Abstract
Aim
The purpose of this study was to analyze the sequence of azurin gene in relation to its expression in Pseudomanas aeruginosa strains isolated from different clinical specimens of burn patients. Moreover, in silico sequence analysis of azurin gene using globally reported sequences was intended.
Materials and Methods
Fifty-nine multidrug-resistant P. aeruginosa isolates were selected from different clinical specimens of patients suffering from burn wound infections in two university hospitals and subjected to antibacterial susceptibility testing. The frequency and genetic diversity of the azurin gene was determined by polymerase chain reaction (PCR) and Sanger sequencing. The azurin gene sequences were compared with the sequence data from other countries. The expression level of azurin gene in P. aeruginosa isolates with different azurin sequences from different clinical specimens was evaluated by real-time PCR.
Results and Conclusion
About 98%–100% of the isolates were resistant to gentamicin, tobramycin, cefoxitin, ciprofloxacin, amikacin, and imipenem, while 100% and 23.9% of the isolates were susceptible to colistin and ceftazidime, respectively. Only eight point mutations were detected with amino acid substitutions in only two positions (81 and 102). In global analysis, 93% of strains showed missense mutation at positions 81 (alanine to threonine). The majority (81%) of Iranian strains were allocated to two major clusters distinct from the rest of world, which may suggest that strains from Iran have made a distinct genetic stockpile through point mutations which has established them separate from the other counties. However, 19% were distributed in different clusters together with the strains from different countries of North and South America, Europe, South and East Asia. The expression level of the azurin gene was statistically higher in the isolates collected from the blood of burns patients with systemic infection compared to the isolates collected from other specimens (wound, catheter and tissue), which shows a positive correlation between azurin gene expression and increased pathogenicity and capability for dissemination. This study may open new insight about azurin genetic variation and significance in P. aeruginosa pathogenesis.
Keywords: azurin, pathogenicity, Pseudomonas aeruginosa, burn, sequence analysis
Highlights
The azurin gene was highly conserved and present in all clinical strains of P. aeruginosa.
Global analysis of azurin gene sequence indicated 97.3% similarity among the 168 sequences.
The expression of the azurin gene was significantly higher in the strains from systemic infections.
Azurin probably affects the pathogenicity and capability of the bacterium for dissemination.
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen.1 Clinical infections of P. aeruginosa are usually related to the immune system compromised situations such as burns, AIDS, cancer and cystic fibrosis.1 P. aeruginosa is frequently recognized as an inhabitant of chronic non-healing wound infections which cause high morbidity and mortality, especially in burn patients, despite antibiotic therapy.2
Several virulence factors are involved in P. aeruginosa infection and pathogenesis. Among them, azurin is a cupredox protein called blue-copper protein which is located in the periplasmic and cytoplasmic space and which has proven different biological functions.3 Azurin is involved in the electron transfer within denitrification process and anaerobic biofilm formation in P. aeruginosa in cystic fibrosis patients.4,5 This protein probably gives the bacterium the capacity to evade the immune system via induction of p53-mediated apoptosis in macrophages and provides bacterial ability to escape from the immune system.6,7 Azurin is also involved in Cu uptake and hemostasis in P. aeruginosa, while the lack of Cu+-ATPase induces an increase in azu transcription.3,8 This protein is involved in cytoplasmic oxidative stress and anaerobical respiration.6
The azurin protein structure has similarity to the variable domains of the immunoglobulin superfamily members (Ig superfamily).9 This protein has antitumor,10 antiparasitic (against Plasmodium falciparum and Toxoplasma gondii) and anti-HIV9,11 properties associated with different domains of the protein. The P18 (amino acids 50–67) is the azurin transport domain (PTD) which is responsible for its penetration into cancerous cells and P28 (amino acids 50–77) is a functional region which interacts with the DNA-binding domain of p53, preventing its proteasomal degradation, enhancing its levels and Bax protein, then resulting in the release of mitochondrial cytochrome c into the cytosol and apoptosis.10 Azurin also can bind to Eph receptor tyrosine kinase and VEGFA (vascular endothelium growth factor A) in cancerous and endothelial cells and inhibits the progression of cell cycle and angiogenesis, respectively.12–15
Azurin and its derivatives can attach to the surface antigen SAG1 in T. gondii, the C-terminal of the merozoite surface protein 1 (MSP1) in P. falciparum, gp120 in HIV-1, and the dendritic cell-specific adhesion receptor DC-SIGN and mimics the function of the intercellular adhesion molecule ICAM-3 and suppresses T. gondii adhesion and P. falciparum and HIV-1 growth in peripheral blood mononuclear cells.9,11 The antibacterial properties of azurin (as an inhibitor of growth, biofilm formation, adhesion and invasion) against different epithelial and intestinal bacterial pathogens was determined in our previous studies.16,17 Therefore, azurin acts like a scaffold protein and possesses high affinity to interact with different molecules using distinct regions.18 These characteristics make it a proper candidate of therapeutic peptides for oncotherapy and antimicrobial purposes. Despite the information available about the effects of azurin against cancerous cells and different microorganisms, there is limited information on its role in P. aeruginosa pathogenesis and virulence.
It was demonstrated that physiological conditions and mutations or deletions in the azurin gene lead to loss of azurin production in some P. aeruginosa isolates.18,19 The main objective of the present study was to determine the genetic variation within the azurin gene sequence of P. aeruginosa from different clinical specimens as a possible factor in the progression of bacterial pathogenesis and to determine the probable correlation between azurin gene frequency, expression and nucleotide sequence variation with the site of infection and geographical origin of strains. Moreover, nucleotide polymorphism analysis of globally reported azurin gene sequences was also intended.
Materials and Methods
Collection and Identification of P. aeruginosa Isolates from Different Clinical Specimens
In this study, 133 P. aeruginosa strains were isolated from burned patients admitted to two major university hospitals in Tehran and Isfahan provinces in Iran from May to October 2016. The strains were isolated from wound, tissue (deep wound), blood (patients with both wound and systemic infections), and genitourinary catheters (patients with both wound and urogenital infections) of burned patients (with Degree III burn) and identified in the hospitals laboratories using biochemical tests. All patients fulfilled the criteria of nosocomial infection with no initial infection prior to admission.
The isolates were transferred to the laboratory of Tarbiat Modares University and subjected to re-identification and confirmation by phenotypic tests including Gram staining, catalase, oxidase, citrate, triple sugar iron agar (TSI), oxidative-fermentative test, growth at 42°C, methyl red/Voges Proskauer (MRVP), sulfur indole motility test (SIM), and pigment production on Muller Hinton agar (MHA)20 and further confirmed by molecular methods using specific polymerase chain reaction (PCR) for 16s rDNA of P. aeruginosa. The primer sequences were species-specific and encompassed variable regions (V2 and V8) of 16S rDNA.21 All culture media were purchased from Merck, Germany, and in all assays, P. aeruginosa ATCC27853 was included as standard control.
Antimicrobial Susceptibility Testing (AST)
Antibiotic susceptibility test was performed by the Kirby–Bauer method on MHA (Merck, Germany), according to the Clinical Laboratory Standard Institute (CLSI) guidelines.22 The antibiotics were chosen from different groups including: ciprofloxacin (5 μg), amikacin (30 μg), gentamicin (10 μg), tobramycin (10 μg), colistin (10 μg), ceftazidime (30 μg), aztreonam (30 μg), imipenem (10 μg), cefotaxime (30 μg), cefoxitin (30 μg), ticarcillin (75 μg), piperacillin (100 μg), and piperacillin/tazobactam (100/10 μg) (Mast, England). Resistance to three or more antibiotic groups was considered as multidrug-resistant (MDR).23
Alignment and In Silico Analysis of Azurin Sequences from GenBank
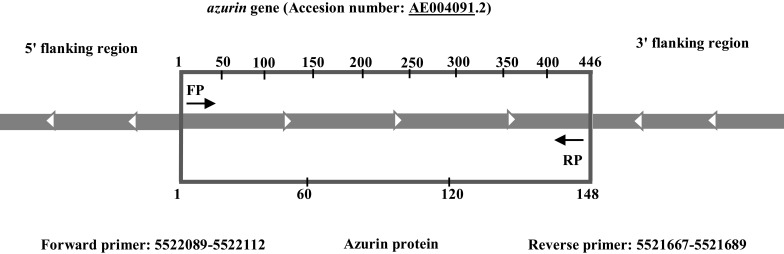
The complete sequences of azurin gene plus flanking regions from whole genome sequence of 108 P. aeruginosa strains (95 strains from clinical specimens such as bacteremia, pneumonia, and burn infections; and 13 strains of environmental origin such as marine, hospital wastewater, and dental clinic wastewater) with defined source and region were selected and acquired from NCBI GenBank (http://www.ncbi.nlm.nih.gov/) (Table 1). Multiple sequence alignment was carried out using CLC Sequence Viewer software Ver. 7.6 (CLC, Denmark). The 3ˈ and 5ˈ flanking regions were composed of very diverse sequences (with 65% similarity), which made it impossible to achieve a conserved sequence for designing a primer pair which could anneal and amplify whole azurin gene sequence in all of the isolates. The aligned sequences were used to design a primer pair which could anneal and amplify a conserved region from the very beginning to the end of azurin gene. The primers were designed by Gene Runner and CLC Sequence Viewer software. The designed primers were analyzed by Primer-BLAST on NCBI (http://www.ncbi.nlm.nih.gov). Schematic representation of azurin gene sequence and primers position are depicted in Figure 1, and the primer pairs’ sequences are displayed in Table 2.
Table 1.
GenBbank Extracted Azurin Gene Sequences Analyzed in This Study
| Origin | Number of Strains | Specimen Source | |
|---|---|---|---|
| Clinical | Environmental | ||
| USA | 49 | 48 | 1 |
| Singapore | 14 | 11 | 3 |
| Colombia | 1 | 1 | – |
| Brazil | 6 | 6 | – |
| Norway | 1 | 1 | – |
| Japan | 3 | 1 | 2 |
| Canada | 2 | 2 | – |
| India | 4 | 4 | – |
| Vietnam | 3 | – | 3 |
| Netherlands | 3 | 3 | – |
| China | 7 | 3 | 4 |
| Korea | 3 | 3 | – |
| Mexico | 7 | 7 | – |
| Germany | 1 | 1 | – |
| Sweden | 1 | 1 | – |
| Switzerland | 3 | 3 | – |
| Total | 108 | 95 | 13 |
Figure 1.
The schematic representation of azurin gene and flanking regions. Position of primers for PCR amplification of azurin gene.
Table 2.
Primer Sequences Used in This Study
| Target Gene | Primer Sequence (5ʹ →3ʹ) | PCR Product Size (bp) | Reference | Amplification Assay |
|---|---|---|---|---|
| azurin | F: CCATGCTACGTAAACTCGCTGCGG R: CTTCAGGGTCAGGGTGCC |
446 | This study | PCR |
| azurin (Blu) | F: GGTGGACATCCAGGGTAACG R: ATGACGTTCTTCGGCAGGTT |
120 | This study | Real time-PCR |
| rpsl (Ribosomal protein S12) | F: GCAAGCGCATGGTCGACAAGA R: CGCTGTGCTCTTGCAGGTTGTGA |
201 | [23, 24] | Real time-PCR |
PCR Amplification and Sequencing of Azurin Gene
All genomic DNA was extracted from the isolates by YTA genomic DNA extraction mini kit (Yekta Tajhiz Azma, Iran) according to the manufacturer’s instructions. DNA isolation procedure was performed in a room physically separated from the room applied for nucleic acid amplification reaction and also from the post-PCR room in order to inhibit or minimize contamination and false positive results. PCR amplification was performed using primers specifically designed for azurin gene (Table 2).
The amplification assay was performed in a total volume of 12.5 μL containing 1 μL of purified DNA (20 ng), 6.5 μL of PCR Master-mix (Ampliqon, Denmark), and 0.5 μL of each primer (10 pM) using Bio Rad Thermal Cycler, Germany. The PCR steps included: an initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 63°C for 45 s, extension at 72°C for 60 s, and a final extension at 72°C for 3 min. The products of PCR were run on electrophoresis by 1% agarose gel and visualized under UV doc apparatus. The amplified fragments of 59 MDR strains were selected (according to the isolate geographical region and clinical specimen type) and subjected to direct sequencing using Applied Biosystems (ABI) capillary sequencer (Macrogen, Korea).
Alignment and Sequence Analysis of Azurin Gene from P. aeruginosa Isolates
Sequences of azurin gene of 59 P. aeruginosa strains and P. aeruginosa ATCC27853 were delivered as Chromas format (Technelysium, Australia). The nucleotide sequence of each strain was deposited in the GenBank database and assigned a GenBank accession number (MK276543– MK276602). The sequences were analyzed and aligned by CLC Sequence Viewer 7 (Qiagen, Denmark) and Gene Runner 6.5.46 (Softpedia, Romania) software. The phylogenetic tree was constructed for the azurin gene using UPGMA (unweighted pair group method with arithmetic mean) algorithm. P. aeruginosa strain PAO1 sequence, which was recorded in GeneBank (accession number AE004091.2), was included in alignment analysis as control. Diversity index was calculated by DI = 1 – n(n – 1)/ƩN(N – 1).
Global Nucleotide Polymorphism of Azurin Gene
A total of 168 azurin gene sequences (59 P. aeruginosa isolates and standard P. aeruginosa ATCC27853 from this study and 108 P. aeruginosa strains from Genbank) (Table 1) were compared and aligned by BioNumerics Ver. 7.6 (Applied Maths company, Belgium). The nucleotide changes, consensus blocks, similarities, amino acid translations and mutations were determined. The circular dendrogram and multidimensional layout were constructed using UPGMA method.
RNA Extraction and cDNA Synthesis
In total, 40 isolates (including 39 clinical isolates and P. aeruginosa ATCC27853 as control) were subjected to total RNA extraction and cDNA synthesis based on the sequence analysis (Sequence Type), isolation location, and clinical sample type (blood, wound, tissue, and catheter). Total RNA was extracted from the isolates by YTA Total RNA Purification Mini kit (Yekta Tajhiz Azma, Iran) according to the manufacturer’s protocol. The removal of genomic DNA was performed by DNase I- RNase free kit (Thermo Scientific, USA). The purity and quality of prepared RNA were assessed by measuring the optical density in 260/280 nm values and electrophoresis on agarose gel.
The cDNA synthesis was performed using cDNA synthesis kit (Yekta Tajhiz Azma, Iran). For this purpose, 100 ng of total RNA, 1 µL oligo (dT) 18 primer (50 µM), 1 µL random hexamer primer (50 µM), and DEPC-treated water up to 13.4 µL were added into a sterile, nuclease-free tube on ice. The mixture was centrifuged briefly and incubated at 70°C for 5 min. In the following cDNA Synthesis Mix, 4 µL 5x-first strand buffer, 1 µL dNTP (10 mM), 0.5 µL RNasin (40 U/µL), and 1 µL M-MLV were added into the microtube, centrifuged, and incubated at 37°C for 60 min. Finally, the reaction was terminated by heating at 70°C for 5 min. The quality of synthesized cDNA was evaluated by gel electrophoresis and reverse transcription PCR. The synthesized cDNAs were subjected to real-time PCR assay.
Real-Time PCR Analysis of Azurin Gene Expression
Relative-real-time PCR was performed to determine the expression level of azurin gene in different isolates of P. aeruginosa using the Light Cycler 96 Real-Time PCR system (Roche Life Science, Germany). Each PCR reaction was performed in a total reaction volume of 20 μL containing 12 μL of real-time PCR Master Mix (Amplliqon, Denmark), 1 μL cDNA template, 1 μL of each primer (Blu), and 5 μL distilled water. Primer designation and sequences are depicted in Table 2. The qPCR was performed according to the following conditions: pre-incubation at 95°C for 5 min, 45 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 30 s. The analysis of melting pick was performed at 95°C for 5 min. In each sample, the same amount of RNA was used, which were converted into the cDNA and pipetted. Ribosomal protein S12 (Rpsl) mRNA expression was applied as the internal control for each sample (Dumas et al., 2006), and the ΔCT (CT target − CT reference) and expression fold change were calculated for each sample according to the comparative CT method (Pfaffl formula).25 Each real-time PCR reaction was performed in duplicate, and the standard deviation was calculated. The efficiency of real-time PCR was determined by amplifying a serial dilution of the template cDNA (10 folds) and calculating E= −1+10 (−1/slope).
Statistical Analysis
The quantity of azurin gene expression in all clinical strains of different locations (Tehran or Isfahan), different clinical samples (wound, blood, tissue and catheter), and different sequence clusters was compared by one-way analysis of variance (ANOVA) statistical analysis test. The t-test analysis was used to compare azurin expression level between the clinical isolates and P. aeruginosa ATCC 27853. A p-value <0.05 was considered as significant in all statistical analysis. The correlation between the sequence type and expression level of azurin gene was analyzed by Pearson test.
Results
Isolates Collection and Identification
In this cross-sectional study, 133 isolates were collected from burned patients, among which 57 and 43% were from the major centers of burn patients in Tehran and Isfahan, respectively. The bacterial strains were isolated from blood [7 (5.3%): 4 (5%) + 3 (5%)], wound [117 (88%): 67 (50%) + 50 (50%)], tissue [1 (0.7%): 1 (1%) + 0], and catheter [8 (6%): 4 (5%) + 4 (7%)] of patients admitted to Tehran and Isfahan university hospitals, respectively. The blood specimens were obtained from patients with both wound and systemic infections, and catheter specimens were from patients with both wound and genitourinary urogenital infections due P. aeruginosa. All isolates (100%) were re-identified as P. aeruginosa by phenotypic tests and confirmed by species-specific 16s rDNA PCR amplification assay.
Antimicrobial Susceptibility Testing
About 100% of the isolates were resistant to gentamicin, tobramycin, and cefoxitin. About 98.5% were resistant to ciprofloxacin, amikacin, and imipenem while 100% and 23.9% of the isolates were susceptible to colistin and ceftazidime, respectively. All strains were determined as MDR according to the criteria determined by CLSI.23
Alignment and In Silico Analysis of Azurin Sequences from GenBank
According to the results of the multiple sequence alignment method, primer pairs were designed to cover the whole azurin gene sequence at the position 5521667–5522112 in P. aeruginosa PAO1 (accession number: AE004091.2). This primer pair amplified the 446 bp amplicon, and the primer blast in NCBI indicated that the primers were specific for P. aeruginosa azurin gene.
Amplification and Sequencing of Azurin Gene
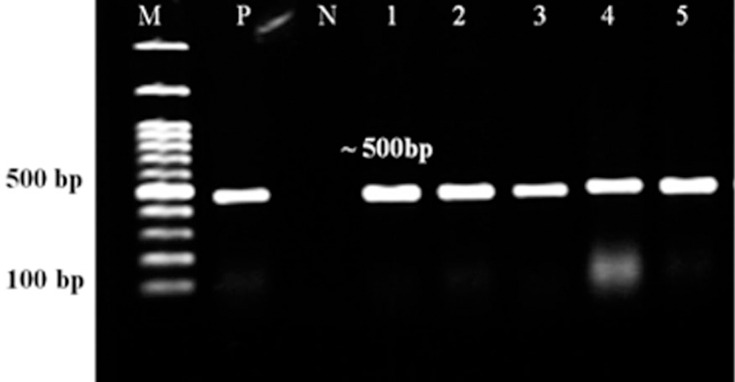
Total isolates (100%) under study contained azurin gene, and it was possible to amplify 446 bp of the azurin gene (Figure 2). In this study, 59 strains isolated from blood (6, 10%), wound (47, 80%), tissue (1, 2%), and catheter (5, 8%) of patients were sequenced. After trimming the low-quality sequences at the ends, a region of 406 bp (which covered 5521700–5522105) was subjected to analysis. The sequence analysis of azurin gene revealed point mutations at eight positions located in nucleotides 51, 241, 243, 282, 297, 298, 304, and 306 (Table 3). Mutation at four locations (241, 243, 304, and 306) resulted in different amino acid substitutions in two positions (threonine to alanine at position 81 and alanine to threonine at position 102 of amino acid sequences) (Table 4). About 62.8% of the isolates from Tehran and Isfahan harbored mutations which resulted in amino acid substitution in two positions (81 and 102). Also, 37.2% of the strains contained mutations in nucleotides 241, 243, 304, and 306, resulting in amino acid substitution at two positions 81 and 102 (missense mutation): A and T at position 81 and T and A at position 102 (Table 4). Generally, in addition, the amino acid sequence type (ST) to P. aeruginosa ATCC27853 and PAO1 there are 3 amino acid STs in our strains (Table 4).
Figure 2.
PCR assay of azurin gene. Lanes 1–5: P. aeruginosa isolates, Lanes N and P: representatives of negative and positive controls. M: 100 bp DNA size marker.
Table 3.
Point Mutations Within the 446-Bp Segment of Azurin Gene in Iranian P. Aeruginosa Isolates in Relation to the Location of Isolation and Accession Numbers
| Isolations | Accession Number | Province | Mutation Position | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 51 | 241 | 243 | 282 | 297 | 298 | 304 | 306 | |||
| PAO1 | AE004091.2 | A | A | C | G | T | G | G | C | |
| ATCC27853 | MK276602 | G | G | A | G | T | G | A | A | |
| 1 | MK276543 | Tehran | G | G | A | A | T | G | A | A |
| 2 | MK276544 | Tehran | G | A | A | A | T | G | A | A |
| 3 | MK276545 | Tehran | G | A | C | A | T | G | A | A |
| 4 | MK276546 | Tehran | G | A | C | G | C | G | A | A |
| 5 | MK276547 | Tehran | G | A | C | G | G | G | A | A |
| 6 | MK276548 | Tehran | G | A | C | A | T | G | A | A |
| 7 | MK276549 | Tehran | G | A | C | A | T | G | A | A |
| 8 | MK276550 | Tehran | G | A | C | A | T | G | A | A |
| 9 | MK276551 | Tehran | G | A | C | A | T | G | A | A |
| 10 | MK276552 | Tehran | G | A | C | A | T | G | A | A |
| 11 | MK276553 | Tehran | G | A | C | A | T | G | A | A |
| 12 | MK276554 | Tehran | G | A | C | G | C | G | A | A |
| 13 | MK276555 | Tehran | G | A | C | A | T | G | A | A |
| 14 | MK276556 | Tehran | G | A | C | A | T | G | A | A |
| 15 | MK276557 | Tehran | G | A | C | A | T | G | A | A |
| 16 | MK276558 | Tehran | G | A | A | A | T | G | A | A |
| 17 | MK276559 | Tehran | G | A | C | G | T | G | A | A |
| 18 | MK276560 | Tehran | G | A | A | G | C | G | A | A |
| 19 | MK276561 | Tehran | G | A | C | A | T | G | A | A |
| 20 | MK276562 | Tehran | G | A | C | A | T | G | G | C |
| 21 | MK276563 | Tehran | G | A | C | G | C | G | G | C |
| 22 | MK276564 | Tehran | G | A | C | A | T | G | A | A |
| 23 | MK276565 | Tehran | G | A | C | G | C | G | A | A |
| 24 | MK276566 | Tehran | G | A | C | A | T | G | G | C |
| 25 | MK276567 | Tehran | G | A | C | A | T | G | G | C |
| 26 | MK276568 | Tehran | G | A | C | G | T | G | G | C |
| 27 | MK276569 | Tehran | G | A | C | G | T | G | G | C |
| 28 | MK276570 | Tehran | G | A | C | G | C | G | G | C |
| 29 | MK276571 | Tehran | G | A | C | G | C | G | G | C |
| 30 | MK276572 | Tehran | G | A | C | A | C | G | G | C |
| 31 | MK276573 | Tehran | G | A | C | A | T | G | G | C |
| 32 | MK276574 | Isfahan | G | A | C | G | C | G | A | A |
| 33 | MK276575 | Isfahan | G | A | C | G | C | G | G | A |
| 34 | MK276576 | Isfahan | G | A | C | G | C | G | A | A |
| 35 | MK276577 | Isfahan | G | A | C | A | T | G | A | A |
| 36 | MK276578 | I Isfahan | A | A | C | G | T | G | A | A |
| 37 | MK276579 | Isfahan | G | A | C | G | C | G | A | A |
| 38 | MK276580 | Isfahan | G | A | A | A | T | G | A | A |
| 39 | MK276581 | Isfahan | G | A | A | A | T | G | A | A |
| 40 | MK276582 | Isfahan | G | A | C | G | C | G | A | A |
| 41 | MK276583 | Isfahan | G | A | A | G | C | G | A | A |
| 42 | MK276584 | Isfahan | G | A | C | A | T | G | A | A |
| 43 | MK276585 | Isfahan | G | G | A | G | C | G | A | A |
| 44 | MK276586 | Isfahan | G | A | C | G | C | G | A | A |
| 45 | MK276588 | Isfahan | G | A | C | A | T | G | A | A |
| 46 | MK276589 | Isfahan | A | A | C | G | T | G | A | A |
| 47 | MK276590 | Isfahan | G | G | C | G | C | G | A | A |
| 48 | MK276591 | Isfahan | G | G | A | G | C | G | A | A |
| 49 | MK276592 | Isfahan | G | G | C | A | T | G | A | A |
| 50 | MK276593 | Isfahan | G | G | C | G | C | G | A | A |
| 51 | MK276594 | Isfahan | G | G | C | A | T | G | A | A |
| 52 | MK276595 | Isfahan | G | G | C | G | C | A | A | A |
| 53 | MK276596 | Isfahan | A | A | C | G | T | G | A | A |
| 54 | MK276597 | Isfahan | G | G | A | A | T | G | A | A |
| 55 | MK276598 | Isfahan | G | G | A | G | C | G | A | A |
| 56 | MK276599 | Isfahan | G | G | A | G | C | G | A | A |
| 57 | MK276600 | Isfahan | G | A | C | G | C | G | A | A |
| 58 | MK276601 | Isfahan | G | A | A | G | C | G | A | A |
| 59 | MK276588 | Isfahan | G | A | A | G | C | G | A | A |
| Total sequenced isolates | 59 isolates and one P. aeruginosa ATCC27853 | |||||||||
Table 4.
The Frequency of Amino Acid Substitutions Among Iranian Azurin Protein Sequences
| Strains | Amino Acids Substitution Site | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 81 | 102 | |||||||||
| PAO1 | . | . | T | . | . | . | . | A | . | . |
| ATCC 27853 | . | . | A | . | . | . | . | T | . | . |
| 37 strains (62.8%) | . | . | T | . | . | . | . | T | . | . |
| 11 strains (18.6%) | . | . | A | . | . | . | . | T | . | . |
| 11 strains (18.6%) | . | . | T | . | . | . | . | A | . | . |
| Total | 59 isolates (100%) | |||||||||
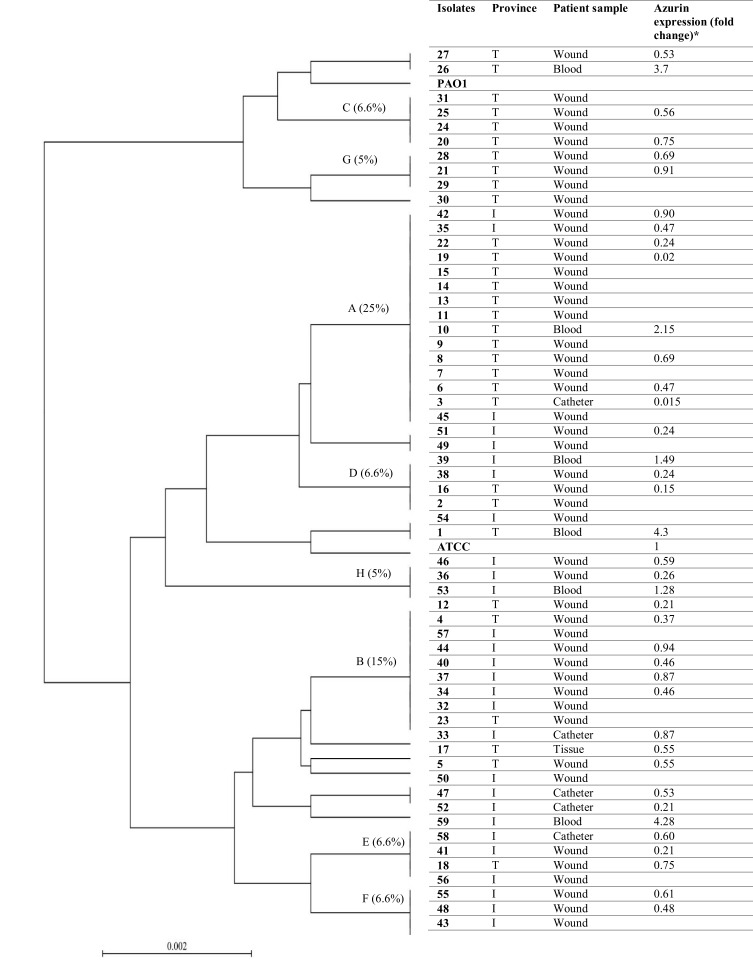
The diversity index was calculated as 0.9 for the population. A dendrogram was constructed based on the sequences of azurin gene from 59 isolated strains, P. aeruginosa ATCC27853, and P. aeruginosa PAO1. The strains were spread in 19 STs (17 STs for Iranian strains and two STs for P. aeruginosa ATCC27853 and PAO1). It was shown that 15 (25%) and nine (15%) clinical isolates allocated in common sequence types (A and B, respectively) (Figure 3). Also, 58 and 50% of the strains from Tehran and Isfahan were distributed in common clusters (A, B, D, and E), while clusters C, G and F, H were specific for the isolates of Isfahan and Tehran, respectively. The strains distributed in cluster A were obtained from the wound (86%), blood (7%), and catheter (7%) samples. On the other hand, 80% and 20% of cluster A strains were collected from Tehran and Isfahan hospitals, respectively. Cluster B included the strains isolated from patients with wound infection (100%). This cluster's strains were isolated in greater numbers from Isfahan (67%) hospital.
Figure 3.
UPGMA dendrogram of P. aeruginosa clinical strains based on the difference in the nucleotides of azurin gene sequences. P. aeruginosa ATCC27853 and PAO1 were used as control. Each strain is presented based on the geographical region of its isolation, clinical sample, and gene expression fold change. T: Tehran, I: Isfahan.
Nucleotide Polymorphism Analysis of Globally Reported Azurin Gene Sequences
The alignment of 168 azurin sequences (59 Iranian clinical strains, 108 strains from GenBank, and P. aeruginosa ATCC27853) displayed 97.3% similarity using BioNumerics Ver. 7.6 (free trial version, Applied Maths Company, Belgium). The point mutations were shown in 23 nucleotides at different positions (51, 81, 114, 117, 124, 126, 169, 201, 241, 243, 255, 273, 282, 297, 298, 304, 306, 336, 342, 378, 418, 432, and 433). The nucleotide sequences were completely conserved in other parts. Only one nucleotide deletion was detected at position 170 in the BH9 strain from India.
The amino acid substitutions (missense mutation) were determined at positions 81 (alanine to threonine) in 93% of isolates, position 99 (valine to isoleucine) in 1% of isolates and position 102 (threonine to alanine or threonine to serine) in 68% and 3% of isolates, respectively. The deletion of nucleotide in BH9 resulted in frameshift and undetermined protein sequence.
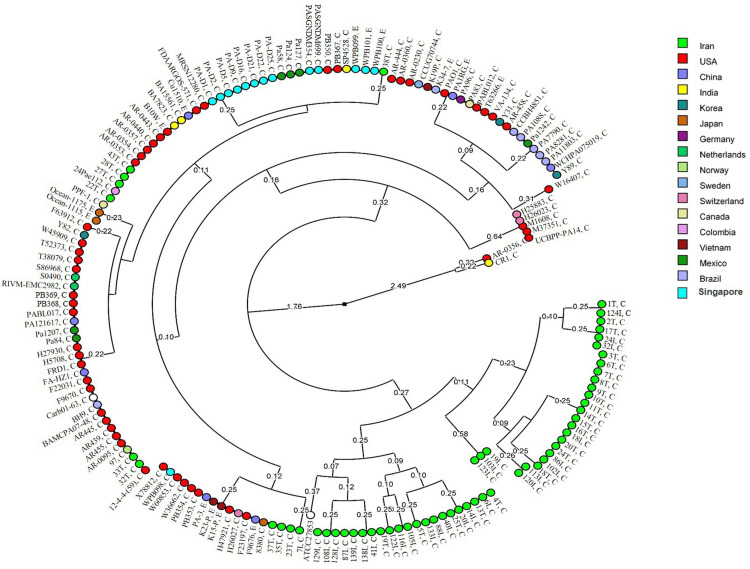
Global analysis of nucleotide polymorphism of azurin gene revealed a total of 26 STs among the P. aeruginosa strains according to the circular dendrogram (Figure 4). The results showed that 59 strains from Iran were distributed in 17 STs, 65 strains from America (USA, Canada, Mexico Brazil, and Colombia) were spread in nine STs, 29 strains from Asia (China, Japan, Korea, Singapore, Vietnam, and India) belonged to seven STs, and nine strains from Europe (Norway, Netherlands, Sweden, Switzerland, and Germany) were distributed in four STs.
Figure 4.
Circular dendrogram according to the global analysis of nucleotide polymorphism of azurin gene among P. aeruginosa strains. C and E next to the ID of the isolates determine the clinical and environmental sources of isolates. The geographical origin of the isolates is displayed using different colors.
Among the Iranian strains, 48 (81%) strains belonged to two major clusters, and 11 (~19%) strains were distributed in different clusters together with the strains from different countries of North and South America, Europe, South and East Asia (Figure 4). Also, 13 environmental strains were located in five STs within the same clusters where the clinical strains were located (Figure 4).
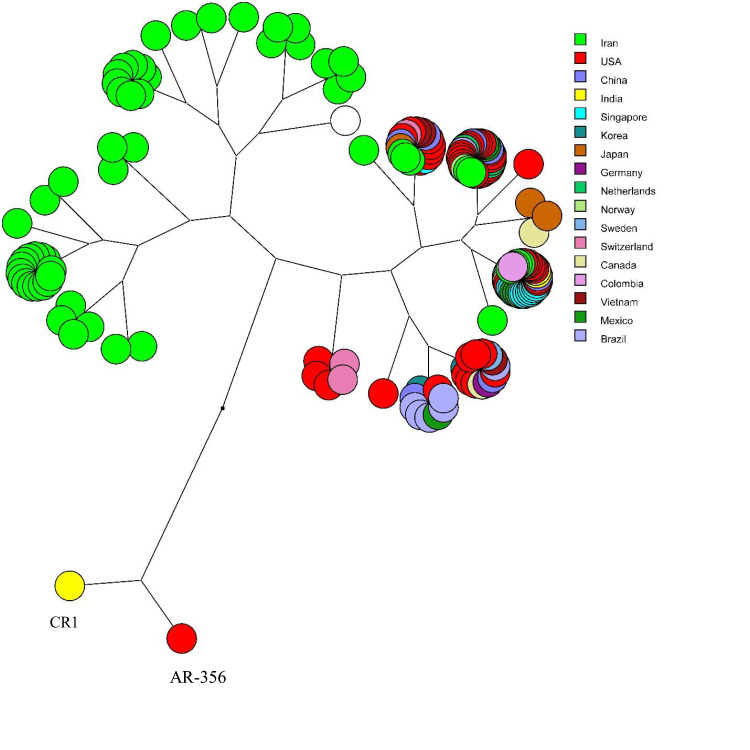
Multidimensional layout of global analysis of azurin gene in different isolates of P. aeruginosa showed that all the isolates were originated from a common ancestor and branched in two main stems, while two strains (CR1 from USA and AR-356 from India) were completely separated from the others (Figure 5).
Figure 5.
The multidimensional layout of global analysis of azurin gene in different isolates of P. aeruginosa according to UPGMA algorithms.
Real-Time PCR Analysis
The purity of each extracted RNA was determined as 1.80–2 by calculating OD260/280 nm, and its quality was confirmed by gel electrophoresis and RT-PCR (data not shown). The amplification picks and melting curve analysis of products in real-time PCR are depicted in Figure S1a and S1b, respectively (supplementary data). No non-specific product and primer dimer were detected at melting temperature of approximately 84°C. Primer efficiency was calculated as 1.96 (Figure 1Sc in supplementary data).
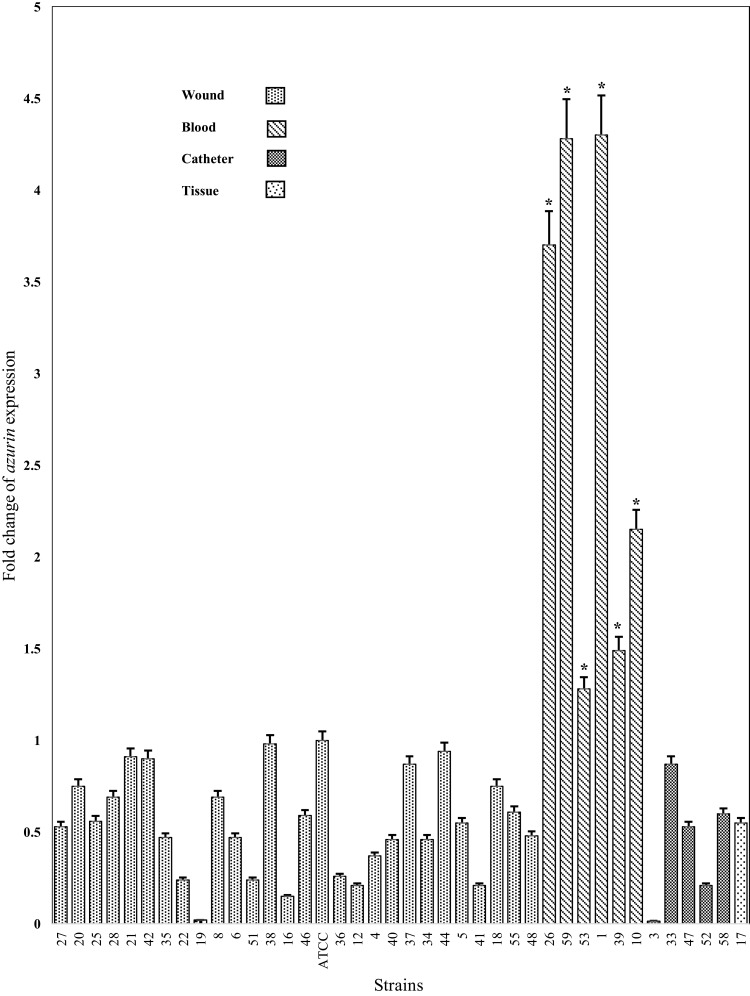
The expression level of azurin gene in different isolates is presented in Figures 3 and 6. The expression level of azurin gene in different Iranian clinical strains was statistically different from its expression level in P. aeruginosa ATCC27853 (p< 0.001) and was 0.015–4.3-fold higher. The azurin gene expression level in the strains isolated from blood was 1.28–4.3-fold higher than in the standard strain, while the expression level in the strains isolated from wound, tissue, and catheter was 0.015–0.91-fold higher (Figure 3). These results showed that the expression level of azurin gene was statistically higher in the strains isolated from blood, compared with the strains of other clinical origin (p< 0.001). There was no significant difference regarding the expression level of azurin between the strains of two provinces (Tehran and Isfahan) (p> 0.05).
Figure 6.
The fold change of azurin gene expression in different P. aeruginosa clinical strains isolated from different specimens (wound, blood, catheter, and tissue). *p< 0.001.
The Pearson correlation test displayed that the expression level of azurin gene was not affected by the nucleotide sequence changes in this gene.
Discussion
In this study, 100% of isolates were susceptible to colistin, which means that this antibiotic is effective for treatment of infections caused by MDR P. aeruginosa. According to CLSI guidelines colistin could be used in antimicrobial assay and probably treatment of MDR P. aeruginosa.22 Moreover, colistin has been reported to be effective against MDR infections in several literatures in this field,26–28 although the use of polymyxins (colistin) should be optimized for dosage administration and indications, in order to maximize effectiveness, prevent the emergence of further polymyxin resistance and reduce adverse effects.29 In the study performed by Sabuda et al. (2008), the intravenous and nebulized colistin was effective for treatment of MDR P. aeruginosa, with mild toxicity in kidneys of two-third of patients.26
In this study, azurin gene was detected in 100% of 133 clinical strains under study, while Sereena and colleagues (2016) reported the presence of this gene in 80% of the strains of environmental origin.14 In another study by Sarwar et al. (2018), the frequency of azurin gene in environmental strains was determined as 25%.30 These findings signify its putative role in the biology of clinical strains and probably disease development in clinical strains.
Sequence alignment and azurin gene analysis in P. aeruginosa strains in this study displayed the presence of eight point mutation sites, of which four resulted in amino acid substitution at two locations (81 alanine to threonine and 102 threonine to alanine) (missense mutation) (Tables 3 and 4). Similarly, the amino acid substitution in azurin sequence were detected in positions 81 and 102 in studies reported from different parts of the world.33–35 Alanine and threonine are structurally different with an extra OH group in threonine (polar amino acid). This is compatible with the preference of alanine (as a hydrophobic amino acid) to form a helical structure and the preference of threonine (as a polar amino acid) to support beta-sheet structures.31 Of course, as the structure of azurin is a complex of alpha helixes and beta sheets, it seems that these amino acid substitutions have no great impact on protein structure and function. In the same study by Nguyen et al. (2019), point mutation and amino acid substitutions were detected, none of which affected the azurin function.32
In the constructed dendrogram based on the sequence analysis of azurin gene in clinical isolates (Figure 3), the isolates were classified in 19 STs and nine (A–H) clusters, considering P. aeruginosa PAO1 and P. aeruginosa ATCC27853 as standard strains. Also, 25% and 15% of the isolates belonged to clusters A and B, respectively. The Diversity Index was calculated as 0.9 for total population. In addition, 58% and 50% of the strains from Tehran and Isfahan were included in common clusters (A, B, D and E), while clusters C, G, and F, H were restricted for Isfahan and Tehran strains, respectively, which may show the dissemination of specific azurin sequence types in each province. According to the distribution of the strains from different clinical specimens (blood, wound, catheter and tissue) in common clusters such as A and B, it was assumed that azurin ST type has no effect on the site of infection. About 80% and 20% of cluster A strains were collected from Tehran and Isfahan hospitals, respectively, and 67% of cluster B strains were isolated from Isfahan. The frequency of distribution of ST types based on the geographical region in different clusters demonstrates the possible role of geographical area on the ST distribution.
Due to limited point mutations and considering the fact that only purine:purine and pyrimidine:pyrimidine conversions were observed in nucleotides, it seems that azurin gene is structurally and functionally conserved in clinical strains and may be considered a useful tool for the detection of clinical isolates of P. aeruginosa. In the study performed by Nguyen et al. (2019), the sequences of azurin gene from the metagenomic DNA sample and P. aeruginosa isolates were analyzed and five point mutations at (51, 282, 297, 305, and 432) were determined. The first mutation was located in the signal peptide region and the other mutations did not affect the three main domains of azurin protein. They concluded that mutations in the mature peptide did not interfere with azurin protein structural properties.32
Kamalakannan et al. (2011) examined the primary and secondary structure of azurin protein and its phylogenetic relatedness in various species of Pseudomonas. Their results showed differences in molecular weight and amino acid composition of azurin in different species; however, phylogenetic analysis indicated that azurin protein of various Pseudomonas species originated from a common ancestor.36 Their study results strengthened our hypothesis about azurin gene conservation in P. aeruginosa species.
The global analysis of azurin gene sequences among the 168 strains of P. aeruginosa (59 sequences from Iranian clinical strains and 108 Genbank registered sequences) displayed point mutations in 22 nucleotides, resulting in amino acid substitution (missense mutations) at three positions: 81 (alanine to threonine), 99 (valine to isoleucine), and 102 (threonine to alanine/serine). The sequence alignment and analysis indicated 97.3% similarity among the 168 sequences with a clear distinct lineage for Iranian sequences (Figure 4). The analysis results are re-emphasizing that the azurin gene is conserved and it may be related the important function of this protein.
The circular dendrogram was constructed according to the sequences alignment and divided the sequences into 26 STs, in which the sequences from Iranian strains were distributed in 17 STs, and the sequences reported from all other countries were spread over nine STs. Among the Iranian sequences, 48 (81%) sequences belonged to two main clusters, but 11 sequences were distributed in other clusters together with the sequences from different countries (Figure 4), emphasizing the probable role of the isolates’ geographical location on azurin gene nucleotide sequence variation. This may suggest that somehow related strains from Iran have made a genetic stockpile through point mutations, which has established them distinct from strains of rest of world.
The sequences of 13 environmental strains (from hospital or dental clinic sewage and marine) were distributed in five STs together with the clinical strains (Figure 4), which may be due to (i) the contamination of hospital or dental clinic sewage by clinical strains or (ii) inefficient disinfection of hospital or dental clinic sewage, which might have caused the contamination to be disseminated to clinics and caused infections.
The multidimensional layout of global analysis of azurin gene in different strains of P. aeruginosa (Figure 5) determined that all isolates were originated from a common ancestor and divided the isolates into two main branches, one of which contained Iranian isolates. Two clinical strains from India and USA (CR1 and AR-356) were absolutely fell apart.
The present study results demonstrated that the strains isolated from blood samples of patients with systemic infections belonged to several clusters and STs; thus, it seems that the azurin gene sequence does not have effect on the infection type.
The results of azurin gene expression among the sequenced isolates demonstrated that the expression of azurin gene was significantly higher in the strains isolated from blood than in those isolated from wound, catheter, and tissue (p< 0.001; Figures 3 and 6). The former isolates were obtained from patients with systemic infections which ultimately resulted in their deaths. This is probably related to the contribution of azurin protein to the pathogenicity and the possibility of bacterial attachment, invasion (our previous study)16,17 and survival in blood. According to the results, it was assumed that the interference or inhibition of azurin function probably led to the destruction of invasion or systematic pathogenicity of P. aeruginosa.
Overall, there are limited studies concerning azurin expression. It was found that azurin is highly expressed in anaerobically constructed biofilms in cystic fibrosis patients.5 This implies the significant role of azurin in P. aeruginosa infection, especially in cystic fibrosis patients. Furthermore, it was previously determined that azurin protein induces apoptosis in macrophages involved in phagocytosis.7 It seems that a high level of azurin expression in blood isolate strains may correlate with its role as an apoptosis inducer in macrophages, which helps its dissemination and increased pathogenesis.
It is worth noting that in this study, no obvious correlation was detected between the azurin sequence type and the level of expression, signifying the importance of clinical source of isolation as the main crucial factor affecting azurin expression level. This increased expression may be due to the presence of stronger azurin promoters in these strains, indisputably explaining why these strains escaped the host immune system and disseminated into the bloodstream.
Conclusion
This study revealed the presence of azurin gene in all clinical isolates of P. aeruginosa, which was highly conserved with point mutations occurring in only eight positions, resulting in only two amino acid substitutions (missense mutation) at positions 81 and 102 (alanine to threonine). It seems that the point mutations have no effect on the virulence potency of P. aeruginosa. The multidimensional layout of global analysis of azurin gene revealed that the majority of Iranian sequences (81%) were allocated in two main clusters distinct from the rest of world, which suggests that Iranian strains have made a distinct genetic stockpile through point mutations which has established them separate from the other counties. However, 19% of Iranian strains were distributed in similar clusters together with strains from different countries of North and South America, Europe, South and East Asia.
The expression of azurin gene was significantly higher in the strains isolated from blood of patients with systemic infections, which may demonstrate their increased pathogenicity and ability to escape the host immune system and disseminate into the bloodstream. This finding may open new insight about azurin genetic variation and significance in P. aeruginosa pathogenesis.
Acknowledgments
I would like to express my special thanks to the laboratory staff of two burns center hospitals, Imam Musa kazem in Isfahan and Shahid Motahhari in Tehran.
Data Sharing Statement
The data sets of the current study are available within article/its supplementary files or can be obtained from corresponding upon request. DNA sequences of genes that have been deposited in GenBank are available in https://pubmed.ncbi.nlm.nih.gov/.
Ethics Approval and Consent to Participate
This study was approved by Medical Ethics Committee of Isfahan University of Medical Sciences Code: 3.094 before the study began. All research was performed in accordance with relevant guidelines/regulations. The consent to participate was obtained from the patients or parents/guardians of the minors included in this study and the data were analyzed anonymously.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Japoni A, Farshad S, Alborzi A. Pseudomonas aeruginosa: burn infection, treatment and antibacterial resistance. Iran Red Crescent Med J IRCMJ ©iranian Red Crescent Med J. 2009;11(3):244–253. [Google Scholar]
- 2.Lau GW, Hassett DJ, Britigan BE. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 2005;13(8):389–397. doi: 10.1016/j.tim.2005.05.011 [DOI] [PubMed] [Google Scholar]
- 3.Han Y, Wang T, Chen G, et al. A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 2019;15(12):e1008198. doi: 10.1371/journal [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada T, Mehta RR, Lekmine F, et al. A peptide fragment of azurin induces a p53-mediated cell cycle arrest in human breast cancer cells. Mol Cancer Ther. 2009;8(10):2947–2958. doi: 10.1158/1535-7163.MCT-09-0444 [DOI] [PubMed] [Google Scholar]
- 5.Yoon SS, Hennigan RF, Hilliard GM, et al. Pseudomonas aeruginosa anaerobic respiration in biofilms. Dev Cell. 2002;3(4):593–603. doi: 10.1016/S1534-5807(02)00295-2 [DOI] [PubMed] [Google Scholar]
- 6.Kamp M, Silvestrini MC, Brunori M, Beeumen J, Hali FC, Canters GW. Involvement of the hydrophobic patch of azurin in the electron-transfer reactions with cytochrome c551 and nitrite reductase. Eur J Biochem. 1990;194(1):109–118. doi: 10.1111/j.1432-1033.1990.tb19434.x [DOI] [PubMed] [Google Scholar]
- 7.Yamada T, Goto M, Punj V, et al. The bacterial redox protein azurin induces apoptosis in J774 macrophages through complex formation and stabilization of the tumor suppressor protein p53. Infect Immun. 2002;70(12):7054–7062. doi: 10.1128/IAI.70.12.7054-7062.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright BW, Kamath KS, Krisp C, et al. Proteome profiling of Pseudomonas aeruginosa PAO1 identifies novel responders to copper stress. BMC Microbiol. 2019;19(1):1–19. doi: 10.1186/s12866-019-1441-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhari A, Fialho AM, Ratner D, et al. Azurin, Plasmodium falciparum malaria and HIV/AIDS: inhibition of parasitic and viral growth by azurin. Cell Cycle. 2006;5(15):1642–1648. doi: 10.4161/cc.5.15.2992 [DOI] [PubMed] [Google Scholar]
- 10.Das Gupta TK, Green A, Shilkaitis A, et al. Noncationic peptides obtained from azurin preferentially enter cancer cells. Cancer Res. 2009;69(2):537–546. doi: 10.1158/0008-5472.can-08-2932 [DOI] [PubMed] [Google Scholar]
- 11.Naguleswaran A, Fialho AM, Chaudhari A, et al. Azurin-like protein blocks invasion of toxoplasma gondii through potential interactions with parasite surface antigen SAG1. Antimicrob Agents Chemother. 2008;52(2):402–408. doi: 10.1128/AAC.01005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apiyo D, Wittung-Stafshede P. Unique complex between bacterial azurin and tumor-suppressor protein p53. Biochem Biophys Res Commun. 2005;332(4):965–968. doi: 10.1016/j.bbrc.2005.05.038 [DOI] [PubMed] [Google Scholar]
- 13.Fialho A, Das Gupta T, Chakrabarty A. Designing promiscuous drugs? Look at what nature made! Lett Drug Des Discov. 2006;4(1):40–43. doi: 10.2174/157018007778992946 [DOI] [Google Scholar]
- 14.Sereena M, Sebastian D. Molecular detection of azurin: a powerful anticancer protein from native pseudomonas isolates. J Adv Med Pharm Sci. 2015;5(1):1–7. doi: 10.9734/jamps/2016/20557 [DOI] [Google Scholar]
- 15.Chen J, Zhuang G, Frieden L, Debinski W. Eph receptors and ephrins in cancer: common themes and controversies. Cancer Res. 2008;68(24):10031–10033. doi: 10.1158/0008-5472.CAN-08-3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammadi Barzelighi H, Nasr-Esfahani B, Bakhshi B, et al. Analysis of antibacterial and antibiofilm activity of purified recombinant Azurin from Pseudomonas aeruginosa. Iran J Microbiol. 2019;11(2):166–176. doi: 10.18502/ijm.v11i2.1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammadi Barzelighi H, Esfahani BN, Bakhshi B, et al. Influence of heterologously expressed azurin from pseudomonas aeruginosa on the adhesion and invasion of pathogenic bacteria to the Caco-2 Cell Line. Probiotics Antimicrob Proteins. 2019. doi: 10.1007/s12602-019-09573-2 [DOI] [PubMed] [Google Scholar]
- 18.Ramachandran S, Sarkar S, Mazumadar A, Mandal M. Azurin synthesis from Pseudomonas aeruginosa MTCC 2453, properties, induction of reactive oxygen species, and p53 stimulated apoptosis in breast carcinoma cells. Journal of Cancer Science & Therapy. 2011;3(5):104–111. doi: 10.4172/1948-5956.1000069 [DOI] [Google Scholar]
- 19.Osman YA, El-Deep D, Younis SA. Azurin: A Powerful Anticancer from “A” Local Pseudomonas aeruginosa Isolate. J Am Sci. 2013;9(12):755–764. [Google Scholar]
- 20.Nanvazadeh F, Khosravi AD, Zolfaghari MR, et al. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns. 2013;39(7):1409–1413. doi: 10.1016/j.burns.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 21.Spilker T, Coenye T, Vandamme P, et al. PCR-based assay for differentiation of pseudomonas aeruginosa from other pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42(5):2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. M07-A10: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition. January 2015. [Google Scholar]
- 23.Falagas ME, Koletsi PK, Bliziotis IA. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J Med Microbiol. 2006;55(12):1619–1629. doi: 10.1099/jmm.0.46747-0 [DOI] [PubMed] [Google Scholar]
- 24.Dumas JL, Van Delden C, Perron K, et al. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett. 2006;254(2):217–225. doi: 10.1111/j.1574-6968.2005.00008.x [DOI] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative Ct method. Nat Prod Rep. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 26.Sabuda DM, Laupland K, Pitout J, et al. Utilization of colistin for treatment of multidrug-resistant Pseudomonas aeruginosa. Can J Infect Dis Med Microbio. 2008;19(6):413–418. doi: 10.1155/2008/743197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martis N, Leroy S, Blanc V. Colistin in multi-drug resistant Pseudomonas aeruginosa blood-stream infections. J Infect. 2014;69(1):1–12. doi: 10.1016/j.jinf.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 28.Cai Y, Yang D, Wang J, Wang R. Activity of colistin alone or in combination with rifampicin or meropenem in a carbapenem-resistant bioluminescent Pseudomonas aeruginosa intraperitoneal murine infection model. J Antimicrob Chemother. 2018;73(2):456–461. doi: 10.1093/jac/dkx399 [DOI] [PubMed] [Google Scholar]
- 29.Bassetti M, Peghin M, Vena A, Giacobbe DR. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front Med. 2019;6:1–10. doi: 10.3389/fmed.2019.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarwar F. Molecular Detection of Anticancer Protein Azurin in Native Pseudomonas Isolates. BRAC University; 2018:31–36. [Google Scholar]
- 31.Malkov S, Zivkovic MV, Beljanski MV, Zaric SD. Correlations of amino acids with secondary structure types: connection with amino acid structure. arxiv.org. 2005;34:1–13. [Google Scholar]
- 32.Nguyen VD, Nguyen TT, Pham TT, Packianather M, Le CH. Molecular screening and genetic diversity analysis of anticancer Azurin-encoding and Azurin-like genes in human gut microbiome deduced through cultivation-dependent and cultivation-independent studies. Int Microbiol. 2019;22(4):437–449. doi: 10.1007/s10123-019-00070-8 [DOI] [PubMed] [Google Scholar]
- 33.Ichise Y, Kosuge T, Uwate M, Nakae T, Maseda H. Complete genome sequence of pseudomonas aeruginosa strain 8380, isolated from the human gut. GenomeAnnounc. 2015;3(3):e00520–e005215. doi: 10.1128/genomeA.00520-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espinosa-Camacho LF, Delgado G, Miranda-Novales G, Soberón-Chávez G, Alcaraz LD, Morales-Espinosa R. Complete genome sequences of two Pseudomonas aeruginosa strains isolated from children with bacteremia. Genome Announc. 2017;5(35):e00927–e00917. doi: 10.1128/genomeA.00927-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magalhães B, Senn L, Blanc DS. High-quality complete genome sequences of three Pseudomonas aeruginosa isolates retrieved from patients hospitalized in intensive care units. Microbiol Resour Announc. 2019;8(9):e01624–18. doi: 10.1128/MRA.01624-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamalakannan T, Rajagopal K, Jothi A, Nirai ST. Analysis of Azurin Protein from Different Pseudomonas sp Using Proteomic Tools. J Commun Comput. 2011;8:727–730. [Google Scholar]