, Atlanta, GA, December 9-12, 2017, and 17th International Myeloma Workshop, Boston, MA, September 12-15, 2019.
SUPPORT
Supported by National Institutes of Health grant R01 CA 205954, a Multiple Myeloma Research Foundation-Perelman Prevention Program grant, a Leukemia and Lymphoma Society Specialized Center of Research grant, a Stand Up To Cancer Dream Team grant, the Adelson Medical Research Foundation, and a Cancer Research UK Early Detection Program grant.
See accompanying editorial on page 2363
AUTHOR CONTRIBUTIONS
Conception and design: Mark Bustoros, Romanos Sklavenitis-Pistofidis, Karma Salem, Jacob Laubach, Tineke Casneuf, Paul Richardson, Meletios-Athanasios Dimopoulos, P. Leif Bergsagel, Salomon Manier, Gad Getz, Irene M. Ghobrial
Financial support: P. Leif Bergsagel, Irene M. Ghobrial
Administrative support: Mark Bustoros, Carl Jannes Neuse, Irene M. Ghobrial
Provision of study material or patients: Mark Bustoros, Efstathis Kastritis, Jacob Laubach, Kenneth C. Anderson, Meletios-Athanasios Dimopoulos, Kwee Yong, Irene M. Ghobrial
Collection and assembly of data: Mark Bustoros, Jihye Park, Karma Salem, Yu-Tzu Tai, Tarek H. Mouhieddine, Cody Boehner, Carl Jannes Neuse, Mahshid Rahmat, Shaji Kumar, Efstathis Kastritis, Elizabeth A. Morgan, Paul Richardson, Kenneth C. Anderson, Meletios-Athanasios Dimopoulos, Kwee Yong, P. Leif Bergsagel, Selina J. Chavda, Salomon Manier, Irene M. Ghobrial
Data analysis and interpretation: Mark Bustoros, Romanos Sklavenitis-Pistofidis, Jihye Park, Robert Redd, Benny Zhitomirsky, Andrew J. Dunford, Shankara Anand, Liudmila Elagina, Carl Jannes Neuse, Justin Cha, Amaro Taylor-Weiner, Eliezer Van Allen, Shaji Kumar, Efstathis Kastritis, Ignaty Leshchiner, Paul Richardson, Nikhil C. Munshi, Kenneth C. Anderson, Lorenzo Trippa, François Aguet, Chip Stewart, P. Leif Bergsagel, Salomon Manier, Gad Getz, Irene M. Ghobrial
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Genomic Profiling of Smoldering Multiple Myeloma Identifies Patients at a High Risk of Disease Progression
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Mark Bustoros
Honoraria: Takeda Pharmaceuticals, DAVA Oncology
Consulting or Advisory Role: Takeda Pharmaceuticals
Travel, Accommodations, Expenses: Takeda Pharmaceuticals, DAVA Oncology
Amaro Taylor-Weiner
Employment: PathAI
Stock and Other Ownership Interests: PathAI
Eliezer Van Allen
Stock and Other Ownership Interests: Syapse, Tango Therapeutics, Genome Medical, Microsoft, Ervaxx
Consulting or Advisory Role: Syapse, Roche, Third Rock Ventures, Takeda Pharmaceuticals, Novartis, Genome Medical, InVitae, Illumina, Tango Therapeutics, Ervaxx, Janssen Pharmaceuticals
Speakers’ Bureau: Illumina
Research Funding: Bristol-Myers Squibb, Novartis
Patents, Royalties, Other Intellectual Property: Patent on discovery of retained intron as source of cancer neoantigens (Inst), patent on discovery of chromatin regulators as biomarkers of response to cancer immunotherapy (Inst), patent on clinical interpretation algorithms using cancer molecular data (Inst)
Travel, Accommodations, Expenses: Roche, Genentech
Shaji Kumar
Consulting or Advisory Role: Takeda Pharmaceuticals (Inst), Janssen Oncology (Inst), Amgen (Inst), AbbVie (Inst), Merck (Inst), Adaptive Biotechnologies, Celgene (Inst), Genentech (Inst), Roche (Inst), Oncopeptides, Kite Pharma (Inst), Genecentrix, Molecular Partners (Inst), Bluebird Bio (Inst)
Research Funding: Celgene (Inst), Takeda Pharmaceuticals (Inst), AbbVie (Inst), Novartis (Inst), Sanofi (Inst), Janssen Oncology (Inst), Merck (Inst), Kite Pharma (Inst), MedImmune (Inst), Roche (Inst), Genentech (Inst), TeneoBio (Inst), Carsgen Therapeutics (Inst)
Efstathis Kastritis
Honoraria: Amgen, Genesis Pharma, Janssen Oncology, Takeda Pharmaceuticals, Prothena, Pfizer
Consulting or Advisory Role: Amgen, Janssen Oncology, Takeda Pharmaceuticals, Genesis Pharma, Prothena, Pfizer
Research Funding: Janssen Oncology (Inst), Amgen (Inst)
Travel, Accommodations, Expenses: Janssen Oncology, Genesis Pharma, Takeda Pharmaceuticals, Pfizer
Ignaty Leshchiner
Consulting or Advisory Role: PACT Pharma
Jacob Laubach
Research Funding: AbbVie (Inst), Bristol-Myers Squibb (Inst), Genentech (Inst), Janssen Research & Development (Inst)
Tineke Casneuf
Employment: Janssen Pharmaceutica NV
Stock and Other Ownership Interests: Johnson & Johnson
Patents, Royalties, Other Intellectual Property: Two patent applications by Johnson & Johnson with a symbolic $1 as compensation
Travel, Accommodations, Expenses: Janssen Pharmaceutica NV
Paul Richardson
Consulting or Advisory Role: Celgene, Janssen Pharmaceuticals, Takeda Pharmaceuticals, Karyopharm Therapeutics, Oncopeptides, Sanofi, Jazz Pharmaceuticals
Research Funding: Celgene (Inst), Takeda Pharmaceuticals (Inst), Bristol-Myers Squibb (Inst), Oncopeptides (Inst),
Nikhil C. Munshi
Stock and Other Ownership Interests: OncoPep
Consulting or Advisory Role: Celgene, Takeda Pharmaceuticals, Janssen Pharmaceuticals, OncoPep, AbbVie, Adaptive Biotechnologies, Amgen, BeiGene, Karyopharm Therapeutics, Bristol-Myers Squibb
Patents, Royalties, Other Intellectual Property: OncoPep
Kenneth C. Anderson
Stock and Other Ownership Interests: C4 Therapeutics, OncoPep
Consulting or Advisory Role: Celgene, Millennium Pharmaceuticals, Gilead Sciences, Bristol-Myers Squibb, Janssen Oncology, Sanofi, Tolero Pharmaceuticals, Precision Biosciences
Patents, Royalties, Other Intellectual Property: C4 Therapeutics, OncoPep
Lorenzo Trippa
Consulting or Advisory Role: Galera Therapeutics
François Aguet
Research Funding: Calico Life Sciences
Patents, Royalties, Other Intellectual Property: Inventor on a patent application related to TensorQTL
Meletios-Athanasios Dimopoulos
Honoraria: Amgen, Celgene, Takeda Pharmaceuticals, Janssen-Cilag, Bristol-Myers Squibb
Consulting or Advisory Role: Amgen, Janssen-Cilag, Takeda Pharmaceuticals, Celgene, Bristol-Myers Squibb
Kwee Yong
Honoraria: Janssen-Cilag, Takeda Pharmaceuticals, Sanofi, Amgen, Roche, Genentech
Consulting or Advisory Role: Janssen-Cilag
Speakers’ Bureau: Takeda Pharmaceuticals, Sanofi
Research Funding: Amgen, Autolus, Takeda Pharmaceuticals (Inst), Janssen-Cilag, Sanofi
Travel, Accommodations, Expenses: Takeda Pharmaceuticals
P. Leif Bergsagel
Consulting or Advisory Role: Isis Pharmaceuticals, Celgene, Janssen Pharmaceuticals, GlaxoSmithKline
Travel, Accommodations, Expenses: Celgene
Salomon Manier
Consulting or Advisory Role: Amgen (Inst), Celgene (Inst), Janssen Oncology (Inst)
Research Funding: Celgene (Inst), Janssen Oncology (Inst)
Gad Getz
Research Funding: IBM (Inst), Pharmacyclics (Inst)
Patents, Royalties, Other Intellectual Property: Patents related to bioinformatic tools (Inst), royalties from licensing tools
Irene M. Ghobrial
Honoraria: Celgene, Bristol-Myers Squibb, Takeda Pharmaceuticals, Amgen, Janssen Pharmaceuticals, Karyopharm Therapeutics, Cellectar, Adaptive Biotechnologies, Sanofi, Medscape
Consulting or Advisory Role: Bristol-Myers Squibb, Novartis, Amgen, Takeda Pharmaceuticals, Noxxon Pharma, Celgene, Sanofi, Genentech, GlaxoSmithKline, GNS Healthcare, Karyopharm Therapeutics, Adaptive Biotechnologies, Janssen Pharmaceuticals, Medscape, AbbVie
Travel, Accommodations, Expenses: Bristol-Myers Squibb, Novartis, Onyx, Millennium Pharmaceuticals, Celgene, Takeda Pharmaceuticals, Janssen Oncology
No other potential conflicts of interest were reported.
REFERENCES
- 1.Weiss BM, Abadie J, Verma P, et al. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009;113:5418–5422. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debes-Marun CS, Dewald GW, Bryant S, et al. Chromosome abnormalities clustering and its implications for pathogenesis and prognosis in myeloma. Leukemia. 2003;17:427–436. doi: 10.1038/sj.leu.2402797. [DOI] [PubMed] [Google Scholar]
- 3.Kyle RA, Remstein ED, Therneau TM, et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N Engl J Med. 2007;356:2582–2590. doi: 10.1056/NEJMoa070389. [DOI] [PubMed] [Google Scholar]
- 4.Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 5.Dispenzieri A, Kyle RA, Katzmann JA, et al. Immunoglobulin free light chain ratio is an independent risk factor for progression of smoldering (asymptomatic) multiple myeloma. Blood. 2008;111:785–789. doi: 10.1182/blood-2007-08-108357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakshman A, Rajkumar SV, Buadi FK, et al. Risk stratification of smoldering multiple myeloma incorporating revised IMWG diagnostic criteria. Blood Cancer J. 2018;8:59. doi: 10.1038/s41408-018-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. doi: 10.1038/s41375-019-0543-4. Misund K, Keane N, Stein CK, et al: MYC dysregulation in the progression of multiple myeloma. Leukemia 34:322-326, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolli N, Maura F, Minvielle S, et al. Genomic patterns of progression in smoldering multiple myeloma. Nat Commun. 2018;9:3363. doi: 10.1038/s41467-018-05058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhodapkar MV, Sexton R, Waheed S, et al. Clinical, genomic, and imaging predictors of myeloma progression from asymptomatic monoclonal gammopathies (SWOG S0120) Blood. 2014;123:78–85. doi: 10.1182/blood-2013-07-515239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uno H, Cai T, Pencina MJ, et al. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]



