Abstract
In the ocean, Bacillariophyta are one of the most successful protistan groups. Due to their considerable biogeochemical implications, diatom diversity, development, and seasonality have been at the center of research, specifically large-sized species. In comparison, nanoplanktonic diatoms are mostly disregarded from routine monitoring and are often underrepresented in genetic reference databases. Here, we identified and investigated the temporal dynamics of relevant nanodiatoms occurring in the Western English Channel (SOMLIT-Astan station). Coupling in situ and laboratory approaches, we revealed that nano-species from the genera Minidiscus and Thalassiosira are key components of the phytoplankton community that thrive in these coastal waters, but they display different seasonal patterns. Some species formed recurrent blooms whilst others were persistent year round. These results raise questions about their regulation in the natural environment. Over a full seasonal cycle at the monitoring station, we succeeded in isolating viruses which infect these minute diatoms, suggesting that these mortality agents may contribute to their control. Overall, our study points out the importance of considering nanodiatom communities within time-series surveys to further understand their role and fate in marine systems.
Subject terms: Microbial ecology, Molecular ecology, Virus-host interactions, Microbial communities, Microbial ecology
Introduction
Within the context of global change, understanding the mechanisms that influence the dynamics of carbon export from the photic layers to the ocean floors is of prime importance. Diatoms, which form massive blooms, have long been recognized as major drivers of the biological pump, especially in productive marine ecosystems [1–3]. Nevertheless, all species do not contribute equally to the export of carbon [4]. Sinking rates depend on diverse parameters such as the cell size and shape, the degree of valve silicification, and also their ability to produce chains [4].
Until recently, studies that aimed at describing and understanding diatoms species dynamics and long-term variability have mainly focused on large-sized species (>20 µm) that belong to the micro-phytoplanktonic community (see for example refs. [5–8] for communities of the English Channel and North Sea). Taking into account the nanoplanktonic diatoms (ranging between 2 and 20 µm) in routine microscopy-based analyses is more challenging, especially for species at the very lower end of the size range 2–5 µm because they are both difficult to detect and identify. Still, several studies, generally involving electron microscopy and in some cases culture experiments, have revealed the importance of nanodiatom species in natural assemblages from different marine regions [9–11]. The genus Minidiscus, that includes the smallest known marine diatom species [12] with cell sizes ranging generally from 2 to 5 µm [13–15] appears to be particularly widespread in the world’s oceans [9, 14–20]. It belongs to the Thalassiosiraceae family, which also encompasses the emblematic genus Thalassiosira (about 170 species described [21]). Minidiscus can even produce intense blooms in turbulent eutrophic environments [9, 22]. Contrarily to common assumptions, these primarily solitary nanodiatoms also contribute to carbon export in the deep ocean most likely due to their ability to form aggregates [9]. This conclusion is supported by recent metabarcoding analyses of the Tara Ocean expedition where sequences assigned to Minidiscus (and other nanodiatoms) were retrieved in abundance both from surface and mesopelagic samples. At a global scale, sequences of Minidiscus were the 21st and 8th most abundant diatom sequences within the Tara Ocean dataset respectively in surface and in mesopelagic samples [9, 19].
Metabarcoding could indeed greatly improve our knowledge of the diversity and dynamics of nanodiatom species in the marine environment. However, the correct assignment of barcodes to species is a prerequisite and highly dependent on the availability of reference sequences. Few nanodiatom representatives have been brought into culture, and the large majority of this group remains uncharacterized genetically to date. To our knowledge, the sequences of the 18S ribosomal RNA gene of 4 of the 11 known Minidiscus species were available in the GenBank sequence database prior to our study. Unfortunately, for most of those sequences, taxonomic annotations to species level or even to genus level were not kept up-to-date. As a consequence, the global significance of these nanodiatoms has been most likely largely underestimated by environmental surveys that rely on automatic taxonomic assignations of barcodes.
In this study, we characterized the nanodiatoms that thrive at the long-term monitoring SOMLIT-Astan station located off Roscoff (Western English Channel, WEC). Using a combination of morphological and molecular approaches, we show that species of the genus Minidiscus, and to a lesser extent nanoplanktonic Thalassiosira species, dominate the diatom community at this sampling station. We investigated their dynamics using metabarcoding data obtained from 2009 to 2016. Minidiscus and Thalassiosira species showed distinct seasonal patterns, suggesting different mechanisms involved in their regulation. A large collection of viruses isolated from these diatom genera indicate that viral pathogens may exert an important biotic pressure at this sampling station. Altogether, these results highlight the necessity of considering nanodiatoms and the mechanisms involved in their dynamics to better understand the functioning of coastal systems.
Material and methods
Environmental isolation and growth conditions of diatom cultures
Diatom strains were isolated from natural seawater samples collected at 1 m depth using a 5 L Niskin bottle at the long-term monitoring SOMLIT-Astan station in the Western English Channel (48:46:18 N, 3:58:6 W) on May 26, 2015 and over a full seasonal cycle (October 2015–October 2016). Strains were isolated either using flow cytometry single cell sorting or dilutions. For isolation using flow cytometry, natural seawater samples were filtered (<50 µm) and diatoms were single cell sorted with a FACS Aria flow cytometer (Becton Dickinson, San Jose, CA, USA), as described in Marie et al. [24]. For strain isolation using dilution, seawater samples were analyzed by flow cytometry to determine the total photosynthetic cell concentration. Samples were diluted into K + Si medium in multiwell plates in order to obtain a final concentration of 5 and 10 cells per well. After 2 weeks of incubation, algal growth was monitored using an inverted microscope (Olympus IX71, Olympus Corporation, Tokyo, Japan). Cultures that appeared monospecific were selected and maintained in sterile condition in K + Si medium [25] at 18 °C, under a 12:12 h light:dark cycle of 100 μmol photons m−2 s−1 provided by a white fluorescent light (Philips Master TL_D 18W/865). Diatom cultures were deposited in the Roscoff Culture Collection (RCC, http://roscoff-culture-collection.org/) (Table 1).
Table 1.
Diatom strains isolated in May 2015 and from October 2015 to October 2016 at the SOMLIT-Astan station. Where indicated, both morphological and genetic identifications were carried out. In bold, fully characterized strains for each taxon. RCC Roscoff culture collection, LM light microscopy, SEM scanning electronic microscopy.
| Species | Strains | Isolation date | Isolation method | Morphological identification | SSU-18S | ITS | LSU-28S D1–D3 region |
|---|---|---|---|---|---|---|---|
| Minidiscus comicus | RCC4660 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete |
| RCC4661 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete | |
| RCC4662 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete | |
| RCC5839 | 20/11/2015 | Dilution | Complete | Complete | |||
| RCC5840 | 04/12/2015 | Dilution | Complete | Complete | |||
| RCC5841 | 04/12/2015 | Dilution | Complete | Complete | |||
| RCC5842 | 04/12/2015 | Dilution | Complete | Complete | |||
| RCC5843 | 04/12/2015 | Dilution | Complete | ||||
| RCC5844 | 04/01/2016 | Dilution | Complete | Complete | |||
| RCC5845 | 04/01/2016 | Dilution | Complete | Complete | |||
| RCC5846 | 02/02/2016 | Dilution | Complete | ||||
| RCC5847 | 02/02/2016 | Dilution | Complete | Complete | |||
| RCC5848 | 02/02/2016 | Dilution | Partial | Complete | |||
| RCC5849 | 04/03/2016 | Dilution | Complete | ||||
| RCC5850 | 04/03/2016 | Dilution | Complete | Complete | |||
| RCC5851 | 04/03/2016 | Dilution | Partial | Complete | |||
| RCC5852 | 01/04/2016 | Dilution | Complete | Complete | |||
| RCC5853 | 01/04/2016 | Dilution | Complete | Complete | |||
| RCC5854 | 01/04/2016 | Dilution | Partial | Complete | |||
| RCC5855 | 01/04/2016 | Dilution | Partial | Complete | |||
| RCC5856 | 15/04/2016 | Dilution | Partial | Complete | |||
| RCC5857 | 15/04/2016 | Dilution | Partial | Complete | |||
| RCC5859 | 13/05/2016 | Dilution | Complete | Complete | |||
| Minidiscus spinulatus | RCC4659 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete |
| RCC5860 | 04/03/2016 | Dilution | Partial | Complete | |||
| RCC5861 | 04/03/2016 | Dilution | Partial | Complete | |||
| Minidiscus variabilis | RCC4657 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete |
| RCC4658 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete | |
| RCC4665 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete | |
| RCC4666 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete | |
| RCC5862 | 06/10/2015 | Dilution | Complete | ||||
| RCC5863 | 04/11/2015 | Dilution | Complete | Complete | |||
| RCC5864 | 04/11/2015 | Dilution | Partial | ||||
| RCC5865 | 04/11/2015 | Dilution | Complete | ||||
| RCC5866 | 04/11/2015 | Dilution | Complete | ||||
| RCC5867 | 04/11/2015 | Dilution | Complete | Complete | |||
| RCC5868 | 20/11/2015 | Dilution | Complete | Complete | |||
| RCC5869 | 20/11/2015 | Dilution | Complete | Complete | |||
| RCC5870 | 04/12/2015 | Dilution | Complete | Complete | |||
| RCC5921 | 04/01/2016 | Dilution | Complete | Complete | |||
| RCC5871 | 02/02/2016 | Dilution | Complete | Complete | |||
| RCC5872 | 04/03/2016 | Dilution | Partial | Complete | |||
| RCC5873 | 01/04/2016 | Dilution | Partial | Complete | |||
| RCC5875 | 15/04/2016 | Dilution | Partial | Complete | |||
| RCC5876 | 30/05/2016 | Dilution | Complete | Complete | |||
| RCC5877 | 13/07/2016 | Dilution | Complete | Complete | |||
| RCC5878 | 09/09/2016 | Dilution | Partial | Complete | |||
| RCC5879 | 24/10/2016 | Dilution | Complete | ||||
| RCC5880 | 24/10/2016 | Dilution | Complete | ||||
| Thalassiosira curviseriata | RCC5154 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete |
| Thalassiosira cf. profunda | RCC4663 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete |
| RCC5881 | 20/11/2015 | Dilution | Partial | ||||
| RCC5882 | 02/02/2016 | Dilution | Partial | ||||
| RCC5883 | 04/03/2016 | Dilution | Complete | Complete | |||
| RCC5884 | 29/04/2016 | Dilution | Partial | Complete | |||
| RCC5885 | 13/05/2016 | Dilution | Partial | Complete | |||
| RCC5886 | 13/07/2016 | Dilution | Partial | Complete | |||
| Thalassiosira sp. | RCC4664 | 26/05/2015 | Flow cytometry | LM, SEM | Complete | Complete | Complete |
| RCC5887 | 02/02/2016 | Dilution | Partial | Complete |
Morphological characterization
Cultures in exponential growth phase were observed using a light microscope (Olympus BX51, Tokyo, Japan) with 40× or 100× objectives and a differential interference contrast. Cultures were imaged with a SPOT RT-slider camera (Diagnostics Instruments, Sterling Heights, MI, USA). Cultures were harvested by gravity on a 0.8 µm polycarbonate filter (Nuclepore, Whatman) and dried for 2 h at 56 °C. The filters were mounted on stubs and adhesive paper, coated with a metallization process and observed using a scanning electronic microscopy (SEM, Phenom G2 Pro, PhenomWorld) operating at 10 kV. Cell diameters were estimated from the SEM pictures using ImageJ software (https://imagej.nih.gov/ij/).
Molecular analysis
The SSU-18S, full ITS, and partial LSU-28S rRNA gene markers were amplified by PCR directly on diatom cultured cells. The primers used were 63F (ACGCTTGTCTCAAAGATTA) and 1818R (ACGGAAACCTTGTTACGA) [26] for the 18S, 329F (GTGAACCTGCRGAAGGATCA) [27] and D1R-R (TATGCTTAAATTCAGCGGGT) [28] for the ITS, and D1R-F (ACCCGCTGAATTTAAGCATA) [28] and D3Ca (ACGAACGATTTGCACGTCAG) [29] for the partial 28S D1-D3 region. For PCR analyses, the aliquots (2.25 µL) of diatom cultures in exponential growth phase were subjected to 95 °C for 5 min for denaturation and cooled to 4 °C. The reaction mixture (30 µL final volume) was then added and included Phusion Master Mix (1× final concentration, Thermo Scientific), 3% DMSO, and 0.25 µM of each primer. PCR amplifications were performed with the following conditions: an initial incubation step at 95 °C for 5 min, followed by 35 (18S-28S) or 40 (ITS) cycles of denaturation at 95 °C for 30 s, annealing step for 30 s at 55°, 52°, and 57° for the amplifications of the 18S, ITS and 28S respectively, and extension at 72 °C for 1 min 30. The cycles were followed by a final extension step at 72 °C for 10 min. PCR products were sent for Sanger Sequencing to GATC Biotech (https://www.gatc-biotech.com/en/index.html, Constance, Germany). Sequences were analyzed using Geneious 9.1.3 and relatives were searched in GenBank using the BLASTn tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic analysis
In order to determine the taxonomic positions of the diatom strains, new DNA sequences of the 18S and of the partial 28S rRNA gene markers were aligned with sequences of other Thalassiosirales (Table S1). The sequence alignments for each gene were generated by the MAFFT version 7 program and with automatic alignment strategy (the L-INS-i iterative refinement method was calculated for both genes) (https://mafft.cbrc.jp/alignment/server/ [30]). For each gene, a phylogenetic reconstruction was performed on the 1590 and 534 aligned nucleotides (18S and 28S respectively) by maximum likelihood with PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/ [31]), with the automatic model selection by SMS [32] and 1000 bootstrap replicates. MEGA7 (ref. [33]) was used to visualize the final tree. For the concatenated tree (18S + 28S), a GTR model was applied to the alignments with 1000 bootstrap replicates.
Temporal dynamics
To study the seasonal dynamics of nanoplanktonic diatoms, we used the SOMLIT-Astan eukaryotic metabarcoding dataset (see Caracciolo et al. [34] for detailed protocols of data acquisition and processing). Briefly, between 2009 and 2016, sea surface water (5 L) was sampled and filtered through 3 µm polycarbonate filter (Whatman). Filters were preserved in a lysis buffer at −80 °C until DNA extraction. Nucleic acids were extracted from the filters using a phenol–chloroform method and DNA was then purified using filter columns from NucleoSpin® PlantII kit (Macherey–Nagel) following a modified protocol. DNA extracts were used as templates for PCR amplification of the V4 region of the 18S rRNA (∼380 bp) using the primers TAReuk454FWD1 and TAReukREV3 (ref. [35]). Following polymerase chain reactions, DNA amplicons were purified, quantified, and sent to Fasteris (https://www.fasteris.com/dna/, Plan-les-Ouates, Switzerland) for high throughput sequencing using paired-end 2 × 250 bp Illumina MiSeq. Sequencing reads were processed following a metabarcoding pipeline available online (https://github.com/frederic-mahe/swarm/wiki). Sequences were grouped into operational taxonomic units (OTUs) using the Swarm approach [36]. Taxonomical assignment of each OTU was performed using the Protist Ribosomal Reference (PR2) database (version 4.7.2) [23]. When taxonomic ranks were too high (above the genus level), representative sequences of each OTU were directly compared with the NCBI nonredundant database using the BLASTn tool. After quality checking, data corresponding to 25 sampling dates were removed from the dataset (total number of reads very low compared to that of other dates). Most of these dates corresponded to a period between 2014 and 2015. In total, the final dataset consisted of 24,795 eukaryotic OTUs (14,356,643 reads) from 163 sampling dates over the period 2009–2016. Read abundances of OTUs for which sequences were 100% similar to those of Minidiscus and Thalassiosira strains isolated from our survey were retrieved from this dataset using the BLASTn tool on Geneious 9.1.3.
Isolations of viruses and morphological features of virions and infected diatoms
In order to detect the presence of viruses infecting nanoplanktonic diatoms at the SOMLIT-Astan monitoring station, we used the established cultures of Minidiscus spp. and Thalassiosira spp. (Table 1, diatoms highlighted in bold from May 2015) both to amplify and isolate potential lytic biological agents (for details, see Arsenieff et al. [37]). Briefly, pre-filtered (150 µm) natural seawater was enriched with F/2 medium and fresh diatom cultures, and incubated for 2 weeks under the hosts culture conditions described above. These enrichment cultures were clarified by successive filtrations (GF/F filters, Whatman and 0.22 μm PES filter, Whatman) and 0.5 mL of the 0.22 µm filtered aliquots were added to 1.5 mL host cultures in 24-multiwell plates. Cultures were inspected by light microscopy 2 weeks after inoculation. When algal lysis was observed, three extinction dilution cycles were carried out to clone the pathogens [38]. New filtrations on 0.22 μm were repeated (at least three times) to verify the transferability of the putative clonal virus isolates. Viral prospection was conducted using samples collected between the end of September 2015 and October 2016.
The morphological features of several viral isolates were determined by transmission electronic microscopy (TEM). Fresh viral lysates were filtered through 0.22 μm pore size filter and concentrated by ultrafiltration (Vivaspin 50 kDa, Sartorius). Concentrated viral suspensions were negative stained for 40 s using uranyl acetate (2% w/v) on a copper grid and observed with a JEOL-JEM 1400 electron microscope (JEOL Ltd., Tokyo, Japan) operating at 80 kV. Appropriate controls (uninfected hosts) were also examined by TEM. To inspect the replication site of these viruses within their hosts, ultrathin sections of exponentially growing culture of M. comicus inoculated with a virus strain were examined (see Arsenieff et al. [37]) for the detailed protocol). Briefly, after fixation, diatoms were embedded in Spurr’s epoxy resin and thin sections were obtained. Sections were mounted on microscopic grids stained with 0.4% uranyl acetate and studied by TEM.
Accession numbers
Sequences obtained from the eukaryotic nuclear rRNA/ITS were deposited in the NCBI database: strains RCC4660-RCC5887 (MN528601–MN528659) for the 18S, strains RCC4660-RCC4664 (MN528660–MN528670) for the 28S and strains RCC4660-RCC5887 (MN528671–MN528718) for the ITS gene marker. 18S sequences of the six diatom species are being submitted to the PR2 reference database (version 4.13.0 in preparation).
Results
Morphogenetic characterization of nanodiatoms of the genera Minidiscus and Thalassiosira from SOMLIT-Astan (WEC)
Three species of Minidiscus (M. comicus, M. spinulatus, and M. variabilis) and three nanoplanktonic Thalassiosira species (T. curviseriata, T. cf. profunda, and Thalassiosira sp.) were identified based on the morphogenetic characterization of 11 strains isolated in May 2015 from the SOMLIT-Astan time-series station (Table 1).
Minidiscus comicus
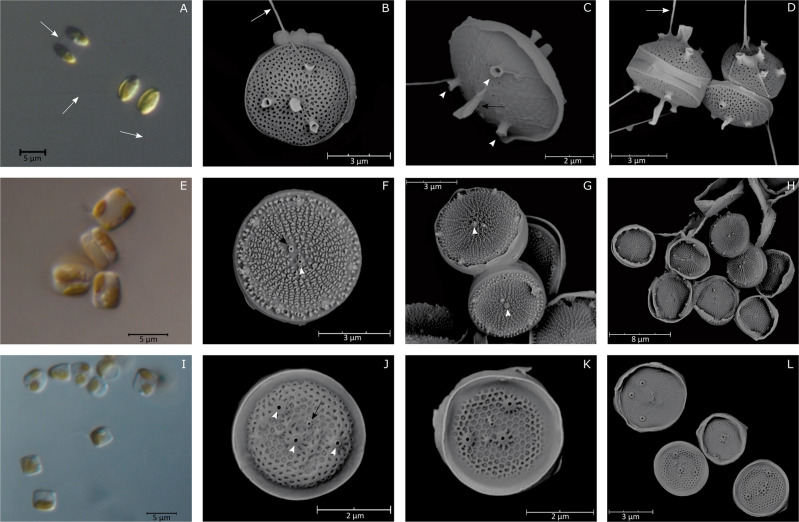
Morphological features of RCC4660, RCC4661, and RCC4662 were identical to that of M. comicus [12, 15]. Cells were solitary or aggregated in pairs (Fig. 1a, b) and had an ovoid shape when observed in girdle view (Fig. 1a, c). The valve was circular and 4.8 ± 0.6 µm (n = 71) in diameter (Fig. 1b). The domed valve face was covered by areolae (Fig. 1b, d). A long and central rimoportula was surrounded by 3–4 fultoportulae (five fultoportulae in few specimens). External tubes of the fultoportulae were shorter than that of the rimoportula. Excretion of mucilaginous threads by the fultoportulae allowed connections between cells (Fig. 1a, d).
Fig. 1. LM and SEM micrographs of Minidiscus species.
M. comicus. a Pairs of cells connected by mucilaginous threads (LM). b, c, d External views of solitary cells (SEM). M. spinulatus. e Aggregated and solitary cells (LM). f, g, h External valve views. Note the Y-shaped ribs and the fultoportulae ring on the margin (SEM). M. variabilis. i Solitary cells (LM). j, k, l External view of valves. White arrows: threads connecting cells. Black arrows: Rimoportula. White arrowheads: Fultoportulae.
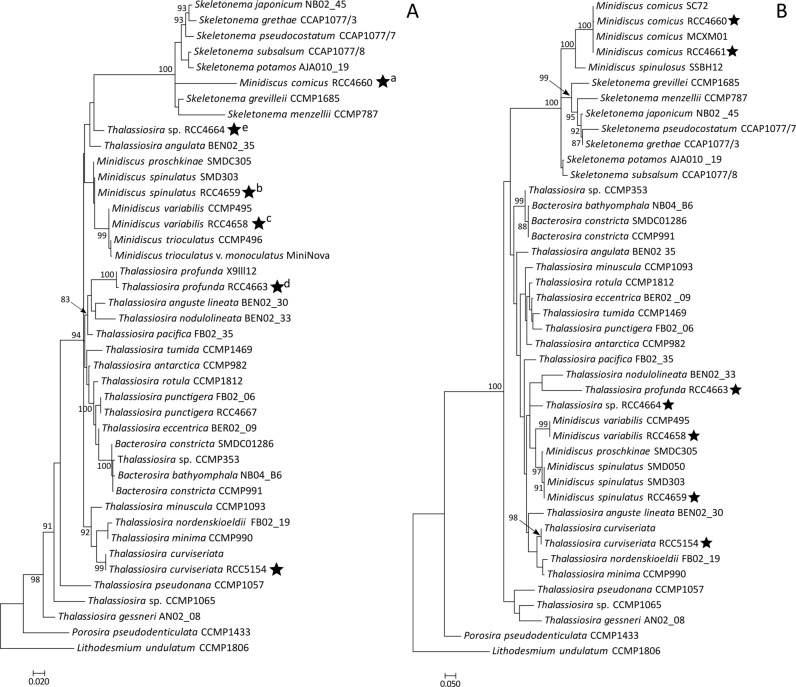
In our phylogenetic analyses of the 18S, partial 28S and concatenated 18S + 28S sequences, the strains RCC4660, RCC4661 and RCC4662 (100% identical 18S sequences) appeared as a distinct branch in a clade that contained ribosomal sequences of Skeletonema (Figs. 3a, b, S1). The partial 28S rRNA gene sequences of these three M. comicus isolates (sequence similarity 99.9%) gathered with M. comicus SC72 and MCXM01 with a high bootstrap (100% value). They formed, together with a sequence belonging to M. spinulosus SSBH12, a highly supported sub-clade (100% bootstrap value) in a clade that included sequences of the genus Skeletonema (Fig. 3b).
Fig. 3. Phylogenetic position of dominant nanodiatoms isolated in the Western English Channel.
Phylogenetic rooted tree based on the 18S (a) and partial 28S (b) sequences of diatoms from the Thalassiosirales order. Porosira pseudodenticulata and Lithodesmium undulatum were taken as outgroups. The black stars indicate the positions of strains for which morphological characterizations were achieved in the frame of this study. Both Maximum Likelihood trees were generated using PhyML 3.0 with 1 000 replicates and a GTR + G + I substitution model according to the SMS analyses. Bootstrap values (%) greater than 80 are shown. Scale bars indicate the number of substitutions per site. Letters in superscript indicate that several strains had identical sequences. a: Minidiscus comicus strains RCC4660, RCC4661, RCC4662, and RCC5839 to RCC5859. b: Minidiscus spinulatus strains RCC4659, RCC5860 and RCC5861. c: Minidiscus variabilis strains RCC4657, RCC4658, RCC4665, RCC4666, and RCC5862 to RCC5880. d: Thalassiosira cf. profunda strains RCC4663 and RCC5881 to RCC5886. e: Thalassiosira sp. RCC4664 and RCC5887.
Minidiscus spinulatus
Morphological features of RCC4649 were identical to those of M. spinulatus [39]. Cells were solitary or aggregated in colonies of 2–3 cells (Fig. 1e). As described in Park et al. [39], the valve face was flat, with a circular shape and was 5.3 ± 0.4 µm (n = 16) in diameter (Fig. 1f). On the valve face, true areolae were absent and were replaced by granules or Y-shaped ribs according to the degree of silicification of the frustule (Fig. 1f–h). A sub-central rimoportula with an ellipsoidal shape was adjacent to a central fultoportula. External tube of the fultoportula was short and surrounded by a hyaline flange. On the valve margin, the cell possessed a ring of 5–8 fultoportulae (Fig. 1f–h).
In phylogenies reconstructed based on the 18 S rRNA, partial 28 S rRNA sequences and on the concatenation of both genes, RCC4659 clustered with two other strains of M. spinulatus (91% bootstrap value) and were closely related to sequences of the species M. proschkinae (97% bootstrap value) and M. variabilis (Figs. 3b, S1).
Minidiscus variabilis
Cells of strains RCC4657, RCC4758, RCC4665, and RCC4666 were solitary and had a cylindrical shape when observed in girdle view (Fig. 1i). The valve (3.4 ± 0.3 µm in diameter, n = 63) presented a sub-central and small rimoportula and a varying number of fultoportulae (2–4 and very rarely 5). For most cells, one fultoportula was located in the central region of the valve while the others were dispersed throughout the valve face. The valves were encircled by a wide and marginal hyaline flange (Fig. 1j–l). Each fultoportula had a ring of silica at the base of the external tube. For the 4 strains examined, the majority of the cells had a tangential-linear areolation, a typical feature of M. trioculatus [13] but few specimens (2 valves out of 147 examined) showed a radial areolation, a typical feature of M. variabilis [13] (Fig. 1l).
The 18S rRNA gene sequences of strains RCC4657, RCC4658, RCC4665, RCC4666 showed 100% of identity with the sequence of M. variabilis CCMP495 and differed from sequences of M. trioculatus at two sites. To be more precise, in the sequence of M. variabilis CCMP495, an ambiguous base (Y) was recorded at a site for which our sequences of M. variabilis and the published sequences of M. trioculatus had a C. Similarly, the 28S rRNA gene sequences of the four strains (that were 100% identical) were also identical to that of M. variabilis CCMP495.
Thalassiosira curviseriata
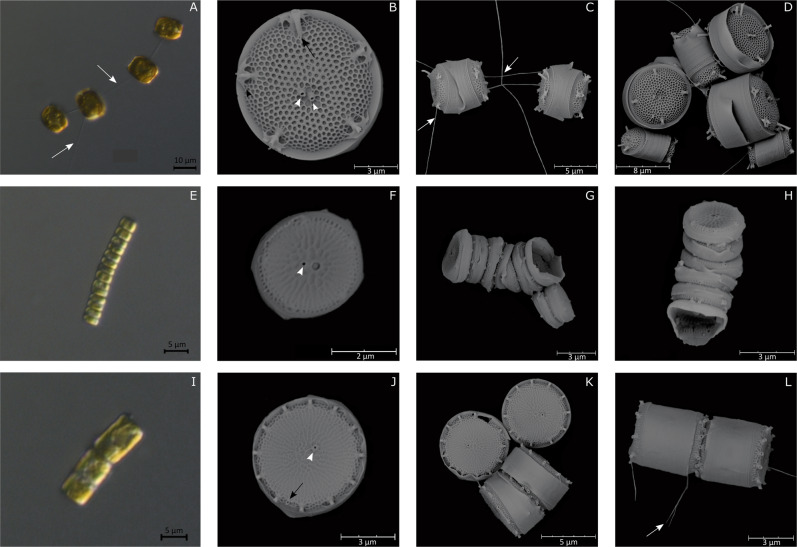
The morphological features of strain RCC5154 fitted with that described by Takano [15] for Thalassiosira curviseriata. Cells (6.7 ± 2.4 µm in diameter, n = 9) were connected in chains by threads (Fig. 2a, c). As described in Takano [15] the circular valve possessed a radial areolation and was encircled by a hyaline mantle. One or two fultoportulae were present in the central region while a ring of 3–5 fultoportulae was disposed on the margin of the valve face. Each marginal fultoportula had two conspicuous wings. A unique rimoportula having a long external process was located close to a marginal fultoportula (Fig. 2b). A considerable variability in cell size and ornamentation was observed in culture conditions (Fig. 2d).
Fig. 2. LM and SEM micrographs of Thalassiosira species.
T. curviseriata. a Chain of cells connected by threads (LM). b External valve view of solitary cells (SEM). Black arrowhead indicates the winged fultoportulae. c Girdle view of short chain of cells. d External view of large and small cells of T. curviseriata (SEM). T. cf. profunda. e Long chain of cells (LM). f Solitary cell in a valve view. Note the large areola adjacent to the central fultoportula (SEM). g, h Girdle views of chains (SEM). Thalassiosira sp. i Chain of cells (LM). j Valve external view of a solitary cell (SEM). k, l External views of solitary cells and of cells associated in pair (SEM). White arrows: threads connecting cells. Black arrows: Rimoportula. White arrowheads: Fultoportulae.
Ribosomal DNA gene sequences of T. curviseriata RCC5154 clustered with other published sequences of Thalassiosira curviseriata in the 18S and 28S rRNA gene phylogenetic trees (99% and 98% bootstrap values respectively) (Fig. 3a, b).
Thalassiosira cf. profunda
The morphological features of strain RCC4663 fitted with that of Thalassiosira profunda [40, 41]. Cells were tightly associated to form long chains and appeared rectangular in girdle view (Fig. 2e). As in Park et al. [41], valve face was flat, 3–3.3 µm (n = 2) in diameter and was covered by radial lines of areolae. A ring of marginal areolae was also present (Fig. 2f). In the central region, a single fultoportula was adjacent to a large areola (F and H). A ring of fultoportulae was disposed on the margin (Fig. 2f–h). No rimoportula could be observed.
The 18S rRNA gene sequence of T. cf. profunda RCC4663 was 99% similar to that of Thalassiosira profunda X9lll12. The two sequences formed a highly supported clade (100% bootstrap support) that emerged in a clade containing T. anguste lineata, T. nodulolineata, and T. pacifica (Fig. 3a).
Thalassiosira sp.
Strain RCC4664 had morphological features that fitted with that of the genus Thalassiosira [42]. Valves of the cylindrical cells (5.6 ± 0.7 µm, n = 16) that were associated in chains (Fig. 2i, l) possessed a sub central fultoportula and one ring of 8–15 marginal fultoportulae (about 1.1 µm apart) with conspicuous external processes. A rimoportula with a short external tube was located midway between two marginal fultoportulae (Fig. 2j–l). Valves had a radial areolation, with 40–45 areolae in 10 µm on valve face and 63–78 areolae in 10 µm on valve mantle.
Our phylogenetic analyses did not provide any additional information on the affiliation of this strain to known species in the Thalassiosirales radiation (Figs. 3a, b, S1).
Prevalence and seasonal dynamics of nanoplanktonic diatoms over the 2009–2016 period
In order to estimate the contribution of the nanoplanktonic diatoms described in this study to the diatom assemblage at the SOMLIT-Astan station, we used two strategies. The first one consisted of analyzing the eukaryotic metabarcoding data obtained for this site over the period 2009–2016 (in situ approach). More precisely we investigated the temporal dynamics of OTUs of which sequences corresponded to those of the Minidiscus spp. and Thalassiosira spp. cited above. Our second strategy was to conduct isolations and genetic characterizations of diatom strains along a full seasonal cycle (October 2015 to October 2016) (culture approach).
In situ approach
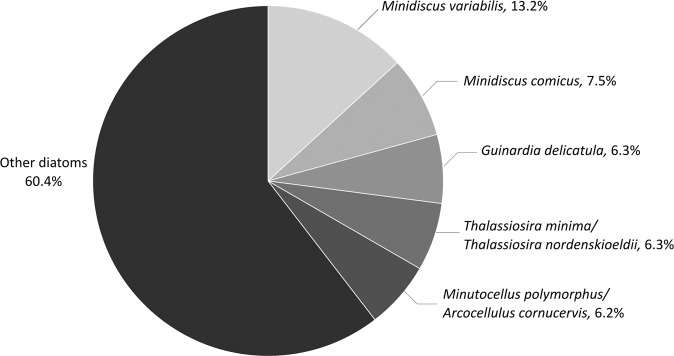
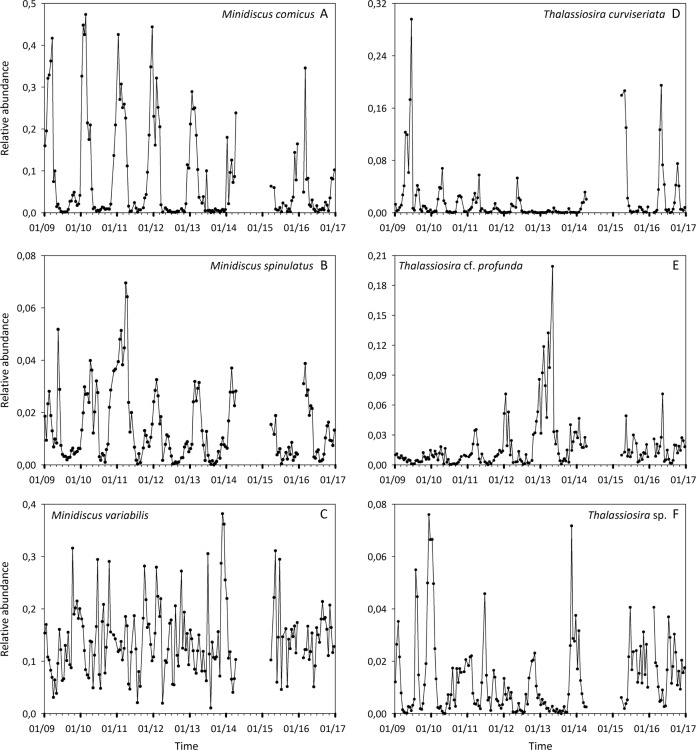
Sequences assigned to the Bacillariophyta accounted for 16.55% of the 14,356,643 eukaryotic reads and 6.46% of the 24,795 eukaryotic OTUs obtained using metabarcoding for the period 2009–2016 at the SOMLIT-Astan time-series. Exact matches of the V4–18S sequences of the Minidiscus and Thalassiosira strains isolated from our time-series were searched in this metabarcoding dataset. OTUs with sequences 100% similar to those of M. variabilis and M. comicus ranked first and second in read abundance when considering all diatom reads (13.2% and 7.5% of diatom reads, respectively) (Fig. 4). The OTUs assigned at 100% to M. spinulatus, T. curviseriata, and Thalassiosira sp. that we isolated contributed to 1.3%, 1.9%, and 1.2%, respectively, of the total diatom reads. No exact match of the T. cf. profunda RCC4663 sequence was retrieved from our metabarcoding dataset. However, a sequence that was 99.7% similar fell in the T. profunda clade in our phylogenetic analyses based upon V4 18S rRNA gene region. This sequence accounted for 1.6% of all diatom reads.
Fig. 4. Relative contributions of the five most abundant OTUs related to Bacillariophyta at the SOMLIT-Astan station (2009–2016).
Taxonomic assignations were based on comparisons with the PR2 or NCBI databases and with the reference diatom strains described in this study.
The relative abundance (to total number of diatom reads) of M. variabilis appeared highly variable at the SOMLIT-Astan station over the studied period and no clear seasonal pattern was detected (Fig. 5c). However, variations in relative abundance of all other studied OTUs that matched with the Minidiscus strains isolated showed clear seasonal patterns. Relative abundances of reads related to M. comicus peaked during winter, generally from January to March, while very low values were recorded during the rest of the year (Fig. 5a). M. spinulatus seemed to develop from autumn to early spring while lower relative abundances were recorded during summer (Fig. 5b).
Fig. 5. Dynamics of the nanodiatoms isolated in the Western English Channel.
Variations in relative abundances of the OTUs related to a. Minidiscus comicus, b Minidiscus spinulatus, c Minidiscus variabilis, d Thalassiosira curviseriata, e Thalassiosira cf. profunda, and f Thalassiosira sp. at the SOMLIT-Astan station during the period 2009–2016. Note that metabarcoding data corresponding to 25 sampling dates (mainly between 2014 and 2015) were removed from the dataset (see “Material and methods”).
According to molecular analyses, T. curviseriata development occurred mainly during spring (late March to early June) (Fig. 5d) while relative abundances of the OTU related to T. cf. profunda peaked principally during winter (Fig. 5e). The seasonal signal of the OTU related to Thalassiosira sp. RCC4664 was less clear. Peaks of relative abundance were generally recorded during winter (January–February) and during summer (end of June-early July) (Fig. 5f).
Overall, all species demonstrated interannual fluctuations in the amplitude of their seasonal peaks. For example, relative abundance of T. curviseriata was relatively low throughout the year 2013 (no spring peak) while exceptionally high values were recorded in 2009 and April 2015 and 2016 (Fig. 5d). Similarly, the OTU related to T. cf. profunda reached exceptionally high reads relative abundances in early spring 2013 (Fig. 5e).
Culture approach
The temporal patterns described above rely on the correct assignment of OTUs to the targeted species, i.e. on the assumption that the chosen barcode is sufficiently variable to distinguish between species. This assumption appears to be true for the studied nanoplanktonic diatoms. However, a second strategy developed within this study consisted of culturing dominant nanodiatoms and sequencing their full 18S rRNA gene and ITS spacer. This strategy was adopted to confirm that the three species of Minidiscus and the three species of Thalassiosira that we identified were present at the sampling station, at least during the periods of read abundance peaks of the corresponding OTUs. It also allowed us to study the intra-specific genetic variability of each species over a seasonal cycle. In total, 48 new nanoplanktonic isolates were obtained, from which the 18S rRNA gene and ITS spacer sequences were analyzed and compared to those of the fully characterized species recorded at SOMLIT-Astan (Table 1).
With this strategy, isolates of the three Minidiscus species as well as of Thalassiosira cf. profunda and Thalassiosira sp. RCC4664 were obtained (Fig. 6). Nineteen new isolates whose 18S rRNA gene sequences were 100% similar to those of M. variabilis RCC4657, RCC4658, RCC4665 and RCC4666 as well as to that of strain CCMP495 were obtained (Table 1, Fig. 3a). Cells were isolated throughout the seasonal cycle, corroborating the dynamics of OTU read abundances (Fig. 6a, b). The 17 ITS sequences obtained for isolates of M. variabilis showed very low variability: only two strains demonstrated one variable nucleotide in the 522 bp alignment. Isolates for which the 18S rRNA gene sequences matched 100% with that of M. comicus RCC4660, RCC4661 and RCC4662 (20 isolates) were obtained between end of November 2015 and May 2016 (Table 1, Figs. 3a, 6b). Similarly, two strains with the 18S sequences matching 100% with that of M. spinulatus RCC4659 were isolated in March 2016 (Table 1, Figs. 3a, 6b). These periods corresponded to blooms of the corresponding species, as suggested by the metabarcoding data (Fig. 6a). The M. comicus strains isolated from our SOMLIT-Astan sampling station (RCC4660, RCC4661, RCC4662, and RCC5839 to RCC5859) showed 99.6% identity between ITS sequences (540 bp), with only two divergent nucleotide sites. ITS sequences of M. spinulatus RCC4659, RCC5860, and RCC5861 were also highly similar (97.6% identity, 537 bp alignment).
Fig. 6. Seasonal dynamics of nanodiatoms and viruses isolated in the Western English Channel.
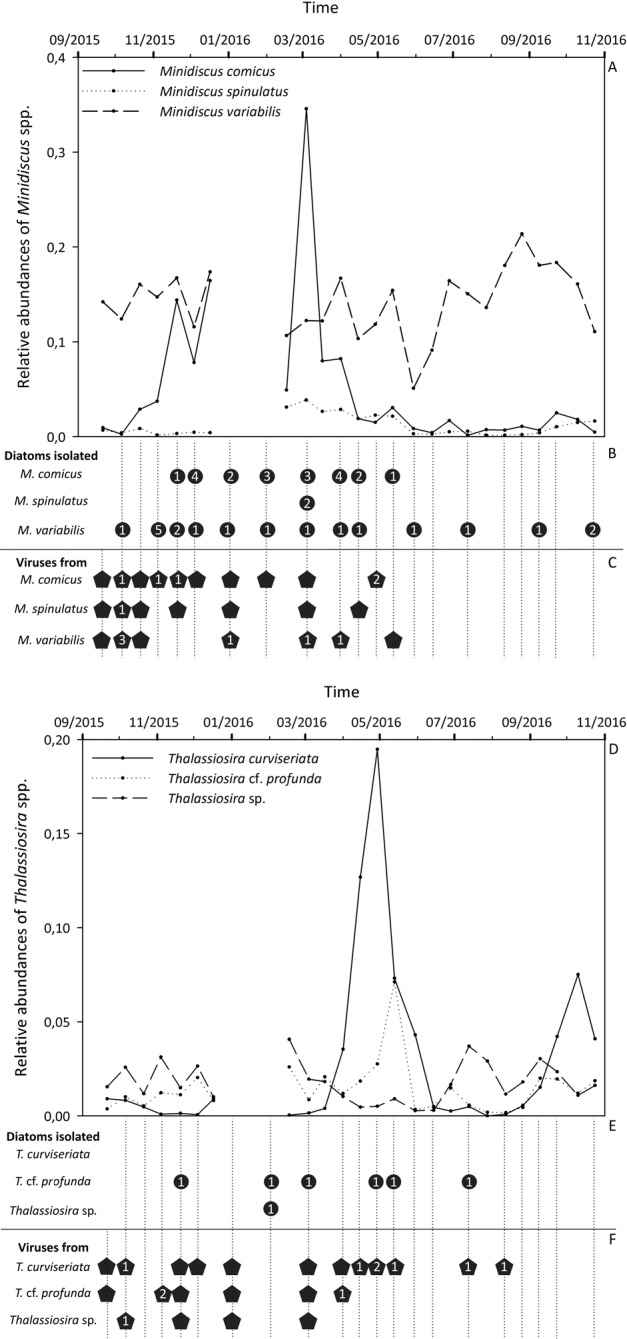
a, d Variations in the read relative abundances of the OTUs related to Minidiscus species (upper panel, a) and to Thalassiosira species (lower panel, d) at the SOMLIT-Astan station during the 2015–2016 period. b, e Respectively, Minidiscus and Thalassiosira isolates obtained during the period of our study. The number of isolated strains is indicated for each species and for each sampling date. c, f Virus isolates obtained respectively from Minidiscus and Thalassiosira cultures during the studied period. The success in the isolation procedure is indicated by pentagons while numbers indicate the number of viral strains still maintained in the laboratory (several strains were lost a few months after isolation). In b, c, e, and f, vertical dashed lines correspond to dates for which dilution series were carried out.
Regarding the genus Thalassiosira, six isolates of T. cf. profunda and one isolate of Thalassiosira sp. RCC4664 were obtained while no culture of T. curviseriata could be established between October 2015 and October 2016 (Fig. 6d, e). The 18S rRNA gene sequences of the six isolates of T. cf. profunda were 100% identical. They were isolated from winter, spring and early July, when OTU related to this species was detected in the metabarcoding dataset (Fig. 6d, e). ITS sequences analysis of strains RCC4663 and RCC5883 to RCC5886 revealed 95.7% of identity in the 571 bp alignment. The culture related to Thalassiosira sp. RCC4664 (18S sequences 100% identical) was isolated in February 2016, a period for which no metabarcoding data are available (Fig. 6d, e). The 614 bp ITS alignment of Thalassiosira sp. RCC4664 and RCC5887 indicated a rather high divergence with only 91.9% of identity, mainly due to an insertion of 46 nucleotides in the ITS sequence from RCC4664.
Virus isolations along the seasonal cycle at the SOMLIT-Astan station
Minidiscus viruses were successfully isolated from our time-series sampling site from the end of September 2015 to May 2016, which roughly corresponded to the blooming periods of M. comicus and M. spinulatus (Fig. 6c). Interestingly, potential viruses of M. variabilis could not be isolated after spring 2016 even if this host species remained abundant throughout the sampling period. Viruses of T. cf. profunda and Thalassiosira sp. were isolated between September 2015 and April 2016 (Fig. 6f) while viruses of T. curviseriata were isolated during the whole seasonal cycle.
All virions examined using TEM appeared to be untailed and showed hexagonal outlines. Viruses isolated from samples collected in October 2015 on M. comicus RCC4662, M. spinulatus RCC4659, M. variabilis RCC4658 and collected in September 2015 on T. cf. profunda RCC4663 and T. curviseriata RCC5154 displayed diameters of 31.8 ± 1.5 nm (n = 39), 30.5 ± 1 nm (n = 51), 26.7 ± 1.5 nm (n = 57), 37.5 ± 2 nm (n = 7), and 37.8 ± 2 nm (n = 43) respectively (Fig. 7a). No virus like particles were observed in the host controls. Thin sections of infected M. comicus showed a clear accumulation of viral particles within its cytoplasm 72 h post-inoculation (Fig. 7b), which suggested that these viruses were likely single-stranded RNA viruses [43].
Fig. 7. TEM micrographs of viruses and infected diatom.
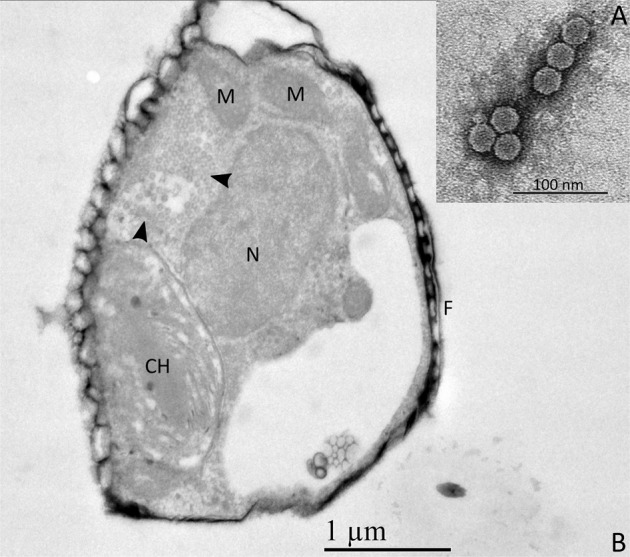
a Negatively stained particles of the viral strain isolated in October 2015 on M. spinulatus RCC4659. As all virions displayed similar morphological features, micrographs of the other viral strains are not shown. b Ultrathin section of M. comicus infected by its associated virus. Arrowheads: viral particles accumulated in the host cytoplasm, F frustule, M mitochondrion, N nucleus, CH chloroplast.
Discussion
Taxonomy and phylogeny of nanoplanktonic diatoms from French coasts of the Western English Channel
Thalassiosirales are important components of phytoplankton communities in the Western English Channel and North Sea [6, 8, 44] and nanoplanktonic species of this group have been recorded from these regions. This study revealed the occurrence of three species of Minidiscus (M. comicus, M. spinulatus and M. variabilis) as well as of three nanoplanktonic species of Thalassiosira (T. curviseriata, T. cf. profunda, and a possibly new species) at SOMLIT-Astan, representative of the WEC waters.
Within the Thalassiosirales, the phylogenetic classification of genera is still under construction. Both new genera and emended descriptions of genera have been published over the last fifteen years [39, 45–50]. Minidiscus is an example of genus for which the description has been recently emended [39]. Originally described to include Thalassiosirales with a rimoportula distant from the margin and non-marginal fultoportulae, this genus is now distinguished from other Thalassiosirales mainly by the size of its valve (<10 µm) and both the position and structure of the rimoportula [39, 51]. However, this genus appears polyphyletic with two clades. M. proschkinae, M. spinulatus and M. variabilis form a monophyletic clade in multigene phylogenies ([39] and this study), while M. comicus and M. spinulosus group together in a separate branch affiliated to species of the genus Skeletonema in phylogenies based on sequences of the partial 28S rRNA gene ([52] and this study).
In our study, a set of strains were assigned to species after examination of valves morphology using SEM. Comparisons of ribosomal sequences to published references served to confirm our conclusions. However, some inconsistencies were recovered between the morphology and genetic sequences of the strains that we assigned to the species M. variabilis (18S sequences 100% similar to that of M. variabilis but areolation pattern of most valves examined similar to that of M. trioculatus). We cannot rule out that our four cultures could contain two lineages (corresponding to the two species M. variabilis and M. trioculatus) that would originate from a pool of cells instead of a single cell sorted using flow cytometry. However, this is improbable given that the inconsistency between morphology and genetic characters was encountered for all examined strains and since the M. trioculatus 18S genotype was not recovered from any of the 49 Minidiscus isolates that we obtained from our time-series station in the frame of this study (while that of M. variabilis was obtained for 23 strains). We thus suggest that the areolation pattern (tangential-linear or radial) should not be used for the distinction of the species M. trioculatus and M. variabilis until more analyses of the morphological and genetic intraspecific variations are conducted. Concerning M. comicus, the morphological and genetic features of the isolated strains corroborated published records [12, 15, 52]. Our phylogenetic analyses, using both the 18S and 28S genes (only 28S gene sequences were available in public database prior to this study), confirmed the position of M. comicus as a sister species of M. spinulosus in a branch within the Skeletonema radiation. This suggests that M. comicus and M. spinulosus should be transferred to the genus Skeletonema. But before proceeding to these taxonomic changes, careful examinations of morphological and genetic features of these Minidiscus species and of all other species (including M. chilensis Rivera, M. decoratus Chrétiennot-Dinet and Quiroga, M. ocellatus Gao, Cheng and Chin, M. subtilis Gao, Cheng and Chin and M. vodyanitskiyi Lyakh and Bedoshvili) are needed to better understand the evolution and further clarify the taxonomy of this important genus. Some of these species may have emerged in different lineages and have evolved convergent morphological features linked to miniaturization.
A few species of the genus Thalassiosira, with sizes in the same range as those of the genus Minidiscus (<10 µm), have been described (for example T. mala Takano, T. profunda (Hendey) Hasle and T. exigua Fryxell and Hasle). Assignation of the isolated strains to the species T. curviseriata and T. cf. profunda was achieved based on the analysis of morphological features of the valve and confirmed by genetic characters. However, to our knowledge, the morphological features and sequences of Thalassiosira sp. RCC4664 and RCC5887 did not match with those of any described species. Thus, we suspect that these strains correspond to a new species or a species not yet sequenced.
These results, and the fact that all nanoplanktonic diatoms identified in the frame of this study are new records for the Atlantic French coasts of the Western English Channel, point to an undersampling and consideration of nanophytoplankton in this region. Given the global importance of nanoplanktonic diatoms in the oceans [9, 19], these new data, and in particular the new sequences that were generated, will help to refine our understanding of the global nanodiatom distribution and to bridge the gap between laboratory identification and environmental studies. To this end, and in order to preserve this important collection for a long-term period, cryopreservation tests were performed for both nanodiatom and virus strains (see supplemental information) and cultures are now available for the scientific community from the RCC.
Nanodiatoms dominate the diatom community at SOMLIT-Astan
The diversity and the seasonal variations of microphytoplanktonic diatoms have been well described in the Western English Channel and North Sea [6, 53–55]. At the SOMLIT-Astan station, Guinardia (especially G. delicatula) and Paralia sp. appear to be key taxa, becoming dominant in spring/summer and winter respectively [6]. Exploration of metabarcoding data provides a more thorough insight into species diversity including small sized organisms that are usually overlooked using traditional microscopy observations. One of the major findings of this study was that nanodiatoms, and more specifically Minidiscus species, largely dominate the diatom community. According to our analyzes of the 8 year metabarcoding survey M. variabilis and M. comicus were dominant species, given the contribution of their read abundances which reached almost 21% of the total diatom reads during the 2009–2016 period. The reads contribution attributed to M. variabilis alone even exceeded G. delicatula read abundance by twofold (see Fig. 4). This minute species ranked in the top five most abundant phytoplankton species at SOMLIT-Astan. Although the contribution of M. spinulatus and nanoplanktonic species of Thalassiosira were less important, they still ranked amongst the top 20 major diatom OTUs. Present results support the hypothesis that nano-sized diatoms are major contributors to the phytoplankton community in temperate coastal waters [9, 11, 13] and emphasize the importance of better understanding their ecology and impacts in nature.
The 8 year monitoring of the six species indicated distinct periods of occurrence. M. comicus, M. spinulatus, and T. cf. profunda formed transient blooms during winter, T. curviseriata during springtime, and Thalassiosira sp. RCC4664 usually peaked twice a year in winter and in summer. Interestingly, M. comicus in the Western English Channel do not develop spring-summer blooms as observed in the Mediterranean Sea [9, 11]. While these species exhibited clear seasonal patterns, M. variabilis persisted year round, with important abundance fluctuations. Few numbers of Thalassiosira strains were isolated between 2015 and 2016 but their ITS analysis demonstrated an important intraspecific variability. Conversely, the genetic characterization of Minidiscus strains isolated between 2015 and 2016 suggested a relatively low intraspecific diversity during the occurrence periods of the studied species based on 18S and ITS sequencing. Genetic markers such as 5.8S + ITS-2 were proposed to be more appropriate to depict diatom diversity [56, 57], especially for species that belong to the Thalassiosirales [58].
Drivers of nanodiatom dynamics: toward a biotic control?
The distinct patterns in species occurrence (bloom forming vs. persistent) and the interannual variability in bloom amplitudes raise the question of whether nanodiatom species are regulated differently. The observed dynamics in read relative abundance most likely result from variable processes affecting either cell growth or cell losses via diverse mechanisms (for example, sedimentation, grazing, programmed cell death, internal clock, or infection by parasites). For decades, researchers have mainly focused their efforts on elucidating the roles of abiotic factors in controlling diatom growth, which are nowadays well established (e.g., [59–63]). The dominance of M. variabilis over the other species throughout the year however, suggests a broad ability to respond to natural environmental variability and thereby a weak influence of physicochemical parameters. Conversely, the marked seasonal development of M. comicus, and of the other nanoplanktonic diatoms, may reflect adaptations to seasonal variations of environmental factors.
Besides the environmental aspect, biotic control is also likely involved in the regulation of nanodiatoms. Pathogens have been described as important mortality agents that may control the dynamics of diatom populations [64–66]. Among them, diatom viruses were reported and were mainly isolated from Chaetoceros species [43]. For the first time, our study provides evidence of viral pathogens that infect the genera Minidiscus and Thalassiosira. A collection of putative viruses was established and preliminary characterization indicates that they possess morphological features similar to those of ssDNA and ssRNA diatom viruses [43, 67]. It is however more likely that our viral strains contain RNA genomes as they accumulate in the cytoplasm of their hosts. We hope you will accept this modification. The period of successful virus isolation (fall to spring) approximately corresponded to the blooming periods of the prospective hosts, except for T. curviseriata for which viruses could be isolated year round. This result may partly explain why we failed to isolate and maintain cultures of T. curviseriata due to a simultaneous isolation of the host and its viruses. More detailed functional and genetic analyses are needed to fully characterize the viral strains that we isolated and study their interplay with nanodiatom species in nature. Yet, our results suggest that virus-driven mortality is involved in the control of the nanodiatom species development.
Concluding remarks
Recently, nanoplanktonic diatoms were proposed as major contributors to phytoplanktonic blooms in coastal as well as offshore regions. Their global ecological significance however, is severely limited by the lack of genetic references and isolates in culture. In this respect, establishing a nanodiatom reference collection was prerequisite for taking a census of these minute organisms and advancing our understanding of their ecology and impacts in nature. Using classic morphological and molecular approaches, our study provides new insights in the diversity and dynamics of relevant nanoplanktonic diatoms (especially Minidiscus) in the Western English Channel. Owing to their prominence in the French coastal waters of the Western English Channel, nanoplanktonic diatoms have undoubtedly important ecological and biogeochemical implications. Persistence and seasonality patterns of Minidiscus and Thalassiosira raise questions about the parameters which contribute to their proliferation and decline. Our study suggests that viruses certainly contribute to the control of these tiny diatoms. Given the global significance of the nanodiatoms, the substantial collection of organisms that were brought into culture should provide biological models of interest in ecological, biogeochemical, and evolutionary studies.
Supplementary information
Acknowledgements
The authors would like to thank the crew of the Neomysis ship for their help during sampling at the SOMLIT-Astan station. We are also grateful to the RCC for the diatom strains provided, to Sarah Romac for her assistance with molecular biology and to Sophie Lepanse for TEM analysis. Christophe Lejeusne who provided advice on phylogenetic analyses, and Mariarita Caracciolo for stimulating discussion, are acknowledged. Lydia White is thanked for her English proofreading. We thank the three anonymous reviewers for their comments on the manuscript.
Funding
This study was supported by PhD fellowships from the Université Pierre et Marie Curie (Sorbonne Université) and the Région Bretagne (ARED), the ANR CALYPSO (ANR-15-CE01-0009) and the CNRS-INSU EC2CO CYCLOBS project.
Author contributions
LA, FLG, ACB, and NS designed the study. LA, FRJ, FLG, ACB, and NS sampled onboard and isolated the nanodiatoms and/or viruses. LA, FLG, and FRJ performed the molecular analyses on the nanodiatoms and/or environmental samples and analyzed the results. LA, FLG, and DS conducted the diatom morphological characterization. FM was in charge of the metabarcoding bioinformatics analyses. LG was in charge of the cryopreservation. LA, ACB, and NS wrote the manuscript. All authors revised the manuscript and approved the final version.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Laure Arsenieff, Florence Le Gall
Contributor Information
Laure Arsenieff, Email: larsenieff@sb-roscoff.fr.
Nathalie Simon, Email: simon@sb-roscoff.fr.
Supplementary information
The online version of this article (10.1038/s41396-020-0659-6) contains supplementary material, which is available to authorized users.
References
- 1.Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob Biogeochem Cycles. 1995;9:359–72. [Google Scholar]
- 2.Leblanc K, Arístegui J, Armand L, Assmy P, Beker B, Bode A, et al. A global diatom database—abundance, biovolume and biomass in the world ocean. Earth Syst Sci Data Discuss. 2012;4:147–85. [Google Scholar]
- 3.Smetacek V. Diatoms and the ocean carbon cycle. Protist. 1999;150:25–32. doi: 10.1016/S1434-4610(99)70006-4. [DOI] [PubMed] [Google Scholar]
- 4.Tréguer P, Bowler C, Moriceau B, Dutkiewicz S, Gehlen M, Aumont O, et al. Influence of diatom diversity on the ocean biological carbon pump. Nat Geosci. 2018;11:27–37. [Google Scholar]
- 5.Gómez F, Souissi S. Unusual diatoms linked to climatic events in the northeastern English Channel. J Sea Res. 2007;58:283–90. [Google Scholar]
- 6.Guilloux L, Rigaut-Jalabert F, Jouenne F, Ristori S, Viprey M, Not F, et al. An annotated checklist of marine phytoplankton taxa at the SOMLIT-Astan time series off Roscoff (Western English Channel, France): data collected from 2000 to 2010. Cah Biol Mar. 2013;54:247–56. [Google Scholar]
- 7.Schlüter MH, Kraberg A, Wiltshire KH. Long-term changes in the seasonality of selected diatoms related to grazers and environmental conditions. J Sea Res. 2012;67:91–7. [Google Scholar]
- 8.Widdicombe CE, Eloire D, Harbour D, Harris RP, Somerfield PJ. Long-term phytoplankton community dynamics in the Western English Channel. J Plankton Res. 2010;32:643–55. [Google Scholar]
- 9.Leblanc K, Quéguiner B, Diaz F, Cornet V, Michel-Rodriguez M, Durrieu de Madron X, et al. Nanoplanktonic diatoms are globally overlooked but play a role in spring blooms and carbon export. Nat Commun. 2018;9:953. doi: 10.1038/s41467-018-03376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Percopo I, Siano R, Cerino F, Sarno D, Zingone A. Phytoplankton diversity during the spring bloom in the northwestern Mediterranean Sea. Bot Mar. 2011;54:243–67. [Google Scholar]
- 11.Ribera d’Alcalà M, Conversano F, Corato F, Licandro P, Mangoni O, Marino D, et al. Seasonal patterns in plankton communities in a pluriannual time series at a coastal Mediterranean site (Gulf of Naples): an attempt to discern recurrences and trends. Sci Mar. 2004;68(S1):65–83. [Google Scholar]
- 12.Jewson D, Kuwata A, Cros L, Fortuno JM, Estrada M. Morphological adaptations to small size in the marine diatom Minidiscus comicus. Sci Mar. 2016;80S1:89–96. [Google Scholar]
- 13.Kaczmarska I, Lovejoy C, Potvin M, Macgillivary M. Morphological and molecular characteristics of selected species of Minidiscus (Bacillariophyta, Thalassiosiraceae) Eur J Phycol. 2009;44:461–75. [Google Scholar]
- 14.Quiroga I, Chretiennot Dinet MJ. A new species of Minidiscus (Diatomophyceae, Thalassiosiraceae) from the eastern English Channel, France. Bot Mar. 2004;47:341–8. [Google Scholar]
- 15.Takano H. New and rare diatoms from Japanese marine waters—VI. Three new species in Thalassiosiraceae. Bull Tokai Reg Fish Res Lab. 1981;105:31–43. [Google Scholar]
- 16.Aké-Castillo JA, Hernandez-Becerril DU, Meave del Castillo ME, Bravo-Sierra E. Species of Minidiscus (Bacillariophyceae) in the Mexican Pacific Ocean. Cryptogam Algol. 2001;22:101–7. [Google Scholar]
- 17.Daniels CJ, Poulton AJ, Esposito M, Paulsen ML, Bellerby R,St, John M, et al. Phytoplankton dynamics in contrasting early stage North Atlantic spring blooms: Composition, succession, and potential drivers. Biogeosciences. 2015;12:2395–409. [Google Scholar]
- 18.Zingone A, Sarno D, Siano R, Marino D. The importance and distinctiveness of small-sized phytoplankton in the Magellan Straits. Polar Biol. 2011;34:1269–84. [Google Scholar]
- 19.Malviya S, Scalco E, Audic S, Vincent F, Veluchamy A, Poulain J, et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc Natl Acad Sci USA. 2016;113:1516–25. doi: 10.1073/pnas.1509523113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang JS, Kang SH, Kim D, Kim DY. Planktonic centric diatom Minidiscus chilensis dominated sediment trap material in eastern Bransfield Strait, Antarctica. Mar Ecol Prog Ser. 2003;255:93–9. [Google Scholar]
- 21.Guiry MD, Guiry GM AlgaeBase [Internet]. World-wide electronic publication, National University of Ireland, Galway. 2018. http://www.algaebase.org.
- 22.Buck KR, Chavez FP, Davis AS. Minidiscus trioculatus, a small diatom with a large presence in the upwelling system of central California. Nov Hedwig, Beih. 2008;133:1–6. [Google Scholar]
- 23.Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marie D, Le Gall F, Edern R, Gourvil P, Vaulot D. Improvement of phytoplankton culture isolation using single cell sorting by flow cytometry. J Phycol. 2017;53:271–82. doi: 10.1111/jpy.12495. [DOI] [PubMed] [Google Scholar]
- 25.Keller MD, Seluin RC, Claus W, Guillard RRL. Media for the culture of oceanic ultraphytoplankton. J Phycol. 1987;23:633–8. [Google Scholar]
- 26.Lepere C, Demura M, Kawachi M, Romac S, Probert I, Vaulot D. Whole-genome amplification (WGA) of marine photosynthetic eukaryote populations. FEMS Microbiol Ecol. 2011;76:513–23. doi: 10.1111/j.1574-6941.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- 27.Moon-Van Der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–10. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 28.Lenaers G, Maroteaux L, Michot B, Herzog M. Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. J Mol Evol. 1989;29:40–51. doi: 10.1007/BF02106180. [DOI] [PubMed] [Google Scholar]
- 29.Orsini L, Sarno D, Procaccini G, Poletti R, Dahlmann J, Montresor M. Toxic Pseudo-nitzschia multistriata (Bacillariophyceae) from the Gulf of Naples: morphology, toxin analysis and phylogenetic relationships with other Pseudo-nitzschia species. Eur J Phycol. 2002;37:247–57. [Google Scholar]
- 30.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017;1–7. [DOI] [PMC free article] [PubMed]
- 31.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 32.Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017;34:2422–4. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caracciolo M, Rigaut-Jalabert F, Romac S, Henry N, Mahe F, Chaffron S, et al. SOMLIT-Astan time-series: a morphogenetic approach to study the seasonal succession of the eukaryotic marine plankton communities. 2020. (In press).
- 35.Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner H-W, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 36.Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arsenieff L, Simon N, Rigaut-Jalabert F, Le Gall F, Chaffron S, Corre E, et al. First viruses infecting the marine diatom Guinardia delicatula. Front Microbiol. 2019;9(Jan 9):3235. doi: 10.3389/fmicb.2018.03235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suttle CA. Enumeration and isolation of viruses. In: Kemp PF, Sherr BF, Sherr EB, Cole JJ, editors. Handbook of methods in aquatic microbial ecology. Boca Raton: Lewis Publisher; 1993. p. 121–37.
- 39.Park JS, Jung SW, Ki JS, Guo R, Kim HJ, Lee KW, et al. Transfer of the small diatoms Thalassiosira proschkinae and T. spinulata to the genus Minidiscus and their taxonomic re-description. PLoS One. 2017;12:1–20. doi: 10.1371/journal.pone.0181980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallegraeff GM. Species of the diatom genus Thalassiosira in Australian waters. Bot Mar. 1984;27:495–513. [Google Scholar]
- 41.Park JS, Jung SW, Lee SD, Yun SM, Lee JH. Species diversity of the genus Thalassiosira (Thalassiosirales, Bacillariophyta) in South Korea and its biogeographical distribution in the world. Phycologia. 2016;55:403–23. [Google Scholar]
- 42.Hasle GR. Some Thalassiosira species with one central process (Bacillariophyceae) Nor J Bot. 1978;25:77–110. [Google Scholar]
- 43.Tomaru Y, Toyoda K, Kimura K. Marine diatom viruses and their hosts: resistance mechanisms and population dynamics. Perspect Phycol. 2015;2:69–81. [Google Scholar]
- 44.Hoppenrath M, Beszteri B, Drebes G, Halliger H, Van Beusekom JEE, Janisch S, et al. Thalassiosira species (Bacillariophyceae, Thalassiosirales) in the North Sea at Helgoland (German Bight) and Sylt (North Frisian Wadden Sea)—a first approach to assessing diversity. Eur J Phycol. 2007;42:271–88. [Google Scholar]
- 45.Park JS, Alverson AJ, Lee JH. A phylogenetic re-definition of the diatom genus Bacterosira (Thalassiosirales, Bacillariophyta), with the transfer of Thalassiosira constricta based on morphological and molecular characters. Phytotaxa. 2016;245:1–16. [Google Scholar]
- 46.Alverson AJ, Beszteri B, Julius ML, Theriot EC. The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella. BMC Evol Biol. 2011;11:125. doi: 10.1186/1471-2148-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alverson AJ, Cannone JJ, Gutell RR, Theriot EC. The evolution of elongate shape in diatoms. J Phycol. 2006;42:655–68. [Google Scholar]
- 48.Stachura-Suchoples K, Williams DM. Description of Conticribra tricircularis, a new genus and species of Thalassiosirales, with a discussion on its relationship to other continuous cribra species of Thalassiosira Cleve (Bacillariophyta) and its freshwater origin. Eur J Phycol. 2009;44:477–86. [Google Scholar]
- 49.Alverson AJ. Molecular systematics and the diatom species. Protist. 2008;159:339–53. doi: 10.1016/j.protis.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaczmarska I, Beaton M, Benoit AC, Medlin LK. Molecular phylogeny of selected members of the order Thalassiosirales (Bacillariophyta) and evolution of the fultoportula. J Phycol. 2005;42:121–38. doi: 10.1111/j.1529-8817.2008.00522.x. [DOI] [PubMed] [Google Scholar]
- 51.Hasle GR. Some marine plankton genera of the diatom family Thalassiosiraceae. Hedwig Beih. 1974;45:1–49. [Google Scholar]
- 52.Gu H, Zhang X, Sun J, Luo Z. Diversity and seasonal occurrence of Skeletonema (Bacillariophyta) species in Xiamen Harbour and surrounding seas, China. Cryptogam Algol. 2012;33:245–63. [Google Scholar]
- 53.Grall JR. Développement “printanier” de la Diatomée Rhizosolenia delicatula près de Roscoff. Mar Biol. 1972;16:41–8. [Google Scholar]
- 54.Martin-Jezequel V. Facteurs hydrologiques et phytoplancton en Baie de Morlaix (Manche Occidentale) Hydrobiologia. 1983;102:131–43. [Google Scholar]
- 55.Jacques G. Variations saisonnières des populations phytoplanctoniques de la région de Roscoff (1962–1963). Thèse Troisième Cycle, Université de Paris, 1963, 88pp.
- 56.Moniz MBJ, Kaczmarska I. Barcoding diatoms: is there a good marker? Mol Ecol Resour. 2009;9:65–74. doi: 10.1111/j.1755-0998.2009.02633.x. [DOI] [PubMed] [Google Scholar]
- 57.Moniz MBJ, Kaczmarska I. Barcoding of diatoms: nuclear encoded ITS revisited. Protis. 2010;161:7–34. doi: 10.1016/j.protis.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Guo L, Sui Z, Zhang S, Ren Y, Liu Y. Comparison of potential diatom ‘barcode’ genes (The 18S rRNA gene and ITS, COI, rbcL) and their effectiveness in discriminating and determining species taxonomy in the Bacillariophyta. Int J Syst Evol Microbiol. 2015;65:1369–80. doi: 10.1099/ijs.0.000076. [DOI] [PubMed] [Google Scholar]
- 59.Sarthou G, Timmermans KR, Blain S, Tréguer P. Growth physiology and fate of diatoms in the ocean: a review. J Sea Res. 2005;53:25–42. [Google Scholar]
- 60.Litchman E, Klausmeier CA. Trait-based community ecology of phytoplankton. Annu Rev Ecol Evol Syst. 2008;39:615–39. [Google Scholar]
- 61.Bowler C, Vardi A, Allen AE. Oceanographic and biogeochemical insights from diatom genomes. Ann Rev Mar Sci. 2010;2:333–65. doi: 10.1146/annurev-marine-120308-081051. [DOI] [PubMed] [Google Scholar]
- 62.Balzano S, Sarno D, Kooistra WHCF. Effects of salinity on the growth rate and morphology of ten Skeletonema strains. J Plankton Res. 2011;33:937–45. [Google Scholar]
- 63.Bidle KD. The molecular ecophysiology of programmed cell death in marine phytoplankton. Ann Rev Mar Sci. 2015;7:341–75. doi: 10.1146/annurev-marine-010213-135014. [DOI] [PubMed] [Google Scholar]
- 64.Gleason FH, Jephcott TG, Küpper FC, Gerphagnon M, Sime-Ngando T, Karpov SA, et al. Potential roles for recently discovered chytrid parasites in the dynamics of harmful algal blooms. Fungal Biol Rev. 2015;29:20–33. [Google Scholar]
- 65.Peacock EE, Olson RJ, Sosik HM. Parasitic infection of the diatom Guinardia delicatula, a recurrent and ecologically important phenomenon on the New England Shelf. Mar Ecol Prog Ser. 2014;503:1–10. [Google Scholar]
- 66.Gutiérrez MH, Jara AM, Pantoja S. Fungal parasites infect marine diatoms in the upwelling ecosystem of the Humboldt Current System off central Chile. Environ Microbiol. 2016;18:1646–53. doi: 10.1111/1462-2920.13257. [DOI] [PubMed] [Google Scholar]
- 67.King AMQ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, et al. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the International Committee on Taxonomy of Viruses (2018) Arch Virol. 2018;163:2601. doi: 10.1007/s00705-018-3847-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.