Abstract
Natural bacterial populations are subjected to constant predation pressure by bacteriophages. Bacteria use a variety of molecular mechanisms to defend themselves from phage predation. However, since phages are nonmotile, perhaps the simplest defense against phage is for bacteria to move faster than phages. In particular, chemotaxis, the active migration of bacteria up attractant gradients, may help the bacteria escape slowly diffusing phages. Here we study phage infection dynamics in migrating bacterial populations driven by chemotaxis through low viscosity agar plates. We find that expanding phage–bacteria populations supports two moving fronts, an outermost bacterial front driven by nutrient uptake and chemotaxis and an inner phage front at which the bacterial population collapses due to phage predation. We show that with increasing adsorption rate and initial phage population, the speed of the moving phage front increases, eventually overtaking the bacterial front and driving the system across a transition from a regime where bacterial front speed exceeds that of the phage front to one where bacteria must evolve phage resistance to survive. Our data support the claim that this process requires phage to hitchhike with moving bacteria. A deterministic model recapitulates the transition under the assumption that phage virulence declines with host growth rate which we confirm experimentally. Finally, near the transition between regimes we observe macroscopic fluctuations in bacterial densities at the phage front. Our work opens a new, spatio-temporal, line of investigation into the eco-evolutionary struggle between bacteria and phage.
Subject terms: Microbial ecology, Population dynamics, Theoretical ecology
Introduction
In natural environments, including oceans and soils, phages and bacteria interact in a spatially structured context. The spatial structure in these environments spans multiple scales from particles of a few micrometers such as marine snow [1] to large scale microbial populations such as blooms [2]. Despite the ubiquity of spatial structure in these natural environments, our understanding of the dynamics of phage infections is largely based on experiments in well-mixed systems [3–8]. For example, decades of studies of phage in chemostats have shed light on the long-term coevolutionary processes in well-mixed conditions [6, 9–11].
Less is known, however, about the role of spatial structure on phage–host interactions. The most detailed accounting of phage–host interactions in space is available in the context of plaque formation. Plaque-forming phages interact with a dense, spatially homogeneous host population. However, unlike well-mixed systems, phages in plaques must infect and propagate locally in space. The dynamics of plaque formation have been studied in detail theoretically and experimentally. For example, several studies have examined the expansion rate of a growing plaque as a function of the microscopic properties of the phage–host interaction [12]. A careful theoretical study showed that the plaque expansion dynamics could be understood quantitatively if the latency period of the phage in the host and the role of bacterial crowding in limiting phage diffusion are accounted for [13]. Experiments have shown that the plaque formation process can favor mutant phages that exhibit lower adsorption rates to their host [14]. Experimental evolution for plaque size also resulted in phage strains with reduced infectivity [15]. A related study showed that low productivity phages can exhibit enhanced fitness in direct competition experiments when the environment is spatially structured [16]. A recent study showed that colonies of susceptible bacteria can survive a phage attack if they are of sufficient size to protect growing cells at the interior of the colony [17]. These studies of plaque formation dynamics have revealed how phage attacks a dense, growing, and nonmotile host population and that these conditions typically favor phage of lower infectivity and productivity.
The ability of phage to attack bacteria present in biofilms has also been explored [18, 19]. In biofilms, bacteria are embedded in dense exopolysaccharide matrices which can be degraded by enzymes carried on the capsids of phage [20] and it is known that phage infection can impact biofilm morphology [21]. In biofilms, as in the plaque formation context, bacterial hosts are not motile but instead remain fixed in space. Complementing these experimental efforts are several theoretical attempts to describe spatially structured phage–bacterial interactions [22–24].
In nature, bacteria in the planktonic state are often motile and exhibit chemotaxis up gradients of attractant. As a result, host motility is an important and unexplored aspect of phage–bacteria interactions. To our knowledge there is no existing study of the dynamics of phage–host interactions in a situation where the host is actively migrating up a gradient of nutrients. Here, we study the interaction between phages and bacterial populations moving through a porous environment. In our experiments, bacterial populations migrate at rates of several millimeters per hour due to chemotaxis and growth driven by a self-organized nutrient gradient [25, 26]. Since chemotaxis is widespread in many natural habitats shared by bacteria and phages, including oceans [27, 28] and soil [29], we expect the process studied here may be relevant to bacteria–phage interactions in environments where motility and chemotaxis are important.
Here we show that the interplay between chemotaxis and phage predation results in rich spatio-temporal dynamics. In this context, a bacterial front composed of the fastest migrating bacteria, which harbor a hitchhiking phage population, is trailed by a secondary phage front where the bacterial population collapses under predation pressure. The rate of migration of this phage front increases with the size of the initial phage population and the rate of infection until it overtakes the migrating bacterial front and forces its collapse. Therefore, as infectivity or initial phage populations increase, the system transitions from a regime where bacterial abundances are regulated by chemotaxis and nutrient consumption to one where they are regulated by phage lysis. Near the transition between these two regimes the phage front becomes rough, exhibiting millimeter scale fluctuations in phage and bacterial densities.
Materials and methods
Experimental methods
All experiments were performed with lysogeny broth (LB) rich medium (10 gL−1 tryptone, 5 gL−1 yeast extract, 5 gL−1 NaCl). To create a swarm plate, LB medium was autoclaved with 0.3% w/v agar after autoclaving 5 mM MgSO4 and an appropriate CaCl2 concentration were added. Prior to cooling 100 mL of medium was measured in a sterile graduated cylinder and poured into a 15 cm diameter sterile plate. The plate was allowed to solidify at room temperature for 3 h. Plates were then transferred to a walk-in incubator 30 °C (Darwin Chambers) and allowed to reach thermal equilibrium for no <24 h prior to inoculation. All experiments reported here were performed at 30 °C.
To initiate an experiment, an overnight culture of Escherichia coli (E. coli) (MG1655, motile, Coli Genetic Stock Center #6300) was initiated in LB from frozen glycerol stocks. 10 μL of saturated overnight culture (≈1 × 106 cells) was taken from this culture and mixed with 10 μL phage P1 lysate and vortexed thoroughly before being inoculated into the center of the swarm plate. P1 lysates were titered by standard serial dilution and plating on top agar and counting plaques. Lysate titers were ∼1 × 1010 PFU/mL. Changing the initial phage population was accomplished by diluting the P1 lysate into LB medium prior to mixing with bacteria. Phage lysates were not used for more than a year and each new phage lysate that was made was titered by serial dilution and plating.
Plates were imaged using computer controlled Logitech webcams housed in light tight boxes with illumination provided by LED strips which encircled the plates and were turned on for 1 s per image acquisition [26]. Imaging occurred in one of two ways: either plates were left in light tight boxes and images were acquired once every 2–4 min (Fig. 1) or plates were housed in the incubator but were manually transferred to the imaging apparatus to acquire images at fixed points in time since inoculation (Fig. 2). All image analysis was performed with custom written Matlab scripts. For plates subjected to continuous imaging (every 2–4 min) the images were processed as described in a previous publication [26]. Briefly, a background image was constructed from the median projection of five images at the beginning of the acquisition before a colony formed. The center of the colony was determined using a circular Hough transform from an image acquired 24 h after inoculation. From this center, a radial line of pixels was extracted, and the image intensity recorded for each frame of the acquisition. The location of the bacterial front was determined by finding the outermost peak in the image intensity at all time points where the front could be detected. The bacterial front migration rate was then determined by linear regression (Fig. S3). The phage front location was determined by finding the point in this radial profile where the image intensity fell to 80% of its maximum (Fig. 1, yellow arrows).
Fig. 1. Spatio-temporal dynamics of bacteria and phage in a soft agar plate with 1mM concentration CaCl2.
The center of the plate (radius 7.5 cm) was inoculated with ~2.3 × 107 phages (PFUs/inoculum) and 1 × 106 bacteria. a Images visualizing the spatial distribution of bacteria (light regions) on the plate at 8, 14, and 19h after inoculation. Scale bar applied to all three panels. b The radial profile of the bacterial population density at 4 h (green), 9 h (blue), 14 h (red), 19 h (orange), and 24 h (purple) after inoculation along a radial direction at angle 90° right from the vertical. c The radial profile of the phage population density measured at 19 h. The phage density reaches a plateau around the same spatial position at which the bacterial population collapses (referred to as the phage front). d–f Are identical to (a–c) except they show results of model simulations in which the phage latency time is inversely proportional to the bacterial growth rate (with offset), and phage burst size is proportional to the growth rate (see “Methods” and Box 1 for details of the model). Phage and bacterial front locations at 19 h are marked with yellow and orange arrows, respectively, in all panels.
Fig. 2. Phages or nutrients regulate bacterial abundances depending on initial phage population size and infectivity.
a Bacterial populations shown 19 h after inoculation with varying phage load quantified by the initial number of phages (PFUs) in the inoculum (y-axis), and CaCl2 concentrations in the agar. Note the logarithmic scale of both axes. The white intensity is proportional to the local density of bacteria with the inner black circle marking the area where bacterial population has collapsed due to phages. Measurements of adsorption rate with CaCl2 concentration show that it varies by a factor 2 over this range (Supplementary Appendix, “Methods”). b The radial position of the expanding outer bacterial front at 19 h. c The width of the region of high bacterial density between the outer bacterial front and the inner phage front. The position of the phage front is defined by the bacterial density dropping to 80% of its peak value. d Model simulations (see Main Text and “Methods”). e–f Bacterial front location and the width of the region of high bacterial density at 19 h in model simulations as in panel (d), but without resistant bacteria.
For plates where intermittent images were acquired manually (e.g., 8, 14, and 19 h after inoculation, Fig. 2), background subtraction could not be performed using the median projection technique described above. Instead, we computed a radial profile of pixel intensities from the colony center at the earliest image acquired when the colony was small (8 h in the case of Fig. 2a). For a radial profile of pixel values, we computed a polynomial fit (fourth order) to the region of the image beyond the outermost bacterial front. We used this fit to construct a synthetic background image by extrapolating to the center of the plate and assuming azimuthal symmetry. The synthetic background image was then subtracted from the 19 h time point. The result is shown in Fig. 2. The bacterial and phage front locations were determined manually for the images in Fig. 2 using the same criteria described above.
To assess the reproducibility of the transition from the regime where nutrients regulate bacterial abundances to the regime where phage infections limit bacterial growth, we performed an additional set of at least five experiments at low (1 mM) and high (≥8 mM) CaCl2 concentrations and low and high initial phage densities. All of these experiments showed colony morphologies and dynamics consistent with the results shown in Fig. 2.
Plate reader experiments were performed in 48-well plates using a BMG Labtech Clariostar plate reader equipped with an incubator maintained at 30 °C. Media volumes in each well were 0.75 mL. The plate reader made automated optical density measurements on 5-min intervals with orbital shaking between measurements. Phage titer, adsorption rate, burst size, and bacterial carrying capacity were assayed using standard techniques, which are described in the Supplementary Appendix.
Model
To interpret experimental observations, we developed a reaction-diffusion model to describe the spatio-temporal dynamics of bacterial and phage densities. The model is described graphically and mathematically in Box 1.
Our model includes susceptible bacteria B(x, t), resistant bacteria Br(x, t), infected bacteria I(x, t), phages P(x, t), nutrient n(x, t), and two chemoattractants c1(x, t) and c2(x, t). These populations are depicted graphically in Box 1a. The model is shown in Box 1b where a description of each term in the model is given. Model parameter values along with the source of values used in our simulations are given in Table 1.
Table 1.
Model parameters.
| Parameters | Description | Value | Source |
|---|---|---|---|
| β | Maximum burst size | 20 | Measured |
| η | Adsorption rate | 1.5–3.0 × 10−8 mL h−1 | Measured |
| τ | Minimum latent period | 1 h | [38] |
| DB | Bacterial diffusion constant | 0.05 mm2 h−1 | Fitted |
| Dn | Nutrient diffusion constant | 3.0 mm2 h−1 | [26] |
| Dp | Phage diffusion constant | 0.04 mm2 h−1 | [39] |
| a− | Low chemoattractant sensing constant | 3.5 μM | [40, 41] |
| a+ | High chemoattractant sensing constant | 1000 μM | [40, 41] |
| χ1 | Chemotactic coefficient for serine | 1.5 mm2 h−1 | Fitted |
| χ2 | Chemotactic coefficient for aspartate | 0.8 mm2 h−1 | Fitted |
| Maximal depletion rate of nutrient | 2.8 h−1 | Fitted | |
| Maximal depletion rate of serine | 2.8 h−1 | [42] | |
| Maximal depletion rate of aspartate | 0.28 h−1 | [29] | |
| Kn | Monod constant of nutrient | 10 mM | Fitted |
| Kc1 | Monod constant of serine | 5 × 10−2 mM | [42] |
| Kc2 | Monod constant of aspartate | 5 × 10−3 mM | [29, 33] |
| Y | Nutrient yield | 108 mL mM−1 | [26] |
| n(0) | Initial nutrient concentration | 10 mM | [30] |
| Initial serine concentration | 0.2 mM | [30] | |
| Initial aspartate concentration | 0.2 mM | [30] | |
| pr | Mutation rate of resistant bacteria | 10−5 | [17, 43, 44] |
| sr | Growth reduction of resistant bacteria | 0.2 | Fitted |
| rb | Ratio of the minimal burst size to its maximum | 0.1 | Fitted |
| r1 | Ratio of the minimal latent period to its maximum | 0.5 | Fitted |
Several assumptions are made in the model and justified below:
Bacterial growth and nutrient depletion: the depletion of the nutrient n leads to bacterial replication where the specific growth rate is assumed to follow Monod’s growth kinetics
Bacterial chemotaxis: the chemotaxis is assumed to be driven by gradients of two chemoattractants. The chemotactic velocity models bacterial chemotaxis up the gradient of c1 and c2 with the log-adaptation range between a– and a+ [30].
Resistant bacteria: the generation of a phage-resistant bacteria (Br) by spontaneous mutations is assumed to occur with probability pr = 10−5 per replication. Resistant bacteria (Br) are assumed to grow 20% slower than the susceptible bacteria B(sr = 0.2). We cannot measure these parameters directly and therefore they are constrained by a qualitative comparison to data and sensitivity is determined by simulations (Fig. S8).
Phage infection: the infection of bacteria by phage follows a simple mass action form η × B × P.
Latent period of infection: latency is modeled by introducing S = 10 auxiliary intermediate infection states of bacteria, I1, I2, …, Is taking place between the adsorption event and the production of phage progeny (Eq. (3), Box 1b). These intermediate states are a well-established mathematical method to model the dispersion of latency periods in the populations in deterministic models [31–33]. In Supplementary Appendix and Fig. S7, we show that multiple states of infection are necessary for our model to recapitulate the spatial patterns we observed in Fig. 1. It is important to note that our conclusions are not sensitive to S so long as it is ~≥10. These intermediate states do not have an explicit microscopic interpretation in terms of the phage infection process. Instead they permit us to capture the fact that the latency period is variable across the population of infected bacteria and avoid the computational complication of simulating time-delayed differential equations.
Bacterial growth-dependent phage physiology: to reproduce the experimental results, we included a reduction of phages’ burst size and extension of their latency time as nutrients are depleted and bacterial growth rates decline. As n declines, we assume that the burst size β(n) decreases and the latency time τ(n) increases as and . Here is the specific growth rate and gn(n = n0) is the specific growth rate when nutrients are not depleted (n0 = 10 mN). We note that this dependence of phage infectivity on host physiology has been observed previously [34, 35]. We are able to constrain these parameters, but we cannot specify them quantitatively directly from our data (see Figs. S2, S4 and Supplementary Appendix). In Fig. S5, we compare our assumed dependence of burst size and latency period on growth rate with that observed in ref. [34] and find that they are similar. Other studies have shown similar dependences of burst size and latency period on growth rate (ref. [36] Fig. 2; ref. [37] Fig. 3).
Fig. 3. Bacterial population dynamics with 5 mM CaCl2 and ∼50,000 phage (PFUs) in the inoculum.
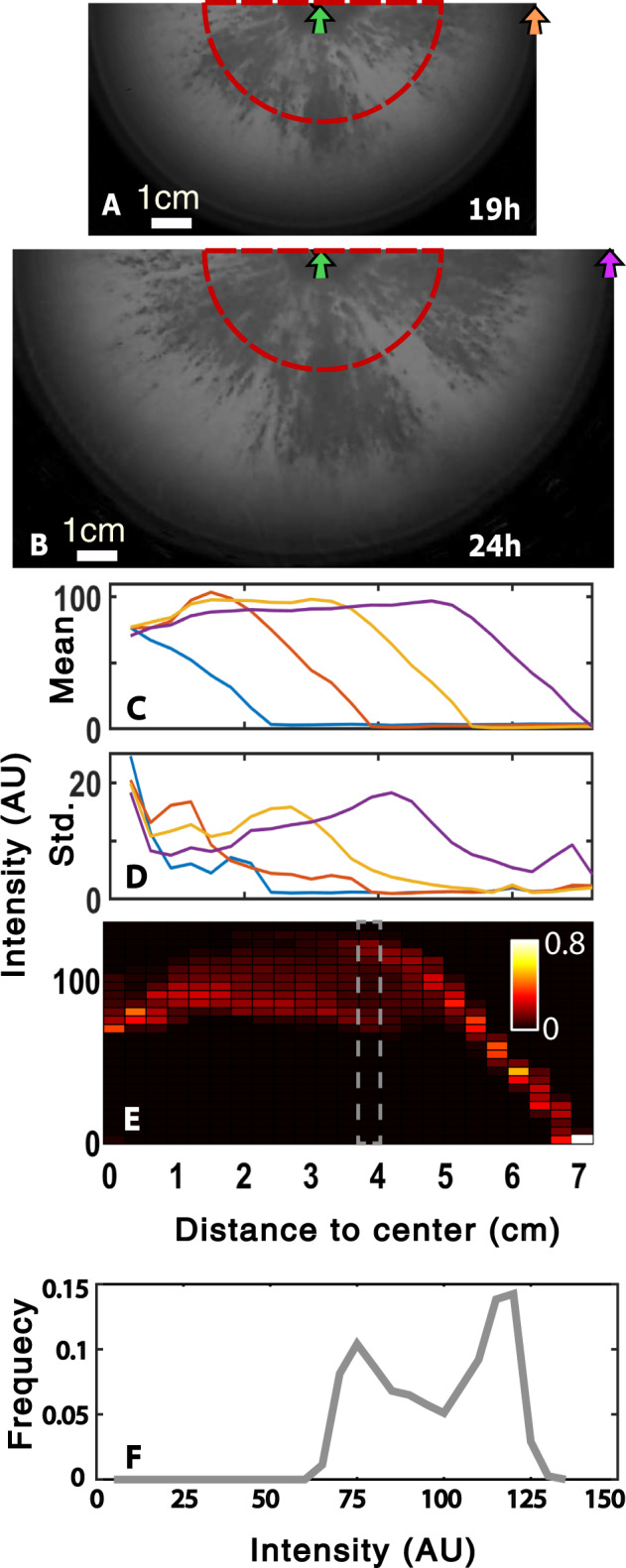
Background-subtracted (“Methods”) half-plate images at a 19 h and b 24 h. c, d show average and standard deviation of pixel intensity as a function of the distance from the center at 9 h (blue), 14 h (red), 19 h (orange), and 24 h (purple). Means and standard deviations in pixel intensities are computed by considering all pixels within a small annulus for radius. e Probability distribution of pixel intensity (y-axis). The distribution is normalized at each radius (x-axis). f Frequency distribution of pixel intensity around 4 cm (position of the dashed box in panel e). Green, purple, and orange arrows in panels (c–d) mark radial locations as indicated in panels (a, b). The red semi-circle in panels (a, b) has a radius of 3 cm.
Sensitivity analysis
The sensitivity of several model assumptions was tested by comparing the fitted models with and without each assumption to well-mixed batch culture experiments, and the result is shown in Fig. S4. The spatial profiles of phage density are shown as a function S in Fig. S7. To further analyze the sensitivity of model parameters related to those assumptions, a Markov Chain Monte Carlo was performed. The posterior distributions for parameters generated by the adaptive Metropolis sensitivity analysis are shown in Fig. S8. Because the posterior distribution can give an approximation of the basin of attraction around a global minimum, the sensitivity of a parameter is reflected by the standard deviation of the posterior distribution (i.e., the smaller the standard deviation, the more sensitive to the variation of the parameter).
Box 1 Description of the model
Panel A shows a graphical description of the modeling formalism developed to describe spatio-temporal phage–bacteria dynamics. Four populations are captured in the model and shown schematically: bacteria (B), infected bacteria (I), phage (P), and bacterial mutants that are resistant to phage infection (Br). These populations all depend explicitly on space and time and interact with three chemical fields: nutrients (n) and two chemoattractants (c1) and (c2). Sketches of the spatial profile of these chemical fields are shown in the plot below. Panel B shows the explicit equations used in our numerical simulations. The interpretation of each term is given.
Results
To study phage–bacterial interactions in a spatially structured context, we used the rich medium (LB) low viscosity (0.3% w/v) agar plates. When E. coli is inoculated at the center of these plates, it rapidly consumes nutrients locally creating a gradient in the primary attractant (serine) and the secondary attractant (aspartate) that drives radially symmetric chemotactic migration of the population (rate ∼0.36 cm h−1, conditions here are identical to those studied in ref. [26] with the addition of phages). We inoculated an agar plate with a mixture of bacteria and phage (P1, virulent strain) at the center of the plate. Phage P1 requires calcium (CaCl2, ranging between 1 and 10 mM in our experiments) and magnesium (MgSO4, 5 mM) to infect E. coli [45]. The dependence of the infection on CaCl2 allows us to modulate the adsorption rate of phage by varying calcium concentration in the plate (MgSO4 concentration is fixed at 5 mM in all experiments shown here). We measured the difference in adsorption rate as a function of CaCl2 and found that it changed by a factor 2 between 1 and 10 mM CaCl2. We refer to the initial phage population as the number of plaque-forming units (PFU) in the inoculum.
Figure 1 shows the spatio-temporal dynamics of populations of bacteria (~1 × 106 cells/inoculum) and phages (~2.3 × 107 PFU/inoculum) for 1 mM CaCl2. At this relatively low calcium concentration, the phage adsorption rate is low. Figure 1a shows an image of the agar plate taken 9, 14, and 19 h after inoculation. The overall radial expansion of the bacterial population (referred to as bacterial front, the bright outer ring of the colony, orange arrow Fig. 1a) proceeds at a constant rate of 0.36 cm h−1. This rate of expansion is identical to bacteria migrating in the plates without phages present [26]. The bacterial front is followed by a deepening and widening black region in the center of the agar plate, whose outer boundary we refer to as the phage front (yellow arrow, Fig. 1a). This central black region is caused by a collapse of the local bacterial population due to phage predation. Radial profiles of the bacterial density were measured by extracting and averaging pixel intensities in background-subtracted images (see “Methods”). Figure 1b shows these radial density profiles of bacteria at the time points shown in Fig. 1a. Here the phage front is visible as a sharp decline in image intensity (bacterial density) near the center of the plate at later times. For example, the orange trace in Fig. 1b shows a radial profile at 19 h where the phage front is evident at a radius of ~3 cm, while that of the bacterial front is present at a radius of 6 cm. For the conditions shown in Fig. 1, the separation between the phage front and the bacterial front increases with time (compare red and orange traces, Fig. 1b). Thus, under these conditions, phages are not able to keep up with the expansion of the bacterial colony.
To confirm that the collapse in bacterial densities at the center of the plate was in fact driven by phage predation and to measure phage densities in the expanding bacterial front, we sampled a plate after 19 h of expansion at several distances from the center and measured the phage density (“Methods”). Figure 1c shows the phage population density (PFU) as a function of the radius where, as expected, we observe very high phage densities in regions where the image intensity is low. Unexpectedly, a small number of phages are present at the outer edge of the expanding bacterial colony.
How could the phage have reached the outer edge of the colony (radius 6 cm) in a period of 19 h? The diffusion constant of phage P1 in water is 4 × 10−8 cm2 s−1 [39] limiting diffusive transport on this timescale to ~0.1 cm. So, it appears passive transport of phage alone is insufficient to explain the presence of phage at distances of 6 cm from the site of inoculation. Previous studies of plaque formation have found that phage plaques expand through lawns of dense bacteria as a traveling wave with speeds up to ∼0.025 cm h−1 [12, 13]. At this rate of expansion, we expect the phage population to travel <0.5 cm over the 19 h duration of the experiment. These quantitative considerations strongly suggest that the phage could not have reached the outer edge of the expanding colony by passive transport alone.
Instead, we suggest that the phage travels along with migrating bacteria either by attaching to the surface of swimming bacterial cells and not injecting DNA or by being transported during the latent period of infection. If phage is hitchhiking with the moving bacterial front, we expect plaques to form behind the front as the hitchhiking phage completes multiple rounds of infection on the host. We observe the formation of these plaques in conditions where the bacterial front is moving quickly relative to the phage front (Fig. 2a, lower-left corner, Fig. 3a, b). Note that in these images the visible plaques are well behind the outermost bacterial front. Visible plaques typically harbor 107–108 phage per plaque [14] and therefore require many (>10) rounds of lysis and infection. Given the latency period of at least 1 h for phage P1, we expect visible plaques take on the order of 10 h to form. In this time the bacterial front travels ~3.5 cm and as a result, the visible plaques arising from hitchhiking phages are present well behind the primary bacterial front.
In the conditions shown in Fig. 1, the phage front is moving at a slower rate than the bacterial front. As a result, we expect that phages are being diluted out of the expanding bacterial front and therefore were the front to expand indefinitely the bacterial population would be free from phage infection. So, while phage must be actively transported by the migrating bacterial front, in the conditions shown in Fig. 1, they are unable to keep up with the traveling bacterial wave.
To describe the spatio-temporal dynamics observed in Fig. 1a–c, we developed a deterministic ordinary differential equation model of the phage–bacteria system, which describes the dynamics of bacterial and phage populations in space and time. Numerical simulations of the model are shown in Fig. 1d–f where we find that the model captures the core features of the data. A schematic of the model is shown in Box 1a and a detailed description in “Methods”, Box 1b, and Supplementary Appendix.
Are bacteria always capable of moving faster than their phage predators, or can phage populations slow down or even stop the advancing bacterial front? To investigate this question, we repeated our experiment for a broad range of parameters characterizing phage infectivity and initial population size. Figure 2a shows snapshots of bacterial populations taken 19 h after inoculation as a function of CaCl2 concentration on agar plates (x-axis) and the initial number of phages in the inoculum (y-axis). Higher concentrations of CaCl2 increase the adsorption rate of phages [45], thereby making it easier for them to spread. This is indeed seen as a systematic increase in the radius of the inner black region (phage front), as one moves from left to right in Fig. 2a, bottom row. Similarly, increasing the initial phage density speeds up the phage front even at fixed CaCl2 concentration (Fig. 2a, left column). Increasing both CaCl2 concentration and initial phage population size above a certain threshold results in a slowed bacterial front (upper-right corner, Fig. 2a). Figure 2a suggests that at low infectivity and initial phage populations the bacterial front is able to move faster than the phage front (lower-left region), whereas at high infectivity or starting phage populations bacteria are overtaken and likely evolve resistance in order to form a migrating front (upper-right region).
To test these conclusions, we made two additional measurements. First, for plates above and below the transition (1 or 8 mM CaCl2 and 1.55 × 105 phages), we tested bacterial populations for phage resistance after the front expansion process had proceeded for 19 h. Indeed, at low infectivity (1 mM CaCl2) the outermost bacterial front is comprised of susceptible bacteria. In contrast, at high infectivity, when a smaller bacterial colony is formed, the entire bacterial population is phage resistant (Fig. S1, Supplementary Appendix). Second, we made time-lapse measurements of a series of plates with varying initial phage population sizes ranging from 1.55 × 104 to 1.55 × 108 and measured the speed of the phage and bacterial fronts (see Movies S1–S4). We found that the bacterial front migration rate did not vary substantially with initial phage population, but that the phage front speed increased more than threefold with a 1000-fold increase in the starting phage population size (Fig. S3, Supplementary Appendix).
Based on these measurements, we conclude that at low initial phage populations and low infectivity the bacterial front manages to escape phage predation. In contrast, at high initial phage populations, or infectivity, the bacterial population fails to begin migration and instead must evolve phage resistance before expanding across the plate. As a result, when the phage population inhibits the bacterial expansion, this inhibition is temporary due to the growth of phage-resistant bacterial strains [46]. For phage P1, these mutant bacteria arise at a rate of about 10−5 per generation [44]. Notice that just above this transition (e.g., 8 mM CaCl2, 1 × 104 phages or 1 mM CaCl2, 1 × 108 phages; Fig. 2a) the bacterial colony exhibits sectors which potentially arise from clonal expansion started from single resistant mutants. These dynamics are reminiscent of spatial antibiotic resistance patterns [47]. Supplementary Movie S1 shows an expanding bacterial front at 10 mM CaCl2 and 1.5 × 105 initial phage population size where we see an expanding front of bacteria which is halted after 12 h of expansion only to resume expanding, presumably with resistant bacteria. However, when the phage load is large, and infectivity high, all susceptible bacteria are eliminated at the outset, resulting in a delayed but spatially symmetric expansion of the resistant bacterial population (Fig. 2a, upper-right corner).
To quantify the speed of both phage and bacterial fronts, we measured their respective locations in space under different experimental conditions. Figure 2b, c shows the position of the outer (bacterial) front and the width of the bright region between bacterial and phage fronts measured 19 h after inoculation. These measurements show the properties of the system on both sides of the transition: the bright area in the lower-left corner of the panel Fig. 2b shows that for low phage loads bacterial chemotaxis proceeds at a rate that is approximately independent of the phage load and CaCl2 concentration. Above the transition, bacterial populations are decimated at the outset of the experiment and the remaining resistant bacteria migrate only a short distance by 19 h. The progressive darkening of Fig. 2b in the upper-right corner quantifies the slowing of the migrating bacterial front by phages under increasing initial phage population sizes and infectivity. Finally, Fig. 2c quantifies the width of the ring where bacteria are abundant (the distance between the bacterial and phage fronts). This distance shrinks to zero as phages overtake bacteria (black region, upper-right corner, Fig. 2c).
Overall Fig. 2a–c shows the existence of two distinct parameter regimes in which bacterial densities are either regulated by migration and access to nutrients or phage lysis. As either the phage load (initial population) or phage adsorption rate (CaCl2 concentration) increases, the speed of the phage front increases, and eventually causes a transition from the regime where bacterial abundances are set by chemotaxis and nutrient availability to one where phage lysis limits total bacterial abundances.
Figure 2d–f shows that our deterministic ODE model reproduces the general properties of the measured phase diagram shown in Fig. 2a–c. The intensity profiles shown in Fig. 2d include the resistant bacteria, which dominate the bacterial populations in the upper-right corner of the diagram but are not significant at or below the transition along the diagonal. As we observe experimentally, the model predicts that increasing the adsorption rate will lead to a transition from a regime where phages are progressively lost from the bacterial front to one where phages overtake the bacterial front resulting in collapse. Figure 2 is the central finding of this study.
To better understand the nature of the transition between the two regimes characterized by low infectivity and low initial phage population or high infectivity and high initial phage density, we studied a simple model of phage reproduction in a bacterial traveling wave (Fig. S6, Supplementary Appendix). In this model, as bacteria are migrating, phages are lost if they do not infect the host. The ability of phages to be retained in the migrating front is therefore controlled by a balance between phage infection and bacterial migration. Using this formalism, we analytically compute a threshold adsorption rate (ηc) which defines the transition between phage populations either growing, and thereby driving bacterial colony collapse, or being lost from the migrating front. We find ηc ∼2 × 10−8 mL/h which agrees well with the adsorption rate at which we observe the transition experimentally and computationally (Fig. 2).
For conditions close to the transition region, where the phage front speed is only slightly slower than that of the bacterial front (e.g., 4 mM CaCl2, 1 × 104 phage, Fig. 2a), the phage front becomes rough, exhibiting a star-like spatial structure with centimeter-long phage-dominated dark regions extending into lighter regions with high bacterial densities. The roughening of the phage front near the transition suggests that there are large differences in bacterial densities for regions of the phage front over length scales of a millimeter or more. In plates where no phages are added the profile of bacterial densities is smooth (Fig. 1b in [19]), ruling out nutrient inhomogeneities. The strong spatial variability in bacterial densities suggests a role for contingencies in infection histories in spatially structuring the phage–bacteria interactions.
To document the roughening of the phage front more carefully, we studied a single plate in detail. Figure 3 shows two snapshots (19 and 24 h after initiation) of a plate containing 5 mM CaCl2 and 4.7 × 104 phage in the inoculum. These images reveal a clear spatial structure in the phage front characterized by alternating black and white regions that are more spatially extended than those shown in Fig. 1a. In this condition, the dark regions (where bacteria are being killed by phage) expand rapidly, at a rate approaching the speed of the bacterial front (compare the width of the light area near protruding black regions at 19 and 24 h in Fig. 3a, b). In contrast, in Fig. 1a well below the transition the black region expands at a steady, but slower, rate than the bacterial front and the rate of expansion is approximately independent of the azimuthal direction.
To better quantify the spatial structure of the dark regions visible in Fig. 3a, b, we computed statistics of image intensity for background-subtracted images in annuli of pixels falling within a fixed range of radii. Figure 3c shows the azimuthally averaged image intensity as a function of the radius at several times after the plate was initiated. Figure 3d shows the standard deviation of all pixel intensities at a fixed radius. Figure 3d clearly shows that the variation in pixel intensities is maximal at intermediate radii (peak around 4 cm for 24 h purple trace). The peak in pixel intensity variability marks the center of the phage front which is comprised of regions where phage (dark) and bacteria (light) dominate. This peak in the variability of pixel intensities migrates ~2 cm between 19 and 24 h. The outermost bacterial front migrates approximately the same distance over this time interval. Therefore, as we expect, the roughened phage front moves at a pace that is similar to that of the outermost bacterial front in conditions near the transition from a regime where bacterial abundances are nutrient regulated to one where they are phage regulated.
To further elaborate on this observation, in Fig. 3e we plotted the normalized histogram of bacterial abundances (quantified by image intensity, y-axis) at 24 h as a function of the distance from the center (x-axis). These histograms were computed for a set of radii from the center of the plate. In Fig. 3e, for distance around 4 cm, one can see that the distribution of bacterial abundances is, in fact, bimodal, corresponding to white and black regions visible in Fig 3a, b and the position of the largest variation in Fig. 3d. The distribution of pixel intensities at a radius of 4 cm is shown in Fig. 3f. Simulations show that this spatial structure could arise from spatially inhomogeneous initial phage densities (Fig. S9).
Therefore, for 5 mM CaCl2 and 4.7 × 104 phage (PFU) in the inoculum, phages keep up with bacterial front expansion in some directions but are left behind in others. This suggests that the local dynamics in our system are potentially driven by positive feedback which amplifies initially small variations into macroscopic differences manifesting themselves at a scale of several centimeters.
Discussion
We have shown how spatial structure in bacterial populations driven by chemotaxis interacts with phage population size and adsorption rate to determine the outcome of bacteria–phage interactions. Our study has three important results.
First, we find that nonmotile phages are able to hitchhike with expanding bacterial populations by repeatedly re-infecting cells in an expanding bacterial front (Fig. 1). This observation suggests that the latent period of phage infection, or another mechanism by which phages attach to swimming bacteria, permit phage to hitchhike with their hosts into new spatial niches. At present, we cannot definitively determine which mechanism of hitchhiking is dominant. However, the latency time is on the order of 1 h [48, 49]. On this timescale, the bacterial front travels 0.36 cm suggesting that phages transport during the latent period would outpace passive transport. In contrast, the time for DNA injection during infection is typically 1–10 min [50, 51], but to our knowledge has not been precisely measured in phage P1. We cannot rule out the possibility that phages are transported while being attached to the cell prior to DNA injection. The fact that hitchhiking occurs suggests that the timescale over which phages attach to, infect, and subsequently lyse bacterial cells can be related to the spatial structure of phage–bacteria populations. To our knowledge, the relationship between the physiology of phage infecting bacteria and the spatial transport of phage by migrating bacteria has not previously been appreciated.
Second, Fig. 2 shows how the outcome of phage–bacteria interactions depends on population size and infectivity. A transition region defines a boundary between regimes, one where phage-susceptible bacteria escape phage infection, and another where the bacteria are forced to evolve phage resistance to survive. This observation has important implications for understanding phage–bacteria interactions in complex environmental contexts such as lakes or the particle-filled water columns in the ocean, where bacterial growth is variable due to transient chemotaxis driven exploitation of patchy resources [27]. In particular, for phages to dominate a migrating and growing bacterial population requires high initial population densities or high adsorption rates. In situations where these conditions are not met, phages remain able to attack smaller localized bacterial populations, which could limit their overall fecundity and possibly their impact on bacterial genome evolution via transduction [52].
It is interesting to note that we observe the phage-regulated regime when infectivity (adsorption rates) is high and the nutrient-regulated regime when infectivity is low (Fig. 2). Therefore, for phage infecting motile host populations, higher infectivity is likely favored. This result stands in contrast to results obtained with expanding phage plaques in bacterial lawns [14] where selection for large plaques favors lower infectivity [15]. It would appear then that the evolution of phage life-history traits may depend qualitatively on the motility of the bacterial host.
Our modeling framework confirmed that a key feature responsible for the spatial structure in phage–bacteria populations is the strong dependence of phage infection on host physiology [19, 34, 36, 37, 53–55]. The decreasing burst size and increasing latency of phage infections with host growth rate interact with the self-organizing nutrient gradients in migrating bacterial populations to give rise to regions of space where phage is excluded. Previous studies suggest this dependence on host physiology arises from limits on the rate of synthesis of new phages and on the size of host cells prior to lysis [34]. If this mechanism is indeed responsible, it suggests that the dependence of phage infectivity on host physiology may be generic. In this case, there is a clear advantage for slow growing bacterial populations in spatial niches with substantial phage predation pressure.
Despite the overall success of the model in describing the dynamics in our phage–bacteria system, we do note that the model fails to capture certain features of the data. In particular, at 8 mM CaCl2 and 104 or 105 phage we observe large, dense, colonies forming (Fig. 2a), whereas the model predicts a dim outer ring and central region of resistant bacteria. We suspect that the reason for this disagreement is the presence of additional chemoattractants in the rich media that are not modeled explicitly, potentially threonine [25], which permit the chemotaxis of resistant bacteria even if the susceptible population (the outer ring) has already consumed serine and aspartate. The presence of such an attractant would give rise to larger colonies since resistant bacteria which arise at the center of the colony could then migrate outward via chemotaxis. We omitted this feature in our model for the sake of simplicity.
Third, there is a role of contingency in determining the spatial structure of the phage front for communities near the transition (Fig. 3a, b), where centimeter-wide spatially extended regions are alternately dominated by phage and bacteria. A full accounting of this phenomenon awaits future work; however, it appears that large differences in bacterial populations are amplified from relatively small fluctuations in the inoculum with a large number of phages (between 104 and 108 in our experiments). At first glance, the alternating black and white regions in the phage front are reminiscent of sectors in microbial range expansion with two fluorescently labeled strains with the same fitness [56]. In our system, the roughening may then result from phage and bacteria having nearly equal rates of expansion near the transition and the fact that phages can be present in the bacterial front at very low concentrations (Fig. 1). Indeed, our results suggest that the roughened phage front near the transition does not arise from phage-resistant bacterial populations. First, the regions of the front which are dominated by bacteria (Fig. 3a, b) are eventually overtaken by dark, phage-dominated regions. Second, a sample taken from the phage front showed bacteria at this spatial location to be phage susceptible (Fig. S1). Our results suggest that resistant bacteria typically arise at a later time and have very different fan-like spatial structure.
To better understand these spatial fluctuations will require a detailed understanding of spatially localized phage fluctuations within the inoculum before and just after the start of migration. Furthermore, exploring any mechanisms of positive feedback present during multiple rounds of infection that might be present and may amplify small fluctuations in phage and bacterial populations. One possible mechanism for the positive feedback is that phage which initially succeeds in killing the bacteria limit nutrient consumption which subsequently drives the growth of bacteria which can then be infected by phage. Weitz et al. [57] used the Lotka–Volterra framework to demonstrate that reduced adsorption rate as bacteria approach stationary state can provide sufficient feedback to give rise to bi-stability, potentially driving the roughening of the phage front we observe.
We have shown that actively moving hosts impose an important set of selection pressures on phages. For example, we expect that phages with infection dynamics inhibiting chemotaxis or with very short latency periods may be at a disadvantage with respect to colonizing new spatial niches. Chemotaxing bacteria thereby add an additional selection pressure on phages with extremely aggressive infection cycles.
Supplementary information
Acknowledgements
This research has received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007 2013)/ERC Grant Agreement No. 740704. DP and SK acknowledge the support from the National Science Foundation Physics Frontiers Center Program (PHY 0822613 and PHY 1430124). This work was performed in part at the Aspen Center for Physics, which is supported by National Science Foundation grant PHY 1607611.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Derek Ping, Tong Wang
Contributor Information
Sergei Maslov, Email: maslov@illinois.edu.
Kim Sneppen, Email: ksneppen@gmail.com.
Seppe Kuehn, Email: seppe.kuehn@gmail.com.
Supplementary information
The online version of this article (10.1038/s41396-020-0664-9) contains supplementary material, which is available to authorized users.
References
- 1.Datta MS, Sliwerska E, Gore J, Polz MF, Cordero OX. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat Commun. 2016;7:11965. doi: 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michalak AM, Anderson EJ, Beletsky D, Boland S, Bosch NS, Bridgeman TB, et al. Record-setting algal bloom in Lake Erie caused by agricultural and meteorological trends consistent with expected future conditions. Proc Natl Acad Sci USA. 2013;110:6448–52. doi: 10.1073/pnas.1216006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell A. Conditions for the existence of bacteriophage. Evolution. 1961;15:153. [Google Scholar]
- 4.Levin BR, Stewart FM, Chao L. Resource-limited growth, competition, and predation—a model and experimental studies with bacteria and bacteriophage. Am Nat. 1977;111:3–24. [Google Scholar]
- 5.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–8. [Google Scholar]
- 6.Bohannan BJ, Lenski RE. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology. 1997;78:2303–15. [Google Scholar]
- 7.Wang Z, Goldenfeld N. Fixed points and limit cycles in the population dynamics of lysogenic viruses and their hosts. Phys Rev E. 2010;82:171–18. doi: 10.1103/PhysRevE.82.011918. [DOI] [PubMed] [Google Scholar]
- 8.Jover LF, Cortez MH, Weitz JS. Mechanisms of multi-strain coexistence in host–phage systems with nested infection networks. J Theor Biol. 2013;332:65–77. doi: 10.1016/j.jtbi.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Chao L, Levin BR, Stewart FM. A complex community in a simple habitat: an experimental study with bacteria and phage. Ecology. 1977;58:369–78. [Google Scholar]
- 10.Mizoguchi K, Morita M, Fischer CR, Yoichi M, Tanji Y, Unno H. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl Environ Microbiol. 2003;69:170–6. doi: 10.1128/AEM.69.1.170-176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterbury JB, Valois FW. Resistance to co-occurring phages enables marine synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–9. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fort J, Méndez V. Time-delayed spread of viruses in growing plaques. Phys Rev Lett. 2002;89:786–4. doi: 10.1103/PhysRevLett.89.178101. [DOI] [PubMed] [Google Scholar]
- 13.Yin J, McCaskill JS. Replication of viruses in a growing plaque—a reaction-diffusion model. Biophys J. 1992;61:1540–9. doi: 10.1016/S0006-3495(92)81958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallet R, Shao Y, Wang I-N. High adsorption rate is detrimental to bacteriophage fitness in a biofilm-like environment. BMC Evolut Biol. 2009;9:241–12. doi: 10.1186/1471-2148-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roychoudhury P, Shrestha N, Wiss VR, Krone SM. Fitness benefits of low infectivity in a spatially structured population of bacteriophages. Proc Biol Sci. 2014;281:20132563–9. doi: 10.1098/rspb.2013.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennehy JJ, Abedon ST, Turner PE. Host density impacts relative fitness of bacteriophage Phi6 genotypes in structured habitats. Evolution. 2007;61:2516–27. doi: 10.1111/j.1558-5646.2007.00205.x. [DOI] [PubMed] [Google Scholar]
- 17.Eriksen RS, Svenningsen SL, Sneppen K, Mitarai N. A growing microcolony can survive and support persistent propagation of virulent phages. Proc Nat Acad Sci. 2018;115:337–42. doi: 10.1073/pnas.1708954115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutherland IW, Hughes KA, Skillman LC, Tait K. The interaction of phage and biofilms. FEMS Microbiol Lett. 2004;232:1–6. doi: 10.1016/S0378-1097(04)00041-2. [DOI] [PubMed] [Google Scholar]
- 19.Vidakovic L, Singh PK, Hartmann R, Nadell CD, Drescher K. Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat Microbiol. 2018;3:26–31. doi: 10.1038/s41564-017-0050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes KA, Sutherland IW, Jones MV. Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology. 1998;144:3039–47. doi: 10.1099/00221287-144-11-3039. [DOI] [PubMed] [Google Scholar]
- 21.Corbin BD, McLean RJ, Aron GM. Bacteriophage T4 multiplication in a glucose-limited Escherichia colibiofilm. Can J Microbiol. 2001;47:680–4. [PubMed] [Google Scholar]
- 22.Abedon ST, editor. Bacteriophage ecology: population growth, evolution, and impact of bacterial viruses. 2008. Cambridge University Press; 2008 May 1.
- 23.Heilmann S, Sneppen K, Krishna S. Sustainability of virulence in a phage-bacterial ecosystem. J Virol. 2010;84:3016–22. doi: 10.1128/JVI.02326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heilmann S, Sneppen K, Krishna S. Coexistence of phage and bacteria on the boundary of self-organized refuges. Proc Nat Acad Sci. 2012;109:12828–33. doi: 10.1073/pnas.1200771109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adler J. Chemotaxis in bacteria. Science. 1966;153:708–16. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 26.Fraebel DT, Mickalide H, Schnitkey D, Merritt J, Kuhlman TE, Kuehn S. Environment determines evolutionary trajectory in a constrained phenotypic space. Elife. 2017;6:e24669. doi: 10.7554/eLife.24669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stocker R, Seymour JR. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev. 2012;76:792–812. doi: 10.1128/MMBR.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allweiss B, Dostal J, Carey KE, Edwards TF, Freter R. The role of chemotaxis in the ecology of bacterial pathogens of mucosal surfaces. Nature. 1977;266:448–50. doi: 10.1038/266448a0. [DOI] [PubMed] [Google Scholar]
- 29.Chet I, Mitchell R. Ecological aspects of microbial chemotactic behavior. Annu Rev Microbiol. 1976;30:221–39. doi: 10.1146/annurev.mi.30.100176.001253. [DOI] [PubMed] [Google Scholar]
- 30.Cremer J, Honda T, Tang Y, Wong-Ng J, Vergassola M, Hwa T. Chemotaxis as a navigation strategy to boost range expansion. Nature. 2019;575:658–63. 10.1038/s41586-019-1733-y. [DOI] [PMC free article] [PubMed]
- 31.Getz WM, Dougherty ER. Discrete stochastic analogs of Erlang epidemic models. J Biol Dyn. 2018;12:16–38. doi: 10.1080/17513758.2017.1401677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurtado PJ, Kirosingh AS. Generalizations of the ‘Linear Chain Trick’: incorporating more flexible dwell time distributions into mean field ODE models. J Math Biol. 2019;79:1831–83. doi: 10.1007/s00285-019-01412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champredon D, Dushoff J, Earn DJD. Equivalence of the Erlang-distributed SEIR epidemic model and the renewal equation. SIAM J Appl Math. 2018;78:3258–78. [Google Scholar]
- 34.Hadas H, Einav M, Fishov I, Zaritsky A. Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology. 1997;143:179–85. doi: 10.1099/00221287-143-1-179. [DOI] [PubMed] [Google Scholar]
- 35.Middelboe M. Bacterial growth rate and marine virus-host dynamics. Microb Ecol. 2000;40:114–24. doi: 10.1007/s002480000050. [DOI] [PubMed] [Google Scholar]
- 36.Golec P, Karczewska-Golec J, Łoś M, Węgrzyn G. Bacteriophage T4 can produce progeny virions in extremely slowly growing Escherichia colihost: comparison of a mathematical model with the experimental data. FEMS Microbiol Lett. 2014;351:156–61. doi: 10.1111/1574-6968.12372. [DOI] [PubMed] [Google Scholar]
- 37.Choua M, Bonachela JA. Ecological and evolutionary consequences of viral plasticity. Am Naturalist. 2019;193:346–58. doi: 10.1086/701668. [DOI] [PubMed] [Google Scholar]
- 38.Fehér T, Karcagi I, Blattner FR, Pósfai G. Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb Biotechnol. 2011;5:466–76. doi: 10.1111/j.1751-7915.2011.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baltus RE, Badireddy AR, Delavari A, Chellam S. Free diffusivity of icosahedral and tailed bacteriophages: experiments, modeling, and implications for virus behavior in media filtration and flocculation. Environ Sci Technol. 2017;51:1433–40. doi: 10.1021/acs.est.6b05323. [DOI] [PubMed] [Google Scholar]
- 40.Fu X, Kato S, Long J, Mattingly HH, He C, Vural, et al. Spatial self-organization resolves conflicts between individuality and collective migration. Nat Commun. 2018;9:1–12. doi: 10.1038/s41467-018-04539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Pollard AM, Höfler C, Poschet G, Wirtz M, Hell R, et al. Relation between chemotaxis and consumption of amino acids in bacteria. Mol Microbiol. 2015;96:1272–82. doi: 10.1111/mmi.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hama H, Shimamoto T, Tsuda M, Tsuchiya T. Characterization of a novel L-serine transport system in Escherichia coli. J Bacteriol. 1988;170:2236–9. doi: 10.1128/jb.170.5.2236-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schellenberg GD, Furlong CE. Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J Biol Chem. 1977;252:9055–64. [PubMed]
- 44.Franklin NC. Mutation in gal U gene of E. coli blocks phage P1 infection. Virology. 1969;38:189–91. doi: 10.1016/0042-6822(69)90144-5. [DOI] [PubMed] [Google Scholar]
- 45.Thomason LC, Costantino N, Court DL. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol. 2007;Chapter 1:Unit 1.17. doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 46.Demerec M, Fano U. Bacteriophage-resistant mutants in Escherichia Coli. Genetics. 1945;30:119–36. doi: 10.1093/genetics/30.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, Yelin I, Kishony R. Spatiotemporal microbial evolution on antibiotic landscapes. Science. 2016;353:1147–51. [DOI] [PMC free article] [PubMed]
- 48.De Paepe M, Taddei F. Viruses’ life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 2006;4:e193–9. doi: 10.1371/journal.pbio.0040193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wall JD, Harriman PD. Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology. 1974;59:532–44. doi: 10.1016/0042-6822(74)90463-2. [DOI] [PubMed] [Google Scholar]
- 50.García LR, Molineux IJ. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J Bacteriol. 1995;177:4066–76. doi: 10.1128/jb.177.14.4066-4076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novick SL, Baldeschwieler JD. Fluorescence measurement of the kinetics of DNA injection by bacteriophage lambda into liposomes. Biochemistry. 1988;27:7919–24. doi: 10.1021/bi00420a050. [DOI] [PubMed] [Google Scholar]
- 52.Dixit PD, Pang TY, Studier FW, Maslov S. Recombinant transfer in the basic genome of Escherichia coli. Proc Nat Acad Sci. 2015;112:9070–5. doi: 10.1073/pnas.1510839112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabinovitch A, Fishov I, Hadas H, Einav M, Zaritsky A. Bacteriophage T4 development in Escherichia coli is growth rate dependent. J Theor Biol. 2002;216:1–4. doi: 10.1006/jtbi.2002.2543. [DOI] [PubMed] [Google Scholar]
- 54.Edwards KF, Steward GF. Host traits drive viral life histories across phytoplankton viruses. Am Nat. 2018;191:566–81. doi: 10.1086/696849. [DOI] [PubMed] [Google Scholar]
- 55.Birch EW, Ruggero NA, Covert MW. Determining host metabolic limitations on viral replication via integrated modeling and experimental perturbation. PLoS Comput Biol. 2012;8:e1002746–12. doi: 10.1371/journal.pcbi.1002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA. 2007;104:19926–30. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weitz JS, Dushoff J. Alternative stable states in host–phage dynamics. Theor Ecol. 2007;1:13–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




