Abstract
Background/Aim: To investigate the function of chromobox 2 (CBX2) in oesophageal squamous cell carcinoma (OSCC). Materials and Methods: We used real-time quantitative reverse transcription PCR (qRT-PCR) and immunohistochemistry to determine CBX2 expression levels in 13 human OSCC cell lines and clinical specimens of two independent cohorts of patients with OSCC. Results: PCR array analysis revealed that CBX2 was co-ordinately expressed with WNT5B in OSCC cell lines. RT-qPCR analysis of clinical samples revealed a high tumour-specific CBX2 expression compared with normal oesophageal tissues. High CBX2 expression was significantly associated with shorter disease-specific survival, hematogenous recurrence, and overall recurrence. Analysis of tissue microarrays of one cohort revealed that patients with higher CBX2 levels tended to have a shorter disease-specific survival. Conclusion: CBX2 overexpression in OSCC tissues may serve as a novel biomarker for predicting survival and hematogenous recurrence.
Keywords: Biomarker, chromobox 2 (CBX2), oesophageal squamous cell carcinoma, recurrence
The main histopathological subtypes of oesophageal carcinoma, the sixth leading cause of cancer-related mortality worldwide (1), are adenocarcinoma and oesophageal squamous cell carcinoma (OSCC). The latter is the predominant subtype in Asia and Africa (2). OSCC is generally associated with poor prognosis, mainly because of its high potential to metastasize, which accounts for its dismal overall survival rate of <40%, despite radical treatment (3).
A primary goal of efforts to improve the management of OSCC and patients’ outcomes is to develop methods that accurately predict the risk of recurrence and survival after curative oesophageal resection (4,5). Specific biomarkers that predict the recurrence pattern of OSCC, particularly for hematogenous recurrence after radical treatment, will likely contribute to improving patients’ outcomes and are therefore the focus of our research.
We the aim to identify novel biomarkers, we first focused on the chromobox 2 (CBX2) gene, a member of the chromobox (CBX) gene family, because of its reported association with certain cancers (6-8). CBX2 encodes a subunit of the polycomb group (PcG), whose members form complexes that mediate homeostasis (9). These proteins regulate the expression of numerous genes that control the maintenance, differentiation and proliferation of adult stem cells and cancer cells (10). Biochemical characterization led to the categorization of PcG complexes into two subtypes: Polycomb repressive complex (PRC) 1 and PRC2 (6,11,12). The CBX gene family comprises CBX2, CBX4, CBX6, CBX7 and CBX8, which encode components of PRC1 (11). PRC1 members are associated with tumourigenesis. For example, CBX4 plays important roles in cellular proliferation and in the repair of DNA damage in certain human tumours (13,14). CBX6 was identified as a susceptibility loci by a genome-wide association study on bladder cancer (15). Numerous studies have shown that CBX7 is specifically expressed in human tumours, including brain cancer (16), colon cancer (17), lung cancer (18) and gastric cancer (19); and CBX8 is involved in the pathogenesis of leukaemia (20). Although CBX2 has been studied in multiple cancers (21-25), its role in OSCC is unknown.
Here, we aimed to assess whether CBX2 will serve as a predictive biomarker for the prognosis of patients with OSCC. For this purpose, we investigated CBX2 expression in cell lines derived from OSCCs and in patients’ tissue samples. To determine whether CBX2 expression correlates with disease recurrence and survival, we performed immunohistochemical analyses of tumour tissue microarrays (TMAs) prepared from an independent cohort of patients with OSCC.
Materials and Methods
Ethics. This study conformed to the ethical guidelines of the World Medical Association Declaration of Helsinki–Ethical Principles for Medical Research Involving Human Subjects and was approved by the Institutional Review Board of Nagoya University, Japan (approval number: 2014-0044). Written informed consent for the use of clinical samples and data, as required by the Institutional Review Board, was granted by all patients.
Sample collection. The human OSCC cell lines TE1, TE2, TE3, TT, TTn, and a non-tumorigenic epithelial cell line Het-1A were obtained from the American Type Culture Collection (Manassas, VA, USA). The KYSE510, KYSE590, KYSE890, KYSE1170, KYSE1260 and KYSE1440 cell lines were obtained from the Japanese Collection of Research Bio Resources Cell Bank (Osaka, Japan). The NUEC2 and WSSC cell lines were established at Nagoya University (26). Cells were cultured in RPMI-1640 medium supplemented with 10% foetal bovine serum and maintained at 37˚C in an atmosphere containing 5% CO2.
We acquired 189 primary OSCC and corresponding normal adjacent tissues from patients who underwent radical oesophageal resection at Nagoya University Hospital between October 2001 and January 2016. The tumour was radically resected when a patient was pathologically diagnosed with stage I, II or III disease. Tissue samples were immediately frozen and stored at −80˚C upon resection, and specimens were confirmed as OSCC according to the histological classification criteria of the TNM Classification of Malignant Tumours, 8th Edition of the Union for International Cancer Control (UICC) (27). Unless contraindicated, fluorouracil combined with platinum-based neoadjuvant chemotherapy has been recommended for patients in Japan with clinical stages II-IV since 2006.
Real-time quantitative reverse transcription PCR (qRT-PCR) and PCR array analyses. The CBX2 mRNA expression levels of cell lines and tissue samples were analysed using RT-PCR with an ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) as previously described (28,29). A 186 base-pair product was amplified using the following primers: sense, 5’-GCAAGCTGGAGTACCTGGTC-3’ in CBX2 exon 2 and antisense 5’-ACATGGCAGTGAGCTTCCTT-3’ in CBX2 exon 4. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was determined as an internal standard, and the expression level of each sample was calculated as the value of CBX2 mRNA divided by that of GAPDH mRNA.
To identify genes co-ordinately expressed with CBX2 in OSCC cell lines, we used the Human Epithelial to Mesenchymal Transition (EMT) RT2 Profiler PCR Array (Qiagen, Hilden, Germany). This array includes 84 key genes that encode proteins with the following functions: transcription factor, extracellular matrix protein, and proteins involved in the EMT, cell differentiation, morphogenesis, growth, proliferation, migration, cytoskeleton, or associated with other signalling pathways (30).
Assessment of the clinical significance of CBX2 expression. Patients were stratified into high and low CBX2 expression groups using the median level of CBX2 mRNA as the cut-off value. We evaluated the correlations between high and low CBX2 mRNA expression; clinicopathological parameters; and outcomes, including disease-specific survival (DSS), disease-free survival (DFS), and recurrence pattern-specific survival.
Analysis of tissue microarrays. To prepare a validation cohort, primary OSCC tissues were collected from 177 patients who underwent oesophageal resection for OSCC at the Department of Thoracic Surgery, Akita University Hospital between 2000 and 2011 (31). These patients were not administered treatment before curative surgery. The tissue specimens were embedded in paraffin, and a TMA prepared at the Pathology Institute (Toyama, Japan) (32-34) was analysed using immunohistochemistry (IHC) as described below. TMA blocks were sectioned and incubated for 16 h at 4˚C with a rabbit anti-CBX2 polyclonal antibody (HPA023083, Atlas Antibodies, Stockholm, Sweden) diluted 1:100 in Antibody Diluent (Dako, Carpinteria, CA, USA). Sections were incubated with a biotinylated secondary antibody [SignalStain Boost IHC Detection Reagent (HRP, Rabbit); Cell Signaling Technology, Beverly, MA, USA] for 30 min. Antigen-antibody complexes were visualized using 3, 3’-diaminobenzidine (Nichirei, Tokyo, Japan) for 1 min. Two independent observers evaluated the specimens and assigned scores as follows: 3+ (intense cytoplasmic or nuclear staining in >30% of cells), 2+ (moderate cytoplasmic or nuclear staining in >10% of cells), 1+ (weak staining in <10% of cells), and 0 (no staining, negative).
Statistical analysis. The quantitative Mann–Whitney and the qualitative χ2 tests were employed to evaluate differences between groups. Correlations between two variables were assessed using Spearman’s rank correlation coefficient. The χ2 test was used to analyse the significance of associations between gene expression and clinicopathological parameters. DSS and DFS were calculated using the Kaplan–Meier method and analysed using a Cox proportional hazards model. Univariate regression analysis of potential prognostic factors was performed using a Cox proportional hazards model, and variables with p<0.05 were included in the final multivariate model (35). All statistical analyses were performed using JMP 14 software (SAS Institute Inc., Cary, NC, USA), and p<0.05 was considered to indicate a significant difference.
Results
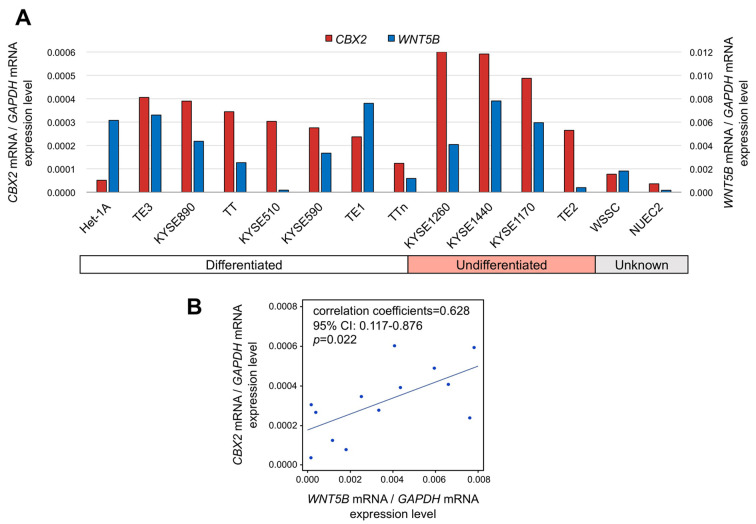
Expression of CBX2 and cancer-related genes in OSCC cell lines. To detect CBX2 mRNA levels in OSCC tissues and to investigate its potential function, we measured CBX2 mRNA expression in 13 human OSCC cell lines. Although absolute levels differed, CBX2 mRNA was present at higher levels in 12 of the OSCC cell lines than in the control non-tumourigenic epithelial cell line (Figure 1A). Significant differences in CBX2 mRNA levels were not detected among OSCC cell lines derived from metastases (TT, TTn, KYSE1170 and KYSE1260), primary tumours (p=0.364), or cell lines with different degrees of differentiation (p=0.454). When we performed PCR array analysis to identify cancer-related genes co-ordinately expressed with CBX2 in the OSCC cell lines, we found that only WNT5B mRNA levels correlated significantly with those of CBX2 (Figure 1B), which suggests a functional relationship.
Figure 1. Expression of CBX2 mRNA in human OSCC cell lines. A) qRT-PCR analysis of CBX2 mRNA expression in 13 OSCC cell lines and a nontumorigenic epithelial cell line (Het-1A). B) PCR array analysis. Correlation coefficients of WNT5B and CBX2 expression levels in OSCC cell lines.
Study subjects and CBX2 mRNA expression in OSCC tissues
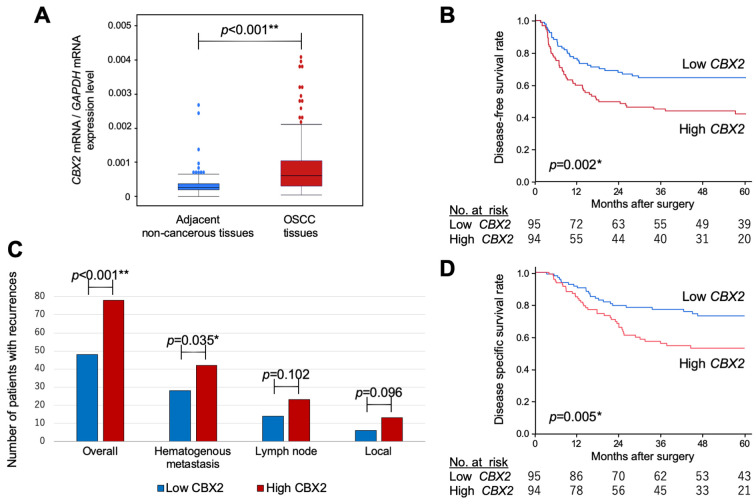
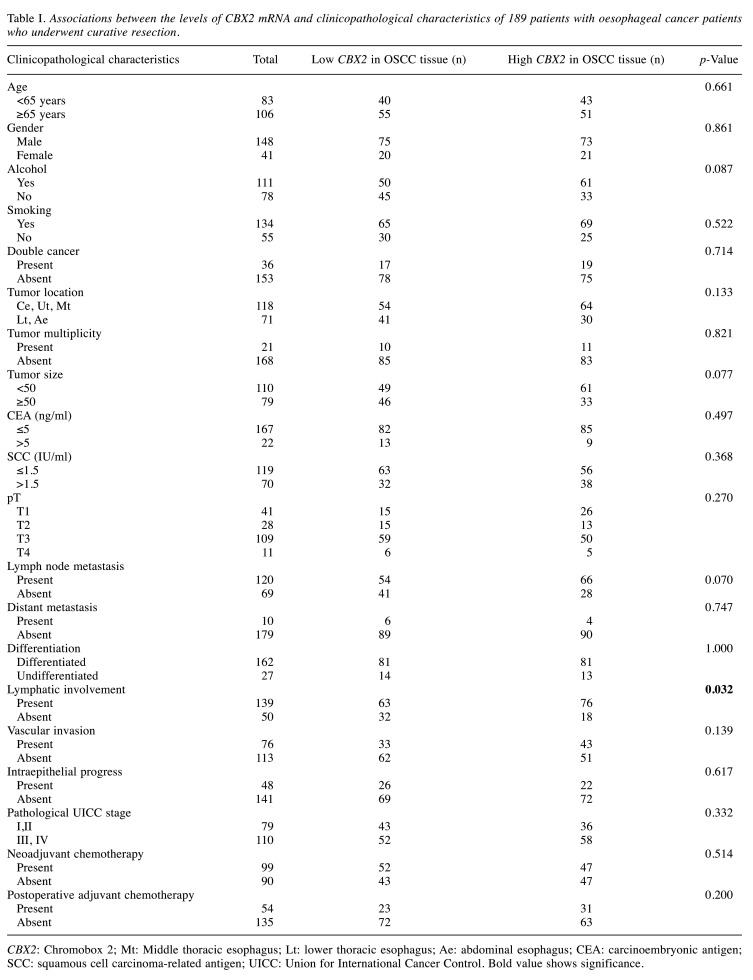
The median age of the entire cohort (n=189) was 66 years (range=44-84 years), and the female:male ratio was 41:148. Most patients (n=162) were diagnosed with differentiated OSCC and the remainder (27) with undifferentiated OSCC. The numbers of patients with OSCC pathological stage I, II, III, or IV were 36, 43, 76 and 34, respectively. Neoadjuvant or adjuvant chemotherapy was administered to 99 (52%) and 54 patients (29%), respectively. The median follow-up was 40.9 months, during which 83 patients (44%) experienced recurrence and 66 patients (35%) died because of OSCC. CBX2 mRNA levels in the 189 paired samples of primary OSCC and adjacent normal tissues were measured using qRT-PCR. CBX2 mRNA levels were significantly higher (p<0.001) in OSCC tissues than in adjacent normal oesophageal tissue in 147/189 (78%) patients (Figure 2A). With respect to lymphatic involvement, CBX2 mRNA levels in 139 (74%) samples with lymphatic involvement were significantly higher than those without [50 (26%); p=0.024]. With respect to vascular invasion, the levels of CBX2 mRNA with vascular invasion, were significantly higher than those without [76 (40%) vs. 113 (60%); p=0.035].
Figure 2. Analysis of CBX2 expression in clinical samples. A) CBX2 mRNA levels in 189 resected oesophageal squamous cell carcinoma tissues and adjacent noncancerous oesophageal mucosa. B) Kaplan–Meier analysis of disease-free survival as a function of high or low CBX2 expression. C) Analysis of recurrence patterns. Numbers of sites of initial recurrence in the high and low CBX2 expression groups. (D) Kaplan–Meier analysis of disease-specific survival as a function of high or low CBX2 expression.
Prognostic significance of CBX2 mRNA levels. To assess the potential prognostic value of CBX2 mRNA expression, patients were divided according to the median value of CBX2 mRNA levels, which served as the cut-off value. Analysis of the correlations between CBX2 expression and clinicopathological factors revealed that high CBX2 mRNA levels were significantly and specifically associated with lymphatic involvement (p=0.032; Table I).
Table I. Associations between the levels of CBX2 mRNA and clinicopathological characteristics of 189 patients with oesophageal cancer patients who underwent curative resection.
CBX2: Chromobox 2; Mt: Middle thoracic esophagus; Lt: lower thoracic esophagus; Ae: abdominal esophagus; CEA: carcinoembryonic antigen; SCC: squamous cell carcinoma-related antigen; UICC: Union for International Cancer Control. Bold value shows significance.
Survival analyses demonstrated a significantly lower 5-year DFS rate for patients with high vs. low CBX2 mRNA levels [hazard ratio (HR)=1.98, 95% confidence interval (CI)=1.28-3.06, p=0.002; Figure 2B]. We therefore determined whether CBX2 expression correlated with recurrence patterns. Of the 189 patients, 86 (46%) experienced postoperative recurrence at 128 initial total recurrence sites. High CBX2 mRNA levels were significantly associated with disease-specific (p<0.001; Figure 2C) and hematogenous recurrences (p=0.035; Figure 2C), but not with lymph node or local recurrence. Moreover, the 5-year DSS rate was lower for patients with high tumour-specific CBX2 mRNA levels (HR=2.01, 95%CI=1.22-3.31, p=0.005; Figure 2D).
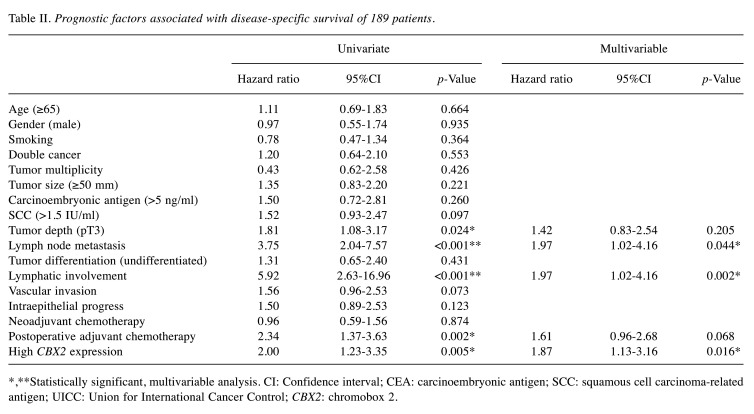
Univariate analysis identified tumour depth (pT3), lymphatic involvement, lymph node metastasis, postoperative adjuvant chemotherapy, and tumour-specific CBX2 expression as significant indicators of poor DSS (Table II). Multivariate analysis revealed that lymph node metastasis (HR=1.97, 95%CI=1.02-4.16; p=0.044), lymphatic involvement (HR=1.97, 95%CI=1.02-4.16; p=0.002), and CBX2 expression (HR=1.87, 95%CI=1.13-3.16; p=0.016) were the only independent prognostic factors.
Table II. Prognostic factors associated with disease-specific survival of 189 patients.
*,**Statistically significant, multivariable analysis. CI: Confidence interval; CEA: carcinoembryonic antigen; SCC: squamous cell carcinoma-related antigen; UICC: Union for International Cancer Control; CBX2: chromobox 2.
IHC analysis of CBX2 expression. To evaluate the significance of CBX2 expression associated with patients’ clinicopathological characteristics, we performed IHC using TMAs of tumour and normal tissue samples collected from 177 patients undergoing curative surgery for OSCC. The patient population comprised 24 women and 153 men (66±8.2 years, mean±SD; range=38-82 years) with pathologically diagnosed differentiated (n=120) or undifferentiated (n=57) OSCC. Disease stages were as follows: 10, 44, 105 and 18 patients with stage I, II, III or IV OSCC, respectively.
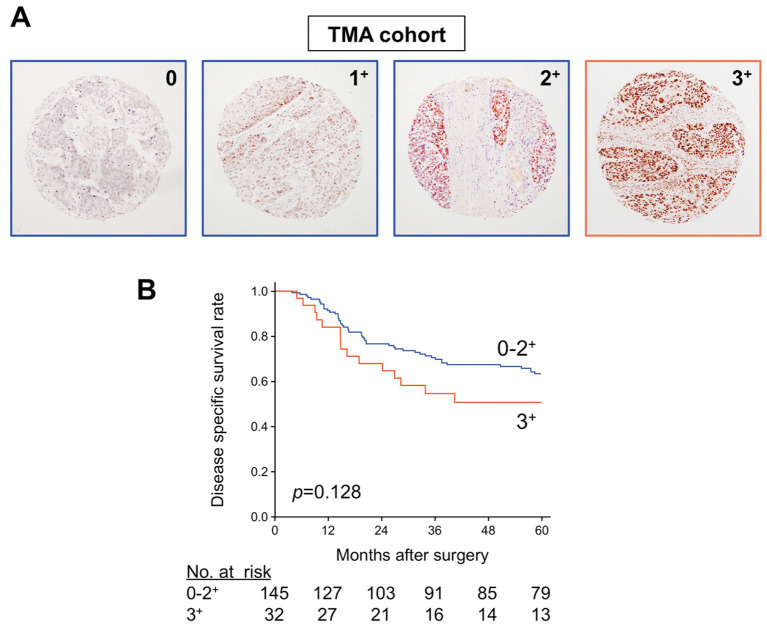
IHC analysis of CBX2 expression was evaluated using a semiquantitative scoring system described in the Methods section that evaluates the localization, intensity and distribution of protein expression. Representative photomicrographs of specimens with negative (0) and positive (3+, 2+, 1+) expression are shown in Figure 3A. Of the 177 samples, 37 (20.9%) scored 0, 58 (32.8%) scored 1+, 50 (28.2%) scored 2+, and 32 (18.1%) scored 3+.
Figure 3. Analysis of CBX2 protein expression in OSCC and correlation with survival outcomes. A) Representative images of immunohistochemical analyses of CBX2 expression using a TMA prepared from the first validation cohort. Examples of negative (0) and positive (1+, 2+, and 3+) expression are shown (100× magnification). B) Kaplan–Meier analysis of disease-specific survival of the validation cohort (n=177) stratified according to strongly positive (3+; n=32) or not strongly positive (0, 1+, and 2+; n=145) CBX2 protein expression.
To assess the prognostic value of CBX2 expression, patients were divided into a CBX2-strongly positive group (scores 3+; n=32) and a CBX2-negative or weakly positive group (score 0, 1+, or 2+; n=145), and DSS was compared using the Kaplan–Meier method. We found that the 5-year DSS rate of the CBX2-strongly positive group was lower than that of the CBX2-negative or weakly positive group, though the difference was not statistically significant (HR=1.56, 95%CI=0.85-2.71, p=0.128; Figure 3B).
Discussion
In the present study, we sought to understand the functions of CBX2 in OSCC and to determine the prognostic value of CBX2 mRNA and protein expression in two independent patient cohorts. Here, our study of the clinical significance of CBX2 expression in OSCC revealed the potential value of CBX2 as a predictive biomarker of hematogenous recurrence after radical esophagectomy.
Specifically, we found that CBX2 mRNA was present at higher levels in 12 OSCC cell lines vs. a non-tumourigenic epithelial cell line. These results are consistent with our findings that CBX2 mRNA was expressed at higher levels in OSCC tissues compared with adjacent normal tissue. Further, high CBX2 expression levels in tumours were significantly associated with poorer DFS and DSS.
Multivariate analysis revealed that lymph node metastasis, lymphatic involvement, and CBX2 expression were independent prognostic factors for DSS and that higher differential tumour-specific CBX2 levels were associated with an increase in overall and hematogenous recurrences. These findings suggest that CBX2 expression was an independent risk factor of OSCC, which supports the conclusion that it serves as a biomarker specific for hematogenous recurrence.
CBX2 encodes a component of the polycomb multiprotein complex. In 2014, Clermont et al. (32) initially identified an oncogenic role for CBX2 through a meta-analysis of gene transcription in human cancers. Aberrantly high CBX2 levels are expressed in phenotypically diverse tumours such as breast cancer, hepatocellular cancer, oral cancer, ovarian cancer and lung cancer, for which it serves as a biomarker (21-25). Conversely, little is known about the expression or function of CBX2 in OSCC. For example, we identified only one study on the relationship between oesophageal cancer and CBX2 mRNA expression (33). This transcriptomic study of OSCC identified CBX2 as one of nine possible diagnostic markers in The Cancer Genome Atlas data (33). Thus, our present study illuminates significant clinical and functional associations between CBX2 expression and OSCC.
PcG complexes, which inhibit the transcription of specific sets of genes through chromatin modification, maintain cell phenotypes during development (5). Certain PcG components contribute to the regulation of cell proliferation and mediate oncogenesis and tumour progression (5). We reasoned therefore that CBX2 regulates the expression of multiple genes involved in these processes, among which the EMT mediates the hematogenous dissemination of carcinomas (36). Moreover, evidence indicates that CBX2 regulates autophagy, apoptosis, and the EMT (8). Overexpression of WNT5B, which was co-ordinately expressed with CBX2 in our mRNA microarray analysis of OSCC, contributes to the EMT and is associated with increased hematogenous metastasis in lung cancer and pancreatic cancer (37). Therefore, CBX2 may interact with the products of other genes (e.g. SOX9, etc.) in the WNT5B cascade to promote hematogenous dissemination of OSCC (38).
We used TMAs to analyse a second cohort to determine if CBX2 expression is associated with DSS. Although TMAs enable the analysis of multiple samples, it can only be evaluated at three sampling times (39,40). Further, there is a risk of bias because of tumour characteristics at the sampling site (e.g. tumour margins, immune cell invasion, necrosis, etc.), which may affect the results. Although our analysis did not detect a statistically significant difference in DSS; CBX2 expression tended to be involved in poorer DSS (Figure 3B).
Our results may have clinical applications in two areas. First, patients with high tumour CBX2 mRNA levels may benefit from postoperative surveillance with a focus on early detection of hematogenous recurrence. Contrast-enhanced computerized tomography, which is the most common imaging technique employed in postoperative follow-up, may not detect early metastasis to the liver and bone, indicating that multimodal imaging surveillance, including gadolinium ethoxybenzyl diethylenetriamine penta-acetic acid-enhanced magnetic resonance imaging and positron emission tomography, may be required to detect early hematogenous recurrence. Second, CBX2 mRNA expression in biopsy samples or surgical specimens may serve as a reference measure in the design of perioperative therapy. For example, in a study of 418 patients with cancer of the oesophagus or gastroesophageal junction, neoadjuvant chemotherapy significantly reduced hematogenous recurrence after radical surgery compared with surgery alone (41), suggesting that active neoadjuvant chemotherapy may improve the prognosis of patients with high CBX2 expression.
Although the application of adjuvant chemotherapy is controversial, it is considered beneficial for specific groups of patients with OSCC (42). Therefore, measurement of CBX2 mRNA levels in clinical samples may assist clinicians to decide whether to administer adjuvant chemotherapy. Chemotherapeutic strategies based on recurrence patterns are currently being established for many cancers (43,44), but they are not yet available for OSCC. In the future, tumour-specific CBX2 expression levels may provide useful information that will guide treatment selection when such strategies become available for oesophageal cancer.
There are several limitations to the present study. For example, the study was retrospective; we used the median value as the cut-off for stratification of patients according to high or low CBX2 mRNA levels, and an optimal cut-off value derived from larger-scale studies will be required for clinical application. Further, the interpretation of the results of the subgroup analyses may have been affected by sample size. Thus, the efficacy of neoadjuvant or adjuvant chemotherapy will require further analysis of more patients.
In conclusion, differential CBX2 mRNA overexpression in OSCC tissues may serve as a biomarker for predicting hematogenous recurrence after radical surgery.
Conflicts of Interest
The Authors declare no conflicts of interest associated with this manuscript.
Authors’ Contributions
SU and MK conceived the study concept and design, analysed data and wrote the manuscript. MK, KS, DS, SU, MK, TN and YK contributed to data acquisition and interpretation. SU contributed to statistical analysis. KS, DS, SU, MK, TN and YK revised the draft. All Authors have read and approved the final version of the manuscript.
Acknowledgements
The Authors thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–386. doi: 10.1002/ijc.29210. PMID: 25220842. DOI: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra GK, Yanala U, Ravipati A, Follet M, Vijayakumar M, Are C. Global trends in esophageal cancer. J Surg Oncol. 2017;115:564–579. doi: 10.1002/jso.24592. PMID: 28320055. DOI: 10.1002/jso.24592. [DOI] [PubMed] [Google Scholar]
- 3.Chapman BC, Weyant M, Hilton S, Hosokawa PW, McCarter MD, Gleisner A, Nader ND, Gajdos C. Analysis of the National Cancer Database Esophageal Squamous Cell Carcinoma in the United States. Ann Thorac Surg. 2019;108:1535–1542. doi: 10.1016/j.athoracsur.2019.05.053. PMID: 15526259. DOI: 10.1016/j.athoracsur.2019.05.053. [DOI] [PubMed] [Google Scholar]
- 4.Kadam PD, Chuan HH. Erratum to: Rectocutaneous fistula with transmigration of the suture: a rare delayed complication of vault fixation with the sacrospinous ligament. Int Urogynecol J. 2016;27:505. doi: 10.1007/s00192-016-2952-5. PMID: 26811110. DOI: 10.1007/s00192-016-2952-5. [DOI] [PubMed] [Google Scholar]
- 5.Cohen C, Tessier W, Gronnier C, Renaud F, Pasquer A, Thereaux J, Gagniere J, Meunier B, Collet D, Piessen G, Mariette C. Salvage surgery for esophageal cancer: how to improve outcomes. Ann Surg Oncol. 2018;25:1277–1286. doi: 10.1245/s10434-018-6365-1. PMID: 29417405. DOI: 10.1245/s10434-018-6365-1. [DOI] [PubMed] [Google Scholar]
- 6.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. PMID: 12435631. DOI: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont PL, Sun L, Crea F, Thu KL, Zhang A, Parolia A, Lam WL, Helgason CD. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: Initial evidence of an oncogenic role. Br J Cancer. 2014;111:1663–1672. doi: 10.1038/bjc.2014.474. PMID: 15321827. DOI: 10.1038/bjc.2014.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler LJ, Watson ZL, Qamar L, Yamamoto TM, Post MD, Berning AA, Spillman MA, Behbakht K, Bitler BG. CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis. 2018;7:1–14. doi: 10.1038/s41389-018-0103-1. PMID: 21579024. DOI: 10.1038/s41389-018-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senthilkumar R, Mishra RK. Novel motifs distinguish multiple homologues of Polycomb in vertebrates: expansion and diversification of the epigenetic toolkit. BMC Genomics. 2009;10:549. doi: 10.1186/1471-2164-10-549. PMID: 19930571. DOI: 10.1186/1471-2164-10-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparmann A, Van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. PMID: 17060944. DOI: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 11.Vincenz C, Kerppola TK. Different polycomb group CBX family proteins associate with distinct regions of chromatin using nonhomologous protein sequences. Proc Natl Acad Sci USA. 2008;105:16572–16577. doi: 10.1073/pnas.0805317105. PMID: 18927235. DOI: 10.1073/pnas.0805317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. PMID: 12167701. DOI: 10.1128/mcb.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. PMID: 22078878. DOI: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Tang J, Liao D, Wang G, Zhang M, Sang Y, Cao J, Wu Y, Zhang R, Li S, Ding W, Zhang G, Kang T. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol. 2013;20:684–692. doi: 10.1245/s10434-013-3171-7. PMID: 23943028. DOI: 10.1245/s10434-013-3171-7. [DOI] [PubMed] [Google Scholar]
- 15.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, Real FX, Van Den Berg D, Matullo G, Baris D, Thun M, Kiemeney LA, Vineis P, De Vivo I, Albanes D, Purdue MP, Rafnar T, Hildebrandt MAT, Kiltie AE, Cussenot O, Golka K, Kumar R, Taylor JA, Mayordomo JI, Jacobs KB, Kogevinas M, Hutchinson A, Wang Z, Fu Y-P, Prokunina-Olsson L, Burdett L, Yeager M, Wheeler W, Tardon A, Serra C, Carrato A, Garcia-Closas R, Lloreta J, Johnson A, Schwenn M, Karagas MR, Schned A, Andriole GJ, Grubb R 3rd, Black A, Jacobs EJ, Diver WR, Gapstur SM, Weinstein SJ, Virtamo J, Cortessis VK, Gago-Dominguez M, Pike MC, Stern MC, Yuan J-M, Hunter DJ, McGrath M, Dinney CP, Czerniak B, Chen M, Yang H, Vermeulen SH, Aben KK, Witjes JA, Makkinje RR, Sulem P, Besenbacher S, Stefansson K, Riboli E, Brennan P, Panico S, Navarro C, Allen NE, Bueno-de-Mesquita HB, Trichopoulos D, Caporaso N, Landi MT, Canzian F, Ljungberg B, Tjonneland A, Clavel-Chapelon F, Bishop DT, Teo MTW, Knowles MA, Guarrera S, Polidoro S, Ricceri F, Sacerdote C, Allione A, Cancel-Tassin G, Selinski S, Hengstler JG, Dietrich H, Fletcher T, Rudnai P, Gurzau E, Koppova K, Bolick SCE, Godfrey A, Xu Z, Sanz-Velez JI, D Garcia-Prats M, Sanchez M, Valdivia G, Porru S, Benhamou S, Hoover RN, Fraumeni JFJ, Silverman DT, Chanock SJ. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–984. doi: 10.1038/ng.687. PMID: 20972438. DOI: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW, Lancini C, van Lohuizen M. In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell. 2013;23:660–676. doi: 10.1016/j.ccr.2013.03.030. PMID: 23680149. DOI: 10.1016/j.ccr.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Pallante P, Terracciano L, Carafa V, Schneider S, Zlobec I, Lugli A, Bianco M, Ferraro A, Sacchetti S, Troncone G, Fusco A, Tornillo L. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur J Cancer. 2010;46:2304–2313. doi: 10.1016/j.ejca.2010.05.011. PMID: 20542683. DOI: 10.1016/j.ejca.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Forzati F, Federico A, Pallante P, Abbate A, Esposito F, Malapelle U, Sepe R, Palma G, Troncone G, Scarfo M, Arra C, Fedele M, Fusco A. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122:612–623. doi: 10.1172/JCI58620. PMID: 22214847. DOI: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni S-J, Zhao L-Q, Wang X-F, Wu Z-H, Hua R-X, Wan C-H, Zhang J-Y, Zhang X-W, Huang M-Z, Gan L, Sun H-L, Dimri GP, Guo W-J. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-kappaB-miR-21 pathways. J Hematol Oncol. 2018;11:17. doi: 10.1186/s13045-018-0562-z. PMID: 29422082. DOI: 10.1186/s13045-018-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. PMID: 22094252. DOI: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen WY, Zhang XY, Liu T, Liu Y, Zhao YS, Pang D. Chromobox homolog 2 protein: A novel biomarker for predicting prognosis and taxol sensitivity in patients with breast cancer. Oncol Lett. 2017;13:1149–1156. doi: 10.3892/ol.2016.5529. PMID: 17921082. DOI: 10.3892/ol.2016.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao J, Tian Y, Wang C, Jiang K, Li R, Yao Y, Zhang R, Sun D, Liang R, Gao Z, Wang Q, Wang L. CBX2 regulates proliferation and apoptosis via the phosphorylation of YAP in hepatocellular carcinoma. J Cancer. 2019;10:2706–2719. doi: 10.7150/jca.31845. PMID: 31258779. DOI: 10.7150/jca.31845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parris TZ, Aziz L, Kovács A, Hajizadeh S, Nemes S, Semaan M, Chen CY, Karlsson P, Helou K. Clinical relevance of breast cancer-related genes as potential biomarkers for oral squamous cell carcinoma. BMC Cancer. 2014;14:1–11. doi: 10.1186/1471-2407-14-324. PMID: 14712407. DOI: 10.1186/1471-2407-14-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gui T, Bai H, Zeng J, Zhong Z, Cao D, Cui Q, Chen J, Yang J, Shen K. Tumor heterogeneity in the recurrence of epithelial ovarian cancer demonstrated by polycomb group proteins. Onco Targets Ther. 2014;7:1705–1716. doi: 10.2147/OTT.S67570. PMID: 11786930. DOI: 10.2147/OTT.S67570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadakis AI, Sun C, Knijnenburg TA, Xue Y, Grernrum W, Hölzel M, Nijkamp W, Wessels LFA, Beijersbergen RL, Bernards R, Huang S. SMARCE1 suppresses EGFR expression and controls responses to MET and ALK inhibitors in lung cancer. Cell Res. 2015;25:445–458. doi: 10.1038/cr.2015.16. PMID: 17487838. DOI: 10.1038/cr.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsunoo H, Komura S, Ohishi N, Yajima H, Akiyama S, Kasai Y, Ito K, Nakao A, Yagi K. Effect of transfection with human interferon-beta gene entrapped in cationic multilamellar liposomes in combination with 5-fluorouracil on the growth of human esophageal cancer cells in vitro. Anticancer Res. 2002;22:1537–1543. PMID: 12168834. [PubMed] [Google Scholar]
- 27.James DB, Mary KG, Christian W Brierley JD, Gospodarowicz MK WC, editors. New York, Wiley-Blackwell. 2016. TNM Classification of Malignant Tumours, 8th Edition. [Google Scholar]
- 28.Nakamura S, Kanda M, Koike M, Shimizu D, Umeda S, Hattori N, Hayashi M, Tanaka C, Kobayashi D, Yamada S, Omae K, Kodera Y. KCNJ15 Expression and malignant behavior of esophageal squamous cell carcinoma. Ann Surg Oncol. 2020;27 (7):2559–2568. doi: 10.1245/s10434-019-08189-8. PMID: 32052303. DOI: 10.1245/s10434-019-08189-8. [DOI] [PubMed] [Google Scholar]
- 29.Kanda M, Shimizu D, Tanaka H, Tanaka C, Kobayashi D, Hayashi M, Iwata N, Niwa Y, Yamada S, Fujii T, Sugimoto H, Murotani K, Fujiwara M, Kodera Y. Significance of SYT8 for the detection, prediction, and treatment of peritoneal metastasis from gastric cancer. Ann Surg. 2018;267:495–503. doi: 10.1097/SLA.0000000000002096. PMID: 28026832. DOI: 10.1097/SLA.0000000000002096. [DOI] [PubMed] [Google Scholar]
- 30.Umeda S, Kanda M. Expression of sushi domain containing two reflects the malignant potential of gastric cancer. Cancer Med. 2018;7:5194–5204. doi: 10.1002/cam4.1793. PMID: 30259711. DOI: 10.1002/cam4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato Y, Motoyama S, Wakita A, Kawakita Y, Liu J, Nagaki Y, Nanjo H, Terata K, Imai K, Saito H. TLR3 expression status predicts prognosis in patients with advanced thoracic esophageal squamous cell carcinoma after esophagectomy. Am J Surg. 2018;216:319–325. doi: 10.1016/j.amjsurg.2018.01.038. PMID: 29395019. DOI: 10.1016/j.amjsurg.2018.01.038. [DOI] [PubMed] [Google Scholar]
- 32.Sato Y, Motoyama S, Nanjo H, Ito S, Yoshino K. REG1A expression status suggests chemosensitivity among advanced thoracic esophageal squamous cell carcinoma patients treated with esophagectomy followed by adjuvant chemotherapy. Ann Surg Oncol. 2013;20:3044–3051. doi: 10.1245/s10434-013-2983-9. PMID: 23645481. DOI: 10.1245/s10434-013-2983-9. [DOI] [PubMed] [Google Scholar]
- 33.Sato Y, Motoyama S, Nanjo H, Wakita A. CXCL10 expression status is prognostic in patients with advanced thoracic esophageal squamous cell carcinoma. Ann Surg Oncol. 2016;23:936–942. doi: 10.1245/s10434-015-4909-1. PMID: 26464192. DOI: 10.1245/s10434-015-4909-1. [DOI] [PubMed] [Google Scholar]
- 34.Sato Y, Motoyama S, Wakita A, Kawakita Y, Liu J, Nagaki Y, Nanjo H, Ito S, Terata K, Imai K, Minamiya Y. High TLR4 expression predicts a poor prognosis after esophagectomy for advanced thoracic esophageal squamous cell carcinoma. Esophagus. 2020 doi: 10.1007/s10388-020-00732-x. PMID: 32170544. DOI: 10.1007/s10388-020-00732-x. [DOI] [PubMed] [Google Scholar]
- 35.Kanda M, Tanaka H, Shimizu D, Miwa T, Umeda S, Tanaka C. SYT7 acts as a driver of hepatic metastasis formation of gastric cancer cells. Oncogene. 2018;37:5355–5366. doi: 10.1038/s41388-018-0335-8. PMID: 29858600. DOI: 10.1038/s41388-018-0335-8. [DOI] [PubMed] [Google Scholar]
- 36.Banyard J, Bielenberg DR. The role of EMT and MET in cancer dissemination. Connect Tissue Res. 2015;56:403–413. doi: 10.3109/03008207.2015.1060970. PMID: 26291767. DOI: 10.3109/03008207.2015.1060970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato S, Hayakawa Y, Sakurai H, Saiki I, Yokoyama S. Mesenchymal-transitioned cancer cells instigate the invasion of epithelial cancer cells through secretion of WNT3 and WNT5B. Cancer Sci. 2014;105:281–289. doi: 10.1111/cas.12336. PMID: 24344732. DOI: 10.1111/cas.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higo N, Okumura H, Uchikado Y, Omoto I, Sasaki K, Kita Y, Ryosuke D, Noda M, Owaki T, Ishigami S, Natsugoe S. Expression of SOX9 is related to prognosis in patients with oesophageal squamous cell carcinoma. In Vivo. 2018;32:835–838. doi: 10.21873/invivo.112316. PMID: 17917549. DOI: 10.21873/invivo.11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tennstedt P, Koster P, Bruchmann A, Mirlacher M, Haese A, Steuber T, Sauter G, Huland H, Graefen M, Schlomm T, Minner S, Simon R. The impact of the number of cores on tissue microarray studies investigating prostate cancer biomarkers. Int J Oncol. 2012;40:261–268. doi: 10.3892/ijo.2011.1216. PMID: 21956230. DOI: 10.3892/ijo.2011.1216. [DOI] [PubMed] [Google Scholar]
- 40.Saremi N, Lam AK. Application of tissue microarray in esophageal adenocarcinoma. Methods Mol Biol. 2018;1756:105–118. doi: 10.1007/978-1-4939-7734-5_10. PMID: 29600364. DOI: 10.1007/978-1-4939-7734-5_10. [DOI] [PubMed] [Google Scholar]
- 41.Oppedijk V, van der Gaast A, van Lanschot JJB, van Hagen P, van Os R, van Rij CM, van der Sangen MJ, Beukema JC, Rutten H, Spruit PH, Reinders JG, Richel DJ, van Berge Henegouwen MI, Hulshof MCCM. Patterns of recurrence after surgery alone versus preoperative chemoradiotherapy and surgery in the CROSS trials. J Clin Oncol. 2014;32:385–391. doi: 10.1200/JCO.2013.51.2186. PMID: 24419108. DOI: 10.1200/JCO.2013.51.2186. [DOI] [PubMed] [Google Scholar]
- 42.Ando N, Iizuka T, Ide H, Ishida K, Shinoda M, Nishimaki T, Takiyama W, Watanabe H, Isono K, Aoyama N, Makuuchi H, Tanaka O, Yamana H, Ikeuchi S, Kabuto T, Nagai K, Shimada Y, Kinjo Y, Fukuda H. Surgery plus chemotherapy compared with surgery alone for localized squamous cell carcinoma of the thoracic esophagus: a Japan Clinical Oncology Group Study—JCOG9204. J Clin Oncol. 2003;21:4592–4596. doi: 10.1200/JCO.2003.12.095. PMID: 14673047. DOI: 10.1200/JCO.2003.12.095. [DOI] [PubMed] [Google Scholar]
- 43.Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, Furukawa H, Nakajima T, Ohashi Y, Imamura H, Higashino M, Yamamura Y, Kurita A, Arai K. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007;357:1810–1820. doi: 10.1056/NEJMoa072252. PMID: 17978289. DOI: 10.1056/NEJMoa072252. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida K, Kodera Y, Kochi M, Ichikawa W, Kakeji Y, Sano T, Nagao N, Takahashi M, Takagane A, Watanabe T, Kaji M, Okitsu H, Nomura T, Matsui T, Yoshikawa T, Matsuyama J, Yamada M, Ito S, Takeuchi M, Fujii M. Addition of docetaxel to oral fluoropyrimidine improves efficacy in patients with stage III gastric cancer: Interim analysis of JACCRO GC-07, a Randomized controlled trial. J Clin Oncol. 2019;37:1296–1304. doi: 10.1200/JCO.18.01138. PMID: 30925125. DOI: 10.1200/JCO.18.01138. [DOI] [PMC free article] [PubMed] [Google Scholar]