Abstract
The global outbreak of COVID-19 (Coronavirus Disease 2019) caused by SARS-CoV-2 (Severe Acute Respiratory Syndrome caused by Coronavirus 2) began in December 2019. Its closest relative, SARS-CoV-1, has a slightly mutated Spike (S) protein, which interacts with ACE2 receptor in human cells to start the infection. So far, there are no vaccines or drugs to treat COVID-19. So, research groups worldwide are seeking new molecules targeting the S protein to prevent infection by SARS-CoV-2 and COVID-19 establishment. We performed molecular docking analysis of eight synthetic peptides against SARS-CoV-2 S protein. All interacted with the protein, but Mo-CBP3-PepII and PepKAA had the highest affinity with it. By binding to the S protein, both peptides led to conformational alterations in the protein, resulting in incorrect interaction with ACE2. Therefore, given the importance of the S protein-ACE2 interaction for SARS-CoV-2 infection, synthetic peptides could block SARS-CoV-2 infection. Moreover, unlike other antiviral drugs, peptides have no toxicity to human cells. Thus, these peptides are potential molecules to be tested against SARS-CoV-2 and to develop new drugs to treat COVID-19.
Keywords: SARS-CoV-2, COVID-19, Synthetic peptides, Molecular docking, ACE2 receptor
Highlights
-
•
Synthetic peptides bind to SARS-CoV-2 Spike protein.
-
•
Synthetic peptides induced conformational changes on SARS-CoV-2 spike protein structure.
-
•
Synthetic peptides bind to ACE2 protein but did not affect its structure.
-
•
Synthetic peptides induced the wrong interaction of SARS-CoV-2 with ACE2 receptor.
1. Introduction
Coronaviruses (CoV) are a group of non-segmented positive single-stranded RNA, with size up to 32 kb, belonging to the Coronaviridae family and Nidovirales order [1,2]. Seventeen coronaviruses can infect humans. Typically, this infection presents slight symptoms, like a common cold. However, among them, three cause the same symptoms but with higher intensity and are also responsible for atypically severe pneumonia complications, which increases their mortality compared to other viruses [[1], [2], [3], [4], [5]].
In the last two decades, three CoV outbreaks have spread widely, becoming world pandemics. The first began in the winter of 2002 in China, where viral pneumonia caused by CoVs resulted in severe acute respiratory syndrome coronavirus (SARS-CoV-1), officially infecting 8906 people in 26 countries and leading to 774 deaths [6,7]. The second was 2012 in Saudi Arabia, called Middle East respiratory syndrome coronavirus (MERS-CoV), which led to death of 37% of infected patients. MERS-CoV spread to 27 countries, infecting 2254 people and causing 800 deaths worldwide [6,8].
The third outbreak is ongoing, a pandemic situation that started in December 2019 in China. Patients developed new severe pneumonia (COVID-19) caused by SARS-CoV-2.COVID-19. The disease spread rapidly to 210 countries, at this writing infecting almost 5 million people and causing around 362 thousand deaths [9,10], figures that are rising steeply. The origin of SARS-CoV-2 is obscure. Although there are similarities between SARS-CoV-2 and bat SARS-CoV-like coronaviruses, the progenitor is hard identify [3,11,12].
Genome sequence data has revealed at least 70% similarity between SARS-CoV-1 and SARS-CoV-2 [3,[12], [13], [14]]. Both coronaviruses share the same entry path in human cells, but with particularities. SARS-CoV-1 and SARS-CoV-2 possess a membrane-bound trimeric spike (S) protein, which has a receptor-binding domain (RBD) able to interact with Angiotensin I Converting Enzyme 2 (ACE2) receptor in human cells [7,15]. Despite the similarities, the S protein from SARS-CoV-2 has modifications in the RBD sequence that enhance the affinity with ACE2 of humans. Indeed, the SARS-CoV-2 S protein has a 20-fold higher affinity with human ACE2 than the S protein from SARS-CoV-1, leading to faster spread from human to human [3,7,[13], [14], [15]].
As yet there are no drugs for clinical treatment or vaccines to prevent human infection by SARS-CoV-2. Drug repositioning seems to be a faster method to find an effective medicine to treat clinical symptoms of COVID-19 [16]. However, in this chaotic scenario, computational screening and molecular docking (MD) are promising alternatives to find a new or existing drug to treat symptoms of COVID-19. In all those MD analyses, a common target has been tested, the SARS-CoV-2 S protein, because it is essential to the entry in cells to start the infection process [[17], [18], [19], [20], [21]].
Here we report a MD study using eight antimicrobial peptides (Mo-CBP3-PepI, Mo-CBP3-PepII, Mo-CBP3-PepIII [22,23], RcAlb-PepI, RcAlb-PepII, RcAlb-PepIII [24], PepGAT and PepKAA [25] (unpublished data)) that target the SARS-CoV-2 S protein. With different affinities, all peptides interact with the RBD from the SARS-CoV-2 S protein. By interacting with SARS-CoV-2 S protein, peptides change its native conformation, inducing incorrect interaction with the ACE2 receptor and thus blocking the entry of SARS-CoV-2 into the cell. The MD results suggest peptides as potential alternative molecules to be used against SARS-CoV-2. To the best of our knowledge, this is the first study to employ synthetic peptides as drugs to target spike protein from SARS-CoV-2.
2. Methodology
2.1. Three-dimensional (3D) structures
The 3D structures of Mo-CBP3-PepI, Mo-CBP3-PepII, and Mo-CBP3-PepIII were the same reported by Oliveira et al. [22]. The 3D structures of RcAlb-PepI, RcAlb-PepII, RcAlb-PepIII were the same as employed by Dias et al. [24]. The 3D structures of PepGAT and PepKAA were the same as employed by Souza et al. [25]. The 3D structure files of SARS-CoV-2 Spike protein (open state, PDB: 6VYB; closed state, PDB: 6VXX) and human ACE2 (ACE2; PDB: 1R42) were downloaded from Protein Data Bank (PDB, https://www.rcsb.org/).
2.2. Bioinformatic analyses of antiviral potential of synthetic peptides
Bioinformatics was employed to evaluate whether the peptides presented any antiviral potential. We used the free server iAMPPred (http://cabgrid.res.in:8080/amppred/).
2.3. Molecular docking (MD) assays
Blind molecular docking assays were carried out in two systems. First, the molecular docking experiments involved synthetic peptides (ligands) against SARS-CoV-2 spike glycoprotein in the open state (6VYB) and closed state (6VXX). Second, SARS-CoV-2 spike-peptide complexes were tested by molecular docking against the ACE2 human receptor. All molecular docking experiments were performed using the ClusPro 2.0 server (https://cluspro.org), which is the best performing server currently available for the CAPRI challenge [26]. It is also used for the study of inhibitor-enzyme, peptide-protein, and inhibitor-protein complexes, additionally allowing the analysis and choice of the most appropriate scoring scheme for the complex in this study [[27], [28], [29]].
The GPU option was selected because it uses more specific computer graphic units of the Massachusetts Green High-Performance Computing Center (MGHPC). The choice of models was standardized and followed the criteria use of the same scoring scheme (Balanced) and balance between the number of members and lowest binding energy. The balanced scoring scheme has the best weight coefficients for the energy used in the structures. The best complexes generated by molecular docking studies were analyzed in terms of interface energy and interaction between surface residues, and for models with a high number of members. The first 12 models of the balanced energy parameter were used for the analysis of waste and interface energy, and models with less than 20 anchored structures were excluded from the analysis. The models were also selected from the analysis of the PIPER energy in the center of the cluster, that is, the structure that has the largest number of neighboring structures in the center of the cluster, as well as from the analysis of the structure that had the lowest energy of the cluster.
2.4. Minimization and equilibration by molecular dynamics simulation
To reach a higher stabilized state, the six systems of interest (S protein closed:: ACE2; S protein open::ACE2; S protein closed::Mo-CBP3-PepII::ACE2; S protein open::Mo-CBP3-PepII::ACE2; S protein closed::PepKAA:: ACE2; and S protein open::PepKAA:: ACE2) were minimized and equilibrated using Gromacs version 2018.4 [30]. Initially, the topology was recorded using the OPLS-AA/L all-atom force field [31,32], after a cubic box was created with 2 nm from the box edge. The box solvation was done with the SPC/E water model. The systems were neutralized and Na+ and Cl− ions were added at a concentration of 0.15 M. The minimization was carried out until reaching negative potential energy and maximum force under 1000 kJ mol−1 nm−1. Finally, the equilibration of temperature and pressure was performed for 100 ps.
2.5. Analysis of the complexes interface
The database for structural analysis of 3D structures PDBdum (EMBL - EBI; http://www.ebi.ac.uk/pdbsum) was used to analyze the general content of the interface region resulting from the molecular docking tests. The Ligplot software was used to generate the 2D figures with the hydrogen bonds and the hydrophobic interactions, and the Pymol software was used to generate the figures with 3D structures [28]. The Pymol software, in addition to being used to visualize the molecular structure of models, was also used to identify molecular interactions between complexes, identify polar and non-polar interactions, and perform the alignment of structures. PyMol was also used to calculate RMSD and generate the score for the overlapping of structures.
2.6. Refinement and validation of structures
The complex structures generated by the molecular docking tests were refined by the Galaxy Refine Complex, a tool belonging to the molecular interaction server GalaxyWEB. All 3D structures were validated for steric impediment by the MolProbity and PROCHECK servers.
3. Results
3.1. Antiviral potential of synthetic peptides
Before molecular docking analyses, the peptides were analyzed in an antiviral sever to evaluate whether or not they had antiviral potential. We only considered peptides that were at least 70% antiviral to be promising. Among all peptides tested, only four that criterion: Mo-CBP3-PepI, Mo-CBP3-PepII, RcAlb-PepIII, and PepKAA, respectively with 70, 85, 84, 74% being antiviral (Supplementary Table 1).
3.2. Interaction of SARS-Cov-2 spike glycoprotein with human ACE2 receptor
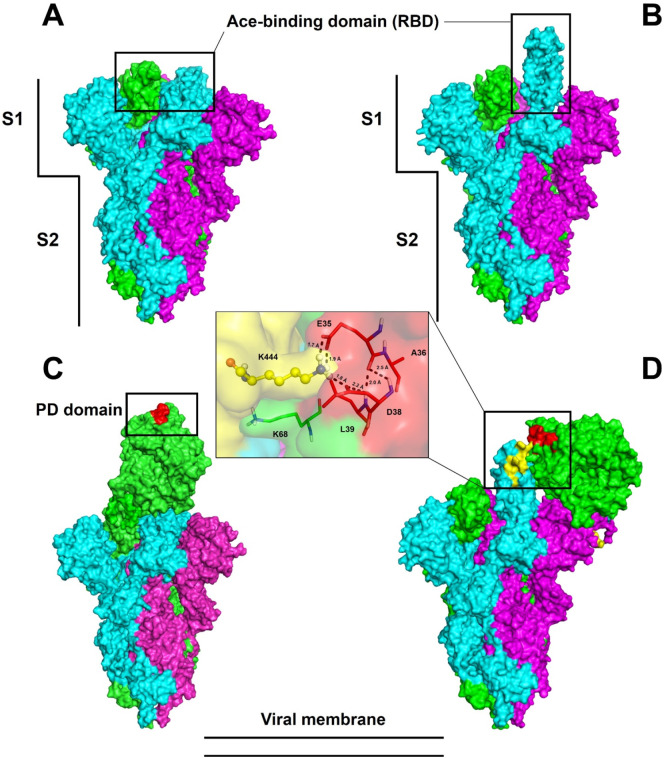
The subunits and domains of spike protein are represented in Fig. 1 , in which their two conformational states are also observed: the closed state (Fig. 1A) and the open state (Fig. 1B). As expected, in the open state (Fig. 1B) the receptor-binding domain (RBD) responsible for interacting with the human ACE2 receptor was positioned outside the structure, favoring interaction with ACE2 (Fig. 1B). Additionally, it the S1 and S2 domains of spike protein, which are respectively responsible for the S1 domain's interaction with human ACE2 and viral and cell membrane fusion before entrance (Fig. 1A and B) [7]. The results of blind molecular docking assays performed by Cluspro 2.0 revealed that in both closed (Fig. 1C) and open (Fig. 1D) state, the spike protein can interact with ACE2. Fig. 1D insert is a zoom view of the interaction between spike protein and ACE2 receptor.
Fig. 1.
Trimeric structure of SARS-CoV-2 spike glycoprotein. The conformational states closed (A) and partially open (B) define the effectiveness of the connection to ACE2. The regions indicated by rectangles correspond to the RBD domains of the glycoprotein spike (A, B) and the PD domain (C, D) of ACE2. The vertical bars define the S1 and S2 subunits that are essential for initial SARS-CoV-2 infection. The enlarged area shows some interaction amino acids according to the crystals in the literature. The trimer chains are highlighted by the colors cyan, magenta, and green.
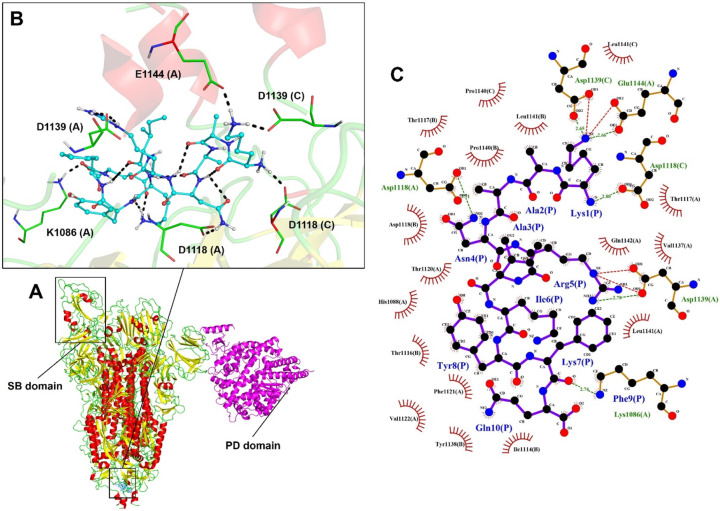
The interface region of the spike protein in the open state with ACE2 had the lowest energy among all the models generated, with the lowest binding energy (LBE), −1032.0 kJ·mol−1. Thus it was used as a control for the analysis of the interaction between peptides and spike protein. In the closed state, spike protein binds to a different region of ACE2 with an LBE of −899.0 kJ·mol−1 and interaction with 33 members. This model correlates with the literature data for the ACE2 region (31–41 interaction with spike protein; Uniprot: Q9BYF1 [33]), demonstrating the importance of open conformational state for binding with ACE2 and onset of infection (Fig. 1C).
These results confirm the predictions identified by [33], who suggested that spike protein must be in the open conformation to interact with the PD domain of ACE2 on the cell surface. The spike glycoprotein must expose the RBD domain for the interaction to occur, because in the closed conformational state, the ACE2 recognition motifs (RBDs) are buried at the interface between the proteins.
3.3. Molecular docking of SARS-Cov-2 S protein with peptides
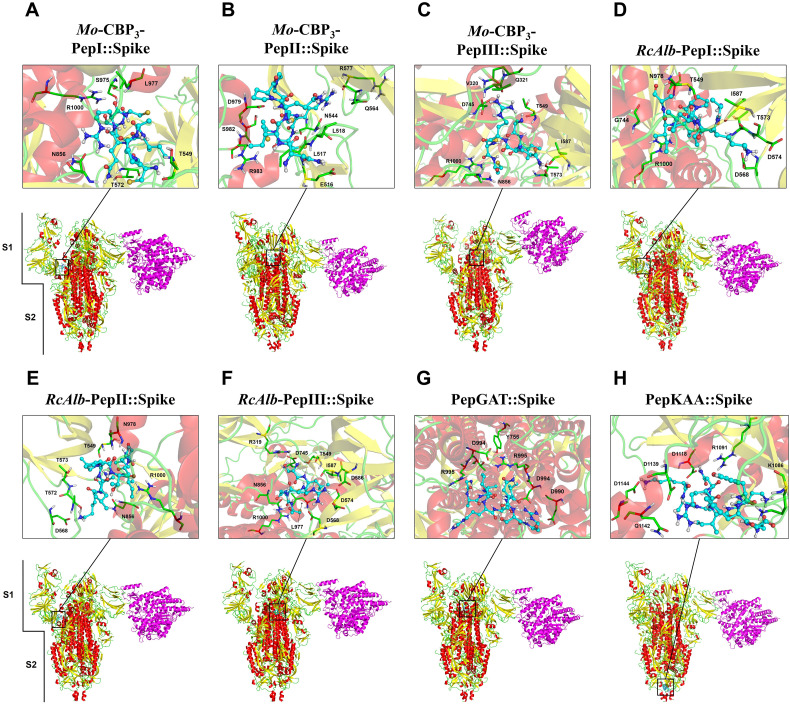
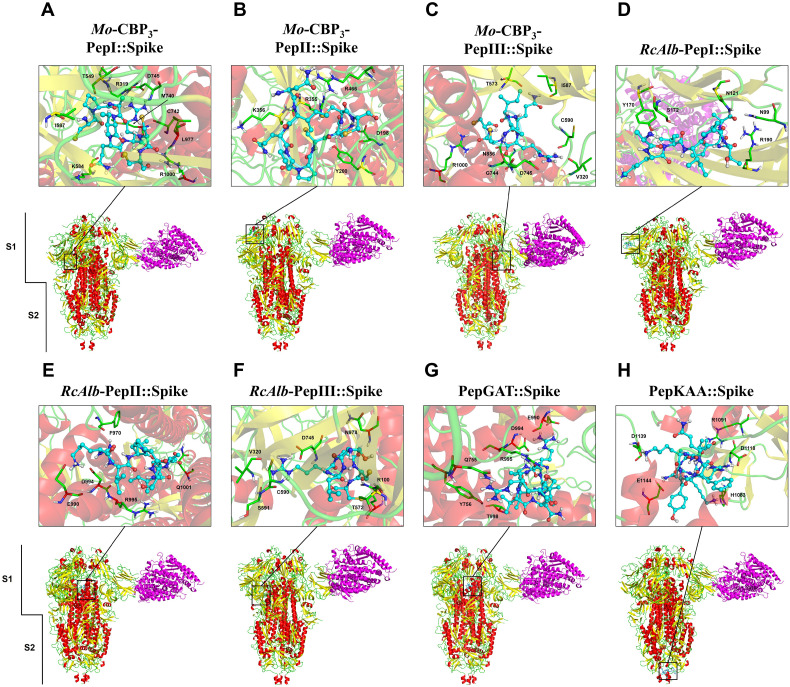
The molecular docking experiments were performed using the eight peptides against spike protein in both open (Fig. 2 ) and closed (Fig. 3 ) states. MD assays with the open state of spike protein showed that Mo-CBP3-PepI, Mo-CBP3-PepII, Mo-CBP3-PepIII, RcAlb-PepI, RcAlb-PepII and PepGAT interact with the S1 domain (Fig. 2A–E, G). In contrast, RcAlb-PepIII and PepKAA interacted with the S2 domain (Fig. 2F and H). Similar results were found out during the MD assays with peptides against the closed state of spike protein (Fig. 3). Only one difference was observed: in this model, RcAlb-PepIII interacted with the S1 domain of the spike protein instead of S2 as in the open state (Fig. 3G).
Fig. 2.
3D structures of the spike glycoprotein in the open state complexed with synthetic peptides and ACE2. The representation shows the viral protein in the partially open state. The RBD and PD domains of ACE2 (magenta) are altered due to the interaction between spike glycoprotein and the peptides. The connection to ACE2 does not occur between the RBD and PD domains. In the S protein, colors are defined by SS: alpha-helix in red, beta-sheets in yellow, and loops in green.
Fig. 3.
3D structures of the spike glycoprotein in the closed state complexed with synthetic peptides and ACE2. The representation shows the viral protein in the partially closed state. The RBD and PD domains of ACE2 (magenta) are altered due to the interaction between spike glycoprotein and the peptides. The connection to ACE2 does not occur between the RBD and PD domains. In the S protein, colors are defined by SS: alpha-helix in red, beta-sheets in yellow, and loops in green.
The number of hydrogen bonds and hydrophobic interactions of the peptides and spike protein suggests their strong interaction, in both open (Table 1 ) and closed (Table 2 ) states. Our results revealed distinct behaviors of peptides when in contact with spike protein. Among the peptides that interacted with the S1 domain, Mo-CBP3-PepII presented the lowest binding energy, of −814.4 kJ·mol−1 (Table 1), and the highest number of interactions (185), of which 170 were hydrophobic interactions and 15 were hydrogen bonds in the open state, and 173 were hydrophobic interactions and 12 were hydrogen bonds in the closed state (Table 1, Table 2).
Table 1.
Molecular interaction docking between synthetic peptides and SARS-CoV-2 spike protein in the open state. The molecular docking assay was performed using the ClusPro server 2.0 and structural analysis was performed by using the Pymol program.
| Complexes | Lowest binding energya | Hydrogen bondsb | Number of hydrophobic interactionsc |
|---|---|---|---|
| SARS-CoV2 spike protein/Mo-CBP3-PepI | −658.0 | THR549 (CYS1); ARG567 (CYS8); THR572 (GLN5); TYR741 (ARG6); ASN856 (GLN5, ARG6); SER975 (CYS7); LEU977 (ARG6) | 111 |
| SARS-CoV2 spike protein/Mo-CBP3-PepII | −814.4 | CYS391 (CYS8); GLU516 (ASN1); LEU517 (CYS6, ASN1); ASN544 (ARG7); GLN564 (ARG7); ARG567 (PRO4, GLN3); ARG577 (ARG7); ASP979 (CYS6); SER982 (CYS9); ARG983 (CYS9). | 170 |
| SARS-CoV2 spike protein/Mo-CBP3-PepIII | −700.8 | ASP745 (ARG4); SER975 (CYS6); ARG1000 (CYS5, CYS6); THR549 (ARG4); THR573 (GLN3); ILE587 (GLN3); CYS590 (ARG4) | 134 |
| SARS-CoV2 spike protein/RcAlb-PepI | −649.8 | ASP568 (LYS2); THR573 (ALA1); ASP574 (LYS2); LEU977 (PRO5); ASN978 (ALA8); ARG1000 (THR6) | 116 |
| SARS-CoV2 spike protein/RcAlb-PepII | −744.8 | THE549 (ALA1); GLY744 (ALA8); ASN856 (LEU3); ASN978 (PRO5); ARG1000 (THR6). | 143 |
| SARS-CoV2 spike protein/RcAlb-PepIII | −703.0 | ASP745 (SER1); ASN856 (GLY4); SER975 (CYS6); ARG1000 (CYS5); THR549 (LEU2, SER1); ASP568 (ARG3); ASP574 (ARG3); ILE587 (ARG3); | 123 |
| SARS-CoV2 spike protein/PepGAT | −593.1 | TYR756 (GLY1); TYR756 (ALA2); GLU990 (ARG5); ASP994 (ARG5); ARG995 (ARG5); ARG995 (SER9); THR998 (THR3); GLU990 (ARG10); ASP994 (ARG10); ARG995 (ASN8) | 141 |
| SARS-CoV2 spike protein/PepKAA | −674.9 | LYS1086 (PHE9); HIS1088 (TYR8); ARG1091 (ASN4); ASP1118 (ASN4); GLN1142 (ARG5); GLU1144 (LYS1); GLN1113 (LYS7); ASP1118 (LYS1); ASP1139 (LYS1); ASP1139 (LYS1) | 161 |
Calculated by the ClusPro server 2.0.
Analyzed in the Pymol program.
Analyzed in the Ligplot program.
Table 2.
Molecular interaction docking between synthetic peptides and SARS-CoV-2 spike protein in the closed state. The molecular docking assay was performed using the ClusPro server 2.0 and structural analysis was performed by using the Pymol program.
| Complexes | Lowest binding energya | Hydrogen bondsb | Number of hydrophobic interactionsc |
|---|---|---|---|
| SARS-CoV2 spike protein/Mo-CBP3-PepI | −703.0 | THR549 (CYS1); ASP568 (GLN5); ASP 571 (CYS 8); THR572 (GLN5); MET740 (ARG 6); GLY744 (CYS 7); ASP745 (ARG6); PHE855 (GLN5); LEU977 (CYS7); ASN978 (CYS7); ARG1000 (ARG6, CYS8, CYS7) | 194 |
| SARS-CoV2 spike protein/Mo-CBP3-PepII | −787.3 | TRP353 (CYS8); ASN354 (ASN1); ARG355 (CYS8); LYS356 (GLN3); ARG357 (GLN3); ARG466 (ASN1), (CYS8); THR167 (CYS9,); ASP198 (ARG7); TYR200 (ARG7); ILE231 (CYS9); GLY232 (CYS9). | 173 |
| SARS-CoV2 spike protein/Mo-CBP3-PepIII | −723.8 | GLY744 (ALA1); ASP745 (ARG4); ASN856 (GLN3); ASN978 (ILE2); ARG1000 (CYS5); THR573 (GLN3); ILE587 (GLN3); CYS590 (ARG4). | 135 |
| SARS-CoV2 spike protein/RcAlb-PepI | −587.8 | ASN121 (ALA8); TYR170 (LEU3); SER172 (ALA1, LEU3); ARG190 (THR6, ILE7, ALA8) | 81 |
| SARS-CoV2 spike protein/RcAlb-PepII | −671.8 | ASP994 (LYS2, LEU3); ARG995 (THR6, PRO5); GLN1002 (LEU9); PHE970 (ALA1); ARG995 (LYS2) | 139 |
| SARS-CoV2 spike protein/RcAlb-PepIII | −674.6 | ASN978 (GLY4); ARG1000 (CYS5); THR573 (SER1); CYS590 (ARG3). | 91 |
| SARS-CoV2 spike protein/PepGAT | −619.0 | GLU990 (ARG5); ASP994 (ALA2, ARG5); ARG995 (ARG5, SER9); GLN755 (ARG10); TYR756 (ARG10); ASP994 (ARG10); ARG995 (ASN8). | 142 |
| SARS-CoV2 spike protein/PepKAA | −715.6 | ASP1118 (LYS7); ASP1139 (LYS7); GLU1144 (ASN4, LYS7); HIS1083 (LYS1); ARG1091 (ALA2); ASP1118 (ARG5); VAL1137 (LYS1) | 175 |
Calculated by the ClusPro server 2.0.
Analyzed in the Pymol program.
Analyzed in the Ligplot program.
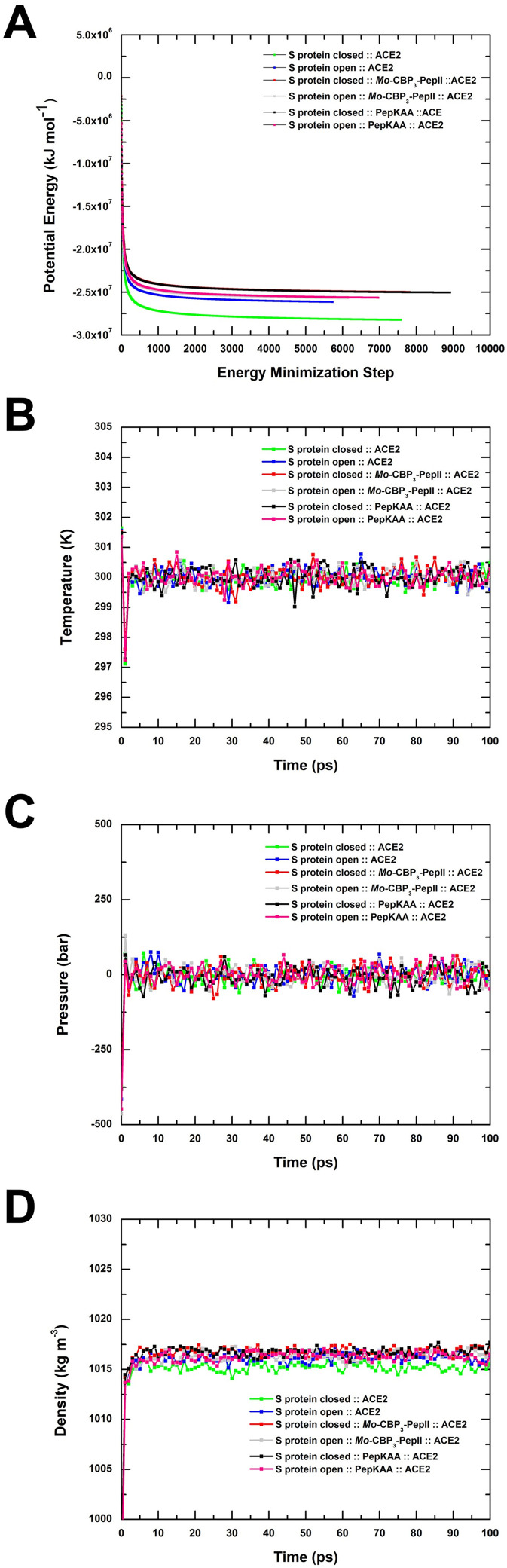
3.4. Stable and balanced structures
All structures were minimized with potential energy of about −2.5 × 10−7 after 5500 energy minimization steps and maximum force of less than 1000 kJ mol−1 nm−1 (Fig. 4A). The minimized structures occurred at the equilibrium of temperature. From 5 to 100 ps, the temperature of all complexes was balanced at 300 K, with small variations (Fig. 4B). Finally, with variable pressure and density it was possible to observe equilibrium after 5 ps and continuing stability until 100 ps. The pressure and density equilibrium were achieved at about 0 bar and 1015 kg m−3, respectively (Fig. 4C and D). The final stable complexes were used in the subsequent analyses.
Fig. 4.
Minimization and equilibrium of the S protein complexed with Mo-CBP3-PepII, PepKAA, and ACE2. The minimizations are observed through potential energy during the energy minimization step (A). The temperature, pressure and density equilibrium along 100 ps (B, C, and D, respectively). The S protein in closed form complexed with ACE2 is represented in green line and square. The open-complexed S protein with ACE2 is represented by the blue line and square. Mo-CBP3-PepII with ACE2 and closed S protein is represented by the red line and square. The open S protein is represented in the gray line and square. PepKAA with ACE2 and closed S protein is represented by the black line and square, and when with open S protein is represented by a pink line and square.
3.5. Interaction of SARS-Cov-2 S protein with peptides
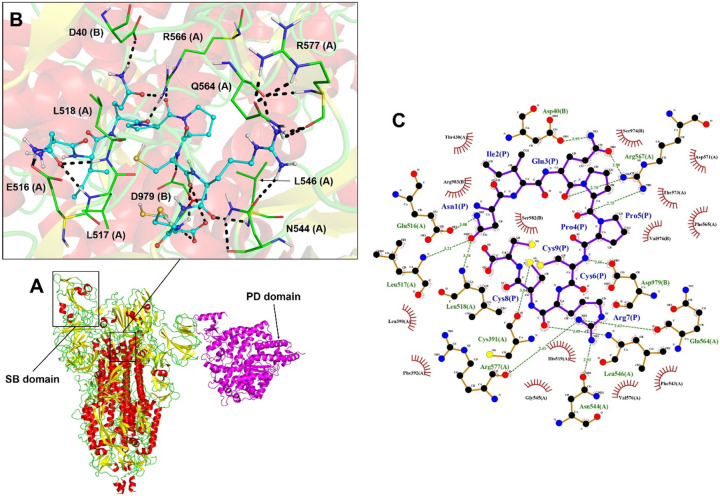
Mo-CBP3-PepII interacted with the α-helices spike S1 domain (Fig. 5A). This interaction is mainly supported by hydrogen bonds formed by Asn1, Gln3, Pro4, and Arg7 from Mo-CBP3-PepII, respectively with Glu516, Asp40, Arg567, Gln564, from the S1 domain of the S protein (Fig. 5B). Moreover, other interactions were also crucial for this interaction, as revealed by Ligplot analyses, such as hydrogen bonds Asn1, Cys6, Arg7, and Cys8, from Mo-CBP3-PepII with, respectively, Leu517, Leu518, Asp979, Arg577, Asn544, Cys391 (Fig. 5C) from the S protein. Several hydrophobic interactions were observed in the residues with spoked red arcs.
Fig. 5.
3D and flat structure of the S protein complexed with Mo-CBP3-PepII and ACE2. Diagram of the interface region showing hydrogen bonds and hydrophobic interactions are visualized in 3D (A, B) and 2D (C). The peptide Mo-CBP3-PepII interacts with the α-helix of the S1 subunit. The images were generated automatically by Lig-Plot and analyzed on PyMol. In B, the peptide is shown in cyan and the viral protein in green. LigPlot images show peptide residues in dark blue and viral residues in dark red.
Regarding peptides that interacted with the S2 domain close to the transmembrane region of the spike protein (Fig. 6A), PepKAA was the best, with 171 interactions, including 10 hydrogen bonds and 161 hydrophobic interactions and an LBE value of −674.9 kJ·mol−1 in the open state (Table 1). The hydrogen bond interactions that support this were Lys1, Asn4, Arg5, and Lys7 from PepKAA, respectively, with Glu1144, Asp1139, Asp1118, and Lys1086 S2 domain of the S protein (Fig. 6B). Moreover, Ligplot analyses revealed several hydrophobic interactions with all PepKAA, represented by residues in spoked red arcs (Fig. 6C).
Fig. 6.
3D and flat structure of the S protein complexed with PepKAA and ACE2. Diagram of the interface region showing hydrogen bonds and hydrophobic interactions are visualized in 3D (A, B) and 2D (C). The peptide PepKAA interacts with the α-helix of the S1 subunit. The images were generated automatically by LigPlot and analyzed on the PyMol. In B, the peptide is shown in cyan and the viral protein in green. LigPlot images show peptide residues in dark blue and viral residues in dark red.
3.6. Peptide-induced conformational changes in SARS-Cov-2 spike
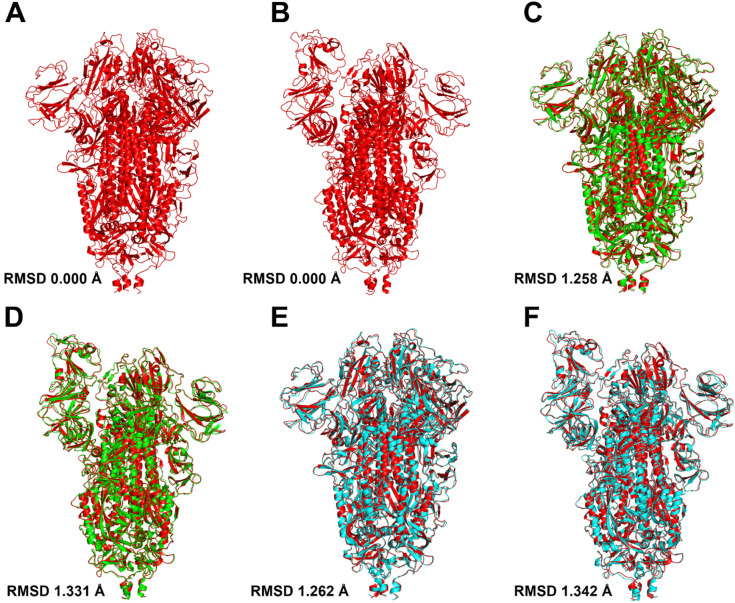
Unfortunately, Mo-CBP3-PepII did not bind to the RBD domain of the spike protein in the S1 subunit. However, a meticulous analysis revealed an interesting point. The analysis of conformational alignment and overlapping of S protein structures, before and after interaction with Mo-CBP3-PepII, revealed that by interacting with the S1 subunit, Mo-CBP3-PepII induced a change in the conformational state of the S protein structure in the closed and open states (Fig. 7C and D) in comparison with the spike protein alone in the closed and open states (Fig. 7A and B). The RMSD of the spike protein in the closed and open state was 0, indicating typical 3D conformation. However, after interaction with Mo-CBP3-PepII, the RMSD values changed from 0 to 1.258 and 1.331, respectively, for the open and closed states. PepKAA also induced conformational changes in the S protein after interacting with the S2 domain (Fig. 7E and F), which was revealed by RMSD values that changed from 0 in the control to 1.262 and 1.342, respectively, in open and closed state of the S protein. These data suggest Mo-CBP3-PepII and PepGAT induce changes in the conformation of the S protein, which may decrease the binding affinity of the SARS-CoV-2 spike protein with human ACE2.
Fig. 7.
3D visualization SARS-CoV-2 S protein:peptides complexes and RMSD calculations. The RMSD calculation of the closed (A) and partially open (B) states of the viral protein before (in red) and after (green and blue) the interaction with the Mo-CBP3-PepII (C, D) peptides and PepKAA (E, F) suggest conformational alteration caused by the peptides. The structure before (red) and after (green) the interaction of Mo-CBP3-PepII and cyan for PepKAA. The structural alignment was carried out by Pymol.
3.7. Peptides do not lead to alterations in the structure of ACE2 receptor
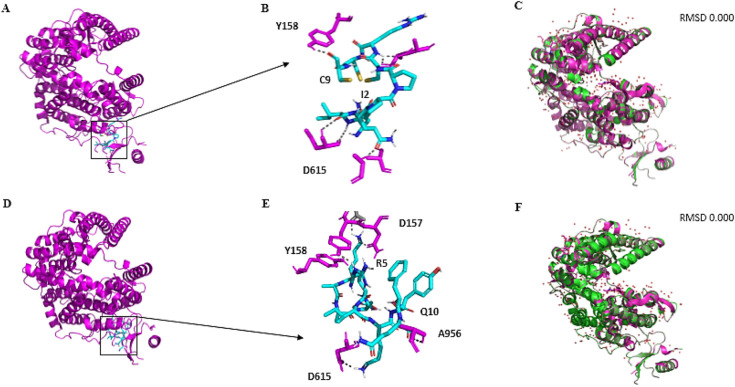
MD assays were also carried out to investigate possible interaction between peptides and ACE2 receptor. The results revealed that both peptides interact with the extracellular region of the ACE2 (Fig. 8A and D). Even though interacting in the same region, the interaction energy and amino acids involved were different. Mo-CBP3-PepII had a slightly lower affinity with ACE2, as shown by an LBE value of −665.5 kJ·mol−1. The amino acids involved in this interaction are Ile2 and C9 from Mo-CBP3-PepII, respectively, with Try158 and Asp615 from ACE2 (Fig. 8B). For PepKAA, the LBE was −700.5 kJ·mol−1 and the amino acids involved were Arg5 and Gln10 with Tyr158, Asp157, Asp615, and Ala956 from the ACE2 receptor (Fig. 8E).
Fig. 8.
Representation of the interface of interaction between ACE2 and Mo-CBP3-PepII and PepKAA. Mo-CBP3-PepII (A, B) and PepKAA (D, E) did not bind to the PD domain of ACE2 and did not interfere with the binding to the RBD domain of the spike glycoprotein. RMSD data displayed of Mo-CBP3-PepII (C) and PepKAA (D) suggest that the peptides do not cause a conformational change in the ACE2 protein. Overlapping of crystals before and after interaction with synthetic peptides is shown in magenta and green, respectively.
Even though both peptides bound to ACE2, none of them caused any conformational change in its structure (Fig. 8C and F). This result suggests that despite binding to ACE2, the peptides did not alter structure, and hence its function.
3.8. By interacting with spike protein, peptides block its interaction with ACE2
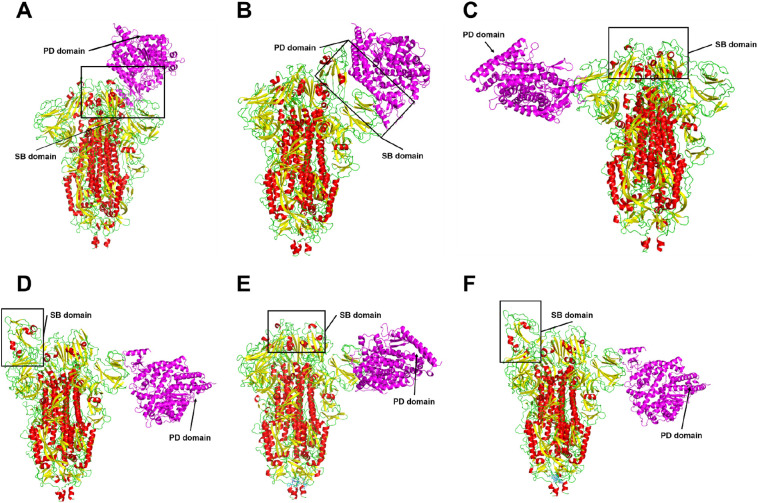
As shown above, the S protein can interact with ACE receptor in both closed and open state (Fig. 9A and B). However, only in the open state of S protein did the interaction happen correctly by the RBD domain of the S protein with the PD domain of ACE2 (Fig. 9B), allowing the entrance of SARS-CoV into cells [33].
Fig. 9.
Effectiveness of the link between SARS-CoV-2 S protein and ACE2 in the presence of peptides. ACE2 binds to the RBD domains in the conformational states closed (A) and open (B) of the spike glycoprotein. Binding to the RBD domains does not occur between the S protein: Mo-CBP3-PepII (C, D) and ACE2 complexes, S protein: PepKAA (E, F) and ACE2. Protein S is colored by SS; in magenta ACE2.
We already found that peptides induced conformational changes in the S protein (Fig. 7). To see if these changes could interfere in the interaction of that protein with ACE2 MD analyses were performed with the S protein:peptides complex with ACE2 (Fig. 9C–F). MD assays carried out with S protein:Mo-CBP3-PeII complexed with ACE2 in both open (Fig. 9C) and closed states (Fig. 9D) revealed that protein S still interacted with ACE2, but this interaction occurred in a different portion rather than the RDB domain. The complex formed by the open state of the S protein and Mo-CBP3-PeII (Fig. 9C) interacted with a portion far from the RBD domain. The same results were found when the complex S protein:PepKAA interacted with ACE2 (Fig. 9E and F). When complexed with PepKAA, the S protein could not correctly interact with ACE2.
4. Discussion
COVID-19 disease is entirely new, and there is no treatment of even vaccine to protect people. Besides respiratory complications, COVID-19 can cause cytokine-mediated inflammatory responses in patients, leading to complications of the heart, kidneys and liver, as well as decreased platelet counts. All these complication are led by the overproduction of response proinflammatory cytokines such as tumor necrosis factor (TNF), interleukin (IL) 6 and 1β [[34], [35], [36]]. The high transmissibility and infectivity of SARS-CoV-2 compared to SARS-CoV-1 and MERS are, in part, the result of a mutation in the spike protein [3,15,20].
The spike protein is a trimeric glycoprotein with two domains, S1 and S2, both critical for SARS-CoV-2 infection. In humans, the interaction between spike protein and ACE2 allows the entrance of SARS-CoV-2 in the lungs. The ability of SARS-CoV-2 to infect many different cells is supported by the interaction between the S protein and ACE2 receptor. ACE2 is expressed in many human tissues, such as lungs, liver, heart, kidneys, gut and brain. Thus, once inside the human body, SARS-CoV-2 can virtually infect all these tissues, leading to multiple infections. Because of that, the spike protein has been the target of many studies worldwide seeking ways to neutralize SARS-CoV-2 infection [7,33,37].
Based on its importance, the spike protein has been targeted by many research groups seeking treatment to COVID-19. Supercomputers and molecular docking assays have been employed to find an existing or develop a new drug that targets this protein and thus prevents SARS-CoV form infecting human cells [18,19,38]. We showed by molecular docking that Mo-CBP3-PepII and PepKAA interact, respectively, with the S1 and S2 domains of the S protein (Fig. 2).
The lowest binding energy values were −814.4 and −674.9 kJ·mol−1, respectively, for Mo-CBP3-PepII and PepKAA. That is 2.71 and 2.24 higher than the energy of arbidol to interact with the S protein [38], suggesting that both peptides have more affinity to bind with the S protein than with arbidol. Despite proven toxicity, some researchers are still analyzing chloroquine and hydroxychloroquine to treat COVID-19. Molecular docking analysis revealed that chloroquine and hydroxychloroquine have binding energies of 100 kJ·mol−1 and 137 kJ·mol−1 to interact with the spike protein. These values are too low compared to the values presented by peptides.
Another study reported molecular docking assays using other drugs against SARS-CoV-2 RNA polymerase (RdRp). The drugs galidesivir, remdesivir, tenofovir, sofosbuvir, and ribavirin bind with SARS-CoV-2 RdRp, with very low binding energies of −7.0, −7.6, −6.9, −7.5, and −7.8 kcal/mol, respectively [39]. Besides the low binding energies, SARS-CoV-2 RpRd might not be a good target because this enzyme can only be assessed during SARS-CoV-2 replication within the cell. By binding to the S protein, peptides prevent cell infection, and thus all the downstream process of SARS-CoV-2 entrance in cells.
Many studies have investigated drug repositioning to an available drug for faster development of treatment for COVID-19 [16,40]. Wu et al. [40] conducted molecular docking screening using many already available drugs such as antihypertensive, antifungal, antibacterial and anticoagulant drugs. Some of them presented low affinity to interact with the S protein, so could not interfere in S protein-ACE2 interaction like the peptides presented here did.
Our experiments revealed that the energy of S protein and ACE2 is 899.0 kJ·mol−1. By showing energies of 814.4 and −674.9 kJ·mol−1, respectively, Mo-CBP3-PepII and PepKAA can interfere in the S protein-ACE2 interaction. This interaction raised two questions: (i) What are the consequences of peptide interaction with the S protein?; and (ii) After interact with peptides, could the S protein bind to the ACE2 protein? The first question was solved by employing RMSD analyses. The RMSD values of the S protein after interaction with peptides were 1.331 and 1.342, respectively, for Mo-CBP3-PepII and PepKAA (Fig. 7), suggesting that by interaction with the S protein, these peptides induce conformation changes in it, which could lead either to no interaction or wrong interaction with ACE2. Vankadari [38] showed that the RMSD value of arbidol after binding to the S protein was 0.82, indicating a slight alteration in that protein's structure.
Indeed, after binding to peptides, the S protein can still bind to ACE2 but with very low affinity and in the wrong place (Fig. 9). The wrong interaction of the S protein with ACE2 has severe consequences for SARS-Cov-2. In humans, the domain (targeted by Mo-CBP3-PepII) is responsible for interacting with ACE2 (angiotensin-converting enzyme 2) receptors in lung cells. After interaction with ACE2, the spike protein is cleaved by a cellular protease, releasing the S2 domain (targeted by PepKAA), responsible for the fusion between viral and cell membranes [7,33,34,41]. These results strongly suggest that both peptides act by targeting the S protein and can block the interaction with ACE2 and thus inhibit the entrance of SARS-CoV-2, first in lung cells and then other cells [2,42]. As happens with other viruses, without a cell to infect, SARS-Cov-2 becomes unstable and thus is naturally degraded.
ACE2 is an essential cellular receptor in many types of cells involved in the conversion of angiotensin (Ang) I and Ang II into Ang 1–9 and Ang 1–7 [37]. Its malfunction can lead to severe multiorgan damages. Based on that, when used in clinical trials, peptides might not show any collateral effects. Molecular docking assays revealed that Mo-CBP3-PepII and PepKAA indeed bind to ACE2, but RMSD analyses showed that no conformational changes occur (Fig. 8). This suggests that although peptides bind to ACE2, its function in cells is not affected. In addition, the binding energy values indicate that peptides have more affinity with the spike protein than ACE2, which means that in the same environment, peptides will prefer to bind to the spike protein instead of ACE2.
Antiviral drugs already available, such as arbidol, chloroquine and hydroxychloroquine, have many side effects, such as diarrhea, nausea, vomiting, liver, and heart damage [12,43,44]. Compared to those drugs, the synthetic peptides presented here have no side effects, no hemolytic activity or toxicity to human cells [23]. This indicates that peptides, based on molecular docking, are potential molecules for use to develop new drugs, given their higher affinity with the spike protein without interfering with ACE2 activity, and absence of toxic effects.
The peptides have high potential against SARS-CoV-2. However, some limitations have to solve: (1) The peptides were only tested against the ACE2 receptor. However, there is an alternative receptor in the human lungs cells called CD209L, a C-type lectin (also called L-SIGN), which could be used by coronaviruses to cause infection [45]. (2) SARS-CoV-2 is an RNA virus that suggests a high mutational rate [45,46], which makes it challenging to find a good target. Indeed, the peptides tested here could not prevent the mutations in SARS-CoV-2 RNA. Because of that, at the same time, peptides could be employed to disrupt the interaction between SARS-CoV-2 and ACE2, they also can be tested against the CD209L receptor by molecular dynamics simulations and against the SARS-CoV-2 RNA polymerase, to see if peptides could inhibit its activity. Further analysis will be carried out to increase the broad spectrum of peptides against SARS-CoV-2.
5. Conclusion
This is the first study to report peptides as potential antiviral molecules that can be used to inhibit SARS-CoV-2 entrance in cells. These peptides can interact with SARS-CoV-2 S protein and block its entrance in human cells and thus inhibit infection and COVID-19 development. Further analyses are needed to prove this, but we can suggest that Mo-CBP3-PepII and PepKAA have potential for development of new drugs against SARS-CoV-2 and perhaps other viruses.
The following is the supplementary data related to this article.
Antiviral potential of synthetic peptides.
Funding and acknowledgments
This work was supported by grants from the following Brazilian agencies: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (process numbers 308107/2013-6 and 306202/2017-4); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES); and Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP).
Author contributions
All authors made substantial contributions in the following steps: (1) conception and design of the study by PFNS; (2) acquisition of data, analysis, interpretation of data by FESL and JLA (3) drafting the article or revising it critically with important intellectual content by FESL, CDTF, JTAO, and PFNS; and (3) final approval of the version to be submitted by PFNS.
Ethical approval
None sought.
Declaration of competing interest
All authors declare no conflict of interest.
References
- 1.Peiris J.S.M. Med. Microbiol. Eighteenth ed. Elsevier Inc.; 2012. Coronaviruses; pp. 587–593. [DOI] [Google Scholar]
- 2.Burrell C.J., Howard C.R., Murphy F.A. Chapter 13 - Coronaviruses. Fifth edition. 2017. Fenner and White’s medical virology; pp. 437–446. [DOI] [Google Scholar]
- 3.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO . WHO; 2020. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [Google Scholar]
- 9.COVIDView weekly summary | CDC, (n.d.). https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html (accessed April 26, 2020).
- 10.Practice B.B. Coronavirus disease 2019. World Heal. Organ. 2019;2020:2633. doi: 10.1001/jama.2020.2633. [DOI] [Google Scholar]
- 11.Almazán F., Sola I., Zuñiga S., Marquez-Jurado S., Morales L., Becares M., Enjuanes L. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Liu S.-M., Yu X.-H., Tang S.-L., Tang C.-K. Coronavirus disease 2019 (COVID-19): current status and future perspective. Int. J. Antimicrob. Agents. 2020;105951 doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8 doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drug repositioning is an alternative for the treatment of coronavirus COVID-19. Int. J. Antimicrob. Agents. 2020:105969. doi: 10.1016/J.IJANTIMICAG.2020.105969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.B. Robson, COVID-19 coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles' heel conserved region to minimize probability of escape mutations and drug resistance, Comput. Biol. Med.. (n.d.). doi: 10.1016/J.COMPBIOMED.2020.103749. [DOI] [PMC free article] [PubMed]
- 18.Hall D.C., Ji H.-F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med. Infect. Dis. 2020:101646. doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujii T., Nakamura T., Iwamoto A. Current concepts in SARS treatment. J. Infect. Chemother. 2004;10(1) doi: 10.1007/s10156-003-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calligari P., Bobone S., Ricci G., Bocedi A. 2020. Molecular Investigation of SARS – CoV-2 Proteins and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira J.T.A., Souza P.F.N., Vasconcelos I.M., Dias L.P., Martins T.F., Van Tilburg M.F., Guedes M.I.F., Sousa D.O.B. Mo-CBP3-PepI, Mo-CBP3-PepII, and Mo-CBP3-PepIII are synthetic antimicrobial peptides active against human pathogens by stimulating ROS generation and increasing plasma membrane permeability. Biochimie. 2019;157:10–21. doi: 10.1016/j.biochi.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Lima P.G., Souza P.F.N., Freitas C.D.T., Oliveira J.T.A., Dias L.P., Neto J.X.S., Vasconcelos I.M., Lopes J.L.S., Sousa D.O.B. Anticandidal activity of synthetic peptides: mechanism of action revealed by scanning electron and fluorescence microscopies and synergism effect with nystatin. J. Pept. Sci. 2020:1–13. doi: 10.1002/psc.3249. [DOI] [PubMed] [Google Scholar]
- 24.Dias L.P., Souza P.F.N., Oliveira J.T.A., Vasconcelos I.M., Araújo N.M.S., Tilburg M.F.V., Guedes M.I.F., Carneiro R.F., Lopes J.L.S., Sousa D.O.B. RcAlb-PepII, a synthetic small peptide bioinspired in the 2S albumin from the seed cake of Ricinus communis, is a potent antimicrobial agent against Klebsiella pneumoniae and Candida parapsilosis. Biochim. Biophys. Acta Biomembr. 2019:183092. doi: 10.1016/j.bbamem.2019.183092. [DOI] [PubMed] [Google Scholar]
- 25.Souza P.F.N., Marques L.S.M., Oliveira J.T.A., Lima P.G., Dias L.P., Neto N.A.S., Lopes F.E.S., Sousa J.S., Silva A.F.B., Caneiro R.F., Lopes J.L.S., Ramos M.V., Freitas C.D.T. Synthetic antimicrobial peptides: from choice of the best sequences to action mechanisms. Biochimie. 2020;175:132–145. doi: 10.1016/j.biochi.2020.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Inbar Y., Schneidman-Duhovny D., Halperin I., Oron A., Nussinov R., Wolfson H.J. Proteins Struct. Funct. Genet., Proteins. 2005. Approaching the CAPRI challenge with an efficient geometry-based docking; pp. 217–223. [DOI] [PubMed] [Google Scholar]
- 27.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moal I.H., Bates P.A. SwarmDock and the use of normal modes in protein-protein docking. Int. J. Mol. Sci. 2010;11:3623–3648. doi: 10.3390/ijms11103623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 31.Robertson M.J., Tirado-Rives J., Jorgensen W.L. Improved peptide and protein torsional energetics with the OPLS-AA force field. J. Chem. Theory Comput. 2015;11:3499–3509. doi: 10.1021/acs.jctc.5b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. doi: 10.1021/ja9621760. [DOI] [Google Scholar]
- 33.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(e6):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. 2019;2019:2019–2020. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ. Res. 2020 doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking the trimerization of viral spike glycoprotein? Int. J. Antimicrob. Agents. 2020:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alifano M., Alifano P., Forgez P., Iannelli A. Renin-angiotensin system at the heart of COVID-19 pandemic. Biochimie. 2020 doi: 10.1016/j.biochi.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A.E. Gorbalenya, S.C. Baker, R.S. Baric, R.J. de Groot, C. Drosten, A.A. Gulyaeva, B.L. Haagmans, C. Lauber, A.M. Leontovich, B.W. Neuman, D. Penzar, S. Perlman, L.L. Poon, D. Samborskiy, I.A. Sidorov, I. Sola, J. Ziebuhr, Severe Acute Respiratory Syndrome-related Coronavirus: The Species and Its Viruses-A Statement of the Coronavirus Study Group, (n.d.). doi: 10.1101/2020.02.07.937862. [DOI]
- 42.Oakes J.M., Fuchs R.M., Gardner J.D., Lazartigues E., Yue X. Nicotine and the renin-angiotensin system. Am. J. Phys. Regul. Integr. Comp. Phys. 2018;315:R895–R906. doi: 10.1152/ajpregu.00099.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q., Xiong H.R., Lu L., Liu Y.Y., Luo F., Hou W., Yang Z.Q. Antiviral and anti-inflammatory activity of arbidol hydrochloride in influenza A (H1N1) virus infection. Acta Pharmacol. Sin. 2013;34:1075–1083. doi: 10.1038/aps.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., DeMartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., Su X., Cao B. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antiviral potential of synthetic peptides.