Abstract
Clues to the evolutionary steps producing innovations in oxygenic photosynthesis may be preserved in the genomes of organisms phylogenetically placed between non-photosynthetic Vampirovibrionia (formerly Melainabacteria) and the thylakoid-containing Cyanobacteria. However, only two species with published genomes are known to occupy this phylogenetic space, both within the genus Gloeobacter. Here, we describe nearly complete, metagenome-assembled genomes (MAGs) of an uncultured organism phylogenetically placed near Gloeobacter, for which we propose the name Candidatus Aurora vandensis {Au’ro.ra. L. fem. n. aurora, the goddess of the dawn in Roman mythology; van.de’nsis. N.L. fem. adj. vandensis of Lake Vanda, Antarctica}. The MAG of A. vandensis contains homologs of most genes necessary for oxygenic photosynthesis including key reaction center proteins. Many accessory subunits associated with the photosystems in other species either are missing from the MAG or are poorly conserved. The MAG also lacks homologs of genes associated with the pigments phycocyanoerethrin, phycoeretherin and several structural parts of the phycobilisome. Additional characterization of this organism is expected to inform models of the evolution of oxygenic photosynthesis.
Subject terms: Bacterial genetics, Phylogenetics
Introduction
Around 2.4 billion years ago, Earth’s surface environments changed dramatically. Atmospheric oxygen rose from <10−5 times present atmospheric level (PAL) to >1% PAL [1–4]. This Great Oxygenation Event (GOE) permanently changed Earth’s surface geochemistry, fundamentally reshaped the cycling of key elements [5] and altered the evolutionary path of life by allowing widespread oxygen respiration [6, 7]. The GOE was enabled by photosynthetic oxygen production by Cyanobacteria, making the evolution of oxygenic photosynthesis one of the most important innovations in Earth’s history [4, 8]. However, the evolutionary processes leading to oxygenic photosynthesis are poorly constrained [9–13]. In one hypothesis, Cyanobacteria acquired photosynthetic genes for both photosystems I and II (PSI and PSII, respectively) via horizontal gene transfer and then combined and refined them to form the photosystems that drive oxygenic photosynthesis in Cyanobacteria [14, 15]. In another hypothesis, the common ancestor of all phototrophic bacteria contained the genes necessary for photosynthesis, which diversified through time and were selectively lost in non-phototrophic portions of those lineages [16–20]. In either scenario, the gene content of photosystems in Cyanobacteria can help elucidate the evolutionary processes leading to oxygenic photosynthesis.
Researchers have attempted to extract evolutionary information by studying the photosynthetic genus Gloeobacter, which is an outgroup to all other Cyanobacteria [21–24]. In these studies, traits absent from Gloeobacter species but common in other Cyanobacteria are interpreted to have evolved after the divergence of Gloeobacter. For example, the Gloeobacter do not contain thylakoid membranes, which host the photosynthesis enzymes in other Cyanobacteria [25, 26]. In Gloeobacter species, photosynthesis and respiration occur in the cytoplasmic membrane rather than in the thylakoid membrane [27]. Gloeobacter spp. also contain structurally simple phycobilisomes, the protein complex responsible for absorbing photons and transferring energy to the PSII reaction center. The six rods of the Gloeobacter phycobilisome form a single bundle whereas many phycobilisomes are hemidiscoidal in many other Cyanobacteria [28]. However, pycobilisome structure can vary within the Cyanobacteria. For example, Acaryochloria marina contains both rod and hemidiscoidal structures [29, 30] Additionally, Gloeobacter spp. lack several PSII proteins, including PsbY, PsbZ and Psb27, and others are poorly conserved, including PsbO, PsbU, and PsbV [31]. The absence of the thylakoid membrane, differences in light harvesting, and missing photosynthesis proteins have been used by some researchers to constrain models for the evolution of oxygenic photosynthesis as well as the ecology and photochemistry of ancestral Cyanobacteria [23, 24, 32].
The Vampirovibrionia are a sister group to all Cyanobacteria, including Gloeobacter [10, 11, 33], and researchers have also interrogated their genomes for insights into the evolution of oxygenic photosynthesis [10–12, 33, 34]. No characterized Vampirovibrionia have genetic content that indicates a potential for phototrophy [10, 11, 33]. The absence of known phototrophic Vampirovibrionia could be due to: (1) incomplete characterization of the clade, which may contain some as yet unidentified phototrophs; (2) the genes necessary for photosynthesis were present in a common ancestor of Vampirovibrionia and Cyanobacteria and then lost in Vampirovibrionia and related lineages [35] or (3) oxygenic photosynthesis evolved after the divergence of Vampirovibrionia and Cyanobacteria [10–12, 33].
An undescribed group of organisms, known only from 16S rRNA gene surveys, is phylogenetically placed near the Gloeobacter and may provide insights into innovations in oxygenic photosynthesis. We recovered two nearly compete metagenome-assembled genomes (MAGs) of this taxon from microbial mats in Lake Vanda, McMurdo Dry Valleys, Antarctica, and we propose to name the taxon Candidatus Aurora vandensis.
Candidatus Aurora (Au’ro.ra. L. fem. n. aurora, the goddess of the dawn in Roman mythology, referring to the organism preference for inhabiting high latitude locations with low light).
Candidatus Aurora vandensis (van.de’nsis. N.L. fem. adj. vandensis of Lake Vanda, Antarctica, referring to the place where the organism was found).
Methods
Site description
Lake Vanda is a perennially ice-covered lake located within Wright Valley, McMurdo Dry Valleys, Antarctica. Lake Vanda has a perennial ice cover of 3.5–4.0 m. The ice cover transmits 15–20% of incident photosynthetically active radiation [36]. Wavelengths shorter than 550 nm dominate the light spectrum because ice transmits little red light and water is particularly transparent to blue-green light [37]. Nutrient concentrations are low, and therefore there is little biomass in the water column [38]. However, benthic mats are abundant [39], covering the lake bottom from the base of the ice to >50 m [40]. The microbial mats contain abundant 0.1–30 cm tall pinnacles [41] that incorporate annual mud laminae. Mat surfaces have brown-purple coloration due to trapped sediment and pigments. The underlying layers are characterized by green and purple pigmentation. The inner sections of large pinnacles are comprised of beige decomposing biomass. The dominant cyanobacterial genera based on morphological and 16S rRNA gene surveys are trichome-forming Leptolyngbya, Pseudanabaena, Wilmottia, Phormidium, Oscillatoria and some unicellular morphotypes [41, 42]. The microbial mats also contain diverse algae, especially diatoms, and other bacteria and archaea [39, 43]. Incident irradiance penetrates millimeters into the mats, and samples from the interior of the mat analyzed here were exposed to low irradiance (<1 µmol photons m−2 s−1) in their natural environment [37].
Sampling and DNA extraction
To obtain samples, SCUBA divers collected intact benthic microbial mats from 9 and 19 m lake depth and brought them to the surface in sterilized plastic containers. Pinnacles were either kept intact as bulk samples or dissected in the field based on color using sterile technique. Subsamples were placed in Zymo Xpedition buffer (Zymo Research, Irvine, CA), and cells were lysed via bead beating in the field. The stabilized samples were then frozen on dry ice and maintained frozen in the field. Samples were transported at −20 °C to UC Davis. DNA was extracted at UC Davis using the QuickDNA Fecal/Soil Microbe kit using the manufacturer’s instructions (Zymo Research, Irvine, CA, USA). The extracted DNAs were quantified using Qubit (Life Technologies) and were concentrated via evaporation until the concentration was ≥ 10 ng/uL. The one green mat and one purple subsample used for this project were sequenced at the US Department of Energy Joint Genome Institute (JGI).
DNA sequencing
The JGI generated sequence data using Illumina technology and reads were quality controlled using their in-house pipeline. Briefly, an Illumina library was constructed and sequenced 2 ×151 bp using the Illumina HiSeq-2500 1TB platform. BBDuk (version 37.36) was used to remove common contaminants (removehuman=t, removedog=t, removecat=t, removemout=t, and removemicrobes=t), trim reads that contained adapter sequence and bases to the right of bases where the quality score drops to 0. BBDuk was also used to remove reads that contained 4 or more ‘N’ bases, had an average quality score across the read less than 3 or had a minimum length ≤ 51 bp or 33% of the full read length. Reads mapped to masked human, cat, dog and mouse references at 93% identity were removed. Reads aligned to common microbial contaminants were also removed.
Bioinformatic analysis
Quality controlled, filtered raw data were retrieved from IMG Gold (JGI Gold ID GP0191362 and Gp0191371). Metagenomes were individually assembled using MEGAHIT 1.0.6 [44] using a minimum contig length of 500 bp and the paired end setting. Reads were mapped back to the assembly using Bowtie2 1.2.2 [45]. A depth file was created using jgi_summarize_bam_contig_depths and the assemblies were binned using MetaBAT with a minimum contig length of 2500 bp [46]. CheckM 1.0.7 assessed the quality of the bins [47], and bins of interest were identified based on phylogenetic placement. Average nucleotide identity (ANI) was calculated using the OrthoANI algorithm [48]. Protein-coding regions were identified by prodigal V2.6.3 [49] within CheckM. GhostKOALA and Prokka 1.12 were used to annotate translated protein sequences [50, 51]. Nucleotide and translated nucleotide sequences from the MAGs are available on OSF.io [52]. The whole-genome shotgun project has been deposited at Genbank under the accessions JAAXLU000000000 and JAAXLT000000000. The versions described in this paper are versions JAAXLU010000000 and JAAXLT010000000.
When homologs of genes from the KEGG photosynthesis module were not present in the bin, they were searched for in assembled, unbinned data by performing a BLASTX search with an E-value cutoff of 1E-5. BLASTP was used to find the best hit for the retrieved sequences and to exclude those that were not the target gene. Any unbinned sequences phylogenetically similar to A. vandensis were identified as belonging to the MAGs based on their position in a phylogenetic gene tree constructed using the methodology described below.
Phylogenetic inference
Aligned, nearly full-length 16S rRNA gene sequences (~1000–1500 bp) of reference Cyanobacteria were collected from the Silva database (v123; [53, 54]). We recovered a single 16S rRNA gene sequence from the green mat MAG and this sequence was added to the overall alignment using MAFFT [54]. A maximum-likelihood tree was constructed in RAxML-HPC2 on XSEDE [55] in the CIPRES Science Gateway [56]. The tree was rooted and visualized in the interactive tree of life [57] (Supplemental File 1). A maximum-likelihood tree based on 16S rRNA gene sequences were separately constructed in MEGA7 [58]. For this tree, sequences were aligned with Muscle [59] and a maximum-likelihood tree was constructed using the Tamura-Nei model [60] with uniform rates among sites and complete deletion and 100 bootstrap replicates. The maximum-likelihood heuristic model was nearest-neighbor interchange (Supplemental File 2).
A maximum-likelihood tree was constructed using translated nucleotide sequences of single-copy marker genes including IF3 C-terminal and L2 proteins (Supplemental Files 3, 4). Sequences were aligned in Muscle [59] in the CIPRES Science Gateway [56] using default parameters. A maximum-likelihood tree was constructed in RAxML-HPC2 on XSEDE [55] in the CIPRES Science Gateway [56] as described above.
The D1 sequence from the green subsample was added to a previously generated alignment in MAFFT [18, 54]. A maximum-likelihood tree was constructed in RAxML-HPC2 on XSEDE [55] in the CIPRES Science Gateway [56] as described above (Supplemental File 5).
A concatenated, single-copy marker gene tree was constructed from the MAGs, published Vampirovibrionia and Sericytochromatia genomes and one genome for each genus of NAG Cyanobacteria available from the Integrated Microbial Genomes and Microbiomes (IMG) database using a custom wrapper script leveraging Phylosift’s search and align functions [61–63]. The alignments were generated by using Phylosift’s search and align functions using lastal and hmm align to identify 37 mostly single-copy marker genes as well as 16S sequences (Table S1). A maximum-likelihood tree was constructed as described above and trees were rooted and visualized in the interactive tree of life [57] (Supplemental File 6). Alignment files are available on osf.io [52].
Results
Assembled metagenomes contained 313–1306 Mbp in 228,837–861,358 contigs with a mean sequence length of 1301–1669 bp. 49.6 and 53.3% of unassembled reads mapped back to the assembly for the green and purple samples, respectively. We recovered two bins of a taxon most closely related to Gloeobacter, one from each sample. Hereafter, we will refer to the taxon represented by the bins as a MAG. The bins were 3.07 and 2.96 Mbp in total length, had a GC content of 55.4% and 55.3%, and contained 3025 and 3123 protein-coding sequences respectively. Bins were 90.1 and 93.2% complete with 1.7 and 0.85% contamination based on marker gene analysis in CheckM. GhostKOALA annotated 41.1 and 41.7% of the predicted protein-coding sequences. Marker gene sequences and key photosynthetic gene sequences from the bins were identical or nearly identical and the genomes were 99.96% similar based on ANI.
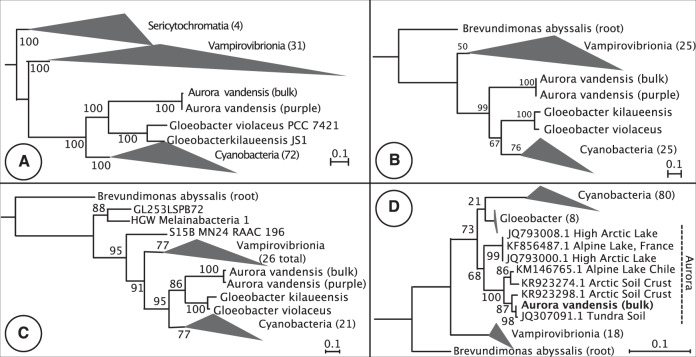
The MAG is most similar to G. violaceous with which it has 69.9% ANI across the genome and G. kilauensis with which it has 69.8% ANI across the genome. The Phylosift concatenated marker gene tree placed the MAG as a sister group to the Gloeobacter (Fig. 1a). The individual marker gene trees differed in their topologies. For example, A. vandensis is a sister group to all other photosynthetic Cyanobacteria in the IF3 C terminal phylogeny and is a sister group to the Gloeobacter in the ribosomal protein L2 phylogeny (Fig. 1b, c). Phylogenetic trees based on 16S rRNA gene sequences varied and placed the MAG either as a sister group to all other Cyanobacteria (Fig. 1d) or as a sister to the Gloeobacter (Figure S1) dependent on the program used for tree building and the16S rRNA gene taxa included in the analyses. The 16S rRNA gene from the MAG was >99% similar to clones from moss pillars in an Antarctic lake (AB630682) and tundra soil (JG307085) and was 91.2% similar to G. violaceous strain PCC 7421 and 90.8% similar to G. kilaueensis (NR_074282; Fig. 1d).
Fig. 1. Phylogeny of A. vandensis.
a Genome phylogeny based on the concatenation of 37 single-copy marker genes showing A. vandensis as a sister group to the Gloeobacter; b 16S rRNA gene phylogeny showing the genus Aurora as a sister group to all other Cyanobacteria; c Ribosomal protein L2 phylogeny with A. vandensis sister to the Gloeobacter; and d IF3 C terminal phylogeny showing A. vandensis as sister to the Gloeobacter.
Based on KEGG annotations, the MAG contained homologs of all the genes necessary for carbon fixation via the Calvin Cycle. It also contained many of the genes necessary for glycolysis via the Embden–Meyerhof–Parnas pathway (EMP; missing pfkABC) and citrate cycle. The MAG contains no genes associated with nitrogen fixation. The MAG contained homologs of chlorophyll biosynthesis genes chlB, chlL,chlN, chlD, and chlM but did not contain homologs of chlH, chlI, chlG, or chlP. The MAG contained homologs of many genes associated with oxygenic photosynthesis, but psbM, psbZ, psbY, psb27, and psbU from photosystem II (PSII) were not found. Similarly, homologs of psbA were absent from the bin, but a BLASTX search of assembled, unbinned data located a psbA sequence that is a sister to the Gloeobacter D1 group 4 sequence (Fig. 2). This sequence is on a contig that was too short to be binned. PSII genes psbJ,psbP, psbO, and psbV were annotated in the MAG but were dissimilar to those found in other Cyanobacteria (Table S2). The MAG lacked homologs of genes encoding phycobilisome proteins ApcD, ApcF, CpcD, RpcG, and CpcG, and any genes associated with phycoerythrocyanin (PEC) or phycoerythrin (PE) (Table S2). The PSI genes psaI, psaJ, psaK, and psaX, and the photosynthetic electron transport gene petJ (cytochrome c6) were also absent. Homologs of chlorophyll biosynthesis genes chlB, chlN, chlM, and chlN were also present in the MAG. For each missing photosynthesis gene, no homologs were found in the assembled, unbinned data that had similar phylogenetic placements to other genes in the MAG, except psbA as described above.
Fig. 2. Phylogenetic position of D1 protein located in the green mat assembly.
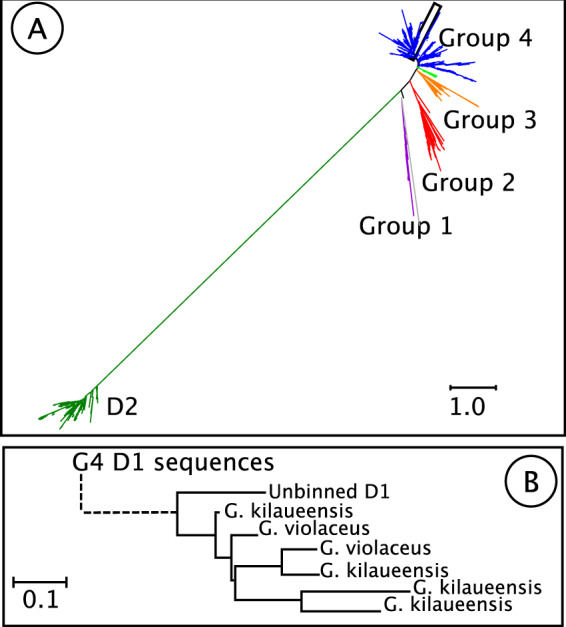
a D1 and D2 phylogeny showing D1 groups as defined by [18] Location of putative A. vandensis D1 highlighted by black box. b Region of tree highlighted by black box in A showing the putative A. vandensis sequence and its closest relatives.
Discussion
Genus and species description
We propose that our MAG is the first genome within a new genus. It has a 66.8% average nucleotide identity (ANI) and approximately 91% similarity of its 16S rRNA gene with G. violaceous strain PCC 7421 and G. kilauensis, the most similar taxa. On average, genera contain taxa that are 96.5% similar based on 16S rRNA genes. Therefore, we propose the creation of a new genus, Aurora, which includes our MAG, Aurora vandensis, and numerous representatives in 16S rRNA gene sequence databases, nearly all from polar or alpine environments (Fig. 1). The candidate genus is named after Aurora, the goddess of the dawn, to reflect its early divergence from other Cyanobacteria and its presence in low light environments. Aurora also refers to the northern and southern lights, aurora borealis and aurora australis, mirroring Aurora’s apparent preference for high latitude locations. The species, A. vandensis, is named after Lake Vanda where the samples originated. Lake Vanda was named after a sled dog used in the British North Greenland Expedition [64].
The phylogenetic placement of A. vandensis varies based on the genes or proteins used to construct the phylogeny and the taxa included in the analysis (e.g., Fig. 1). In most analyses it was placed as a sister to the Gloeobacter, but phylogenies of the IC3 C terminal and a 16S rRNA gene phylogeny placed it also as a sister group to all other cyanobacterial lineages, e.g., non-Aurora, non-Gloeobacter Cyanobacteria (hereafter NAG Cyanobacteria). Additionally, A. vandensis and Gloeobacter are located in a portion of the phylogenetic tree that is (a) at the end of a long branch and (b) does not contain many sequenced genomes. Therefore, we cannot be confident in the phylogenetic placement of A. vandensis. Additional genomes may help to better resolve Aurora’s family-level classification within the Cyanobacteria.
To date, Aurora is composed of taxa from high altitude or high latitude regions including Arctic microbial mats [65], Patagonian Andes [66], Nunavut, Canada [67], the French Alps [68], and perennially ice-covered lakes in Antarctica [69] and the current study (Fig. 1d), with a single taxon from modern stromatolites in Tasmania [70]. Based on this geographic distribution, Aurora may be a cold-adapted clade [65, 71].
Metabolic characterization of the uncultured Aurora genome
Based on its genome content, A. vandensis is predicted to be capable of both carbon fixation via the Calvin Cycle and glycolysis via the EMP. Many Cyanobacteria use the EMP pathway to ferment glycogen under dark conditions [72, 73], and A. vandensis may do so in Lake Vanda during the four months of darkness over the Antarctic winter.
A. vandensis is likely capable of performing oxygenic photosynthesis, but the MAG does not contain homologs to proteins that act as accessory subunits present in most Cyanobacterial photosystems. The lack of these genes may result from the incompleteness of the MAG or because they are actually missing from the A. vandensis genome. Here, we provide an analysis of the potential metabolic consequences if these proteins are actually absent in the A. vandensis genome rather than missing due to bin completeness.
Thylakoid membrane
In NAG Cyanobacteria, most photosynthetic processes occur in the thylakoid membrane, but this membrane is absent in Gloeobacter species, and photosynthesis and respiration occur in the cellular membrane [25, 26]. Few genes are known to be associated with the construction of the thylakoid membrane in Cyanobacteria. The VIPP1 gene is important [74] or essential to building a thylakoid membrane in organisms capable of oxygenic photosynthesis and is absent from Gleobacter [75]. It is also not present in the MAGs. Therefore A. vandensis may not contain thylakoid membranes.
Light harvesting
A. vandensis is predicted to be capable of collecting light energy for use in photosynthesis despite lacking homologs of several phycobiliproteins used for light-harvesting proteins in some other Cyanobacteria. Phycobilisomes harvest photons for use in PSII. These structures contain stacks of pigment proteins (biliproteins) connected by linker proteins, which are anchored into the thylakoid membrane in NAG Cyanobacteria or the cell membrane in the Gloeobacter. The pigments in many phycobilisomes include a core of allophycocyanin which best captures photons at ~650 nm, surrounded by rods of phycocyanin (~620 nm), phycoerythrin (maxima between 495–560 nm) and phycoerythrocyanin (575 nm). The A. vandensis MAG lacks homologs for genes associated with phycoerythrin and phycoerythrocyanin. Therefore, A. vandensis may have either lost those genes or they may have evolved after the divergence of A. vandensis from other Cyanobacteria. Gloeobacter spp. also lack genes for phycoerythrocyanin, so if the last common ancestor for Aurora and Gloeobacter had phycoerythrocyanin genes, they were lost in both lineages. In contrast, phycoerythrin-associated genes are present in Gloeobacter spp. so these may have been present in a common ancestor of A. vandensis and Gloeobacter but lost in A. vandensis. Cyanobacteria can lose biliproteins in response to their light environment. For example, Leptolyngbya sp. BC1307, isolated from the McMurdo Dry Valley Lake Hoare, Antarctica, lacks phycoerythrin, which may prevent it from being photoinhibited [76]. Alternately, the lack of these proteins may be an adaptive strategy that allows it to reduce the number of resources directed to the phycobilisome as has been proposed for other polar phototrophs [76–79].
Biliprotein linker proteins transfer energy from the biliproteins to the reaction center [31, 80] and neither A. vandensis nor Gloeobacter encode homologs of some linker proteins that are present in other Cyanobacteria. Homologs for the phycobilisome rod-core linker protein for PEC, cpcG, are absent in Gloeobacter (and A. vandensis). Instead, Gloeobacter uses cpcJ (Glr2806), which connects phycocyanin and allophycocyanin, and cpeG (Glr1268), which connects phycocyanin and PE. The A. vandensis MAG also lacks the phycoerythrocyanin linker proteins. In addition, the sequences that may be homologous to cpcJ and cpeG, in the A. vandensis MAG are not known to serve similar functions. The linker proteins that are absent from the A. vandensis MAG are not essential to oxygenic photosynthesis. Knockouts of these genes in other Cyanobacteria do not inhibit photosynthesis, but mutants have lower energy transfer to the reaction center than wildtype strains [81].
Overall, A. vandensis is predicted to capture irradiance for photosynthesis using allophycocyanin and phycocyanin and to transfer that energy to PSII via the associated linker proteins.
Photosystem II reaction center
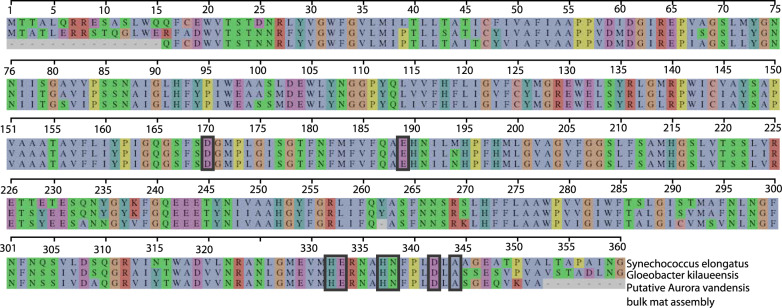
To perform oxygenic photosynthesis, A. vandensis is expected to need a functional PSII reaction center, including the D1and D2 proteins and other proteins structuring the oxygen-evolving center (OEC). The A. vandensis MAG did not contain psbA, which encodes the D1 protein. However, we located a D1 sequence from each of the assemblies that is phylogenetically located at the same place as A. vandensis (Fig. 2), is 91% similar to that of G. violaceus (WP_023172020), and is located on a contig that is too short to be binned using our methods (<2500 bp). Based on these attributes, we hypothesize that these psbA sequences belong to the A. vandensis MAG. The translated psbA sequence produces a D1 protein within Group 4, the group that is functional for oxygen production and present in all known photosynthetic Cyanobacteria [18]. It contains all of the previously described amino acid ligands necessary to support the Mn4CaO5 cluster in the OEC (Asp170, Glu189, His332, Glu333, His 337, Asp342, and Ala344) [82, 83] Fig. 3). Thus, this D1 protein is likely fully functional.
Fig. 3.
Alignment of D1 protein sequences from Synechococcus elongatus and Gloeobacter kilaueensis and the translated D1 sequence from the green mat assembly.
Most other critical PSII reaction center subunits have homologs in the A. vandensis MAG (Table S2), which suggests it is capable of oxygenic photosynthesis. However, the MAG lacks homologs to a number of genes involved in supporting the PSII reaction center (Table S2). The minimum suite of genes necessary for a functional PSII is not known. Gloeobacter spp. perform oxygenic photosynthesis without psbY, psbZ, and psb27 [31] which are also absent from the A. vandensis MAG, suggesting that they are not essential. Other PSII homologs that are absent from the A. vandensis MAG (psbM and psbU) or are poorly conserved (psbJ and psbV) have been individually knocked out in NAG Cyanobacteria. Mutants lacking psbM, which helps stabilize the PSII D1/D2 dimer, still form functional D1/D2 dimers [84]. Those lacking the stabilizing psbU or psbV still perform oxygenic photosynthesis, but they have decreased energy transfer between the allophycocyanin phycobilisome and PSII [85], are highly susceptible to photoinhibition, have decreased light utilization under low-light conditions, and have lowered oxygen evolution and electron donation rates than the wildtype [86–88]. Mutants lacking psbJ have less stable D1/D2 dimers but still photosynthesize with lower rates of oxygen production than the wildtype [89]. Finally, the gene encoding PsbO is poorly conserved in A. vandensis relative to Gloeobacter (46%) and NAG Cyanobacteria (≤36%), compared with ≥55% similarity among NAG Cyanobacteria. Despite this, PsbO in A. vandensis contains all the features previously identified as necessary to interact with the D1, D2, CP43 and C47 subunits [90]. Therefore, the A. vandensis PsbO subunit likely helps stabilize the Mn4CaO5 cluster and support the OEC as it does in other Cyanobacteria.
PsbO, PsbU, PsbV, a region of the D1, and other extrinsic proteins help control the concentration of Cl−, Ca2+, and H+ and create an environment that is amenable to water oxidation [84, 91–93]. Although PsbU is missing in the A. vandensis MAG, other PSII proteins conserve residues that have been identified as important for Cl− and Ca2+ regulation. For example, the D1 chloride ligand site Asn338 is conserved in the translated psbA that we attribute to A. vandensis (Fig. 3). Similarly, the translated psbO contains Glu54, Glu114, and His231 residues that bind with Ca2+ [94], suggesting that A. vandensis is capable of some Cl− and Ca2+ regulation.
Individually, each of the missing or poorly conserved PSII reaction center genes reduce energy transfer, growth rate, or oxygen evolution of oxygenic photosynthesis in mutated NAG Cyanobacteria. If the genes missing in the A. vandensis MAG are missing in the organism, this taxon may have a less stable OEC and lower oxygen evolution potential than other Cyanobacteria. However, none of the missing genes are individually necessary to perform oxygenic photosynthesis, suggesting that, if A. vandensis is missing all of the genes absent from the MAG, it could still be capable of oxygenic photosynthesis.
Cytochrome b6f
Once through PSII, the electrons pass through the cytochrome b6f complex, which is composed of eight subunits in NAG Cyanobacteria. The A. vandensis MAG contains homologs of genes encoding six of these subunits, including PetA, PetB, PetC, PetD, PetM and PetG. However, the MAG is missing petL and petN. Gloeobacter is also missing petL, and a Synechocystis mutant was able to grow photoautotrophically without petL, although it produced oxygen at a reduced rate compared to the wildtype [95]. Therefore, an absence of petL should not inhibit the functionality of cytochrome b6f. The effect of an absence of petN is less clear. Deletion of petN prevented plants from photosynthesizing [96, 97]. These results have been interpreted to mean that petN is necessary for photosynthesis in both plants and Cyanobacteria [95, 98], but attempts to delete petN in Cyanobacteria have been unsuccessful [95]. Therefore, it is not possible to determine what effect its absence may have on electron transport if it is not present in A. vandensis. Overall, even if these genes are absent from A. vandensis, oxygenic photosynthesis and aerobic respiration should not be inhibited.
Cytochrome b6f passes electrons to either plastocyanin or cytochrome c6. Aurora vandensis contains homologs of genes necessary to produce plastocyanin, but the MAG lacks homologs of petJ, which codes for cytochrome c6. petJ knockout Cyanobacteria grow at the same rate as wildtype [99]. Therefore, A. vandensis would not be affected by the lack of cytochrome c6 even if petJ is absent from the organism because it could use plastocyanin as the final electron carrier delivering electrons to PSI.
Photosystem I reaction center
The genetic makeup of PSI in A. vandensis MAG is similar to that in Gloeobacter genomes. Both contain the main subunits for PSI, but lack homologs of several genes including psaI, psaJ, and psaK that are present in NAG Cyanobacteria. However, the A. vandensis MAG also lacks homologs of psaM, which is present in Gloeobacter. PsaM is involved in forming stable PSI trimers, and Cyanobacteria mutants lacking this gene perform photosynthesis and grow at a normal rate [100]. The A. vandensis MAG also contains the genes encoding for a homolog of PsaZ, a protein only found in Gloeobacter species. This protein may stabilize the PSI structure in the absence of PsaI, PsaJ, and PsaK [32]. Therefore, PSI in A. vandensis likely functions similarly to PSI in Gloeobacter, even if psaM is absent from the organism.
Photoprotection
Cyanobacteria can experience photoinhibition under high light conditions when photon absorption outstrips the ability to dissipate electrons through photochemical pathways, and reactive oxygen species accumulate at the PSII reaction center. These reactive species damage photosynthetic machinery, especially the D1 protein, which then requires reassembly [101]. Cyanobacteria protect themselves from photoinhibition in two key ways. First, they use orange carotenoid proteins (OCP) as receptors to reduce the amount of energy transferred from the phycobilisome to PSII and PSI [101]. The A. vandensis genome contains homologs of a gene coding for a protein 68% similar to the OCP in G. violaceous. Because the OCP interacts directly with the phycobilisome [101] the sequence differences may reflect structural differences in the phycobilisomes of A. vandensis and G. violaceous. Alternately, the OCP in A. vandensis may be belong to recently described paralogs of OCPs called helical carotenoid proteins [102–104]. Some of these proteins are capable of antennae or singlet oxygen quenching activity, but the function of others is unknown [102].
Cyanobacteria also protect themselves from photoinhibition using high light-inducible proteins (HLIP) to dissipate energy. A. vandensis contains homologs of genes for three proteins that are 69–85% similar to HLIP in G. violaceous. We hypothesize that these genes act as HLIP and protect A. vandensis against photoinhibition.
Importance of Aurora vandensis
A. vandensis, G. violaceous and G. kilauensis are the only species identified to date occupying the phylogenetic space between the NAG Cyanobacteria and the Vampirovibrionia. The addition of the genus Aurora to this portion of the phylogenetic tree will provide key insights into evolutionary events associated with the divergence of two lineages with very different ecological distributions and dominant metabolisms, e.g., the NAG Cyanobacteria, which are ubiquitous in illuminated environments and which are nearly universally capable of oxygenic photosynthesis, and the Vampirovibrionia, none of which are known to be capable of phototrophy [10–12, 33].
Supplementary information
Acknowledgements
Sequencing was provided by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, and is supported under Contract No. CSP502867. Samples used in this project were collected during a field season supported by the New Zealand Foundation for Research, Science and Technology (grant CO1X0306) with field logistics provided by Antarctica New Zealand (project K-081). Salary support for CG was provided by the Massachusetts Institute of Technology node of the NASA Astrobiology Institute and the National Science Foundation (grant 1745341). We would also like to thank Dr Tanai Cardona for providing his alignment of D1 and D2 sequences and four anonymous reviewers for their help in improving this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0668-5) contains supplementary material, which is available to authorized users.
References
- 1.Karhu JA, Holland HD. Carbon isotopes and the rise of atmospheric oxygen. Geology. 1996;24:867–70. [Google Scholar]
- 2.Kump LR. The rise of atmospheric oxygen. Nature. 2008;451:277–8. doi: 10.1038/nature06587. [DOI] [PubMed] [Google Scholar]
- 3.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506:307. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 4.Planavsky NJ, Reinhard CT, Wang X, Thomson D, McGoldrick P, Rainbird RH, et al. Low Mid-Proterozoic atmospheric oxygen levels and the delayed rise of animals. Science. 2014;346:635–8. doi: 10.1126/science.1258410. [DOI] [PubMed] [Google Scholar]
- 5.Reinhard CT, Planavsky NJ, Robbins LJ, Partin CA, Gill BC, Lalonde SV, et al. Proterozoic ocean redox and biogeochemical stasis. Proc Natl Acad Sci USA. 2013;110:5357–62. doi: 10.1073/pnas.1208622110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Summons RE, Bradley AS, Jahnke LL, Waldbauer JR. Steroids, triterpenoids and molecular oxygen. Philos Trans R Soc B Biol Sci. 2006;361:951–68. doi: 10.1098/rstb.2006.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blankenship RE. Early evolution of photosynthesis. Plant Physiol. 2010;154:434–8. doi: 10.1104/pp.110.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowe SA, Døssing LN, Beukes NJ, Bau M, Kruger SJ, Frei R, et al. Atmospheric oxygenation three billion years ago. Nature. 2013;501:535. doi: 10.1038/nature12426. [DOI] [PubMed] [Google Scholar]
- 9.Mulkidjanian AY, Koonin EV, Makarova KS, Mekhedov SL, Sorokin A, Wolf YI, et al. The cyanobacterial genome core and the origin of photosynthesis. Proc Natl Acad Sci USA. 2006;103:13126–31. doi: 10.1073/pnas.0605709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Rienzi SC, Rienzi SCD, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife. 2013;2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soo RM, Skennerton CT, Sekiguchi Y, Imelfort M, Paech SJ, Dennis PG, et al. An expanded genomic representation of the phylum cyanobacteria. Genome Biol Evol. 2014;6:1031–45. doi: 10.1093/gbe/evu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soo RM, Hemp J, Parks DH, Fischer WW, Hugenholtz P. On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science. 2017;355:1436–40. doi: 10.1126/science.aal3794. [DOI] [PubMed] [Google Scholar]
- 13.Magnabosco C, Moore KR, Wolfe JM, Fournier GP. Dating phototrophic microbial lineages with reticulate gene histories. Geobiology. 2018;16:179–89. doi: 10.1111/gbi.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathis P. Compared structure of plant and bacterial photosynthetic reaction centers. Evolutionary implications. Biochimica Et Biophysica Acta Bba - Bioenerg. 1990;1018:163–7. [Google Scholar]
- 15.Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Plant Biol. 2011;62:515–48. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- 16.Olson JM. The evolution of photosynthesis. Science. 1970;168:438–46. doi: 10.1126/science.168.3930.438. [DOI] [PubMed] [Google Scholar]
- 17.Olson JM. ‘Evolution of photosynthesis’ (1970), re-examined thirty years later. Photosynth Res. 2001;68:95–112. doi: 10.1023/A:1011807229154. [DOI] [PubMed] [Google Scholar]
- 18.Cardona T. A fresh look at the evolution and diversification of photochemical reaction centers. Photosynth Res. 2015;126:111–34. doi: 10.1007/s11120-014-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sousa FL, Shavit-Grievink L, Allen JF, Martin WF. Chlorophyll biosynthesis gene evolution indicates photosystem gene duplication, not photosystem merger, at the origin of oxygenic photosynthesis. Genome Biol Evol. 2013;5:200–16. doi: 10.1093/gbe/evs127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson JM. Evolution of photosynthetic reaction centers. Biosystems. 1981;14:89–94. doi: 10.1016/0303-2647(81)90024-1. [DOI] [PubMed] [Google Scholar]
- 21.Turner S, Pryer KM, Miao VPW, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis1. J Eukaryot Microbiol. 1999;46:327–38. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 22.Gupta RS. Protein signatures (molecular synapomorphies) that are distinctive characteristics of the major cyanobacterial clades. Int J Syst Evol Micr. 2009;59:2510–26. doi: 10.1099/ijs.0.005678-0. [DOI] [PubMed] [Google Scholar]
- 23.Montejano G, Becerra-Absalón I, Gold-Morgan M, Osorio-Santos K. Gloeobacter violaceus: primitive reproductive scheme and its significance. Plant Syst Evol. 2018;304:1221–9. [Google Scholar]
- 24.Mimuro M, Tomo T, Tsuchiya T. Two unique cyanobacteria lead to a traceable approach of the first appearance of oxygenic photosynthesis. Photosynth Res. 2008;97:167. doi: 10.1007/s11120-008-9311-4. [DOI] [PubMed] [Google Scholar]
- 25.Rippka R, Waterbury J, Cohen-Bazire G. A cyanobacterium which lacks thylakoids. Arch Microbiol. 1974;100:419–36. [Google Scholar]
- 26.Saw JHW, Schatz M, Brown MV, Kunkel DD, Foster JS, Shick H, et al. Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a Lava Cave in Kīlauea Caldera, Hawai’i. Plos One. 2013;8:e76376. doi: 10.1371/journal.pone.0076376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rexroth S, Mullineaux CW, Ellinger D, Sendtko E, Rögner M, Koenig F. The plasma membrane of the cyanobacterium gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell. 2011;23:2379–90. doi: 10.1105/tpc.111.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guglielmi G, Cohen-Bazire G, Bryant DA. The structure of Gloeobacter violaceus and its phycobilisomes. Arch Microbiol. 1981;129:181–9. [Google Scholar]
- 29.Marquardt J, Senger H, Miyashita H, Miyachi S, Mörschel E. Isolation and characterization of biliprotein aggregates from Acaryochloris marina, a Prochloron‐like prokaryote containing mainly chlorophyll d. Febs Lett. 1997;410:428–32. doi: 10.1016/s0014-5793(97)00631-5. [DOI] [PubMed] [Google Scholar]
- 30.Hu Q, Marquardt J, Iwasaki I, Miyashita H, Kurano N, Mörschel E, et al. Molecular structure, localization and function of biliproteins in the chlorophyll a/d containing oxygenic photosynthetic prokaryote Acaryochloris marina. Biochimica Et Biophysica Acta Bba - Bioenerg. 1999;1412:250–61. doi: 10.1016/s0005-2728(99)00067-5. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura Y, Kaneko T, Sato S, Mimuro M, Miyashita H, Tsuchiya T, et al. Complete genome structure of gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. Dna Res. 2003;10:137–45. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- 32.Inoue H, Tsuchiya T, Satoh S, Miyashita H, Kaneko T, Tabata S, et al. Unique constitution of photosystem I with a novel subunit in the cyanobacterium Gloeobacter violaceus PCC 7421. Febs Lett. 2004;578:275–9. doi: 10.1016/j.febslet.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Soo RM, Woodcroft BJ, Parks DH, Tyson GW, Hugenholtz P. Back from the dead; the curious tale of the predatory cyanobacterium Vampirovibrio chlorellavorus. Peerj. 2015;3:e968. doi: 10.7717/peerj.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soo RM, Hemp J, Hugenholtz P. The evolution of photosynthesis and aerobic respiration in the cyanobacteria. Free Radical Bio Med. 2019;140:200–5. doi: 10.1016/j.freeradbiomed.2019.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Cardona T. Reconstructing the origin of oxygenic photosynthesis: do assembly and photoactivation recapitulate evolution? Front Plant Sci. 2016;7:257. doi: 10.3389/fpls.2016.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard-Williams C, Schwarz A-M, Hawes I, Priscu JC. Optical properties of the McMurdo Dry Valley Lakes, Antarctica. In: Priscu JC editor. Ecosystem Dynamics in a Polar Desert: the McMurdo Dry Valleys. Vol. 72 . Antarctica: AGU Publications; 1998. p 189–3.
- 37.Hawes I, Schwarz AJ. Absorption and utilization of irradiance by cyanobacterial mats in two ice‐covered antarctic lakes with contrasting light climates. J Phycol. 2001;37:5–15. [Google Scholar]
- 38.Vincent WF, Vincent CL. Factors controlling phytoplankton production in Lake Vanda (77S) Can J Fish Aquat Sci. 1982;39:1602–9. [Google Scholar]
- 39.Love FG, Jr, GMS, Parker BC, Jr, RAW, Seaburg KG. Modern conophyton‐like microbial mats discovered in Lake Vanda, Antarctica. Geomicrobiol J. 2009;3:33–48. [Google Scholar]
- 40.Mackey TJ, Sumner DY, Hawes I, Jungblut AD. Morphological signatures of microbial activity across sediment and light microenvironments of Lake Vanda, Antarctica. Sediment Geol. 2017;361:82–92. [Google Scholar]
- 41.Sumner DY, Jungblut AD, Hawes I, Andersen DT, Mackey TJ, Wall K. Growth of elaborate microbial pinnacles in Lake Vanda, Antarctica. Geobiology. 2016;14:556–74. doi: 10.1111/gbi.12188. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Jungblut AD, Hawes I, Andersen DT, Sumner DY, Mackey TJ. Cyanobacterial diversity in benthic mats of the McMurdo Dry Valley lakes, Antarctica. Polar Biol. 2015;38:1097–110. [Google Scholar]
- 43.Kaspar M, Simmons GM, Parker BC, Seaburg KG, Wharton RA, Smith RIL. Bryum Hedw. Collected from Lake Vanda, Antarctica. Bryologist. 1982;85:424. [Google Scholar]
- 44.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. Peerj. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon S-H, Ha S, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–6. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 49.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. Bmc Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 52.Grettenberger CL, Sumner DY, Wall K, Brown CT, Eisen JA, Mackey TJ, et al. Aurora vandensis supplemental files [Internet]. 2020 [cited May 13 2020]. Available from https://www.osf.io/jbctp
- 53.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 56.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. in proc Gatew Comput Environ Work (GCE). New Orleans, LA: 2010. p 1–8.
- 57.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245.. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 61.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. PhyloSift: phylogenetic analysis of genomes and metagenomes. Peerj. 2014;2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–93. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.HMMER 3.1b2.
- 64.Chinn T, Mason P. The first 25 years of the hydrology of the Onyx River, Wright Valley, Dry Valleys, Antarctica. Polar Rec. 2016;52:16–65. [Google Scholar]
- 65.Jungblut AD, Hawes I, Mackey TJ, Krusor M, Doran PT, Sumner DY, et al. Microbial mat communities along an oxygen gradient in a perennially ice-covered Antarctic Lake. Appl Environ Micro. 2016;82:620–30. doi: 10.1128/AEM.02699-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elser JJ, Navarro MB, Corman JR, Emick H, Kellom M, Laspoumaderes C, et al. Community structure and biogeochemical impacts of microbial life on floating pumice. Appl Environ Micro. 2015;81:1542–9. doi: 10.1128/AEM.03160-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lynch MDJ, Bartram AK, Neufeld JD. Targeted recovery of novel phylogenetic diversity from next-generation sequence data. Isme J. 2012;6:2067. doi: 10.1038/ismej.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Billard E, Domaizon I, Tissot N, Arnaud F, Lyautey E. Multi-scale phylogenetic heterogeneity of archaea, bacteria, methanogens and methanotrophs in lake sediments. Hydrobiologia. 2015;751:159–73. [Google Scholar]
- 69.Nakai R, Abe T, Baba T, Imura S, Kagoshima H, Kanda H, et al. Microflorae of aquatic moss pillars in a freshwater lake, East Antarctica, based on fatty acid and 16S rRNA gene analyses. Polar Biol. 2012;35:425–33. [Google Scholar]
- 70.Proemse BC, Eberhard RS, Sharples C, Bowman JP, Richards K, Comfort M, et al. Stromatolites on the rise in peat-bound karstic wetlands. Sci Rep. 2017;7:15384. doi: 10.1038/s41598-017-15507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chrismas NAM, Anesio AM, Sánchez-Baracaldo P. Multiple adaptations to polar and alpine environments within cyanobacteria: a phylogenomic and Bayesian approach. Front Microbiol. 2015;6:1070. doi: 10.3389/fmicb.2015.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moezelaar R, Stal LJ. Fermentation in the unicellular cyanobacterium Microcystis PCC7806. Arch Microbiol. 1994;162:63–69. [Google Scholar]
- 73.Moezelaar R, Bijvank SM, Stal LJ. Fermentation and sulfur reduction in the mat-building cyanobacterium microcoleus chthonoplastes. Appl Environ Micro. 1996;62:1752–8. doi: 10.1128/aem.62.5.1752-1758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Awai K. Lipids in plant and algae. Dev Sub-Cell Biochem. 2016;86:85–101. doi: 10.1007/978-3-319-25979-6_4. [DOI] [PubMed] [Google Scholar]
- 75.Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, et al. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA. 2001;98:4238–42. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chrismas NAM, Williamson CJ, Yallop ML, Anesio AM, Sánchez‐Baracaldo P. Photoecology of the Antarctic cyanobacterium Leptolyngbya sp. BC1307 brought to light through community analysis, comparative genomics and in vitro photophysiology. Mol Ecol. 2018;27:5279–93. doi: 10.1111/mec.14953. [DOI] [PubMed] [Google Scholar]
- 77.Davison IR. Environmental effects on algal photosynthesis: temperature. J Phycol. 1991;27:2–8. [Google Scholar]
- 78.Geide RK. Light and temperature dependence of the carbon to chlorophyll a ratio in microalgae and Cyanobacteria: Implications for physiology and growth of phytoplankton. N. Phytol. 1987;106:1–34. [Google Scholar]
- 79.Tang EPY, VincentT WF. Strategies of thermal adaptation by high‐latitude cyanobacteria. N. Phytol. 1999;142:315–23. [Google Scholar]
- 80.Koyama K, Tsuchiya T, Akimoto S, Yokono M, Miyashita H, Mimuro M. New linker proteins in phycobilisomes isolated from the cyanobacterium Gloeobacter violaceus PCC 7421. Febs Lett. 2006;580:3457–61. doi: 10.1016/j.febslet.2006.04.098. [DOI] [PubMed] [Google Scholar]
- 81.Ashby MK, Mullineaux CW. The role of ApcD and ApcF in energy transfer from phycobilisomes to PS I and PS II in a cyanobacterium. Photosynth Res. 1999;61:169–79. [Google Scholar]
- 82.Murray JW. Sequence variation at the oxygen-evolving centre of photosystem II: a new class of ‘rogue’ cyanobacterial D1 proteins. Photosynth Res. 2012;110:177–84. doi: 10.1007/s11120-011-9714-5. [DOI] [PubMed] [Google Scholar]
- 83.Vermaas W, Charite J, Shen G. Glu-69 of the D2 protein in photosystem II is a potential ligand to Mn involved in photosynthetic oxygen evolution. Biochemistry. 1990;29:5325–32. doi: 10.1021/bi00474a017. [DOI] [PubMed] [Google Scholar]
- 84.Umena Y, Kawakami K, Shen J-R, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9Å. Nature. 2011;473:55. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 85.Veerman J, Bentley FK, Eaton-Rye JJ, Mullineaux CW, Vasil’ev S, Bruce D. The PsbU subunit of photosystem ii stabilizes energy transfer and primary photochemistry in the phycobilisome−photosystem ii assembly of synechocystis sp. PCC 6803 †. Biochemistry. 2005;44:16939–48. doi: 10.1021/bi051137a. [DOI] [PubMed] [Google Scholar]
- 86.Shen J-R, Burnap RL, Inoue Y. An independent role of cytochrome c-550 in cyanobacterial photosystem ii as revealed by double-deletion mutagenesis of the psbO and psbV genes in synechocystis sp. PCC 6803. Biochemistry. 1995;34:12661–8. doi: 10.1021/bi00039a023. [DOI] [PubMed] [Google Scholar]
- 87.Shen JR, Qian M, Inoue Y, Burnap RL. Functional characterization of Synechocystis sp. PCC 6803 delta psbU and delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry. 1998;37:1551–8. doi: 10.1021/bi971676i. [DOI] [PubMed] [Google Scholar]
- 88.Inoue-Kashino N, Kashino Y, Satoh K, Terashima I, Pakrasi HB. PsbU provides a stable architecture for the oxygen-evolving system in cyanobacterial photosystem II. Biochemistry. 2005;44:12214–28. doi: 10.1021/bi047539k. [DOI] [PubMed] [Google Scholar]
- 89.Sugiura M, Iwai E, Hayashi H, Boussac A. Differences in the interactions between the subunits of photosystem II dependent on D1 protein variants in the thermophilic cyanobacterium thermosynechococcus elongatus. J Biol Chem. 2010;285:30008–18. doi: 10.1074/jbc.M110.136945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koyama K, Suzuki H, Noguchi T, Akimoto S, Tsuchiya T, Mimuro M. Oxygen evolution in the thylakoid-lacking cyanobacterium Gloeobacter violaceus PCC 7421. Biochimica Et Biophysica Acta Bba - Bioenerg. 2008;1777:369–78. doi: 10.1016/j.bbabio.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 91.Yocum CF. The calcium and chloride requirements of the O2 evolving complex. Coord Chem Rev. 2008;252:296–305. [Google Scholar]
- 92.Chu H-A, Nguyen AP, Debus RJ. Amino acid residues that influence the binding of manganese or calcium to photosystem II. 2. The carboxy-terminal domain of the D1 polypeptide. Biochemistry. 1995;34:5859–82. doi: 10.1021/bi00017a017. [DOI] [PubMed] [Google Scholar]
- 93.Guskov A, Gabdulkhakov A, Broser M, Glöckner C, Hellmich J, Kern J, et al. Recent progress in the crystallographic studies of photosystem II. Chemphyschem. 2010;11:1160–71. doi: 10.1002/cphc.200900901. [DOI] [PubMed] [Google Scholar]
- 94.Murray JW, Barber J. Identification of a calcium-binding site in the PsbO protein of photosystem II. Biochemistry. 2006;45:4128–30. doi: 10.1021/bi052503t. [DOI] [PubMed] [Google Scholar]
- 95.Schneider D, Volkmer T. Rögner M. PetG and PetN, but not PetL, are essential subunits of the cytochrome b6f complex from Synechocystis PCC 6803. Res Microbiol. 2007;158:45–50. doi: 10.1016/j.resmic.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 96.Hager M, Biehler K, Illerhaus J, Ruf S, Bock R. Targeted inactivation of the smallest plastid genome‐encoded open reading frame reveals a novel and essential subunit of the cytochrome b6f complex. Embo J. 1999;18:5834–42. doi: 10.1093/emboj/18.21.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwenkert S, Legen J, Takami T, Shikanai T, Herrmann RG, Meurer J. Role of the low-molecular-weight subunits PetL, PetG, and PetN in Assembly, Stability, and Dimerization of the Cytochrome b6f Complex in Tobacco. Plant Physiol. 2007;144:1924–35. doi: 10.1104/pp.107.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bernát G, Rögner M. Center of the Cyanobacterial Electron Transport Network: The Cytochrome b6f Complex. In: Peschek A, Obinger C, Renger, G editors. Bioenergetic processes of cyanobacteria, from evolutionary singularity to ecological diversity Dordrecht: Springer; 2011. p 573–606.
- 99.Durán RV, Hervás M, Rosa MAD, la, Navarro JA. The efficient functioning of photosynthesis and respiration in synechocystis sp. PCC 6803 strictly requires the presence of either cytochrome c6 or plastocyanin. J Biol Chem. 2004;279:7229–33. doi: 10.1074/jbc.M311565200. [DOI] [PubMed] [Google Scholar]
- 100.Naithani S, Hou J-M, Chitnis PR. Targeted inactivation of the psaK1, psaK2 and psaM genes encoding subunits of Photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res. 2000;63:225–36. doi: 10.1023/A:1006463932538. [DOI] [PubMed] [Google Scholar]
- 101.Kirilovsky D. Photoprotection in cyanobacteria: the orange carotenoid protein (OCP)-related non-photochemical-quenching mechanism. Photosynth Res. 2007;93:7. doi: 10.1007/s11120-007-9168-y. [DOI] [PubMed] [Google Scholar]
- 102.López-Igual R, Wilson A, Leverenz RL, Melnicki MR, Carbon CB, de, Sutter M, et al. Different functions of the paralogs to the N-terminal domain of the orange carotenoid protein in the cyanobacterium anabaena sp. PCC 7120. Plant Physiol. 2016;171:1852–66. doi: 10.1104/pp.16.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Melnicki MR, Leverenz RL, Sutter M, López-Igual R, Wilson A, Pawlowski EG, et al. Structure, diversity, and evolution of a new family of soluble carotenoid-binding proteins in cyanobacteria. Mol Plant. 2016;9:1379–94. doi: 10.1016/j.molp.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 104.Bao H, Melnicki MR, Kerfeld CA. Structure and functions of Orange Carotenoid Protein homologs in cyanobacteria. Curr Opin Plant Biol. 2017;37:1–9. doi: 10.1016/j.pbi.2017.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.