To the Editor,
We read with interest the work by Pasomsub et al. [1] on saliva as a non-invasive specimen for the diagnosis of coronavirus disease-2019 (COVID-19). We think their work timely and useful in addressing a key sampling question for community assessment of acute COVID-19 cases. While this sample type is attractive for both its reduced invasiveness and absent need for swabs and trained personnel, in our prospective UK cohort of mild community cases, comparing reverse transcription polymerase chain reaction (RT-PCR) results from combined oropharyngeal/nasopharyngeal (OP/NP) swabs with saliva, we find saliva provides inferior sample adequacy with reduced sensitivity and negative predictive value (NPV).
We conducted a prospective cross-sectional study among symptomatic healthcare workers (HCWs) and household contacts presenting to a dedicated COVID-19 outpatient clinic in London, between 28 April and 7 May 2020. Those with acute (<7-day duration) symptoms meeting the Public Health England case definition were included.
One hundred and thirty-two patients underwent combined OP/NP swab and saliva collection during the same clinic visit. Combined swabs were performed by a trained HCW swabbing both sides of the oropharynx, then nasopharynx, with collection in 4.3 mL of Roche cobas® PCR medium. Saliva was self-collected by the patient spitting into a container, without preceding coughing. On receipt in the laboratory 4.3 mL of cobas® PCR medium was added to saliva. All samples were processed utilizing one of the assays in use for routine diagnostics (Roche, AusDiagnostics, ThermoFisher and Abbott), approved for detection of SARS-CoV-2, in the North West London Pathology laboratory. Where swab and/or saliva detected SARS-CoV-2, samples were re-run using the AusDiagnostics platform for standardization and comparison of semi-quantitative cycle threshold (cT) values, inversely proportional to viral load. This assay comprises a multiplex tandem RT-PCR, targeting open reading frame (ORF) 1ab and 8 (annotated as target A and B, respectively) and includes a sample adequacy control (non-POU domain-containing octamer-binding protein, ‘NONO’).
Sensitivity, specificity, positive predictive value (PPV) and NPV were calculated to assess diagnostic performance of saliva, using OP/NP swab as a reference standard.
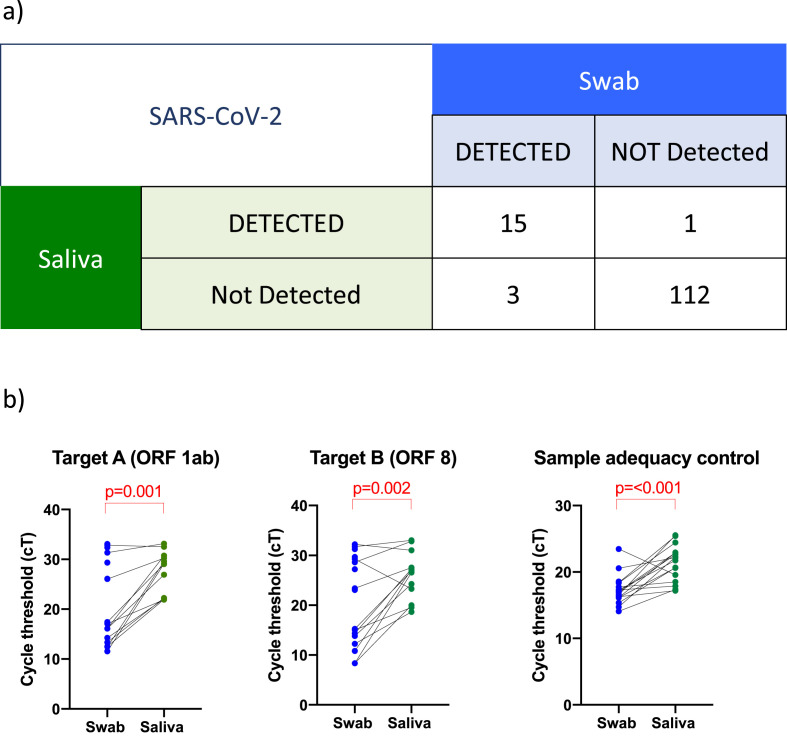
A total of 132 paired OP/NP and saliva specimens were collected; 89/132 (67.4%) patients were female. The median age was 39 years (interquartile range 30–51 years). There were five paediatric patients (<18 years) in the cohort. A total of 18/132 (13.6%) patients had SARS-CoV-2 detected by OP/NP swab (all adults >18 years). One saliva specimen was inhibited on testing, leaving 131 paired samples for comparison.
Using OP/NP results as the reference standard, saliva was positive for 15/18 patients (Fig. 1 a), with a sensitivity and specificity of 83.3% (95% CI 60.8–94.2%) and 99.1% (95% CI 95.2–100%), respectively. The PPV and NPV were 93.8% (95% CI 71.7%-99.7%) and 97.4% (95% CI 92–99.3%), respectively. cT values for SARS-CoV-2 target A, target B and sample adequacy control from saliva were significantly higher than swabs, reflecting lower viral load (Fig. 1b).
Fig. 1.
Patient-matched saliva and swab a) raw data table b) SARS-CoV-2 PCR cT values for target A (ORF 1ab), target B (ORF 8) and sample adequacy control (NONO) for 18 patients with SARS-CoV-2 detected by swab. Where SARS-CoV-2 was not detected by saliva, there is no paired cT value. p values calculated by Wilcoxon matched-pairs signed rank test.
We agree with Pasomsub et al. [1] that saliva offers a non-invasive, low-cost diagnostic specimen that avoids the need for specialist consumables (swabs and PPE) and trained personnel, removing exposure risks to HCW, advantageous in any setting, but particularly in low- and middle-income countries and community screening programmes. Published reports comparing saliva to NP swab for SARS-CoV-2 detection have shown conflicting results. In 38 inpatients admitted to Yale New Haven Hospital with severe disease, Wyllie et al. [2] found saliva more sensitive, with higher SARS-CoV-2 titres, than patient-matched NP swabs. Similarly, 25 inpatients with severe disease in Italy all had SARS-CoV-2 detected in saliva [3]. However, in an outpatient setting in Melbourne, Australia, of 662 patients screened, 33/39 (84.6%) confirmed COVID-19 patients had SARS-CoV-2 detected in saliva [4]. The median cT value was significantly higher in saliva, suggestive of lower viral loads. Our findings, and those of Pasomsub et al. [1], both testing symptomatic outpatients with mild disease, are similar. We found saliva had 83.3% sensitivity and 99.1% specificity and cT values for both SARS-CoV-2 gene targets in saliva were significantly higher than patient-matched swabs, reflecting lower levels of viral nucleic acid. This may be explained, in part, by suboptimal sample adequacy, given we found higher cT values for the housekeeping gene in saliva. Further investigation to optimize saliva collection methodology is required, which might improve sample adequacy and sensitivity.
It has been well documented that patients with more severe disease have higher viral loads [5], which may explain the acceptable performance of saliva in inpatient populations. However, if the aim for use of saliva is in a lower prevalence community setting, particularly as the background rate of infection reduces with time, this reduction in sensitivity becomes extremely important, as false negative results carry substantial risk for public health. Rather than concluding saliva is a suitable alternative to NP swab, we caution reliance on its use as a diagnostic test in this setting, without further robust evidence from a larger cohort of patients, to support its use.
Transparency declaration
L.S.P.M. has consulted for bioMerieux (2013), DNAelectronics (2015–2018), Dairy Crest (2017–2018), Umovis Lab (2020) and Pfizer (2018–2020), received speaker fees from Profile Pharma (2018), received research grants from the National Institute for Health Research (2013–2020), CW+ Charity (2018–2019) and Leo Pharma (2016), and received educational support from Eumedica (2016–2018). R.J. has received honoraria, speaker fees, travel support and/or research grant funding from Gilead, ViiV Healthcare and MSD. All other authors have no conflicts of interest to declare. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. This study was deemed a verification of a ce marked in vitro diagnostic test at North West London Pathology, Imperial College Healthcare NHS Trust. The data analysed during the current study and further details on the assays are available from the corresponding author (K.S. on reasonable request, as long as this meets local ethical and research governance criteria.
Author contributions
P.R. and L.S.P.M. designed the study methodology. M.R. and R.J. collected samples and P.M. conducted the assays. All authors reviewed the results and data analysis and contributed comments. K.S. drafted the initial manuscript with all authors contributing significantly to revising this for submission. All authors agreed on the final version for submission to the journal.
Acknowledgements
L.S.P.M. acknowledges support from the National Institute of Health Research (NIHR) Imperial Biomedical Research Centre (BRC) and the National Institute for Health Research Health Protection Research Unit (HPRU) in Healthcare Associated Infection and Antimicrobial Resistance at Imperial College London in partnership with Public Health England. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, or the UK Department of Health.
Editor: F. Allerberger
References
- 1.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease-2019 (COVID-19): a cross-sectional study. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.05.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. BMJ Yale. 2020 doi: 10.1101/2020.04.16.20067835. In press. [DOI] [Google Scholar]
- 3.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020 doi: 10.1016/j.jinf.2020.04.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/JCM.00776-20. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Yan L.-M., Wan L., Xiang T.-X., Le A., Liu J.-M. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30232-2. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]