Abstract
Species of Gnomoniaceae are commonly associated with leaf spot diseases of a wide range of plant hosts worldwide. During our investigation of fungi associated with tree diseases in China, several gnomoniaceous isolates were recovered from symptomatic branches and leaves on different woody plants in the Fagaceae, Pinaceae, and Salicaceae families. These isolates were studied by applying a polyphasic approach including morphological, cultural data, and phylogenetic analyses of partial ITS, LSU, tef1, rpb2 and tub2 gene sequences. As a result, three species were identified with characters fitting into the family Gnomoniaceae. One of these species is described herein as Cryphognomonia pinigen. et sp. nov., characterized by developed pseudostromata and ascospores with obvious hyaline sheath; Gnomoniopsis xunwuensissp. nov. is illustrated showing sympodially branched conidiophore, oval or fusiform conidia; and one known species, Plagiostoma populinum. The current study improves the understanding of gnomoniaceous species causing diebacks and leaf spot on ecological and economic forest trees.
Keywords: forest trees, Gnomoniaceae , new genus, phylogeny, systematics
Introduction
The Gnomoniaceae (Diaporthales, Sordariomycetes, Ascomycota) is a family of perithecial ascomycetes that occur as endophytes, pathogens, or saprobes on growing and overwintered leaves of hardwood trees, shrubs, and herbaceous plants (Walker 2012). Many species in the Gnomoniaceae cause serious tree diseases such as cherry leaf scorch (Apiognomonia erythrostoma (Pers.) Höhn.), oak dieback (A. errabunda (Roberge) Höhn), sycamore canker (A. veneta (Sacc. & Speg.) Höhn), and chestnut dieback (Gnomoniopsis daii Tian & Jiang) (Sogonov et al. 2008; Walker et al. 2010; Jiang et al. 2019).
The sexual morph of Gnomoniaceae is characterized by ascomata that are generally immersed, solitary or aggregated in an undeveloped stroma (Rossman et al. 2007; Sogonov et al. 2008). The perithecia are dark brown to black and pseudoparenchymatous with central, eccentric, or lateral necks (Rossman et al. 2007; Sogonov et al. 2008). Asci usually have an inconspicuous or distinct apical ring. Ascospores are generally small, hyaline, uniseptate. The asexual morph is characterized by acervular or pycnidial, phialidic, with non-septate conidia (Monod 1983).
The generic concepts of Gnomoniaceae were recently revised based on a survey of leaf-inhabiting diaporthalean fungi (Sogonov et al. 2008). Phylogenetic analyses of molecular markers is the primary methodology for systematic studies of the Gnomoniaceae, however, host specificity and morphology can also be useful for species identification. Recent phylogenetic studies have shown that species of Gnomoniaceae often have a narrow host range associating with a single host genus or species (Mejía et al. 2008, 2011a, b, 2012; Sogonov et al. 2008; Walker et al. 2010, 2012, 2013). For example, Cryptosporella is a well-defined genus which was frequently limited to a single host species, especially in the host family Betulaceae, except for C. wehmeyeriana on Tilia spp. and type species C. hypodermia on Ulmus spp. (Mejía et al. 2008, 2011b).
Several fungal species of Gnomoniaceae, Cryptosporella platyphylla from Betula platyphylla, Flavignomonia rhoigena from Rhus chinensis, Gnomoniopsis daii and Ophiognomonia castaneae from Castanea mollissima, have been reported from China (Fan et al. 2016; Gong et al. 2017; Jiang and Tian 2019; Jiang et al. 2019). In the present study, tree inhabiting gnomoniaceous species, mainly on cankered branches and leaves, were surveyed in China. The aim of the present study was to identify these fungi via morphology and multi-locus phylogeny based on modern taxonomic concepts.
Materials and methods
Isolates
Fresh specimens of Gnomoniaceae-related fungi were collected from branches and leaves of hosts in Beijing, Jiangxi and Shaanxi provinces (Tables 1–3). Isolates from host material were obtained by removing a mucoid spores mass from perithecia and pycnidia-like conidiomata, spreading the suspension on the surface of 1.8% potato dextrose agar (PDA), and incubating at 25 °C for up to 24 h. Single germinating conidia/ascospore was removed and plated on to fresh PDA plates. Specimens are deposited in the Museum of the Beijing Forestry University (BJFC). Axenic cultures are maintained in the China Forestry Culture Collection Centre (CFCC).
Table 1.
Strains and GenBank accession numbers used in the phylogenetic analyses of Gnomoniaceae
| Species | Strains | Genbank accession number | |||
|---|---|---|---|---|---|
| ITS | LSU | tef1 | rpb2 | ||
| Alnecium auctum | CBS 124263 | KF570154 | KF570154 | KF570200 | KF570170 |
| Ambarignomonia petiolorum | CBS 116866 | EU199193 | AY818963 | NA | EU199151 |
| CBS 121227 | EU254748 | EU255070 | EU221898 | EU219307 | |
| Amphiporthe tiliae | CBS 119289 | EU199178 | EU199122 | NA | EU199137 |
| Anisogramma anomala | 529478 | EU683064 | EU683066 | NA | NA |
| Anisogramma virgultorum | 529479 | EU683062 | EU683065 | NA | NA |
| Apiognomonia errabunda | AR 2813 | DQ313525 | NG027592 | DQ313565 | DQ862014 |
| Apiognomonia veneta | MFLUCC 16-1193 | MF190114 | MF190056 | NA | NA |
| Apioplagiostoma populi | 858501 | KP637024 | NA | NA | NA |
| Asteroma alneum | CBS 109840 | EU167609 | EU167609 | NA | NA |
| Asteroma sp. | Masuya 8Ah9-1 | NA | AB669035 | NA | NA |
| Cryphognomonia pini | CFCC 53020 | MK432672 | MK429915 | MK578144 | MK578100 |
| CFCC 53021 | MK432673 | MK429916 | MK578145 | MK578101 | |
| Cryptosporella hypodermia | CBS 116866 | EU199181 | AF408346 | NA | EU199140 |
| Discula destructiva | MD 254 | AF429741 | AF429721 | AF429732 | NA |
| Ditopella biseptata | MFLU 15-2661 | MF190147 | MF190091 | NA | MF377616 |
| Ditopella ditopa | CBS 109748 | DQ323526 | EU199126 | NA | EU199145 |
| Ditopellopsis sp. | CBS 121471 | EU254763 | EU255088 | EU221936 | EU219254 |
| Flavignomonia rhoigena | CFCC 53118 | MK432674 | MK429917 | NA | MK578102 |
| CFCC 53119 | MK432675 | MK429918 | NA | MK578103 | |
| CFCC 53120 | MK432676 | MK429919 | NA | MK578104 | |
| Gnomonia gnomon | CBS 199.53 | DQ491518 | AF408361 | EU221885 | EU219295 |
| CBS 829.79 | AY818957 | AY818964 | EU221905 | NA | |
| Gnomoniopsis alderdunensis | CBS 125680 | GU320825 | NA | NA | NA |
| Gnomoniopsis chamaemori | CBS 803.79 | EU254808 | EU255107 | NA | NA |
| Gnomoniopsis racemula | AR 3892 | EU254841 | EU255122 | EU221889 | EU219241 |
| Mamianiella coryli | BPI 877578 | EU254862 | NA | NA | NA |
| Marsupiomyces quercina | MFLUCC 13-0664 | MF190116 | MF190061 | NA | NA |
| Marsupiomyces epidermoidea | MFLU 15-2921 | NA | MF190058 | NA | NA |
| Melanconis marginalis | CBS 109744 | EU199197 | AF408373 | EU221991 | EU219301 |
| Neognomoniopsis quercina | CBS 145575 | MK876399 | MK876440 | NA | NA |
| Occultocarpon ailaoshanense | LCM 524.01 | JF779849 | JF779853 | NA | JF779856 |
| LCM 522.01 | JF779848 | JF779852 | JF779862 | JF779857 | |
| Ophiognomonia melanostyla | LCM 389.01 | JF779850 | JF779854 | NA | JF779858 |
| Ophiognomonia vasiljevae | AR 4298 | EU254977 | EU255162 | EU221999 | EU219331 |
| Plagiostoma aesculi | AR 3640 | EU254994 | EU255164 | NA | EU219269 |
| Linospora capreae | CBS 372.69 | NA | AF277143 | NA | NA |
| Pleuroceras oregonense | AR 4333 | EU255060 | EU255196 | EU221931 | EU219313 |
| Pleuroceras pleurostylum | CBS 906.79 | EU255061 | EU255197 | EU221962 | EU219311 |
| Phragmoporthe conformis | AR 3632 | NA | AF408377 | NA | NA |
| Valsalnicola oxystoma | AR 5137 | JX519561 | NA | NA | NA |
| AR 4833 | JX519559 | JX519563 | NA | NA | |
| Sirococcus tsugae | AR 4010 | EF512478 | EU255207 | EU221928 | EU219289 |
| CBS 119626 | EU199203 | EU199136 | EF512534 | EU199159 | |
| Tenuignomonia styracis | BPI 89278 | NA | LC379288 | LC379282 | LC379294 |
Note: NA, not applicable. Strains in this study are marked in bold.
Table 3.
Strains and GenBank accession numbers used in the phylogenetic analyses of Gnomoniopsis.
| Species | Strain | Genbank accession number | ||
| ITS | tef1 | tub2 | ||
| Apiognomonia errabunda | AR 4182 | DQ313543 | KJ509937 | KJ509947 |
| Plagiostoma aceris-palmati | CBS 137265 | KJ509959 | KJ509938 | KJ509949 |
| Plagiostoma aesculi | CBS 121905 | EU254994 | GU367022 | GU354005 |
| Plagiostoma amygdalinae | CBS 791.79 | EU254995 | GU367030 | GU354012 |
| Plagiostoma apiculatum | CBS 109775 | DQ323529 | GU367008 | GU353990 |
| CBS 126126 | GU367066 | GU367009 | GU353991 | |
| Plagiostoma barriae | LCM 601.01 | GU367054 | GU366997 | GU353980 |
| LCM 484.01 | GU367053 | GU366995 | GU353979 | |
| Plagiostoma convexum | CBS 123206 | EU255047 | EU219112 | GU353994 |
| Plagiostoma devexum | CBS 123201 | EU255001 | GU367027 | GU354010 |
| Plagiostoma dilatatum | LCM 403.02 | GU367069 | GU367012 | GU353995 |
| CBS 124976 | GU367070 | GU367014 | GU353996 | |
| Plagiostoma euphorbiaceum | CBS 816.79 | EU255003 | EU219158 | GU354013 |
| Plagiostoma euphorbiae | CBS 340.78 | DQ323532 | GU367034 | GU354016 |
| CBS 817.79 | KJ509960 | GU367028 | KJ509950 | |
| Plagiostoma exstocollum | CBS 127662 | GU367046 | GU366988 | GU353972 |
| LCM 422.01 | GU367043 | GU366989 | GU353969 | |
| Plagiostoma fraxini | CBS 121258 | EU255008 | KJ509939 | KJ509951 |
| CBS 109498 | AY455810 | GU367033 | GU354015 | |
| Plagiostoma geranii | CBS 824.79 | EU255009 | GU367032 | GU354014 |
| Plagiostoma imperceptibile | LCM 456.01 | GU367059 | GU367002 | GU353984 |
| Plagiostoma jonesii | MFLUCC 16–1189 | MF190159 | NA | MF377589 |
| Plagiostoma mejianum | CBS 137266 | KJ509961 | KJ509940 | KJ509952 |
| Plagiostoma oregonense | CBS 126124 | GU367073 | GU367016 | GU353999 |
| Plagiostoma ovalisporum | CBS 124977 | GU367072 | GU367015 | GU353998 |
| Plagiostoma petiolophilum | AR 3821 | EU255039 | GU367025 | GU354008 |
| CBS 126123 | GU367078 | GU367023 | GU354006 | |
| Plagiostoma populinum | CFCC 53016 | MK432677 | MK578070 | MK578146 |
| CFCC 53017 | MK432678 | MK578071 | MK578147 | |
| Plagiostoma populinum | CBS 174.58 | GU367074 | GU367017 | GU354000 |
| CBS 144.57 | GU367075 | GU367018 | GU354001 | |
| Plagiostoma pulchellum | CBS 170.69 | EU255043 | KJ509941 | GU353989 |
| CBS 126653 | GU367063 | GU367006 | GU353987 | |
| Plagiostoma rhododendri | CBS 847.79 | EU255044 | GU367026 | GU354009 |
| Plagiostoma robergeanum | CBS 121472 | EU255046 | GU367029 | GU354011 |
| Plagiostoma rubrosporum | CBS 137267 | KJ509962 | KJ509942 | KJ509953 |
| Plagiostoma salicellum | CBS 126121 | GU367037 | GU366977 | GU353961 |
| CBS 121466 | EU254996 | GU366978 | GU353962 | |
| Plagiostoma salicicola | MFLUCC 13–0656 | MF190161 | NA | NA |
| Plagiostoma samuelsii | CBS 125668 | GU367051 | GU366993 | GU353977 |
| LCM 596.01 | GU367052 | GU366994 | GU353978 | |
| Plagiostoma triseptatum | CBS 137268 | KJ509963 | KJ509943 | KJ509954 |
| Plagiostoma tsukubense | CBS 137269 | KJ509964 | KJ509944 | KJ509955 |
| CBS 137270 | KJ509965 | KJ509945 | KJ509956 | |
| Plagiostoma versatile | CBS 124978 | GU367038 | GU366979 | GU393963 |
| LCM 598.01 | GU367040 | GU366981 | GU393965 | |
| Plagiostoma yunnanense | LCM 513.02 | GU367036 | GU366976 | GU353960 |
| CBS 124979 | GU367035 | GU366975 | GU353959 | |
Note: NA, not applicable. Strains in this study are marked in bold.
Morphological analysis
Morphological observations of the asexual/sexual morph in the natural environment were based on features of the conidiomata or ascomata on infected plant tissues and micromorphology, supplemented by cultural characteristics. Ascomata and conidiomata from tree barks were sectioned by hand, using a double-edged blade and structures were observed under a dissecting microscope. The gross morphology of conidiomata or ascomata was recorded using a Leica stereomicroscope (M205 FA). Fungal structures were mounted in clear lactic acid and micromorphological characteristics were examined using a Leica compound microscope (DM 2500) with differential interference contrast (DIC) optics. Thirty measurements of each structure were determined for each collection. Colony characters and pigment production on PDA were noted after 10 d. Colony colors were described according to Rayner (1970).
DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from fresh mycelium grown on PDA using a cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1990). PCR amplifications were performed in a DNA Engine Peltier Thermal Cycler (PTC-200; Bio-Rad Laboratories, Hercules, CA, USA). The primer sets ITS1 and ITS4 (White et al. 1990) were used to amplify the ITS region. The primer sets LR0R and LR7 (Vilgalys and Hester 1990; Vilgalys and Sun 1994) were used to amplify the nuclear ribosomal large subunit (LSU) region. The primer sets EF1-728F (Carbone and Kohn 1999) and EF1-1567R (Rehner 2001) were used to amplify a partial fragment of the translation elongation factor 1-α gene (tef1-α). The primer sets RPB2-5F and fRPB2-7cR (Liu et al. 1999) were used to amplify the partial RNA polymerase II subunit (rpb2) region. The primer sets T1 (O’Donnell and Cigelnik 1997) and Bt2b (Glass and Donaldson 1995) were used to amplify the beta-tubulin gene (tub2). The PCR conditions were: an initial denaturation step of 5 min at 94 °C followed by 35 cycles of 30 sec at 94 °C, 50 sec at 48 °C (ITS, LSU) or 54 °C (tef-1α) or 55 °C (rpb2, tub2) and 1 min at 72 °C, and a final elongation step of 7 min at 72 °C. PCR amplification products were assayed via electrophoresis in 2% agarose gels. DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyser with a BigDye Terminater Kit v.3.1 (Inv-itrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
Phylogenetic analyses
The quality of our amplified nucleotide sequences was checked and combined by SeqMan v.7.1.0 and reference sequences were retrieved from the National Center for Biotechnology Information (NCBI), based on Mejía et al. (2011a), Senanayake et al. (2018), Jiang and Tian (2019), and Jiang et al. (2019), supplemented by sequences of Tenuignomonia styracis and Neognomoniopsis quercina from Crous et al. (2019) and Minoshima et al. (2019). Sequences were aligned using MAFFT v. 6 (Katoh and Toh 2010) and manually corrected using Bioedit 7.0.9.0 (Hall 1999).
The phylogenetical analyses were conducted using Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian inference (BI). MP was performed with PAUP v. 4.0b10 (Swofford 2003) using tree-bisection-reconnection (TBR) as the branch-swapping algorithm. Other calculated parsimony scores were tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency (RC). ML was performed with RAxML (Stamatakis 2006) as implemented in raxmlGUI 1.3 (Silvestro and Michalak 2012), using the ML + rapid bootstrap setting and the GTRGAMMA substitution model with 1000 bootstrap replicates. BI was performed using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v. 3.0b4 (Ronquist and Huelsenbeck 2003). Two MCMC chains, started from random trees for 1,000,000 generations and trees, were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25% of trees were discarded as burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BPP) were estimated in the remaining 7500 trees. Phylogenetic trees were viewed with FigTree v.1.4.3 (Rambaut 2016) and processed by Adobe Illustrator CS5. Alignment and trees were deposited in TreeBASE (submission ID: S26271). The nucleotide sequence data of the new taxa have been deposited in GenBank (Tables 1–3).
Results
Phylogenetic analyses
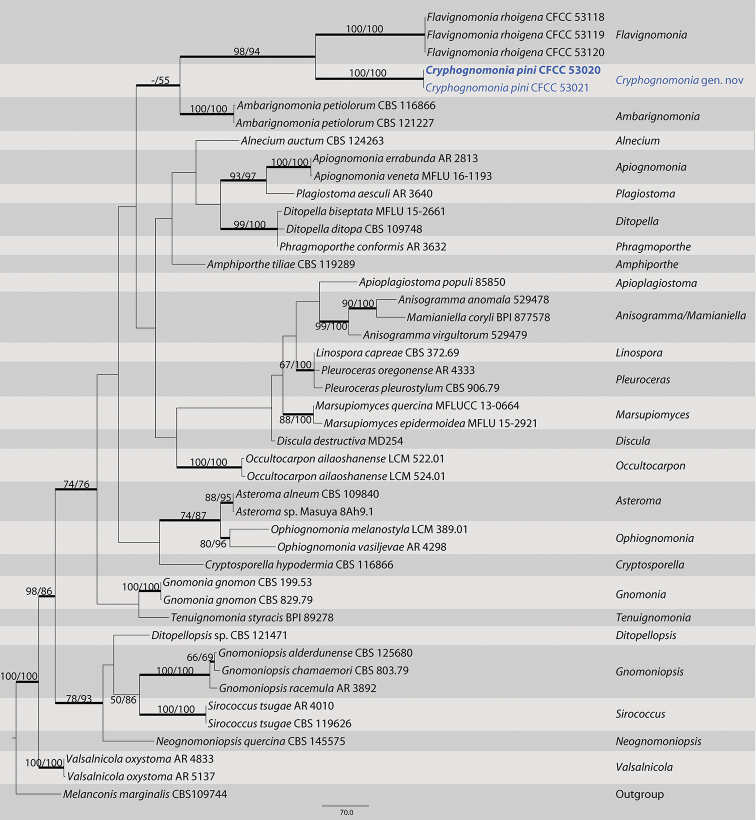
The first sequences dataset for the ITS, LSU, tef1, and rpb2 was analyzed to focus on Gnomoniaceae. The alignment included 45 taxa, including the outgroup sequences of Melanconis marginalis (Table 1). The aligned four-locus datasets included 3388 characters. Of these, 2180 characters were constant, 198 variable characters were par-simony-uninformative and 1010 characters were parsimony informative. The heuristic search using maximum parsimony (MP) generated 4 parsimonious trees (TL = 3241, CI = 0.539, RI = 0.672, RC = 0.362), from which one was selected (Fig. 1). In the phylogenetic tree, two strains form a well-supported clade (MP/ML/BI=100/100/1) sister to the species Flavignomonia rhoigena from Rhus chinensis.
Figure 1.
Maximum parsimony phylogram of Gnomoniaceae based on a combined matrix of ITS, LSU, tef1 and rpb2 genes. The MP and ML bootstrap support values above 50% are shown at the first and second position, respectively. Thickened branches represent posterior probabilities above 0.90 from BI. Scale bar: 80 nucleotide substitutions. Strains in this study are in blue and ex-type strains are in blod.
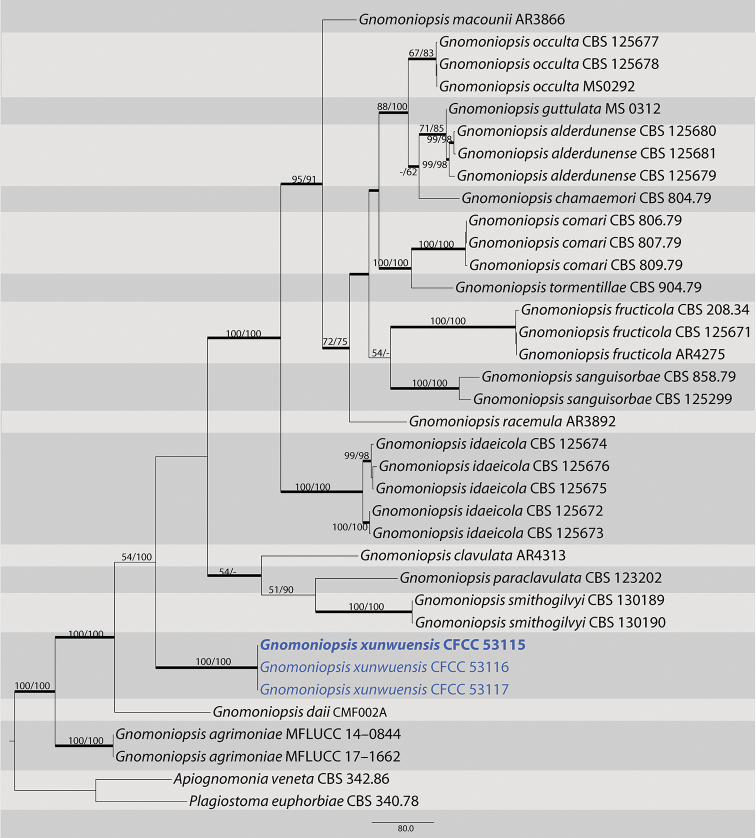
The second dataset with ITS, tef1 and tub2 sequences were analyzed in combination to infer the interspecific relationships within Gnomoniopsis. The alignment included 36 taxa, including the outgroup sequences of Apiognomonia veneta and Plagiostoma euphorbiae (Table 2). The aligned three-locus datasets included 2481 characters. Of these, 1443 characters were constant, 186 variable characters were par-simony-uninformative and 852 characters were parsimony informative. The heuristic search using maximum parsimony (MP) generated one parsimonious tree (TL = 2644, CI = 0.620, RI = 0.781, RC = 0.485), which is shown in Fig. 2. In the phylogenetic tree, three strains form a well-supported clade (MP/ML/BI=100/100/1) that does not include any previously described species.
Table 2.
Strains and GenBank accession numbers used in the phylogenetic analyses of Gnomoniopsis
| Species | Strain | Genbank accession number | ||
|---|---|---|---|---|
| ITS | tef1 | tub2 | ||
| Apiognomonia veneta | CBS 342.86 | DQ313531 | DQ318036 | EU219235 |
| Gnomoniopsis alderdunensis | CBS 125679 | GU320826 | GU320813 | GU320788 |
| CBS 125680 | GU320825 | GU320801 | GU320787 | |
| CBS 125681 | GU320827 | GU320802 | GU320789 | |
| Gnomoniopsis chamaemori | CBS 804.79 | GU320817 | GU320809 | GU320777 |
| Gnomoniopsis chinensis | CFCC 52286 | MG866032 | MH545370 | MH545366 |
| CFCC 52287 | MG866033 | MH545371 | MH545367 | |
| CFCC 52288 | MG866034 | MH545372 | MH545368 | |
| CFCC 52289 | MG866035 | MH545373 | MH545369 | |
| Gnomoniopsis clavulata | CBS 121255 | EU254818 | GU320807 | EU219211 |
| Gnomoniopsis comari | CBS 806.79 | EU254821 | GU320810 | EU219156 |
| CBS 807.79 | EU254822 | GU320814 | GU320779 | |
| CBS 809.79 | EU254823 | GU320794 | GU320778 | |
| Gnomoniopsis daii | CFCC 54043 | MN598671 | MN605519 | MN605517 |
| CMF002B | MN598672 | MN605520 | MN605518 | |
| Gnomoniopsis fructicola | CBS 121226 | EU254824 | GU320792 | EU219144 |
| CBS 208.34 | EU254826 | GU320808 | EU219149 | |
| CBS 125671 | GU320816 | GU320793 | GU320776 | |
| Gnomoniopsis guttulata | MS 0312 | EU254812 | NA | NA |
| Gnomoniopsis idaeicola | CBS 125672 | GU320823 | GU320797 | GU320781 |
| CBS 125673 | GU320824 | GU320798 | GU320782 | |
| CBS 125674 | GU320820 | GU320796 | GU320780 | |
| CBS 125675 | GU320822 | GU320799 | GU320783 | |
| CBS 125676 | GU320821 | GU320811 | GU320784 | |
| Gnomoniopsis macounii | CBS 121468 | EU254762 | GU320804 | EU219126 |
| Gnomoniopsis occulta | CBS 125677 | GU320828 | GU320812 | GU320785 |
| CBS 125678 | GU320829 | GU320800 | GU320786 | |
| Gnomoniopsis paraclavulata | CBS 123202 | GU320830 | GU320815 | GU320775 |
| Gnomoniopsis racemula | CBS 121469 | EU254841 | GU320803 | EU219125 |
| Gnomoniopsis sanguisorbae | CBS 858.79 | GU320818 | GU320805 | GU320790 |
| Gnomoniopsis smithogilvyi | CBS 130190 | JQ910642 | KR072534 | JQ910639 |
| CBS 130189 | JQ910644 | KR072535 | JQ910641 | |
| CBS 130188 | JQ910643 | KR072536 | JQ910640 | |
| MUT 401 | HM142946 | KR072537 | KR072532 | |
| MUT 411 | HM142948 | KR072538 | KR072533 | |
| Gnomoniopsis tormentillae | CBS 904.79 | EU254856 | GU320795 | EU219165 |
| Gnomoniopsis xunwuensis | CFCC 53115 | MK432667 | MK578067 | MK578141 |
| CFCC 53116 | MK432668 | MK578068 | MK578142 | |
| CFCC 53117 | MK432669 | MK578069 | MK578143 | |
| Plagiostoma euphorbiae | CBS 340.78 | DQ323532 | GU354016 | GU367034 |
Note: NA, not applicable. Strains in this study are marked in bold.
Figure 2.
Maximum parsimony phylogram of Gnomoniosis based on a combined matrix of ITS, tef1-α and tub2 genes. The MP and ML bootstrap support values above 50% are shown at the first and second position, respectively. Thickened branches represent posterior probabilities above 0.90 from BI. Scale bar: 80 nucleotide substitutions. Strains in this study are in blue and ex-type strains are in blod.
The third dataset with ITS, tef1 and tub2 sequences were analyzed in combination to infer the interspecific relationships within Plagiostoma. The alignment included 48 taxa, including the outgroup sequences of Apiognomonia errabunda (Table 3). The aligned three-locus datasets included 2311 characters. Of these, 1556 characters were constant, 204 variable characters were parsimony-uninformative and 551 characters were parsimony informative. The heuristic search using maximum parsimony (MP) generated 6 parsimonious trees (TL = 1462, CI = 0.685, RI = 0.779, RC = 0.534), from which one was selected (Fig. 3). In the phylogenetic tree, four strains from this study group in a well-supported clade with Plagiostoma populinum. The topologies resulting from MP, ML and BI analyses of the concatenated dataset were congruent.
Figure 3.
Maximum parsimony phylogram of Plagiostoma based on a combined matrix of ITS, tef1-α and tub2 genes. The MP and ML bootstrap support values above 50% are shown at the first and second position, respectively. Thickened branches represent posterior probabilities above 0.90 from BI. Scale bar: 30 nucleotide substitutions. Strains in this study are in blue.
Taxonomy
Cryphognomonia
C.M. Tian & N. Jiang gen. nov.
C9CCE66F-0558-5DBF-BB0E-433C484F1A90
829509
Etymology.
Crypho + gnomonia, referring to the cryptic stromata on hosts.
Type species.
Cryphognomonia pini C.M. Tian & N. Jiang
Description.
Pseudostromata erumpent, causing a pustulate bark surface. Central column yellowish to brownish. Stromatic zones lacking. Perithecia conspicuous, flask-shaped to spherical, umber to fuscous black, regularly scattered. Paraphyses deliquescent. Asci fusoid, 8-spored, biseriate, with an apical ring. Ascospores hyaline, clavate to cylindrical, smooth, multi-guttulate, symmetrical to asymmetrical, straight to slightly curved, bicellular, with a median septum distinctly constricted, with distinct hyaline sheath. Asexual morph: not observed.
Notes.
Cryphognomonia was classified as a new genus in Gnomoniaceae throughout molecular data and the characteristics of sexual morph. Morphologically, Cryphognomonia can be distinguished from the other genera by pseudostromata and ascospores with obvious hyaline sheath.
Cryphognomonia pini
C.M. Tian & N. Jiang sp. nov.
DDB2516A-8F2F-5985-8344-22CAEA479CB2
829510
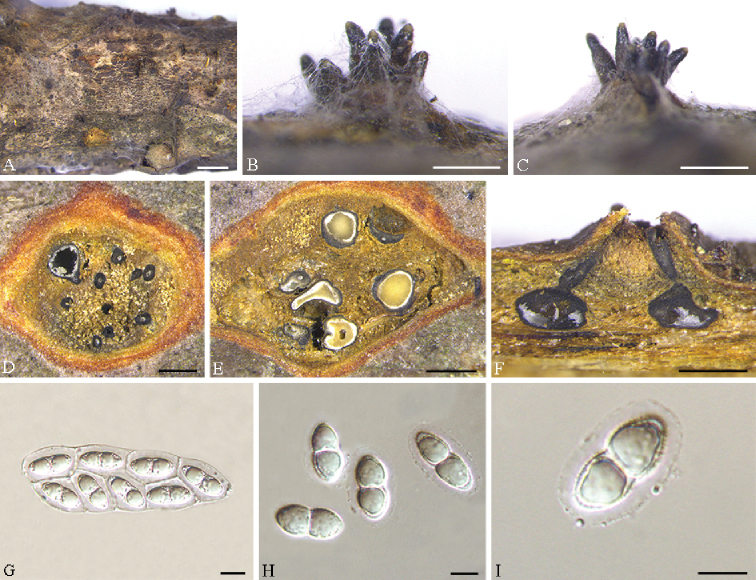
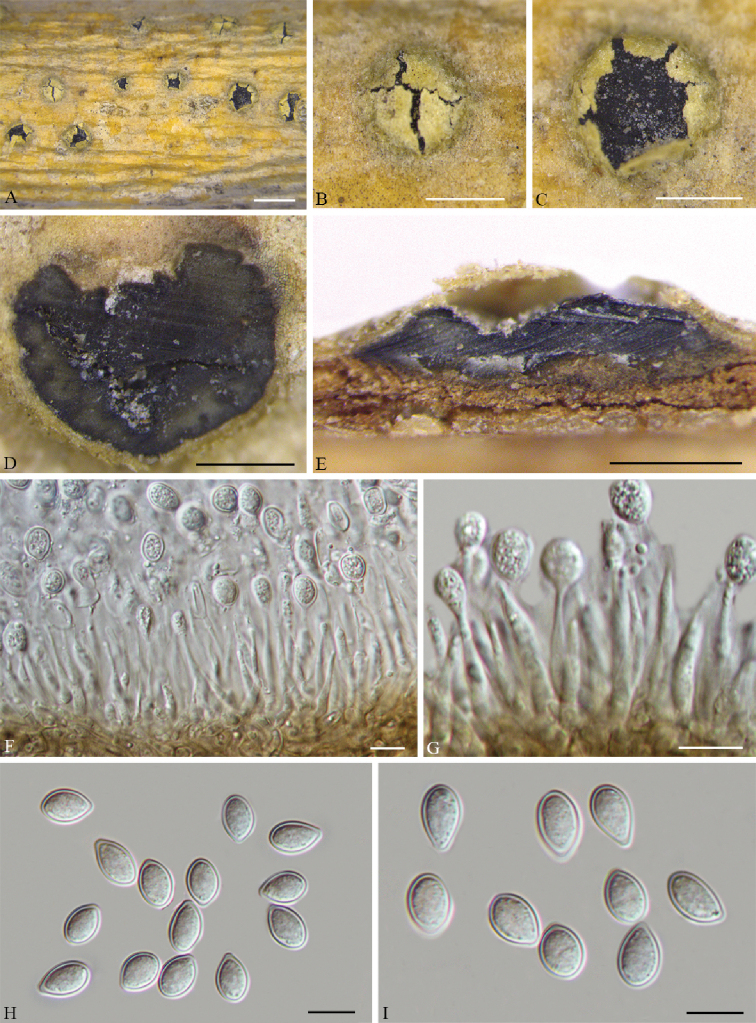
Figure 4.
Cryphognomonia pini on Pinus armandii (BJFC-S1725) A–C habit of ascomata on twigs D, E transverse section of ascomata F longitudinal section through ascomata G asci H, I ascospores. Scale bars: 2 mm (A); 500 μm (B–F); 10 μm (G–I).
Diagnosis.
Cryphognomonia pini differs from its closest phylogenetic neighbor, F. rhoigena, in ITS, LSU, tef1 and rpb2 loci based on the alignments deposited in TreeBASE.
Etymology.
Named after the genus of the host plant from which the holotype was collected, Pinus.
Description.
Pseudostromata erumpent, causing a pustulate bark surface, 650–1200 µm diam., containing up to 12 perithecia. Central column yellowish to brownish. Stromatic zones lacking. Perithecia conspicuous, flask-shaped to spherical, umber to fuscous black, regularly scattered, 350–600 µm diam. Paraphyses deliquescent. Asci fusoid, 8-spored, biseriate, with an apical ring, (60–)65–80(–90) × (21–)22–31(–35) µm. Ascospores hyaline, clavate to cylindrical, smooth, multi-guttulate, symmetrical to asymmetrical, straight to slightly curved, bicellular, with a median septum distinctly constricted, with distinct hyaline sheath, (15.5–)18–25(–27) × (8.5–)9.5–11.5(–12) µm. Asexual morph: not observed.
Culture characters.
Cultures incubated on PDA at 25 °C in the dark, initially pale white, becoming olive-green after 3 wk. The colonies are flat, with regular margins; texture initially uniform, becoming compact after 1 month.
Specimens examined.
China. Shaanxi Province: Ankang City, Huoditang forest farm, 33°26'7"N, 108°26'48"E, on branches of Pinus armandii, 8 June 2018, N. Jiang & C.M. Tian (holotype BJFC-S1725; ex-type living culture: CFCC 53020); 33°26'7"N, 108°26'48"E, on branches of Pinus armandii, 8 June 2018, N. Jiang & C.M. Tian (BJFC-S1726; living culture: CFCC 53021).
Notes.
Cryphognomonia pini is the type species of Cryphognomonia, and occurs on Pinus armandii in China. Morphologically, Cryphognomonia pini is characterized based on bicellular ascospores with obvious hyaline sheath. In the phylogenetic tree, this species is most closely related to F. rhoigena (Fig. 1). However, Cryphognomonia pini can be distinguished from F. rhoigena based on ITS, LSU, tef1 and rpb2 loci (73/512 in ITS, 4/775 in LSU, 186/437 in tef1 and 90/1064 in rpb2).
Gnomoniopsis xunwuensis
C.M. Tian & Q. Yang sp. nov.
C1F2771E-F584-598A-B96A-C7DF66F49514
829529
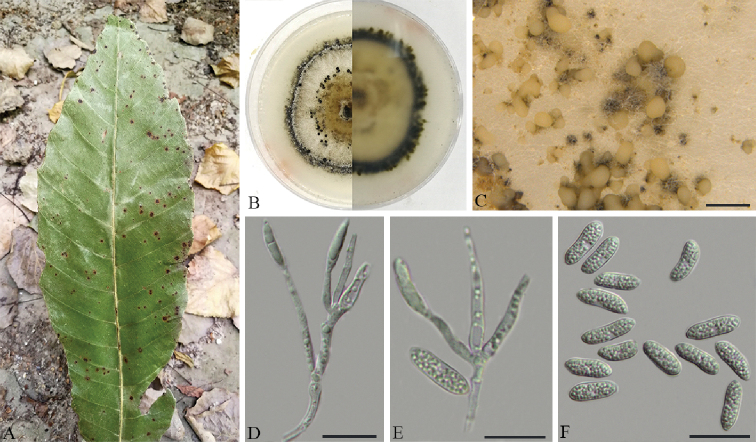
Figure 5.
Gnomoniopsis xunwuensis on Castanopsis fissa (BJFC-S1688) A symptoms on leaves of host plant B the colony on PDAC conidiomata on PDAD, E conidiophores attached with condia F conidia. Scale bars: 500 μm (C); 20 μm (D–F).
Diagnosis.
Gnomoniopsis xunwuensis differs from its closest phylogenetic neighbor, G. daii, in ITS, tef1 and tub2 loci based on the alignments deposited in TreeBASE.
Etymology.
Named after the County (Xunwu), where the species was first collected.
Description.
On PDA: Conidiomata pycnidial, (115–)130–210(–250) μm diam., globose, solitary to gregarious, or occasionally coalescing, deeply embedded in the medium, erumpent, brown to dark black. White to cream conidial drops exuding from the ostioles. Conidiophores (40–)43–58(–60.5) × 2–2.5(–3) μm, cylindrical, hyaline, phiailidic, branched or sympodially branched, straight or slightly curved. Conidia oval or fusiform, straight to slightly curved, hyaline, multiguttules, (14–)16.5–20 × 4–5.5 µm.
Culture characters.
Cultures incubated on PDA at 25 °C in the dark. Colony originally compact and flat with white aerial mycelium, then developing pale brown aerial mycelium at the center and blackish green mycelium at the marginal area, zonate with 2 well defined zones with regular edge; conidiomata dense, regularly distributed over agar surface.
Specimens examined.
China. Jiangxi Province: Ganzhou City, Xunwu County, 24°40'50"N, 115°34'37"E, on leaves of Castanopsis fissa, 12 May 2018, Q. Yang, Y. Liu & Y.M. Liang (holotype BJFC-S1688; ex-type living culture: CFCC 53115); 24°52'20"N, 115°35'25"E, on leaves of Castanopsis fissa, 12 May 2018, Q. Yang, Y. Liu & Y.M. Liang (BJFC-S1689; living culture: CFCC 53116 and CFCC 53117).
Notes.
Gnomoniopsis xunwuensis is associated with leaf spot of Castanopsis fissa, representing the first report from this host in China. It is characterized by sympodially branched conidiophore and oval or fusiform conidia. Morphologically, G. xunwuensis differs from G. daii in having bigger conidia (16.5–20 × 4–5.5 vs. 5.5–7 × 2–3.5 µm) (Jiang and Tian 2019). The phylogenetic inferences indicated this species as an individual well-supported clade (MP/ML/BI=100/100/1) in the genus Gnomoniopsis (Fig. 2).
Plagiostoma populinum
(Fuckel) L.C. Mejía. Stud. Mycol. 68: 225. 2011.
ACBB0FE2-EF21-5D11-B50D-8F58DF82058D
Figure 6.
Plagiostoma populinum on Populus tomentosa (BJFC-S1724) A–C habit of conidiomata on twigs D transverse section through conidiomata E longitudinal section through conidiomata F, G conidiogenous cells attached with conidia H, I condia. Scale bars: 2 mm (A); 1 mm (B, C); 500 μm (D, E); 10 μm (F–I).
Description.
See Butin (1958)
Specimens examined.
China. Beijing: Haidian district, 40°31'55"N, 116°20'24"E, on branches of Populus tomentosa, 12 November 2017, N. Jiang (BJFC-S1724; living culture: CFCC 53016 and CFCC 53017).
Notes.
Plagiostoma populinum is a common plant pathogenic fungus causing poplar canker in China. The current identification follows previous descriptions and records (Butin 1958). In the present study, two isolates (CFCC 53016 and CFCC 53017) from symptomatic branches of Populus tomentosa were congruent with P. populinum based on morphology and DNA sequences data (Fig. 3). We therefore describe P. populinum as a known species for this clade.
Discussion
In this study, three gnomoniaceous species were identified based on morphological and molecular phylogenetic analyses. As a result, Cryphognomonia typified with C. pini is proposed as a new genus in Gnomoniaceae for its distinct phylogenic position and distinctive sexual morphs. Also, Gnomoniopsis xunwuensis strains were successfully isolated from leaf spot of Castanopsis fissa, and were identified as a new species in Gnomoniopsis, which was typified by Gnomoniopsis chamaemori having pycnidia with hyaline, oval, one-celled conidia (Walker et al. 2010).
The type species of Cryphognomonia, C. pini, is unique through its developed pseudostromata and ascospores with distinct hyaline sheath. In the molecular phylogeny, C. pini is closely related to species of F. rhoigena. Flavignomonia rhoigena is characterized by the formation of synnemata and no sexual morph is known for this species (Jiang et al. 2019). However, C. pini can be easily distinguished from F. rhoigena based on ITS, LSU, tef1 and rpb2 loci. Therefore, the unique morphology in combination with an isolated phylogenetic position within Gnomoniaceae warrant the establishment of a new genus.
Most species of Gnomoniopsis show host preference or potentially limited host specificity to genera in the Fagaceae, Onagraceae and Rosaceae (Sogonov et al. 2008). In the present study, isolates were collected from leaf spot of Castanopsis fissa, and described as a novel pathogen depending on its asexual state, G. xunwuensis. Four taxa, G. clavulata, G. daii, G. paraclavulata, and G. smithogilvyi, have been found on Fagaceae host plants. However, Gnomoniopsis xunwuensis can be easily distinguished from the four species in conidial size (16.5–20 × 4–5.5 µm in G. xunwuensis vs. 5.0–8.0 × 2.0–4.0 µm in G. clavulata vs. 5.0–8.0 × 2.0–3.5 µm in G. daii vs. 6.0–9.5 × 2.0–3.5 µm in G. paraclavulata vs. 4.9–9.8 × 2.9–4.9 µm in G. smithogilvyi), as well as supported by molecular data (Walker et al. 2010; Crous et al. 2012; Visentin et al. 2012).
Plagiostoma populinum is regarded as the pathogen responsible for poplar canker. Butin (1958) presented a full description with illustrations of this species as Cryptodiaporthe populea. Mejía et al. (2011a) treated C. populea as a synonym of P. populinum based on analyses of cultural and DNA sequence data. In this paper, P. populinum forms a highly supported monophyletic group (Fig. 3) characterized by having conidia with obvious hyaline sheath. It is the first time that we have been able to provide detailed morphological diagrams in China.
Supplementary Material
Acknowledgements
This study is financed by the Research Foundation of Education Bureau of Hunan Province, China (Project No.: 19B608), the introduction of talent research start-up fund project of CSUFT (Project No.: 2019YJ025) and National Natural Science Foundation of China (Project No.: 31670647). We are grateful to Chungen Piao, Minwei Guo (China Forestry Culture Collection Center (CFCC), Chinese Academy of Forestry, Beijing.
Citation
Yang Q, Jiang N, Tian C-M (2020) Tree inhabiting gnomoniaceous species from China, with Cryphogonomonia gen. nov. proposed. MycoKeys 69: 71–89. https://doi.org/10.3897/mycokeys.69.54012
References
- Butin H. (1958) Über die auf Salix und Populus vorkommenden Arten der Gattung Cryptodiaporthe Petrak. Phytopathologische Zeitschrift 32: 399–415. 10.1111/j.1439-0434.1958.tb01783.x [DOI] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 3: 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JDP, Dima B, Guarnaccia V, Imrefi I, Jurjević Ž, Knapp DG, Kovács GM, Magistà D, Perrone G, Rämä T, Rebriev YA, Shivas RG, Singh SM, Souza-Motta CM, Thangavel R, Adhapure NN, Alexandrova AV, Alfenas AC, Alfenas RF, Alvarado P, Alves AL, Andrade DA, Andrade JP, Barbosa RN, Barili A, Barnes CW, Baseia IG, Bellanger J-M, Berlanas C, Bessette AE, Bessette AR, Biketova AYu, Bomfim FS, Brandrud TE, Bransgrove K, Brito ACQ, Cano-Lira JF, Cantillo T, Cavalcanti AD, Cheewangkoon R, Chikowski RS, Conforto C, Cordeiro TRL, Craine JD, Cruz R, Damm U, de Oliveira RJV, de Souza JT, de Souza HG, Dearnaley JDW, Dimitrov RA, Dovana F, Erhard A, Esteve-Raventós F, Félix CR, Ferisin G, Fernandes RA, Ferreira RJ, Ferro LO, Figueiredo CN, Frank JL, Freire KTLS, García D, Gené J, Gęsiorska A, Gibertoni TB, Gondra RAG, Gouliamova DE, Gramaje D, Guard F, Gusmão LFP, Haitook S, Hirooka Y, Houbraken J, Hubka V, Inamdar A, Iturriaga T, Iturrieta-González I, Jadan M, Jiang N, Justo A, Kachalkin AV, Kapitonov VI, Karadelev M, Karakehian J, Kasuya T, Kautmanová I, Kruse J, Kušan I, Kuznetsova TA, Landell MF, Larsson K-H, Lee HB, Lima DX, Lira CRS, Machado AR, Madrid H, Magalhães OMC, Majerova H, Malysheva EF, Mapperson RR, Marbach PAS, Martín MP, Martín-Sanz A, Matočec N, McTaggart AR, Mello JF, Melo RFR, Me&353;ič A, Michereff SJ, Miller AN, Minoshima A, Molinero-Ruiz L, Morozova OV, Mosoh D, Nabe M, Naik R, Nara K, Nascimento SS, Neves RP, Olariaga I, Oliveira RL, Oliveira TGL, Ono T, Ordoñez ME, de M Ottoni A, Paiva LM, Pancorbo F, Pant B, Pawłowska J, Peterson SW, Raudabaugh DB, Rodríguez-Andrade E, Rubio E, Rusevska K, Santiago ALCMA, Santos ACS, Santos C, Sazanova NA, Shah S, Sharma J, Silva BDB, Siquier JL, Sonawane MS, Stchigel AM, Svetasheva T, Tamakeaw N, Telleria MT, Tiago PV, Tian CM, Tkalčec Z, Tomashevskaya MA, Truong HH, Vecherskii MV, Visagie CM, Vizzini A, Yilmaz N, Zmitrovich IV, Zvyagina EA, Boekhout T, Kehlet T, Læssøe T, Groenewald JZ. (2019) Fungal Planet description sheets: 868–950. Persoonia 42: 291–473. 10.3767/persoonia.2019.42.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GESTJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ. (2012) Fungal Planet description sheets: 107–127. Persoonia 28: 138–182. 10.3767/003158512X652633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. 10.2307/2419362 [DOI] [Google Scholar]
- Fan XL, Du Z, Hyde KD, Liang YM, Pan YP, Tian CM. (2016) Cryptosporella platyphylla, a new species associated with Betula platyphylla in China. Phytotaxa 253: 285–292. 10.11646/phytotaxa.253.4.4 [DOI] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. 10.1128/AEM.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zhang X, Jiang S, Chen C, Ma HB, Nie Y. (2017) A new species of Ophiognomonia from Northern China inhabiting the lesions of chestnut leaves infected with Diaporthe eres. Mycological progress 16: 83–91. 10.1007/s11557-016-1255-z [DOI] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hall T. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Jiang N, Tian CM. (2019) An Emerging Pathogen from Rotted Chestnut in China: Gnomoniopsis daii sp. nov. Forests 10: 1016. 10.3390/f10111016 [DOI]
- Jiang N, Yang Q, Liang YM, Tian CM. (2019) Taxonomy of two synnematal fungal species from Rhus chinensis, with Flavignomonia gen. nov. described. MycoKeys 60: 17–29. 10.3897/mycokeys.60.46395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Mejía LC, Castlebury LA, Rossman AY, Sogonov MV, White JF. (2008) Phylogenetic placement and taxonomic review of the genus Cryptosporella and its synonyms Ophiovalsa and Winterella (Gnomoniaceae, Diaporthales). Mycological Research 112: 23–35. 10.1016/j.mycres.2007.03.021 [DOI] [PubMed] [Google Scholar]
- Mejía LC, Castlebury LA, Rossman AY, Sogonov MV, White JF. (2011a) A systematic account of the genus Plagiostoma (Gnomoniaceae, Diaporthales) based on morphology, host-associations, and a four-gene phylogeny. Studies in Mycology 68: 211–235. 10.3114/sim.2011.68.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía LC, Rossman AY, Castlebury LA, White JF. (2011b) New species, phylogeny, host-associations and geographic distribution of genus Cryptosporella (Gnomoniaceae, Diaporthales). Mycologia 103: 379–399. 10.3852/10-134 [DOI] [PubMed] [Google Scholar]
- Mejía LC, Rossman AY, Castlebury LA, Yang ZL, White JF. (2012) Occultocarpon, a new monotypic genus of Gnomoniaceae on Alnus nepalensis from China. Fungal Diversity 52: 99–105. 10.1007/s13225-011-0108-y [DOI] [Google Scholar]
- Minoshima A, Walker DM, Takemoto S, Hosoya T, Walker AK, Ishikawa S, Hirooka Y. (2019) Pathogenicity and taxonomy of Tenuignomonia styracis gen. et sp. nov., a new monotypic genus of Gnomoniaceae on Styrax obassia in Japan. Mycoscience 60: 31–39. 10.1016/j.myc.2018.08.001 [DOI] [Google Scholar]
- Monod M. (1983) Monographie taxonomique des Gnomoniaceae (Ascomycétes de l’ordre des Diaporthales I). Beihefte zur Sydowia 9: 1–315. [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2016) FigTree, version 1.4.3. University of Edinburgh, Edinburgh.
- Rayner RW. (1970) A mycological colour chart. Commonwealth Mycological Institute, Kew.
- Rehner SA. (2001) EF1 alpha primers. http//ocid.nacse.org/research/deephyphae/EF1primer.pdf
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48: 135–144. 10.1007/S10267-007-0347-7 [DOI] [Google Scholar]
- Senanayake IC, Jeewon R, Chomnunti P, Wanasinghe DN, Norphanphoun C, Karunarathna A, Pem D, Perera RH, Camporesi E, McKenzie EHC, Hyde KD, Karunarathna SC. (2018) Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93: 241–443. 10.1007/s13225-018-0410-z [DOI] [Google Scholar]
- Silvestro D, Michalak I. (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution 12: 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Sogonov MV, Castlebury LA, Rossman AY, Mejía LC, White JF. (2008) Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Studies in Mycology 62: 1–77. 10.3114/sim.2008.62.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis E. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*: Phylogenetic Analyses Using Parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland.
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Sun BL. (1994) Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proceedings of the National Academy of Science USA 91: 4599–4603. 10.1073/pnas.91.10.4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visentin I, Gentile S, Valentino D, Gonthier P, Cardinale F. (2012) Gnomoniopsis castanea sp. nov. (Gnomoniaceae, Diaporthales) as the causal agent of nut rot in sweet chestnu. Journal of Plant Pathology 94: 411–419. [Google Scholar]
- Walker DM. (2012) Taxonomy, systematics, ecology, and evolutionary biology of the Gnomoniaceae (Diaporthales), with emphasis on Gnomoniopsis and Ophiognomonia. Rutgers The State University of New Jersey-New Brunswick.
- Walker DM, Castlebury LA, Rossman AY, Mejía LC, White JF. (2012) Phylogeny and taxonomy of Ophiognomonia (Gnomoniaceae, Diaporthales), including twenty-five new species in this highly diverse genus. Fungal Diversity 57: 85–147. 10.1007/s13225-012-0200-y [DOI] [Google Scholar]
- Walker DM, Castlebury LA, Rossman AY, Sogonov MV, White JF. (2010) Systematics of genus Gnomoniopsis (Gnomoniaceae, Diaporthales) based on a three gene phylogeny, host associations and morphology. Mycologia 102: 1479–1496. 10.3852/10-002 [DOI] [PubMed] [Google Scholar]
- Walker DM, Castlebury LA, Rossman AY, Struwe L. (2013) Host conservatism or host specialization? Patterns of fungal diversification are influenced by host plant specificity in Ophiognomonia (Gnomoniaceae: Diaporthales). Biological Journal of the Linnean Society 111: 1–16. 10.1111/bij.12189 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JM. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.