Abstract
In Saccharomyces cerevisiae, the mitoribosomal RNA of the minor subunit,15S rRNA, is transcribed as a bicistronic transcript along with tRNAW. 5’ and 3’ sequences flanking the mature transcript must be removed by cleavage at the respective junctions before incorporating it into the mitoribosome. An in vivo dose-response triphasic system was created to elucidate the role of Ccm1p in the processing of 15S rRNA: Ccm1p supply (“On”), deprivation (“Off”), and resupply (“Back on”). After 72 h under “Off” status, the cells started to exhibit a complete mutant phenotype as assessed by their lack of growth in glycerol medium, while keeping their mitochondrial DNA integrity (ρ+). Full functionality of mitochondria was reacquired upon “Back on.” 15S rRNA levels and phenotype followed the Ccm1p intramitochondrial concentrations throughout the “On-Off-Back on” course. Under “Off” status, cells gradually accumulated unprocessed 5’ and 3’ junctions, which reached significant levels at 72–96 h, probably due to a saturation of the mitochondrial degradosome (mtEXO). The Ccm1p/mtEXO mutant (Δccm1/Δdss1) showed a copious accumulation of 15S rRNA primary transcript forms, which were cleaved upon Ccm1p resupply. The gene that codes for the RNA component of RNase P was conserved in wild-type and mutant strains. Our results indicate that Ccm1p is crucial in processing the 15S rRNA primary transcript and does not stabilize the already mature 15S rRNA. Consequently, failure of this function in Δccm1 cells results, as it happens to any other unprocessed primary transcripts, in total degradation of 15S rRNA by mtEXO, whose mechanism of action is discussed.
Keywords: Ccm1p, 15S rRNA, processing, mtEXO, mitoribosome, mitochondria
Introduction
During the course of evolution, mitochondrial DNA (mtDNA) underwent a significant reduction by the migration of genes from mtDNA to the nuclear genome, which has been a universal phenomenon in eukaryotes (Keeling and Palmer 2008). This phenomenon also involves the loss of transcription initiation signals that results in the production of polycistronic primary transcripts, which must be processed before becoming fully functional (Anderson et al. 1981). Failures in the processing of primary transcripts cause several disorders (Van Haute et al. 2015). Specifically, problems in the nucleolytic processing of mitochondrial RNA precursors reduce the levels of the electron transport chain components and consequently may cause progressive loss of cognitive and motor function, epilepsy, retinal degeneration (Deutschmann et al. 2014; Falk et al. 2016), cardiomyopathy (Haack et al. 2013; Deutschmann et al. 2014), lactic acidosis, hypotonia, feeding difficulties, and deafness (Metodiev et al. 2016).
Although mtDNA exhibits a remarkable variation in size (Gray 2012), a shared set of genes remain in it, COB, COX1, COX2, COX3, ATP6, and ATP8, which are part of higher-order complexes (Gray 2012), and those coding for ribosomal RNAs, and transfer RNAs (Anderson et al. 1981; Foury et al. 1998). All these genes are essential for mitochondrial functionality.
The need for intramitochondrial expression of these proteins is due to their high content of hydrophobic domains, which is the cause of their mistargeting to the endoplasmic reticulum when expressed in the cytoplasm (Björkholm et al. 2015). This observation supports the notion that such proteins must be synthesized, folded, and incorporated into their corresponding complexes inside the organelle during mitochondria biogenesis. Hence the limit of migration to the nucleus imposed over mitochondrial genes. Thus, nearly 250 nuclear-encoded factors are required to produce proteins inside the organelle (Fox 2012). Interestingly, the cytoplasmic protein Puf3p regulates the translational fluctuation during metabolic adaptation to substrate availability by balancing mitochondrial and cytosolic ribosome biogenesis (Wang et al. 2019). In addition, the co-regulation of the overall network is carried out by Abf1p, an expression factor for ribosomal proteins whose binding motif is also present in mitochondrial ribosomal protein genes (de la Cruz et al. 2018).
While in humans, two initial transcripts are generated (Montoya et al. 1982), in Saccharomyces cerevisiae, a minimum of 11 primary transcripts has been proposed to be synthesized (Turk et al. 2013). All of them must be processed by nucleases to produce functional RNAs. Therefore, the reduction in mtDNA genes has to be compensated by a set of nuclear-encoded proteins that cleave primary transcripts at precise sites. Once released, RNA byproducts like introns must be degraded to prevent their toxicity (Margossian et al. 1996). When not processed, the S. cerevisiae COB, VAR1, and 15S rRNA primary transcripts undergo degradation as well (Dziembowski et al. 2003). The system involved in RNA degradative quality control is the mitochondrial degradosome (mtEXO). Two enzymes, a 3’→5’ exoribonuclease, and an RNA helicase, coded by the DSS1 and SUV3 nuclear genes respectively, compose the mtEXO (Dziembowski et al. 2003).
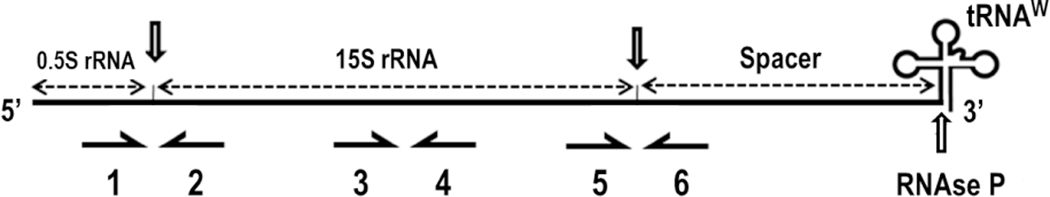
The 15S rRNA primary transcript (15S rRNA, tRNAW; Turk et al. 2013) consists of 76 nucleotides (0.5S rRNA) at the 5’end (Osinga et al. 1981), followed by mature 15S rRNA (1646 nucleotides), a spacer or 3’-precursor (Dziembowski et al. 2003) of 1308 nucleotides and tRNAW at the 3’ end, which encompasses 74 nucleotides (Naquin et al. 2018) (Fig. 1). Thus, three joints must be processed to generate the authentic 15S rRNA and tRNAW. The deletion of either mtEXO subunit results in the accumulation of 3’ end unprocessed forms and a subsequent reduction in processed 15S rRNA levels (Dziembowski et al. 2003). Insufficient amounts of rRNA cause the malfunction of the mitochondrial gene expression system, which results in loss of mtDNA (Merz and Westermann 2009; Guo et al. 2011). Once the tRNAW has been removed from the primary transcript by RNAse P (Hollingsworth and Martin 1986) and RNAse Z (Mörl and Marchfelder 2001), the precursor 15S rRNA may follow two routes that seem to take place simultaneously but are mutually exclusive: This molecule can be processed and integrated into the minor subunit of the mitoribosome or degraded by the quality control (QC) mtEXO. The notion that mtEXO (Gavin et al. 2002; Dziembowski et al. 2003) and Ccm1p (Möller-Hergt et al. 2018) are most probably physically associated with the mitoribosome supports this theory. Processing of the 15S rRNA primary transcript 5’ terminal end is mainly controlled by Pet127p and carried out by a putative 5’→3’ exonuclease (Fekete at al. 2008). Curiously, the S. cerevisiae 62R1 (Δpet127 [Cox 3–662]) double mutant is unable to cleave the 76 nucleotides at the 5’ end, but it grows weakly in the presence of glycerol, which suggests that mitoribosome assembly takes place to some extent (Wiesenberger and Fox 1997).
Fig. 1.
Scheme of the 15S rRNA initial transcript. Downward pointing arrows indicate the processing sites studied in this paper. Horizontal arrows (1 to 6) show the location of the primers used to assess the levels of unprocessed joints and total 15S rRNA
Pentatricopeptide-repeat (PPR) proteins (Small and Peeters 2000) have been reported to be involved, among other activities, in nucleolytic processing of mitochondrial RNA (for a thorough review, see Manna 2015). PPR domains are not considered to be catalytic; instead, they mediate interactions between RNA substrates and intra- or intermolecular nuclease activity (Manna 2015). However, a subset of PPR proteins known as PRORP (protein-only RNase P) have two-domains: An N-terminal domain, composed of several PPR motifs, that confers substrate specificity (Kobayashi et al. 2012) and a C- terminal domain that harbors nucleolytic activity (Pinker et al. 2013). Examples of PRORPs are MRPP3, PRORP2, and PRORP1. These proteins participate in 5’ tRNA nucleolytic processing in the mitochondria of humans, Arabidopsis thaliana, and Trypanosoma brucei, respectively (Holzmann et al. 2008; Gobert et al. 2010; Taschner et al. 2012). In S. cerevisiae, the participation of Ccm1p, a bifunctional PPR protein (Moreno et al. 2012) in the nucleolytic processing of only the 15.5S rRNA has been addressed (De Silva et al. 2015), albeit not yet demonstrated.
The present paper reports a novel in vivo triphasic expression system consisting of Ccm1p supply/deprivation /resupply (“On”/ “Off”/ “Back on”) that causes wild-type/mutant/wild-type phenotypes. Because this system is reversible, it is amenable to conduct dual dose-response experiments that validate one another. It correlates intramitochondrial concentrations of Ccm1p and events on the 15S rRNA primary transcript that define this PPR protein as an essential processing factor. Studies involving double mutant cells (Δccm1/Δdss1) showed that mtEXO and Ccm1p act concertedly but in opposition to determine the fate of the molecule. Thus, the absence of Ccm1p leads to the accumulation of unprocessed forms, which are recognized and degraded by the QC-surveillance mtEXO.
Materials and Methods
Media, strains, and DNA constructs
Media, yeast manipulation, including the selection of heterozygous strains, transformation, sporulation, tetrad dissection, and further analysis of meiotic segregants were carried out as described previously (Moreno et al. 2009). S. cerevisiae harboring intronless mtDNA (I0) (MATa ade1Δ0 lys103940 ura3Δ0) (Séraphin et al. 1987) was kindly provided by Dr. Alan M. Lambowitz (Institute for Cellular and Molecular Biology, Departments of Chemistry, Biochemistry and Microbiology, University of Texas at Austin, TX, USA. The S. cerevisiae Δdss1 strain (MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 dss1Δ0::kanMX) was purchased from Dharmacon (Lafayette, CO, USA). pCCM1ZZLC is a low-copy number vector that expresses the full- length CCM1 gene fused to a ZZ tag (two IgG-binding domains in tandem) at the C-terminus as Ccm1pZZ (Moreno et al. 2012). pCCM1LC, a low-copy number vector that expresses the naturally occurring CCM1 gene without any modifications (authentic protein construct, the gene product is Ccm1pAPC) was prepared as previously described (Moreno et al. 2009). The second bona fide PPR domain (PPR2) of CCM1 open reading frame (ORF) in pCCM1ZZLC was deleted between residues 356 and 390 to generate ΔPPR2ZZ (Moreno et al. 2009). The LguI DNA fragment containing ΔPPR2ZZ ORF and all expression elements were inserted in the SmaI site of the low-copy vector pRS316 (ATCC, Manassas, VA, USA). The resulting vector, pΔPPR2ZZLC that expresses ΔPPR2pZZ. The constitutive expression of Ccm1p was achieved by inserting the CCM1 ORF at the NotI site of the 2μ-vector pDB20 (a generous gift from Dr. Leonard Guarente, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA; Becker et al. 1991) under the control of the ADH1 promoter (pDB20-CCM1).
“On-Off-Back on” time-course experiment
Complemented meiotic segregants harboring pCCM1LC, pCCM1ZZLC, or pΔPPR2ZZLC were initially isolated on YEPG. Protein expression was induced by growing them in synthetic define medium minus uracil (SD) with galactose (SDGal) for 24 h (“On”). Cells were then washed three times with sterile deionized water at room temperature. Repression of CCM1 expression (“Off”) was carried out by adding cells (Abs600 =1.0) to SD with dextrose (SDD) at a ratio of 1:20 (v/v) and incubating them for 12 h and 24 h at 30°C, 300 rpm. Subsequent “Off” times were started by adding a 24 h-culture to SDD at a ratio of 1:100 (v/v) and incubating it for an additional 24 h period (i.e., 48h). This procedure was repeated twice to achieve incubation times of 72 and 96 h. Cells from the 96 h-SDD culture (“Off”) were washed as indicated above and induced to express Ccm1pZZ or Ccm1p by adding the inoculum (Abs600 =1.0) to SDGal (“Back on”) at a ratio of 1:20 (v/v), for 12 and 24 h, or 1:100 (v/v), for 48h. A 48 h-culture inoculum grown in SDGal (Abs600 =1.0) was added to the same medium at a ratio of 1:100 (v/v) and incubated for an additional 24 h for a total of 72 h. The same procedure was carried out again for the SDD control. Samples, representing all conditions and incubation times, were collected and aliquoted. Most of the cells were stored at −70°C for further analysis, while small aliquots were collected for drop assays. They were carried out at the end of each incubation time by spotting 3 μL of 1:10 serially diluted cell suspensions (at an initial concentration of 1 × 108 cells/ mL) onto YEPG and YEPD solid media containing geneticin (Thermo Fisher Scientific, Waltham, MA, USA). Plates were further incubated at 30°C for 48 h and photographed.
Δccm1/ Δdss1 haploid mutant and incubation conditions
I0 haploids complemented with pCCM1ZZLC and carrying functional mitochondria were mated with the Δdss1 strain. The mating mixture was incubated for 3 hours at 30°C and spread on SSD to allow the microscopic identification of young diploids by their characteristic morphology (shmoos) (Amberg et al. 2005). Diploids were separated from the rest of the cell population by micromanipulation and allowed to form colonies, which were tested for pCCM1ZZLC content by growth on SSD. They were pooled and induced to sporulate. Tetrads were dissected on YEPG. Normal-sized and petite colonies were screened for the presence of pCCM1ZZLC, as indicated above. Segregants that harbored the expression vector were studied by colony PCR to determine whether they harbored no deletion, one (Δdss1 or Δccm1), or both deletions (Δccm1/Δdss1). This screening used the primers indicated by the Saccharomyces Genome Deletion Project (Table 1) and the Phire Plant Direct PCR Master Mix (Thermo Fisher Scientific). For mtDNA and mtRNA hybridization analyses, suitable segregants were harvested from the tetrad dissection plate, pooled, and propagated in YEPD for 48 h. The rich medium was used to generate enough cells for the hybridization experiments. For the Δccm1/Δdss1 “Back on” assay, mutants that harbored pCCM1ZZLC were harvested from the tetrad dissection plate and incubated in SDD (“Off”) for 48 h. At the end of the incubation period, a third of each culture was collected for reverse transcription (RT)-quantitative (q)PCR analysis using ALG9 mRNA as the reference transcript (Teste et al. 2009). The rest of the cells were washed as mentioned above, incubated for two more days in SDGal (“Back on”), collected, and analyzed as indicated.
Table 1:
Primers used in this study
| Template | Forward primer | Reverse primer | Amplicon size (bp) | Annealing position (a) | Reference |
|---|---|---|---|---|---|
| 15S_RRNA 5’unprocessed junction | AGTTATATAATAAGGAAAAG (n°1) | AGCTTATTCTATAGTTCATT (n°2) | 173 | 6527–6546, 6680–6699 |
This article |
| Total 15S_RRNA |
GTTAAACCTAGCCAACGATCCA (n°3) | TGTCCAATATTCCTCACTGCTG (n°4) | 108 | 270–291, 356–377 |
Moreno et al. 2009 |
| 15S_RRNA 3’unprocessed junction | GCGAAGTTGAAATACAGTTACCG (n°5) | GGGCCCCGGAACTATTAATA (n°6) | 123 | 8126–8148, 8229–8248 |
This article |
| ALG9 | CACGGATAGTGGCTTTGGTGAACAATTAC | TATGATTATCTGGCAGCAGGAAAGAACTTGGG | 161 | 236545–236573 236413–236443 (Chromosome XIV) |
Teste et al. 2009 |
| CCM1 | CCAAACCTGAGACCACGGA | TAGCGGCTTTCATCACCAGCTC | 509 | 793380–793404 792871–792895 (Chromosome VII) |
Moreno et al. 2012 |
| 21S rRNA | CGGGTCCCGGAACTTAAATA | CGAGGTGGCAAACATAGCTT | 221 | 2548–2567, 2553–2768 |
Moreno et al. 2012 |
| DSS1 | GTTTACAAATTGAATCGGATGACTC | TCAATATCAGGCTCTAATCCTTTTG | 517 | 845573–845597, 845080–845104 (Chromosome XIII) |
Saccharomyces Genome Deletion Project |
| kanMX4 module | N / A | CTGCAGCGAGGAGCCGTAAT | 576 (DSS1) 675 (CCM1) |
250 bp downstream of 5 ‘-end of the kanMX4 module | Saccharomyces Genome Deletion Project |
Mitochondrial genome unless otherwise indicated
mtRNA, mtDNA, preparation of mitochondria, qPCR, antibodies, and protein analysis
All probes were biotinylated by PCR (Biotin PCR Labeling Core Kit, Jena Bioscience, Jena, Germany) and subsequently purified from primers and unincorporated nucleotides (QIAEX II, Qiagen, Valencia, CA, USA). RNA isolation and hybridization were performed as previously described (Moreno et al. 2009). The signals were visualized and recorded with a chemiluminescent blot scanner (C-DiGit) (Li-Cor, Lincoln, NE, USA). DNA probes that detect 15S rRNA spanned either 204 bp (Moreno et al. 2009), or 588 bp from positions 6785–7369 of the mitochondrial genome.
mtDNA was prepared and digested with EcoRV, as indicated previously (Defontaine et al. 1991). DNA fragments were then separated by 1% agarose gel electrophoresis, ethidium bromide-stained, photographed, and transferred to a nylon membrane. This membrane was hybridized with a biotinylated 290-bp COB probe (Moreno et al. 2009) or a 209-bp DNA fragment located within the 3405 bp-EcoRV - EcoRV segment that contains the full-length RPM1 gene (9S RNA) and its promoter (Naquin et al. 2018). DNA hybridization was carried out with the North2South® Chemiluminescent Hybridization and Detection Kit (Thermo Scientific, Rockford, IL. USA), following the manufacturer’s instructions. Signals were visualized and recorded, as indicated above. qPCRs were carried out in a Smart Cycler II thermal cycler (Cepheid, Sunnyvale, CA, USA) using the SensiFAST™ SYBR No-ROX reagent (Bioline, Memphis, TN, USA). Primer sequences, annealing positions, and amplicon sizes are indicated in Fig. 1 and Table 1. RT-qPCR and qPCR products were identified by their melting temperature and endonuclease restriction patterns (Table 2). Calibration curves converted Ct (threshold cycle value) into starting amounts of template for each qPCR template–primers combination. A serial 1/10 dilution of yeast nuclear and mitochondrial DNA was used as the template for all calibration curves. Values that fit within the linear range were chosen for further calculation. The slope and R2 of each calibration curve were determined with the SigmaStat statistical software (SPSS, Chicago, IL, USA). R2 values were at least 0.998. mtDNA levels were assessed by qPCR using 15S_ RRNA and ALG9 as the target and reference genes, respectively. Levels of 15S rRNA, 5’, and 3’ unprocessed junctions (UJs) were assessed by RT-qPCR using 21S rRNA as the reference transcript, as previously indicated (Moreno et al. 2012). The percentage of processed 15S rRNA was calculated by subtracting the sum of 5’ and 3’ UJs from total 15S rRNA, which was considered 100 %. The amount of 3’ processed junctions was determined by subtracting 3’ UJ from total 15S rRNA levels. ALG9 and CCM1 mRNAs levels were measured by RT-qPCR, as mentioned above. Cytoplasm-free mitochondrial fractions were prepared as previously reported (Meisinger et al. 2006).
Table 2:
Characteristics of qPCR products
| qPCR amplicon | Melting temperature (°C) | Restriction endonuclease | Cleavage position |
|---|---|---|---|
| 15S_RRNA 5’unprocessed junction | 77.0 | HinfI | 85 |
| 15S_RRNA | 78.0 | HinfI | 55 |
| 15S_RRNA 3’unprocessed junction | 78.5 | SspI | 60 |
| 21S_RRNA | 79.0 | BfaI | 161 |
| ALG9 | 83.0 | AgeI | 89 |
| CCM1 | 77.0 | HinfI | 58 |
Two different sets of antibodies were used in this study: (i) Intramitochondrial Ccm1p was detected by immunoblot with specific polyclonal antibodies against the full-length protein produced in E. coli as previously described (Moreno et al. 2009). The assay limit of detection was approximately 2.5 ng of Ccm1p (Ccm1pZZ form) per lane. As no signal was displayed in mitochondrial extracts isolated from the Δccm1 strain (Moreno et al. 2009), these antibodies proved to be specific; (ii) the intramitochondrial concentration of Ccm1p was measured by ELISA with affinity-purified polyclonal antibodies against a synthetic peptide (VNKKSHAKALKWEEQELN), which constitutes a potent epitope (E3) located at the C-terminus of Ccm1p. The peptide and the antibodies were obtained as previously described (Moreno et al. 2012). As 5 μg of mitochondrial proteins prepared from the Δccm1 strain displayed no signal, the polyclonal antibodies showed to be specific.
For SDS/PAGE and immunoblotting, cytosol-free mitochondria samples were solubilized in cold 0.1 N NaOH (Nandakumar et al. 2003) and kept in ice for less than one hour for protein determination. Protein concentration was determined by Bradford assay and BSA as standard (Thermo Fisher Scientific).
Quantitation of intramitochondrial Ccm1pZZ levels by ELISA
The Ccm1pZZ calibrator, used for molarity measurement, is a truncated form of Ccm1pZZ that consists of the last 43 residues of Ccm1p. This peptide contains one of the strongest epitopes, VNKKSHAKALKWEEQELN (E3), and is fused at the C-terminus to the ZZ affinity tag (E3ZZ). The E3ZZ was prepared by subcloning the corresponding DNA fragment in pET28b and expressing it in Escherichia coli Rosetta (DE3) pLysS (Novagen, Madison, WI, USA) according to the supplier’s recommendations. The bacterial cell mass was lysed by sonication in PBS, 0.5% Triton X-100, 0.1% Tween, 100 mM EDTA, 100 mM EGTA plus one tablet of Complete™ protease inhibitor (Santa Cruz Biotechnology, Dallas, TX, USA) per 10 mL of lysis buffer. E3ZZ (25.50 kDa) was affinity-purified with IgG-Sepharose (GE Healthcare, Chicago, IL, USA), as previously described (Moreno 1996). The protein concentration was measured using the Bradford assay with BSA as standard (Thermo Fisher Scientific). E3ZZ was then aliquoted, kept frozen at −70 °C and thawed at the moment of use.
The sandwich ELISA was standardized as follows: After measuring the protein concentration in the alkaline mitochondria crude extracts, the samples were kept in ice until the moment of use, but no longer than 1h. Purified porcine IgG (Equitech-Bio, Kerrville, TX, USA) was used as the capture antibody at 2 μg per well. The E3ZZ calibrator was 1:10 diluted in 0.1 N NaOH. 5 μL of E3ZZ or mitochondria alkaline lysates were loaded on ELISA plate wells (Corning, NY, USA), which already contained 200 μL of dilution buffer (DB: 2X PBS, 130 mM EDTA, 130 mM EGTA, 1% Triton X-100, 0.1% Tween 20, 0.5% BSA and one tablet of Complete protease inhibitor per 50 mL of solution). The samples were then 1:3 serially diluted with DB and incubated for 18 h at 4°C. Plates were then washed three times (Wellwash Microplate Washer, Thermo Fisher Scientific) with washing buffer (WB: PBS, 0.5% Triton X-100, and 0.1% Tween 20). Wells were then incubated with purified polyclonal antibodies against the E3 epitope (21st Century Biochemicals, Marlboro, MA, USA), diluted 1:1000 in DB. Plates were allowed to stand at 4° C for 18 h. Wells were washed three times and incubated with 100 μL of 1:10,000 diluted horseradish peroxidase-conjugated goat anti-rabbit IgG (Thermo Fisher Scientific) in DB. Plates were incubated at room temperature for 1 h, washed with WB three times, and processed as indicated previously (Moreno et al. 2012). The limit of detection for this assay was approximately 0.14 nM.
Statistical analysis
All qPCR values represent the means ± S.E.M. of duplicates for the number of independent experiments indicated in each figure legend. When necessary, measured values were transformed into logarithms to normalize the distribution or equalize the variances. mtDNA qPCR data were analyzed by a two-sample t-test. RT-qPCR results were analyzed by one-way ANOVA, followed by Student-Newman-Keuls’ post hoc test for multiple comparisons (SPSS).
Results
Deletion of CCM1 buildups unprocessed 15S rRNA transcripts
Established Δccm1 mutants lose their mtDNA (Merz and Westermann 2009) and, consequently, all the corresponding mitochondrial transcription products. To visualize the effects of the deletion on the 15S rRNA, we analyzed fresh CCM1 and Δccm1 intronless nascent meiotic segregants since the latter had inherited mtDNA, transcripts, and all the cytosolic factors from the heterozygous diploid (Moreno et al. 2012). Wild-type cells exhibited only processed 15S rRNA in predictable quantities, whereas Δccm1 cells had severely diminished amounts of it. Surprisingly, the mutants also showed a longer form of approximately 3000 nucleotides of similar intensity (Fig. 2), which corresponds to the initial full-length 15S rRNA transcript (Fig. 1). The heavier transcript was the first indication that Ccm1p could be involved in a post-transcriptional modification activity that converts this RNA to its final and functional form. This observation and previous reports (Dziembowski et al. 2003; De Silva et al. 2015) prompted us to focus our study on the processing of the 15S rRNA primary transcript by Ccm1p (Fig. 1).
Fig. 2.
Non-complemented Δccm1 nascent meiotic segregants exhibit unprocessed 15S rRNA. Hybridization analysis of total RNA of 10 and 15 μg per well from CCM1 and Δccm1 strains, respectively. The filter was probed with a 204 bp-biotinylated 15S R_RNA DNA probe that spans positions 7167–7370 of the mitochondrial genome, within the mature transcript. Filled and hollow arrowheads indicate the position of processed and non-processed forms, respectively
Mitochondria in Δccm1 non-complemented segregants keep their capability to import Ccm1p.
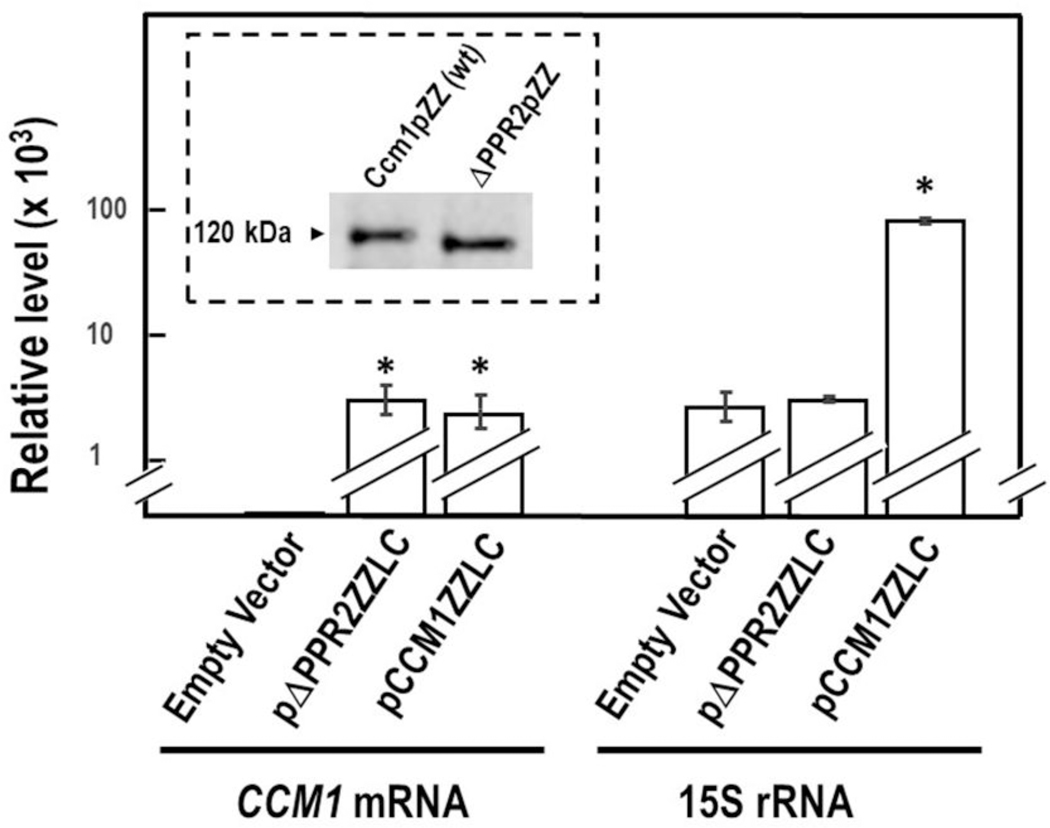
Non-complemented Δccm1 meiotic segregants displayed, in addition to mitochondrial respiratory failure, abnormal morphology (Moreno et al. 2009). Their capacity to revert to full organelle functionality was assessed (see below) by their ability to import cytoplasmic proteins into mitochondria (Kritsiligkou et al. 2017). We have previously reported that, by deleting the second canonical PPR domain of Ccm1p, the protein loses the ability to complement Δccm1 meiotic segregants (Moreno et al. 2009). Consequently, purified mitochondrial fractions from Δccm1 segregants harboring the empty vector, pCCM1ZZLC, or pΔPPR2ZZLC were compared in regards to their relative levels of 15S rRNA and CCM1 mRNA along with protein expression and import into the organelle. Cells harboring pΔPPR2ZZLC and non-complemented Δccm1 (empty vector) segregants had comparable levels of 15S rRNA (Fig. 3). However, nascent Δccm1 segregants harboring pΔPPR2ZZLC produced the corresponding deletant protein without apparently reduced fitness and imported it into mitochondria as efficiently as the wild- type cells (Fig. 3, insert). This observation indicated that mitochondria of non-complemented nascent segregants could sustain the import of a Ccm1p-like protein.
Fig. 3.
Ccm1p is imported by non-functional mitochondria. Δccm1 cells harboring the empty vector, pΔPPR2ZZLC, or pCCM1ZZLC were grown in SDGalfthe ones previously reported or 24h. Their relative levels of 15S rRNA and CCM1 mRNA were measured by RT-qPCR. Columns in association with vertical bars represent the means ± SEM of three independent clones measured in duplicate. ALG9 mRNA was the reference transcript. *, p < 0.05; one-way analysis of variance, Student-Newman- Keuls’ post hoc test. Insert: Crude extracts of purified mitochondria from strains expressing wild-type Ccm1pZZ and Ccm1ΔPPR2p (ΔPPR2pZZ) were electrophoresed in a 5–20 % gradient SDS-PAGE until the 35 kDa marker ran off. Proteins were transferred to a nitrocellulose membrane and probed with polyclonal antibodies against Ccm1p produced in E. coli
The mutant phenotype caused by the sudden mitochondrial deprivation of Ccm1p (SMDC) is reversible
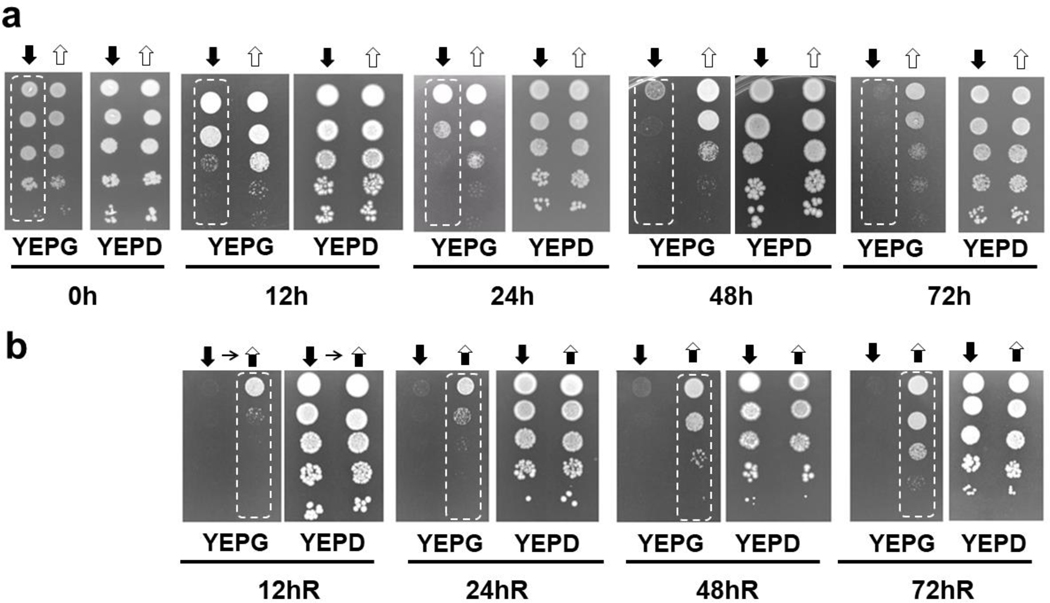
We had previously complemented Δccm1 cells by ectopic expression of CCM1 ORF under the control of the GAL1 promoter. Downregulation of CCM1 expression ended in respiratory failure, as shown by their lack of growth on glycerol medium (Moreno et al. 2009). The “Off” (SMDC) status was enhanced by fusing a ZZ tag to the Ccm1p C-terminus (Moreno et al. 2012) since it constraints en route its mitochondrial import (Schülke et al. 1997). To set up our system of study, we evaluated the cells’ ability to grow again on non-fermentable substrates upon Ccm1p resupply after they had acquired the mutant phenotype. Therefore, cells were initially induced by galactose (“On”), then were incubated in glucose medium for several days (“Off”), before switching them back to galactose (“Back on”). The entire progression of events, induction/repression/reinduction, was monitored by the ability of the cells to grow on medium with glycerol. Cells in “Off” reached the full mutant phenotype after 72 h (Fig. 4a), which was consistent with our previously reported results (Moreno et al. 2012). We further extended the “Off” time up to 96 h and obtained the same results (data not shown). A complete reversion to the wild-type phenotype was observed when the cells were reinduced by switching back to the galactose medium (Fig. 4b). Growth in glycerol medium for all conditions was recorded after 48 h of incubation.Interestingly, after 72 h of additional incubation time, cells under “Off” status resumed growth and reached comparable levels to the ones that were permanently in the “On” phase, since the lack of glucose in YEPG derepressed Ccm1p expression.
Fig. 4.
Progression of the phenotype throughout the “On-Off-Back on” time-course. a A inoculum was taken from a master plate of YEPG-geneticin and grown in SDGal for 48 h (“On”; t = 0 h). Cultures in SSD (“Off” [ ]) and SDGal (“On” [
]) and SDGal (“On” [ ]). b From the 72h-SDD culture (“Off”), cells were either grown in SDD (“Off” [
]). b From the 72h-SDD culture (“Off”), cells were either grown in SDD (“Off” [ ] or SDGal (“Back on” [
] or SDGal (“Back on” [ ]), as shown by the small horizontal arrow. White-dashed rectangles indicate the acquired transient mutant and wild- type phenotype during the entire experimental course
]), as shown by the small horizontal arrow. White-dashed rectangles indicate the acquired transient mutant and wild- type phenotype during the entire experimental course
Cells maintain ρ+ status throughout the entire “On-Off-Back on” time-course experiment
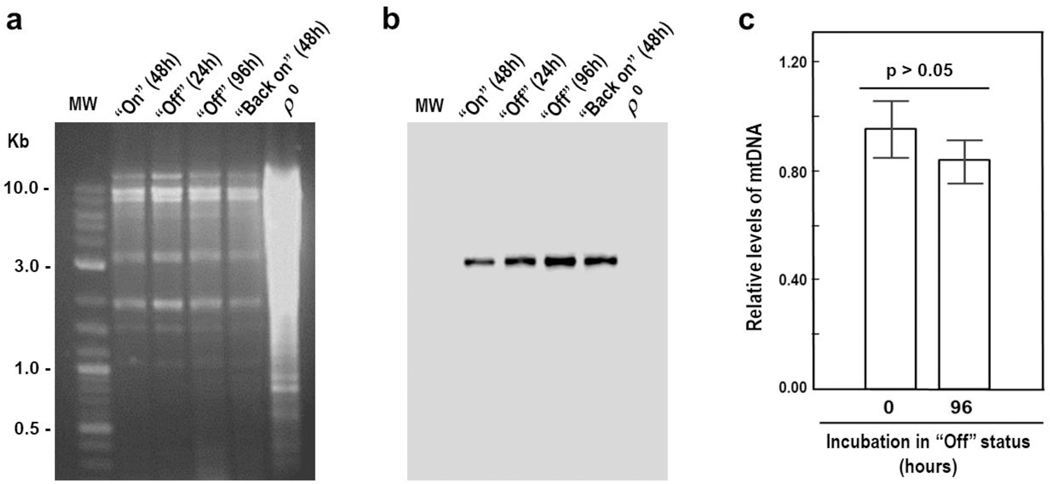
The length of the most critical phase (“Off”) and subsequent reversion to wild-type phenotype prompted us to determine whether the mtDNA underwent modifications. Fig. 5a depicts representative samples of mtDNA from cells subjected to the “On-Off-Back on” conditions. COB was used as the gene marker for genetic rearrangements (Fritsch et al. 2014), deletions (Heude et al. 1979) or depletion (Ulery et al. 1994) (Fig. 5b). All samples showed precisely the same pattern of restriction fragments without smears. Thus, neither ρ0 nor ρ− status, no mtDNA rearrangements, or alteration of mtDNA levels were observed throughout the entire “On-Off-Back on” experiment.
Fig. 5.
Mitochondrial DNA (mtDNA) does not undergo modifications during the “On-Off-Back on” time-course experiment. a Ethidium bromide-stained agarose gel electrophoresis of EcoRV- digested mtDNA isolated from clones grown under “On,” “Off,” and “Back on” conditions. MW: 2-log DNA ladder (New England Biolabs). ρ0 lane: total DNA was purified from an established Δccm1 strain. b DNA hybridization analysis of a using a 290 bp-biotinylated COB DNA probe that spans positions 28033–28322 of the mitochondrial genome. c Relative levels of mtDNA were assessed by qPCR using 15S R_RNA and ALG9 as target and reference genes, respectively. Columns in association with vertical bars represent the means ± SEM of three independent Ccm1pZZ-expressing clones measured in duplicate. The values were not found significantly different by a two-sample t-test.
In addition to repressing the GAL1 promoter, glucose also decreases the mtDNA copy number (Ulery et al. 1994). We had observed a transient-moderate depression of 15S rRNA and 21S rRNA levels upon the same sugar change (Moreno et al. 2012). Therefore, to factor in this swap on the mtDNA copy- number, we measured mtDNA levels in Δccm1 cells complemented with a vector expressing Ccm1p under the control of a constitutive promoter (pDB20-CCM1). Subsequently, mtDNA levels in cells with the regulable expression vector were divided by mtDNA levels in cells with the constitutive expression vector, both in the presence of galactose (t = 0 h). The procedure was also carried out for cells grown in the presence of glucose (t = 96 h). The two ratios were then compared with one another: mtDNA levels in galactose did not differ from those in glucose (p > 0.05, Fig. 5c). Thus, differences in Ccm1p expression did not affect the mtDNA copy number.
So far, our results indicated that even though the total population of mitochondria reached a dysfunctional status, they kept the capacity to uptake Ccm1p and resume their physiological activity. Moreover, the cells re-acquired the wild-type phenotype at approximately the same rate they lost it (Fig. 4a, b). Thus, the idea of a few, still active mitochondria generating 100% of the functional population by fusion and fission (Mishra and Chan 2014) upon Ccm1p resupply could be ruled out; instead, a “stand by” condition appeared to take place during the “Off” phase.
Based on the present work, we have produced and validated a tri-phasic in vivo system that yielded dual dose-response results suitable to study changes on the initial 15S rRNA transcript upon modifications of the Ccm1p intramitochondrial concentrations.
The levels of 5’ and 3’ 15S rRNA unprocessed junctions inversely follow that of intramitochondrial concentration of Ccm1p
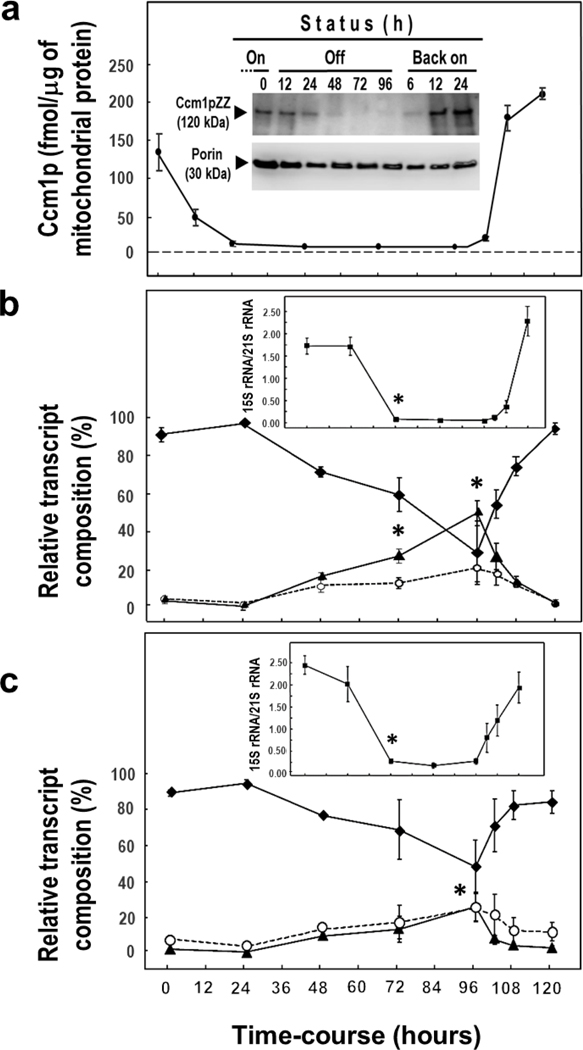
We then assessed how the in vivo system responded to the variation of the intramitochondrial Ccm1p concentration by measuring the levels of total 15S rRNA. The “Back on” status was a confirmatory second dose-response counterpart. Ccm1pZZ was used in this part of the study because the ZZ moiety was essential to measure the intramitochondrial protein concentration by ELISA, increase the sensitivity of the immunoblot analysis, and enhance the “Off” effect (Fig. 6a). Levels of 5’ and 3’ UJs were measured for both Ccm1pZZ (Fig. 6b) and Ccm1pAPC (Fig. 6c). After 24 h under “Off” status, segregants harboring pCCM1ZZLC presented a Ccm1pZZ concentration of approximately 5 fmol per μg of mitochondrial protein while the total 15S rRNA levels remained unchanged (Fig. 6b, insert). Between 48 and 96 h of repression, the Ccm1pZZ concentration never reached 0, but rather maintained a constant value of approximately 1 fmol per μg of mitochondrial protein. These low levels of Ccm1p may account for the fact that the 15S rRNA transcript was never found 100% unprocessed. After 48 h under “Off” status, the total 15S rRNA levels abruptly fell more than 20-fold (Fig. 6b, c, inserts), while the cells’ growth on YEPG was 100 times lower than that of the wild type (Fig. 4a). Additionally, the Ccm1pZZ clones presented less 15S rRNA than those harboring Ccm1pAPC over the entire “Off” period (p < 0.05; Fig. 6b, c), but the amount of CCM1 transcript was similar in both cases (p > 0.05, data not shown). This effect agreed with the en route delay in the protein import caused by the ZZ tag. Remarkably, the percentages of 5’ and 3’ UJs that represent the unprocessed 15S rRNA progressively increased several- fold during the “Off” phase, reaching, at 96 h, their maximum value between 20 (Ccm1pAPC 3’ and 5’ UJs; Ccm1pZZ 5’UJ) to 50 times (Ccm1pZZ 3’UJ) over that of 0 or 24 h (p < 0.05). Once the Ccm1p supply was restored (right after 96 h under “Off” status) for both Ccm1p forms (Fig. 6a, insert “Back on”), the levels of both UJs diminished to negligible values (second dose-response) with a concomitant increase of processed 15S rRNA. This observation strongly suggests that the peaks at 96 h belong to authentic unprocessed UJs rather than abnormal forms of 15S rRNA.
Fig. 6.
Accumulation of unprocessed 15S rRNA transcripts during the “Off” status reaches significant levels at 96 h. “Time course (hours)” at the bottom of the chart represents the total span of the “On-Off-Back on” experiment. a Intramitochondrial Ccm1p levels at “On” (24 h in SDGal medium is t = 0 h), “Off” (t =12 to 96 h), and “Back on” (t =102 to 120 h) during the time-course experiment. Quantitation was carried out by ELISA. Insert: Ccmp1 was visualized by 5–20% PAGE and immunoblot analysis with an anti-Ccm1p polyclonal antibody. Times for each status are designated at the top of the immunoblot. Porin was used as the loading control. b Percentage of 3’ unprocessed junctions (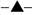 ), 5’ unprocessed junctions (
), 5’ unprocessed junctions ( ), and mature 15S rRNA [i.e. total 15S rRNA – (3’ unprocessed junctions + 5’ unprocessed junctions)] (
), and mature 15S rRNA [i.e. total 15S rRNA – (3’ unprocessed junctions + 5’ unprocessed junctions)] (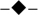 ) was measured by RT-qPCR as the levels of Ccm1pZZ or (c) Ccm1pAPC changed. Inserts in
b
and
c Relative levels of total 15S rRNA, 21S rRNA was used as the reference transcript. Samples for analyses in a, b, and c came from a common set of cultures. Symbols in association with vertical bars represent the means ± SEM of three independent clones measured in duplicate. For statistical analysis, data were log-transformed when necessary to normalize the variables or homogenize the variances. *, p < 0.05, one-way ANOVA, Student-Newman-Keuls post hoc test. Downward pointing arrowheads indicate the last “On” time point and the initiation of the “Back on” status
) was measured by RT-qPCR as the levels of Ccm1pZZ or (c) Ccm1pAPC changed. Inserts in
b
and
c Relative levels of total 15S rRNA, 21S rRNA was used as the reference transcript. Samples for analyses in a, b, and c came from a common set of cultures. Symbols in association with vertical bars represent the means ± SEM of three independent clones measured in duplicate. For statistical analysis, data were log-transformed when necessary to normalize the variables or homogenize the variances. *, p < 0.05, one-way ANOVA, Student-Newman-Keuls post hoc test. Downward pointing arrowheads indicate the last “On” time point and the initiation of the “Back on” status
In addition to the higher percentage of 3’ UJ, the total 15S rRNA decay appears to be more abrupt for the ZZ form-expressing clones. Yet again, these results concur with others of SMDC, which allowed for a faster 15S rRNA primary transcript accumulation. This buildup of the unprocessed molecule might be due to a saturation of the mtEXO activity. Nevertheless, the profiles of 15S rRNA and UJs levels over the “On-Off-Back on” time-course were in agreement for both CCM1 constructs. We did not extend the “Off” time beyond 96 hours due to the risk of damaging the mtDNA.
Ccm1p and mtEXO act concertedly but oppositely in the processing/degradation of 15S rRNA
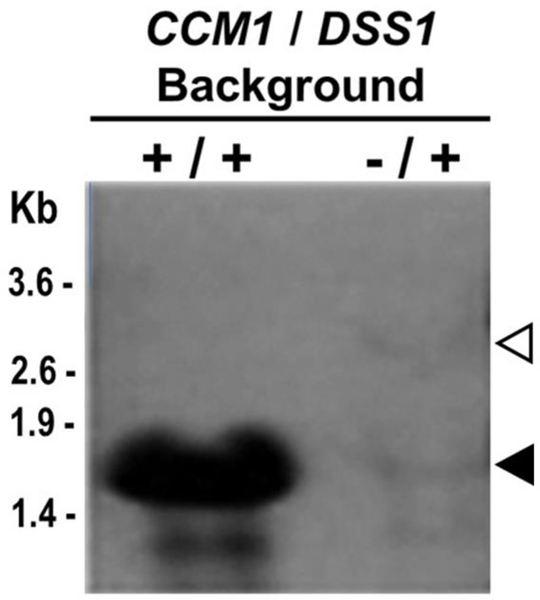
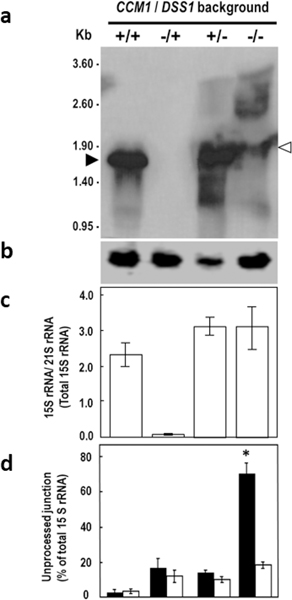
The significant accumulation of 3’ and 5’ UJs (Fig. 6b, c) was investigated in more detail with a study based on Ccm1p/mtEXO double deletant cells. mtEXO is composed of DSS1 and SUV3 gene products (Dziembowski et al. 1998). The deletion of either gene caused substantial accumulation of not only unprocessed 15S rRNA transcripts but also of the processed form, demonstrating that mtEXO is responsible for surveying and degrading the initial 15S rRNA transcript (Dziembowski et al. 2003). RNA hybridization analysis showed that CCM1/DSS1 (+/+) cells exhibited only processed 15S rRNA, but the Δccm1/DSS1 (−/+) mutants did not display any form of 15S rRNA, not even degradation products. Single CCM1/ Δdss1 (+/−) mutants exhibited abundant processed 15S rRNA and limited amounts of non- processed transcripts (Fig. 7a, c), indicating that, as previously reported, mtEXO does not participate in the removal of intervening or flanking sequences (Dziembowski et al. 2003). Remarkably, Δccm1 / Δdss1 (−/ −) cells dramatically accumulated unprocessed transcripts, as evidenced by the abundant heavier forms. Specifically, a form that is approximately 100 nucleotides longer than the processed 15S rRNA was detected in the double mutant (Fig. 7a, 4th lane, hollow arrowhead). As the sum of the independent measurements of 3’ and 5’ UJ levels equaled 100% of the total unprocessed 15S rRNA (Fig. 7d, 4th set of bars), every species displayed in Δccm1 / Δdss1 (−/ −) has at least one UJ intact. Therefore, we concluded that such a form was another degradation product from the initial transcript. Furthermore, we compared our results to the ones previously reported (see Fig. 6b, 15S rRNA panel; Dziembowski et al. 2003) in which both independent mutants, Δdss1 and Δsuv3, were wildtype for CCM1. Their patterns of degradation were strikingly similar to the one displayed in the fourth lane of Fig. 7a, which had a Δccm1 background. These observations indicate that Ccm1p does not participate in the stabilization of the 15S rRNA primary transcript per se.
Fig. 7.
The Δccm1 / Δdss1 (−/ −) mutant displays unprocessed, rather than mature 15S rRNA. a Hybridization analysis of mitochondrial RNA. Total RNA (10–15 μg per well) was isolated from strains of CCM1/DSS1 (+/+), Δccm1/ DSS1 (−/+), CCM1/ Δdss1 (+/−), and Δccm1 / Δdss1 (−/ −) genetic backgrounds and probed with a 585 bp-biotinylated 15S R_RNA DNA fragment that spans positions 6785–7369 of the mitochondrial genome, inside the mature transcript. b Hybridization analysis of EcoRV-digested mtDNA (4 μg per well) with a 209 bp-DNA probe that anneals to a fragment that contains the full-length RPM1 gene and its promoter. RT-qPCR assessment of (c) relative levels of total 15S rRNA and (d) 3’ UJ (black columns) and 5’ UJ (white columns). 21S rRNA was the reference transcript. The assays depicted in b, c, and d correspond to samples from the set of mutants shown in a. Measurements in c and d for each strain were independent. Columns in association with vertical bars represent the means ± SEM of three independent clones measured in duplicate. *, p < 0.05, one-way ANOVA, Student-Newman-Keuls post hoc test
Importantly, the Δccm1 / Δdss1 (−/ −) mitochondrial genome contained RPM1, which encodes for the RNA component (9S RNA) of the RNAse P (Stribinskis et al. 1996), as evidenced by DNA hybridization analysis. An EcoRV DNA fragment of approximately 3400 bp, which includes RPM1 along with its promoter region was detected in all the genetic backgrounds examined in this experiment. (Fig. 7b). Therefore, we ruled out that the substantial accumulation of unprocessed 15S rRNA was on account of the absence of 9S RNA activity.
Quantitation by RT-qPCR detected over 30 times more total 15S rRNA in Δccm1 / Δdss1 (−/ −) cells than in Δccm1/DSS1 (−/+) mutants (p < 0.01) (Fig. 7c, 4th bar); the most abundant RNA was the 3’ UJ-containing transcripts (p < 0.05, Fig. 7d, black bar). Consequently, the results due to the lack of mtEXO agree with the peaks of 3’ and 5’ UJs levels (Fig. 6b, c) in which the surveillance system appears to be saturated, and 100 % of the cells present the mutant phenotype (Fig. 4a).
Ccm1p resupply restores the processing of the initial 15S rRNA transcript in Δccm1/Δdss1 cells
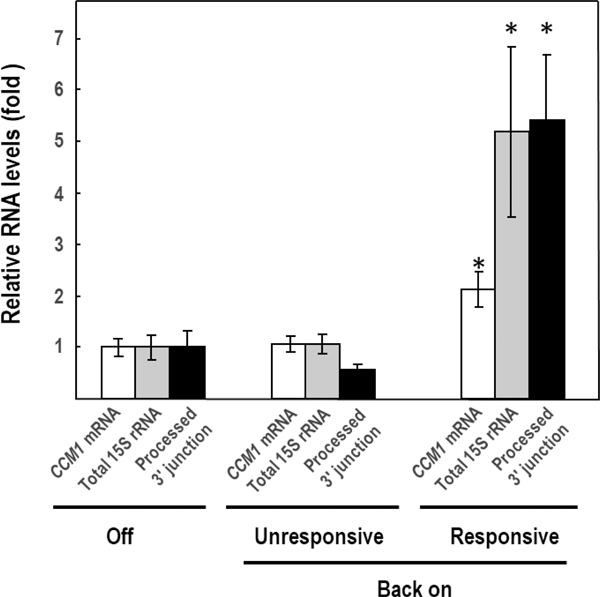
The Δccm1/ Δdss1 diploid mutant was prepared with cells that harbored pCCM1ZZLC at the moment of mating, to produce Δccm1/ Δdss1 meiotic segregants that would be competent for the “On-Off-Back on” experiment. The “Back on” approach aimed to discern whether the longer transcripts observed in the Δccm1/Δdss1 cells (Fig. 7a, 4th lane), were the result of the lack of Ccm1p or just aberrant RNA forms. We obtained seven Δccm1/Δdss1 meiotic segregants harboring pCCM1ZZLC, that under “Off” conditions, showed similar baseline levels of CCM1 mRNA, total 15S rRNA, and processed 3’ junction (Fig. 8). When transferred to SDGal and incubated for 48h (“Back on”), we found that three of them were unresponsive to induction by galactose as they did not differ from the ones under “Off” conditions. These clones were considered as a second negative control. These results were foreseeable since Δccm1/ Δdss1 nascent segregants harboring pCCM1ZZ in “Off” status would have a borderline reduced fitness. It is noteworthy to mention that the established Δccm1 mutant (ρ0/ρ−) is unable to operate the GAL1 promoter due to reduced fitness (unpublished data), but nascent non-complemented segregants still conserved that capacity (Fig. 3, insert). However, four clones were responsive to galactose, as shown by the significant increase in CCM1 mRNA (p < 0.05), and were able to process the 15S rRNA transcript in substantial quantities (p < 0.05). These results confirmed that the abundant heavier species observed in the Δccm1/ Δdss1 cells (Fig. 7a, 4th lane) were authentic unprocessed forms, which could be cleaved under “Back on” conditions.
Fig. 8.
Processing of 3’ UJ is restored in Δccm1Δdss1 (−/ −) segregants upon Ccm1pZZ resupply (Back on). Relative levels of CCM1 mRNA, total 15S rRNA, and 3’ processed junction (total 15S rRNA – 3’ UJ) were assessed by RT-qPCR. “Off” represents the baseline values obtained under SDD conditions (n = 7). ALG9 mRNA was the reference transcript. Columns in association with vertical bars represent the means SEM of three independent clones measured in duplicate. Data were log-transformed to normalize the variables or homogenize the variances.*, p < 0.05, one-way ANOVA, Student-Newman-Keuls post hoc test
Discussion
Several studies reported that PPR proteins are implicated in RNA processing (Manna 2015). PTCD1, a human protein that contains eight PPR motifs (Rackham et al. 2009), is involved in processing the mitochondrion heavy strand polycistronic transcript (Sánchez et al., 2011; Perks et al. 2016). This molecule includes RNA19, a precursor that consists of 16S rRNA (a component of the large mitoribosome subunit) linked at the 3’ end to mt-tRNAL(UUR) and ND1 mRNA (Bindoff et al. 1993). When PTCD1 is not expressed, levels of 16S rRNA decrease dramatically, but heavier precursor molecules (including RNA19) become detectable by RNA hybridization analysis, albeit in much lower concentration than those of the mature rRNA (Perks et al. 2016) as it happens with 15S rRNA in baker’s yeast (this study).
Similarly, another mammalian PPR protein, PTCD2, participates in the processing of the ND5- COB bicistronic transcript (Xu et al. 2008), which includes an intervening sequence of 600 nucleotides, not flanked by tRNAs (Anderson et al. 1981). Mice expressing a deficient PTCD2 are unable to process the bicistronic transcript; instead, they accumulate large amounts of the precursor molecule (Xu et al. 2008). However, it is not degraded by the RNA surveillance system (Szczesny et al. 2010).
Yeast Pet309p and Aep3p participate in the processing of COX1 mRNA and the ATP8/6 bicistronic transcript, respectively. Mutations that inactivate or delete the proteins lead to the complete disappearance of the COX1 transcripts (Manthey and McEwen 1995) or the processed ATP8/6 mRNA with the concomitant accumulation of trace levels of its unprocessed precursor (Ellis et al. 2004). Likewise, suppression of Ccm1p expression reduces mature COB and COX1 mRNAs to undetectable levels (Moreno et al. 2009; 2012). However, the precursor remains visible in minimal amounts throughout the entire “Off” time course at approximately 5 % of the initial levels of the processed transcript (Moreno et al. 2012).
In vitro splicing experiments in the absence of Ccm1p showed that only minute amounts of the full-sized bI4 intron (1417 nucleotides) could be removed. Moreover, under such conditions, the bI4 intron had to be minimized down to 380 nucleotides for efficient splicing (Boniecki et al. 2009). We had proposed that Ccm1p would be able to act on long stretches of RNA that are fated to be removed, thus yielding a bI4 intron structure that is competent to undergo excision (Moreno et al. 2009). Accordingly, Ccm1p acts on the removal of long stretches of nucleotides from the 15S rRNA primary transcript (this report).
The “On-Off” approach was used by an earlier study to determine that the first activity of Ccm1p was essential to the splicing of COB and COX1 pre-mRNAs (Moreno et al. 2009). Follow-up research provided clear insights into the molecular events that took place during SDMC (Moreno et al. 2012), which is the “Off” phase in the present study. During the entire “On-Off-Back on” time course, we have demonstrated that: (i) the mutant phenotype (i.e., failure to grow in the presence of glycerol as the sole source of carbon and energy) acquired during the “Off” phase was reversible upon Ccm1p resupply; (ii) the recovery of the wild-type phenotype was achieved at approximately the same rate as the preceding “Off” phase; (iii) the phenotype changes followed the intramitochondrial Ccm1p levels; (iv) the capacity of mitochondria to import Ccm1p was maintained; (v) no ρ−or ρ0 cells were observed during the entire “On-Off-Back on” course which agrees with points (i) and (sii); (vi) total 15S rRNA levels directly followed the intramitochondrial concentrations of Ccm1p during the complete “On-Off-Back on” course. Consequently, we concluded that this approach was a suitable new triphasic in vivo dose-response system for experimentation. We performed preliminary experiments in which the “Back on” phase was started from the “Off” state at earlier times (65 h and 72 h) upon replacing glucose by galactose, thus reinducing the GAL1 promoter. In all cases, including 96 h under “Off” status, we consistently obtained the same pattern of phenotype loss and recovery (data non-shown). Therefore, there was no credible possibility of suppressor mutation events during the off-state period.
mtEXO is a physiological complex of the RNA metabolism that is required to eliminate unprocessed and aberrant RNAs (Dziembowski et al. 2003). This present work addresses the axiom “any unprocessed RNA molecule is destined to be degraded.” Once our in vivo system “On-Off-Back on” was validated, we investigated how the primary15S rRNA transcript underwent processing as the intramitochondrial concentration of Ccm1p varied. Levels of both 5’ and 3’ UJs gradually increased (probably due to mtEXO saturation) reaching their highest values when the cells fully acquired the mutant phenotype at a Ccm1p concentration of approximately 1 fmol per μg of mitochondrial protein. The last “Off” time point (96 h) showed the maximal accumulation of both 3’ and 5’ UJs. Conversely, the proportion of unprocessed 15S rRNA forms fell upon Ccm1p resupply with a simultaneous increase of total levels of 15S rRNA and recovery of the wild-type phenotype. The 15S recovery in the case of Ccm1pZZ was slower than that of Ccm1pAPC (Fig 6b and c, inner panels, respectively). This result is in agreement with the import delay scenario caused by the ZZ tag. In the case of Ccm1pAPC (no addition of extra sequences), no lag was observed as no hindrance in the import process occurred. There was a six hour-interval between measurements, long enough for the production and degradation of other proteins that may be involved in this process. Nevertheless, we do not discard the possibility of a direct action of Ccm1p on the 15S rRNA initial transcript. Furthermore, the abrupt switch “Off” using Ccm1pZZ caused a higher accumulation of 3’ UJ than of 5’UJ, indicating that the 3’ end processing appears to be more Ccm1p dependent than its 5’ counterpart. These results suggest that Ccm1p acts on both UJs likely associated with different sets of factors.
To confirm these findings independently, we focused on the unprocessed transcripts observed in the “Off” phase. The presence of 15S rRNA and related precursors were examined in Δdss1 and Δdss1/Δccm1 mutants. The Δdss1 mutant exhibited mostly processed 15S rRNA and low levels of longer forms. These results confirmed that mtEXO does not participate in the removal of the 5’ or 3’ flanking sequences (Dziembowski et al. 2003). In agreement with our “On-Off-Back on” results, the Δdss1/ Δccm1 mutant exhibited a significantly higher abundance of unprocessed 15S rRNA species in which the 3’ UJ was predominant. That the absence of Ccm1p causes the accumulation of such abundant quantities of precursors, which contain mature 15S rRNA, is a clear indication that Ccm1p is neither a stabilizer nor a factor to maintain the integrity of this molecule. Likewise, the absence of mtEXO activity allowed us to visualize such unprocessed forms.
Additionally, by resupplying Ccm1p to this double mutant, the amount of processed 3’ junction increased significantly. These results demonstrated that mtEXO is responsible for the degradation of unprocessed 15S rRNA in the Δccm1 mutant strain. Therefore, in addition to being a splicing factor (Moreno et al. 2009; 2012), this protein is essential to process the 15S rRNA primary transcript. Consequently, Ccm1p may be ruled out as a stabilizer, proper assembler, or chaperon to the mature 15S rRNA before it can be fully integrated into the mitoribosome, neither it has a direct role in preventing the degradation of mitochondrial rRNA.
The elimination of the 5’ 0.5S rRNA is accomplished by a purportedly 5’→3’ exonuclease activity associated with Pet127p (Fekete et al. 2008) that trims the 5’ terminal end of the 15.5S rRNA (Wiesenberger and Fox 1997). According to our results, Ccm1p could be a molecular partner of Pet127p to control such exonucleolytic activity; it could act as a signal to anchor the enzyme associated with Pet127p or promote a putative-Ccm1p dependent endoribonuclease.
The processing of the 3’ junction has some remarkable features. When released from the initial transcript, the 1308-nucleotide intervening sequence was not degraded by mtEXO since a probe that specifically hybridized to that region failed to detect a signal in RNA isolated from Δdss1 mutants (Dziembowski et al. 2003). A possible candidate to degrade this sequence is Dis3p, which has 3’ → 5’ exonuclease activity and localizes to the mitochondrion (Turk et al. 2013). However, no data regarding the elimination of the free intervening sequence has been reported so far, and no specific or consensus signal for endonucleolytic activity has been found at the 3’ end of it (Turk et al. 2013). An heptakaidecamer cleavage site has been located approximately 3kb downstream of the end of the 15S rRNA- tRNAW transcript (Turk et al. 2013), too far to yield the mature 15S rRNA. In this case, as it was proposed earlier regarding the removal of the COX1 aI4 and COB bI4 introns (Moreno et al. 2009), Ccm1p acts on long stretches of RNA, at the boundaries of the 3’ junction, to assist the 15S rRNA precursor in acquiring a competent structure for a precise cleavage. This situation resembles that of mammalian ND6 mRNA (Van Haute et al. 2015). The mature ND6 transcript lacks a consensus sequence at the 3’ end (Slomovic et al. 2005), there is a 5700-nucleotide spacer between it and closest tRNA (Anderson et al. 1981), and the processing of the 3’ junction involves an RNA-binding protein, FASTK (Jourdain et al. 2015).
Like any bona fide PPR protein, processing the 15S rRNA primary transcript and splicing the pre- mRNAs obviously imply in vivo Ccm1p-RNA interactions. However, both activities reside at different PPR2 amino acids (Moreno et al. 2012). Along the same lines, replacement of S. cerevisiae Ccm1p by that of S. bayanus also dissects both functions since it reduces 15S rRNA levels without affecting the removal of introns (Jhuang et al. 2017). Interestingly, mutations of two residues in the PPR2 domain rescue the incompatibility.
In S. cerevisiae, the nuclear equivalent version of the cytosolic ribosome consists of one primary transcript (35S pre-RNA) that bears three of the four required rRNAs. The processing of this molecule follows a well-known pathway involving many factors and ribonucleases (Fang et al. 2005). The primary transcript has two internal spacers, ITS1 and ITS2. Particularly, removal of ITS2 is initiated through a specific cleavage inside the spacer by the complex Las1/Crc3 followed by exoribonucleases that eliminate the ITS2 producing the 5.8S and 25S of the 60S ribosomal subunit (Pillon and Stanley 2018). In the mitochondrial case, the scenario seemed somehow related due to the presence of an intervening sequence between an rRNA and a tRNA, but the complete mechanism and identity of the factors involved are still unclear. Based on our findings, particularly the ones obtained with the Δdss1/Δccm1 mutant (Fig. 7), we propose that once the tRNAW has been excised by RNase P and RNase Z (Daoud et al. 2012), the spacer 3’ end is exposed. When it remains bound to 15S rRNA, due to the lack of Ccm1p, this region acts as the signal recognized by mtEXO to anchor and commence the degradation of the unprocessed 15S rRNA-spacer transcript until the entire molecule is degraded. Conversely, if the spacer is removed with the assistance of Ccm1p, mtEXO cannot reach the 15S rRNA moiety, which is protected by primary binding ribosomal proteins. In fact, Nam9p, one of the first ribosomal proteins to bind 15S rRNA (Gan et al. 2002; Shajani et al. 2011), protects it from degradation (Biswas and Getz 1999). Thermosensitive NAM9 strains lose their mature 15S rRNA at the nonpermissive temperature, which proves that the actual 15S rRNA stabilizers are the primary proteins bound to it during the biogenesis of the mitoribosome minor subunit.
By two independent approaches, the “On-Off-Back on” time course and the Δccm1/ Δdss1 double mutant, we have demonstrated that the 15S RNA primary transcript has only two alternatives: Either it is processed and incorporated into the mitoribosome minor subunit, or it is degraded by the mitochondrial RNA surveillance system as a consequence of processing failure in Δccm1 strains.
Acknowledgments
This work was supported by the Mississippi INBRE funded by grants from the National Center for Research Resources [5P20RR016476] and the National Institute of General Medical Sciences (NIGMS) [8P20GM103476] from the National Institutes of Health (NIH), and by grants number [5SC3GM087169] and [W911NF-13-1-0174] from NIGMS, NIH, and DoD, respectively. We thank Mrs. Kimberley S. Buie for her contributions. The support from Dr. Sandra Barnes, Department of Chemistry and Physics, and the office of the Dean of the School of Art and Sciences are honestly appreciated. Finally, we acknowledge the valuable input from the reviewers.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Amberg DC, Burke DJ, Strathern JN (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Becker DM, Fikes JD, Guarente L (1991) A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci USA 88:1968–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindoff LA, Howell N, Poulton J, McCullough DA, Morten KJ, Lightowlers RN, Turnbull DM, Weber K (1993) Abnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA. A potential disease mechanism. J Biol Chem 268:19559–19564 [PubMed] [Google Scholar]
- Biswas TK and Getz GS (1999) The single amino acid changes in the yeast mitochondrial S4 ribosomal protein cause temperature-sensitive defect in the accumulation of mitochondrial 15S rRNA. Biochemistry 38:13042–13054 [DOI] [PubMed] [Google Scholar]
- Björkholm P, Harish A, Hagström E, Ernst AM, Andersson SGE (2015) Mitochondrial genomes are retained by selective constraints on protein targeting. Proc Natl Acad Sci USA 112:10154–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniecki MT, Rho SB, Tukalo M, Hsu JL, Romero EP, Martinis SA (2009) Leucyl-tRNA synthetase- dependent and-independent activation of a group I intron. J Biol Chem 284:26243–26250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud R, Forget L, Lang BF (2012) Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res 4:1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Gómez-Herreros F, Rodríguez-Galán O, Begley V, de la Cruz Muñoz-Centeno M, Chávez S (2018) Feedback regulation of ribosome assembly. Curr Genet 64: 393–404 [DOI] [PubMed] [Google Scholar]
- De Silva D, Tu YT, Amunts A, Fontanesi F, Barrientos A (2015) Mitochondrial ribosome assembly in health and disease. Cell Cycle 14:2226–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defontaine A, Lecocq FM, Hallet JN (1991) A rapid miniprep method for the preparation of yeast mitochondrial DNA. Nucleic Acids Res 19:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschmann AJ, Amberger A, Zavadil C, Steinbeisser H, Mayr JA, Feichtinger RG, Oerum S, Yue WW, Zschocke J (2014) Mutation or knock-down of 17β-hydroxysteroid dehydrogenase type 10 cause loss of MRPP1 and impaired processing of mitochondrial heavy strand transcripts. Hum Mol Genet 23:3618–3628 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Malewicz M, Minczuk M, Golik P, Dmochowska A, Stepien PP (1998) The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol Gen Genet 260:108–114 [DOI] [PubMed] [Google Scholar]
- Dziembowski AJ, Piwowarski R, Hoser M, Minczuk A, Dmochowska M, Siep H, van der Spek LG, Stepien PP (2003) The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem 278:1603–1611 [DOI] [PubMed] [Google Scholar]
- Ellis TP, Helfenbein KG, Tzagoloff A, Dieckmann CL (2004) Aep3p stabilizes the mitochondrial bicistronic mRNA encoding subunits 6 and 8 of the H+-translocating ATP synthase of Saccharomyces cerevisiae. J Biol Chem 16:15728–15733 [DOI] [PubMed] [Google Scholar]
- Falk MJ, Gai X, Shigematsu M, Vilardo E, Takase R, McCormick E, Christian T, Place E, Pierce EA, Consugar M, Gamper HB, Rossmanith W, Hou YM (2016) A novel HSD17B10 mutation impairing the activities of the mitochondrial RNase P complex causes X-linked intractable epilepsy and neurodevelopmental regression. RNA Biol 13:477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Phillips S, Butler JS (2005) Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA 11:1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete Z, Ellis TP, Schonauer MS, Dieckmann CL (2008) Pet127 governs a 5’→ 3’-exonuclease important in maturation of apocytochrome b mRNA in Saccharomyces cerevisiae. J Biol Chem 7:3767–3772 [DOI] [PubMed] [Google Scholar]
- Foury F, Roganti T, Lecrenier N, Purnelle B (1998) The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 440:325–331 [DOI] [PubMed] [Google Scholar]
- Fox TD (2012) Mitochondrial protein synthesis, import, and assembly. Genetics 192:1203–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch ES, Chabbert CD, Klaus B, Steinmetz LM (2014) A genome-wide map of mitochondrial DNA recombination in yeast. Genetics, 198:755–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Kitakawa M, Yoshino KI, Oshiro N, Yonezawa K, Isono K (2002) Tag‐mediated isolation of yeast mitochondrial ribosome and mass spectrometric identification of its new components. Eur J Biochem 269:5203–5214 [DOI] [PubMed] [Google Scholar]
- Gavin AC et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147 [DOI] [PubMed] [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gössringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P (2010) A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17:740–744 [DOI] [PubMed] [Google Scholar]
- Gray MW (2012) Mitochondrial evolution. Cold Spring Harb Perspect Biol 4:a011403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XE, Chen CF, Wang DD, Modrek AS, Phan VH, Lee WH, Chen PL (2011) Uncoupling the roles of the SUV3 helicase in maintenance of mitochondrial genome stability and RNA degradation. J Biol Chem 286:38783–38794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack TB, Kopajtich R, Freisinger P, Wieland T, Rorbach J, Nicholls TJ, Baruffini E, Walther A, Danhauser K, Zimmermann FA, Husain RA, Schum J, Mundy H, Ferrero I, Strom TM, Meitinger T, Taylor RW, Minczuk M, Mayr JA, Prokisch H (2013) ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am J Hum Genet 93:211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heude M, Fukuhara H, Moustacchi E (1979) Spontaneous and induced rho mutants of Saccharomyces cerevisiae: patterns of loss of mitochondrial genetic markers. J Bacteriol 139:460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth MJ, Martin NC (1986) RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol 6:1058–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W (2008) RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135:462–474 [DOI] [PubMed] [Google Scholar]
- Jhuang HY, Lee HY, Leu JY (2017) Mitochondrial-nuclear co-evolution leads to hybrid incompatibility through pentatricopeptide repeat proteins. EMBO Rep 1:87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain AA, Koppen M, Rodley CD, Maundrell K, Gueguen N, Reynier P, Guaras AM, Enriquez JA, Anderson P, Simarro M, Martinou JC (2015) A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Rep 10:1110–1121 [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD (2008) Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9: 605–618 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kawabata M, Hisano K, Kazama T, Matsuoka K, Sugita M, Nakamura T (2012) Identification and characterization of the RNA binding surface of the pentatricopeptide repeat protein. Nucleic Acids Res 40:2712–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritsiligkou P, Chatzi A, Charalampous G, Mironov A Jr., Grant CM, Tokatlidis K (2017) Unconventional targeting of a thiol peroxidase to the mitochondrial intermembrane space facilitates oxidative protein folding. Cell Rep 18:2729–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S (2015) An overview of pentatricopeptide repeat proteins and their applications. Biochimie 113:93–99. [DOI] [PubMed] [Google Scholar]
- Manthey GM, McEwen JE (1995) The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron‐containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J 14:4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margossian SP, Li H, Zassenhaus HP, Butow RA (1996) The DExH box protein Suv3p is a component of a yeast mitochondrial 3’-to-5’ exoribonuclease that suppresses group I intron toxicity. Cell 84:199–209 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. (2006) Methods Mol Biol 313:33–39 [DOI] [PubMed] [Google Scholar]
- Merz S, Westermann B (2009) Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol 10, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Thompson K, Alston CL, Morris AA, He L, Assouline Z, Rio M, Bahi-Buisson N, Pyle A, Griffin H, Siira S, Filipovska A, Munnich A, Chinnery PF, McFarland R, Rötig A, Taylor RW (2016) Recessive mutations in TRMT10C cause defects in mitochondrial RNA processing and multiple respiratory chain deficiencies. Am J Hum Genet 98:993–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC (2014). Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol 15:634–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller-Hergt BV, Carlström A, Stephan K, Imhof A, Ott M. (2018) The ribosome receptors Mrx15 and Mba1 jointly organize cotranslational insertion and protein biogenesis in mitochondria. Mol Biol Cell 29:2386–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G (1982) Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA 79:7195–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JI (1996) A Trypanosoma cruzi polyantigen obtained by gene fusion: its expression in Staphylococcus aureus and rapid purification. Protein Expr Purif 8:332–340 [DOI] [PubMed] [Google Scholar]
- Moreno JI, Buie KS, Price RE, Piva MA (2009) Ccm1p/Ygr150cp, a pentatricopeptide repeat protein, is essential to remove the fourth intron of both COB and COX1 pre-mRNAs in Saccharomyces cerevisiae. Curr Genet 55:475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JI, Patlolla B, Belton KR, Jenkins BC, Radchenkova PV, Piva MA (2012) Two independent activities define Ccm1p as a moonlighting protein in Saccharomyces cerevisiae. Biosci Rep 32:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörl M, Marchfelder A (2001) The final cut. The importance of tRNA 3’-processing. EMBO Rep 2:17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar MP, Shen J, Raman B, Marten MR (2003) Solubilization of trichloroacetic acid (TCA) precipitated microbial proteins via NaOH for two-dimensional electrophoresis. J Proteome Res 2:89–93 [DOI] [PubMed] [Google Scholar]
- Naquin D, Panozzo C, Dujardin G, van Dijk E, d’Aubenton-Carafa Y, Thermes C (2018) Complete sequence of the intronless mitochondrial genome of the Saccharomyces cerevisiae strain CW252. Genome Announc 6:e00219–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinga KA, Evers RF, Van der Laan JC, Tabak HF (1981) A putative precursor for the small ribosomal RNA from mitochondria of Saccharomyces cerevisiae. Nucleic Acids Res. 9:1351–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R (2012) RNA Degradation in Saccharomyces cerevisiae. Genetics 3:671–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perks KL, Rossetti G, Kuznetsova I, Hughes LA, Ermer JA, Ferreira N, Busch JD, Rudler DL, Spahr H, Schöndorf T, Shearwood AM (2018). PTCD1 is required for 16S rRNA maturation complex stability and mitochondrial ribosome assembly. Cell Rep 23:127–142 [DOI] [PubMed] [Google Scholar]
- Pillon MC, Stanley RE (2018) Nuclease integrated kinase super assemblies (NiKs) and their role in RNA processing. Curr Genet 64, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinker F, Bonnard G, Gobert A, Gutmann B, Hammani K, Sauter C, Gegenheimer PA, Giegé P (2013) PPR proteins shed a new light on RNase P biology. RNA Biol 10:1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Davies SM, Shearwood AM, Hamilton KL, Whelan J, Filipovska A (2009) Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res 17:5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez MI, Mercer TR, Davies SM, Shearwood AM, Nygard KK, Richman TR, Mattick JS, Rackham O, Filipovska A (2011) RNA processing in human mitochondria. Cell Cycle 10:2904–2916 [DOI] [PubMed] [Google Scholar]
- Schülke N, Sepuri NB, Pain D (1997) In vivo zippering of inner and outer mitochondrial membranes by a stable translocation intermediate. Proc. Natl. Acad. Sci. U.S.A. 94, 7314–7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B, Boulet A, Simon M, Faye G (1987) Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci USA 84:6810–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR (2011) Assembly of bacterial ribosomes. Annu Rev Biochem 80:501–526 [DOI] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G (2005) Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol Cell Biol 25:6427–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25:46–47 [DOI] [PubMed] [Google Scholar]
- Stribinskis V, Gao GJ, Sulo P, Dang YL, Martin NC. (1996) Yeast mitochondrial RNase P RNA synthesis is altered in an RNase P protein subunit mutant: insights into the biogenesis of a mitochondrial RNA- processing enzyme. Mol Cell Biol 16:3429–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP (2010) Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res 38:279–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner A, Weber C, Buzet A, Hartmann RK, Hartig A, Rossmanith W (2012) Nuclear RNase P of Trypanosoma brucei: a single protein in place of the multicomponent RNA-protein complex. Cell Rep 2:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teste MA, Duquenne M, François JM, Parrou JL (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Das V, Seibert RD, Andrulis ED (2013) The mitochondrial RNA landscape of Saccharomyces cerevisiae. PLoS One 8:e78105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulery TL, Jang SH, Jaehning JA (1994) Glucose repression of yeast mitochondrial transcription: kinetics of derepression and role of nuclear genes. Mol Cell Biol 14:1160–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute L, Pearce SF, Powell CA, D’Souza AR, Nicholls TJ, Minczuk M (2015) Mitochondrial transcript maturation and its disorders. J Inherit Metab Dis 38:655–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sun X, Wee J, Gu Z (2019) Novel insights into global translational regulation through Pumilio family RNA-binding protein Puf3p revealed by ribosomal profiling. Curr Genet 65, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger G, Fox TD (1997) Pet127p, a membrane-associated protein involved in stability and processing of Saccharomyces cerevisiae mitochondrial RNAs. Mol Cell Biol. 17:2816–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Ackerley C, Maj MC, Addis JB, Levandovskiy V, Lee J, Mackay N, Cameron JM, Robinson BH (2008) Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem J 416:15–26 [DOI] [PubMed] [Google Scholar]