Abstract
Proteasomes are highly abundant protein complexes that are responsible for most regulated protein degradation in cells under favorable growth conditions. When yeast cells are under nutritional stress, most proteasomes exit the nucleus and either accumulate in cytoplasmic condensates called proteasome storage granules (PSGs) or are directed to the vacuole by autophagy. Nitrogen starvation does not cause PSG formation but leads to degradation of proteasomes through the classical macroautophagy pathway. By contrast, carbon starvation or extended incubation in stationary phase results in both PSG formation and macroautophagy of proteasomes. Unexpectedly, we found that glucose limitation also causes proteasomes to be taken up directly into vacuoles by a microautophagy mechanism. Macro- and micro-autophagy occur in parallel in glucose-starved cells, and microautophagy appears biased toward aberrant or inactive proteasomes, leaving functional proteasomes to accumulate in PSGs. PSGs dissolve and proteasomes remobilize to the nucleus within minutes after glucose refeeding. We showed that AMP-activated protein kinase (AMPK) and endosomal-sorting-complex-required-for-transport (ESCRT) factors are required for proteasome microautophagy and also impact PSG dissipation and nuclear reimport of proteasomes after glucose refeeding. The insoluble protein deposit (IPOD) compartment provides an alternative means of proteasome homeostasis, including when microautophagy is impaired. Our findings reveal a surprising diversity of mechanisms for proteasome quality and quantity control during starvation. A mechanistic understanding of the AMPK-regulated ESCRT-mediated microautophagy pathway could provide new avenues for manipulating proteasome homeostasis and treating human disease.
Keywords: proteasome storage granule (PSG), microautophagy, proteasome, AMPK, ESCRT
The eukaryotic 26S proteasome is a highly conserved ~2.5 MDa multisubunit complex capable of degrading a vast array of different cellular proteins, usually after they have been covalently modified by ubiquitin (Tomko and Hochstrasser 2013). It is assembled from two major subcomplexes called the 20S proteasome or core particle (CP), which houses the proteolytic active sites, and the regulatory particle (RP). At least 33 different polypeptides constitute the 26S proteasome, and in proliferating cells, between ~104 to 105 particles typically accumulate per cell, often concentrating in the nucleus (Budenholzer, et al. 2017, Enenkel 2014, Ho, et al. 2018).
Proteasome relocalization during glucose starvation
Under specific nutritional stress conditions such as extended carbon starvation, budding yeast proteasomes undergo a massive relocalization from the nucleus to the cytoplasm (Laporte, et al. 2008). In the cytoplasm, they accrete into one or two large foci called proteasome storage granules (PSGs). In early stages of stationary phase, nuclear proteasomes migrate to the nuclear periphery where they form small foci close to the inner nuclear membrane. These proteasomes eventually move into the cytoplasm and assemble into PSGs, which preferentially accumulate in proximity to the vacuole (the yeast lysosome). Strikingly, PSGs dissolve and proteasomes are reimported into the nucleus within minutes of glucose add-back (Laporte, et al. 2008, Peters, et al. 2016).
Yeast PSGs formed after long-term growth in stationary phase are largely devoid of nonproteasomal proteins; the known exceptions are Blm10, a large protein that promotes incorporation of the CP, but not the RP, into PSGs and also is required for re-import of the CP into the nucleus upon glucose refeeding (Weberruss, et al. 2013), and free ubiquitin (Enenkel 2018). PSG assembly can be regulated by a variety of distinct factors, such as carbon source (Laporte, et al. 2008), proteasome subunit integrity (Li, et al. 2019, Saunier, et al. 2013), intracellular pH (Peters, et al. 2013), and other cellular proteins (van Deventer, et al. 2015, Weberruss, et al. 2013). Moreover, PSG formation is conserved in different eukaryotes, including yeast (Laporte, et al. 2008), plants (Marshall and Vierstra 2018), and mammalian cells (Enenkel 2014). PSGs allow cells to regulate proteasome availability and enhance cell fitness during stress conditions and aging (Saunier, et al. 2013, van Deventer, et al. 2015).
Proteasome degradation through autophagy
Not all nutritional stress conditions lead to PSG formation. Most notably, nitrogen starvation in mammalian cells (Cuervo, et al. 1995), plants (Marshall, et al. 2015), and yeast (Marshall, et al. 2016, Nemec, et al. 2017, Waite, et al. 2016) leads instead to degradation of proteasomes in the lysosome/vacuole through the macroautophagy pathway. In macroautophagy, segments of cytoplasm are engulfed by a developing double membrane structure, the phagophore, which seals into an autophagosome. Hemifusion of the autophagosome with the vacuole release the single-membrane enclosed autophagic body into the vacuole interior where resident hydrolases break down its contents (Wen and Klionsky 2016). This type of bulk autophagy is nonselective; however, specific, often defective proteins or organelles can also be targeted by so-called selective macroautophagy. Selectivity is frequently conferred by receptor proteins that bind ubiquitylated substrate on the one hand and the lipidated ubiquitin-like protein ATG8 in the developing phagophore membrane on the other (Gatica, et al. 2018). Interestingly, if proteasomes are inactivated by a covalent active-site inhibitor, they become hyper-ubiquitylated and targeted by specific receptor proteins for selective macroautophagy (Marshall, et al. 2015, Marshall, et al. 2016).
An inverse relationship between PSG formation and proteasome macroautophagy has been observed when yeast or plants are grown without a carbon source (Marshall and Vierstra 2018). In particular, PSGs appear to protect proteasomes from autophagy-mediated destruction. For instance, when yeast cells lack Blm10, which prevents CP assembly into PSGs, CPs are more rapidly consumed by autophagy. It remains unknown how the partitioning of proteasomes between assembling PSGs in the cytoplasm and proteolysis in the vacuole is normally regulated.
Recently, we identified a novel AMPK (yeast Snf1 complex)-regulated and ESCRT-dependent microautophagy pathway that targets proteasomes under limiting glucose conditions. This pathway appears biased toward aberrant or inactive proteasomes, allowing functional proteasomes to accumulate in PSGs (Li, et al. 2019). Upon glucose refeeding, this enables rapid nuclear reimport of working proteasomes following PSG dissolution.
Microautophagy is not nearly as well characterized mechanistically as is macroautophagy, but new examples continue to emerge (Li, et al. 2012, Oku and Sakai 2018). Proteasome microautophagy is unusual in that it involves uptake of a soluble protein complex from the cytoplasm, but this is not unprecedented. Fission yeast have been shown to deliver several vacuolar hydrolases via selective ESCRT-dependent uptake into endosomes. The resulting multivesicular bodies (MVBs) then fuse with the vacuole. This pathway is similar to the chaperone-mediated endosomal microautophagy pathway described in mammalian cells (Tekirdag and Cuervo 2018). Although proteasome uptake into endosomes in budding yeast has not been ruled out, microscopic observation of proteasomes budding directly into the vacuole indicates that an MVB intermediate is not essential (Li, et al. 2019). Microautophagy of other cytoplasmic components in glucose-limited S. cerevisiae cultures has also recently been reported (Iwama and Ohsumi 2019), as has ESCRT-dependent microautophagy of ER membranes under ER stress conditions (Schäfer, et al. 2019).
Distinct roles for proteasome autophagy pathways during glucose starvation
Under limiting glucose conditions, proteasome macroautophagy and microautophagy are both induced (Li, et al. 2019). The two pathways require distinct cellular components and have differential effects on PSG dynamics and proteasome trafficking (Fig. 1). Specifically, proteasome macroautophagy relies on the core macroautophagy-related proteins such as Atg8 and certain non-selective macroautophagy-related scaffold proteins such as Atg17 (Li, et al. 2019, Marshall and Vierstra 2018).
Figure 1. Proteasome trafficking and degradation under nutrient starvation conditions.
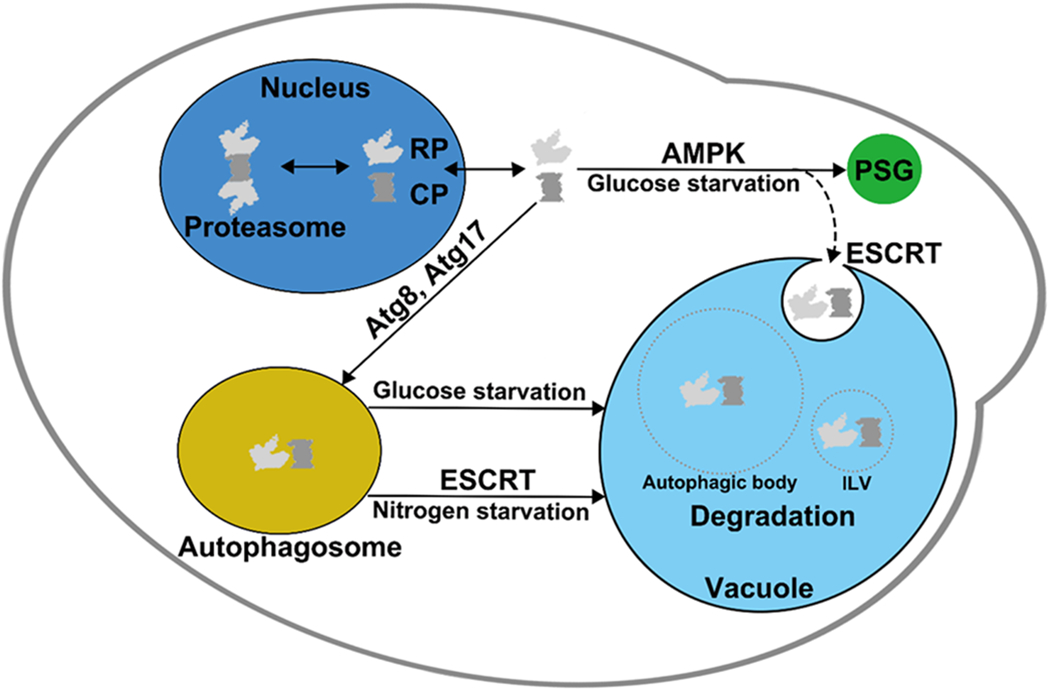
Proteasomes, which in yeast are concentrated in the nucleus, separate into core particle (CP) and regulatory particle (RP) during starvation for nutrients such as nitrogen or carbon. Both CP and RP relocate from the nucleus to the cytoplasm under these conditions. Nitrogen starvation induces degradation of proteasomal particles through the macroautophagy pathway, and this requires ESCRT factors. Under low glucose conditions, AMP-activated protein kinase (AMPK) is activated, and proteasomes can either move into proteasome storage granules (PSGs) or get degraded by one of two autophagic mechanisms, macroautophagy or microautophagy; only the latter is ESCRT-dependent under these conditions. The microautophagy pathway appears to preferentially remove aberrant proteasomes. ILV: Intralumenal vesicle.
PSG formation and dissipation, as well as nuclear reimport of proteasomes, are normal in macroautophagy mutants, arguing that macroautophagy does not alter PSG dynamics or proteasome nucleocytoplasmic trafficking. It is most likely part a non-selective bulk degradation pathway to recycle cellular materials in response to glucose deprivation. Proteasome macroautophagy under conditions of proteasome inhibition, by contrast, appears to be a selective process driven by ubiquitylation of proteasome subunits and recognition of the modified particles by specific autophagic receptors (Marshall, et al. 2016). Proteasome microautophagy during glucose deprivation also relies on ESCRT factors, unlike macroautophagy under these conditions. AMPK appears to regulate ESCRT-dependent microautophagic vesicle induction at the vacuolar membrane (Li, et al. 2019). Proteasome microautophagy alters PSG dynamics insofar as it sorts aberrant proteasomes away from assembling PSGs. This pathway may function as a surveillance system for proteasome quality control during changes of carbon availability.
Integration of IPODs in proteasome homeostasis mechanisms
Assembly of the insoluble protein deposit (IPOD) compartment by small heat shock protein chaperones serves as an intermediary in several of the pathways regulating proteasome levels during nutritional or proteotoxic stress (Karmon and Ben Aroya 2020). When proteasomes are damaged or chemically inactivated, they transit through an IPOD-like compartment, based on the colocalization of the Hsp42 chaperone, before being consumed by macroautophagy. Similarly, proteasomes transiently localize in or near IPODs during early-stage PSG formation during carbon starvation before separating to spatially distinct PSGs (Peters, et al. 2016). Proteasome microautophagy, on the other hand, does not require Hsp42 (J.L. and M.H., unpublished observations).
Interestingly, in the absence of AMPK, which blocks proteasome microautophagy, proteasomes accumulate in irreversible aggregates in the cytoplasm under limiting glucose conditions. CP-containing aggregates in particular gradually convert into IPODs based on their co-localization with Hsp42 and thus become sequestered in the cytoplasm (Li, et al. 2019). Moreover, in cells lacking AMPK, PSGs lose their reversibility as the cells age (J.L. and M.H., unpublished data); that is, even after glucose refeeding, the PSG structures do not dissipate rapidly. This change in PSG dynamics is intriguing, and it will be interesting to determine if the composition of PSGs or post-translational modifications of PSG-localized proteasomes might be altered under these conditions. Such changes might limit proteasome exchange between PSGs and free proteasomes in the cytoplasm.
Perspective
The newly described AMPK-controlled ESCRT-dependent microautophagy pathway for proteasome degradation makes important contributions to proteasome levels and quality control in cells under limiting glucose conditions. Notably, this pathway is much more strongly induced in cells grown in low glucose (0.025%) medium compare to glucose-free medium (Li, et al. 2019). Low glucose levels and gradual glucose loss may more closely mimic physiological carbon starvation, as happens frequently in natural environments. Abrupt changes to a glucose-free state might be less common and could trigger distinct cellular responses. A recent study showed that yeast cells in low glucose medium go through three distinct growth stages that correspond to glucose-utilizing, ethanol-utilizing, and ethanol-depleted phases and that different types of autophagy, including microautophagy, are induced at specific growth stages (Iwama and Ohsumi 2019). Post-transcriptional regulation of autophagy-related genes might play a role in autophagy induction at the specific growth stages and thus promote cell survive under nutritional stress conditions (Gatica and Klionsky 2019, Kuang, et al. 2018). It will be useful to determine how proteasome autophagy is differentially induced in low-glucose versus glucose-free medium. A clear physiological rationale for utilizing distinct autophagic mechanisms remains to be elucidated as well. Proteasome quality control might be more stringent in the microautophagy pathway, for example.
In yeast cells, AMPK is also involved in macroautophagy induction in glucose-limiting conditions (Adachi, et al. 2017), and ESCRT-dependent microautophagy of a vacuolar membrane protein has also recently been described (Oku, et al. 2017). It would be of a great interest to understand precisely how AMPK regulates ESCRT-dependent microautophagy of proteasomes. The kinase may directly target proteasome subunits or ESCRT components and thereby regulate proteasome-ESCRT interactions, or it may separately regulate vacuolar membrane lipid domains to promote membrane invagination or ESCRT recruitment. Alternatively, AMPK may be indirectly involved in the ESCRT-dependent microautophagy pathway through crosstalk with other cellular signaling pathways such as TORC1 or regulators of the AMPK pathway such as Std1 (Simpson-Lavy and Kupiec 2018). AMPK may co-regulate several processes during starvation, such as formation of other cellular granules (e.g., P bodies) involved in the regulation of cell signaling (Zhang and Herman 2019). Proteasome microautophagy could also require proteasome ubiquitylation. If so, specific E3 ligases might be activated, potentially by AMPK.
Cells have evolved multiple pathways to regulate proteasome homeostasis during changes of nutrient availability, suggesting the importance of tuning proteasome levels and activity to different nutritional or growth states. There is still much that is uncertain about the intersections of macroautophagy and microautophagy of proteasomes, the precise roles of the IPOD, and how PSG formation and dissolution and proteasome trafficking across the nuclear envelope are regulated. A better understanding of these pathways could provide potential cellular targets for pharmacological intervention in proteasome homeostasis and trafficking, and might also yield insights into aging and age-related disease.
Acknowledgements
Work from our laboratory in this area was supported by NIH grants GM083050 and GM046904 to M.H.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Adachi A, Koizumi M, Ohsumi Y (2017) Autophagy induction under carbon starvation conditions is negatively regulated by carbon catabolite repression. J Biol Chem 292: 19905–19918 doi: 10.1074/jbc.M117.817510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budenholzer L, Cheng CL, Li Y, Hochstrasser M (2017) Proteasome structure and assembly. J Mol Biol 429: 3500–3524 doi: 10.1016/j.jmb.2017.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, Knecht E (1995) Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem 227: 792–800 doi: 10.1111/j.1432-1033.1995.0792p.x [DOI] [PubMed] [Google Scholar]
- Enenkel C (2014) Proteasome dynamics. BBA-Mol Cell Res 1843: 39–46 doi: 10.1016/j.bbamcr.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Enenkel C (2018) The paradox of proteasome granules. Curr Genet 64: 137–140 doi: 10.1007/s00294-017-0739-y [DOI] [PubMed] [Google Scholar]
- Gatica D, Klionsky DJ (2019) Towards understanding mRNA-binding protein specificity: lessons from post-transcriptional regulation of ATG mRNA during nitrogen starvation-induced autophagy. Curr Genet 65: 847–849 doi: 10.1007/s00294-019-00943-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, Klionsky DJ (2018) Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20: 233–242 doi: 10.1038/s41556-018-0037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, Brown GW (2018) Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Systems 6: 192–205.e193 doi: 10.1016/j.cels.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Iwama R, Ohsumi Y (2019) Analysis of autophagy activated during changes in carbon source availability in yeast cells. J Biol Chem 294: 5590–5603 doi: 10.1074/jbc.RA118.005698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmon O, Ben Aroya S (2020) Spatial organization of proteasome aggregates in the regulation of proteasome homeostasis. Front Mol Biosci 610.3389/fmolb.2019.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Ji H, Boeke JD (2018) Stress response factors drive regrowth of quiescent cells. Curr Genet 64: 807–810 doi: 10.1007/s00294-018-0813-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Salin B, Daignan-Fornier B, Sagot I (2008) Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 181: 737–745 doi: 10.1083/jcb.200711154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Breker M, Graham M, Schuldiner M, Hochstrasser M (2019) AMPK regulates ESCRT-dependent microautophagy of proteasomes concomitant with proteasome storage granule assembly during glucose starvation. PLoS Genet 15: e1008387 doi: 10.1371/journal.pgen.1008387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-w, Li J, Bao J-k (2012) Microautophagy: lesser-known self-eating. Cell Mol Life Sci 69: 1125–1136 doi: 10.1007/s00018-011-0865-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Richard S, Li F, Gemperline David C, Book Adam J, Vierstra Richard D (2015) Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell 58: 1053–1066 doi: 10.1016/j.molcel.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, McLoughlin F, Vierstra RD (2016) Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep 16: 1717–1732 doi: 10.1016/j.celrep.2016.07.015 [DOI] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD (2018) Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. eLife 7: e34532 doi: 10.7554/eLife.34532 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nemec AA, Howell LA, Peterson AK, Murray MA, Tomko RJ (2017) Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J Biol Chem 292: 21466–21480 doi: 10.1074/jbc.M117.817999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M, Maeda Y, Kagohashi Y, Kondo T, Yamada M, Fujimoto T, Sakai Y (2017) Evidence for ESCRT- and clathrin-dependent microautophagy. J Cell Biol 216: 3263–3274 doi: 10.1083/jcb.201611029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M, Sakai Y (2018) Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays 40: 1800008 doi: 10.1002/bies.201800008 [DOI] [PubMed] [Google Scholar]
- Peters LZ, Hazan R, Breker M, Schuldiner M, Ben-Aroya S (2013) Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J Cell Biol 201: 663–671 doi: 10.1083/jcb.201211146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LZ, Karmon O, Miodownik S, Ben-Aroya S (2016) Proteasome storage granules are transiently associated with the insoluble protein deposit in Saccharomyces cerevisiae. J Cell Sci 129: 1190–1197 doi: 10.1242/jcs.179648 [DOI] [PubMed] [Google Scholar]
- Saunier R, Esposito M, Dassa EP, Delahodde A (2013) Integrity of the Saccharomyces cerevisiae Rpn11 protein is critical for formation of proteasome storage granules (PSG) and survival in stationary phase. PLoS One 8: e70357 doi: 10.1371/journal.pone.0070357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer JA, Schessner JP, Bircham PW, Tsuji T, Funaya C, Pajonk O, Schaeff K, Ruffini G, Papagiannidis D, Knop M, Fujimoto T, Schuck S (2019) ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J n/a: e102586 doi: 10.15252/embj.2019102586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy K, Kupiec M (2018) A reversible liquid drop aggregation controls glucose response in yeast. Curr Genet 64: 785–788 doi: 10.1007/s00294-018-0805-0 [DOI] [PubMed] [Google Scholar]
- Tekirdag K, Cuervo AM (2018) Chaperone-mediated autophagy and endosomal microautophagy: Jointed by a chaperone. J Biol Chem 293: 5414–5424 doi: 10.1074/jbc.R117.818237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Hochstrasser M (2013) Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem 82: 415–445 doi: 10.1146/annurev-biochem-060410-150257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deventer S, Menendez-Benito V, van Leeuwen F, Neefjes J (2015) N-terminal acetylation and replicative age affect proteasome localization and cell fitness during aging. J Cell Sci 128: 109–117 doi: 10.1242/jcs.157354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite KA, Mota-Peynado AD-L, Vontz G, Roelofs J (2016) Starvation induces proteasome autophagy with different pathways for core and regulatory particles. J Biol Chem 291: 3239–3253 doi: 10.1074/jbc.M115.699124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberruss MH, Savulescu AF, Jando J, Bissinger T, Harel A, Glickman MH, Enenkel C (2013) Blm10 facilitates nuclear import of proteasome core particles. EMBO J 32: 2697–2707 doi: 10.1038/emboj.2013.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Klionsky DJ (2016) An overview of macroautophagy in yeast. J Mol Biol 428: 1681–1699 doi: 10.1016/j.jmb.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Herman PK (2019) It is all about the process(ing): P-body granules and the regulation of signal transduction. Curr Genet 10.1007/s00294-019-01016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]


