Abstract
Insects feeding on the nutrient-poor diet of xylem plant sap generally bear two microbial symbionts that are localized to different organs (bacteriomes) and provide complementary sets of essential amino acids (EAAs). Here, we investigate the metabolic basis for the apparent paradox that xylem-feeding insects are under intense selection for metabolic efficiency but incur the cost of maintaining two symbionts for functions mediated by one symbiont in other associations. Using stable isotope analysis of central carbon metabolism and metabolic modeling, we provide evidence that the bacteriomes of the spittlebug Clastoptera proteus display high rates of aerobic glycolysis, with syntrophic splitting of glucose oxidation. Specifically, our data suggest that one bacteriome (containing the bacterium Sulcia, which synthesizes seven EAAs) predominantly processes glucose glycolytically, producing pyruvate and lactate, and the exported pyruvate and lactate is assimilated by the second bacteriome (containing the bacterium Zinderia, which synthesizes three energetically costly EAAs) and channeled through the TCA cycle for energy generation by oxidative phosphorylation. We, furthermore, calculate that this metabolic arrangement supports the high ATP demand in Zinderia bacteriomes for Zinderia-mediated synthesis of energy-intensive EAAs. We predict that metabolite cross-feeding among host cells may be widespread in animal–microbe symbioses utilizing low-nutrient diets.
Subject terms: Microbial communities, Microbial ecology
Introduction
Many symbiotic associations between animals and microbes are nutrition based, enabling the animal host to survive on nutrient-poor diets [1, 2]. The dominant focus of research on these beneficial systems is the genetic and metabolic capacity of the microbial partner to produce nutrients of value to the host [3], while the metabolic traits of the host cell that sustain the microbial functions has been relatively neglected.
In various animals with intracellular bacteria, the bacterial symbionts are housed in host cells that are morphologically and functionally distinctive, and often aggregated as an organ. For example, various insects possess bacteriome(s) containing beneficial bacteria which, generally, have very small genomes and are transmitted vertically from mother insect to offspring [1, 4]. Bacteriomes have evolved in insects feeding through the life cycle on nutritionally inadequate diets, including vertebrate blood (e.g., cimicid bedbugs, Glossina tsetse flies), plant phloem and xylem sap (e.g., aphids, whiteflies, cicadas, spittlebugs), and sound wood (e.g., anobiid and bostrychid beetles). Where investigated, the bacterial partners overproduce and release either or both essential amino acids (EAAs) and B vitamins, supplementing the inadequate dietary supply of these nutrients for the host [3].
The purpose of this study was to investigate how the bacteriomes of an insect support the metabolic functions of the bacterial symbionts. The symbiosis in xylem-feeding insects is particularly suitable for this topic for two reasons. First, xylem sap is exceptionally nutrient-poor and insects that utilize this diet as sole food source display remarkable metabolic efficiency [5–7]. Evidence from ancestral state reconstruction studies shows that xylem feeding has apparently evolved once among animals, in the ancestor of hemipteran insects of the suborder Auchenorrhyncha, which likely bore bacterial symbionts [8]. This suggests that the nutritional role of the symbionts is crucial for xylem-feeding insects and, presumably, supported by metabolic adaptations of the bacteriomes. Second, in many xylem-feeding insects, EAA biosynthesis is partitioned between a primary symbiont Candidatus Sulcia muelleri (Bacteroidetes, henceforth Sulcia) which produces seven or eight EAAs, and different co-primary symbionts that produce the complementary and energetically costly set of three or eight EAAs (methionine, histidine, and tryptophan) [9–13]. This dual symbiosis is likely the ancestral condition [8], and both this and the multiple replacements of the co-primary symbiont have been attributed to genomic deterioration and loss of function [4, 10, 14]. Metabolic modeling indicates that this arrangement is metabolically inefficient because it is more costly for a host to maintain populations of two symbionts, each of which provides a subset of EAAs [15], compared with supporting a single symbiont that provides the full complement of EAAs (as occurs in many other insects). Research on other systems, however, suggests that partitioning of function can promote metabolic efficiency in an energy-limiting environment [16–18] raising the possibility that the division of EAA production between two bacterial symbionts in xylem-feeding insects may confer metabolic advantages. However, this alternative scenario has not been investigated.
We used the spittlebug Clastoptera proteus because large numbers of insects of uniform developmental age can be collected from natural populations and dissected in a realistic timeframe to isolate metabolically active bacteriomes for study. In addition, Clastoptera symbionts Sulcia and Candidatus Zinderia insecticola (β-proteobacteria, henceforth Zinderia) are localized to different, closely associated bacteriomes that can be distinguished by pigmentation and can be readily separated with minimal cross contamination of the partners (Fig. 1). These qualities make the spittlebug Clastoptera proteus an experimentally tractable system to compare metabolic function between bacteriomes. Genome analysis of the bacterial symbionts in the congeneric insect C. arizonana has revealed the capacity for complementary nutritional functions: Zinderia can synthesize three EAAs (methionine, histidine, and tryptophan) and riboflavin (vitamin B2), while Sulcia can synthesize the remaining seven EAAs [12, 19]. The symbionts of C. proteus are predicted to have the same metabolic traits, in the light of very limited variation in gene content and especially metabolism-related genes, among vertically transmitted symbionts in conspecific and congeneric insect hosts [20–22].
Fig. 1. Images of insect used in this study and their dissected bacteriomes.
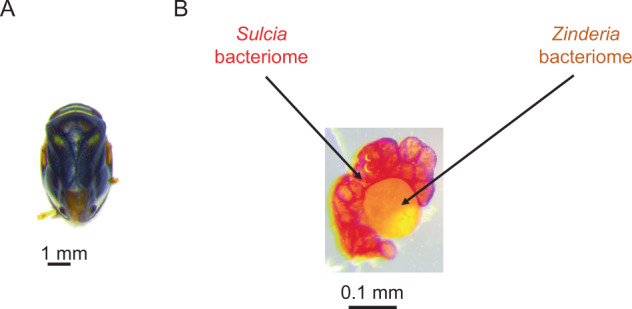
a Adult spittlebug Clastoptera proteus. b Clastoptera bacteriomes showing Sulcia (red) and Zinderia (orange) specific bacteriomes. Each paired bacteriome is readily isolated, and the Sulcia bacteriome and Zinderia bacteriome can then be separated.
We tested two hypotheses: first, that the metabolism of the C. proteus bacteriomes is programed to produce substrates utilized by the bacterial symbionts; and, second, that bacteriomes bearing Zinderia display additional metabolic traits that facilitate the production of energetically expensive EAAs. To test these hypotheses, we applied a stable isotope-assisted metabolomics approach using 13C-glucose and high-resolution liquid chromatography–mass spectrometry. Previous metabolic models of the bacterial symbioses in xylem-feeding insects have presented glucose as the major carbon source for the bacteriome [15] because the glucose disaccharide trehalose is the dominant blood sugar in the insect hemolymph (blood), the location of the bacteriomes [23]. Spittlebug bacteriomes are enriched in transcripts coding the enzyme trehalase, which mediates trehalose hydrolysis to glucose [15]. Furthermore, our experiments were conducted under aerobic conditions, following the observation that spittlebug bacteriomes are supplied with tracheae (the oxygen-delivery system of insects) in vivo.
We extend our analysis of the empirical results using flux balance analysis (FBA) [24] to generate predictions about the mechanisms underlying the differences in metabolic phenotypes observed in the Clastoptera bacteriomes. Where reactions for processing central carbon metabolites are duplicated between the host and symbionts, metabolic modeling predicts the quantitative contributions of host and symbiont reactions to observed bacteriome metabolite profiles. Constraint-based metabolic modeling approaches such as FBA are now widely used in silico tools for investigating species interactions in complex biological systems from humans to soils, and are particularly valuable for systems where traditional empirical approaches are infeasible or have a limited scope [25–29].
Our data support the presence of aerobic glycolysis in the bacteriomes, and further reveal that metabolic routing through the pentose-phosphate, glycolytic, and oxidative phosphorylation pathways differ between the Sulcia-containing and Zinderia-containing bacteriomes. These metabolic traits serve to facilitate the production of EAAs by the bacterial symbionts and promote the overall metabolic efficiency of the insect bacteriome.
Methods
Insect collection
Nymphs of Clastoptera proteus informally known as the dogwood spittlebug were collected from dogwood tree branches in Ithaca, NY in June 2017 and June 2018. Insects were collected from plants ~2 h before dissection. Insects for each biological replicate were collected from different dogwood plants. Species identification was carried out at Cornell University Insect Collection (voucher specimen CU1268).
Metabolite pool sample collection
Spittlebug nymphs were dissected with fine pins and forceps in sterile phosphate-buffered saline on agar plates. Bacteriomes containing both Sulcia and Zinderia (paired bacteriome samples) were isolated from the insect abdomen. For some experiments, the Sulcia-containing (red pigmentation) and Zinderia-containing (yellow pigmentation) bacteriomes (separated bacteriome samples) were separated with fine pins. The identity of symbionts in each bacteriome type was confirmed by 16S rRNA gene amplification and subsequent sequencing of the amplified product. Thirty spittlebugs were dissected to generate bacteriomes for each of three biological replicates, immediately flash-frozen in liquid nitrogen and stored at −80 °C until further processing.
Bacteriome incubation
Bacteriomes were incubated aerobically in filter-sterilized base medium: (8.6 mM NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 50 mM NaH2PO4, 13 mM K2HPO4 (trihydrate)) [pH 7.5] supplemented with 2 mM NH4Cl and 20 mM [U-13C]-glucose (13C6 99%, Sigma-Aldrich, St. Louis, MO, USA) at the room temperature with 10 rpm agitation. The base medium is modified from a medium developed for metabolic analysis of the bacteriomes and symbionts of aphids [30]. This medium supports in vitro culture of bacteriomes for extended periods with minimal cell lysis or culture medium induced cell stress. Bacteriomes were sacrificed for every replicate and timepoint and no non-13C bacteriome incubations were set up for this study.
13C isotopic profiling experiments
Dissected bacteriomes were incubated for up to 3 h in 20 mM 13C-glucose. At each timepoint (5, 30, 60, 120, and 180 min), samples from three replicate sets of pooled bacteriomes from 30 insects were either sacrificed (paired bacteriomes) or 100 µL incubation medium (separated bacteriomes) was collected. All samples were centrifuged at 2348 × g for 2 min. Each of the bacteriome pellet and supernatant fraction per sample was flash-frozen and stored at −80 °C until further processing.
Metabolite extractions from bacteriomes and extracellular medium
Flash-frozen insect bacteriomes were hand homogenized (~10 s) on ice and 100 µL ice-cold filter-sterilized base medium added to each homogenate and briefly vortexed. Metabolite extractions were conducted on 70 µL homogenate and the remainder was used for protein quantification.
Each 70 µL sample for metabolite analysis was extracted with 1 mL extraction solvent (40:40:20 methanol:acetonitrile:water) at −20 °C, and incubated on ice for 2 min, followed by three cycles of centrifugation at 19,090 × g for 2 min at 4 °C, homogenization and vortexing. The supernatant (first extract) was transferred to a new 1.5 mL Eppendorf tube and the remaining pellet resuspended in 100 µL fresh extraction solvent followed by an additional round of vortexing and centrifugation at 19,090 × g for 2 min at 4 °C. The supernatant (second extract) was combined with the first extract and dried under N2 gas. The dried samples were then resuspended in 100 µL HPLC grade water for mass spectrometry analysis. For extracellular metabolite quantification, 100 µL flash-frozen bacteriome incubation medium was thawed on ice and used for metabolite analysis.
Metabolite analysis
Cellular metabolites were identified by reversed-phase ion-pairing liquid chromatography coupled with high-resolution mass spectrometry as detailed previously [31]. Briefly, samples were run through an Acquity UPLC C18 column (Waters, Milford, MA) at a flow rate of 0.180 mL min−1 and an injection volume of 10 µL. Column temperature was maintained at 25 °C. The MS was operated in full scan negative mode. Metabolite identification was based on accurate mass and matches with standard retention time. Metabolites from the isotopic labeling experiments were analyzed on the Metabolomic Analysis and Visualization Engine software [32, 33]. Peak area top counts from extracted ion chromatograms of each metabolite were used as a surrogate for metabolite abundance and were normalized to the protein content of the bacteriome samples (Datasets S1, S2). For each sample of homogenized bacteriomes, protein was quantified by a colorimetric assay using the DC Protein Assay Kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. Absorbance was measured at 750 nm on a Bio-Rad xMarkTM Microplate reader with a standard curve of BSA (0.05–2 mg mL−1).
Data analysis and visualization
Statistical differences were investigated using two-tailed t-tests with FDR correction or one-way ANOVA followed by Tukey’s HSD post hoc test and FDR correction for two-treatment and more than two-treatment analyses, respectively, using MATLAB (The MathWorks, Inc., Natick, MA) and MetaboAnalyst [34]. Boxplots of metabolite abundances, fold changes, and percent label were generated using MATLAB.
Metabolic model reconstruction
Genome-scale metabolic models were generated for the symbiotic bacteria of Clastoptera proteus, using the annotated gene content of publicly available genomes for the bacteria in the related species C. arizonana [S. muelleri (CP002163.1) and Zinderia insecticola (CP002161.1)]. To obtain the bacteriome metabolic model, we collated published metabolic models of three xylem-feeding insects (spittlebugs, cicadas, and sharpshooters) [15] and identified a core set of reactions (~99% of the reactions in each bacteriome model) present in all xylem-feeding insect bacteriomes regardless of insect identity or phylogeny. The genes encoding the core reactions are highly conserved and to our knowledge general among annotated insect genomes. This core set of reactions was used to create the bacteriome metabolic model with default reaction bounds (upper bound of 1000, lower bound of −1000), as is commonly done for genome-scale metabolic models [35]. Individual bacterial and insect metabolic models were integrated into a three-compartment model using previously described methods [15]. Model testing was conducted in COBRA Toolbox version 3.0 [35] run in Matlab 2015b (The MathWorks Inc., Natick, MA), using the Gurobi solver (Gurobi Optimization 2016).
Model constraints and analysis
All model simulations applied aerobic conditions (maximum oxygen uptake flux of 20 mmol gDW−1 h−1), and a minimal external medium comprising glucose, ammonia, cystathionine, and sulfate as sole carbon, nitrogen, and sulfur sources. The constituents of the external medium were selected to match the growth medium used for our in vitro incubations and the nutrient auxotrophies of each symbiont–host model. The maximum uptake fluxes for ammonia and cystathionine were capped at 5 and 0.5 mmol gDW−1 h−1, respectively. Additional metabolites such as B vitamins required in small quantities were added to the external medium, the uptake fluxes of these metabolites were left unbounded (Table S1), and the lower bound of the biomass reaction for each bacterium was fixed at 0.01 mmol gDW−1 h−1. For all model simulations, we used a single objective function derived from published xylem-feeding insect metabolic models [15].
Pyruvate and lactate export simulations
Pyruvate and lactate export simulations were performed using a three-compartment model comprising Sulcia, Zinderia, and the spittlebug host. To simulate changing extracellular nutrient availability, extracellular glucose concentrations between 0.1 and 50 mM were assumed to allow uptake fluxes between 0.1 and 50 mmol gDW−1 h−1. These constraints were applied to glucose exchange reactions. Amino acids were excluded as nutrient sources in all model simulations. Quantitative predictions for the onset of pyruvate and lactate release from spittlebug bacteriomes and ATP synthase flux were investigated by obtaining unique optimal flux vectors by FBA [24, 35]. The proportion of carbon released was calculated by dividing total carbon efflux fluxes by total carbon influx fluxes and multiplying by 100. Total carbon efflux and influx were calculated by multiplying glucose, pyruvate, and lactate transport reaction fluxes by the number of carbon molecules in each transported compound.
Glucose and lactate preference assay simulations
Glucose and lactate preference simulations were performed using two-compartment models comprising Sulcia and the spittlebug host or Zinderia and the spittlebug host. The two-compartment models were generated from the three-compartment models by removing all reactions associated with Sulcia or Zinderia and providing any essential nutrients provided by the removed bacterium in the extracellular medium. Sulcia-spittlebug models were provided with histidine, methionine, tryptophan, and riboflavin. Zinderia-spittlebug models were provided with arginine, isoleucine, leucine, lysine, phenylalanine, threonine, valine, 2-oxobutanoate, homoserine, shikimate, and pantoate. To simulate changing extracellular nutrient availability, extracellular glucose and lactate concentrations between 1 and 50 mM were assumed to allow uptake fluxes between 1 and 50 mmol gDW−1 h−1. Glucose and lactate transport fluxes were allowed to be reversible for all glucose and lactate preference assay simulations. Quantitative predictions of glucose and lactate uptake from the extracellular medium and ATP synthase flux were investigated by FBA [24].
Results
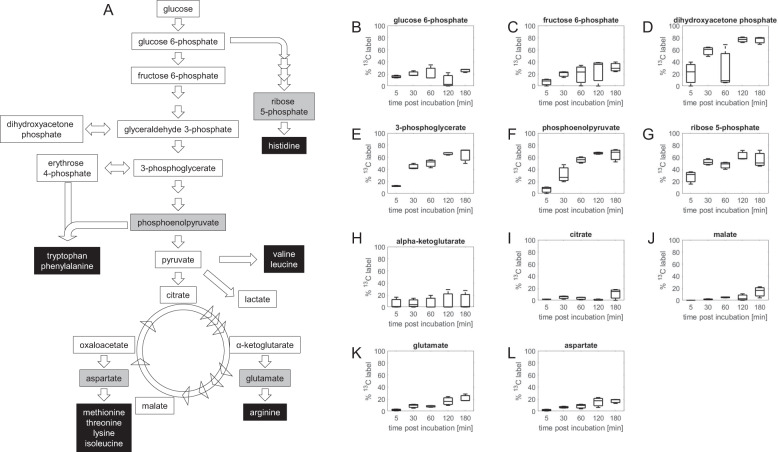
To investigate the metabolic traits of spittlebug bacteriomes that support symbiont EAA production on the extremely nutrient limited diet of xylem sap, we first monitored kinetic 13C profiling of central carbon metabolites in spittlebug bacteriomes incubated in a base medium supplemented with 13C-glucose (Fig. 2a). This substrate is predicted to be metabolized by the insect cells of the bacteriome but not by the symbiotic bacteria, which lack the capacity to utilize glucose as a carbon source (Fig. S1). The first experiments were conducted on bacteriome preparations containing both symbionts, which we term “paired bacteriomes”, and subsequent experiments used symbiont-specific bacteriome preparations, which we refer to as “separated bacteriomes”.
Fig. 2. Glucose-derived 13C incorporation into essential amino acid precursor metabolites.
a Schematic representation of central carbon metabolism. Essential amino acids are shown in black boxes and essential amino acid precursors monitored in this study are shown in gray boxes. Kinetic incorporation of glucose-derived 13C into b–f glycolytic, g pentose-phosphate, h–j TCA cycle metabolites. Kinetic incorporation of glucose-derived 13C into nonessential amino acid precursors, glutamate (k) and aspartate (l). Box-plots show the percentage of the spittlebug intracellular metabolite pool that has accumulated 13C-labeled fraction.
Spittlebug bacteriomes are metabolically active
Monitoring of kinetic isotopic enrichment of intracellular metabolites demonstrated that the isolated paired bacteriomes were metabolically active (Fig. 2). We observed a steady increase in 13C incorporation into the monitored glycolytic metabolites [i.e., glucose 6-phosphate, fructose 6-phosphate, dihydroxyacetone phosphate, 3-phosphoglycerate, phosphoenolpyruvate (PEP)], and the pentose-phosphate pathway (i.e., ribose 5-phosphate (R5P)), reaching 30–80% in 13C-labeled fractions (Fig. 2b–g). The metabolites PEP and R5P are precursors to EAAs (Fig. 2a). Metabolite labeling in the TCA cycle was variable, alpha-ketoglutarate did not display an increasing trend in 13C-labeling (Fig. 2h), but both citrate and malate displayed a steady increase in 13C incorporation, reaching 11–14% in 13C-labeled fractions (Fig. 2i, j). We also observed up to 30% 13C enrichment in two TCA cycle-derived amino acids, glutamate and aspartate, which are precursors to EAAs (Fig. 2k–l).
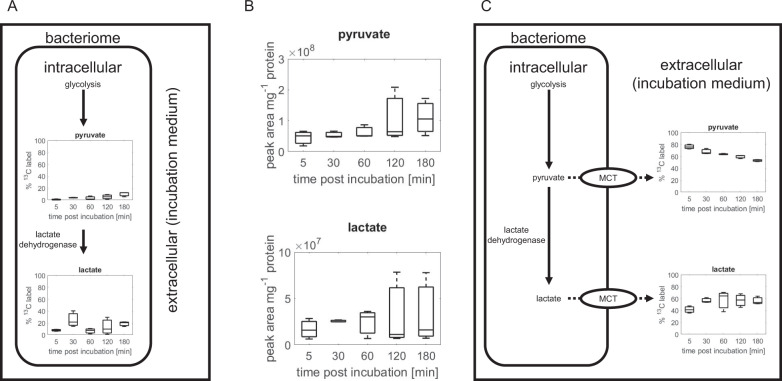
Pyruvate and lactate exported from paired spittlebug bacteriomes
The 13C enrichment of pyruvate was only 10% (Fig. 3a), despite 60% 13C-labeling of its metabolic precursor PEP after 3 h (Fig. 2a). Interestingly, the 13C fraction in lactate, which is derived from pyruvate, was 20% after 3 h (Fig. 3a) suggesting a rapid conversion of pyruvate to lactate by the bacteriomes. The 13C-labeled fraction in lactate (~20%), however, was substantially lower than PEP’s (~60%), implying that rapid conversion of pyruvate to lactate could not fully account for the minimal label incorporated into pyruvate.
Fig. 3. Comparison of intracellular and extracellular pyruvate and lactate 13C-label incorporation during 3-h incubation.
a Intracellular pyruvate and lactate 13C-label incorporation. b Peak area count of extracellular pyruvate and lactate. c Extracellular pyruvate and lactate 13C-label incorporation. Monocarboxylate transporter (MCT) mediates bi-directional transport of pyruvate and lactate. Box-plots show the percentage of the intracellular metabolite pool that has accumulated 13C-label.
We measured the metabolites in the extracellular medium in which the paired bacteriomes were incubated (Fig. 3b). Compared with the 5-min incubation, the extracellular abundance of pyruvate and lactate were both increased by up to approximately threefold by the end of the 3-h incubation, indicating that the bacteriomes exported a fraction of the assimilated glucose as pyruvate and lactate (Fig. 3b). The 13C fraction of the extracellular pyruvate steadily decreased from 80% after 5-min incubation, to 50% at 3-h incubation, but the 13C-labeled fraction of lactate, a pyruvate derivative, started at ~40% and increased to ~65% (Fig. 3c). The latter trend was consistent with the activity of lactate dehydrogenase to produce lactate from pyruvate (Fig. 3a). However, the trend in the labeling of the extracellular pyruvate implied contribution of a non-labeled precursor metabolite to pyruvate (Fig. 3c). Compared with the extracellular labeling scheme of pyruvate and lactate, the intracellular labeling 13C fractions were relatively lower (Fig. 3a). This large discrepancy between the extracellular and intracellular 13C fractions of pyruvate and lactate can be attributed to the high extracellular levels of these metabolites, which can interfere with the accurate determination of intracellular 13C enrichment (Fig. 3a–c). Nevertheless, the relative abundance and labeling schemes of the extracellular metabolites highlighted both rapid generation of lactate from pyruvate and export of both pyruvate and lactate into the extracellular environment (Fig. 3a–c).
High glucose uptake flux triggers overflow pyruvate and lactate export
We employed metabolic modeling of the spittlebug and its symbiotic partners Sulcia and Zinderia to investigate how the export of pyruvate and lactate by the spittlebug bacteriomes varies with extracellular glucose concentrations. Specifically, we used FBA in a three-compartment genome-scale metabolic model, with the extracellular glucose concentration varied between 0.1 and 50 mM and the extracellular ammonia concentration maintained at 5 mM. Our flux simulations identified an abrupt change in the metabolism of spittlebug bacteriomes in response to glucose availability (Fig. S2). At relatively low glucose uptake flux (<9 mmol g dry weight−1 h−1), the bacteriomes exhibited glycolytic flux with minimal export of pyruvate or lactate (<10 mmol g dry weight−1 h−1) but, at higher glucose uptake flux (≥9 mmol g dry weight−1 h−1), the bacteriomes switched to aerobic glycolysis, in which export flux of lactate or pyruvate was substantial, ranging from 11 to 70 mmol g dry weight−1 h−1 (Fig. S2a). Using our flux simulations, we predicted that spittlebug bacteriomes exported up to 90% of their total glucose-derived carbon influx as pyruvate and lactate at the highest uptake of glucose (Fig. S2b). Increasing glucose uptake flux also compromised oxidative phosphorylation activity in spittlebug bacteriomes, ATP synthase flux steadily decreased with increased glucose uptake (Fig. S2c).
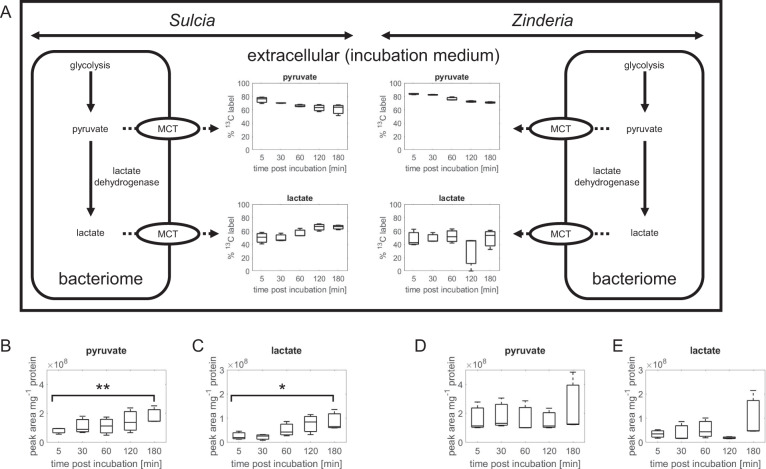
Separated Sulcia and Zinderia bacteriomes exhibit different cellular metabolomes
We tested whether separated bacteriomes displayed pyruvate and lactate export when incubated with exogenous 13C-labeled glucose (Fig. 3). As observed with the paired bacteriomes after 3-h incubations, we also obtained appreciable 13C incorporation into pyruvate (50–80%) and lactate (30–70%) in the extracellular medium of the separated Sulcia and Zinderia bacteriomes (Fig. 4a). Relative to the 5-min incubation, extracellular abundance of pyruvate and lactate significantly (P < 0.1) increased by approximately twofold and fourfold, respectively, in the Sulcia-bacteriome medium by the end of the 3-h incubation (Fig. 4b, c). In the Zinderia-bacteriome medium, the extracellular abundances of pyruvate and lactate increased by ~1.4-fold and 3-fold, respectively, by the end of the 3-h incubation (Fig. 4d, e) but these increases were not statistically significant. In addition, the median of the extracellular abundance of both pyruvate and lactate was higher for Sulcia bacteriomes compared with Zinderia bacteriomes for three of the five timepoints sampled (Fig. S3). These data highlight a possible metabolic programming that may facilitate greater export of pyruvate and lactate in Sulcia bacteriomes relative to Zinderia bacteriome in response to exogenous glucose.
Fig. 4. Comparison of pyruvate and lactate release from Sulcia and Zinderia-spittlebug bacteriomes.
Extracellular pyruvate and lactate, expressed as percentage of 13C-label in Sulcia and Zinderia (a) and expressed as area count, for pyruvate and lactate in Sulcia (b, c) and Zinderia (d, e). Asterisks indicate significant differences between specific timepoints compared with the 5 min timepoint. *p < 0.1; **p < 0.05.
To further investigate possible differences in metabolism between bacteriomes containing Sulcia and Zinderia, we obtained the relative intracellular abundances of 75 metabolites associated with central carbon metabolism, amino acid biosynthesis, and nucleotide biosynthesis (Fig. S4, Dataset S2). The Sulcia bacteriome had elevated levels of metabolites involved in the pentose-phosphate pathway, glycolysis, and the reduced equivalent NADH, which is generated in surplus in glycolysis (Fig. S5a). In contrast, metabolites involved in the TCA cycle were elevated in the Zinderia bacteriome (Fig. S5b). Due to high-sample variability, not all metabolites accumulated in specific bacteriomes were statistically significant (Fig. S5). Approximately 20% (15 out of 75) of the measured metabolites differed significantly (P < 0.1 and >2-fold change) between the Sulcia and Zinderia bacteriomes after false discovery rate (FDR) correction (Fig. S6).
The significantly different metabolites included products from pathways in amino acid biosynthesis (Fig. S6a–c), central carbon metabolism (Fig. S6d, e), B vitamin biosynthesis (Fig. S6f–g), nucleotide metabolism (Fig. S6h–m), and glutathione metabolism (Fig. S6n–o). The EAAs methionine and tryptophan, which are produced by Zinderia, were significantly enriched in the Zinderia bacteriome and aspartate, a precursor for three Sulcia-produced EAAs (threonine, lysine, and isoleucine), was enriched in the Sulcia bacteriome (Fig. S6a–c). Riboflavin, the sole B vitamin produced by Zinderia, and GTP the precursor for riboflavin biosynthesis were depleted in Zinderia bacteriomes compared with Sulcia bacteriomes (Fig. S6g, h). In contrast, the riboflavin derivative FAD (an important cofactor of the TCA cycle) was enriched in Zinderia bacteriomes compared with Sulcia bacteriomes (Fig. S6f), suggesting differences in FAD utilization and the availability of FAD precursors between the Sulcia and Zinderia bacteriomes.
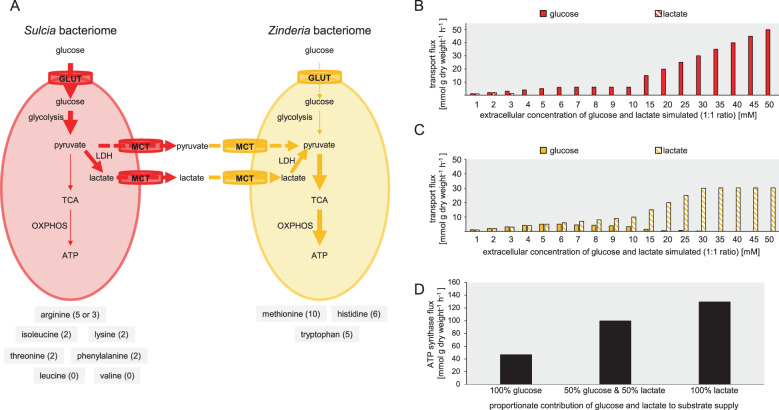
A final and striking difference in the datasets was the enrichment in intracellular lactate abundance in Zinderia bacteriomes, compared with Sulcia bacteriomes (Fig. S5a). The enrichment of lactate in Zinderia bacteriomes could be attributed to three mechanisms (1) increased lactate dehydrogenase activity in Zinderia bacteriomes, (2) decreased utilization of lactate pools in Zinderia bacteriomes, or (3) increased assimilation of exogenous lactate by Zinderia bacteriomes. We decided to prioritize testing of the last mechanism because this mechanism would increase the metabolic efficiency of energy production, a trait of selective advantage to insects feeding on the extremely nutrient-poor diet of xylem sap. Specifically, lactate requires fewer enzymatic steps than glucose to enter the TCA cycle for energy generation, thereby minimizing the protein costs associated with glycolytic enzyme synthesis and maintenance [36] for the Zinderia bacteriomes. These considerations led to the specific prediction that the Sulcia and Zinderia bacteriomes are metabolically linked, such that host-derived glucose is metabolized by the Sulcia bacteriome, via aerobic glycolysis, to lactate; and this lactate is imported into Zinderia bacteriomes to enhance energy production by oxidative phosphorylation (Fig. 5a).
Fig. 5. Metabolic specialization in spittlebug bacteriomes.
a Model for metabolite transfer in bacteriomes. Lactate released as an end product of aerobic glycolysis from Sulcia bacteriomes is imported by Zinderia bacteriomes and oxidized to produce energy. Essential amino acids produced in each bacteriome are listed in gray boxes and energy costs (μmol ATP per μmol amino acid synthesized) for the synthesis of each amino acid are provided in parentheses. Energy costs data obtained from ref. [9]. Reaction fluxes (solid arrows) and transport fluxes (dashed arrows) are shown. Arrow thickness illustrate the magnitude of flux size, thicker arrows represent higher fluxes. GLUT glucose transporter, LDH lactate dehydrogenase, MCT monocarboxylate transporter, TCA tricarboxylic acid, OXPHOS oxidative phosphorylation, ATP adenosine triphosphate. In silico glucose and lactate choice assay for (b) Sulcia bacteriomes and (c) Zinderia bacteriomes. d In silico predictions of ATP synthase flux in Zinderia bacteriomes when provided with glucose, a mixture of glucose and lactate or lactate as carbon sources.
Cross-feeding of Sulcia-bacteriome derived lactate enhances Zinderia-bacteriome energy production
A key element to the rationale for our hypothesis that the assimilation by Zinderia bacteriomes of lactate derived from Sulcia bacteriomes (Fig. 5a) is that this metabolic link enhances the oxidative phosphorylation capacity of Zinderia bacteriomes. To test this prediction, we applied FBA to two-compartment metabolic models of the bacteriomes.
Our first analysis concerned the uptake fluxes of glucose and lactate provided in silico to isolated Sulcia bacteriomes. Our simulations reveal that Sulcia bacteriomes utilize both carbon sources, but they utilize glucose preferentially as a bioenergetic substrate with increasing availability of glucose and lactate (Fig. 5b). Our simulations also showed that ATP synthase flux steadily decreased in Sulcia bacteriomes with increased glucose uptake (Fig. S7a). We then investigated the uptake fluxes of glucose and lactate provided in silico to isolated Zinderia bacteriomes. These simulations reveal that Zinderia bacteriomes also utilize both carbon sources. However, in contrast to Sulcia bacteriomes, Zinderia bacteriomes utilize lactate preferentially as a bioenergetic substrate with increasing availability of glucose and lactate (Fig. 5c). ATP synthase flux also steadily increased in Zinderia bacteriomes with increased lactate uptake (Fig. S7b).
We then compared the energy-generating potential of lactate and glucose assimilated by the Zinderia bacteriomes. These analyses reveal a progressive increase in ATP synthase flux with increasing contribution of lactate (Fig. 5d). When the Zinderia bacteriome is simulated to receive glucose as a sole carbon source, ATP synthase flux was 47 mmol g dry weight−1 h−1 (Fig. 5d), increasing to 99 mmol g dry weight−1 h−1 when lactate was provided and assimilated by Zinderia bacteriomes in addition to glucose (Fig. 5d), and increasing even further to 129 mmol g dry weight−1 h−1 when lactate was provided as a sole source of carbon (Fig. 5d).
In summary, our simulations show that lactate released from Sulcia bacteriomes diverts pyruvate away from the TCA cycle and compromises oxidative phosphorylation activity in Sulcia bacteriomes. At the same time, lactate released from Sulcia bacteriomes when assimilated by Zinderia bacteriomes into the TCA cycle enhances energy production by increased oxidative phosphorylation activity.
Discussion
This study of the spittlebug Clastoptera proteus symbiosis with the bacteria Sulcia and Zinderia demonstrates the metabolic feasibility and energetic advantage of the syntrophic splitting of the central carbon metabolism between the symbiosis organs (bacteriomes): glycolysis in the Sulcia bacteriomes and the TCA cycle in Zinderia bacteriomes, linked by lactate transfer. These metabolic traits are predicted to facilitate the metabolic function of the bacterial symbionts. In particular, the capacity for aerobic glycolysis, where cells preferentially use glycolysis for energy generation when oxygen is not limiting [37, 38] is adopted by many rapidly proliferating cells, e.g., some cancer cells and rapidly growing insect larvae [39, 40], as well as metabolically active nonproliferating cells, e.g., activated cells in the innate immune system [41, 42]. Aerobic glycolysis of the cells of the spittlebug bacteriomes may provide precursors for both EAA synthesis by the bacterial symbionts and, more generally, precursors for biomass increase of the bacteriomes. Indeed, our data suggest that aerobic glycolysis in spittlebug bacteriomes supports the production of biosynthetic intermediates upstream of pyruvate. Nevertheless, our observation that most assimilated carbon is eventually exported from spittlebug bacteriomes as pyruvate and lactate raises the possibility that a subpopulation of cells in some bacteriomes utilize the exported pyruvate and lactate as bioenergetic substrates thereby increasing the overall metabolic efficiency of spittlebug bacteriomes.
How might pyruvate and lactate export from spittlebug bacteriomes impact EAA production and overall bacteriome metabolic efficiency? Two complementary processes may be involved. First, exporting pyruvate and lactate may eliminate bottlenecks in maintaining high glycolytic flux caused by accumulation of glycolytic intermediates, thereby maintaining rapid EAA precursor production in Sulcia. Additionally or alternatively, the exported pyruvate and lactate may be re-assimilated for use as bioenergetic substrates by a subset of the bacteriome cells. Of note, our in silico simulations indicate that Zinderia-containing bacteriomes utilize lactate preferentially as a carbon source and generate more energy from lactate compared with glucose. These results also raise the possibility that, as in other systems [43, 44], lactate secreted from spittlebug bacteriomes may function as a signal or mediate acidification of the bacteriome extracellular microenvironment, with consequences for bacteriome metabolism and the regulation of symbiont function. While Zinderia bacteriomes can in principle produce, export and re-assimilate exported lactate for use as a carbon source, a few conditions make the simultaneous production and uptake of lactate by the same bacteriome type unlikely. First, high rates of glycolysis deplete cellular NAD+ availability and thereby reduce glycolytic flux [38]. To maintain high glycolytic activity, cells alter their metabolism to regenerate NAD+ through lactate dehydrogenase activity which converts pyruvate to lactate and regenerates NAD+ from NADH. Second, measurements of the direction of lactate transport between metabolically heterogeneous eukaryotic cell populations suggest lactate is transported from glycolytic cells to oxidative cells [45, 46].
Our demonstration that Sulcia- and Zinderia-containing bacteriomes exhibit distinct glycolytic and oxidative metabolic profiles further supports our prediction that pyruvate and lactate produced by the Sulcia bacteriome are shuttled to the Zinderia bacteriomes, where they can be assimilated into the TCA cycle for biosynthetic intermediate production and energy generation through oxidative phosphorylation. One potential caveat to this interpretation is that contribution of different partners to metabolic reactions mediated by both the host and symbionts could not be quantified experimentally. Nevertheless, two lines of evidence indicate that the host is the primary driver of bacteriome central carbon metabolism. First, neither Sulcia nor Zinderia has the genetic capacity to assimilate and process glucose as a carbon source or produce and export lactate, indicating that the host mediates glucose consumption and the production of released lactate. Second, our model simulations predict that symbiont contributions to metabolic reactions also present in the host are up to 200-fold lower than in the host.
This use of multistep glucose oxidation is best documented in cells of tumors [47]. However, it is also known in nonmalignant cells, including glycolytic astrocytes which shuttle lactate to oxidative neurons in the brain [48, 49] and fast ‘white’ glycolytic fibers and slow ‘red’ oxidative fibers which exchange lactate in skeletal muscle [50, 51]. These findings along with those observed in our insect bacteriomes indicate that the function of multistep glucose oxidation extends beyond cell proliferation and is the hallmark of healthy metabolism in various tissues.
A key inference from our research is that the metabolism of bacteriomes containing Zinderia is structured to generate the required energy needed to synthesize expensive EAAs using products released from Sulcia-containing bacteriomes. This trait may contribute to the prevalence of multi-partner symbiosis among insects feeding on extremely nutrient-poor diets [9]. The occurrence of multi-partner symbioses has largely been attributed to genomic decay of a symbiont, requiring compensatory responses by either recruitment of the lost metabolic function by the host or by the recruitment of additional symbionts [52]. Our metabolic modeling data demonstrate that partitioning of metabolic tasks between multiple symbionts and metabolic specialization by the host cells harboring the symbionts may be of selective advantage to the host by increasing resource use proficiency and metabolic efficiency through cooperative cross-feeding of metabolites between metabolically heterogeneous bacteriome populations.
How general is the incidence of metabolic differentiation and coupling in carbon metabolism between the host cells bearing nutritional symbionts? In the context of evidence that a dual symbiosis including Sulcia is both the ancestral condition and general to xylem-feeding insects [8], we argue that this metabolic arrangement may be widespread among insects bearing Sulcia and Zinderia, and possibly relevant to other xylem-feeding insects that harbor different co-symbionts with Sulcia. The evolutionary supplementation of a single symbiont with a second (or more) symbiont(s) partitioned to different bacteriocytes has also occurred multiple times in plant phloem sap hemipteran insects [1] and the metabolic functions outsourced to the additional symbiont generally comprise synthesis of energetically expensive EAAs [9]. This raises the possibility that the integration of the second symbiont into the symbiosis has been accompanied by the syntrophic splitting of host cell metabolic function. A further consideration is the potential for functional heterogeneity among individual host cells of bacteriomes bearing a single symbiont. As in other systems, metabolic efficiency may be promoted by the phenotypic specialization of a subset of host cells and their complement of bacterial symbionts for the synthesis of costly EAAs, supported by the products of neighboring cells displaying aerobic glycolysis. Overall, a greater research focus on the metabolic function of the host cell will greatly facilitate our understanding of the evolution and function of intracellular symbioses.
Supplementary information
Acknowledgements
We thank Seung Ho Chung, Lu Liu, Frances Blow and Alyssa Bost for assistance with insect dissections, Jason Dombroskie (Cornell University Insect Collection) for assistance with insect identification, and Brandon Barker (Aristotle Cloud Federation) for assistance with virtual machine image development, made possible by National Science Foundation grant ACI-1541215. This study was funded by NSF grant IOS-1354743 awarded to A.E.D., and a NSF CAREER grant (CBET-1653092) awarded to L.A. Graduate support for RAW was also provided by the NSF Graduate Research Fellowship Program (DGE-1650441).
Author contributions
NYDA, LA, and AED designed research; NYDA, FZ, DZ, and TK performed research; RAW and LA performed mass spectrometry analysis; NYDA created the models and performed model analysis, NYDA analyzed data; NYDA and AED wrote the first draft of the paper; paper revisions made by all authors.
Data availability
All models have been provided in three formats—SBML (.xml), MATLAB (.mat), and Excel (.xls)—and deposited in GitHub (https://github.com/na423/symbiosis_Warburg). SBML files of the models have also been submitted to the BioModels database [53] with the following identifiers: MODEL1908040002, MODEL1908040003, and MODEL1908040004.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ludmilla Aristilde, Email: ludmilla.aristilde@northwestern.edu.
Angela E. Douglas, Email: aes326@cornell.edu
Supplementary information
The online version of this article (10.1038/s41396-020-0661-z) contains supplementary material, which is available to authorized users.
References
- 1.Buchner P. Endoymbiosis of animals with plant microorganisms. Chichester, United Kingdom: John Wiley and Sons; 1965.
- 2.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–36. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douglas AE. Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol. 2015;60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 5.Mattson WJ., Jr Herbivory in relation to plant nitrogen content. Annu Rev Ecol Syst. 1980;11:119–61. [Google Scholar]
- 6.Brodbeck BV, Mizell RF, Andersen PC. Physiological and behavioral adaptations of three species of leafhoppers in response to the dilute nutrient content of xylem fluid. J Insect Physiol. 1993;39:73–81. [Google Scholar]
- 7.Redak RA, Purcell AH, Lopes JRS, Blua MJ, Mizell RF, Andersen PC. The biology of xylem fluid feeding insect vectors of xylella fastidiosa and their relation to disease epidemiology. Annu Rev Entomol. 2004;49:243–70. doi: 10.1146/annurev.ento.49.061802.123403. [DOI] [PubMed] [Google Scholar]
- 8.Bell-Roberts L, Douglas AE, Werner GD. Match and mismatch between dietary switches and microbial partners in plant sap-feeding insects. Proc R Soc B. 2019;286:20190065. doi: 10.1098/rspb.2019.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas AE. How multi-partner endosymbioses function. Nat Rev Microbiol. 2016;14:731–43. doi: 10.1038/nrmicro.2016.151. [DOI] [PubMed] [Google Scholar]
- 10.Koga R, Bennett GM, Cryan JR, Moran NA. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol. 2013;15:2073–81. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 11.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC–rich genome of a bacterial symbiont. PLos Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–18. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–8. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ankrah NY, Chouaia B, Douglas AE. The cost of metabolic interactions in symbioses between insects and bacteria with reduced genomes. mBio. 2018;9:e01433–18. doi: 10.1128/mBio.01433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey E, Heys J, Gedeon T. Quantifying the effects of the division of labor in metabolic pathways. J Theo Biol. 2014;360:222–42. doi: 10.1016/j.jtbi.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roell GW, Zha J, Carr RR, Koffas MA, Fong SS, Tang YJ. Engineering microbial consortia by division of labor. Micro Cell Fact. 2019;18:35. doi: 10.1186/s12934-019-1083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thommes M, Wang T, Zhao Q, Paschalidis IC, Segrè D. Designing metabolic division of labor in microbial communities. mSyst. 2019;4:e00263–18. doi: 10.1128/mSystems.00263-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–10. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Z, Jones DH, Khuri S, Tsinoremas NF, Wyss T, Jander G, Wilson AC. Comparative analysis of genome sequences from four strains of the Buchnera aphidicola Mp endosymbion of the green peach aphid, Myzus persicae. BMC Genom. 2013;14:917. doi: 10.1186/1471-2164-14-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang ZF, Xia F, Johnson KW, Brown CD, Bartom E, Tuteja JH, Stevens R, Grossman RL, Brumin M, White KP, Ghanim M. Comparison of the genome sequences of “Candidatus Portiera aleyrodidarum” primary endosymbionts of the whitefly Bemisia tabaci B and Q biotypes. Appl Environ Microbiol. 2013;79:1757–9. doi: 10.1128/AEM.02976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–82. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 23.Thompson SN. Trehalose—the insect ‘blood’sugar. Adv Insect Physiol. 2003;31:85. [Google Scholar]
- 24.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotech. 2010;28:245–8. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall RJ, Flanagan LA, Bottery MJ, Springthorpe V, Thorpe S, Darby AC, et al. A tale of three species: adaptation of Sodalis glossinidius to tsetse biology, Wigglesworthia metabolism, and host diet. MBio. 2019;10:e02106–18. doi: 10.1128/mBio.02106-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Zarecki R, Medina S, Ofaim S, Liu X, Chen C, et al. Modeling microbial communities from atrazine contaminated soils promotes the development of biostimulation solutions. ISME J. 2019;13:494–508. doi: 10.1038/s41396-018-0288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medlock GL, Carey MA, McDuffie DG, Mundy MB, Giallourou N, Swann JR, et al. Inferring metabolic mechanisms of interaction within a defined gut microbiota. Cell Syst. 2018;7:245–7. doi: 10.1016/j.cels.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann J, Obeng N, Yang W, Pees B, Petersen C, Waschina S, et al. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. ISME J. 2020;14:26–38. doi: 10.1038/s41396-019-0504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnúsdóttir S, Thiele I. Modeling metabolism of the human gut microbiome. Curr Opin Biotechnol. 2018;51:90–6. doi: 10.1016/j.copbio.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Russell CW, Bouvaine S, Newell PD, Douglas AE. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 2013;79:6117–23. doi: 10.1128/AEM.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aristilde L, Reed ML, Wilkes RA, Youngster T, Kukurugya MA, Katz V, et al. Glyphosate-induced specific and widespread perturbations in the metabolome of soil Pseudomonas species. Front Environ Sci. 2017;5:34. [Google Scholar]
- 32.Clasquin MF, Melamud E, Rabinowitz JD. LC‐MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinf. 2012;37:14.11. 11–14.11. 23. doi: 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC− MS data. Anal Chem. 2010;82:9818–26. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nuc Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v. 3.0. Nat Protoc. 2019;14:639. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noor E, Flamholz A, Bar-Even A, Davidi D, Milo R, Liebermeister W. The protein cost of metabolic fluxes: prediction from enzymatic rate laws and cost minimization. PLoS Comp Biol. 2016;12:e1005167. doi: 10.1371/journal.pcbi.1005167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noor E, Flamholz A, Bar-Even A, Davidi D, Milo R, Liebermeister W. The protein cost of metabolic fluxes: prediction from enzymatic rate laws and cost minimization. PLoS Comp Biol. 2016;12:e1005167. doi: 10.1371/journal.pcbi.1005167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13:139–48. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tennessen JM, Bertagnolli NM, Evans J, Sieber MH, Cox J, Thummel CS. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3. 2014;4:839–50. doi: 10.1534/g3.114.010652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison JF, Greenlee KJ, Verberk WC. Functional hypoxia in insects: definition, assessment, and consequences for physiology, ecology, and evolution. Annu Rev Entomol. 2018;63:303–25. doi: 10.1146/annurev-ento-020117-043145. [DOI] [PubMed] [Google Scholar]
- 42.O’Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–65. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–43. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–8. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40:323–32. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clasadonte J, Scemes E, Wang Z, Boison D, Haydon PG. Connexin 43-mediated astroglial metabolic networks contribute to the regulation of the sleep-wake cycle. Neuron. 2017;95:1365–80. doi: 10.1016/j.neuron.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mächler P, Wyss MT, Elsayed M, Stobart J, Gutierrez R, von Faber-Castell A, Baeza-Lehnert F. In vivo evidence for a lactate gradient from astrocytes to neurons. Cell Metab. 2016;23:94–102. doi: 10.1016/j.cmet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Magistretti PJ, Allaman I. A cellular perspective on brain energy metabolism and functional imaging. Neuron. 2015;86:883–901. doi: 10.1016/j.neuron.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 49.Pellerin L, Pellegri G, Bittar PG, Charnay Y, Bouras C, Martin J-L, et al. Evidence supporting the existence of an activity-dependent astrocyte-neuron lactate shuttle. Dev Neurosci. 1998;20:291–9. doi: 10.1159/000017324. [DOI] [PubMed] [Google Scholar]
- 50.Dubouchaud H, Butterfield GE, Wolfel EE, Bergman BC, Brooks GA. Endurance training, expression, and physiology of LDH, MCT1, and MCT4 in human skeletal muscle. Am J Physiol-Endocrin Metab. 2000;278:E571–9. doi: 10.1152/ajpendo.2000.278.4.E571. [DOI] [PubMed] [Google Scholar]
- 51.Gladden L. Lactate metabolism: a new paradigm for the third millennium. J Physiol. 2004;558:5–30. doi: 10.1113/jphysiol.2003.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–76. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chelliah V, Juty N, Ajmera I, Ali R, Dumousseau M, Glont M, et al. BioModels: ten-year anniversary. Nuc Acids Res. 2015;43:D542–8. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All models have been provided in three formats—SBML (.xml), MATLAB (.mat), and Excel (.xls)—and deposited in GitHub (https://github.com/na423/symbiosis_Warburg). SBML files of the models have also been submitted to the BioModels database [53] with the following identifiers: MODEL1908040002, MODEL1908040003, and MODEL1908040004.