Abstract
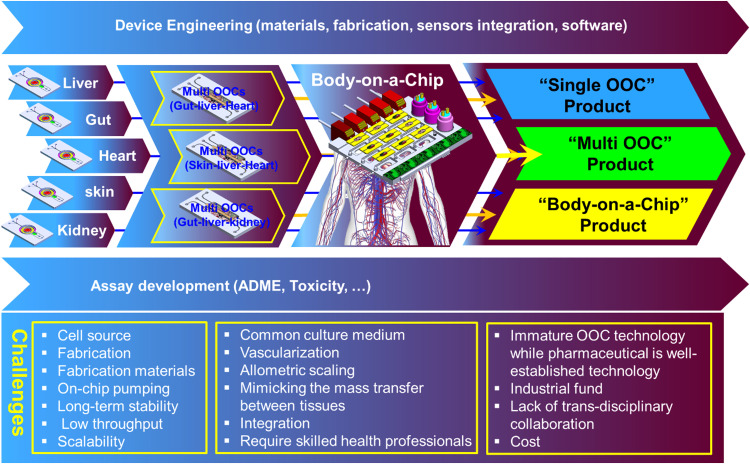
Organ-on-a-chip (OOC) is a very ambitious emerging technology with a high potential to revolutionize many medical and industrial sectors, particularly in preclinical-to-clinical translation in the pharmaceutical arena. In vivo, the function of the organ(s) is orchestrated by a complex cellular structure and physiochemical factors within the extracellular matrix and secreted by various types of cells. The trend in in vitro modeling is to simplify the complex anatomy of the human organ(s) to the minimal essential cellular structure “micro-anatomy” instead of recapitulating the full cellular milieu that enables studying the absorption, metabolism, as well as the mechanistic investigation of drug compounds in a “systemic manner.” However, in order to reflect the human physiology in vitro and hence to be able to bridge the gap between the in vivo and in vitro data, simplification should not compromise the physiological relevance. Engineering principles have long been applied to solve medical challenges, and at this stage of organ-on-a-chip technology development, the work of biomedical engineers, focusing on device engineering, is more important than ever to accelerate the technology transfer from the academic lab bench to specialized product development institutions and to the increasingly demanding market. In this paper, instead of presenting a narrative review of the literature, we systemically present a synthesis of the best available organ-on-a-chip technology from what is found, what has been achieved, and what yet needs to be done. We emphasized mainly on the requirements of a “good in vitro model that meets the industrial need” in terms of the structure (micro-anatomy), functions (micro-physiology), and characteristics of the device that hosts the biological model. Finally, we discuss the biological model–device integration supported by an example and the major challenges that delay the OOC technology transfer to the industry and recommended possible options to realize a functional organ-on-a-chip system.
I. INTRODUCTION
Pre-clinical drug screening, along with the Toxicity Testing for the 21st Century (TT21C), aims to transform toxicity testing from a system reliant on high-dose animal studies to one based primarily on human-relevant in vitro models. The phylogenetic distance between laboratory animals and humans,1–4 the discrepancy between current in vitro systems and the human body,5 and the limitations of in silico modeling6 have generated the need for new solutions to the ever-increasing demand for safety screening of new substances. The inherent complexity of interconnected tissues in animal models makes it difficult to elucidate and track the physiological events that characterize the interaction between an animal's organs and exogenic factors. Therefore, translational medical research should be focused more on complex human factors and conditions, rather than relying on animal models. On the other hand, while the simplicity of the traditional in vitro models makes them robust and suitable for high throughput research,7 unfortunately, only little biological relevance is provided to the complex biological tissues of the human body.
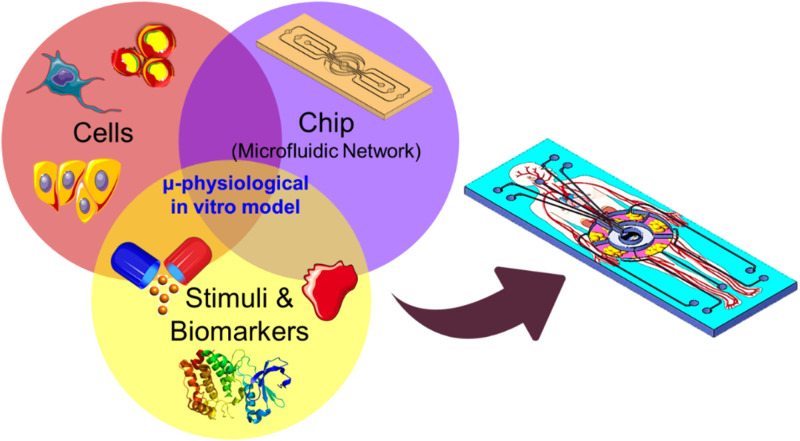
Organ-on-a-chip (OOC) is an emerging trans-disciplinary technology as a result of the recent advances in microtechnology (particularly microfluidics), cell biology, physiology, and tissue engineering and is driven by the need for low cost and reliable animal-alternative in vitro models for drug screening in most lengthy and costly product developments (Fig. 1). The target of the OOC technology is to develop effective and translatable integrated microphysiological models for investigating the physiological events that characterize the interaction between organs, immune system, and exogenic (e.g., pharmaceutics and nutraceutics) stimuli in health and disease states. This would be achieved by recapitulating the key structure and functions of a specific human tissue or a network of functional organs in vitro.
FIG. 1.
The advancement of organ-on-a-chip technology relies on the development of three main components: the cell source, chip technology, and biomarker discovery. The drawings of the biological items, cells, drug, and circulation, are available online from https://smart.servier.com, licensed under a Creative Commons Attribution 3.0 Unported License.
Cell patterning, with defined spatial positioning and stable cell growth, in microfluidic chips have been widely used for fundamental cell biology8,9 and various applications such as tissue engineering,10,11 neuron networks,12 cell-based biosensors,13,14 and drug screening.15 However, the concept of long-term culture of heterogeneous cells in perfusion microfluidic chips, i.e., OOC, was triggered by the pioneering work of Schuler’s group, termed as micro-cell culture analog (μCCA), in 2004,16 and Ingber’s group17,18 in 2010, which aim to accelerate drug discovery processes and ultimately replacing animal testing as a more accurate and affordable in vitro platform for drug development and personalized medicine.
OOC systems would represent powerful tools for providing physiologically relevant in vitro disease models that faithfully reproduce the key physiological aspects of the complex human. However, achieving this goal is still beyond the capability of the currently available microfluidic technology used in research labs. Despite the tremendous effort by various research groups all over the world on building in vitro microphysiological models or OOCs of various tissues or organs, for example, liver,19–26 lung,18,27–33 gut,18,34–39 kidney,40–44 skin,45–52 bone,53–56 adipose,57–59 heart,60,61 brain/blood–brain barrier,62–66 vasculature,67–71 and diseases such as cancer,72–75 diabetes,59 infection,76,77 and thrombosis,78 the technology gap between the lab models and industry/clinic adoptable models is still dramatically wide. The vast majority of the current OOC devices rely on simple microfluidic chips that consist of either planner or double-layered channels with a porous membrane. Such devices were implemented in academia labs or by start-up companies that lack the capability to invest in technological development of OOC engineering systems (hardware). Therefore, despite the large number of OOC studies published during the last decade, only scattered biological data that mainly demonstrate the co-culture of two or three cell types as a model of a specific human organ and characterization of cell–cell interaction and simple functional assays are available. Several excellent reviews on the OOC technology were recently published,79–83 which surveyed the landscape of OOC technology and presented the recent development in OOC. Bhatia and Ingber79 discussed the technical challenges that must be overcome to develop organ-on-a-chip models into acceptable predictive models of human physiology and sketched the possible directions in future related research. Skardal et al.81 described the progress that has been made to generate complex multi-organoid body-on-a-chip platforms and their applications and discussed the impact of this technology on drug and toxicology screening, disease modeling, and personalized medicine. Mencattini et al.82 emphasized the role of time-lapse microscopy and machine learning approaches on the advances of OOC technology, while van den Berg et al.83 discussed the potential use of OOC in personalized and precision medicine.
A careful look at the current OOC research landscape reveals that the state of the art of this technology is still dominated by proof-of-concept studies that aim to reproduce a specific tissue or organ-like structure in vitro “X-on-a-chip.” “X” here is an organ or tissue. Most of these studies share a similar chip structure and methodology by co-culturing relevant cells together in close proximity by varying the cell types in different studies. With only a few exceptions, there is still a severe lack in deep and focused accumulated research on a single organ. To develop multi-organ-based models, it is necessary to gain a deep understanding of the cell–cell interaction and overall tissue structure and function of a single organ before integrating with another functional organ.
A successful OOC in vitro model relies on device engineering, cell biology, and biomarker discovery (Fig. 1). The lack of development in these areas would slow the advances toward the realization of reliable in vitro models. In this paper, we focus on the engineering development aspect by surveying the literature landscape and highlighting the state-of-art OOC engineering in both academia and industry. We emphasized the requirements of a “good in vitro model that meets the industrial need” in terms of the structure (micro-anatomy), functions (micro-physiology), and characteristics of the device that host the biological model. We will discuss the current manufacturing technologies used in OOC and, finally, will sketch the possible pathways that may push the boundaries of this technology toward transferring the technology from the lab to fabrication and hopefully to market.
II. FROM LAB-ON-A-CHIP TO ORGAN-ON-A-CHIP
A. What is organ-on-a-chip?
OOC is an emerging trans-disciplinary technology that overlaps with tissue engineering and lab-on-a-chip technologies, benefited from recent advances in microtechnology (particularly microfluidics), cell biology, physiology, and tissue engineering and driven by the need of low cost and reliable animal-alternative in vitro models for drug screening. An OOC device can be defined as a microfluidic-based perfusion device that hosts a (co-)culture of cells in vitro and aims to recapitulate specific structure(s), function(s), and key aspects of human metabolism of a certain tissue or an organ in normal and pathological physiology. Thanks to the well-defined and precise features produced by microfabrication, OOC would enable key aspects of living organs, including physiologically relevant tissue microarchitecture, spatiotemporal cell–cell interaction, and extracellular microenvironments. Currently, OOC devices are fabricated by soft-lithographic processes, with polydimethylsiloxane (PDMS) and glass representing the common fabrication materials owing to their optical properties, which enables live-cell imaging.
In vivo, it is challenging to isolate the interactions between two organs because they are embedded within the complex whole-body environment; signals released by each organ are diluted into the bloodstream and delivered to many other tissues. OOC would enable tracking the cell–cell/tissue–tissue signals in isolation in a simple structure or within a complex structure with precise spatiotemporal control, therefore, providing a better understanding of the contribution of a specific cell type within the tissue milieu or organ.
B. OOC's microfluidics: From the flat “lab-on-a-chip” to the 3D “organ-on-a-chip”
The fabrication of micro-electro-mechanical systems (MEMS) has benefited from the well-established semiconductor microfabrication technology, namely, lithography and etching, employing a silicon substrate, the material used to fabricate integrated circuits (ICs), for creating planar miniaturized features with unprecedented high precision and high throughput capabilities. MEMS and microfabrication technologies have pushed the boundaries of miniaturization beyond electrical and optical systems to explore new applications such as microfluidics. The prominent microfluidics technology enables handling of minute amounts of fluids as low as a few picoliters in a network of microchannels and manipulation of various biochemical reactions at very small volumes. In the early 1990s, Manz et al. proposed that MEMS/microfluidic technologies could be applied to biological analysis, and by using tiny planar channels, many laboratory processes including sample preparation, separation, and detection can be carried out using very small volumes.84 This emerging “flat-microfluidic” technology, which was enabled by the surface and bulk micromachining techniques, has shown a spectacular growth over the last three decades and sparked the development of a wide spectrum of on-chip assays for molecular and cellular analyses ranging from in vitro diagnostic, personalized medicine, and infectious disease, among others. However, advances in cell-based applied research (e.g., cell culture) in miniaturized device were much slower with limited diffusion in biomedical practice compared to other biological applications such as DNA microarray technology.
Since its introduction by Harrison,85 the process of growing eukaryotic cells in vitro remained basically unchanged for almost a century. This is partially due to the dynamic nature of the biological processes in cell growth that require continuous monitoring of the cell environment. With a plethora of recent publications, it has been shown that the utilization of microfabrication and microfluidics in cell biology practice enables high spatial resolution of cell positioning and patterning and opens a new avenue to increase the resolution of analysis and sampling throughput that push the boundaries of cell biology toward more advances in “cellomics” science.
C. From monoculture to co-culture and organ-on-a-chip
Cell co-culture is a cell cultivation setup, in which two or more different type of cells are grown with some degree of contact between them86 to synthesize a physiologically relevant multicellular system. The motivations for creating such systems include studying the interaction of heterogeneous population of cells and creating human-based biomimetic tissue models for pre-clinical drug screening. It becomes evident that the cellular phenotype is produced by complex interactions between genotypes and strongly affected by the extracellular environment. For example, the crosstalk between cancer cells and accessory cells fuels and shapes tumor development,87 and it was demonstrated that that inflammatory immune cells are essential players of cancer-related inflammation.88 The organ structure is characterized by the intricate composition of various specialized cell types arranged in precise geometries that lead to a complex interaction between cells and the microenvironment to enable the organ function that is achieved by a well-defined tissue interface and geometrical orientation. Mimicking the in vivo cellular heterostructure within the microfluidic system requires designing complex microfluidic systems. Cell positioning and cell–cell separation distance in the co-culture system needs to be carefully chosen to ensure relevance to the ultimate application and to ensure relevant substance exchange between the two cell types. Such cell–cell separation can be achieved in microfluidic devices using compartmentalization. Using microfabrication technology allows creating a unique structure with the desirable scaling and precise control of chemical and physical and flow conditions with cellular scale spatial resolution in three-dimensional space. For example, the inter-compartment fluid exchange or diffusion can be achieved by placing a semi-porous barrier between the compartments with porosity that is custom designed to enable the desired permeability and diffusion and thickness that allows cell–cell communication and some degree of contact.
III. OOC ENGINEERING: REQUIREMENTS OF THE MICROFLUIDIC SYSTEMS
OOC functions are different from other lab-on-chip (LOC) systems because they host a viable cell co-culture in an artificial milieu for a relatively long period of time and recreates in vivo conditions in a micro-scale bioreactor. The optimum function of the OOC system is to provide the tools and environment that enable studying of the cellular assembly in vitro that mimics their counterpart in vivo and capturing the spatiotemporal cellular behavior when exposed to exogenic stimuli and substances. Furthermore, miniaturization would enable the integration of process control, sensors, imaging systems, and other analytical components. Therefore, the design of an OOC system needs to take into account all these functional parameters; however, these may vary depending on the in vitro model of interest, e.g., liver, skin, heart, etc. The following are the general requirements of OOC system engineering that would facilitate achieving the major functions and enabling translation of the in vivo milieu into a physiologically relevant in vitro system.
A. Steady, continuous supply of cell nutrients and waste removal in a stable perfusion circuit
The cellular system in the OOC recapitulates the interaction of cells/tissue with blood and circulating substances. To enable this, a reliable fluidic circuit needs to be designed that ensures stable fluid flow at the designated flow rate with possible flow modulation as well as media dilution. Due to their small volume, microfluidic channels are prone to bubble generation, which severely impacts the experimental conditions; therefore, the fluidic circuit must include the bubble removing/prevention mechanism. Over the course of the culture, cell population density varies (in general, cases increase), and in the case of co-culture, different cell populations may have different proliferation profiles that require nutrient supply adjustment accordingly. Perfusion is a major differentiating factor of OOC, which is accomplished by retaining cells within a channel/chamber, while exchanging the nutrient-carrying medium to sustain cell growth and viability. OOC systems maintain cells over much longer periods by continuously feeding the cells with fresh media and removing cell waste while keeping the cells live in culture. In general, the following are the major advantages of perfusion (Fig. 2):
-
(i)
By continuous nutrient supply and waste removal, the nutrient levels are maintained for optimal growing conditions while the cell waste product is removed to avoid cell toxicity;
-
(ii)
During the culture process, cells secrete the protein of interest into the flowing media, which can be sampled and analyzed either in situ or off the chip.
-
(iii)
Preventing the exposure of the drug/stimuli of interest to excessive waste that may cause deviation of the planned conditions (e.g., drift in the pH value).
-
(iv)
By optimizing the cell-to-liquid ratio within the OOC micro-fluidic chip, less culture media will be used thereby reducing the overall cost of culture.
-
(v)
Providing physiologically relevant shear stress on the cell membrane. In vivo endothelial cells, in particular, develop and differentiate under flow-induced shear stress. Therefore, to mimic the action of such shear stress in vitro, endothelial cells should be cultured under steady flow with a calculated flow rate for prolonged time to induce the physiological mechanical force.
-
(vi)
Inducing liquid exchange through the compartmentalized fluidic system and enabling time-dependent sampling for downstream analysis.
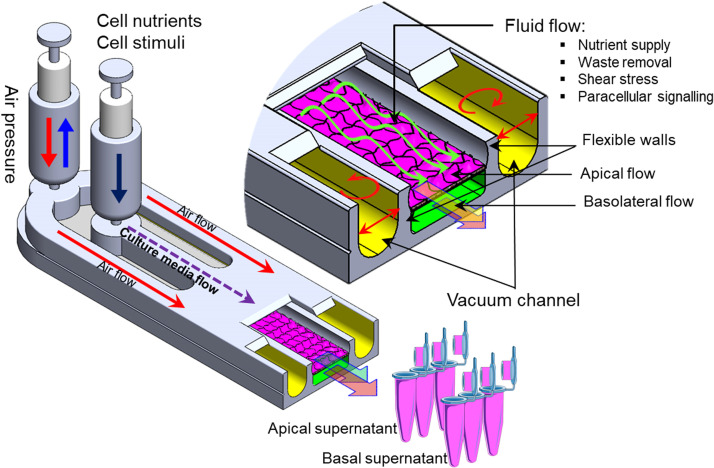
FIG. 2.
Schematic drawing of a simple OOC device with major functions: fluid perfusion, nutrient supply, waste removal, mechanical force application, and downstream sampling.
Figure 2 schematically depicts a simple OOC chip with major functional components. Cell–cell interaction in a heterogeneous cellular culture system can be achieved by employing a semi-porous membrane that separates two vertically stacked fluidic channels where two different cell types can be grown. The two cell types are physically separated but fluidically connected due to the diffusion through the porous membrane. For instance, epithelial cells (e.g., intestinal,36 epidermal,46 and lung alveolar17) can be cultured on the upper surface of the porous membrane (apical chamber), and other parenchymal cells are grown in the lower (basolateral) chamber. Besides maintaining in vivo-like biochemical conditions and continuous nutrients and waste removal, microfluidic technology also enables mimicking the mechanical forces on cells and tissue such as shear stresses and stretching. For example, using two side hollow channels adjacent to the elastic porous membrane (Fig. 2) and cyclic vacuum suction induced cyclic mechanical stretching of the adherent cell layers, which recapitulates the breathing action of lung.17
B. Device fabrication materials
The main problem that still hinders the progress of OOC device development is the material that is used for fabrication and the lack of standards that govern possible mass production of chips similar to the well-established semiconductor industry. Currently, the vast majority of LOC and OOC devices are fabricated by PDMS using the soft lithography technique. PDMS is a great rapid prototyping material that enabled the research community to use it in a plethora of applications and produce high impact results due to the ease of fabrication and rapid molding process, thanks to its elasticity, gas permeability, biocompatibility, and good optical clarity. However, PDMS absorbs small biomolecules and other organic compounds and drugs, which limit its applications as an OOC material. In addition, despite the ease of rapid fabrication in the lab, PDMS may not be suitable to be mass produced due to the lack of industrial standards. Recently, a number of other materials were used for the fabrication of microfluidic/LOC/OOC devices that aim to overcome the shortcoming of PDMS such as polystyrene (PS)89 and polymethylmethacrylate (PMMA),90,91 which is reviewed elsewhere.92,93 Table I shows a list of recently used materials in LOC applications and their advantages and disadvantages. The materials are compared in terms of the key OOC-relevant properties, namely, biocompatibility, transparency, elasticity, and manufacturability.
TABLE I.
List of common materials used in the fabrication of LOC and OOC devices.
| Materials | Biocompatibility | Optical property (transparency) | Mechanical property (elasticity) | Chemical resistance | Manufacturability | Cost (Ref. 16) | Reference |
|---|---|---|---|---|---|---|---|
| PDMS | Good | Good | Good elasticity | Poor | Lab-based soft lithography No scale up production |
High | 94–96 |
| Polymethylmethacrylate (PMMA) | Good | Good | Rigid | Good | Microinjection molding, hot embossing, casting, reactive ion etching, mechanical milling (CNC), laser micromachining | Low | 90, 91, 97, and 98 |
| Polystyrene (PS) | Good | Good | Rigid | Poor | Injection molding, hot embossing | Low | 89 and 99 |
| Polyimide (PI) | Good | Poor | Poor | Good | Photosensitive, lithography | High | 100–102 |
| Polycarbonate (PC) | Good | Poor | Rigid | Good | Injection molding, hot embossing | Low | 99 and 103 |
| Cyclic olefin copolymer (COC) | Good | Poor | Rigid | Good | Injection molding, hot embossing | Moderate | 104–105 |
C. Tissue architecture: Cell–cell and tissue–tissue interface
Cellular organization reflects the tissue function as cells/tissue and organs communicate by secreting various soluble factors and extracellular vesicles that mediate peripheral crosstalk with the circulatory system.106 The connection of different organ modules in vitro can greatly affect their functionality and effectiveness. Inter-organ communication is mainly studied through a systemic common medium that interconnects different tissue/organ modules to mimic the circulating blood that can transport nutrients, soluble factors, cell metabolites, and drugs and mediates organ crosstalk. Developing a common cell culture medium for the maintenance of phenotypes and functions of all organs in the OOC system is a challenging task that demands innovative solutions. Due to the central role of liver in metabolism, several multi-organ systems, with a liver module, employed liver culture media as the common medium to ensure hepatic functions in the co-culture system.107,108
OOC is a sophisticated form of cell culture architecture that ensures precise cellular positioning and in vivo-like cell polarization by providing a template on which cells can reproduce a complex assembly and mimic the actual tissue organization. To construct an in vitro organotypic cellular structure with fluidic/bio/chemical exchange, microfluidic systems are fabricated to enable organizing of various types of cells with the appropriate tissue architecture. Three-dimensional (3D) cell culture is utilized to create multicellular structures or spheroids. However, the tissue architectures in vivo, in many cases, do not contain spheroidal tissue. Therefore, cell culture methods need to force the formation of cell assembly and their extracellular milieu to enable the relevant physiological structure before using the in vitro model for analysis. For instance, polarity is an inevitable architectural feature of organs, which is created by the asymmetrical distribution of proteins in the cell membrane and determined by the formation of cell–cell tight junctions (TJs) that separate the basolateral and apical membrane.109 It is the major characteristic of the epithelial, endothelial, and liver structure and function, and it can be produced in a prober extracellular matrix (ECM) or cell culture process.110
Microfluidic technology has enabled the integration of various cell types and/or organs in a single fluidic circuit that allow organ–organ crosstalk while preserving individual organ functionality and mimics the in vivo role of vascular perfusion. A straightforward approach is to use common cell culture medium to support all the cell types/organs within the integrated system. However, this approach is limited to tissues that are already mature and phenotypically stable.111 In microfluidic systems, multi-tissue structures could be hosted within a compartmentalized system that is separated by an endothelial barrier to mimic the tissue–blood interfacing in the body such that tissue-specific media could be maintained in each compartment to support and mature each tissue in an optimal manner, while enabling the crosstalk among the tissue units via vascular flow.106 The design of the compartmentalized fluidic system for OOC, which ensures a relevant in vitro system, involves several critical parameters such as the relative size of individual compartment (i.e., the size of the hosted organ), the order in which the organs are connected, the tissue orientation, and the perfusion rate within each compartment. Since the cell metabolism varies from organ to organ and during cell maturity, a carefully designed OOC system could provide a deeper understanding of cell metabolism and organ–organ interaction. The design of OOC is mainly driven by the physiological parameters that need to be recapitulated in vitro. This could be achieved by a minimal functional structure of an individual organ or integrated organs by selecting the key cell models [from either cell lines or the induced pluripotent stem cell (iPSC) source], biochemical stimuli (e.g., drugs and toxins), and physical stimuli (e.g., hydrodynamic, mechanical, and electrical).
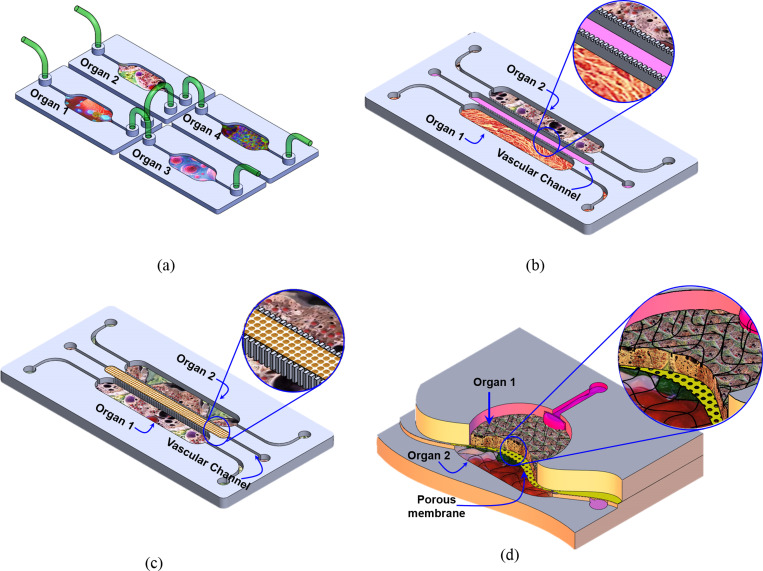
To achieve a physiologically relevant organotypic structure and functional coupling that enable cell nutrients supply, chemical, and paracrine signaling, OOCs are integrated in various connection strategies such as
-
(i)
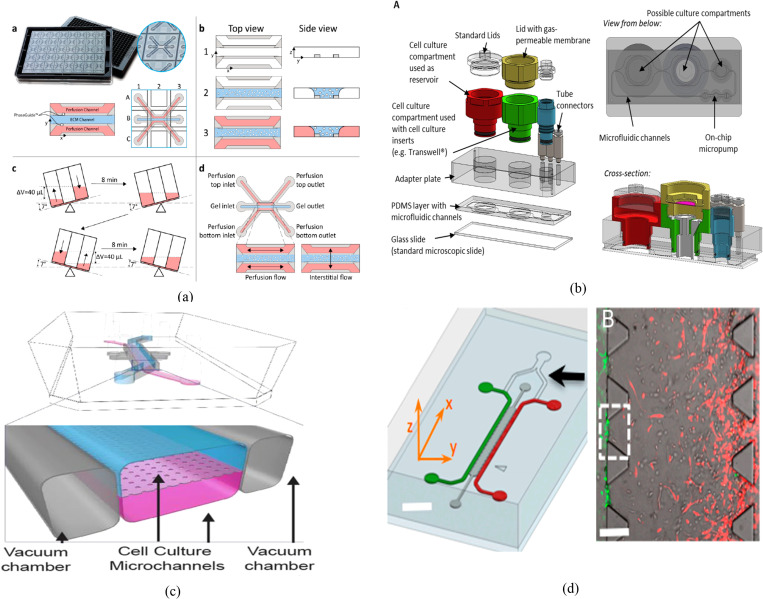
Convection-based fluidic transfer (manual pipetting or through tubing): This simple connection method does not require a micro-fabricated channel to connect the fluidic chambers but provides flexibility for integration of several individual organs and enables organ–organ via secreted factors.111–118 However, it fails to recapitulate the physiological flow between organs and is limited to the use of organs that can be supported by the same culture media [Fig. 3(a)].
-
(ii)
Connection through porous barriers/gel with planar orientation: In this configuration, cells are assembled/cultured in a two-dimensional (2D) organization within a planar compartmentalized microfluidic structure. The multi-compartments in such structures are physically separated by semi-porous vertical barriers [Fig. 3(b)]. So, cells of different types are cultured in close proximity to each other such that they are physically isolated but fluidically and chemically connected.119,120 The semi-porous barriers are characterized by an array of small pores that allow only liquid but not cells to be exchanged through the adjacent compartments. The size of these pores could be adjusted to specific requirement (e.g., type/size of the cells) to control the mass transfer between the different compartments. The porous channels can be filled with porous materials (e.g., hydrogel) to enable biomimetic vascular structures between different organs [Fig. 3(c)]. In sequence, this would enable chemical/biological interactions (paracrine signaling) between the cells in different neighboring compartments. The flow profile, rate, and direction of the inter-compartment flow can be controlled using either internal or external pumps or valves. Taking advantage of the porous barriers structure, multi-cell type cultures can be arranged in the desired order where the interaction between the cells/tissue within the various compartments is defined by the direction of the perfusion flow. The perfusion micro-pores with the combination of micro-flow in the adjacent compartment can be utilized to generate chemical gradients when desired;121 hence, the composition and concentration of chemicals within the side compartment can be selectively adjusted. The horizontal order of organs is a common structure due to the ease of fabrication and suitability to study the interaction of specific organ/cells with the immune system. Table II lists examples of OOC modes that are demonstrated in planar order.
-
(iii)
Connection through porous barriers with vertical orientation: In this configuration, two-layer, or more, fluidic compartments are vertically stacked and interfaced through a porous membrane, which is sandwiched between them such that two or more cell types can be co-cultured in close proximity to each other in two vertical orientations. In this structure, a porous membrane with optimized thickness and pore size can be used to physically separate the two cellular structures. In addition, the porous membrane also can be used as a substrate for culturing two types of cell on both sides. When epithelial/endothelial/epidermal cells are cultured on the upper surface of the membrane and the corresponding parenchymal tissue on the lower side, this structure would mimic the structure and function of important parts of the human body, which are biological barriers such as the small intestine, lung parenchyma, skin, and blood vessels that generally control the interaction of the body with drugs, food, and the environment. The lower compartment can host a model of a specific organ such as the liver, adipose, muscle, bone, etc. This structure can be used to recapitulate the transport (absorption and distribution) of bioactive materials or drug through the epithelial and measuring the bioavailability of these substances in the target organs and its metabolic profile. Furthermore, the lower compartment/channel can be used to host circulating immune cells to model the activation of the immune system after the transport of a foreign substance through the epithelia. Table III lists examples of OOC modes that are demonstrated in a vertical orientation.
FIG. 3.
Fluidic exchange configuration between cell culture compartments in the in vitro models. (a) Various microfluidic devices are externally connected by tubing. (b) The fluidic compartments are separated by permeable thin solid barriers. (c) The fluidic compartments are separated by a narrow channel filled with porous materials (e.g., gel). (d) Two vertically stacked compartments separated by a porous membrane.
TABLE II.
List of OOC examples that are demonstrated in a planar organization/order.
| Organ model(s) | Used cells | Key studied parameter | Physiological parameters/readout | Reference |
|---|---|---|---|---|
| Liver, bone marrow, uterine cancer | HepG2/C3A, MEG-01, MES-SA, MESSA/DX-5 | Drug mixtures for potential efficacy in treating multidrug resistant cancers | Inhibiting MES-SA/DX-5 cell proliferation | 122 |
| Intestine, liver, cancer, and connective tissue | HCT-116, HepaRG, Caco-2, TIG-121 fibroblasts | Pneumatic pressure driven medium circulation platform with a microplate-sized format | Evaluation of the effects of prodrugs on multiple organ models | 116 |
| Breast cancer, bone, muscle, and microvascular network | hBM-MSCs, HUVECs, osteoblast-differentiated cells (OBs), C2C12 | Bone- and muscle-mimicking microenvironments through a microvascular network concentrically wrapped with mural cells | Cancer cell extravasation; extravasation rates and microvasculature permeabilities | 123 |
| Adipose-immune cells | Human primary adipocytes and U937 | Simultaneous multiplexed measurements of pro-inflammatory cytokines The immune-metabolic correlation |
Pro-inflammatory cytokines and glucose uptake | 120 |
TABLE III.
List of OOC examples that are demonstrated in the vertical organization/order.
| Organ model(s) | Used cells | Key studied parameter | Physiological parameters/readout | Reference |
|---|---|---|---|---|
| Gut-immune cells | Caco-2 and U937 | Caco-2 as a protection layer | TEER and inflammatory cytokines | 37 |
| Skin-immune cells | HaCaT and U937 | HaCaT as a protection layer | TEER and inflammatory cytokines | 46 |
| Gut-liver and skin-liver | Human primary cryopreserved hepatocytes and Kupffer cells, Caco2 and HT29 cells | Integration barrier tissue with parenchymal for pharmacokinetics | Permeability | 107 and 124 |
| Blood–brain barrier | Human microvascular endothelial (BMVECs) from the iPS cell line IMR90-4 cells and primary human astrocytes | Differentiation under hypoxic conditions | Permeability and drug transport | 125 |
| Lung airway | Primary airway epithelial hAECs and human lung microvascular endothelial cells | Lung inflammatory | Cytokine secretion | 126 |
D. Allometric scaling
Very little research has been carried out on the rational design of cell–cell or tissue–tissue interaction in in vitro models that faithfully reflect that in vivo. A good OOC design needs to be taken into account for the relationship between cells, cell metabolism, and exchange in the human body using allometric rules in order to establish appropriate cell ratios in the system in a rational manner.127 Current design approaches are based on relative sizes and not functions.128 Two different allometric scaling models (i.e., the cell number scaling model and the metabolic scaling model) need to be considered127 to enable the predictive potential. For example, Ucciferri et al.127 proposed connecting hepatocytes with endothelial cells before adding other cells or to construct an in vitro model of biotransformation and distribution in liver. Therefore, the hepatocytes in the model are scaled with reference to the basal metabolism, whereas the endothelium is scaled using the surface area of the human vascular system. As cells are usually plated in monolayers, the allometric design process begins by considering the metabolism of a two-dimensional culture of human hepatocytes in a single module. By integrating in vivo and in vitro data allometric scaling would enable more accurate prediction of human pharmacokinetics/pharmacodynamics.
E. Stable performance over extended periods of time
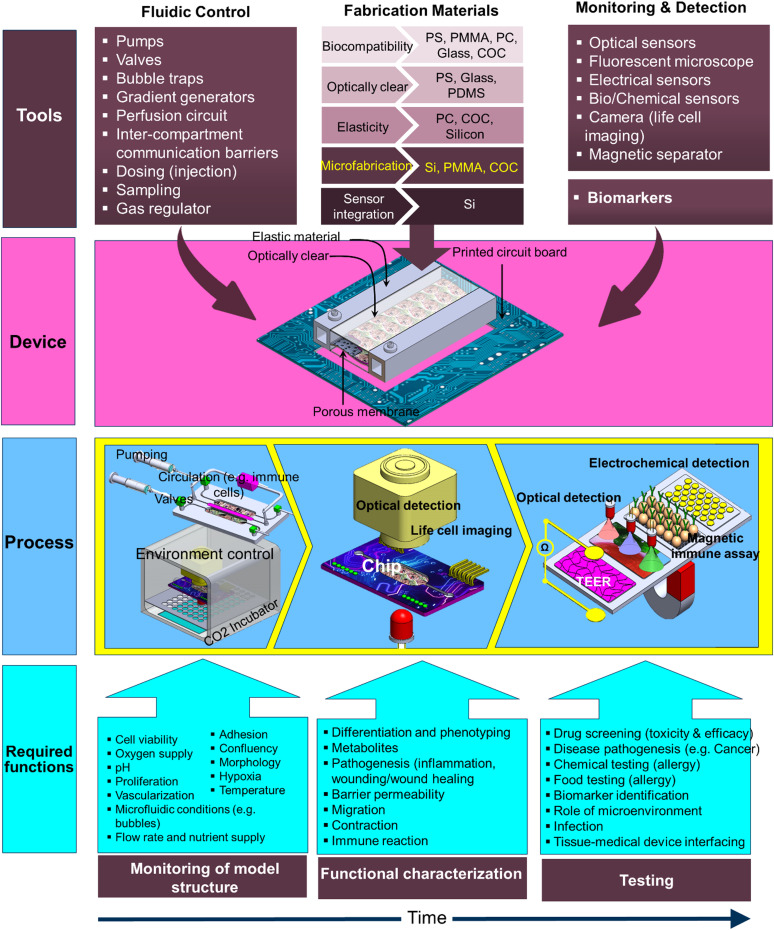
The OOC system is a miniaturized bioreactor that hosts the hybrid cellular structure for extended periods of time for studying the time-dependent cellular behavior that depends on the type of cells/tissue and the downstream analysis. To achieve reliable in vitro results, it is important to maintain stable cell viability and control the phenotypic stability in their artificial milieu during the relatively long-life span of the OOC model. The life cycle of the OOC system can be divided into three stages: (i) creating the model structure, (ii) model's functional characterization, and (iii) model validation and testing (Fig. 4). These processes and functionalities should drive the engineering design of the OOC system when transferring the prototype to product development. Such a methodology is not common in the bioengineering product within the LOC domain, but, despite the lack of standards, the industrial design applied to mechanical and electrical products can be adopted in the OOC development.129 In general, regardless of the type of the in vitro model under study, the longevity of use of the model is crucial to enable reliable and consistent results and at the same time to allow repeatable and consistent experiments on the same set of cells/tissues. The following summarizes the required process control during the stages of the OOC model:
FIG. 4.
General presentation of the integrated OOC tools, device, process, and the required functions through the development of the in vitro model stages (model building, functional characterization, and testing).
1. Model structure (micro-anatomy)
During the stage of building the structure of the in vitro model, it is important to control the environment and monitor the cell growth and phenotyping. Depending on the in vitro model under study, the controlled components in the system need to be aligned to the intended structure and functional parameters. In general, the following parameters are important in most of the OOC in vitro models:
-
(1)
The microfluidic dynamic properties: The dynamic of the perfusion flow is a crucial parameter in the dynamic cell culture. Therefore, it is important to take the fluid dynamic within the OOC system into consideration during the design of the chip. These include the geometry and dimensions of the channels/chambers and the time-dependent pressure gradient, which need to be optimized based on the required cell–liquid/nutrients interaction and the human physiological data. In addition, the flow/pump mode needs to be initially defined to configure the type of flow such as steady or pulsatile flow. In the second case, the driving frequency should be set within a physiologically relevant range (i.e., in the range of then human heartbeat of 60–180 beats per minute or a frequency of 1–3 Hz). The fluid flow within the microfluidic channel imposes mechanical shear stresses and results in strain on cells and tissues, which are integral parts of the cellular microenvironment that modulates the proliferation, differentiation, phenotype, and migration. These actions in sequence affect the function of multicellular systems in organ-level health and disease.130
-
(2)
The physicochemical parameters (temperature, pH, oxygen, CO2): The optimum micro-bioreactor could supply cells with dynamic profiles of nutrients, oxygen, and growth factors and, at the same time, control the physical and chemical environments of the cell culture with in-line monitoring modality. Maintaining ideal culture temperatures is vital for optimal cell growth, where mammalian cells thrive at around 37 °C. The pH value is also critical to cellular function. The cell culture pH may deviate from the optimum ranges due to the build-up of acidic metabolites by cultures that have grown too dense or grown too long in that medium and the low oxygen levels and contamination by fast-growing bacteria or fungi. The vast majority of cell culture media contain carbonate-based buffers that work with elevated CO2 levels in the incubator to stabilize the cell culture pH131 as they are also present in vivo. In a recent study, Zhang et al.111 demonstrated an integrated pH and oxygen sensor within a compact micro-bioreactor. The pH sensor detects the changes in the light absorption of phenol red in the culture medium to translate into a voltage change, while the oxygen-quenchable luminescent dye [Ru(dpp)3]2 + Cl2 – tris(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) chloride was used for the oxygen sensor. A comprehensive review of various instrumented in vitro microphysiological systems has been presented by Soucy et al.132 Recently, Kirkstall133 developed the Quasi Vivo® System, which consists of interconnected cell culture chambers and a peristaltic pump to create a continuous flow of media over cells. The Quasi Vivo® system is available with three different culture chambers (QV500, QV600, and QV900), which enable monitoring of variables during the cell culture experiment. The system also fits with the standard cell culture inserts (Transwell).
-
(3)
Mass transfer at the tissue–fluid interface: To mimic in vivo cross-organ interactions, reliable fluidic connections between organ modules (fluidic chambers) are essential. The architecture of the fluidic networks in multi-organ systems can have a large impact on tissue/organ crosstalk. The perfusion circuit in recirculating microfluidic systems allows mimicking blood circulation and facilitates communications among organs. One of the key challenges in OOC systems, particularly when involving 3D cell culture or a multi-organ model, is to maintain acceptable cell viability during the course of cell culture. Therefore, nutrients, oxygen, growth factors, and other regulatory molecules have to be efficiently transported from the bulk of culture media and through the multi-tissue structure (internal mass transfer).134 The organization of the cellular system and the order or orientation of the tissue in the multi-organ system have a significant impact on the inter-tissue mass transfer, which is generally dependent on the combination of diffusion and convection mechanisms, the porosity and size of the microfluidic system, and the diffusion rate through the biomaterial.134 Taking into account the mass transfer parameters is an essential consideration to achieve long-term viability of the model. For example, the fluidic barrier between various cell type chambers needs to be designed and numerically simulated prior to designing the compartmentalized microfluidic system. The most important parameters are the porosity, the thickness, and the material (e.g., hydrogel) of the structure. Improving the microfluidic design will result in efficient mass transfer. In particular, it is critical to maintain a balance between oxygen delivery to cells and their oxygen consumption. Oxygen transfer is also a key parameter in the cell culture setup that requires monitoring due to the poor solubility of oxygen in the culture medium135 and taking into consideration the diffusion distance of oxygen in tissue of 100–200 μm.136
2. Functional characterization (micro-physiology)
In the downstream stages, to gain confidence in the structured in vitro model, it is necessary to evaluate it by demonstrating the relevant in vivo physiology and assess the analogy between the in vitro model and its in vivo counterpart. The type of functional components depends on the organ(s) under study. The following are some of major characteristic parameters that are commonly studied:
a. Differentiation
One of the key challenges in the OOC technology is to obtain functional, human, organ-specific parenchymal cells that can produce specific human physiological characteristics. Various human-based cell types are currently commercially available, which assisted to push the boundaries of this technology. In addition, induced pluripotent stem cells (iPSCs) have also been used to create specialized organ-specific tissues. For example, the structure of the in vitro model representing the small intestine for pharmacokinetics screening/drug absorption should include the major cell types: enterocytes, Paneth cells, and goblet cells with a relevant in vivo ratio. However, since it is difficult to include all the necessary features in a single chip, the complexity of the model varies depending on the specific desired function. Monitoring of cell differentiation within the OOC system is essential to ensure the model relevancy during the course of culture. For example, a rapid loss of “liver” phenotype during culture was observed, which makes the culture system not amenable to repeat-dose studies.137 In fact, OOC technology could be also used to enhance stem cell differentiation in vitro and to study patient-specific developmental responses for personalized medicine.138
b. Nutrient and metabolite profile in cell culture (e.g., glucose and lactate)
Assessing the metabolic activity of a cell culture provides a detailed understanding of the physiological state of cell culture. In general, in large-scale bioreactors, glucose and lactate concentrations are usually measured using colorimetric assays and bulky electrochemical detection systems, which require manual and tedious sample preparation that is not suitable for monitoring over extended periods of time. Prill et al.139 demonstrated an automated detection of drug-induced changes in cellular viability by continuous monitoring of glucose consumption and lactate secretion of a hepatic tumor cell line. Using microfluidically addressed electrochemical sensors, the system probes the mitochondrial apoptotic pathway by exposing the cells to the complex I inhibitor rotenone, which reduces OXPHOS that in turn upregulates other catabolic mechanisms, like anaerobic glycolysis. Bavli et al.140 reported real-time monitoring of mitochondrial respiration using two-frequency phase modulation of tissue-embedded phosphorescent microprobes in a liver-on-chip device. The computer-controlled microfluidic switchboard allowed contiguous electrochemical measurements of glucose and lactate, providing real-time analysis of minute shifts from oxidative phosphorylation to anaerobic glycolysis, an early indication of mitochondrial stress.
c. Permeability of epithelial tissue barriers
A key characteristic of epithelial tissue structures, such as gastrointestinal (GI) tract epithelium, blood vessel endothelium, and skin epidermis is the tight arrangement of cells that strongly express intercellular tight junctions (TJs), which play an important role in modulating the transport of nutrients/drugs from the apical side to the basolateral blood stream. This barrier is not static but can be modulated by specific stimuli. For example, the increased permeability of intestinal epithelium has been linked to the pathogenesis of inflammatory bowel disease, ulcerative colitis, Crohn’s disease, and food allergies. On the other hand, a transient increase in paracellular transport could improve the bioavailability of desirable bioactive compounds, which normally are poorly absorbed.141
One of the common methods of monitoring epithelial integrity is based on the measurement of transepithelial electrical resistance (TEER) of barrier forming cells grown on porous membranes using two sets of electrodes that are connected to a volt-ohmmeter. This non-invasive method can be applied to living cells without markers and allows them to be monitored during growth and differentiation. TEER reflects the resistance to the passage of ions through the physiological epithelial barrier and is recognized as one of the most accurate and sensitive measures of epithelial integrity and permeability. TEER can also be used to measure the effect of different stimuli on the barrier permeability and any biological response of the underlying cells within the lower chamber. These quantitative data provide an effective tool to predict the influence of tissue exposure to chemical, biological, electrical, thermal, and mechanical stimuli related to organ inflammation (e.g., skin and gut), toxicity, and immune modulation. The electrical behavior of a cell/cell layer is frequency dependent. The cell membrane is a poorly conducting lipid bilayer, and at low frequencies, the cell behaves as an insulator. As the frequency increases, the electric field lines penetrate the cell so that the cytoplasmic resistance of the cell can be probed.142 Therefore, frequency-dependent impedance measurements would provide a wealth of information not only about the cell layer integrity but also about the molecular pathway of transported bio/chemical substances through the epithelial monolayer because impedance measurements allow for the separation of paracellular resistance (governed by TJ properties) from transcellular resistance (determined by conductive structures residing in the cell membranes).
d. Response to mechanical stimulation
Among the earlier OOC models is the lung-on-a-chip model,17 a biomimetic microsystem that recapitulates the microarchitecture and dynamic microenvironment of the alveolar–capillary unit of the human lung. The alveolar epithelial cells were exposed to air to induce differentiation and the formation of the alveolar–epithelium barrier tissue. To recapitulate the breathing action of lung, cyclic vacuum suction was applied to two side hollow chambers adjacent to the cell culture channels to induce cyclic mechanical stretching of the adherent cell layers. The mechanical strain also showed to enhance the uptake of the epithelial layer of nanoparticles and stimulates their transport into the underlying microvascular channel. Besides the lung, a number of tissues in the human body are exposed to mechanical strain, such as the heart, bone, skin, gut, and urothelium.143 Recent research on the role of mechanical forces in wound healing and repair opens the possibility of targeting mechano-transduction pathways to reduce scar formation.144 Epidermal keratinocytes and dermal fibroblasts can sense the mechanical force, which, together with soluble and immobilized signal cues, are responsible for the observed wound healing response.143 The ability to reproduce in vivo-like strain on tissue culture in vitro would enable the investigation of the mechanical–biochemical signal interaction and target specific pathways associated with mechanical strain.145
e. Migration
Motility is an essential feature of live cells during all the stages of cell life. Monitoring cell migration is important for understanding a variety of physiological and pathological processes in the tissue in vitro models such as organogenesis, cancer metastasis, and inflammation. Data obtained from these tests may allow for an understanding of how a particular cell type migrates or responds to a chemical stimulation and migrates toward it. The most common cell migration assays are wound healing and the transwell migration and invasion assays.146,147 Most of microfluidic-based cell migration studies have focused on single cell types rather than heterogeneous cellular structure, which dramatically simplifies the in vivo conditions. More sophisticated tools may be required to study the tissue-level cell migration for clinical applications. As an example of OOC-based cell migration study is the cancer metastasis. During metastasis, cancer cells migrate from their initial location to distant organs that lead to the formation of new tumor, which involves cell–cell interaction and chemotaxis.148,149 Zervantonakis et al.72 developed a microfluidic-based assay to recreate the tumor–vascular interface to test the effect of biochemical factors from the interacting cells on carcinoma cell intravasation. The study showed that signaling with macrophages via secretion of TNFα results in endothelial barrier impairment and the intravasation rates increase as validated with live imaging. It was also found that endothelial barrier impairment is associated with a higher number and faster dynamics of tumor cell–endothelial cell interactions. In another study, Lei et al.150 developed a compartmentalized microfluidic chip to examine the interaction between neurons and cancer cells. It was observed that the nerve bundles to guide a directional migration of cancer cells. The on-chip model allows the screening of compounds that inhibit cancer cell migration along neurites in vitro.
f. Immune competence: Response to toxic or pathogenic substances
The communication between parenchymal cells and immune cells is a key characteristic in any biologically active in vitro model, which takes place via either soluble factors or direct cell–cell interaction at the crossroad of drug/nutrient metabolism and immune modulation. Inflammation is an immunological mechanism that assists in the removal of infectious and other foreign substances, which consists of a cascade of immunological, physiological, and behavioral processes, and is orchestrated by soluble immune signaling molecules, called cytokines.151,152 This also leads to a variety of cellular responses in tissues including elevated permeability in microvessels, attachment of circulating cells to the vessels in the vicinity of the injury site, migration of several cell types, cell apoptosis, and growth of new tissue and blood vessels.152,153 The response of various epithelial in vitro models to allergen exposure was studied using OOC systems such as the small intestine,36–37 skin,46,154 and lung/airway.17,155
3. Testing, validation, and screening
The use of OOC devices for screening depends on successful device validation to ensure that the biological functions reproduced in vitro are representative of the native tissues in vivo, which is crucial to enable adopting this technology in industrial applications. A direct comparison against the current industry standards as well as between different technologies might be necessary for the translational OOC technology and to increase the confidence in the outputs of OOC systems.156 However, to date, validating studies of OOC systems are very limited and the vast majority of OOC studies focus on the demonstration of the micro-anatomy of specific tissue or organ and characterization of simple functions. Ewart et al.157 describe the main steps and key considerations that need to be taken in order to enable OOC technology adoption into the industrial laboratory. Written by scientists from 13 pharmaceutical companies and partners at the National Institutes of Health (NIH), this article captures a consensus view on the progression strategy to facilitate and accelerate the adoption of this valuable technology.
In general, there are three primary uses of OOC systems that provide initial value for industry:157
-
(1)
Mechanistic investigation: The OOC system will enable rapid assessment of new drug mechanisms in pharmacology or toxicity. The technical standards for use in mechanistic investigation would be defined by the biology or pathobiology of the site of action to be investigated.157
-
(2)
Preclinical safety screening: The key functionality of in vitro models is to provide in vitro datasets that can accurately predict the adverse effects in vivo. Such compounds with high risk profile to be deprioritized, while those with a lower risk profile are brought forward.158
-
(3)
Drug absorption, distribution, metabolism, and excretion (ADME): ADME assays are critical in gaining insights into the metabolism and potential drug interactions. Robust in vivo predictions can be achieved with well-characterized in vitro models with the combination of computational methods (physiology-based pharmacokinetics/toxicokinetics).
IV. BIOLOGICAL INTEGRATION (CELL/TISSUE HETEROGENEITY)
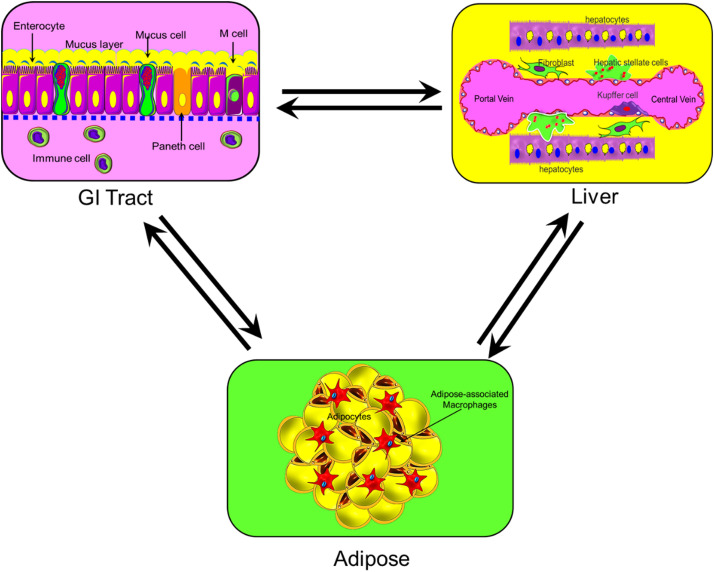
The function of the organ(s) is orchestrated by complex cellular structures and physiochemical factors within the ECM and secreted by various types of cells. Each organ in the human body contains a diverse type of cells that are necessary to perform its function.159–164 Knowing the cellular structure (microanatomy) of the target organ is essential to determine the accurate biological model of the OOC system. Various OOC systems with multiple cell types have been or are being constructed, but none have yet to capture the full diversity of cell types within an organ that is highly complex, therefore, posing a tremendous challenge to mimic. It is unrealistic to expect an OOC system that recapitulates the full cellular structure of a healthy/diseased organ in the near future. Nonetheless, it is still important to study cellular heterogeneity of target tissue/organs and identify the best subset of cells that provide a sufficient physiological representation that models the structure of that organ. By creating the minimal structure and analyzing its function, the response of the organ(s) model can be extrapolated, which enables capturing the specific functions of interest. For example, integration of the GI tract, liver, and adipose represent organ-level heterogeneity of the “the gut–liver–adipose axis” as illustrated in Fig. 5. The GI tract is a complex environment where human epithelial cells, commensal bacteria, and opportunistic pathogens constantly interact with each other and are faced with a continuous flow of nutrients and metabolites. The microarchitecture of the GI tract barrier (epithelium) includes a single layered or pseudostratified columnar epithelium enterocytes, Paneth cells, mucus-secreting goblet cells, hormone-secreting enteroendocrine cells, and antigen-sampling microfold (M) cells165 (Fig. 5). On the other hand, the microarchitecture of the liver forms a 3D structure that consists of epithelial and mesenchymal cells arranged in repetitive microscopic units. The major cell types that constitute the minimal structure of the liver are hepatocytes, hepatic stellate cells, fibroblasts, and Kupffer cells, which also include vessels and biliary duct parenchymal distributions.166 An engineered system can include the GI tract module in close vicinity to a liver module to emulate the drugs/nutrients metabolization by the liver immediately after GI tract absorption before reaching the systemic circulation. Furthermore, to study the obesity-associated metabolic pathophysiology, adipose tissue can be integrated into the system, which further increases the complexity of the model. This complex interconnected model would enable studying the absorption, metabolism, and mechanistic investigation of drug compounds in a systemic manner (Fig. 5). A recent review by Mertz et al.167 addresses the advances in characterization of cell heterogeneity and cell plasticity and how it impacts tissue and organ function.
FIG. 5.
An example of biological model integration: interconnected GI tract, liver, and adipose represent organ-level heterogeneity of the “the gut–liver–adipose axis.”
V. ENGINEERING SYSTEM INTEGRATION
Some of the obvious advantages of OOC compared to conventional multi-well plates are the dynamic interactions between several organs that enable reproducing specific functions in vitro and the ability to perform time resolved measurements at different check points. This would require in situ, real-time, non-destructive detection assays, rather than end point assays. OOCs are highly integrated and complex systems by nature as they reflect human physiology. To construct a sufficiently physiologically relevant in vitro model, the minimal number of cell subsets of cells must be included in the co-culture system. Integration of additional cell type/tissue/organ into the system adds another level of unavoidable complexity. In consequence, the complexity of the biological model would require engineering a complex device. Different cell types may require different culture medium, nutrients, and growth factors and may not be compatible to co-culture before differentiation/maturation. In addition, cell organization/order and interfacing vary in different tissue and organs as discussed earlier. Such complexity puts the OOC technology at risk of delay of market penetration and acceptance by the targeted end users. Furthermore, new medical technologies require competent health professionals who are well-trained to adopt such advanced technology. In consequence, this would pose a high assay cost that is prone to a high rate of use-related errors, which raises an important question “Can this complex technology be commercialized?”
OOC technology is progressing through two parallel development tasks: cell models (biological model) and device engineering; however, the recent significant development in cell biology (e.g., iPSCs) does not accompany the same speed of OOC device engineering. To accelerate this translational technology, reducing the gap between the academic research and industry and, therefore, enabling smoother adaptation by the end user, many challenges must yet be overcome to meet the requirements listed above to realize market-driven products. The ability to perform a morphological non-invasive in-line assessment is advantageous for assessing the structural integrity and drug- or disease-induced structural changes. Sensors and imaging modalities integration and enabling real-time sampling of the cellular products and multi-channel monitoring would also be attractive features.
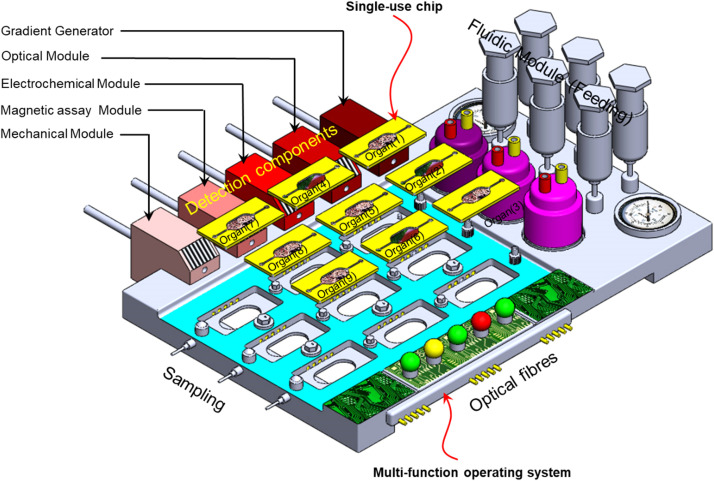
Engineering OOC devices may be advantageous to the well-established semiconductor industry, as it is initially the case of the fabrication of the single device, such as integrated circuits (ICs), printed circuit boards (PCBs), and field-programmable gate arrays (FPGAs). OOC chips are single-use devices that need not be expensive to be adopted by the end user; therefore, one approach is to minimize the complexity of the fluidic chip that hosts the organotypic culture while maximizing the functionality of the operating unit. Figure 6 depicts an integrated OOC system with disposable organ chips (OCs) and a multi-function operating system (OS).
FIG. 6.
Depicted integrated OOC system with disposable organ chips (OOCs) and multi-function operating system (OS).
VI. OOC IN THE INDUSTRY
OOC technology is still in its beginning stage and has considerable interest among researchers in academia with increasing number of publications that cover various aspects of physiology. However, as we emphasized earlier, tremendous effort is needed to tackle the technological challenges to enable transferring this technology from the laboratory to the industry. A rough estimation of the highest technology readiness level (TRL), which describes the maturity of a technology, among current industry players can be positioned at TRL4, which refers to technology validation in the laboratory environment,168 while the TRL for technology that is tested in the application environment is TRL9. OOC technology has gained a lot of momentum toward commercialization fuelled by support from governmental funders as well as the industry, in particular, the pharmaceutical industry. Two major initiatives actively working in this domain are listed in Table IV.
TABLE IV.
The two major initiatives actively working toward the advancement of OOC technology.
| Initiative (where) | Funding agencies and organizers | Academia involved | Industry partners | Activities |
|---|---|---|---|---|
| Tissue Chip (USA) (https://ncats.nih.gov/tissuechip) | National Institute of Health (NIH), National Center for advancing Translational Science (NCATS), Food and Drug Administration (FDA), Defense Advanced Research Projects Agency (DARPA) | Columbia University, Duke University, Harvard University, Massachusetts Institute of Technology, University of Wisconsin–Madison, Northwestern University, University of California (Berkeley), University of Pittsburgh, University of Washington (Seattle), Vanderbilt University, Washington University (St. Louis), Cornell University, Duke University, University of California (Irvine), Johns Hopkins University, University of Florida, Stanford University | GlaxoSmithKline (GSK), Pfizer, Inc. AstraZeneca, Children's Hospital of Philadelphia, Emulate, Inc., Boston |
|
| ORCHID (Europe) (https://h2020-orchid.eu/) | European Union's Horizon 2020 research and innovation program | The Institute for human Organ and Disease Model technologies (hDMT), the Netherlands University of Twente, Eindhoven University, University of Delft, Leiden University, Erasmus Medical Center, the Hubrecht Institute, Fraunhofer, IMEC, and Universidad Zaragoza |
Genmab and Galapagos, Leiden University medical center, TNO, Amsterdam UMC, Hubrecht Organoid Technology (HUB), and Philips |
|
In 2012, the National Institutes of Health (NIH), led by the National Center for Advancing Translational Sciences (NCATS), launched the Tissue Chip program. The program is a joint collaboration with the U.S. Food and Drug Administration (FDA) and the Defense Advanced Research Projects Agency (DARPA), which awarded 12 projects that supported the development OOC systems that represent human organs and 7 projects that explored the use of stem and progenitor cells to differentiate into multiple cell types that represent the cellular architecture within organ systems. In 2014, NIH funded the next phase of the tissue chip program to improve ways of predicting drug safety and effectiveness. The next phase aimed to link individual organs on chips to develop a human multi-organ model system to replicate the complex human response to drug exposure. Ultimately, linking a major human organ model in one system to form a human-body-on-a-chip model intends to accelerate the translation of these basic discoveries into the clinic and industry. In 2016, NCATS with a partnership with the Center for the Advancement of Science in Space (CASIS), launched the Tissue Chips in Space initiative, which aims to help refine the Tissue Chip platforms for in-flight experiments at the International Space Station U.S. National Laboratory (ISS National Lab), so that scientists can better understand diseases and translate their findings to affect human health on Earth. In 2017, NCATS announced 13 additional awards to develop 3D Tissue Chip research platforms that model diseases and test drug efficacy before clinical trials. In 2018, NCATS announced the Tissue Chip Consortium awardees to develop tissue chip models for pain, opioid addiction, and type 2 diabetes. In order to provide a way for independent testing and validation of platforms developed by Tissue Chip for drug screening, NCATS awarded three testing centers to assess the chips’ robustness and provide inputs for further improvement of the devices.
Over the past few years, several companies and start-ups have lunched OOC-based devices or tools for tissue/organ modeling (Table V). Among these products, MIMETAS OrganoPlate® has shown many uses in the literature. The MIMETAS OrganoPlate® features 96 (with two-lane) or 40 (with three-lane) independent culture cells, each supporting one in-gel culture and one/two perfusion channels [Fig. 7(a)]. The system employs the Phaseguides™ technology as meniscus pinning barriers that enable precise, barrier-free definition of culture matrices and cells in three dimensions.169 The pump-less media flow is achieved by using a gravity-driven leveling technique. TissUse GmbH (Berlin, Germany) introduced a variety of HUMIMIC Chips, such as HUMIMIC Chip2, HUMIMIC Chip3, HUMIMIC Chip4, and HUMIMIC Chip XX/XY, for different in vitro modeling purposes [Fig. 7(b)]. The organ models are connected by microfluidic channels, which are covered with human dermal microvascular endothelial cells.178 A peristaltic on-chip pump enables an in vivo-like nutrient and oxygen supply to the cells and generates pulsatile shear stress in an adjustable range. In 2010, the lung-on-a-chip system was developed by Ingber's group17 at the Wyss Institute, which mimics the function of a lung alveolus, including the breathing mechanism. Emulate Inc. (Boston, USA) is currently commercializing a variety of organ chips based on this platform [Fig. 7(c)]. The basic chip comprises two microfluidic chambers separated by a semi-porous membrane with two side vacuum channels. Applying a cyclic vacuum through the side channels causes the cell layer on the membrane to stretch, mimicking natural stretching during inhalation of the lung. Another interesting OOC chip is provided by AIMBiotech, which comprises a gel region with adjoining media channels separated from the gel channel by trapezoidal posts [Fig. 7(d)]. The chip allows 3D cell culture in the gel channel and immune cell (e.g., T cell) circulation in the adjacent channels.
TABLE V.
Companies and start-ups actively working in the area of OOC.
| Company (founding year), website | Technology | Product(s) | Structure and characteristics | Applications (examples) | Reference |
|---|---|---|---|---|---|
| Hμrel Corp. (2006) www.hurelcorp.com |
|
8 species and 10 models, e.g., HμRELhuman™ HμRELhumanPool™ HμRELrat™ HμRELratWH™ HμRELprimate™ HμRELminipig™ HμRELdog™ HμRELmouse™ HμRELrabbit™ HμRELcat™ |
|
Studying the cellular response, metabolic competency, and pharmacokinetic interactions among multiple tissues and organs Viral chronicity in liver disease models |
169 and 170 |
| Mimetas (2013), www.mimetas.com |
|
OrganoPlate® PhaseGuide™ OrganoPlate® 2-lane, OrganoPlate® 3-lane, OrganoPlate® Graft, OrganoPlate® Caco-2, OrganoTEER®, OrganoFlow® L |
|
Pancreatic cancer, blood vessel, kidney proximal tube, CNS toxicology, blood–brain barrier, gut, angiogenesis, liver, breast cancer | 171–177 |
| TissUse (2010) www.tissuse.com |
|
HUMIMIC Chip HUMIMIC Starter HUMIMIC AutoLab HUMIMIC AutoPlant |
Organ compartments of varying size On-chip pump |
Combination of multi-organs, e.g., intestine, liver, kidney, and neuronal tissue Human skin |
178–180 |
| Emulate (2013) www.emulatebio.com | Human Emulation System for multi-organ culture Chip-lab equipment interfacing |
Organ-Chips Pod™ Portable Module Zoë™ Culture Module Orb™ Hub Module Bio-kit for Kidney, liver and intestine |
Stretchable plastic chip with porous membrane Allow organ chips to be transported and placed on standard microscopes for imaging Up to 12 Organ Chip combinations |
lung, liver, intestine, and kidney | 181–184 |
| AIM Biotech (2012) www.aimbiotech.com |
|
Microscope slide format chips | Compatible with all polymerizable gels Enables the control of interstitial flow across the 3D gel region Rapid media exchange through vacuum aspiration |
Immunotherapy, neurobiology, and vascular biology | 185–186 |
| 4DBio (2014) www.4designbiosciences.com |
|
Vascularized 96-well microfluidic plate | Channels are lined with endothelial cells and act as arteriole and venule, connected by a network of living capillaries within a physiologic extracellular matrix | Tumor, metastasis, and vascular pathology | |
| AxoSim (2014) www.axosim.com |
|
NERVESIM™ BrainSim™ |
iPSC derived spheroids in a 3D culture environment | Neurotoxicity neurodegenerative diseases |
|
| CNBio (2009) www.cn-bio.com |
|
PhysioMimix: Multi-organ interactions, ADME, and toxicity | Benchtop fluidic control Up to six MPS plates can be run for multiple independent experiments |
Toxicology, drug metabolism, and disease modeling | |
| AlveoliX AG (2015), www.alveolix.com | Lung-on-chip model | AXLung-on-Chip System | Ultrathin membrane breathing motion | Toxicity | |
| Hesperos Inc. (2015) www.hesperosinc.com | Multi-organ fluidic chip | Organ models: heart–liver, heart–liver–skeletal muscle–neuron, neuromuscular junction, heart–liver–cancer | Serum-free cell medium Gravity flow system |
Customized human-on-a-chip systems | |
| Kirkstall (2006) www.kirkstall.com |
Interconnected cell culture flow system | QV500 QV600 QV900 |
Produced from medical-grade silicone Compatible with commercially available transwells and inserts |
Liver, brain, cardio, respiratory, kidney, and gut | |
| Nortisbio (2015) https://www.nortisbio.com |
|
ParVivo™ perfusion system | Plug and play Chips with pre-established tissues | Toxicity and efficacy testing | |
| Synvivo https://www.synvivobio.com/ |
|
SynBBB (blood–brain barrier model) SynTumor (various cancer models) SynRAM (inflammation model for rolling adhesion and migration assays) SynTox (Toxicology) |
Air liquid interface (ALI) In vivo-like airway structure Real-time visualization |
Toxicity assays Biomarker analysis Therapeutic screening |
FIG. 7.
(a) The microfluidic microtiter plate OrganoPlate from MIMETAS with three-line 40 integrated microfluidic chips. A gel channel (blue) holds the extracellular matrix (ECM) in place through the PhaseGuide's pressure barrier function. Cells can be introduced in the middle lane (gel channel) to create a 3D cell culture while medium flow through the adjacent channels. Reproduced with permission from Kramer et al., J. Mol. Sci. 20, 4647 (2019), Copyright 2019 MDPI.171 (b) The multi-organ chip platform (Chip3) from TissUse GmbH. The chip comprises five assembled layers: glass slide, PDMS layer with microfluidic channels, adapter layer, cell culture insert layer, and reservoir layer. Reproduced with permission from Schimek et al., Sci. Rep. 10, 7865 (2020). Copyright 2020 Nature Research. (c) The organ chip from Emulate. The chip was used to construct an in vitro model of the human lung alveolus. Epithelial cells are cultivated on top of a semi-porous membrane, and human pulmonary microvascular endothelial cells are cultivated on the bottom of the membrane. Applying a cyclic vacuum through the side chambers causes the cell layer to stretch, mimicking natural stretching during inhalation. Reproduced with permission from Kasendra et al., Sci. Rep. 8, 2871 (2018). Copyright 2018 Nature Research.182–184 (d) AIM Bioterch cell culture chip comprises a gel region with adjoining media channels separated from the gel channel by trapezoidal posts. Reproduced with permission from Zervantonakis et al., Proc. Natl. Acad. Sci. U.S.A. 109(34), 13515–13520 (2012). Copyright 2012 the National Academy of Sciences of the United States of America.186
VII. INDUSTRY TRANSFER HURDLES
The ultimate goal of OOC technology is to emulate human organs and eventually the whole body in health and disease states and to enable mimicking the interaction of drugs with body. The majority of OOC studies focus mainly on the biological characterization of the cellular components with limited effort toward device engineering. The main hurdle delaying the maturation of OOC device engineering is the fact that every OOC study uses different sets of cells to investigate different biological/medical effects. Most of these studies share a similar simple device structure that relies on various peripheral tools to provide the necessary cell culture environment or capture cell signaling. To develop a reliable single-organ OOC system, a key cellular structure needs to be present. Also, a precise cellular manipulation within the OOC system and a detailed understanding of the human body's complex response to xenobiotics are necessary. This requires engineering a robust device that enables a long-term cell manipulation and signal capturing without compromising the cellular system viability. Increasing the biological complexity, i.e., employing more cell types or adding additional organ(s) to the model, would require more complex fluidic networks, hence increasing the complexity of the OOC device.
The drug industry is well established, which involves complex, costly, and regulated process in a highly competitive environment.187 Therefore, adopting a new technology within this environment may involve risking the drug production. On the other hand, OOC is a relatively young technology with most of its related research implemented as exploratory studies outside the mainstream pharmaceutical development and is not mature enough to be adopted as a part of ongoing drug development programs. Despite the tremendous effort focused on OOC device prototyping among many start-ups around the world, penetrating of the OOC technology into the pharmaceutical industry is still facing many scientific (as described above), manufacturing, and industrial challenges. OOC chips need to be reliable, affordable, scalable, and mass manufacturable. Current OOC devices require skilled scientific personnel to operate and rely on various peripheral equipment during the cell culture process, such as incubators, pumps, automated dispensers, sensors, and actuators, as well as model characterization and readout, such as optical imaging devices. It becomes crucial to enable the OOC devices beyond the research laboratory bench by either developing robust instrumentations particularly for OOCs or enabling the interface of OOC devices with the classical analytical tools.
Manufacturing of OOC chips for industrial use is a major hurdle that the OOC technology faces as the adopted fabrication methodologies are originated and extended from academic labs. OOC fabrication requires a multidisciplinary approach of engineers, fabrication expertise, material scientists, and biologists. In this respect, it is important to apply mature manufacturing expertise to accelerate device engineering and prototyping processes. Industry aligned funds, with strong collaboration with the pharma industry, need to be directed toward solving the major challenges facing the industry transfer including device engineering, fabrication material, cell–material interface, biological complexity, and sensors integration, among others, and enabling chips prototyping beyond the academic labs. Figure 8 depicts a possible road map of OOC production with some of the associated challenges. One possible approach is to develop a modular universal OOC platform that enables hosting a variety of organ-on-a-chip or body-on-a-chip models. Early communication with stakeholders (researcher, regulators, suppliers, and end users) would help identify the industry needs in early stages of development. Depending on the nature of the assay, OOC systems (products) can be designed to provide a single or multi-organ in vitro model. Ultimately, body-on-a-chip can be realized by integrating all the key organs in one system.
FIG. 8.
A road map of OOC production and the associated challenges.
Although there are only a few young companies developing OOC devices such as those listed in Table V, they are, in fact, showing great progress in providing exploratory platforms for feasibility studies with exciting innovations at increasing speed. Additionally, many major pharmaceutical and consumer care companies are involved, in collaboration with the companies mentioned above, in feasibility studies and have started to build their own OOC R&D investigational units. For example, Emulate Inc. has started a collaboration program with Johnson & Johnson company (NJ, USA)188 for drug candidate screening. TissUse GmbH is also collaborating with Bayer AG (Leverkusen, Germany)189 and AstrZeneca190 to explore the potential use of OOC models in drug screening. Other start-ups also announce similar collaboration with unannounced industry partners.
VIII. ADDITIVE MANUFACTURING FOR ORGAN-ON-A-CHIP
The vast majority of microfluidic, LOC, and OOC chips are realized using the conventional fabrication techniques including microfabrication (i.e., silicon fabrication process such as lithography, etching, and deposition), soft lithography, injection molding, laser cutting, embossing, etc. These techniques belong to the subtractive manufacturing (SM) technology by which 3D objects are constructed by cutting material away from a solid block of material (e.g., silicon wafer, polymer sheet). The advent of the additive manufacturing (AM), which is the process of joining materials, through a layer-by-layer addition to create a 3D object from a CAD model, such as 3D printing, would trigger a manufacturing revolution in a wide industry sectors and microfluidics is not an exception. More recently, 3D printing has been adopted to utilize life cells and gels as printing materials (bioinks) to create in vitro tissue models in the macro-scale.191 Such technology would push the boundaries of the in vitro testing including OOC technology in different ways:
-
•
3D printing of the OOC's microfluidic devices: Adoption of 3D printing in microfluidics would overcome many constrains of the current fabrication methods such as enabling 3D microfluidic structures (microfluidic fabrication conventionally fabricated in 2D), elimination of the back-end assembly process (e.g., bonding of multilayers), and integration of the necessary fluidic components (e.g., valves), sensors, and actuators in a precise and automated manner. This new generation of microfluidic devices would be more autonomously flexible to use in many LOC and OOC applications. A comprehensive review on 3D printed microfluidics can be found elsewhere.192
-
•
3D printing of the tissue scaffolds: complex tissue scaffolds can be produced based on computer designs obtained from patient-specific anatomical data to construct a scaffold on which living cells might be seeded to grow an approximation of a tissue. The ability to create these complex 3D shapes is very attractive for tissue engineering, which enables precise control of both microstructures and microarchitecture. The pre-fabrication of 3D scaffolds from biocompatible materials as a top–down approach would be followed by seeding the cells onto the scaffolds to create the miniaturized 3D tissue structure that emulates the structure and morphology of the human organ.193,194 Recent work by Tan et al.195 describes the use of the cryogenic 3D printing method to produce 3D structures that mimic the mechanical properties of the softest tissues in the human body by utilizing the liquid to solid phase change of a composite hydrogel ink. This was achieved by rapidly cooling the ink solution below its freezing point using solid carbon dioxide (CO2) in an isopropanol bath.
-
•
3D bio-printing of tissue/organ models: Another option is to follow a bottom–up approach to spatially immobilize various types of living cells to generate heterogeneous functional structures within a prefabricated chip and scaffold. This would enable the creation of a heterogeneous tissue structure, i.e., linked multi-organs. Recently, various printed organ models were realized including liver,194 heart,196 vasculature,197 and kidney.198 A recent strategy, termed 4D printing, is to add the time factor, which enables the fabrication tissue structure with flexible shapes that can be actuated to reproduce specific functionality.199
IX. CONCLUSIONS AND FUTURE OUTLOOK
In vitro microphysiological models, or the so-called “tissue/organ/human-on-a-chip,” is a very ambitious technology which could revolutionize many aspects in medicine and industry, particularly for preclinical-to-clinical translation in the pharmaceutical arena. However, many technical challenges, as well as standardization and regulatory endorsement, are still ahead and need to be overcome before adopting this technology as an alternative to animal models and transferring them from the laboratory to industry. To realize physiologically relevant in vitro models, a complex functional tissue structure “micro-anatomy” needs to be established, and this would require devising tools for cell and reagent manipulating, monitoring, sensing, and readout capturing. Many significant works were recently demonstrated for inter-connected multi-organ systems for drug screening, which show a high momentum toward the realization of the “human-on-a-chip” concept. However, individual organ models need to be established to increase their reliability and maximize the key physiological robustness. Research in iPSCs is actively mobilized for transferring to many applications including OOC technology, and these cells form an attractive option to be adopted for organ and disease modeling as well as personalized medicine. Trans-disciplinary research involving stem cell biologists, engineers, physiologists, and pharmaceutical industry partners is necessary to enable crossing the boundary between the lab and industry. Furthermore, the last few years witnessed the emergence of several partnerships between pharmaceutical corporations, start-ups, and academia that demonstrate the importance of this technology that carries a tremendous potential in mitigating the current business needs in the drug discovery and promise commercial success.
OOC is an engineering driven technology and is benefited from well-established multi-engineering disciplines including microfabrication, microfluidics, microscopy, sensors, etc. Recently, the convergence of 3D (bio) printing with microfluidics, LOC, and OOCs adds an extra momentum toward the enabling the technology and its commercialization. OOCs would represent a paradigm shift in physiology research as they would enable continuous monitoring of living cells and rapid detection of mode of action, allowing an unprecedented advance in our understanding of living cells and how they respond to stimuli and providing a wealth of information at low cost and in a short time. But OOC technology goes far beyond the pharmaceutical industry and holds great promise to deepen our understanding of the complex interactions between the body and exogenic stimuli and provides a more accurate assessment of the unknown effects of drugs, food, chemicals, pathogens, and environmental toxins.
DATA AVAILABILITY
The data that support the findings of this study are available within the article.
REFERENCES
- 1.Curry S. H., Ann. N. Y. Acad. Sci. 993, 69–74 (2003). 10.1111/j.1749-6632.2003.tb07512.x [DOI] [PubMed] [Google Scholar]
- 2.Knight A., Bailey J., and Balcombe J., Altern. Lab. Anim. 34, 19–27 (2006). 10.1177/026119290603400117 [DOI] [PubMed] [Google Scholar]
- 3.Shanks N., Greek R., and Greek J., Philos. Ethics Humanit. Med. 4(2), (published online 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seok J., Warren H. S., Cuenca A. G., Mindrinos M. N., Baker H. V., Xu W., Richards D. R., McDonald-Smith G. P., Gao H., Hennessy L., Finnerty C. C., López C. M., Honari S., Moore E. E., Minei J. P., Cuschieri J., Bankey P. E., Johnson J. L., Sperry J., Nathens A. B., Billiar T. R., West M. A., Jeschke M. G., Klein M. B., Gamelli R. L., Gibran N. S., Brownstein B. H., Miller-Graziano C., Calvano S. E., Mason P. H., Cobb J. P., Rahme L. G., Lowry S. F., Maier R. V., Moldawer L. L., Herndon D. N., Davis R. W., Xiao W., and Tompkins R. G., Proc. Natl. Acad. Sci. U.S.A. 110, 3507–3512 (2013). 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentile C., Curr. Stem Cell. Res. Ther. 11, 652–665 (2016). 10.2174/1574888X10666151001114848 [DOI] [PubMed] [Google Scholar]
- 6.Geris L., Guyot Y., Schrooten J., and Papantoniou I., Interface Focus 6, 20150105 (2016). 10.1098/rsfs.2015.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astashkina A., Mann B., and Grainger D. W., Pharmacol. Ther. 134, 82–106 (2102). 10.1016/j.pharmthera.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 8.Xia N., Thodeti C. K., Hunt T. P., Xu Q. B., Ho M., Whitesides G. M., Westervelt R., and Ingber D. E., FASEB J. 22, 1649–1659 (2008). 10.1096/fj.07-090571 [DOI] [PubMed] [Google Scholar]
- 9.Desai R. A., Gao L., Raghavan S., Liu W. F., and Chen C. S., J. Cell Sci. 122, 905–911 (2009). 10.1242/jcs.028183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho C. T., Lin R. Z., Chen R. J., Chin C. K., Gong S. E., Chang H. Y., Peng H. L., Hsu L., Yew T. R., Chang S. F., and Liu C. F., Lab Chip 13, 3578–3587 (2013). 10.1039/c3lc50402f [DOI] [PubMed] [Google Scholar]
- 11.Khademhosseini A., Langer R., Borenstein J., and Vacanti J. P., Proc. Natl. Acad. Sci. U.S.A. 103, 2480–2487 (2006). 10.1073/pnas.0507681102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takayama Y., Kotake N., Haga T., Suzuki T., and Mabuchi K., J. Biosci. Bioeng. 114, 92–95 (2012). 10.1016/j.jbiosc.2012.02.011 [DOI] [PubMed] [Google Scholar]
- 13.Hynes W. F., Doty N. J., Zarembinski T. I., Schwartz M. P., Toepke M. W., Murphy W. L., Atzet S. K., Clark R., Melendez J. A., and Cady N. C., Biosensors 4, 28–44 (2014). 10.3390/bios4010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H., Jiang D., Zhu P., Pi F., Ji J., Sun C., Sun J., and Sun X., Biosens. Bioelectron. 3, 126–133 (2016). 10.1016/j.bios.2016.04.028 [DOI] [PubMed] [Google Scholar]
- 15.Liu Z. B., Zhang Y., Yu J. J., Mak A. F. T., Li Y., and Yang M., Sens. Actuators B Chem. 143, 776–783 (2010). 10.1016/j.snb.2009.10.020 [DOI] [Google Scholar]
- 16.Sin A., Chin K. C., Jamil M. F., Kostov Y., Rao G., and Shuler M. L., Biotechnol. Prog. 20(1), 338–345 (2004). 10.1021/bp034077d [DOI] [PubMed] [Google Scholar]
- 17.Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328(5986), 1662–1668 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H. J., Huh D., Hamilton G., and Ingber D. E., Lab Chip 12, 2165–2174 (2012). 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 19.Carraro A., Hsu W. M., Kulig K. M., Cheung W. S., Miller M. L., Weinberg E. J., Swart E. F., Kaazempur-Mofrad M., Borenstein J. T., Vacanti J. P., and Neville C., Biomed. Microdevices 10, 795–805 (2008). 10.1007/s10544-008-9194-3 [DOI] [PubMed] [Google Scholar]
- 20.Lee P. J., Hung P. J., and Lee L. P., Biotechnol. Bioeng. 97, 1340–1346 (2007). 10.1002/bit.21360 [DOI] [PubMed] [Google Scholar]
- 21.Kane B. J., Zinner M. J., Yarmush M. L., and Toner M., Anal. Chem. 78, 4291–4298 (2006). 10.1021/ac051856v [DOI] [PubMed] [Google Scholar]
- 22.Chao P., Maguire T., Novik E., Cheng K. C., and Yarmush M. L., Biochem. Pharmacol. 78, 625–632 (2009). 10.1016/j.bcp.2009.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legendre A., Baudoin R., Alberto G., Paullier P., Naudot M., Bricks T., Brocheton J., Jacques S., Cotton J., and Leclerc E., J. Pharm. Sci. 102, 3264–3276 (2013). 10.1002/jps.23466 [DOI] [PubMed] [Google Scholar]
- 24.Novik E., Maguire T. J., Chao P., Cheng K. C., and Yarmush M. L., Biochem. Pharmacol. 79, 1036–1044 (2010). 10.1016/j.bcp.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delalat B., Cozzi C., Ghaemi S. R., Polito G., Kriel F. H., Michl T. D., Harding F. J., Priest C., Barillaro G., and Voelcker N. H., Adv. Funct. Mater. 28, 1801825 (2018). 10.1002/adfm.201801825 [DOI] [Google Scholar]
- 26.Lee J. B., Park J. S., Shin Y. M., Lee D. H., Yoon J. K., Kim D. H., Ko U. H., Kim Y. T., Bae S. H., and Sung H. J., Adv. Funct. Mat. 29, 1900075 (2019). 10.1002/adfm.201900075 [DOI] [Google Scholar]
- 27.Huh D., Fujioka H., Tung Y. C., Futai N., Paine R., Grotberg J. B., and Takayama S., Proc. Natl. Acad. Sci. U.S.A. 104, 18886–18891 (2007). 10.1073/pnas.0610868104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huh D., Leslie D. C., Matthews B. D., Fraser J. P., Jurek S., Hamilton G. A., Thorneloe K. S., McAlexander M. A., and Ingber D. E., Sci. Transl. Med. 4, 159ra147 (2012). 10.1126/scitranslmed.3004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fritsche C. S., Simsch O., Weinberg E. J., Orrick B., Stamm C., Kaazempur-Mofrad M. R., Borenstein J. T., Hetzer R., and Vacanti J. P., Int. J. Artif. Organs 32, 701–710 (2009). 10.1177/039139880903201001 [DOI] [PubMed] [Google Scholar]
- 30.Tavana H., Zamankhan P., Christensen P. J., Grotberg J. B., and Takayama S., Biomed. Microdevices 13, 731–742 (2011). 10.1007/s10544-011-9543-5 [DOI] [PubMed] [Google Scholar]
- 31.Felder M., Stucki A. O., Stucki J. D., Geiser T., and Guenat O. T., Integr. Biol. 6, 1132–1140 (2014). 10.1039/C4IB00149D [DOI] [PubMed] [Google Scholar]
- 32.Felder M., Trueeb B., Stucki A. O., Borcard S., Stucki J. D., Schnyder B., Geiser T., and Guenat O. T., Front. Bioeng. Biotechnol. 22, 3 (2019). 10.3389/fbioe.2019.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stucki A. O., Stucki J. D., Hall S. R. R., Felder M., Mermoud Y., Schmid R. A., Geiserce T., and Guenat O. T., Lab Chip. 15, 1302–1310 (2015). 10.1039/C4LC01252F [DOI] [PubMed] [Google Scholar]
- 34.Shin W. and Kim H. J., Proc. Natl. Acad. Sci. U.S.A. 115, E10539–E10547 (2018). 10.1073/pnas.1810819115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H. J. and Ingber D. E., Integr. Biol. 5, 1130–1140 (2013). 10.1039/c3ib40126j [DOI] [PubMed] [Google Scholar]
- 36.Ramadan Q., Jafarpoorchekab H., Huang C., Silacci P., Carrara S., Koklu G., Ghaye J., Ramsden J., Ruffert C., Vergeres G., and Gijs M. A. M., Lab Chip 13, 196 (2013). 10.1039/C2LC40845G [DOI] [PubMed] [Google Scholar]
- 37.Ramadan Q. and Jing L., Biomed. Microdevices 18(1), 11 (2016). 10.1007/s10544-016-0035-5 [DOI] [PubMed] [Google Scholar]
- 38.Gourikutty S. B. N., Chiam S. Y., and Ramadan Q., J. Micromech. Microeng. 27(12) 124004 (2017). 10.1088/1361-6439/aa96bd [DOI] [Google Scholar]
- 39.Shin W., Wu A., Massidda M. W., Foster C., Thomas N., Lee D. W., Koh H., Ju Y. W., Kim J. H., and Kim H. J., Front. Bioeng. Biotechnol. 7, 13 (2019). 10.3389/fbioe.2019.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang K. J. and Suh K. Y., Lab Chip 10, 36–42 (2010). 10.1039/B907515A [DOI] [PubMed] [Google Scholar]
- 41.Baudoin R., Griscom L., Monge M., Legallais C., and Leclerc E., Biotechnol. Prog. 23, 1245–1253 (2007). 10.1021/bp0603513 [DOI] [PubMed] [Google Scholar]
- 42.Jang K. J., Mehr A. P., Hamilton G. A., McPartlin L. A., Chung S., Suh K. Y., and Ingber D. E., Integr. Biol. 5, 1119–1129 (2013). 10.1039/c3ib40049b [DOI] [PubMed] [Google Scholar]
- 43.Astashkina A. I., Mann B. K., Prestwich G. D., and Grainger D. W., Biomaterials 33, 4700–4711 (2012). 10.1016/j.biomaterials.2012.02.063 [DOI] [PubMed] [Google Scholar]
- 44.Desrochers T. M., Palma E., and Kaplan D. L., Adv. Drug Deliv. Rev. 69–70, 67–80 (2014). 10.1016/j.addr.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ataç B., Wagner I., Horland R., Lauster R., Marx U., Tonevitsky A. G., Azarc R. P., and Lindner G., Lab Chip 13, 3555–3561 (2013). 10.1039/c3lc50227a [DOI] [PubMed] [Google Scholar]
- 46.Ramadan Q. and Ting C. W., Lab Chip 16, 1899 (2016). 10.1039/C6LC00229C [DOI] [PubMed] [Google Scholar]
- 47.Alberti M., Dancik Y., Sriram G., Wu B., Teo Y. L., Feng Z., Bigliardi M., Wu R. G., Wang Z. P., and Bigliardi P. L., Lab Chip 17, 1625–1634 (2017). 10.1039/C6LC01574C [DOI] [PubMed] [Google Scholar]
- 48.Lee S., Jin S. P., Kim Y. K., Sung G. Y., Chung J. H., and Sung J. H., Biomed. Microdevices 19, 22–35 (2017). 10.1007/s10544-017-0156-5 [DOI] [PubMed] [Google Scholar]
- 49.Song H. J., Lim H. Y., Chun W., Choi K. C., Sung J. H., and Sung G. Y., J. Ind. Eng. Chem. 56, 375–381 (2017). 10.1016/j.jiec.2017.07.034 [DOI] [Google Scholar]
- 50.Song H. J., Lim H. Y., Chun W., Choi K. C., Lee T. Y., Sun J. H., and Sun G. Y., J. Ind. Eng. Chem. 60, 355–359 (2018). 10.1016/j.jiec.2017.11.022 [DOI] [Google Scholar]
- 51.Sriram G., Alberti M., Dancik Y., Wu B., Wu R., Feng Z., Ramasamy S., Bigliardi P. L., Bigliardi M., and Wang Z. P., Mater. Today 21(4), 326–340 (2018). 10.1016/j.mattod.2017.11.002 [DOI] [Google Scholar]
- 52.Abaci H. E., Guo Z., Coffman A., Gillette B., Lee W. H., Sia S. K., and Christiano A. M., Adv. Health Mater. 5, 1800–1807 (2016). 10.1002/adhm.201500936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park S. H., Sim W. Y., Min B. H., Yang S. S., Khademhosseini A., and Kaplan D. L., PLoS ONE 7, e46689 (2012). 10.1371/journal.pone.0046689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., Gazit Z., Pelled G., Gazit D., and Vunjak-Novakovic G., Integr. Biol. 3, 39–47 (2011). 10.1039/C0IB00053A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hao S., Ha L., Cheng G., Wan Y., Xia Y., Sosnoski D. M., Mastro A. M., and Zheng S. Y., Small 14(12), 1702787 (2018). 10.1002/smll.201702787 [DOI] [PubMed] [Google Scholar]
- 56.Marturano-Kruik A., Nava M. M., Yeager K., Chramiec A., Hao L., Robinson S., Guo E., Raimondi M. T., and Vunjak-Novakovic G., Proc. Natl. Acad. Sci. U.S.A. 115(6), 1256–1261 (2018). 10.1073/pnas.1714282115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y., Kongsuphol P., Gourikutty S. B. N., and Ramadan Q., Biomed. Microdevices 19(3), 18 (2017). 10.1007/s10544-017-0164-5 [DOI] [PubMed] [Google Scholar]
- 58.Loskill P., Sezhian T., Tharp K. M., Lee-Montiel F. T., Jeeawoody S., Reese W. M., Zushin P. H., Stahl A., and Healy K. E., Lab Chip 7(9), 1645–1654 (2017). 10.1039/C6LC01590E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kongsuphol P., Gupta S., Liu Y. X., Gourikutt S. B. N., Biswas S. K., and Ramadan Q., Sci. Rep. 9, 4887 (2019). 10.1038/s41598-019-41338-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agarwal A., Goss J. A., Cho A., McCain M. L., and Parker K. K., Lab Chip 3, 3599–3608 (2013). 10.1039/c3lc50350j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qian F., Huang C., Lin Y. D., Ivanovskaya A. N., O'Hara T. J., Booth R. H., Creek C. J., Enright H. A., Soscia D. A., Belle A. M., Liao R., Lightstone F. C., Kulp K. S., and Wheeler E. K., Lab Chip 17, 1732–1739 (2017). 10.1039/C7LC00210F [DOI] [PubMed] [Google Scholar]
- 62.Booth R. and Kim H., Lab Chip 12, 1784–1792 (2012). 10.1039/c2lc40094d [DOI] [PubMed] [Google Scholar]
- 63.Kilic O., Pamies D., Lavell E., Schiapparelli P., Feng Y., Hartung T., Quinones-Hinojosa A., Guerrero-Cazares H., and Levchenko A., Lab Chip 16, 4152–4162 (2016). 10.1039/C6LC00946H [DOI] [PubMed] [Google Scholar]
- 64.Griep L. M., Wolbers F., de Wagenaar B., Braak P. M., Weksler B. B., Romero I. A., Couraud P. O., Vermes I., van der Meer A. D., and van den Berg A., Biomed. Microdevices 15, 145–150 (2013). 10.1007/s10544-012-9699-7 [DOI] [PubMed] [Google Scholar]
- 65.Wevers N. R., Kasi D. G., Gray T., Wilschut K. J., Smith B., van Vught R., Shimizu F., Sano Y., Kanda T., Marsh G., Trietsch S. J., Vulto P., Lanz H. L., and Obermeier B., Fluids Barriers CNS 15, 23 (2018). 10.1186/s12987-018-0108-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeong S., Kim S., Buonocore J., Park J., Welsh C. J., Li J., and Han A., IEEE Trans. Biomed. Eng. 65(2), 431–439 (2018). 10.1109/TBME.2017.2773463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng Y., Chen J., Craven M., Choi N. W., Totorica S., Diaz-Santana A., Kermani P., Hempstead B., Fischbach-Teschl C., López J. A., and Stroock A. D., Proc. Natl. Acad. Sci. U.S.A. 109, 9342–9347 (2012). 10.1073/pnas.1201240109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schimek K., Busek M., Brincker S., Groth B., Hoffmann S., Lauster R., Lindner G., Lorenz A., Menzel U., Sonntag F., Walles H., Marx U., and Horland R., Lab Chip 13, 3588–3598 (2013). 10.1039/c3lc50217a [DOI] [PubMed] [Google Scholar]
- 69.Jeon J. S., Bersini S., Gilardi M., Dubini G., Charest J. L., Moretti M., and Kamm R. D., Proc. Natl. Acad. Sci. U.S.A. 112, E818–E818 (2015). 10.1073/pnas.1501426112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandian N. K. R., Mannino R. G., Lam W. A., and Jain A., Curr. Opin. Biomed. Eng. 5, 29–34 (2018). 10.1016/j.cobme.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mannino R. G., Pandian N. K., Jain A., and Lam W. A., Curr. Opin. Biomed. Eng. 5, 13–20 (2017). 10.1016/j.cobme.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zervantonakis I. K., Hughes-Alford S. K., Charest J. L., Condeelis J. S., Gertler F. B., and Kamm R. D., Proc. Natl. Acad. Sci. U.S.A. 109, 13515–13520 (2012). 10.1073/pnas.1210182109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shirure V. S., Bi Y., Curtis M. B., Lezia A., Goedegebuure M. M., Goedegebuure S. P., Aft R., Fieldsbd R. C., and George S. C., Lab Chip 18, 3687–3702 (2018). 10.1039/C8LC00596F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsai H. F., Trubelja A., Shen A. Q., and Bao G., J. R. Soc. Interface 14(131), 20170137 (2017). 10.1098/rsif.2017.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carvalho M. R., Barata D., Teixeira L. M., Giselbrecht S., Reis R. L., Oliveira J. M., Truckenmüller R., and Habibovic P., Sci. Adv. 5(5), eaaw1317 (2019). 10.1126/sciadv.aaw1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y., Warrick J. W., Haubert K., Beebe D. J., and Yin J., Biomed. Microdevices 11(3), 565–570 (2009). 10.1007/s10544-008-9263-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ortega-Prieto A. M., Skelton J. K., Wai S. N., Large E., Lussignol M., Vizcay-Barrena G., Hughes D., Fleck R. A., Thursz M., Catanese M. T., and Dorner M., Nat. Commun. 9, 682 (2018). 10.1038/s41467-018-02969-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y. S., Davoudi F., Walch P., Manbachi A., Luo X., Dell’Erba V., Miri A. K., Albadawi H., Arneri A., Li X., Wang X., Dokmeci M. R., Khademhosseini A., and Oklua R., Lab Chip 16(21), 4097–4105 (2016). 10.1039/C6LC00380J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhatia N. S. and Ingber D. E., Nat. Biotechnol. 32(8), 760–772 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 80.Perestrelo A. R., Águas A. C. P., Rainer A., and Forte G., Sensors 15, 31142–31170 (2015). 10.3390/s151229848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skardal A., Shup T., and Atala A., Drug Discovery Today 21(9), 1399–1411 (2016). 10.1016/j.drudis.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mencattini A., Mattei F., Schiavoni G., Gerardino A., Businaro L., Di Natale C., and Martinelli E., Front. Pharmacol. 10, 100 (2019). 10.3389/fphar.2019.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van den Berg A., Mummery C. L., Passier R., and van der Meer A. D., Lab Chip 19, 198–205 (2019). 10.1039/C8LC00827B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manz A., Graber N., and Widmer H. M., Sens. Actuators B Chem. 1(1–6), 244–248 (1990). 10.1016/0925-4005(90)80209-I [DOI] [Google Scholar]
- 85.Harrison R. G., Greenman M. J., Mall F. B., and Jackson C. M., Anat. Rec. 1, 116–128 (2005). 10.1002/ar.1090010503 [DOI] [Google Scholar]
- 86.Goers L., Freemont P., and Polizzi K. M., J. R. Soc. Interface 11, 20140065 (2014). 10.1098/rsif.2014.0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gonzalez H., Hagerling C., and Werb Z., Genes Dev. 32, 1267–1284 (2018). 10.1101/gad.314617.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hanahan D. and Weinberg R. A., Cell 144, 646–674 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 89.Li H., Fan Y., Kodzius R., Kodzius R., and Foulds I. G., Microsyst. Technol. 18, 373–379 (2012). 10.1007/s00542-011-1410-z [DOI] [Google Scholar]
- 90.Matellan C. and Hernández A. E. R., Sci. Rep. 8, 6971. (2018). 10.1038/s41598-018-25202-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liga A., Morton J. A. S., and Kersaudy-Kerhoas M., Microfluid. Nanofluid. 20, 164 (2016). 10.1007/s10404-016-1823-1 [DOI] [Google Scholar]
- 92.Tsao C. W., Micromachines 7, 225 (2016). 10.3390/mi7120225 [DOI] [Google Scholar]
- 93.Gale B. K., Jafek A. R., Lambert C. J., Goenner B. L., Moghimifam H., Nze U. C., and Kamarapu S. K., Inventions 3, 60 (2018). 10.3390/inventions3030060 [DOI] [Google Scholar]
- 94.Xia Y. and Whitesides G. M., Angew. Chem. Int. Ed. Engl. 37, 550–575 (1998). [DOI] [PubMed] [Google Scholar]
- 95.Eddings M. A., Johnson M. A., and Gale B. K., J. Micromech. Microeng. 18, 067001 (2008). 10.1088/0960-1317/18/6/067001 [DOI] [Google Scholar]
- 96.Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., Science 288, 113–116 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- 97.Guckenberger D. J., de Groot T. E., Wan A. D. M., Beebe D. J., and Young E. W. K., Lab Chip 15, 2364–2378 (2015). 10.1039/C5LC00234F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rahmanian O. and DeVoe D. L., Lab Chip 13, 1102–1108 (2013). 10.1039/c2lc41057e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang J. Y., Zhan J., Yue W., Yang M., Yi C., and Li C. W., RSC Adv. 5, 36036 (2015). 10.1039/C5RA02220G [DOI] [Google Scholar]
- 100.Constantin P. C., Aflori M., Damian R. F., and Rusu R. D., Materials 12, 3166 (2019). 10.3390/ma12193166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hopp B., Smausz T., Papdi B., Bor Z., Szabó A., Kolozsvári L., Fotakis C., and Nógrádi A., Appl. Phys. A 93, 45–49 (2008). 10.1007/s00339-008-4648-2 [DOI] [Google Scholar]
- 102.Moon K. H., Chae B., Kim K. S., Lee S. W., and Jung Y. M., Polymers 11, 489 (2019). 10.3390/polym11030489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Y., Ganser D., Schneider A., Liu R., Grodzinski P., and Kroutchinina N., Anal. Chem. 73(17), 4196–4201 (2001). 10.1021/ac010343v [DOI] [PubMed] [Google Scholar]
- 104.Nunes P. S., Ohlsson P. D., Ordeig O., and Kutter J. P., Microfluid. Nanofluid. 9, 145–161 (2010). 10.1007/s10404-010-0605-4 [DOI] [Google Scholar]
- 105.Keller N., Nargang T. M., Runck M., Kotz F., Striegel A., Sachsenheimer K., Klemm D., Länge K., Worgull M., Richter C., Helmer D., and Rapp B. E., Lab Chip 16, 1561–1564 (2016). 10.1039/C5LC01498K [DOI] [PubMed] [Google Scholar]
- 106.Ronaldson-Bouchard K. and Vunjak-Novakovic G., Cell Stem Cell 22, 310 (2018). 10.1016/j.stem.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maschmeyer I., Lorenz A. K., Schimek K., Hasenberg T., Ramme A. P., Hübner J., Lindner M., Drewell C., Bauer S., Thomas A., Sambo N. S., Sonntag F., Lauster R., and Marx U., Lab Chip 15(12), 2688–2699 (2015). 10.1039/C5LC00392J [DOI] [PubMed] [Google Scholar]
- 108.Materne E. M., Ramme A. P., Terrasso A. P., Serra M., Alves P. M., Brito C., Sakharov D. A., Tonevitsky A. G., Lauster R., and Marx U., J. Biotechnol. 205, 36–46 (2015). 10.1016/j.jbiotec.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 109.Wodarz A. and Nathke I., Nat. Cell Biol. 9, 1016–1024 (2007). 10.1038/ncb433 [DOI] [PubMed] [Google Scholar]
- 110.Lelièvre S. A., Kwok T., and Chittiboyina S., Toxicol. In Vitro 45(Pt 3), 287–295 (2017). 10.1016/j.tiv.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Y. S., Alemana J., Shina S. R., Kilica T., Kima D., Shaegh S. A. M., Massa S., Riahi R., Chaea S., Hu N., Avci H., Zhang W., Silvestria A., Nezhad A. S., Manbohi A., De Ferrari F., Polini A., Calzone G., Shaikh N., Alerasool P., Budina E., Kang J., Bhise N., Ribasa J., Pourmand A., Skardald A., Shupe T., Bishop C. E., Dokmeci M. R., Atala A., and Khademhosseini A., Proc. Natl. Acad. Sci. U.S.A. 14(12), E2293–E2302 (2017). 10.1073/pnas.1612906114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mahler G. J., Esch M. B., Glahn R. P., and Shuler M. L., Biotechnol. Bioeng. 104(1), 193–205 (2009). 10.1002/bit.22366 [DOI] [PubMed] [Google Scholar]
- 113.Sonntag F., Schilling N., Mader K., Gruchow M., Klotzbach U., Lindner G., Horland R., Wagner I., Lauster R., Howitz S., Hoffmann S., and Marx U., J. Biotechnol. 148, 70–75 (2010). 10.1016/j.jbiotec.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 114.Yu Z. T., Kamei K., Takahashi H., Shu C. J., Wang X., He G. W., Silverman R., Radu C. G., Witte O. N., Lee K. M., and Tseng H. R., Biomed. Microdevices 11, 547–555 (2009). 10.1007/s10544-008-9260-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vernetti L., Gough A., Baetz N., Blutt S., Broughman J. R., Brown J. A., Foulke-Abel J., Hasan N., In J., Kelly E., Kovbasnjuk O., Repper J., Senutovitch N., Stabb J., Yeung C., Zachos N. C., Donowitz M., Estes M., Himmelfarb J., Truskey G., Wikswo J. P., and Taylor D. L., Sci. Rep. 7, 42296 (2017). 10.1038/srep42296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Satoh T., Sugiura S., Shin K., Onuki-Nagasaki R., Ishida S., Kikuchi K., Kakiki M., and Kanamori T., Lab Chip 18, 115–125 (2018). 10.1039/C7LC00952F [DOI] [PubMed] [Google Scholar]
- 117.van Duinen V., van den Heuvel A., Trietsch S. J., Lanz H. L., van Gils J. M., van Zonneveld A. J., Vulto P., and Hankemeier T., Sci. Rep. 7, 18071 (2017). 10.1038/s41598-017-14716-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oleaga C., Bernabini C., Smith A. S. T., Srinivasan B., Jackson M., McLam W., Platt V., Bridges R., Cai Y., Santhanam N., Berry B., Najjar S., Akanda N., Guo X., Martin C., Ekman G., Esch M. B., Langer J., Ouedraogo G., Cotovio J., Breton L., Shuler M. L., and Hickman J. J., Sci. Rep. 6, 20030 (2016). 10.1038/srep20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chan C. Y., Gora Michael V. N., DeRosa E., Huang T. J., and Yuen P. K., Biomicrofluidics 8, 046505 (2014). 10.1063/1.4894409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Y. X., Kongsuphol P., Chiam S. Y., Zhang Q. X., Gourikutty S. B. N., Saha S., Biswas S. K., and Ramadan Q., Lab Chip 18, 3550–3560 (2018). 10.1039/C8LC00605A [DOI] [PubMed] [Google Scholar]
- 121.Parittotokkaporn S., Dravid A., Bansal M., Aqrawe Z., Svirskis D., Suresh V., and O’Carroll S. J., Biomed. Microdevices 21, 77 (2019). 10.1007/s10544-019-0427-4 [DOI] [PubMed] [Google Scholar]
- 122.Tatosian D. A. and Shuler M. L., Bioengineering 103(1), 187–198 (2009). 10.1002/bit.22219 [DOI] [PubMed] [Google Scholar]
- 123.Jeona J. S., Bersinib S., Gilardic M., Charestf G. D. J. L., Morettic M., and Kamm R. D., Proc. Natl. Acad. Sci. U.S.A. 12(1), 214–219 (2015). 10.1073/pnas.1417115112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maschmeyer I., Hasenberg T., Jaenicke A., Lindner M., Lorenz A. K., Zech J., Garbe L. A., Sonntag F., Hayden P., Ayehunie S., Lauster R., Marx U., and Materne E. M., Eur. J. Pharm. Biopharm. 95(Pt. A), 77–87 (2015). 10.1016/j.ejpb.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park T. E., Mustafaoglu N., Herland A., Hasselkus R., Mannix R., FitzGerald E. A., Prantil-Baun R., Watters A., Henry O., Benz M., Sanchez H., McCrea H. J., Goumnerova L. C., Song H. W., Palecek S. P., Shusta E., and Ingber D. E., Nat. Commun. 10, 2621 (2019). 10.1038/s41467-019-10588-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Benam K. H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H. H., Alves S. E., Salmon M., Ferrante T. C., Weaver J. C., Bahinski A., Hamilton G. A., and Ingber D. E., Nat. Methods 13, 151–157 (2016). 10.1038/nmeth.3697 [DOI] [PubMed] [Google Scholar]
- 127.Ucciferri N., Sbrana L. T., and Ahluwalia A., Front. Bioeng. Biotechnol. 2, 74 (2014). 10.3389/fbioe.2014.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maass C., Stokes C. L., Griffith L. G., and Cirit M., Integr. Biol. 9(4), 290–302 (2017). 10.1039/C6IB00243A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mandenius C. F., Bioengineering 5, 56 (2018). 10.3390/bioengineering5030056 [DOI] [Google Scholar]
- 130.Kaarj K. and Yoon J. Y., Micromachines 10, 700 (2019). 10.3390/mi10100700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Baicu S. C. and Taylor M. J., Cryobiology 45(1), 33–48 (2002). 10.1016/S0011-2240(02)00104-9 [DOI] [PubMed] [Google Scholar]
- 132.Soucy J. R., Bindas A. J., Koppes A. N., and Koppes R. A., iScience 21, 521–548 (2019). 10.1016/j.isci.2019.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.See https://www.kirkstall.com/why-quasi-vivo for more info on the Quasi Vivo® System.
- 134.Salehi-Nik N., Amoabediny G., Pouran B., Tabesh H., Ali Shokrgozar M., Haghighipour N., Khatibi N., Anisi F., Mottaghy K., and Zandieh-Doulabi B., Biomed. Res. Int. 2013, 762132 10.1155/2013/762132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martin I., Wendt D., and Heberer M., Trends Biotechnol. 22(2), 80–86 (2004). 10.1016/j.tibtech.2003.12.001 [DOI] [PubMed] [Google Scholar]
- 136.Muschler G. F., Nakamoto C., and Griffith L. G., J. Bone Jt. Surg. A 86(7), 1541–1558 (2004). 10.2106/00004623-200407000-00029 [DOI] [PubMed] [Google Scholar]
- 137.Sutherland J. J., Jolly R. A., Goldstein K. M., and Stevens J. L., PLoS Comput. Biol. 12, e1004847 (2016). 10.1371/journal.pcbi.1004847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ingber D. E., Development 145, 156125 (2018). 10.1242/dev.156125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Prill S., Jaeger M. S., and Duschl C., Biomicrofluidics 8(3), 034102 (2014). 10.1063/1.4876639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bavli D., Prill S., Ezra E., Levy G., Cohen M., Vinken M., Vanfleteren J., Jaeger M., and Nahmias Y., Proc. Natl. Acad. Sci. U.S.A. 113(16), E2231–E2240 (2016). 10.1073/pnas.1522556113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kosińska A. and Andlauer W., Food Chem. 135(3), 999–1005 (2012). 10.1016/j.foodchem.2012.05.101 [DOI] [PubMed] [Google Scholar]
- 142.Sun T., Swindle E. J., Collins J. E., Holloway J. A., Davies D. E., and Morgan H., Lab Chip 10, 1611–1617 (2010). 10.1039/c000699h [DOI] [PubMed] [Google Scholar]
- 143.Guenat O. T. and Berthiaume F., Biomicrofluidics 12, 042207 (2018). 10.1063/1.5024895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barnes L. A., Marshall C. D., Leavitt T., Hu M. S., Moore A. L., Gonzalez J. G., Longaker M. T., and Gurtner G. C., Adv. Wound Care 7(2), 47–56 (2018). 10.1089/wound.2016.0709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Krishnan R., Park J., Seow C. Y., Lee P. V., and Stewart A. G., Trends Pharmacol. Sci. 37(2), 87–100 (2016). 10.1016/j.tips.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Castellone R. D., Leffler N. R., Dong L., and Yang L. V., Cancer Lett. 312(2), 197–208 (2011). 10.1016/j.canlet.2011.08.013 [DOI] [PubMed] [Google Scholar]
- 147.Albini A., Pathol. Oncol. Res. 4(3), 230–241 (1998). 10.1007/BF02905254 [DOI] [PubMed] [Google Scholar]
- 148.Nguyen D. X., Bos P. D., and Massagué J., Nat. Rev. Cancer 9, 274 (2009). 10.1038/nrc2622 [DOI] [PubMed] [Google Scholar]
- 149.Calvo F. and Sahai E., Curr. Opin. Cell Biol. 23, 621–629 (2011). 10.1016/j.ceb.2011.04.010 [DOI] [PubMed] [Google Scholar]
- 150.Lei Y., Li J., Wang N., Yang X., Hamada Y., Li Q., Zheng W., and Jiang X., Integr. Biol. 8, 359–367 (2016). 10.1039/c5ib00309a [DOI] [PubMed] [Google Scholar]
- 151.Ashley N. T., Weil Z. M., and Nelson R. J., Annu. Rev. Ecol. Evol. Syst. 43, 385–406 (2012). 10.1146/annurev-ecolsys-040212-092530 [DOI] [Google Scholar]
- 152.Ramadan Q. and Gijs M. A. M., Lab Chip 15, 614 (2015). 10.1039/C4LC01271B [DOI] [PubMed] [Google Scholar]
- 153.Schmid-Schonbein G. W., Annu. Rev. Biomed. Eng. 8, 93–151 (2006). 10.1146/annurev.bioeng.8.061505.095708 [DOI] [PubMed] [Google Scholar]
- 154.Pupovac A., Senturk B., Griffoni C., Maniura-Weber K., Rottmar M., and McArthur S. L., Adv. Healthcare Mater. 7(12), e1701405 (2018). 10.1002/adhm.201701405 [DOI] [PubMed] [Google Scholar]
- 155.Esch M. B., Sung J. H., Yang J., Yu C., Yu J., March J. C., and Shuler M. L., Biomed. Microdevices 14(5), 895–906 (2012). 10.1007/s10544-012-9669-0 [DOI] [PubMed] [Google Scholar]
- 156.Zhang B., Korolj A., Fook B., Lai L., and Radisic M., Nat. Rev. Mater. 3, 257 (2018). 10.1038/s41578-018-0034-7 [DOI] [Google Scholar]
- 157.Ewart L., Fabre K., Chakilam A., Dragan Y., Eswaraka D. B. D. J., Gan J., Guzzie-Peck P., Otieno M., Jeong C. G., Keller D. A., de Morais S. M., Phillips J. A., Proctor W., Sura R., Van Vleet T., Watson D., Will Y., Tagle D., and Berridge B., Exp. Biol. Med. 242, 1579–1585 (2017). 10.1177/1535370217715441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McKim J. M., Comb. Chem. High Throughput Screening 3(2), 188–206 (2010). 10.2174/138620710790596736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Griffith L. G. and Swartz M. A., Nat. Rev. Mol. Cell Biol. 7, 211–224 (2006). 10.1038/nrm1858 [DOI] [PubMed] [Google Scholar]
- 160.Yamada K. M. and Cukierman E., Cell 130(4), 601–610 (2007). 10.1016/j.cell.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 161.Sung J. H., Esch M. B., Prot J. M., Long C. J., Smith A. et al. , Lab Chip 13, 1201–1212 (2013). 10.1039/c3lc41017j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang C. P., Lu J., Seon H., Lee A. P., Flanagan L. A. et al. , Lab Chip 9, 1740–1748 (2009). 10.1039/b818401a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zervantonakis I. K., Kothapalli C. R., Chung S., Sudo R., and Kamm R. D., Biomicrofluidics 5, 13406 (2011). 10.1063/1.3553237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ronnov-Jessen L., Petersen O. W., and Bissell M. J., Physiol. Rev. 76, 69–125 (1996). 10.1152/physrev.1996.76.1.69 [DOI] [PubMed] [Google Scholar]
- 165.Sato T., Vries R. G., Snippert H. J., Van De Wetering M., Barker N., Stange D. E., Van Es J. H., Abo A., Kujala P., Peters P. J., and Clevers H., Nature 459, 262–265 (2009). 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 166.Didio L. J. A., Gaudio E., and Correr S., in Biopathology of the Liver, edited by Motta P. M. (Springer, Dordrecht, 1988). [Google Scholar]
- 167.Mertz D. R., Ahmed T., and Takayama S., Lab Chip 18(16), 2378–2395 (2018). 10.1039/c8lc00413g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Straub J., Aerosp. Sci. Technol. 46, 312–320 (2015). 10.1016/j.ast.2015.07.007 [DOI] [Google Scholar]
- 169.Maguire T. J., Novik E., Chao P., Barminko J., Nahmias Y., Yarmush M. L., and Cheng K.-C., Curr. Drug Metab. 10(10), 1192–1199 (2009). 10.2174/138920009790820093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Oh T., Sung J. H., Tatosian D. A., Shuler M. L., Kim D., Cytometry Part A 71A, 857–865 (2007). 10.1002/cyto.a.20427 [DOI] [PubMed] [Google Scholar]
- 171.Kramer B., de Haan L., Vermeer M., Olivier T., Hankemeier T., Vulto P., Joore J., and Lanz H. L., Int. J. Mol. Sci. 20, 4647 (2019). 10.3390/ijms20184647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu L., Koo Y., Russell T., Gay E., Li Y., and Yun Y., PLoS One 15(3), e0230335 (2020). 10.1371/journal.pone.0230335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Petrosyan A., Cravedi P., Villani V., Angeletti A., Manrique J., Renieri A., De Filippo R. E., Perin L., and Sacco S. D., Nat. Commun. 10, 3656. (2019). 10.1038/s41467-019-11577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Beaurivage C., Naumovska E., Chang Y. X., Elstak E. D., Nicolas A., Wouters H., van Moolenbroek G., Lanz H. L., Trietsch S. J., Joore J., Vulto P., Janssen R. A. J., Erdmann K. S., Stallen J., and Kurek D., Int. J. Mol. Sci. 20, 5661 (2019). 10.3390/ijms20225661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.van Duinen V., Zhu D., Ramakers C., van Zonneveld A. J., Vulto P., and Hankemeier T., Angiogenesis 22, 157–165 (2019). 10.1007/s10456-018-9647-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.van Duinen V., Stam W., Borgdorff V., Reijerkerk A., Orlova V., Vulto P., Hankemeier T., and Jan van Zonneveld A., J. Vis. Exp. 153, e59678 (2019). 10.3791/59678 [DOI] [PubMed] [Google Scholar]
- 177.Liu L., Koo Y., Russell T., Gay E., Li Y., and Yun Y., PLoS One 15(3), e0230335 (2020). 10.1371/journal.pone.0230335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schimek K., Frentzel S., Luettich K., Bovard D., Rütschle I., Boden L., Rambo F., Erfurth H., Dehne E. M., Winter A., Marx U., and Hoeng J., Sci. Rep. 10, 7865 (2020). 10.1038/s41598-020-64219-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hasenberg T., Mühleder S., Dotzler A., Bauer S., Labuda K., Holnthoner W., Redl H., Lauster R., and Marx U., J. Biotechnol. 216, 1–10 (2015). 10.1016/j.jbiotec.2015.09.038 [DOI] [PubMed] [Google Scholar]
- 180.Vahav I., van den Broek L. J., Thon M., Monsuur H. N., Spiekstra S. W., Atac B., Scheper R. J., Lauster R., Lindner G., and Marx U., J. Tissue Eng. Regener. Med. 14, 761–773 (2020). 10.1002/term.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kasendra M., Tovaglieri A., Sontheimer-Phelps A., Jalili-Firoozinezhad S., Bein A., Chalkiadaki A., Scholl W., Zhang C., Rickner H., Richmond C. A., Li H., Breault D. T., and Ingber D. E., Sci. Rep. 8, 2871 (2018). 10.1038/s41598-018-21201-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sheyn D., Cohn-Yakubovich D., Ben-David S., De Mel S., Chan V., Hinojosa C., Wen N., Hamilton G. A., Gazit D., and Gazit Z., Microfluid. Nanofluid. 23, 99 (2019). 10.1007/s10404-019-2261-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kasendra M., Luc R., Yin J., Manatakis D. V., Kulkarni G., Lucchesi C., Sliz J., Apostolou A., Sunuwar L., Obrigewitch J., Jang K. J., Hamilton G. A., Donowitz M., and Karalis K., eLife 9, e50135 (2020). 10.7554/eLife.50135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Peel S., Corrigan A. M., Ehrhardt B., Jang K. J., Caetano-Pinto P., Boeckeler M., Rubins J. E., Kodella K., Petropolis D. B., Ronxhi J., Kulkarni G., Foster A. J., Williams D., Hamiltonb G. A., and Ewart L., Lab Chip 19, 410–421 (2019). 10.1039/C8LC00829A [DOI] [PubMed] [Google Scholar]
- 185.Park J. H., Kim M. J., Kim W. J., Kwon K.-D., Choi K.-T. H. B. T., Lee S.-Y., and Shin H. K., Cancer Lett. 478, 71–81 (2020). 10.1016/j.canlet.2020.03.009 [DOI] [PubMed] [Google Scholar]
- 186.Campisi M., Shin Y., Osaki T., Hajal C., Chiono V., and Kamm R. D., Biomaterials 180, 117–129 (2018). 10.1016/j.biomaterials.2018.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Marx U., Akabane T., Andersson T. B., Elizabeth B., Beilmann M., Beken S., Brendler-Schwaab S., Cirit M. et al. , Altern. Anim. Exp. (published online 2020). 10.14573/altex.2001241 [DOI] [Google Scholar]
- 188.See https://www.emulatebio.com/press/strategic-collaboration for the press release on the collaboration between Emulate Inc. and Janssen Biotech, Inc.
- 189.See https://www.tissuse.com/en/news/press-releases/ for the press release on the collaboration between TissUse GmbH and Bayer AG.
- 190.Bauer S., Huldt C. W., Kanebratt K. P., Durieux I., Gunne D., Andersson S., Ewart L., Haynes W. G., Maschmeyer I., Winter A., Ämmälä C., Marx U., and Andersson T. B., Sci. Rep. 7, 14620 (2017). 10.1038/s41598-017-14815-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Peng W., Unutmaz D., and Ozbolat I. T., Trends Biotechnol. 34(9), 722–732 (2016). 10.1016/j.tibtech.2016.05.013 [DOI] [PubMed] [Google Scholar]
- 192.Weisgrab G., Ovsianikov A., and Costa P. F., Adv. Mater. Technol. 4(10), 1900275 (2019). 10.1002/admt.201900275 [DOI] [Google Scholar]
- 193.Hinton T. J., Jallerat Q., Palchesko R. N., Park J. H., Grodzicki M. S., Shue H. J., Ramadan M. H., Hudson A. R., and Feinberg A. W., Sci. Adv. 1, e1500758 (2015). 10.1126/sciadv.1500758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lee J. W., Choi Y. J., Yong W. J., Pati F., Shim J. H., Kang K. S., Kang I. H., Park J., and Cho D. W., Biofabrication 8, 015007 (2016). 10.1088/1758-5090/8/1/015007 [DOI] [PubMed] [Google Scholar]
- 195.Tan Z. C., Parisi C., Di Silvio L., Dini D., and Forte A. E., Sci. Rep. 7, 16293 (2017). 10.1038/s41598-017-16668-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Guerzoni L. P. B., Tsukamoto Y., Gehlen D. B., Rommel D., Haraszti T., Akashi M., and De Laporte L., Biomacromolecules 20, 3746–3754 (2019). 10.1021/acs.biomac.9b00786 [DOI] [PubMed] [Google Scholar]
- 197.Grigoryan B., Paulsen S. J., Corbett D. C., Sazer D. W., Fortin C. L., Zaita A. J., Greenfield P. T., Calafat N. J., Gounley J. P., Ta A. H., Johansson F., Randles A., Rosenkrantz J. E., Louis-Rosenberg J. D., Galie P. A., Stevens K. R., and Mille J. S., Science 364, 458–464 (2019). 10.1126/science.aav9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lin N. Y. C., Homan K. A., Robinson S. S., Kolesky D. B., Duarte N., Moisan A., and Lewis J. A., Proc. Natl. Acad. Sci. U.S.A. 116, 5399–5404 (2019). 10.1073/pnas.1815208116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gao B., Yang Q., Zhao X., Jin G., Ma Y., and Xu F., Trends Biotechnol. 34, 746–756 (2016). 10.1016/j.tibtech.2016.03.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available within the article.