Abstract
Tissues- and organs-on-chips are microphysiological systems (MPS)s that model the architectural and functional complexity of human tissues and organs that is lacking in conventional cell monolayer cultures. While substantial progress has been made in a variety of tissues and organs, chips recapitulating immune responses have not advanced as rapidly. This review discusses recent progress in MPSs for investigation of immune responses. To illustrate recent developments, we focus on two cases in point:) immunocompetent tumor microenvironment-on-a-chip devices that incorporate stromal and immune cell components, and pathomimetic modeling of human mucosal immunity and inflammatory crosstalk. More broadly, we discuss the development of systems immunology-on-a-chip devices that integrate microfluidic engineering approaches with high-throughput omics measurements, and emerging immunological applications of MPSs.
Keywords: microphysiological systems, engineering immune system responses, immune system-on-a-chip, tumor immune microenvironment, intestinal inflammation, systems immunology
Modeling human tissues and organs with microphysiological systems
Microphysiological systems (MPSs) (see Glossary) were developed to complement animal models, which often do not translate well to clinical practice [1]. Moreover, conventional two-dimensional (2D) cell cultures fail to account for important properties including substrate stiffness and topographical cues found in the in vivo extracellular matrix (ECM). For example, culturing cells on substrates with nanoscale ridges and grooves reminiscent of the native ECM topography enhances the structural and functional properties of engineered cardiomyocyte tissues [2, 3] and enhances cell migration [4–6]. When used in combination with primary patient tissue or induced pluripotent stem cells, MPSs can be used for disease modeling and personalized testing of candidate drugs and therapies. These devices allow precise control over physiological parameters including ECM composition, biochemical gradients, oxygen levels, biomechanical stress, and electrical stimulation. To date, substantial progress has been made in developing chips modeling the physiology of the heart, liver, lung, kidney, skin, muscle, blood vessels, blood-brain barrier, gastrointestinal system, nervous system, reproductive system, adipose tissue, tumor angiogenesis, and cancer metastasis [7, 8]. These multiple MPSs have also been linked together to form interconnected tissue and organ systems [9–11]. However, only recently have efforts expanded to incorporate immune system components. Understanding the physiology of the immune system is critical for clinical management and new treatments for conditions such as pathogen responses, autoimmune diseases, and cancer. Furthermore, it is also important to understand how the immune system is compromised in disease states such as impaired wound healing in patients with diabetes. While there has been recent progress in developing inflammation-on-a-chip models [12], there is still a paucity of MPSs for immune system organs and tissues such as the spleen, lymph nodes, and thymus, as well as MPSs for modeling wound healing and autoimmune disorders. This review discusses recent progress in developing human tissue- and organ-on-a-chip models for investigation of immune responses.
MPSs for modeling immune responses in the tumor microenvironment
For in vivo studies, human tumor cells are implanted as xenografts in immunodeficient mice, precluding assessment of the endogenous immune system. Likewise, xenograft models are used to study the infiltration and cytotoxicity of ex vivo cultured/engineered T cells, natural killer (NK) cells, and immunomodulatory agents. However, animal testing is time-consuming and expensive. Interactions of therapeutic cells and agents with human immune, stromal, and vascular endothelial cells cannot be assessed, and it is not conducive to rapidly evaluating the efficacy of therapeutics in a precision medicine context. Recent advances in MPS modeling the tumor microenvironment have established physiologically relevant platforms for in vitro testing [13]. These platforms are now being engineered to facilitate the investigation of immune cell responses.
3D microfluidic models of immune cell infiltration and antitumor cytotoxicity
There has been tremendous recent success in the immunotherapy of hematological malignancies with autologous T-cell receptor (TCR)-engineered T cells. For example, chimeric antigen receptor (CAR) T cells engineered to recognize the cluster of differentiation 19 (CD19) protein on the surface of B cells have demonstrated remarkable antitumor activity in refractory CD19+ B-cell leukemias and lymphomas [14]. However, robust results for this approach have yet to be achieved in solid tumor (non-hematological) malignancies. Major barriers to effective cellular immunotherapy in solid tumors include the physical ECM and endothelial barriers and the harsh immunosuppressive solid tumor microenvironment influenced by hypoxia, metabolic conditions, waste products, pH gradients, and the enrichment of immunosuppressive cells. Therefore, human MPSs have recently been developed to model these factors.
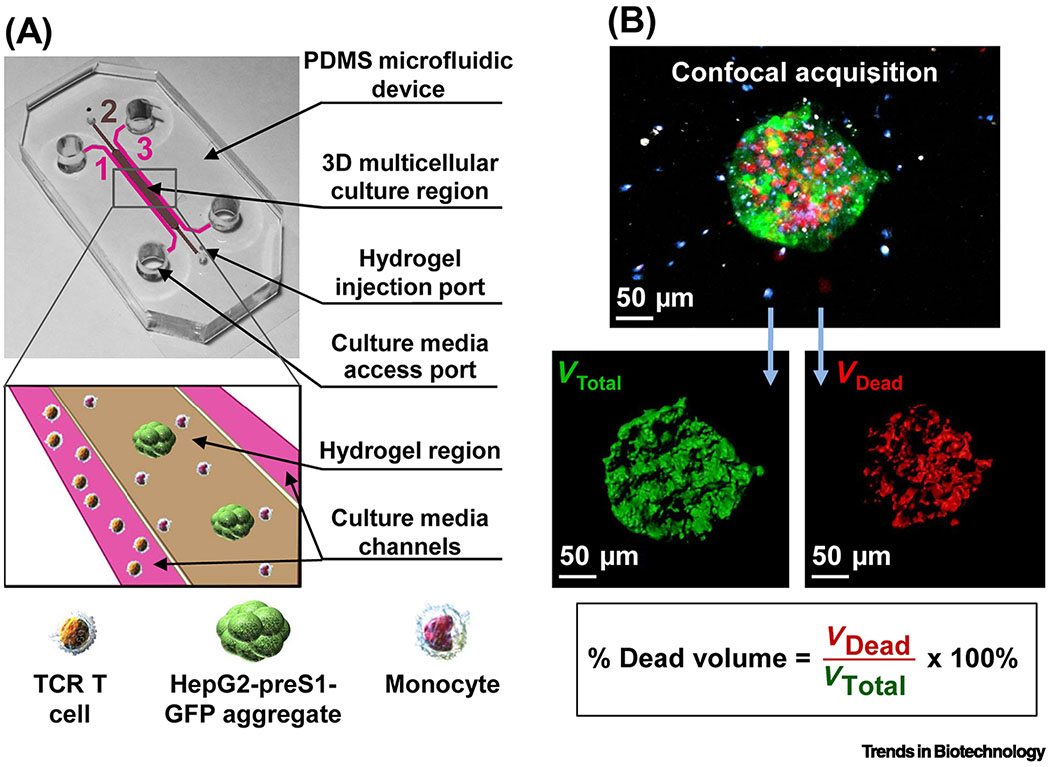
A simple and customizable microfluidic model was developed for investigating the ability of human TCR-engineered T cells to overcome physical and metabolic barriers in the tumor microenvironment [15] (Table 1). The 3D system consisted of a confined central type I collagen hydrogel in which target liver tumor cells were suspended as single cells or aggregates adjacent to a media channel in which tumor-specific TCR-engineered T cells were injected. The T cells were pre-labeled with CellTracker dye to track migration and interactions between tumor cells and T cells, while the live/dead cell discrimination dye DRAQ7 was included to track tumor cell killing by time-lapse confocal imaging. The microfluidic device revealed tumor cell-dependent T-cell migration and tumor cell killing and the importance of oxygen levels and inflammatory cytokines for optimal activity. To increase the cellular diversity, human primary monocytes were co-cultured with the target tumor cells (Figure 1) (Table 1) [16]. By expressing programmed death-ligand 1 (PD-L1) on their surface, monocytes can inhibit T cells by activating the checkpoint protein programmed death-1 receptor (PD-1). This system uncovered that the ability of the monocytes to suppress the cytotoxicity of T cells was dependent on only one of two types of methods that was used to engineer the T cells to equip them with the tumor-specific TCR and expand them, and could be reversed by blocking the PD-1/PD-L1 pathway with monoclonal antibodies. This result was not observed in 2D cultures, demonstrating the importance of the 3D culture model for preclinical prediction of therapeutic T-cell efficacy.
Table 1.
Engineering Tumor Immune Microenvironments
| Design of microphysiological device | Key cell types | Biological findings | Refs |
|---|---|---|---|
| 3D immune cell infiltration and cytotoxicity | |||
| Tumor cells and monocytes in collagen hydrogel; T cells delivered in microfluidic channel | Hepatitis B virus surface antigen (HBsAg)+ HepG2 liver tumor cells; Anti-HBsAg T cells; primary monocytes |
Importance of oxygen levels and inflammatory cytokines for optimal T-cell activity; Monocyte suppression of retrovirally-transduced but not mRNA-electroporated T cells by PD-1/PD-L1 pathway |
[15, 16] |
| Porcine collagen scaffold with intact basement membrane for establishing 3D tumor mass with static or dynamic flow conditions | Receptor tyrosine kinase-like orphan receptor 1 (ROR1)+ A549 lung and ROR1+ MDA-MB-231 breast tumor cells; Anti-ROR1 CAR T cells |
Scalable system revealed the dynamics of T-cell cytolytic activity, cytokine secretion, proliferation, and exhaustion; Differential efficacy of anti-ROR1 CAR T constructs |
[17] |
| Tumor cells in gelatin methacryloyl hydrogel between two oxygen diffusion barriers; CAR T cells delivered in microfluidic channel | HER2+ SKOV3 ovarian tumor cells; anti-HER2 CAR T cells |
Importance of matrix stiffness and oxygen concentration on CAR T-cell infiltration, cytotoxicity, and contact-independent mechanisms of CAR T-cell killing | [20] |
| Tumor cells in collagen hydrogel in multiwell injection molded plastic array; NK cells delivered in microfluidic channel | HeLa cervical tumor cells; primary NK cells |
Importance of greater effector to target cell ratio in 3D compared to 2D for tumor cell killing; Influence of collagen stiffness on NK-cell migration and contact time with target |
[21] |
| Tumor cells and NK cells in collagen hydrogel flanked by two lateral vascular lumens | MCF7 breast tumor spheroids; NK-92CD16V NK cells; HUVEC endothelial cells |
Endothelial barrier delays diffusion of anti-EpCAM-IL-2 antibody-cytokine conjugate; Anti-EpCAM-IL-2 enhances NK-cell cytotoxicity and ADCC at the periphery |
[24] |
| Tumor cells and monocytes Encapsulated in inner layer of gelatin methacryloyl hydrogel; endothelial cell layer at periphery; T cells delivered in perfusate to the bilayer | MCF7 and MDA-MB-231 tumor spheroids; THP-1 monocytes; TALL-104 T cells |
Enhanced recruitment of T cells by tumor spheroids compared to dispersed cells; Monocyte enhancement of T-cell recruitment; Upregulation of T-cell activating chemokines CCL4, CCL5, and CCL11 in monocyte/spheroid co-cultures; Upregulation of endothelial cell-cell junction destabilizing chemokine CXCL8 |
[25] |
| Engineering complex 3D tumor immune microenvironments | |||
| Organotypic tumor spheroids in a collagen hydrogel flanked by antiparallel microfluidic channels | Murine or patient organotypic tumor spheroids containing endogenous lymphoid and myeloid immune cells | Spheroids retained immune cell profile of the original bulk tumor and displayed differential sensitivity to PD-1 blockade; PD-1 blockade induced spheroids to produce the chemokines CCL19 and CXCL13; Identified cytokine secretion profiles correlating with ineffective response to PD-1 blockade |
[27, 28] |
| Central vascular-lined microchamber surrounded by two parallel collagen hydrogels containing tumor cells and immune cells flanked by microfluidic channels | HER2+ BT474 breast tumor cells; Hs578T CAF; Primary human PBMC |
Trastuzumab (anti-HER2 monoclonal antibody) stimulates long-term cancer immune cell interactions and ADCC in an immune cell density-dependent manner, an effect antagonized by CAF | [30] |
Figure 1. MPS modeling of the infiltration and cytotoxicity of therapeutic lymphocytes.
(A) To study the influence of monocytes on tumor cell killing by TCR-engineered T cells in the tumor microenvironment, a simple and customizable microfluidic device was developed for the 3D culture of HepG2 hepatocellular carcinoma tumor spheroids expressing the preS1 portion of the hepatitis B virus envelope protein linked to green fluorescent protein (preS1-GFP) and monocytes in a central collagen hydrogel. This was flanked by two media channels with or without TCR-engineered T cells specific for the hepatitis B virus antigen present on the tumor cells. (B) Tumor cell killing was assessed by confocal imaging of pre-labeled tumor spheroids (green) surrounded by monocytes (blue) and T cells (white). Dead tumor target cells were labeled by the live/dead cell exclusion dye DRAQ7 (red), and the volume of dead cells was quantified relative to the total spheroid volume. Reproduced from [16] under a Creative Commons license.
In another approach for modeling T-cell infiltration and cytotoxicity, human lung and breast cancer 3D cultures were established in which the small intestine submucosa and mucosa (sisMUC) from decellularized porcine jejunum were used as a collagen scaffold with intact basement membrane for the engraftment and expansion of tumor cells [17] (Table 1). The resulting tumor masses with an invasive phenotype were challenged with CAR T cells over 3-5 days under static or dynamic conditions providing continuous flow and shear stress with a flow bioreactor. Even under the more challenging condition of arterial flow, the CAR T cells adhered to and robustly penetrated the sisMUC tumor matrix and exhibited potent antitumor activity as revealed by levels of caspase-cleaved keratin 18 in the culture media as a measure of tumor cell apoptosis. This system profiled the time-course of important immunological parameters including cytolytic activity, cytokine secretion, proliferation, and acquisition of an exhausted phenotype of the T cells and could prioritize the efficacy of different CAR constructs. Disassembling the scaffolds at the end of the assay allowed detailed histology and phenotyping of the T cells, tumor cells, and stromal cells. While laborious to set up, the ability to generate 130 sisMUC scaffolds per porcine jejunum makes it amenable to medium- to high-throughput testing.
The increased collagen stiffness characteristic of the tumor ECM causes changes in gene expression that reduce the cytotoxicity of T cells and can instruct cells such as tumor-associated macrophages to acquire an immunosuppressive phenotype [18, 19]. In addition, the hypoxic tumor microenvironment promotes immunosuppression. Thus, 3D MPS that model these features would be useful for elucidating mechanisms. A 3D device was developed in which CAR T cells were delivered in microfluidic channels to human ovarian cancer cells embedded in gelatin methacryloyl in the presence of micromilled oxygen diffusion barrier pillars to create an oxygen gradient without complex external oxygen control [20] (Table 1). This allowed the study of the influence of matrix stiffness and heterogenous oxygen concentration on CAR T-cell infiltration, cytotoxicity, and contact-independent mechanisms of CAR T-cell killing. To increase the throughput of these 3D infiltration/cytotoxicity approaches, a multiwell cytotoxicity assay of cytotoxic lymphocytes in an injection molded plastic array 3D culture platform was developed [21] (Table 1). Hydrophilic rail-based microstructures allowed spatially compartmented collagen hydrogel patterning by simple pipetting, eliminating time intensive polydimethylsiloxane (PDMS) batch fabrication. The enhanced throughput allowed investigation of multiple parameters influencing the infiltration and cytotoxicity of human primary NK cells towards tumor cell targets including effector to target cell ratio and collagen matrix stiffness.
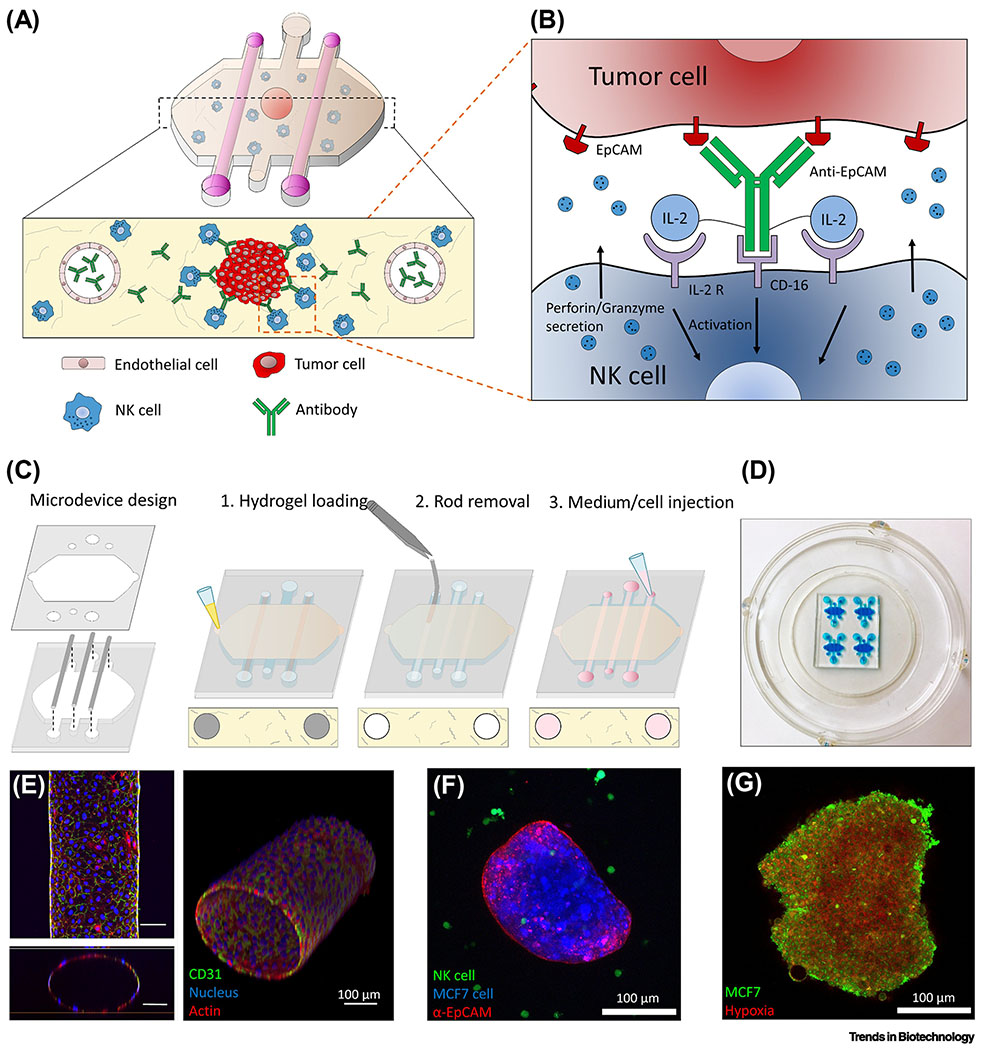
To reach a solid tumor target in vivo, therapeutic immune cells must overcome the physical and biochemical barrier of the tumor vasculature, which is known to inhibit immune cells through multiple mechanisms [22]. Combining a vascularized tumor MPS such as the human vascularized renal cell carcinoma-on-a-chip [23] with the approaches described above, it is now possible to assess how the endothelium influences the function of immunomodulatory cells and agents in the tumor microenvironment. A microfluidic model was developed consisting of human breast spheroids with hypoxic cores embedded with NK cells in a 3D collagen hydrogel flanked by two endothelial cell vascular lumens [24] (Figure 2) (Table 1). The device allowed perfusion of antibodies through the lateral vessels or direct inclusion in the hydrogel and the study of NK-cell cytotoxicity and antibody-dependent cell-mediated cytotoxicity (ADCC). This study focused on a modified antibody known as an immunocytokine, anti-EpCAM-IL-2, that simultaneously binds to epithelial cell adhesion molecule (EpCAM) protein on the surface of tumor cells and CD16 protein on the surface of NK cells and uses the costimulatory molecule Interleukin-2 (IL-2) to stimulate NK-cell cytotoxicity. Perfused anti-EpCAM-IL-2 antibody penetration into the collagen matrix was delayed by the inclusion of an endothelial cell barrier in the lumen. Once extravasated out of the vessels, the antibodies diffused through the matrix, but their penetration into tumor cell spheroids was inhibited by tumor cell-cell contacts. In contrast, NK cells penetrated the spheroids faster and destroyed them at the periphery and innermost layers, an effect that was enhanced by anti-EpCAM-IL-2 at the periphery. To study T-cell infiltration across an endothelial barrier, a bilayer microfluidic platform was developed in which tumor cells and monocytes were encapsulated in an inner gelatin methacryloyl hydrogel layer surrounded by an outer peripheral endothelial layer [25] (Table 1). This approach revealed pro-inflammatory chemokine secretion profiles associated with enhanced T-cell recruitment in tumor spheroid/monocyte co-cultures as well as enhanced secretion of a chemokine promoting vascular permeability. To date, most models of the tumor vasculature have used human umbilical vein endothelial cells (HUVECs). Ideally, future models should incorporate organotypic endothelial cells derived from the specific organ the tumor originated from or endothelial cells isolated directly from the tumor, which differ significantly from HUVECs [26].
Figure 2. MPS modeling of the ADCC of NK cells in the tumor microenvironment.
(A) Tumor spheroids were embedded in a collagen hydrogel containing endothelial-lined lateral lumens and antibodies and NK cells were administered via the lumens or by direct inclusion in the collagen. (B) The inset shows how an immunocytokine combining an epithelial-directed antibody (anti-EpCAM) with a co-stimulating cytokine (IL-2) via IL-2 receptor (IL-2 R) enhances NK-cell cytotoxicity. (C) The steps in fabrication and cell seeding of the microfluidic device. (D) Photograph of actual microarray device labeled with a blue dye for visualization. (E) Confocal (top), the orthogonal cross-sectional view (bottom) and 3D reconstruction (right) of an endothelial lumen fixed and stained with an antibody for the endothelial marker CD31 (green), Hoechst (nucleus, blue), and phalloidin (actin, red). (F) The microfluidic device allowed the simultaneous detection of prelabeled human breast cancer MCF7 cells in a spheroid (blue), the epithelial marker EpCAM on the surface (red), and NK cells (green). (G) A hypoxia-sensing dye (red) confirmed increased hypoxia in the central region of a GFP-labeled MCF7 cell spheroid. Reproduced from [24] with permission.
Engineering complex 3D tumor immune microenvironments
To more fully capture the complexity of cell types in the human tumor immune microenvironment, a 3D microfluidic device (DAX-1, Aim Biotech, USA) was adapted for the culture of murine- and patient-derived multicellular organotypic tumor spheroids containing autologous stromal and immune cells in collagen hydrogels [27] (Figure 3A) (Table 1). Immune profiling of these organoids demonstrated patient heterogeneity in the range of relevant immune cell types including B cells, CD4+ and CD8+ T cells, granulocytes, monocytes, and dendritic cells as well as heterogeneity of expression of exhaustion markers (PD-1, CTLA4, TIM3) on T cells and PD-1 receptor ligands (PD-L1, PD-L2) on dendritic cells, myeloid-derived suppressor cells, and tumor-associated macrophages. The system was compatible with serial microscopy for live/dead cell analysis and dynamic multiplex cytokine secretion profiling from conditioned media. The spheroids expanded in the microfluidic devices, retained the immune cell profile of the original bulk tumors, and recapitulated differential sensitivity to PD-1 blockade. In patient tumor spheroids, the microfluidic device was required for robust generation of the chemokines CCL19 and CXCL13 that serve as chemoattractants for immune cell recruitment in response to PD-1 blockade, which occurs in patients. The device also facilitated identification of cytokine secretion profiles predictive of an ineffective response to PD-1 blockade. In a follow-up study, RNA sequencing (RNA-seq) of patient tumor spheroids exposed to PD-1 blockade in the device was performed in order to infer the relative number of immune cells using computational methods [28] (Table 1). Similarly, a microfluidic droplet-based method of simultaneously monitoring RNA-seq profiles and T-cell receptor and B-cell immunoglobulin repertoires from single cells isolated from primary patient-derived tumor organoids was recently introduced [29]. This method successfully modeled immune checkpoint blockade with anti-PD-1 and anti-PD-L1 antibodies, activation of antigen-specific tumor-infiltrating lymphocytes, and tumor cytotoxicity. Taken together, these studies demonstrate that it is possible to use an engineered patient tumor organoid-based MPSs to investigate the mechanisms of immune checkpoint blockade and personalized immunotherapy testing.
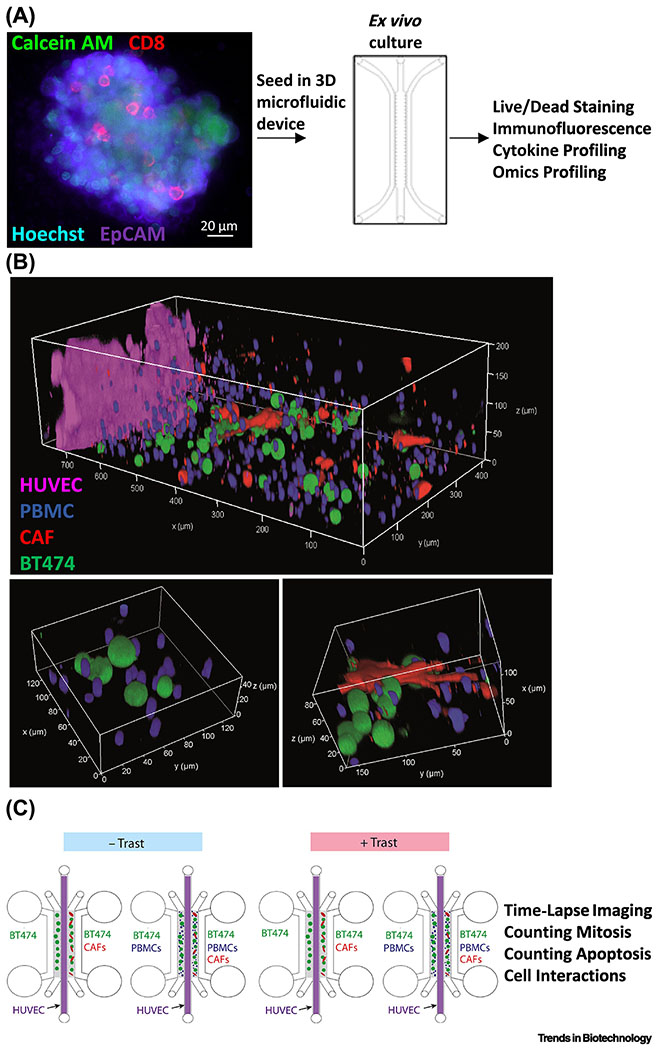
Figure 3. MPS modeling of complex cell types in the tumor immune microenvironment.
(A) In one approach, patient tumor organoids can be harvested to retain the autologous stromal cells and immune cells prior to embedding in 3D microfluidic hydrogel devices. Confocal imaging of a patient high-grade serous ovarian carcinoma organoid containing viable (calcein acetoxymethyl ester (AM), green), EpCAM+ epithelial cells (purple) demonstrated the presence of CD8+ T cells (red) and all nucleated cells (Hoechst, blue). Microfluidic devices have facilitated the identification of cytokine profiles predictive of differential sensitivity to PD-1 blockade and exogenous agents that enhance sensitivity. Adapted from [28] under a Creative Commons license. (B) To provide for tunable control over cell density, type, ratio, and orientation, a “tumor ecosystem-on-a-chip” was developed (top). Confocal imaging of one half of the device demonstrated the position of pre-labeled human BT474 breast cancer cells (green), PBMC including immune cells (blue), and human breast Hs578T CAF (red) parallel to a central HUVEC monolayer (magenta). Magnified 3D images show physical interactions between various cell types (bottom). (C) Experimental design of the microfluidic devices containing two hydrogel compartments (gray) containing the indicated cell type configurations to allow for the study of how ADCC stimulated by the therapeutic antibody trastuzumab (Trast) is antagonized by CAF. Adapted from [30] under a Creative Commons license.
While primary patient tumor organoids allow for the assessment of endogenous tumor immune and stromal cell populations, there is limited experimental control over cell density, type, ratio, or orientation. To address this, a “tumor ecosystem-on-a-chip” was developed for co-culture of vascular endothelial cells, cancer cells, cancer-associated fibroblasts (CAF), and immune cells [30] (Figure 3 B,C) (Table 1). The microfluidic device consisted of a central endothelial microchamber surrounded by two parallel collagen hydrogel microchambers containing combinations of cancer, CAF, and immune cells. Bilateral outer chambers served as media and drug administration reservoirs, or as channels for perfusion. Different cell types were labeled with unique dyes before incorporation in the chip to facilitate the identification and motility tracking by high content video microscopy. Nuclear and active caspase staining provided an assessment of mitosis and apoptosis, while confocal live-cell imaging in combination with cell localization and cell tracking software allowed for assessment of cancer-immune cell interactions. To test the device, a human breast tumor cell line expressing Human epidermal growth factor receptor 2 (HER2) known to be sensitive to the therapeutic anti-HER2 antibody trastuzumab was seeded in various combinations with breast CAF and/or primary human peripheral blood mononuclear cells (PBMCs), a source of T, B, and NK cells and monocytes. The device revealed that trastuzumab stimulates long term cancer-immune cell interactions and ADCC in an immune cell-density dependent manner, an effect antagonized by CAF. Taken together, these studies demonstrate the utility of immune MPSs for understanding the factors that influence the efficacy of therapeutic immune cells and immunomodulatory agents for improving cancer immunotherapy.
Modeling mucosal immunity and inflammatory crosstalk on chips
The human mucosal barrier, including tight junctions and mucus, is a physical microarchitecture that protects the body from external harmful pathogens and allergens. The human gastrointestinal (GI) tract is one of the most representative mucosal interfaces in which tissue-specific local immune cells constitutively exert homeostatic intercellular crosstalk with the host epithelium as well as the gut microbiome [31]. In this complex milieu, intestinal immunity in the “epithelium-microbiome-immune (EMI) axis” orchestrates intestinal homeostasis by regulating immune responses against various foreign antigens [32]. Cells in the EMI axis continuously communicate and coordinate with each other by producing essential microbial metabolites (e.g., short-chain fatty acids [33, 34]) or protecting (e.g., epithelial junctional barrier and mucus [35]) and surveilling exogenous antigens (e.g., gut-associated lymphoid tissues, GALT [36]). When this intercellular crosstalk is compromised by a pathogenic infection [32], barrier dysfunction [35], dysbiosis [37], or immune dysregulation [38], inflammatory immune responses germane to intestinal inflammatory diseases (e.g., inflammatory bowel disease, IBD) may be triggered. However, animal models that are genetically [39], chemically [40], or immunologically [41] manipulated often lack physiological relevance with humans in terms of the genetic heterogeneity [42], microbiome [43], etiological factors [40], and pharmacological validation (e.g., drug induction [44], first-pass effect [45, 46], and microbial drug metabolism [47–49]). These technical challenges in animal models have stimulated the development of in vitro and ex vivo models to simulate immune-associated inflammatory responses. A representative in vitro model includes a multiwell plate that contains a microporous insert (i.e., Transwell) to reconstruct the polarized EMI axis [50, 51], where an intestinal epithelial layer grown on a porous membrane forms a tissue barrier and provides luminal and abluminal compartments to harbor microbial and immune cells, respectively. The addition of immune cells on the basolateral side of the Transwell lined by an intestinal epithelial layer [50] or in the culture medium during the intestinal organoid culture [52] rapidly induces a simple immune response. However, conventional static cell culture models lack physiological biomechanics, cytodifferentiation, 3D histogenesis, and controllable protocols for a longitudinal host-microbiome co-culture.
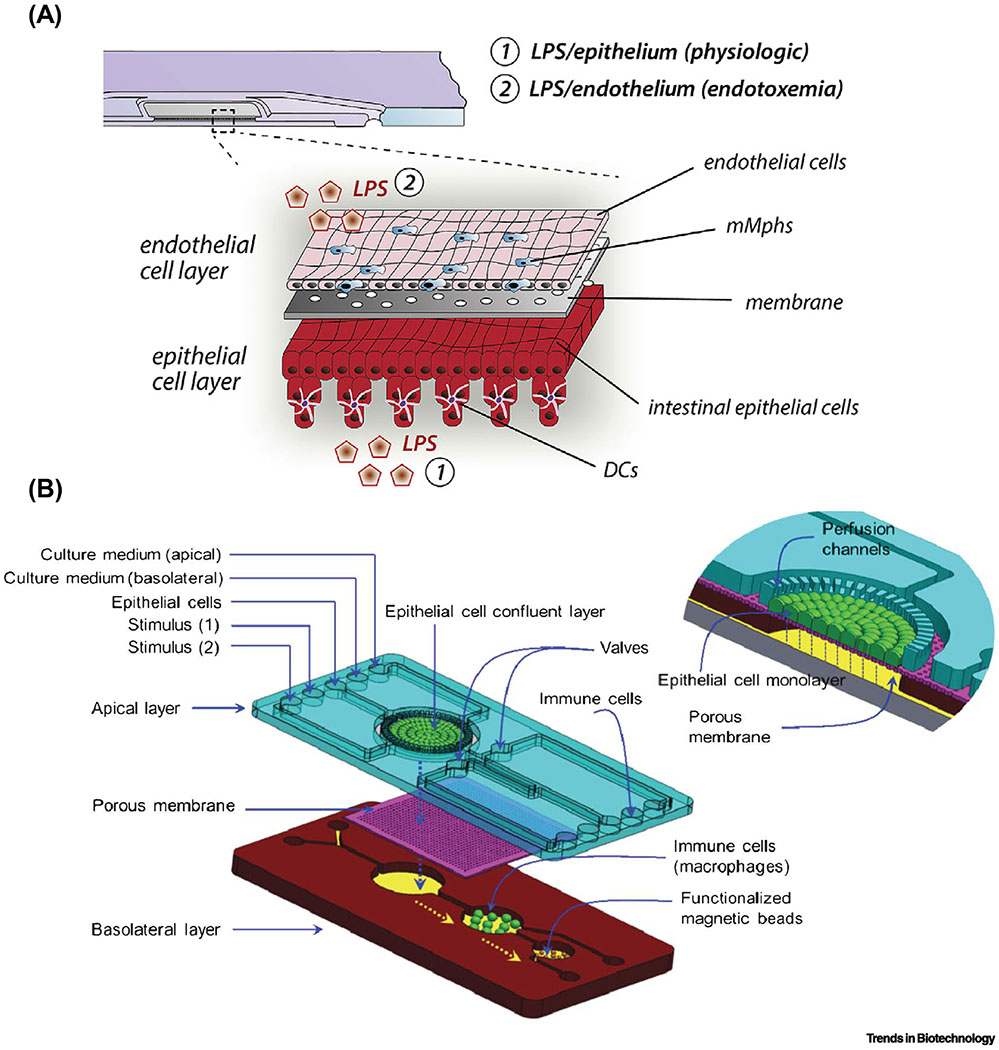
To improve these caveats in the conventional cell culture models, a limited number of novel MPS models have been suggested to emulate the immune-mediated inflammatory reactions in the mucosal interface (Table 2). A major advance includes spatiotemporal induction and monitoring of immune cells as well as living microbial cells in the model. Precision control of hydrodynamic flow and mechanical deformation precludes a factitious outgrowth of microbial cells while it emulates the physiological biomechanics in the gut. For instance, the gut-on-a-chip that recreates the intestinal mucosal interface allows the formation of differentiated 3D villus epithelium [53–56], intestinal lumen-capillary interface [57–60], a luminal stimulation with a beneficial (e.g., co-culture with probiotic bacteria [57, 58, 60]) or a detrimental factor (e.g., lipopolysaccharide, LPS), and immune cell-mediated interactions [58, 60] (Figure 4A). By switching over the immune cells associated with a particular immune milieu (e.g., pro- vs. anti-inflammatory cascade, parenchymal vs. peripheral), the gut-on-a-chip can be manipulated to trigger various tissue-specific immune responses in the EMI axis. By introducing PBMCs into the vascular microchannel, the “switch-on” effect of the immune components can be accurately controlled in the “gut inflammation-on-a-chip”, by which the temporal effect of immunological stimulation in response to the luminal component can be independently investigated (Figure 4 B,C) [58, 60]. Alternatively, the differentiated immune subsets such as monocyte-derived phagocyte subsets, including macrophages or dendritic cells, can be added to the vascular microenvironment to induce tissue-specific crosstalk or physiological immune tolerance in the intestine [59, 61]. For instance, a microfluidic device made of polystyrol was leveraged to induce immune responses between probiotic or pathogenic bacteria and tissue-resident mucosal macrophages and dendritic cells in an immunocompetent environment (Figure 5A). This MPS model specifically demonstrated a normal physiological condition as well as endotoxemia by introducing the bacterial endotoxin LPS [62] in the epithelial or endothelial layer, respectively. This compelling study accurately demonstrated the fungal infection by Candida albicans and the anti-fungal effect of a probiotic Lactobacillus rhamnosus strain via multimodal imaging as well as biochemical assays to monitor the extracellular proinflammatory cytokines. A conceptual idea contemplated in the “NutriChip” also suggested a modular immune microenvironment that induces proinflammatory stimuli (e.g., LPS) and a consequent response by differentiated macrophages or lymphoma-derived macrophage-like immune cells (Figure 5B) [61]. Interestingly, the NutriChip model proposes to validate the physiological contribution of anti-inflammatory dairy food to the defined immune-mediated inflammation, in which the “nutrikinetics” that studies the bioavailability of nutrient components on inflammation is an emerging discipline. One representative ex vivo MPS model utilized the beauty of the microfluidics system, where dissected mouse guts were connected to a devised platform enabling luminal flow and components could be applied with the original intestinal architecture and differentiated cells, including intestinal epithelium, immune cells, and enteric neuronal cells [63]. This approach possessed the EMI axis and a controlled access to the inside and outside of the lumen, independently. However, the limited longevity of ex vivo tissue (<30-40 h) and integration of human tissues remain challenging.
Table 2.
Modeling Mucosal Immunity and Inflammatory Crosstalk in the EMI axis
| Design of microphysiological device | Key cell types | Biological findings | Refs |
|---|---|---|---|
| The gut-on-a-chip that is configured with dual cell microchannels separated by a flexible porous membrane and vacuum chambers that induce mechanical deformations | Caco-2 intestinal epithelium; PBMC; Capillary or lymphatic human microvascular endothelium; Non-pathogenic E. coli, enteroinvasive E. coli, or probiotic VSL#3 |
Demonstrated an intestinal inflammation under complex interactions between luminal and abluminal components; Anti-inflammatory effects of probiotic or antibiotic treatment and the effects of the intestinal peristalsis on the microbial overgrowth were studied |
[58] |
| The gut inflammation-on-a-chip that modifies the basic configuration of the gut-on-a-chip microdevice | Caco-2 intestinal epithelium; PBMC; Non-pathogenic E. coli or probiotic VSL#3 |
The first mechanistic study using an MPS inflammation model to investigate the triggering factor of intestinal inflammation; Independent coupling of inflammatory factors revealed that the maintenance of epithelial barrier function is necessary and sufficient to suppress the initiation of gut inflammation |
[60] |
| A microfluidic device that has dual channels separated by a non-flexible porous membrane | Caco-2 intestinal epithelium; PBMC or tissue-resident mucosal macrophages; HUVEC; L. rhamnosus or C. albicans |
Demonstrated the effect of LPS during physiological condition or endotoxemia in the presence of tissue-resident mucosal macrophages; Anti-fungal effect of a probiotic L. rhamnosus against C. albicans was evaluated |
[59] |
| NutriChip that contains dual channels separated by a porous polyester membrane | Caco-2 intestinal epithelium; U937 macrophage cell line; Microbial LPS |
Suggested a new concept of “nutrikinetics” to assess the contribution of anti-inflammatory effects of dairy food products | [61] |
| A new microfluidic device with a medium chamber with ports for lumen input to connect dissected mouse gut |
Ex vivo mouse gut; Acinetobacter baumannii, Bacteroides dorei, Bacteroides fragilis, Bacteroides ovatus, Clostridia ramosum, Fusobacterium varium, Peptostreptococcus magnus, Candidatus Arthromitus |
Integrated ex vivo mouse gut to MPS maintaining its structure and multiple cells in the tissue, while applying luminal component by flow; Activation of the enteric nervous system by gut microbiome via regulatory T-cell induction was demonstrated |
[63] |
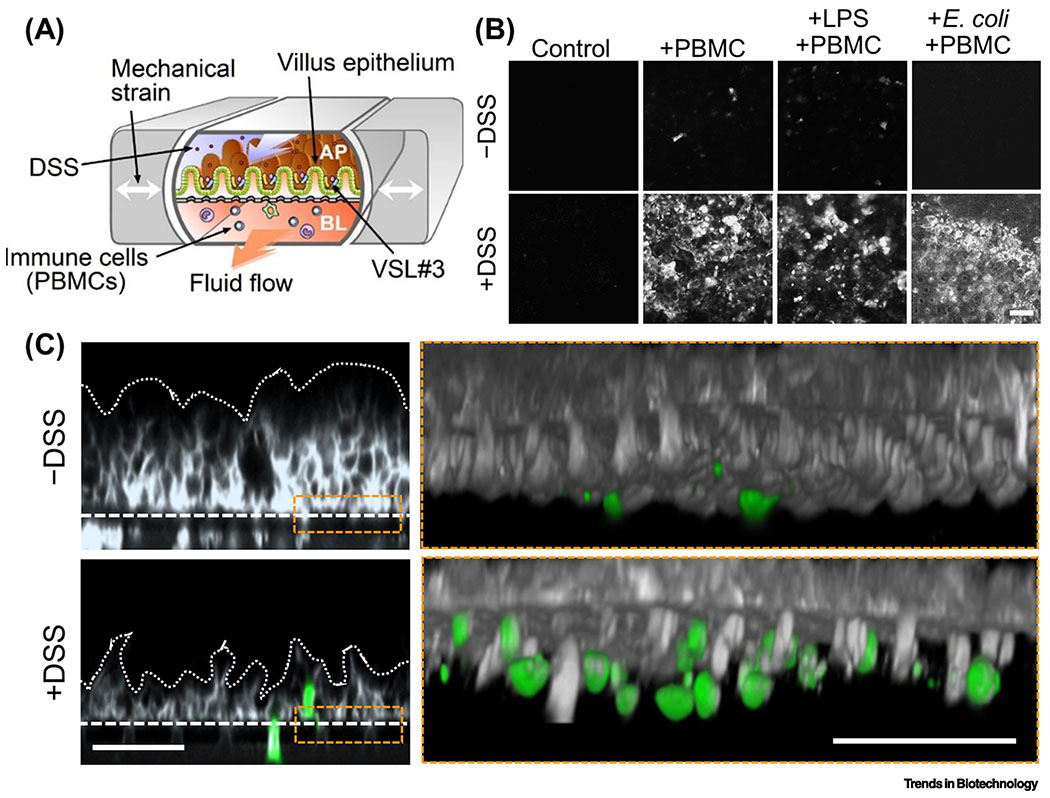
Figure 4. MPS modeling of intestinal inflammation.
(A) A schematic of the human gut inflammation-on-a-chip that provides the accessibility and versatility to manipulate the complex inflammatory milieu. Arrows in the central microchannels and the vacuum chambers indicate the directions of fluid flow and the peristalsis-like mechanical distortions, respectively. AP, apical; BL, basolateral. (B) Top-down views of the fluorescently labeled villous epithelium under the oxidative stress in the presence or absence of dextran sodium sulfate at various combinations of inflammatory triggers (PBMC, LPS, and E. coli) in the human gut inflammation-on-a-chip. Bar, 50 μm. (C) Cross-sectional views of the immune cell recruitment visualized in situ in the gut inflammation-on-a-chip. Gray, microengineered villous epithelium; green, PBMC. Dotted lines indicate the contour of the 3D villi. Dashed lines show the location of the basement membrane in the gut inflammation-on-a-chip. Bars, 50 μm. B and C reproduced from [60] with permission.
Figure 5. Immunocompetent intestinal MPS models that incorporate differentiated immune subsets.
(A) An MPS model that induces inflammatory responses in the EMI axis consisting of the intestinal epithelium, endothelium, LPS or bacterial cells, and tissue-resident mucosal macrophages (mMphs) and dendritic cells (DCs). Reproduced from [59] with permission. (B) A NutriChip that assesses the effects of dairy products by reconstituting intestinal epithelium, LPS or dairy food products, and macrophages. Bars, 50 μm. Reproduced from [61] with permission.
One of the most significant features of the mucosal MPS model is the experimental modularity, where the independent uncoupling of the individual contributing factors allows simulating complex immune responses via combinatorial recoupling. For instance, when epithelial cells were simultaneously exposed to both luminal (i.e., LPS or bacteria) and immune elements (i.e., PBMCs) in the lumen and capillary microchannels, respectively, in the gut inflammation-on-a-chip, aggressive inflammatory responses, elevated oxidative stress, loss of villous microarchitecture, and epithelial barrier dysfunction were triggered (Figure 4C). However, when each factor was independently applied, no inflammatory outcomes were observed [58]. Notably, animal models cannot demonstrate such experimental modularity. Furthermore, directional secretion of proinflammatory cytokines in the gut inflammation-on-a-chip is reminiscent of the pathophysiology germane to the chronic inflammation that occurs in IBD patients [58, 60] in which ensuing recruitment of activated immune cells also emulated the pathology of gut inflammation (Figure 4C). It is also amenable to investigate the effect of intestinal epithelial barrier on the regulation of intestinal immunity on “physiological tolerance” [60]. For instance, dextran sodium sulfate (DSS) can be applied in the gut inflammation-on-a-chip as a negative regulator of the intestinal barrier function, by which the tight junction barrier can be manipulated to discover which specific component orchestrates the epithelial oxidative stress (Figure 4B) and immune recruitment and activation (Figure 4C) pertinent to gut inflammation. This landmark study effectively demonstrated how a microengineered disease model and the reverse engineering principle could contribute to the interrogation of the disease initiating factor. In this context, the microengineered gut inflammation-on-a-chip model can validate the impact of the intercellular crosstalk on the immune responses triggered by the luminally stimulating components such as commercially available probiotic strains (e.g., VSL#3) or clinically relevant enteric pathogens (e.g., enteroinvasive Escherichia coli).
To better model the tissue-specific immunity of various GI diseases, it is critical to have reliable and reproducible models that closely recapitulate the patient-oriented immune microenvironment of the intestine. However, there have been numerous challenges, including limited sources of immune cells besides PBMCs, a lack of a protocol to induce in situ differentiation of immune cells inside the GI MPS models, difficulty in recreating a local 3D microenvironmental niche that secures the intercellular signaling, and a lack of a patient-specific personalized model for the immune system. Regardless of the scientific consensus of the in vitro immune systems, it is not yet easy to recreate the physiology of the immunity in a defined model system with a controlled spatiotemporal management including the recreation of the GALT, parenchymal-mesenchymal transition of immune cells, differentiation and polarization of naïve immune cells, professional antigen presentation, or the robust establishment of the specialized local immune cells (e.g., innate lymphoid cells, intraepithelial lymphocytes). The scalability of the current GI MPS immune models can be improved by incorporating patient-derived cells such as 3D organoid-derived epithelium, advanced enrichment of local immune cells depending on the type of disease (e.g., CD4+ T helper cell subsets in IBD, CD8+ cytotoxic T lymphocytes as well as regulatory T cells in cancer), or a sophisticated 3D design of a microenvironment to induce intercellular crosstalk in the EMI axis.
Facilitating systems immunology with MPSs
By integrating microfluidic approaches with high-throughput omics measurements, it is now possible to envision systems immunology-on-a-chip. Cell subset or single-cell measurements, facilitated by microfluidic droplet technology, are critical to account for cellular heterogeneity in immune cell responses in a population, as opposed to measurements that are typically averaged from bulk populations of immune cells. To this end, a multiplexed system combining microfluidics and multi-omics has been developed to enrich low abundance subsets from low input human PBMC with on-chip sequence library construction for genome-wide RNA-seq [64]. The approach demonstrated subset-specific responses to immune challenges in vitro and subset-specific transcriptional profiles in samples isolated from healthy donors or those with systemic lupus erythematosus. This approach overcomes the limitations of established methodologies for enriching PBMC subsets (fluorescence- and automated magnetic-activated cell sorting), which are labor- and cost-intensive and have high starting cell number requirements. Another multiparameter system combining tunable droplet-based microfluidics and single-cell RNA-seq was used to study the Toll-like receptor (TLR)-dependent secretion of interferon (IFN) and transcriptional profiles of plasmacytoid dendritic cells (pDC) [65], which are important for anti-viral and anti-cancer immunity and are implicated in autoimmunity. This approach revealed that IFN production is limited to a minor subpopulation of pDC cells and is controlled by stochastic gene regulation that is amplified by environmental signals (TLR signaling, cell density, IFN priming) rather than the existence of a pre-existing IFN-secreting pDC subset, providing an understanding of the heterogeneity of pDC responses to therapies that target them and insights towards optimizing DC-based immunotherapy. Finally, a microfluidic device was developed for isolating peptide-specific CD8+ T cells labeled with magnetic nanoparticles containing peptide-MHC tetramers and directly amplifying the T-cell receptor (TCR) genes from individual cells for sequencing [66]. The system provided 1000-fold increased sensitivity compared to bulk cell population analysis for identifying the specific TCR genes encoding antigen recognition. These pioneering studies predict an exciting future for combining microfluidic chips with omics approaches for enhancing our understanding of immune responses and developing new immunomodulatory therapies.
Concluding Remarks and Future Perspectives
These MPSs that recapitulate the tumor immune microenvironment and intestinal inflammatory immune responses, and the systems immunology applications highlighted here, demonstrate the promise of MPSs for modeling the architectural and functional complexity of human immune responses. Some emerging immunological applications of MPSs include incorporating immune cells in existing tissues- and organs-on-chips modeling a variety of diseases. For example, a human vascularized liver acinus MPS was developed to model the inflammatory phenotypes in liver diseases such as nonalcoholic fatty liver disease and diabetes [67]. This system recapitulated the role of the vascular inflammatory response in promoting the binding and transmigration of immune cells across a vascular endothelial barrier to a hepatic chamber. The incorporation of wound healing assays in MPSs as demonstrated in a breathing lung-on-a-chip [68] will be useful to study the role of immune cells in impaired wound healing in disease states such as repetitive lung microinjury or diabetes. MPSs can also be used to model how environmental toxins influence immune cells as recently shown in a human placenta-on-a-chip model of placental impairment [69]. These and other emerging immune MPS models will increase our understanding of human immune physiology and provide physiologically relevant platforms for the development of novel immunomodulatory approaches. The ability to precisely control the composition, stiffness, topography, and density of the ECM used in these 3D models will also help determine the effects of these important properties on the migration and mechanobiology of immune cells [70] as well as cytokine and chemokine secretion [71]. Several gaps and challenges remain in the broad adoption of this technology (see Outstanding Questions).
Outstanding Questions.
What are the existing challenges to recapitulate tissue-specific immunity in MPSs and how can these be overcome?
How can a disease-specific immune system and its responses be recreated in vitro?
Can an immune system-on-a-chip be utilized to study unknown disease mechanisms or develop or test a new therapeutic intervention of auto-immune diseases and immunodeficiency disorders?
Can incorporating immune cell components such as effector and regulatory cells expand the physiological relevance of existing tissues-and-organs-on-chips and emerging body-on-a-chip approaches?
Can immune system-on-a-chip models incorporate primary human or iPSC-derived cell inputs or tissue-specfic microbiota in order to understand patient heterogeneity in responses to immunotherapeutic approaches and develop precision medicine screening applications?
Can organ-on-chip approaches be developed to model immune organs such as the spleen, lymph node, and thymus?
How can immune system-on-a-chip technologies be designed to improve our understanding of impaired wound healing and repair in conditions such as diabetes?
Can immune system response-on-a-chip technologies that are mainly developed in highly specialized engineering labs be simplified and commercially distributed at large scale to facilitate widespread adoption in conventional cellular and molecular immunology labs and the broader biotechnology community?
Can immune MPSs be designed to be compatible with dynamic, miltiparameter monitoring of cellular responses and omics approaches to facilitate systems level immunology investigations?
Finally, we envision that tissue- and organ-on-a-chip technology and patient-specific organoid cultures and cell sources will synergize to build patient-specific immunocompetent MPS models as a new route to fulfilling the Precision Medicine Initiative [72]. Demographic, environmental, and epigenetic factors that can influence the development of diseases such as cancer or chronic inflammation will be more accurately depicted in the immunocompetent patient MPS avatars. Collaborative studies with pharmaceutical companies will contribute to compile patient-specific database in terms of pharmacokinetics, pharmacodynamics, and pharmacogenomics [73]. Combination with other advanced clinical and biomedical technologies in liquid biopsies [74], spatial transcriptomics [75], or cryopreservation will also contribute to the development of comprehensive human immune system-on-a-chip models. Overall, further development of human immune system responses on chips will increase our understanding of the role of the immune system in disease and lead to the development of new immunotherapeutic approaches.
Highlights.
Human tumor immune microenvironment-on-a-chip models have been developed to emulate cell type-dependent interactions, physical and chemical perturbations, and the infiltration and cytotoxicity of therapeutic anti-tumor lymphocytes and clinically relevant immunomodulatory agents.
Intestinal inflammation-on-a-chip models recapitulating the 3D intestinal transmural interface have been developed to discover how pathophysiological factors impair the intercellular crosstalk in the epithelial-microbiome-immune axis and trigger chronic inflammatory immune responses.
Combining engineered microphysiological immune system responses with high-throughput multi-omics measurements at the single-cell level facilitates a systems immunology-on-a-chip approach to gain novel insights into immune disorders.
Immune cells are being incorporated in tissues- and organs-on-chips modeling a variety of diseases.
Acknowledgements
This work was supported by the National Institutes of Health (R01HL146436, UG3EB028094, R01NS094388, and R01HL94388 to D.H.K.), the Human Frontier Science Program (RGP0038/2018 to D.H.K.), the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (HI19C0642 to D.H.K), the Bio & Medical Technology Development Program of the National Research Foundation funded by the Ministry of Science and ICT, Republic of Korea (2018M3A9H3025030 to H.J.K.), the Technology Impact Award of the Cancer Research Institute (UTA18-000889 to H.J.K.), the Alternative Research and Development Foundation (UTA18-001198 to H.J.K.), the Leona M. & Harry B. Helmsley Charitable Trust (Grant #1912-03604 to H.J.K.), the National Cancer Institute of the National Institutes of Health (R21CA236690 to H.J.K. and R21CA220111 to E.H.A.) and the F99/K00 Predoctoral to Postdoctoral Transition Award (1F99CA245801-01 to W.S.).
Glossary
- Antibody-dependent cell-mediated cytotoxicity (ADCC)
A cell-mediated immune response in which the fragment crystallizable (Fc) region of antibodies coating a target cell bind to Fc receptors on an effector cell, typically a natural killer cell, stimulating the release of cytotoxic factors
- Autologous T-cell receptor (TCR)-engineered T cells
T cells that are harvested from patients and engineered to express a T-cell receptor or a chimeric antigen receptor with specificity for an antigen expressed on tumor cells
- Chimeric antigen receptor (CAR) T cells
A type of T-cell receptor-engineered T cell expressing a chimeric antigen receptor consisting of an extracellular tumor antigen recognition domain, an extracellular hinge region, a transmembrane domain, and an intracellular domain for potently activating signaling pathways that promote T-cell activation and antitumor activity
- Collagen hydrogel
A semisolid medium used to create 3D cell culture environments consisting of type 1 collagen, the most abundant protein in the extracellular matrix
- Dextran sodium sulfate (DSS)
A polyanionic dextran derivative used to induce colitis in murine models. DSS can specifically disrupt the tight junction barrier in the gut
- Dysbiosis
An imbalanced population of the gut microbiome in the pathological stage that compromises host-microbiome crosstalk in the gut
- Gelatin methacryloyl
A photo-crosslinkable hydrogel derived from collagen with tunable stiffness properties
- Gut-associated lymphoid tissue (GALT)
A diffusive immune system of the gut including Peyer’s patches, mesenteric lymph nodes, and other local immune elements in the lamina propria
- Inflammatory bowel disease (IBD)
Chronic inflammation with autoimmunity in the human GI tract that includes Crohn’s disease and ulcerative colitis
- Lipopolysaccharide (LPS)
A bacterial endotoxin, a ligand of the toll-like receptor 4 (TLR4) and a component of the cell membrane of Gram-negative bacteria
- Microphysiological systems (MPSs)
Human biomimetic platforms based on microfluidics and microfabrication technologies to create a modular controllable microenvironment with physiologically relevant hydrodynamics and biomechanics for co-culturing different cell types in 3D to model the architectural and functional complexity of human tissues and organs
- Peripheral blood mononuclear cells (PBMCs)
A whole blood-derived mixed population of circulating immune cells including lymphocytes, monocytes, and granulocytes isolated using a density gradient separation
- Polydimethylsiloxane (PDMS)
An optically transparent, soft, biocompatible elastomer often used in the fabrication of microfluidic devices
- Programmed death-1 receptor (PD-1) blockade
A therapeutic approach in which monoclonal antibodies are used to disrupt interactions between the programmed death-1 receptor checkpoint receptor on the surface of cytotoxic T cells and programmed death-ligand 1 on the surface of tumor cells in order to promote anti-tumor responses
- Programmed death-ligands (PD-L1,2)
Proteins expressed on the surface of tumor cells and other immune cells that interact with programmed death-1 receptor on T cells to promote T-cell exhaustion which prevents antitumor responses
- Spheroids
A 3D cell modeling structure that better mimics live cell conditions in the extracellular matrix compared to 2D monolayer cultures including the presence of a hypoxic core with reduced oxygen levels
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
D.H.K. is a scientific founder and equity holder of NanoSurface Biomedical Inc. H.J.K. is a founder of 3D Health Solutions Inc. and holds an equity interest in the company.
References
- 1.Arrowsmith J and Miller P (2013) Trial watch: phase II and phase III attrition rates 2011-2012. Nat Rev Drug Discov 12 (8), 569. [DOI] [PubMed] [Google Scholar]
- 2.Tsui JH et al. (2018) Conductive silk-polypyrrole composite scaffolds with bioinspired nanotopographic cues for cardiac tissue engineering. J Mater Chem B 6 (44), 7185–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith AST et al. (2019) NanoMEA: A tool for high-throughput, electrophysiological phenotyping of patterned excitable cells. Nano Letters doi: 10.1021/acs.nanolett.9b04152. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nam KH et al. (2016) Multiscale cues drive collective cell migration. Sci Rep 6, 29749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J et al. (2019) Switch-like enhancement of epithelial-mesenchymal transition by YAP through feedback regulation of WT1 and Rho-family GTPases. Nat Commun 10 (1), 2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CL et al. (2016) migration phenotype of brain-cancer cells predicts patient outcomes. Cell Rep 15 (12), 2616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudmann DG (2019) The emergence of microphysiological systems (organs-on-chips) as paradigm-changing tools for toxicologic pathology. Toxicol Pathol 47 (1), 4–10. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B et al. (2018) Advances in organ-on-a-chip engineering. Nature Reviews Materials 3 (8), 257–278. [Google Scholar]
- 9.Chang SY et al. (2017) Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight 2 (22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edington CD et al. (2018) Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci Rep 8 (1), 4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernetti L et al. (2017) Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep 7, 42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irimia D and Wang X (2018) Inflammation-on-a-chip: probing the immune system ex vivo. Trends Biotechnol 36 (9), 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HN et al. (2019) Microphysiological systems as enabling tools for modeling complexity in the tumor microenvironment and accelerating cancer drug development. Advanced Functional Materials 29 (22), 1807553. [Google Scholar]
- 14.Salter AI et al. (2018) Chimeric antigen receptor-modified T cells: CD19 and the road beyond. Blood 131 (24), 2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavesi A et al. (2017) A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight 2 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SWL et al. (2018) Characterizing the role of monocytes in T cell cancer immunotherapy using a 3D microfluidic model. Front Immunol 9, 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallstabe L et al. (2019) ROR1-CAR T cells are effective against lung and breast cancer in advanced microphysiologic 3D tumor models. JCI Insight 4 (18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuczek DE et al. (2019) Collagen density regulates the activity of tumor-infiltrating T cells. J Immunother Cancer 7 (1), 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen AMH et al. (2019) Collagen density modulates the immunosuppressive functions of tumor-associated macrophages. bioRxiv, 513986. [Google Scholar]
- 20.Ando Y et al. (2019) Evaluating CAR-T cell therapy in a hypoxic 3D tumor model. Adv Healthc Mater 8 (5), e1900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park D et al. (2019) High-throughput microfluidic 3D cytotoxicity assay for cancer immunotherapy (CaCI-IMPACT platform). Front Immunol 10, 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanitis E et al. (2015) Targeting the tumor vasculature to enhance T cell activity. Curr Opin Immunol 33, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller CP et al. (2018) A 3D human renal cell carcinoma-on-a-chip for the study of tumor angiogenesis. Neoplasia 20 (6), 610–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayuso JM et al. (2019) Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. Oncoimmunology 8 (3), 1553477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aung A et al. (2019) An engineered tumor-on-a-chip device with breast cancer-immune cell interactions for assessing T-cell recruitment. Cancer Res doi: 10.1158/0008-5472.CAN-19-0342. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ligresti G et al. (2016) A novel three-dimensional human peritubular microvascular system. J Am Soc Nephrol 27 (8), 2370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins RW et al. (2018) Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov 8 (2), 196–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aref AR et al. (2018) 3D microfluidic ex vivo culture of organotypic tumor spheroids to model immune checkpoint blockade. Lab Chip 18 (20), 3129–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neal JT et al. (2018) Organoid modeling of the tumor immune microenvironment. Cell 175 (7), 1972–1988 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen M et al. (2018) Dissecting effects of anti-cancer drugs and cancer-associated fibroblasts by on-chip reconstitution of immunocompetent tumor microenvironments. Cell Rep 25 (13), 3884–3893 e3. [DOI] [PubMed] [Google Scholar]
- 31.Kinross JM et al. (2011) Gut microbiome-host interactions in health and disease. Genome Med 3 (3), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mowat AM and Agace WW (2014) Regional specialization within the intestinal immune system. Nat Rev Immunol 14 (10), 667–85. [DOI] [PubMed] [Google Scholar]
- 33.Rooks MG and Garrett WS (2016) Gut microbiota, metabolites and host immunity. Nat Rev Immunol 16 (6), 341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith PM et al. (2013) The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341 (6145), 569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson LW and Artis D (2014) Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14 (3), 141–53. [DOI] [PubMed] [Google Scholar]
- 36.Rhee KJ et al. (2004) Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J Immunol 172 (2), 1118–24. [DOI] [PubMed] [Google Scholar]
- 37.Carding S et al. (2015) Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 26, 26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown EM et al. (2013) The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol 14 (7), 660–7. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn R et al. (1993) Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75 (2), 263–74. [DOI] [PubMed] [Google Scholar]
- 40.Perse M and Cerar A (2012) Dextran sodium sulphate colitis mouse model: traps and tricks. J Biomed Biotechnol 2012, 718617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostanin DV et al. (2009) T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol 296 (2), G135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seok J et al. (2013) Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110 (9), 3507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen TL et al. (2015) How informative is the mouse for human gut microbiota research? Dis Model Mech 8 (1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martignoni M et al. (2006) Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol 2 (6), 875–94. [DOI] [PubMed] [Google Scholar]
- 45.Musther H et al. (2014) Animal versus human oral drug bioavailability: do they correlate? Eur J Pharm Sci 57, 280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi M et al. (2009) The species differences of intestinal drug absorption and first-pass metabolism between cynomolgus monkeys and humans. J Pharm Sci 98 (11), 4343–53. [DOI] [PubMed] [Google Scholar]
- 47.Carmody RN and Turnbaugh PJ (2014) Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest 124 (10), 4173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koppel N et al. (2017) Chemical transformation of xenobiotics by the human gut microbiota. Science 356 (6344). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace BD et al. (2010) Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330 (6005), 831–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haller D et al. (2000) Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut 47 (1), 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noel G et al. (2017) A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci Rep 7, 45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnalzger TE et al. (2019) 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J 38 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HJ and Ingber DE (2013) Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb) 5 (9), 1130–40. [DOI] [PubMed] [Google Scholar]
- 54.Shin W et al. (2019) Human intestinal morphogenesis controlled by transepithelial morphogen gradient and flow-dependent physical cues in a microengineered gut-on-a-chip. iScience 15, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sung JH et al. (2011) Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 11 (3), 389–92. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y et al. (2017) A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HJ et al. (2012) Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12 (12), 2165–74. [DOI] [PubMed] [Google Scholar]
- 58.Kim HJ et al. (2016) Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A 113 (1), E7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maurer M et al. (2019) A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 220, 119396. [DOI] [PubMed] [Google Scholar]
- 60.Shin W and Kim HJ (2018) Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc Natl Acad Sci U S A 115 (45), E10539–E10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramadan Q et al. (2013) NutriChip: nutrition analysis meets microfluidics. Lab Chip 13 (2), 196–203. [DOI] [PubMed] [Google Scholar]
- 62.Park BS and Lee JO (2013) Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yissachar N et al. (2017) An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 168 (6), 1135–1148 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reyes M et al. (2019) Multiplexed enrichment and genomic profiling of peripheral blood cells reveal subset-specific immune signatures. Sci Adv 5 (1), eaau9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wimmers F et al. (2018) Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat Commun 9 (1), 3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng AHC et al. (2019) MATE-Seq: microfluidic antigen-TCR engagement sequencing. Lab Chip 19 (18), 3011–3021. [DOI] [PubMed] [Google Scholar]
- 67.Li X et al. (2018) A glass-based, continuously zonated and vascularized human liver acinus microphysiological system (vLAMPS) designed for experimental modeling of diseases and ADME/TOX. Lab Chip 18 (17), 2614–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Felder M et al. (2019) Impaired wound healing of alveolar lung epithelial cells in a breathing lung-on-a-chip. Front Bioeng Biotechnol 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yin F et al. (2019) A 3D human placenta-on-a-chip model to probe nanoparticle exposure at the placental barrier. Toxicol In Vitro 54, 105–113. [DOI] [PubMed] [Google Scholar]
- 70.Kim JK et al. (2019) Unraveling the mechanobiology of the immune system. Adv Healthc Mater 8 (4), e1801332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeon H et al. (2015) Combined effects of substrate topography and stiffness on endothelial cytokine and chemokine secretion. ACS Appl Mater Interfaces 7 (8), 4525–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collins FS and Varmus H (2015) A new initiative on precision medicine. N Engl J Med 372 (9), 793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whirl-Carrillo M et al. (2012) Pharmacogenomics knowledge for personalized medicine. Clinical Pharmacology & Therapeutics 92 (4), 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Di Meo A et al. (2017) Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer 16 (1), 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stahl PL et al. (2016) Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353 (6294), 78–82. [DOI] [PubMed] [Google Scholar]