Abstract
CRISPR/Cas9 is a programmable genome editing tool widely used for biological applications and engineered Cas9s have increased discrimination against off-target cleavage compared with wild-type Streptococcus pyogenes (SpCas9) in vivo. To understand the basis for improved discrimination against off-target DNA containing important mismatches at the distal end of the guide RNA, we performed kinetic analyses on the high-fidelity (Cas9-HF1) and hyper-accurate (HypaCas9) engineered Cas9 variants. We show that DNA cleavage is impaired by more than 100- fold for the high-fidelity variants. The high-fidelity variants improve discrimination by slowing the observed rate of cleavage without increasing the rate of DNA rewinding and release. The kinetic partitioning favors release rather than cleavage of a bound off-target substrate only because the cleavage rate is so low. Further improvement in discrimination may require engineering increased rates of dissociation of off-target DNA.
Subject terms: Enzyme mechanisms, CRISPR-Cas9 genome editing
Engineered high-fidelity Cas9s have increased discrimination against off-targets. Kinetic analyses of Cas9-HF1 and HypaCas9 engineered Cas9 variants show that their DNA cleavage is impaired by more than 100- fold, which leads to release rather than cleavage of a bound off-target substrate.
Introduction
CRISPR/Cas9 has become widely used for precise genome editing applications in basic research and represents a promising tool for future applications1–6. However, Cas9 endonucleases show unintended off-target cleavage events that pose serious limitations to Cas9-based gene therapies. Several Cas9 enzyme variants have been developed that lead to significant improvements in discrimination in vivo, including: high-fidelity (Cas9-HF1: N467A, R661A, Q695A, and Q926A mutations)7, enhanced specificity (eSpCas9(1.1): K848A, K1003A, and R1060A mutations)8 and hyper-accurate (HypaCas9: N692A, M694A, Q695A, and H698A mutations)9. However, substrate discrimination is a kinetic phenomenon, and the basis for target discrimination and the guidelines for further improving fidelity remain unclear10–14. Recently, we showed that DNA cleavage is fast and DNA unwinding constitutes the rate-limiting and specificity-determining step for on-target cleavage by SpCas915. Later single molecule studies confirmed that DNA unwinding appeared to be the rate-limiting step for SpCas9 but further suggested that DNA unwinding is the determinant for specificity with both wild-type and high-fidelity Cas9 variants, Cas9-HF1 and eCas9(1.1), for 3 bp PAM-distal mismatches16. However, single molecule kinetic studies that lack a parallel measurement of the rates of DNA cleavage provide limited information to establish the mechanistic basis for DNA cleavage specificity, which is a function of all steps leading up to the first largely irreversible reaction17.
Because the PAM-distal mismatches (Fig. 1) have been shown to play an important role in selectivity as observed in single molecule experiments of both DNA unwinding and conformational dynamics within the enzyme18–22, we chose to continue our study of the effects of this off-target activity using single turnover kinetic analysis. These studies include parallel measurements of conformational changes and DNA cleavage and are interpreted globally to yield a single unifying model defining intrinsic rate constants governing specificity. We show that high-fidelity variants increase discrimination largely through kinetic partitioning, favoring release rather than cleavage of off-target substrates.
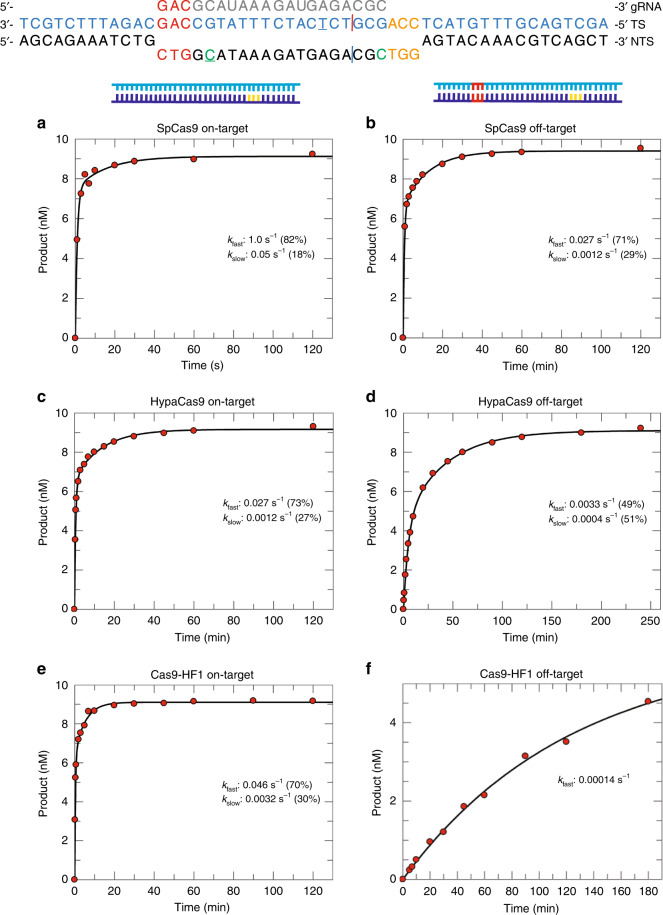
Fig. 1. PAM-distal mismatches dramatically slow cleavage by HypaCas9 and Cas9-HF1.
Schematic of target DNA substrate. PAM site is orange; target sequence is blue; mismatches in the PAM distal region are red; gRNA is colored gray; cleavage sites are shown as red and blue vertical lines for the HNH and RuvC domains, respectively; tCo labeled site at position −16 nt or −1 nt in the non-target strand is green. Cy3 and Cy5 labeled sites at position −6 nt and −16 nt in the target strand and non-target strand, respectively, are underlined. A time course of cleavage of on-target DNA (10 nM) was monitored with 28 nM of each enzyme. Data were fit to a double-exponential equation and the percentage of the total signal occurring with each decay rate is indicated. SpCas9 cleavage of on-target (a) (from Gong et al.15) and off-target (b) DNA. c, d HypaCas9 cleavage with on-target (c) and off-target (d) DNA. e, f Cas9-HF1 cleavage of on-target (e) and off-target (f) DNA. Data in f were fit to a single-exponential function.
Results
HypaCas9 and Cas9-HF1 show slow observed rates of cleavage
We begin our kinetic analyses by measuring the enzyme active site concentration for each of the Cas9 variants23–25. Measuring the amount of product formed in a titration of enzyme with increasing concentrations of DNA revealed active site concentrations of 31 nM, 26 nM, 23 nM for SpCas9, HypaCas9, and Cas9-HF1, respectively, for enzyme samples with a 100 nM nominal concentration based on absorbance at 280 nm (Supplementary Fig. 1a–d). We also measured the active site concentrations of SpCas9 and HiFiCas925 from Integrated DNA Technologies (IDT) and observed similar concentrations of active enzyme (Supplementary Fig. 1e–f). It is important to note that, in each case, the concentration of DNA required to saturate the signal was equal to the concentration of product formed, which eliminates concerns that some of the enzyme might bind DNA but not react. All subsequent experiments were set up using the concentration of active enzyme determined in the active site titration.
To compare the kinetics of on- or off-target DNA substrates of SpCas9 with the engineered variants, we first examined the time course of target strand (HNH) cleavage for each enzyme (Fig. 1 and Supplementary Fig. 2). Data were fit with either a single- or double-exponential function using Eq. (1) or Eq. (2), respectively.
| 1 |
where Y represents concentration of cleavage product, A1 represents the amplitude, and λ1 represents the observed decay rate (eigenvalue)17.
| 2 |
where Y represents concentration of cleavage product, A1 represents the amplitude and λ1 represents the observed rate for the first phase. A2 represents the amplitude and λ2 represents the observed rate for the second phase.
Comparison of the observed cleavage decay rates of on- and off-target substrates by SpCas9 shows that the 3 bp PAM-distal mismatch slows the enzyme 13-fold (from 1 s−1 to 0.076 s−1). Both high-fidelity Cas9 variants dramatically decrease the observed decay rate for cleavage of on-target DNA substrates 21- to 35-fold compared to SpCas9 (0.028 s−1 for HypaCas9 and 0.047 s−1 for Cas9-HF1 vs 1 s−1 for SpCas9). Moreover, HypaCas9 and Cas9-HF1 further reduce the decay rates of off-target DNA cleavage 8- to 290-fold (rates of 0.0033 s−1 and 0.00016 s−1, respectively) relative to their respective rates with on-target DNA. These data demonstrate dramatic changes in the observed rates of cleavage by the engineered variants with on- and off-target DNA. However, these measurements alone do not define changes in specificity, which is a function of the kinetic partitioning of DNA cleavage versus dissociation, encompassing all steps leading up to the first irreversible step in the pathway17, including reversible DNA binding, R-loop formation, HNH domain docking, and DNA cleavage.
DNA unwinding is largely unchanged with the Cas9 variants
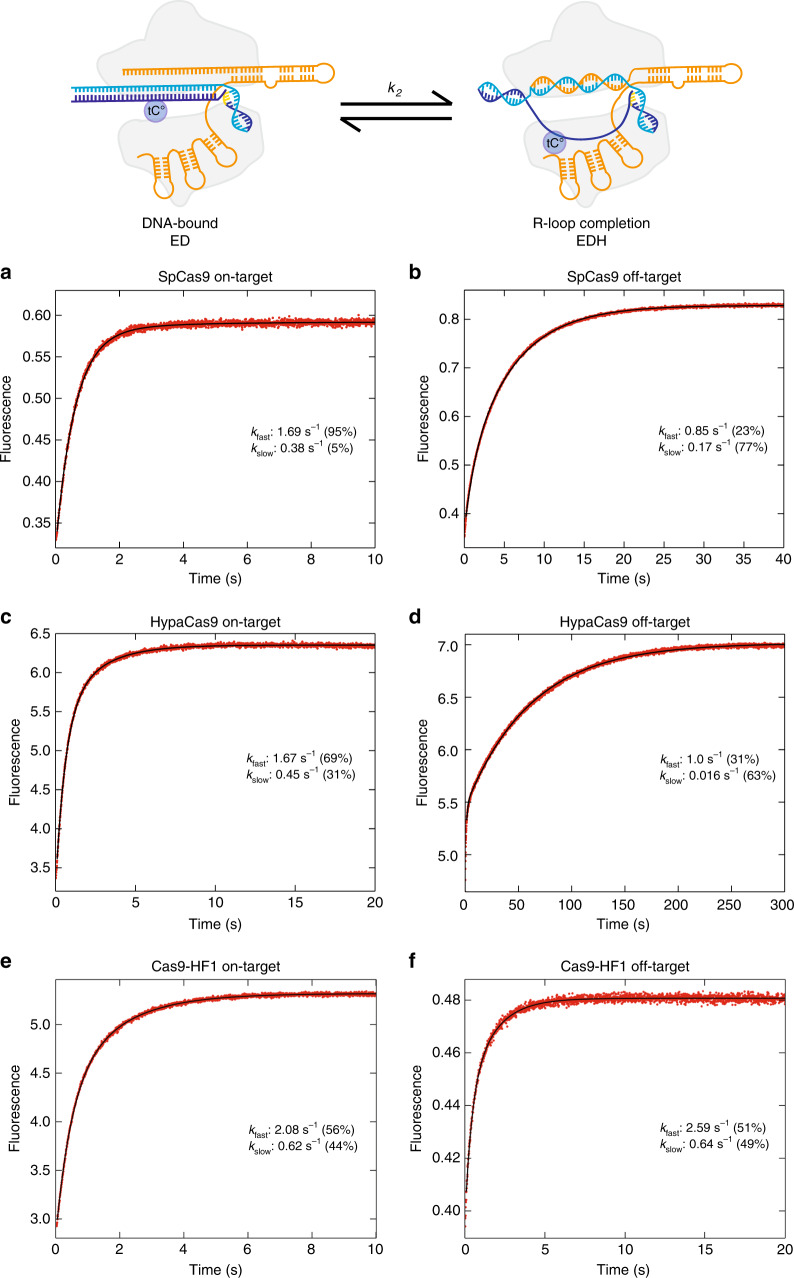
Since our previous work identified R-loop formation as rate-limiting for on-target cleavage and others subsequently suggested that R-loop formation and rewinding rates may dictate enzyme specificity for SpCas9 and Cas9-HF116, we tested whether HypaCas9 would display similar kinetics. To directly measure the rates of R-loop formation for all enzymes, we used a stopped-flow assay based on measuring fluorescence of tCo at −16 nt, a fluorescent tricyclic cytosine analog that is quenched by base stacking in dsDNA so that opening of the duplex results in a large increase in fluorescence. Our control experiments using both our tCo- and 2-AP-labeled base analog substrates at positions −16, −9, and −1 nt, respectively (Supplementary Fig. 3), show that neither of the analogs affect the observed decay rate for DNA cleavage. The chemical structures of tCo and 2AP are much less bulky than larger Cy3 and Cy5 labels and less likely to interfere with enzyme kinetics (Supplementary Fig. 4). In the presence of Mg2+, SpCas9, HypaCas9, and Cas9-HF1 unwind the on-target DNA substrate with nearly identical decay rates (~2 s−1) (Fig. 2). Surprisingly, the decay rate of R-loop formation for off-target DNA substrates for all Cas9 variants was also largely unchanged (between 0.85 s−1 and 2.59 s−1). Therefore, DNA unwinding is not rate-limiting and is not correlated with observed rates of cleavage for the high-fidelity variants, unlike SpCas9.
Fig. 2. DNA unwinding rates are nearly identical for on-targets and off-targets.
R-loop formation decay rates were measured by the monitoring the fluorescence increase from tCo (position −16 in the non-target strand) as a function of time after mixing enzyme (28 nM) with DNA (10 nM) in the presence of 10 mM Mg2+. Data were fit to a double exponential function to get the decay rates shown. a, b SpCas9 (28 nm) with on-target (a) and off-target (b) DNA, c, d HypaCas9 with on-target (c) and off-target (d) DNA. e, f Cas9-HF1 with on-target (e) and off-target (f) DNA.
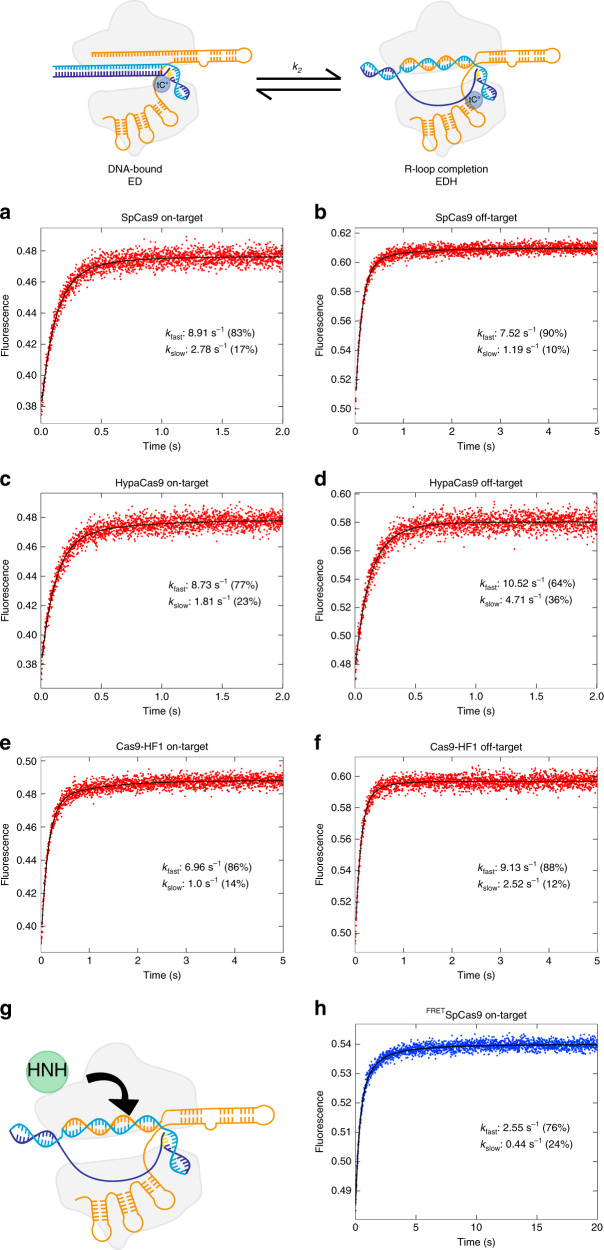
Given the location of the tCo at −16 nt, it is likely that our measurements reflected a step late in the unwinding process. For comparison, we measured the decay rate for R-loop formation using tCo labeled at a position immediately proximal to the PAM (position −1 nt). After rapidly mixing Cas9-RNA with the on-target DNA substrate, we observed a marked increase in fluorescence as the DNA unwinds the tCo base analog. We fit the data to a double-exponential to determine the major decay rate of 10 s−1 (Fig. 3a–f). Intriguingly, these results indicate that once a PAM site is engaged with the enzyme, initial unwinding of the DNA is faster than observed at −16 nt. These data suggest that the net observed decay rate of unwinding observed at −16 nt may be a function of several fast unwinding step leading to complete unwinding. However, because no lag was observed in the kinetics, it appears that the faster, earlier partial unwinding steps lead to a final rate-limiting step of full unwinding, measured at the −16 nt position. Although further studies are needed to examine the effects of mismatches at earlier stages of unwinding, our current measurement provides the best estimate of the net rate of R-loop formation. The decay rate measured for R-loop formation using this label is indistinguishable for both SpCas9 and the high-fidelity variants on all substrates tested, so the rates for DNA unwinding appear to be unchanged by the engineered Cas9 enzymes.
Fig. 3. DNA unwinding occurs more rapidly near the PAM.
R-loop formation decay rates were measured by the monitoring the fluorescence increase from tCo (position −1 in the non-target strand) as a function of time after mixing enzyme (28 nM) with DNA (10 nM) in the presence of 10 mM Mg2+. Data were fit to a double exponential function to get the decay rates shown. a, b SpCas9 (28 nm) with on-target (a) and off-target (b) DNA, c, d HypaCas9 with on-target (c) and off-target (d) DNA. e, f Cas9-HF1 with on-target (e) and off-target (f) DNA. g Cartoon of the HNH domain rearrangement. h The FRETSpCas9 labeled with Cy3 and Cy5 pair at C867 and C355, respectively, was used to measure the HNH domain motion during cleavage of on-target substrates.
To correlate the rates of R-loop formation with steps involved in HNH-domain docking onto the target strand, we measured the enzyme conformational change using stopped-flow analysis on a Cas9 which was labeled with Cy3 and Cy5 as described previously18. Briefly, a Cysteine-light version of Cas9 was labeled with Cy3 at amino acid 355 and Cy5 at amino acid 867. The FRET efficiency increases when the HNH domain rearranges to the catalytically active state (Fig. 3g). After rapidly mixing the FRET-pair-labeled Cas9 with a perfectly matched on-target DNA, we observed an increase in FRET efficiency, indicating that the HNH domain was rearranging to a catalytically active state. We measured the decay rate for HNH docking to be ~2.5 s−1 (Fig. 3h), which is similar to single molecule FRET measurements. Surprisingly, the observed rates of R-loop formation (1.5 s−1) and HNH domain docking are very similar, indicating that these steps may be kinetically linked. The slightly faster observed decay rate for HNH domain movement could be due to the reverse reaction as the domain movement comes to equilibrium after or coincident with R-loop formation.
The decay rates of DNA unwinding have also been measured using single molecule methods using a FRET-pair labeled DNA, Cy3 and Cy5 were labeled on position −6 nt of the target strand and -16 nt on the non-target strand, respectively16. Therefore, we also tested the FRET-paired DNA substrates in an attempt to correlate the FRET signal with the observed rates of DNA cleavage. First, we tested the substrate previously used in single molecule studies without the Cy3/Cy5 labels16, which has a two-nucleotide difference with our substrate, specifically, a T substitution at positions −16 and −18. The decay rate of target strand (HNH) cleavage is similar to that of our substrate (0.7 s−1 vs 1 s−1, Supplementary Fig. 5a), so sequence context at this position does not have a significant affect. We also tested cleavage with Cy3/Cy5 labeled DNA with this other sequence, and it showed a similar decay rate as our sequence (0.05 s−1 vs 0.06 s−1, Supplementary Figs. 5b and 6a). Later, we examined the time course of target strand (HNH) cleavage of the Cy3/Cy5 labeled DNA with our sequence for each enzyme (Supplementary Fig. 6). With SpCas9, the reaction of the Cy3/Cy5 labeled DNA follows a single exponential with a markedly reduced decay rate (0.06 s−1 vs 1 s−1) showing a ~17-fold decrease in the observed decay rate for DNA cleavage compared to unlabeled substrate. The engineered HypaCas9 and Cas9-HF1 also exhibited decreased DNA cleavage rates of 0.0046 s−1 and 0.0016 s−1, respectively, which are 5.9-fold and 28.75-fold slower than unlabeled substrates measured under identical conditions. It is clear that interference by the Cy3/Cy5 labels alters the effect of the high-fidelity variants on the DNA cleavage rates. Taken together, labeling of the DNA with bulky Cy3 and Cy5 labels dramatically impacted the enzyme. For these reasons, no further studies were performed using the FRET-pair labeled DNA.
Cas9 specificity is governed by kinetic partitioning
Since enzyme specificity is a function of all steps leading up to the first largely irreversible step, all events prior to DNA cleavage must be considered17. To measure the intrinsic cleavage rate, we bypassed the normally rate-limiting R-loop formation step by preincubating SpCas9 with off-target DNA in the absence of Mg2+ to allow binding and conformational changes to come to equilibrium to form an SpCas9.DNA complex. We then initiated the chemical reaction by the addition of 10 mM Mg2+. In our previous studies, R-loop formation was rate-limiting with SpCas9 reacting with on-target DNA, and the intrinsic cleavage decay rate was faster when measured using the preincubation protocol. Here, with off-target DNA, the rates of HNH and RuvC cleavage were measured to be 0.12 s−1 and 0.14 s−1, respectively (Supplementary Fig. 7c, d, and Supplementary Fig. 8), which are much slower than the rates of R-loop formation. Intriguingly, these results show that the rate-limiting step in the enzyme pathway of SpCas9 with an off-target is DNA cleavage since the decay rate for R-loop formation we measured was 0.85 s−1 (Fig. 2b). These data indicate that discrimination is based, at least in part, on a change in the identity of the rate-limiting step in comparing on- and off-target DNA.
We repeated these experiments with HypaCas9 and Cas9-HF1 with on- or off-target DNA. These results define observed rates for HNH cleavage of 0.035 s−1 and 0.0023 s−1 for HypaCas9 with on- and off-target substrates, respectively (Supplementary Fig. 7g, k). Observed cleavage rates of Cas9-HF1 for on- and off-target substrates were measured as 0.038 s−1 and 0.00014 s−1, respectively (Supplementary Fig. 7o, q). These observed cleavage rates are somewhat faster than those measured with the simultaneous addition of DNA and Mg2+, indicating that some step other than R-loop formation but preceding DNA cleavage may slow the observed decay rate. Nonetheless, these results show that the observed cleavage rates for on-target DNA are reduced ~100-fold for both HypaCas9 and Cas9-HF1 relative to SpCas9. For off-target DNA, the observed cleavage rates are reduced 50- or 860-fold for HypaCas9 or Cas9-HF1, respectively, relative to SpCas9.
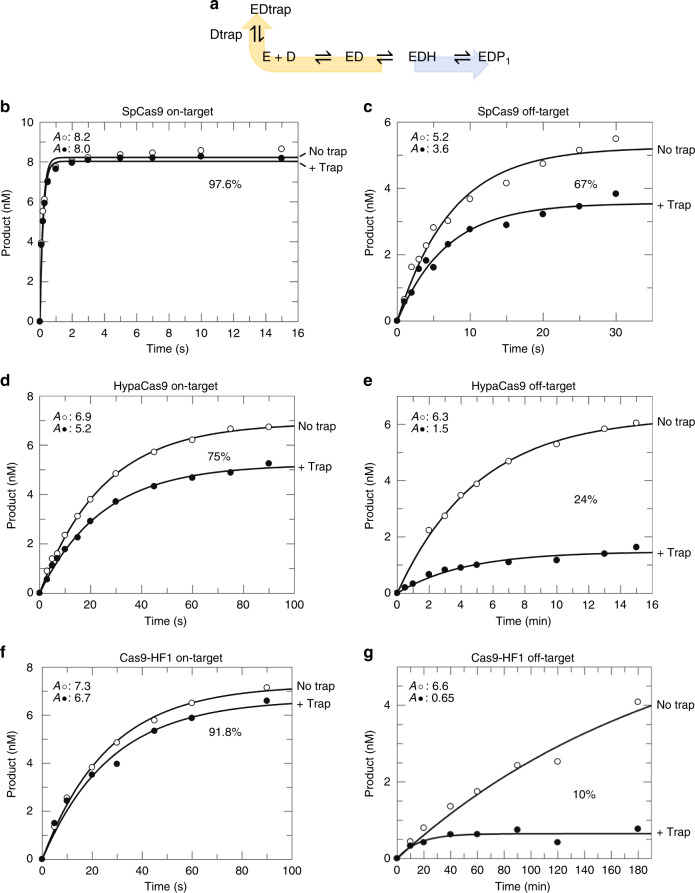
Discrimination is not defined solely by the relative rates of DNA cleavage. Rather, because R-loop formation is fast, discrimination is a function of the kinetic partitioning between the rates of DNA release versus cleavage17,26,27. In order to quantify the kinetic partitioning, we incubated enzyme and labeled DNA in the absence of Mg2+, which allows for R-loop formation to come to equilibrium, but prevents catalysis15. We then added Mg2+ to initiate the reaction in the presence of a large excess of identical, unlabeled on-target DNA to serve as a trap. Comparison between parallel experiments performed in the presence and absence of the DNA trap provides an estimate for the fractional kinetic partitioning for dissociation versus cleavage of bound DNA (Fig. 4a). Once SpCas9 was bound to on-target DNA, it was cleaved rapidly, and the DNA trap had little effect (Fig. 4b). In contrast, 33% of the off-target DNA disassociated from the enzyme, while ~67% of the DNA was committed to cleavage (Fig. 4c and Supplementary Fig. 9a). These results show that SpCas9 discriminates against the PAM-distal mismatched DNA by decreasing the rate of cleavage, which increases the fraction of DNA that is released rather than cleaved. However, the effect is small so the dissociation rate appears to be slower than cleavage.
Fig. 4. Engineered Cas9s improve specificity through a decreased rate of DNA cleavage.
a Schematic of DNA trap experiment. b, c SpCas9 cleavage of on-target (from Gong et al.15) (b) or off-target (c) DNA. d, e HypaCas9 cleavage of on-target (d) or off-target (e) DNA. f, g Cas9-HF1 cleavage of on-target (f) or off-target (g) DNA. Enzyme (28 nM) and labeled DNA (10 nM) were incubated in the absence of Mg2+ and the reaction was initiated by adding Mg2+ in the presence or absence of an excess of unlabeled DNA trap. A• and A◦ represent the amplitude with (filled circles) and without trap (open circles), respectively. The percentage indicates the fraction of pre-bound DNA committed to going forward for cleavage relative to the reaction in the absence of trap.
Next, we examined the kinetic partitioning for HypaCas9 and Cas9-HF1 bound to on-target DNA (Fig. 4d, f, Supplementary Fig. 9b, d). Our results with HypaCas9 and Cas9-HF1 show that ~75% and ~92% of the on-target DNA was cleaved in the presence of the trap, respectively. The percentage cleaved is smaller than for wild-type SpCas9 because the observed intrinsic cleavage decay rate for on-target DNA by HypaCas9 (0.035 s−1) and Cas9-HF1 (0.038 s−1) was 100-fold slower than with SpCas9 (4.3 s−1). This slower cleavage rate gives time for a small fraction (8 to 25%) of the on-target DNA to dissociate before it is cleaved.
Kinetic partitioning to favor dissociation was enhanced when HypaCas9 and Cas9-HF1 react with off-target DNA because the cleavage rates were further reduced to 0.0023 s−1 and 0.00014 s−1, respectively (Fig. 4e, g, Supplementary Fig. 9c, e). These rates are 50- to 860-fold slower, respectively, compared to SpCas9 on an off-target substrate. Accordingly, only ~24% and ~10% of the bound off-target DNA was committed to going forward for cleavage by HypaCas9 and Cas9-HF1, respectively, in the presence of trap DNA. Taken together, these results show that the engineered high-fidelity variants acquired improved specificity against the PAM-distal mismatched DNA through a markedly decreased rate of cleavage, which alters kinetic partitioning to favor release rather than cleavage of the bound substrate for both on- and off-target DNA, but the effect is larger for off-target DNA. Calculation of the apparent dissociation rate constants using Eq. (3) shows that the high-fidelity variants do not increase the apparent rate of DNA release (Supplementary Table 1).
| 3 |
Rather, the increased discrimination is entirely attributed to decreases in the apparent rate constant for cleavage.
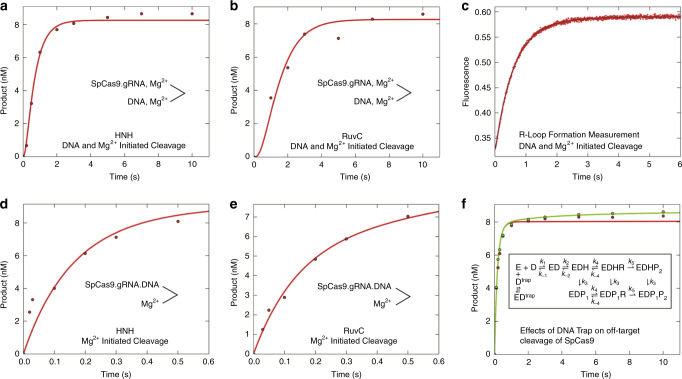
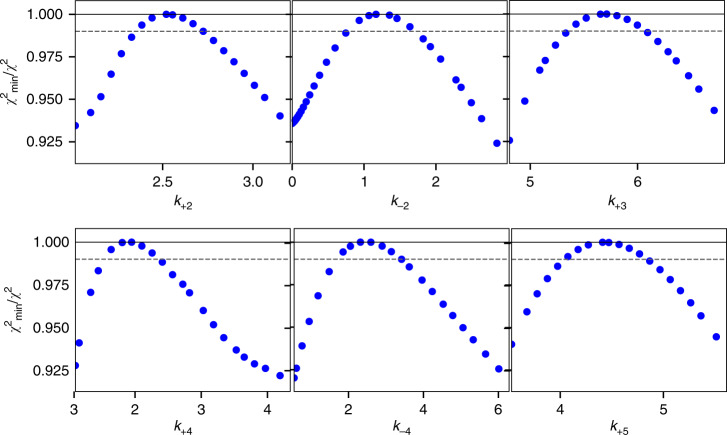
In our descriptions of the kinetics of Cas9 enzymes thus far, the observed decay rates of reactions were estimated based on fitting data to exponential functions, which yield eigenvalues that are usually complex functions of multiple rate constants17. To fully understand the kinetics and mechanism, all of the experiments for each enzyme and DNA substrate were fit globally based on numerical integration of the rate equations using a single unified model (Fig. 5 and Supplementary Figs. 10–14). Global data fitting shows that our minimal model (Fig. 5f, inset) accounts for our data and each of the rate constants is well defined based on confidence contour analysis testing the extent to which each parameter is constrained by the data (Fig. 6)17,28. Note in particular that our model accounts for the biphasic traces seen in some experiments without having to invoke heterogeneity, and it provides intrinsic rate constants for each step in the pathway including R-loop formation and HNH and RuvC cleavage events. Although it is likely that additional, unresolved structural rearrangements may be required for alignment of catalytic residues for DNA cleavage, these have not yet been resolved as kinetically distinct events. The current model represents a minimal pathway necessary and sufficient to account for all available data, and can be easily expanded as new information becomes available to define additional steps in the pathway.
Fig. 5. Global fitting of experiments performed to interrogate on-target activity of SpCas9.
a DNA and Mg2+ initiated cleavage by the HNH domain. b DNA and Mg2+ initiated cleavage by the RuvC domain. c R-loop formation rate. d Mg2+ initiated cleavage by the HNH domain. e Mg2+ initiated cleavage by the RuvC domain. f Effect of DNA trap on kinetic partitioning of Cas9 cleavage. All curves were calculated based on the global fit to the data according to the kinetic model (inset), with rate constants shown in Table 1. Error estimates were based on confidence contour analysis as shown in Fig. 6. E is Cas9.gRNA, D is target DNA, ED is Cas9.gRNA.DNA, EDH is R-loop formation with docking of target strand to HNH, EDHR and EDP1R are R-loop formation with docking of non-target strand to RuvC, EDHP2 is RuvC cleavage of non-target strand, EDP1 is HNH cleavage of the target-strand, EDP1P2 is cleavage of both strands, Dtrap is excess unlabeled, perfect-matched DNA. EDtrap is Cas9.gRNA.DNAtrap.
Fig. 6. Confidence contour analysis for global data fitting.
Confidence contours are shown for fitting data in Fig. 5. Numbered rate constants are defined in our kinetic model (Fig. 5f, inset). The χ2 threshold (dashed line) was set to 0.99 to define upper and lower limits for each rate constant as described17,32. Because the contours were symmetrical with parameters well constrained, we report ± error limits in Table 1. For this analysis a χ2 threshold was calculated using an F-distribution and was used to define the lower and upper limits for each parameter.
To understand enzyme specificity, rate constants must be interpreted in the context of all kinetically relevant steps as illustrated in the free energy profile (calculated using Eq. (4) from rate constants in Table 1).
| 4 |
The transmission coefficient A = 0.001
Table 1.
Kinetic parameters for Cas9 enzymes.
| DNA binding and R-loop formation | HNH cleavage | RuvC alignment | RuvC cleavage | |||||
|---|---|---|---|---|---|---|---|---|
| Enzyme | 1/K11 (nM) | Kd,net2 (nM) | k2 (s−1) | k-2 (s−1) | k3 (s−1) | k4 (s−1) | k-4 (s−1) | k5 (s−1) |
| SpCas9 on-target | [3]3 | [1]3 | 2.5 ± 0.2 | 1.2 ± 0.4 | 5.6 ± 0.4 | 1.8 ± 0.3 | 2.3 ± 0.6 | 4.4 ± 0.4 |
| SpCas9 off-target | 24 | 8.1 | 1.1 ± 0.1 | 0.5 ± 0.1 | 0.17 ± 0.01 | 0.7 ± 0.1 | 0.30 ± 0.05 | 0.38 ± 0.02 |
| Hypa on-target | 4.3 | 1.6 | 0.7 ± 0.03 | 0.4 ± 0.02 | 0.042 ± 0.0007 | 1.0 ± 0.02 | 0.08 ± 0.006 | 0.054 ± 0.001 |
| Hypa off-target | 52 | 17 | 0.8 ± 0.1 | 0.4 ± 0.01 | 0.005 ± 0.001 | 0.04 ± 0.001 | 0.0004 ± 0.0003 | 0.008 ± 0.003 |
| Cas9HF1 on-target | 1.6 | 0.12 | 2.2 ± 0.2 | 0.2 ± 0.08 | 0.038 ± 0.002 | 0.71 ± 0.04 | 0.02 ± 0.01 | 0.032 ± 0.002 |
| Cas9HF1 off-target | 33 | 9.8 | 2.5 ± 2 | 0.7 ± 0.3 | 0.0002 ± 0.00002 | 1.1 ± 0.8 | 0.4 ± 0.2 | 0.0002 ± 0.00005 |
1We used a nominal value of k1 = 1 nM-1s-1 and fit data to derive k-1 to compute K1 = k-1k1.
2Kd,net = 1/(K1(1 + K2)), where K2 = k2/k-2
3Values for DNA dissociation rates are a function of k-1 and k-2, and for SpCas9 on-target DNA these values are not well defined by the data because of the low kinetic partitioning for DNA dissociation (Fig. 4a). For global data fitting, we locked rate constants k-1 = 3 s−1 at a nominal upper limit. Because this rate constant was not well-defined by the data, locking it at a reasonable upper limit had little effect on the values for other rate constants.
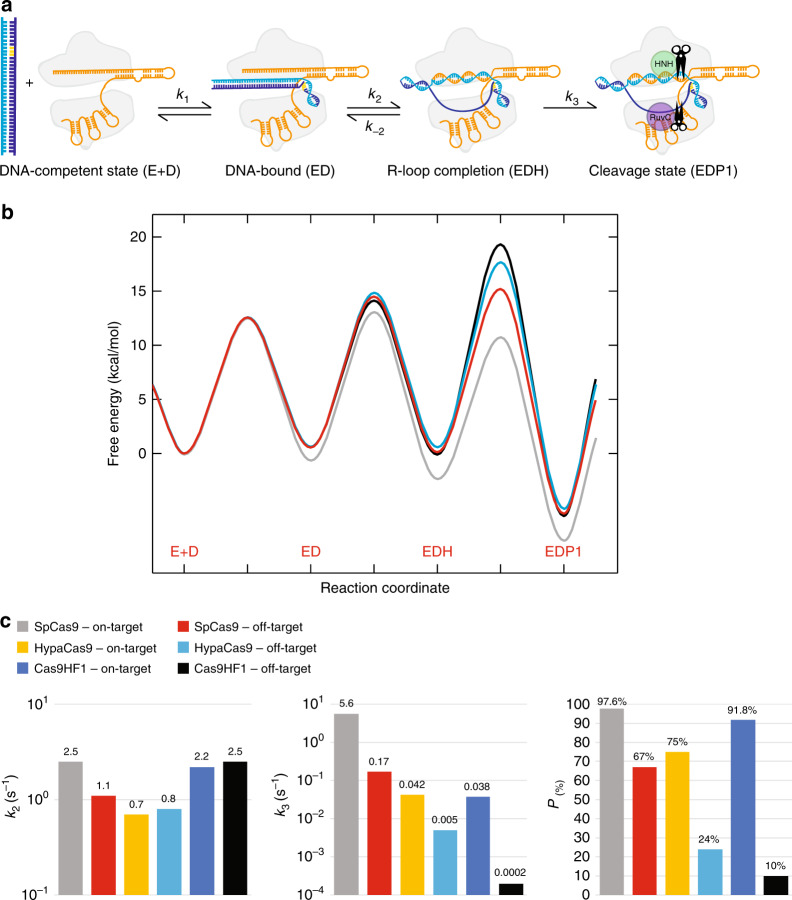
Because global data fitting provides the rate constants for each relevant step (Figs. 5 and 6), we can construct a bona fide free energy profile (Fig. 7b, and Supplementary Figs. 10–14). The free energy profiles comparing SpCas9, HypaCas9, and Cas9-HF1 show a change in rate-limiting and specificity-determining steps. Enzyme specificity is defined by kcat/Km and is given by the highest overall barrier relative to the starting state, while the maximum rate, kcat, is defined by the highest local barrier relative to the preceding state, attenuated by any preceding rapid-equilibrium steps. Because the rate constants for R-loop formation do not change significantly with different substrates and enzyme variants, specificity is governed largely by the kinetic partitioning of the Cas9 R-loop, EDH state in our kinetic model (Fig. 5f, inset), to either go forward resulting in irreversible cleavage versus release via re-annealing of the DNA and ejection from the enzyme. The higher overall barriers for cleavage seen with the high-fidelity variants increase the kinetic partitioning probability to favor dissociation of the DNA (Fig. 6c).
Fig. 7. Specificity is governed by the kinetic partitioning of Cas9.
a Cartoon representation of Cas9 enzymes reaction pathway. b Free energy profiles for SpCas9 cleavage of an on-target (gray line) from Gong et al.15 and off-target (red line), respectively; HypaCas9 cleavage of an off-target (blue line); and Cas9-HF1 cleavage of an off-target (black line). Each profile was calculated using transition state theory using rate and equilibrium constants that were derived from globally fitting each set of experiments (Table 1). Note the higher barriers for DNA cleavage relative to the lower barrier for the preceding reverse reaction increase the probability of DNA release. c Summary bar graph for k2, k3, and kinetic partitioning (P).
Discussion
Our results are important to rationalize in relation to single molecule studies. Previous studies have measured the rate of transition of the HNH domain from the inactive to the active state on an on-target using single-molecule FRET to be 1.5 s−120. This is consistent with our measurements of R-loop formation and HNH domain docking (1.5–2.5 s−1), suggesting that HNH docking is limited by the rate-limiting step of R-loop formation. Since SpCas9 has an intrinsic cleavage rate of 4 s−1, our calculations demonstrate kinetic partitioning to favor cleavage rather than DNA release so that that R-loop formation and coincident HNH docking constitute the specificity-determining steps for wild-type enzyme with on-target DNA.
Interestingly, the previously determined decay rate for the HNH domain transition for the off-target DNA is 0.05 s−1 20. This is consistent with our measurements of the rate of cleavage of an off-target DNA when Mg2+ and DNA are simultaneously mixed with SpCas9 of 0.076 s−1. Our measurement of the intrinsic cleavage decay rate for SpCas9 on an off-target (0.12 s−1) suggests that there may be another conformational change or structural rearrangement that occurs before cleavage, as this transition is not rate-limiting. This also suggests that our measurements using pre-equilibration of Cas9 and DNA in the absence of Mg2+ has no effect on the ability of the HNH domain to sample the active state as this intrinsic cleavage rate is faster than that measured with simultaneous addition of Mg2+ and DNA.
Previous studies have also measured the decay rate for the conversion of the HNH domain of Cas9-HF1 from the inactive to active state (0.2 s−1)9. Our data show that the decay rates for cleavage with either simultaneous addition of DNA and Mg2+ or pre-equilibration followed by initiation with Mg2+ are 0.037 s−1. This would again suggest that while HNH domain movement is slower than for SpCas9, it is faster than the observed rate of DNA cleavage and not rate-limiting.
In this study, we provide a more complete understanding of enhanced specificity of high-fidelity Cas9 variants. HypaCas9 and Cas9-HF1 are seriously impaired in terms of enzyme efficiency of on-target DNA cleavage. For each variant, the cleavage rate was 100-fold slower for on-target cleavage as compared to SpCas9. HypaCas9 and Cas9-HF1 gain discrimination mainly through further slowing of the rate of DNA cleavage with off-target DNA, which shifts kinetic partitioning to favor release rather than cleavage of the bound DNA26,27. We propose that Cas9 uses an induced-fit mechanism analogous to DNA polymerases, where a conformational change after initial substrate binding (HNH domain movement) is an important determinant of enzyme specificity. For SpCas9, the conformational change is rate-limiting and determines specificity because R-loop formation is largely irreversible and is followed by fast DNA cleavage15. The higher-fidelity variants have extraordinarily slow rates of DNA cleavage, allowing for release of the substrate from the enzyme before the irreversible cleavage reaction even though the dissociation rate is slow. Although the high-fidelity variants have increased fidelity, calculated from the kcat/Km values for on- versus off-target DNA, they achieved this through markedly reduced enzyme efficiency (kcat/Km for on-target DNA). The slower rates of DNA cleavage may be the result of a slower rate of HNH domain movement, an unfavorable equilibrium for the conformational change, and/or a suboptimal alignment of catalytic residues for DNA cleavage. DNA polymerases have evolved much higher fidelity by dramatically increasing the rate of dissociation of mismatched nucleotides in addition to decreasing the rate of catalysis26,27. Improvements in Cas9 specificity without sacrificing efficiency may require engineering an enzyme that leads to more rapid release of off-target DNA.
Engineered Cas9 enzymes achieve improved discrimination against mismatches at the distal end of the guide sequence by decreasing the rate of catalysis. Our results are consistent with the model that the REC3 domain senses whether or not the final 3-nt of the target strand are base-paired with the guide RNA, and that this signal is communicated to the tethered REC2 domain. In this model, the REC2 domain is evicted from its normal position, which allows the HNH domain to dock on the target strand for cleavage9. This is likely due to REC3 domain sensing the shape of the substrate, and we hypothesize that duplex formation between the gRNA and target strand is the trigger for the catalytical rearrangement29,30. This moderate improvement in discrimination may be sufficient in the context of the full recognition sequence to disfavor off-target cleavage in vivo. However, further improvements to enable gene therapy may require engineered enzymes that increase rates of off-target DNA dissociation without requiring such drastic reductions in efficiency of on-target cleavage.
Methods
Expression and purification of Cas9s
Plasmid pMJ806 containing Streptococcus pyogenes Cas9 and plasmid pSHS306 containing the HNH FRET variant of Streptococcus pyogenes Cas9 were obtained from Addgene (Cambridge, MA). Plasmid pHypaCas9 was a gift from Dr. Ilya Finkelstein at the University of Texas at Austin. For bacterial expression of the recombinant SpCas9 and HypaCas9, plasmids were transformed into BL21-Rosetta 2 (DE3)-competent cells (Millipore). The E. coli cells were cultured at 37 °C in LB media (containing working concentration 50 μg/mL kanamycin for SpCas9 and HypaCas9 and 100 μg/mL ampicillin for FRETSpCas9) until the OD600 reached to 0.5–0.8, and then Cas9 expression was induced by the addition of 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 20 h at 18 °C. The His6-MBP tagged Cas9 were purified by a combination of affinity, cation exchange, and size exclusion chromatographic steps, essentially as described previously15 with following modifications. Briefly, bacterial cells were lysed by sonication in buffer containing 50 mM Tris-HCl, pH 7.5, 10% glycerol, 500 mM NaCl, 2 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM TCEP and Pearce protease inhibitor cocktails (Thermo Scientific). Clarified lysate was applied to HisPur TM Ni-NTA resin (Thermo Scientific) and the resin was washed extensively with buffer containing 50 mM Tris-HCl, pH 7.5, 10% glycerol, 500 mM NaCl, 1 mM PMSF, 0.5 mM TCEP, and 25 mM Imidazole. The bound protein was eluted in 50 mM Tris-HCl, pH 7.5, 10% glycerol, 500 mM NaCl, 1 mM PMSF, 0.5 mM TCEP, and 200 mM Imidazole. The His6-MBP-Cas9 fusion protein was cleaved to remove the His6-MBP tag by adding the TEV protease to the dialysis tubing (Spectrum labs) during dialysis against buffer containing 20 mM HEPES pH 7.5, 5% glycerol, 150 mM KCl, 2 mM PMSF, and 0.5 mM TCEP. After further purification by SP cation exchange chromatography, the cleaved Cas9 was concentrated and loaded onto Superdex 200 g 16/600 in 20 mM HEPES pH 7.5, 5% glycerol, 150 mM KCl, 2 mM PMSF, and 0.5 mM TCEP. The eluent proteins were concentrated to ~5 mg/ml, snap frozen, and store at −80 °C. Alt-R®SpCas9 and Alt-R®SpHiFiCas925 are purchased from Integrated DNA Technologies. Alt-R®HiFiCas9 contains a single substitution at R691A. Here, we named Alt-R®SpCas9 and Alt-R®SpHiFiCas9 as IDTSpCas9 and IDTHiFiCas9 in our article. Fluorescent labeling of FRETSpCas9 was performed as described in Sternberg et al. 201518. FRETSpCas9 was mixed with 20-fold excess of Cy3- and Cy5-maleimide (GE Healthcare) dissolved in DMSO, the final concentration of which did not exceed 5%. The reaction was quenched with 10 mM DTT following incubation for 2 h at room temperature and overnight at 4 °C. Labeling reactions were performed in the dark. Labeled FRETSpCas9 was separated via size-exclusion chromatography, concentrated, snap frozen, and stored at −80 °C.
In vitro sgRNA transcription and refolding
In vitro sgRNA transcription and refolding were performed as described previously15,31. The primers used for the templates of sgRNA transcription are listed (Supplementary Table 2). Equimolar concentrations of complementary oligos were mixed in 10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA and heated to 95 °C for 5 min, then slowly cooled to room temperature in about 60 minutes. The sgRNA was in vitro transcribed using the HiScribe Qiuk T7 RNA synthesis kit (New England Biolab) following the manufactory protocol. The transcribed sgRNA was further purified using a PureLink column (Thermo Scientific) and refolded by heating to 95 °C and then slowly cooled to room temperature in 10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA. We purchased the gRNA for the Singh et al. substrate from Synthego.
DNA duplex formation and probe labeling
55-nt DNA duplexes were prepared from unmodified DNA oligonucleotides synthesized, and PAGE gel purified by Integrated DNA Technologies. FRET paired-labeled (Cy3/Cy5) DNA oligonucleotides were synthesized, and PAGE gel purified by Gene LinkTM. A thymine modified with an amine group through a C6 linker (amino dT) was used to label DNA with Cy3 N-hydroxysuccinimido (NHS) or Cy5 NHS. A cytosine modified with an amine group through a C6 linker (amino dC) was used to label DNA with Cy5 NHS. The detailed sequence information is listed in Supplementary Table 2. The tricyclic fluorescent cytidine analog (tCo)-labeled DNA oligonucleotides were synthesized, and PAGE gel purified by Bio-Synthesis, Inc. The synthesized oligonucleotides and the position of FRET paired and tCo labeling are listed in the Supplemental Experimental Methods. The DNA duplex used for RuvC cleavage assays was prepared by γ-32P labeling or 6-FAM the non-target strand before annealing with the cold complementary strand at a 1:1.15 molar ratio. The DNA duplex used for HNH cleavage assays was prepared by γ-32P or 6-FAM labeling the target strand before annealing with cold non-target strand at a 1:1.15 molar ratio.
Buffer composition for kinetic reactions
Prepared 5X cleavage buffer (100 mM Tris-Cl, pH 7.5, 500 mM KCl, 25% glycerol, 5 mM DTT). After allowing Cas9–gRNA to react with substrate in 1X cleavage buffer (20 mM Tris-Cl, pH 7.5, 100 mM KCl, 5% glycerol, 1 mM DTT) under the conditions for the stopped-flow kinetic, Active-site titration, DNA cleavage and DNA trap cleavage assays.
Stopped-flow kinetic assay
Stopped-flow experiment was performed as previously described15,31. Briefly, Cas9-gRNA complex (1:1 ratio) was mixed with tCo-labeled 55/55 nt DNA substrate at 37°C using AutoSF-120 stopped-flow instrument (KinTek Corporation, Austin, TX). The fluorophore was excited at 367 nm, and emission was monitored at 445 nm using a single band-pass filter with 20 nm bandwidth (Semrock) (data in Figs. 2a–f and 3a–f). For the Cy3/Cy5 FRET signal, samples were excited at 550 nm, and the time-dependent fluorescence change was monitored at 670 nm using a single band-pass filter with a 30-nm bandwidth (Semrock) (data in Fig. 3h).
Global analysis of kinetic data
The kinetic data defining CRISPER-Cas9 cleavage were globally fit using the models shown in our kinetic model (Fig. 5f, inset) by KinTek Explorer software (KinTek Corporation, Austin, TX) to obtain rate constants (Figs. 5 and 6, and Supplementary Figs. 10–14). The output for experiments to measure HNH and RuvC DNA cleavage were modeled simply as the sum of products for each cleavage site. For example, for HNH cleavage the products were defined according to our kinetic model (Fig. 5f, inset) as the sum of all species containing P1: EDP1 + EDP1R + EDP1R.Mg +EDP1P2. RuvC cleavage products were defined as the sum of all species containing P2: EDHP2 + EDHP2.Mg+EDP1P2. The fluorescence signal for R-loop formation was modeled as a weighted sum of all species contributing to net fluorescence, with an enhancement in fluorescence in HNH alignment in forming EDH (and subsequent products) and a smaller enhancement in RuvC alignment in forming EDHR (and subsequent products). Specifically, the fluorescence output was modeled as a*(D + ED + b*(EDH + EDP1 + EDH.Mg) +c*(EDHR + EDHR.Mg +EDP1R + EDP1R.Mg +EDHP2 + EDHP2.Mg +EDP1P2)). The scaling factors, a, b, and c were derived as variable parameters in globally fitting the data. FitSpace confidence contour analysis was performed to define the lower and upper limits for each kinetic parameter and to establish that all parameters, including scaling factors, were well constrained according to this minimal model. This model can be readily expanded to include additional information from new experiments.
Active-site titration assay
To measure the active-site concentration of SpCas9 in off-target cleavage, a fixed concentration of enzyme (100 nM, estimated from absorbance at 280 nm) of SpCas9.gRNA was allowed to react with various concentrations of off-target DNA (5’ end labeled on the target strand) in the presence of 10 mM Mg2+. According to a previous study9 and our current results, the intrinsic cleavage rates for SpCas9 off-target cleavage, HypaCas9 and Cas9-HF1 on-target cleavage are slower than SpCas9 on-target cleavage. After 1 hour at 37°C, the products were quenched and resolved on a sequencing gel, quantified, and plotted as a function of DNA concentration (Supplementary Fig. 1b–d). The results showed an active enzyme concentration of 31 nM. Because we used a new preparation of Cas9 enzyme, we also measured the active-site concentration of SpCas9 in on-target cleavage (Supplementary Fig. 1a). The results of IDT Alt-R®SpCas9 and Alt-R®SpHiFiCas9 are shown in Supplementary Fig. 1e, f).
DNA cleavage kinetics
To better understand the reaction on the timescale of a single turnover, it is important to design experiments to accurately measure the rate of catalysis. For these measurements, we employed two complementary approaches. In the first case, we examined the time courses of Cas9s on- and off-target cleavage by simultaneously mixing Cas9.gRNA (28 nM active-site concentration) with 10 nM radiolabeled DNA target in the presence of Mg2+. After quenching by adding EDTA, we resolved the products on a sequencing gel and quantified the products using a phosphor imager (data in Fig. 1, Supplementary Figs. 2, 3, 5a and 7a, b, e, f, i, j, m, and n). In some experiments we used 6-FAM-labeled DNA and resolved and quantified products using an Applied Biosystems DNA sequencer (ABI 3130xl). By using the first case method, we also can directly measure Cy3/Cy5-labeled DNA cleavage to detect Cy3 signal for target strand (HNH) cleavage from Applied Biosystems DNA sequencer (data in Supplementary Figs. 5b, c and 6). In the second case, we first allow the formation of the Cas9.gRNA.DNA (28 nM active-site concentration with 10 nM radiolabeled or 6-FAM label DNA target) in the absence of Mg2+, and then directly measure the cleavage after the addition of Mg2+ (data in Supplementary Fig. 7c, d, g, h, k, l, o, p–r). All the detailed protocol information can be found in Liu et al. as described previously31. Control experiments using radiolabeled DNA established that the 6-FAM label did not alter the kinetics.
DNA trap cleavage kinetics
To measure the forward commitment for kinetic partitioning, we employed perfectly matched on-target cold DNA serving as DNA trap. Firstly, Cas9.gRNA (28 nM active-site concentration) and labeled DNA target (10 nM) were incubated in the absence of Mg2+ and the reaction was initiated by simultaneously adding Mg2+ in the presence or absence of an excess of unlabeled DNA trap (200 nM), and quenched each reaction by the addition of 0.5 M EDTA (data in Fig. 4 and Supplementary Fig. 9).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank members of the Johnson and Taylor labs for helpful discussions. This work was supported in part by Welch Foundation grants F-1604 (to K.A.J.) and F-1938 (to D.W.T.), Army Research Office Grant W911NF-15-1-0120 (to D.W.T.), and a Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation Medical Research Award (to D.W.T.). D.W.T is a CPRIT Scholar supported by the Cancer Prevention and Research Institute of Texas (RR160088) and an Army Young Investigator supported by the Army Research Office (W911NF-19-1-0021).
Author contributions
M.L. performed the kinetic studies. H.H.Y. and K.J. purified and reconstituted the Cas9–gRNA complexes and assisted with the kinetic studies. M.L., S.G. and K.A.J. analyzed the kinetic data. M.L., K.A.J., and D.W.T. interpreted the results and wrote the manuscript. K.A.J. and D.W.T. conceived the experiments, supervised the research, and secured funding for the project.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
Competing interests
K.A.J. is the President of KinTek, Corp., which provided the stopped-flow and chemical-quench flow instruments and the KinTek Explorer software used in this study. D.W.T. is an inventor on a Cas9-related patent. The remaining authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Chirlmin Joo and Nils Walter for their contributions to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kenneth A. Johnson, Email: kajohnson@mail.utexas.edu
David W. Taylor, Email: dtaylor@utexas.edu
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-17411-1.
References
- 1.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1077. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 4.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strutt SC, et al. RNA-dependent RNA targeting by CRISPR-Cas9. Elife. 2018;7:e32724. doi: 10.7554/eLife.32724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang HF, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 7.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Geen H, Yu AS, Segal DJ. How specific is CRISPR/Cas9 really? Curr. Opin. Chem. Biol. 2015;29:72–78. doi: 10.1016/j.cbpa.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Kriz AJ, Sharp PA. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014;2:59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong S, Yu HH, Johnson KA, Taylor DW. DNA unwinding is the primary determinant of CRISPR-Cas9 activity. Cell Rep. 2018;22:359–371. doi: 10.1016/j.celrep.2017.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh D, et al. Mechanisms of improved specificity of engineered Cas9s revealed by single-molecule FRET analysis. Nat. Struct. Mol. Biol. 2018;25:347. doi: 10.1038/s41594-018-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson KA. Kinetic Analysis for the New Enzymology: Using Computer Simulation to Learn Kinetics and Solve Mechanisms., 484. Austin, USA: KinTek Corporation; 2019. [Google Scholar]
- 18.Sternberg SH, LaFrance B, Kaplan M, Doucina JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh D, Sternberg SH, Fei JY, Doudna JA, Ha T. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat. Commun. 2016;1:12778. doi: 10.1038/ncomms12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dagdas YS, Chen JS, Sternberg SH, Doudna JA, Yildiz A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017;3:eaao0027. doi: 10.1126/sciadv.aao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim Y, et al. Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat. Commun. 2016;7:13350. doi: 10.1038/ncomms13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 24.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 25.Vakulskas CA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellinger MW, Johnson KA. Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc. Natl. Acad. Sci. USA. 2010;107:7734–7739. doi: 10.1073/pnas.0913946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirmizialtin S, Nguyen V, Johnson KA, Elber R. How conformational dynamics of DNA polymerase select correct substrates: experiments and simulations. Structure. 2012;20:618–627. doi: 10.1016/j.str.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Press WH, Vetterling WT, Flannery BP. Numerical Recipies in C++ New York: Cambridge University Press; 2007. [Google Scholar]
- 29.Zhu X, et al. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat. Struct. Mol. Biol. 2019;26:679–685. doi: 10.1038/s41594-019-0258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor DW. The final cut: Cas9 editing. Nat. Struct. Mol. Biol. 2019;26:669–670. doi: 10.1038/s41594-019-0267-1. [DOI] [PubMed] [Google Scholar]
- 31.Liu MS, Gong S, Yu HH, Taylor DW, Johnson KA. Kinetic characterization of Cas9 enzymes. Methods Enzymol. 2019;616:289–311. doi: 10.1016/bs.mie.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson KA, Simpson ZB, Blom T. FitSpace Explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.