Abstract
Background
Highly pathogenic avian influenza (HPAI)-H7N9 virus arising from low pathogenic avian influenza (LPAI)-H7N9 virus with polybasic amino acid substitutions in the hemagglutinin was detected in 2017.
Methods
We compared the tropism, replication competence, and cytokine induction of HPAI-H7N9, LPAI-H7N9, and HPAI-H5N1 in ex vivo human respiratory tract explants, in vitro culture of human alveolar epithelial cells (AECs) and pulmonary microvascular endothelial cells (HMVEC-L).
Results
Replication competence of HPAI- and LPAI-H7N9 were comparable in ex vivo cultures of bronchus and lung. HPAI-H7N9 predominantly infected AECs, whereas limited infection was observed in bronchus. The reduced tropism of HPAI-H7N9 in bronchial epithelium may explain the lack of human-to-human transmission despite a number of mammalian adaptation markers. Apical and basolateral release of virus was observed only in HPAI-H7N9- and H5N1-infected AECs regardless of infection route. HPAI-H7N9, but not LPAI-H7N9 efficiently replicated in HMVEC-L.
Conclusions
Our findings demonstrate that a HPAI-H7N9 virus efficiently replicating in ex vivo cultures of human bronchus and lung. The HPAI-H7N9 was more efficient at replicating in human AECs and HMVEC-L than LPAI-H7N9 implying that endothelial tropism may involve in pathogenesis of HPAI-H7N9 disease.
Keywords: alveolar epithelial cells, H7N9, HPAI, influenza, risk assessment
We compared the replication competences of HPAI- and LPAI-H7N9, which were comparable in ex vivo cultures of human bronchus and lung. HPAI-H7N9 and H5N1 replicated to higher titers than LPAI-H7N9 in both human alveolar epithelial and pulmonary microvascular endothelial cells.
Since the first detection of human disease caused by low pathogenic avian influenza (LPAI) A H7N9 in February 2013 in Eastern China, this virus has caused 5 epidemics [1, 2]. The fifth epidemic in 2016–2017 was associated with the largest number of reported human cases and was of particular concern [3]. As of October 3, 2018, 1567 laboratory-confirmed cases of H7N9 disease and 615 deaths were reported, with a case-fatality rate of ~39%. An HPAI-H7N9 with an insertion of multibasic amino acids at the hemagglutinin (HA) cleavage site was first detected in environmental samples collected from poultry cages in November 2016 [4]. In January 2017, HPAI-H7N9 viruses were identified in Guangdong, and subsequently 32 human cases were identified to be HPAI-H7N9 infections, as of October 3, 2018 [5]. During the fifth wave in September 2017, there was an introduction of an H5/H7 bivalent influenza vaccine in chickens and it was highly effective. The H7N9 virus isolation rate in poultry was reduced by 93.3%, and there were only 3 H7N9 human cases between October 2017 and February 2019 [5, 6].
Phylogenetic analysis revealed that HPAI-H7N9 viruses were derived from LPAI-H7N9 of the Yangtze lineage currently circulating in domestic poultry [7]. The most important risk factor for human infection of the HPAI-H7N9 virus was exposure to sick or dead poultry. Although HPAI-H7N9 viruses were able to replicate efficiently in mammals and transmit via respiratory droplets among ferrets [8], there is currently no evidence of sustained human-to-human transmission [9]. Clinical outcomes and fatality risk of HPAI- H7N9 and LPAI-H7N9 infections appeared to be comparable; infected patients primarily developed viral pneumonia with some progressing to acute respiratory distress syndrome, which is a type of respiratory failure characterized by rapid onset of widespread inflammation in the lungs and increased alveolar-capillary permeability leading to alveolar edema with high mortality rate. However, HPAI-H7N9 infection usually resulted in earlier symptoms onset, hospitalization, and death [7, 9, 10].
We have previously shown that LPAI-H7N9 isolated from the first wave of infection efficiently replicated and infected human bronchus and lung explant cultures [11]. To better understand the pathogenesis of the HPAI-H7N9 infection in humans, we compared the tissue tropism and viral replication competence of an HPAI-H7N9 strain (A/Guangdong/17SF006/2017) isolated from a fatal human case in Guangdong, with an LPAI-H7N9 chicken isolate isolated during the fifth wave, 3 LPAI-H7N9 viruses isolated from human during the first 2 waves, an HPAI-H5N1 virus and a 2009 pandemic H1N1 virus in ex vivo cultures of human bronchus and lung and in vitro cultures of human alveolar epithelial cells (AECs), and microvascular endothelial cell cultures (HMVEC-L). The innate host responses induced by HPAI- and LPAI-H7N9 infection were studied in human AECs and in HMVEC-L.
METHODS
Viruses
A/Guangdong/17SF006/2017 (QY/HPAI-H7N9) (GISAID accession nos. EPI919592–EPI919599) was isolated from the endotracheal aspirate of a 56-year-old male patient on day 6 of disease onset. The patient was treated with oseltamivir and peramivir and died on day 43 [7]. A/chicken/HK/YO84/2016 (ck/YO84/H7N9) was isolated from chicken carcass in Hong Kong (GISAID accession nos. EPI1345893–EPI1345900). A/Shanghai/2/2013 (Sh2/H7N9) (GISAID accession nos. EPI439495–EPI439502) and A/Anhui/1/H7N9 (Ah1/H7N9) (GISAID accession nos. EPI439503–EPI439510) were isolated from fatal human cases during the first wave of the H7N9 epidemic. A/HK/734/2014 (734/H7N9) (GISAID accession nos. EPI498793–EPI498800) was isolated from a fatal human case during the second wave. A/Hong Kong/483/1997 (483/H5N1), a HPAI-H5N1 isolated from a fatal human case in Hong Kong, and A/Hong Kong/415742/2009 (H1N1pdm), a pandemic H1N1 virus, were also used for comparison. Alignment and analysis of viral sequences were carried out using MEGA6. QY/HPAI-H7N9 and ck/YO84/H7N9 were first isolated in pathogen-free embryonated chicken eggs, whereas other viruses were isolated in Madin-Darby canine kidney (MDCK) cells. Virus stocks used were prepared in MDCK cells, and viral titers were determined by 50% tissue culture infectious dose (TCID50) titrations. All experiments were performed in a biosafety level-3 facility.
Virus Infection of Human Ex Vivo Cultures
Fresh human bronchus and lung were obtained from tissue removed as part of the routine clinical care of patients undergoing surgical resection. After diagnostic requirements were met, any residual excess tissue was used in this study. Informed consent was obtained from all subjects, and approval was granted by the Institutional Review Board of the University of Hong Kong and the Hospital Authority (Hong Kong West) (approval no. UW 14-119). The bronchus and lung tissues were cultured, infected, and analyzed as previously described [11–13]. Supernatant of the infected tissues were harvested for virus titration by TCID50 assay at 1, 24, and 48 hours postinfection (hpi). Area under the curve (AUC) was calculated from the viral titers (see Supplementary Material). Tissues were fixed in 10% neutral-buffered formalin at 24 and 48 hpi for immunohistochemical staining.
Isolation of Primary Human Alveolar Epithelial Cells
Primary human AECs were isolated and used for infection as previously described (see Supplementary Material) [11, 13].
Isolation of Primary Human Pulmonary Microvascular Endothelial Cells
The HMVEC-L were isolated from human nontumor lung tissue using the method described previously with modifications (see Supplementary Material) [14, 15].
Virus Infection of In Vitro Cultures
The AECs and HMVEC-L were infected at a multiplicity of infection (MOI) of 0.01 or 2 for 1 hour at 37°C [11, 13]. Cells were washed with phosphate-buffered saline and replenished with saline-adenine-glucose-mannitol (SAGM) for AECs and EGM-2MV for HMVEC-L. The viral titer of supernatants collected at 1, 24, 48, and 72 hpi was determined by TCID50 assay. The AUC was calculated from the viral titers (see Supplementary Material). Cell lysates and supernatants were collected at 24 hpi for measurement of messenger ribonucleic acid (mRNA) and protein expression of cytokine and chemokine.
Polarity of Virus Release Upon Infection
To study the polarity of HPAI-H7N9 infection in AECs and HMVEC-L, cells were seeded on the apical compartment of a 0.4-µm pore size Transwell (Corning) with a density of 3 × 105 cells/cm2. Polarized AECs and HMVEC-L were infected at an MOI of 0.01 from the apical or basolateral surface for 1 hour at 37°C after 1 and 10 days of seeding, respectively, when the transepithelial electrical resistance reached 25 ohm.cm2. Cells were washed and culture supernatants were harvested at 48 hpi from apical and basolateral chambers. Viral titers were determined by TCID50 assay.
Cytokine and Chemokine Measurement
The mRNA expression and protein concentrations of cytokine and chemokine in AECs after infection were measured by real-time polymerase chain reaction and cytometric bead array, respectively, as previously described (see Supplementary Material) [12, 13].
Desialylation-Hemagglutination Assay
The effect of desialylation of Turkey red blood cells (TRBC) (Lampire) on virus hemagglutination was investigated as previously described [11, 13]. The TRBCs treated with Glyko Sialidase S (Prozyme), which removed only α-2,3-linked sialic acids (SAs), or sialidase DAS181 (NexBio), which removed both α-2,3-linked and α-2,6-linked SAs, were compared with untreated TRBCs as controls (see Supplementary Material).
Immunohistochemical Staining
Human tissues were paraffin-embedded and stained for influenza viral protein (see Supplementary Material). In brief, the sectioned and deparaffinized tissues were incubated with pronase and blocked with an avidin/biotin kit (Vector Laboratories). The sections were incubated with influenza A virus nucleoprotein (NP)-specific antibody (HB65; EVL) and a biotinylated secondary antibody. Visualization of the bound antibodies was performed with a Strep-ABC complex and an AEC substrate kit (Vector).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism, version 7. Viral replication kinetics between experimental groups at different time points were compared by two-way analysis of variance (ANOVA), whereas mRNA and protein expressions of cytokines and chemokines were compared by one-way ANOVA.
RESULTS
Molecular Markers Associated With Pathogenicity, Replication, and Antiviral Resistance of A/Guangdong/17SF006/2017 (QY/HPAI-H7N9)
The viral sequences of QY/HPAI-H7N9 were compared with ck/YO84/H7N9, a LPAI-H7N9, Sh2/H7N9, Ah1/H7N9, and 734/H7N9. An insertion of multiple basic amino acids (PEVPKRKRTARGL) was found in the HA cleavage site of QY/HPAI-H7N9, indicating that this virus would be highly pathogenic to chickens (Supplementary Table 1). This motif was not observed in ck/YO84/H7N9 and all H7N9 isolates from the previous waves. A L226Q mutation that potentially associated with lower binding to human-like α-2,6-linked SA receptors was identified in QY/HPAI-H7N9 HA. NA-R292K mutation conferring resistance to oseltamivir and zanamivir was observed in QY/HPAI-H7N9 but not in other H7N9 viruses in this study. A signature mammalian adaptation marker E627K was identified in the PB2 genes of QY/HPAI-H7N9 and other human isolates, whereas ck/YO84/H7N9 retained the avian phenotype 627E. Moreover, QY/HPAI-H7N9 was the sole isolate here found to possess an additional A588V substitution in PB2, which was associated with enhanced polymerase activity, replication efficiency, and virulence in mice. S31N substitution in the matrix protein conferring to amantadine resistance was found in all H7N9 isolates.
Virus Replication Competence and Tissue Tropism of HPAI-H7N9
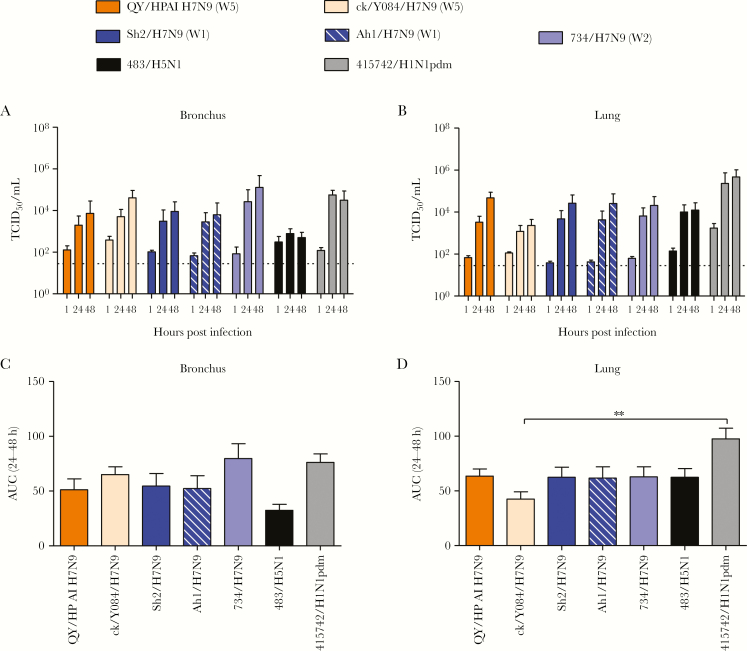
QY/HPAI-H7N9 replicated efficiently in ex vivo cultures of human bronchus and lung (Figure 1A–D). In the bronchus, QY/HPAI-H7N9 replicated to similar viral titers as H1N1pdm and all LPAI-H7N9 viruses tested (Figure 1A and C and Supplementary Table 2). There was a trend of higher replication efficiency when compared with H5N1, although the difference was not statistically significant. In lung tissue cultures, QY/HPAI-H7N9 replicated to significantly higher viral titers than ck/YO28/H7N9 at 48 hpi but similarly to Sh2/H7N9, Ah1/H7N9, 734/H7N9, and H5N1 at both 24 and 48 hpi (Figure 1B and D and Supplementary Table 3). However, its replication efficiency was significantly lower than H1N1pdm at all time points. The titers of H1N1pdm at 1 hour in lung tissue explants were higher than the other viruses; however, by using thermal inactivation assay, we found that H1N1pdm at 103 dropped to a detection limit of TCID50 assay after 24 hours incubation (Supplementary Figure 1). These results suggest that the titers of H1N1pdm at 24 and 48 hpi come from the progeny virus but not from the residual virus after washing.
Figure 1.
Viral replication kinetics of influenza A virus in ex vivo cultures of human bronchus (A) and lung (B) infected with 106 50% tissue culture infectious dose (TCID50)/mL at 37oC. The viral titer in culture supernatant was determined by TCID50 assay, and data were pooled from at least 3 independent experiments and shown as mean ± standard error of the mean (SEM). The horizontal dashed lines denote the detection limit of TCID50 assay. The area under the curve (AUC) over 24–48 hours postinfection is shown for both bronchus and lung (C and D). Data were pooled from at least 3 independent experiments and shown as mean ± SEM, and statistical analysis was calculated by one-way analysis of variance. **, P < .01.
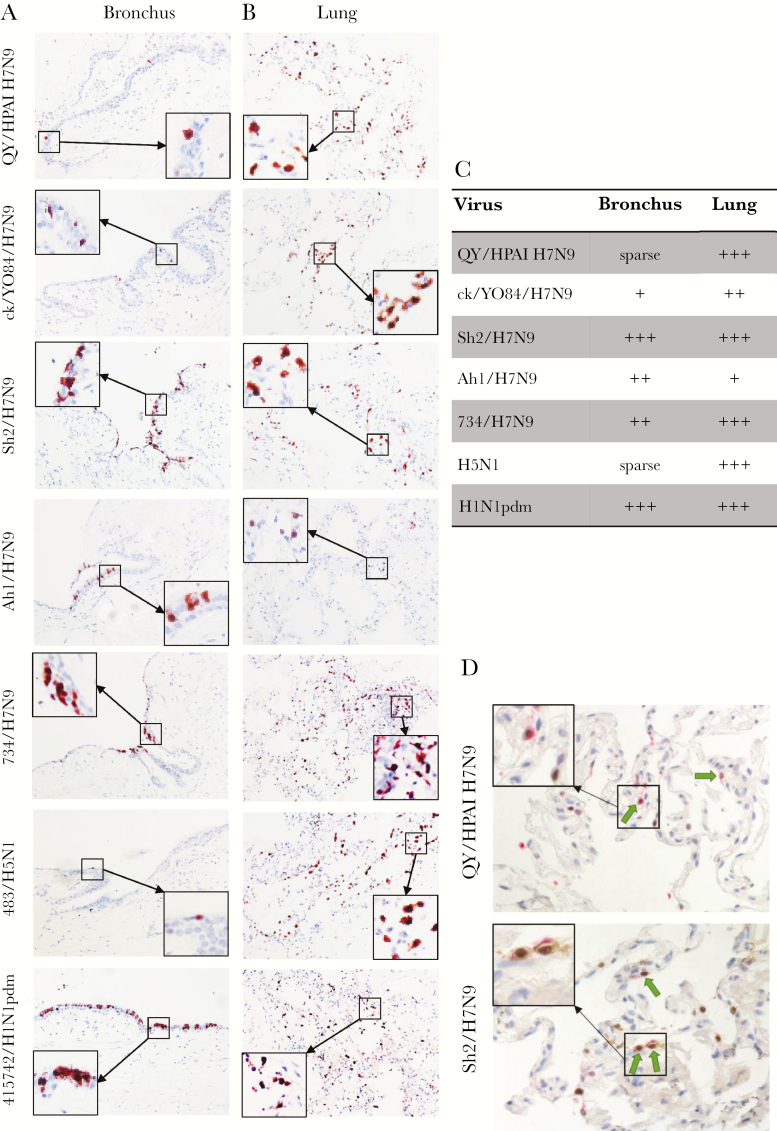
Immunohistochemical staining for viral NP revealed only sparse positive epithelial cells in QY/HPAI-H7N9- and H5N1-infected bronchi compared with the more extensive positive staining in bronchi infected by other tested viruses, especially Sh2/H7N9 and H1N1pdm (Figure 2A and C). In the lung tissue cultures, QY/HPAI-H7N9 showed extensive infection of alveolar cells, comparable to those of Sh2/H7N9, ck/YO84/H7N9, 734/H7N9, H5N1, and H1N1pdm and higher than that of Ah1/H7N9 (Figure 2B and C). QY/HPAI-H7N9 mainly targeted type-II pneumocytes (Figure 2D) and alveolar macrophages, having similar preference with Sh2/H7N9, ck/YO84/H7N9, and 734/H7N9 [11].
Figure 2.
Tissue tropism of influenza A viruses in ex vivo cultures of human bronchus (A) and lung (B). Human bronchus and lung tissue sections infected with QY/HPAI H7N9, ck/YO28/H7N9, Sh2/H7N9, Ah1/H7N9, 734/H7N9, 483/H5N1, and 415742/H1N1pdm were formalin-fixed and paraffin-embedded and subjected to immunohistochemical staining at 24 hours postinfection. Sections were stained with a monoclonal antibody against the influenza nucleoprotein with positive cells identified by red color. Tissue tropism of influenza virus infection in ex vivo cultures of human bronchus and lung was also assessed by immunohistochemical staining. Magnification, ×200. (C) Quantification of tissue tropism of influenza virus infection of human bronchus and lung in immunohistochemistry IHC photos. Sparse: <1%; +, <10%; ++, 10%–50%; +++, >50% cells with positive staining. (D) Lung tissue sections were doubled stained with a monoclonal antibody against the influenza nucleoprotein with positive cells identified by brown color and a monoclonal antibody against prosurfactant-C with positive cells identified by pink color. The arrow indicated the type-II alveolar epithelial cells infected with influenza virus. Magnification, ×400.
Receptor Binding Specificity of HPAI-H7N9
The TRBCs treated with Sialidase S, which specifically cleaved the α-2,3-linkages of SA with galactose, abolished hemagglutination with H5N1 as expected because it predominantly binds α-2,3-linkages of SA. It did not affect hemagglutination of the other viruses, but it marginally decreased the hemagglutination of QY/HPAI-H7N9 by 1-fold when compared with untreated TRBCs (Supplementary Table 4). As a control, TRBCs treated with sialidase DAS181, which cleaved both α-2,3- and α-2,6-linkages of SA with galactose, prevented the hemagglutination of all influenza viruses. Our findings suggested that ck/YO84/H7N9, Sh2/H7N9, Ah1/H7N9, 734/H7N9, QY/HPAI-H7N9, and H1N1pdm can bind glycans with α-2,6-links to SA, whereas H5N1 predominantly binds α-2,3-linked glycans. The marginal reduction in hemagglutination of QY/HPAI-H7N9 observed with Sialidase S-treated TRBCs suggested that QY/HPAI-H7N9 virus may have slightly more dependence on α-2,3-linked glycans than the other H7N9 viruses.
Proinflammatory Cytokine Induction of HPAI-H7N9 in Human Alveolar Epithelial Cells
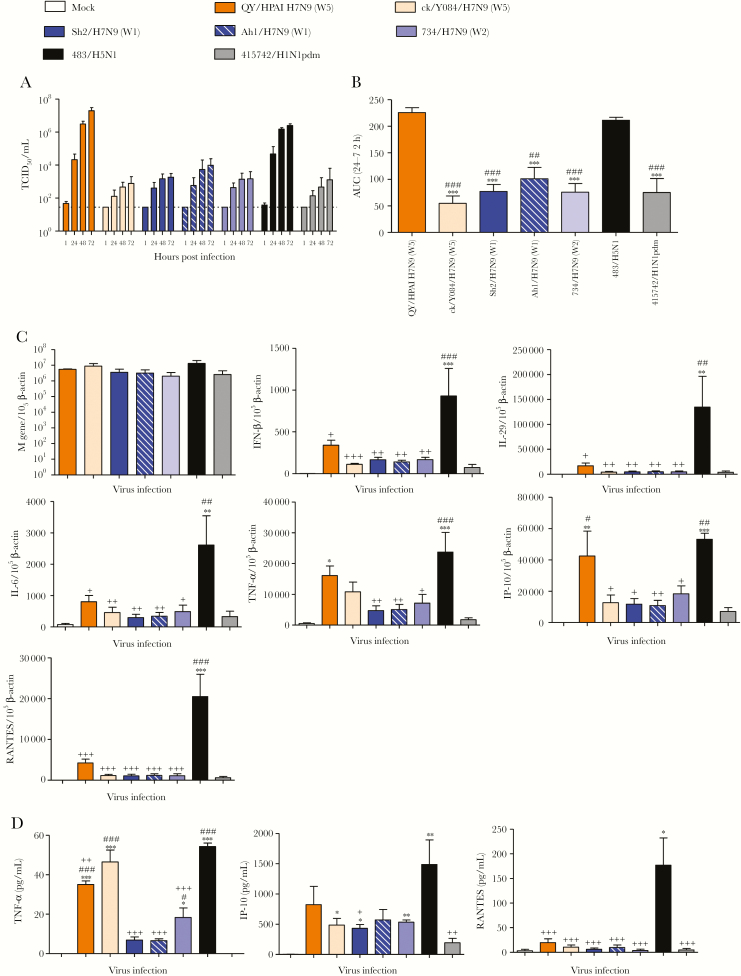
In human AECs, QY/HPAI-H7N9 replicated to significantly higher titers than all LPAI-H7N9 viruses tested and H1N1pdm at an MOI of 0.01, while similar to H5N1 (Figure 3A and B). At an MOI of 2, mRNA levels of the influenza matrix (M) gene were comparable among all viruses at 24 hpi (Figure 3C). However, QY/HPAI-H7N9 induced significantly more interferon gamma-induced protein 10 (IP-10) mRNA than H1N1pdm, with a level comparable to that induced by the high cytokine inducer H5N1. There were trends showing greater induction of proinflammatory cytokine interferon (IFN)-β, interleukin (IL)-29, IL-6, tumor necrosis factor (TNF)-α, IP-10, and RANTES mRNA by QY/HPAI-H7N9 than the LPAI-H7N9 and H1N1pdm viruses, although differences were not statistically significant. However, the QY/HPAI-H7N9-induced mRNA levels of these cytokines, except for TNF-α, were significantly lower than those of H5N1. In culture supernatant, QY/HPAI-H7N9 induced significantly more TNF-α protein than H1N1pdm (Figure 3D). Similar induction of TNF-α, IP-10, and RANTES proteins were observed between QY/HPAI-H7N9 and the LPAI-H7N9 viruses although the trends mostly showed higher induction by QY/HPAI-H7N9. Compared to H5N1, QY/HPAI-H7N9 induced significantly less TNF-α and RANTES proteins, with no significant difference in the level of IP-10 protein.
Figure 3.
Viral replication kinetics of influenza A viruses in in vitro cultures of human alveolar epithelial cells (AECs). The AECs were infected with indicated viruses at a multiplicity of infection of 0.01 and cultured at 37oC for 72 hours. Culture supernatants were harvested at 1, 24, 48, and 72 hours postinfection, and virus titers were determined by 50% tissue culture infectious dose (TCID50) assay. Results (A) were pooled from at least 3 independent experiments and shown as mean ± standard error of the mean (SEM). The horizontal dashed lines denote the detection limit of TCID50 assay. The area under curve (AUC) over 24 to 72 hours postinfection is pooled from at least 3 independent experiments and shown as mean ± SEM (B). ***, P < .001 (compared with QY/HPAI H7N9); ###, P < .001 (compared with 483/H5N1). Statistical significances were calculated by one-way analysis of variance (ANOVA). (C) Cytokine and chemokine expression profiles in human AECs infected with influenza A virus. The expression of messenger ribonucleic acid (mRNA copies per 105 β-actin copies) for influenza matrix (M) gene, interferon (IFN)-β, interleukin (IL)-29, IL-6, tumor necrosis factor (TNF)-α, IFN gamma-induced protein 10 (IP-10), and chemokine ligand 5 (RANTES) is shown for various viruses at 24 hours postinfection. Data are pooled from at least 3 independent experiments and presented as mean ± SEM. (D) Protein concentration in culture supernatants for TNF-α, IP-10, and RANTES were measured at 24 hours postinfection by cytometric bead array immunoassay. The mean concentrations were pooled from at least 3 independent experiments, and data are presented as mean ± SEM. *, P < .05; **, P < .01; and ***, P < .001 (compared with mock). #, P < .05; ##, P < .01; and ###, P < .001 (compared with 415742/H1N1pdm). +, P < .05; ++, P < .01; and +++, P < .001 (compared with 483/H5N1). Statistical significance was calculated by one-way ANOVA.
HPAI-H7N9 Replicated Productively in Human Pulmonary Microvascular Endothelial Cells
Endothelial cell tropism was reported to be a determinant of HPAI-H5N1 pathogenesis in ferrets and mice [16], and influenza viruses targeting endothelial cells might contribute to vascular leakage and severely progressing viral pneumonia [17, 18]. It may also contribute to dissemination of virus beyond the respiratory tract. At an MOI of 0.01, QY/HPAI-H7N9 replicated to similar titers as H5N1 in HMVEC-L (mean titers >105 TCID50/mL) (Figure 4A and B). Both viruses replicated to significantly higher titers than all LPAI-H7N9 viruses tested and H1N1pdm, which displayed no or limited replication (mean titers <102.5 TCID50/mL). However, at an MOI of 2, M gene mRNA levels were similar among all viruses at 24 hpi (Figure 4C), indicating that both HPAI and LPAI viruses were able to infect HMVEC-L, yet only HPAI viruses resulted in productive replication. In terms of host responses, QY/HPAI-H7N9 induced similar mRNA levels of IFN-β, IL-29, IL-6, TNF-α, IP-10, RANTES, and monocyte chemoattractant protein-1 (MCP-1) as the LPAI-H7N9 and H1N1pdm viruses. The H5N1 virus showed a trend of higher induction of these proinflammatory cytokines and chemokines, and the difference in IP-10 level compared with QY/HPAI-H7N9 was statistically significant (Figure 4C).
Figure 4.
Viral replication kinetics of influenza A viruses in in vitro cultures of human lung microvascular endothelial cells (HMVEC-L). The HMVEC-L were infected with indicated viruses at a multiplicity of infection of 0.01 and cultured at 37oC for 72 hours. Culture supernatants were harvested at 1, 24, 48, and 72 hours postinfection, and virus titers were determined by 50% tissue culture infectious dose (TCID50) assay. Results (A) were pooled from at least 3 independent experiments and shown as mean ± standard error of the mean (SEM). The horizontal dashed lines denote the detection limit of TCID50 assay. The area under curve (AUC) over 24 to 72 hours postinfection is pooled from at least 3 independent experiments and shown as mean ± SEM (B). *, P < .05; **, P < .01 (compared with QY/HPAI H7N9). ###, P < .001 (compared with 483/H5N1). Statistical significances were calculated by one-way analysis of variance (ANOVA). (C) Cytokine and chemokine messenger ribonucleic acid (mRNA) expression profiles in HMVEC-L infected with influenza A virus. The expression of mRNA (mRNA copies per 105 β-actin copies) for influenza matrix (M) gene, interferon (IFN)-β, interleukin (IL)-29, IL-6, tumor necrosis factor (TNF)-α, IFN gamma-induced protein 10 (IP-10), chemokine ligand 5 (RANTES), and monocyte chemoattractant protein-1 (MCP-1) is shown for various viruses at 24 hours postinfection. Data are pooled from at least 3 independent experiments and presented as mean ± SEM. *, P < .05 (compared with mock); #, P < .05 (compared with 415742/H1N1pdm); +, P < .05 (compared with 483/H5N1). Statistical significance was calculated by one-way ANOVA.
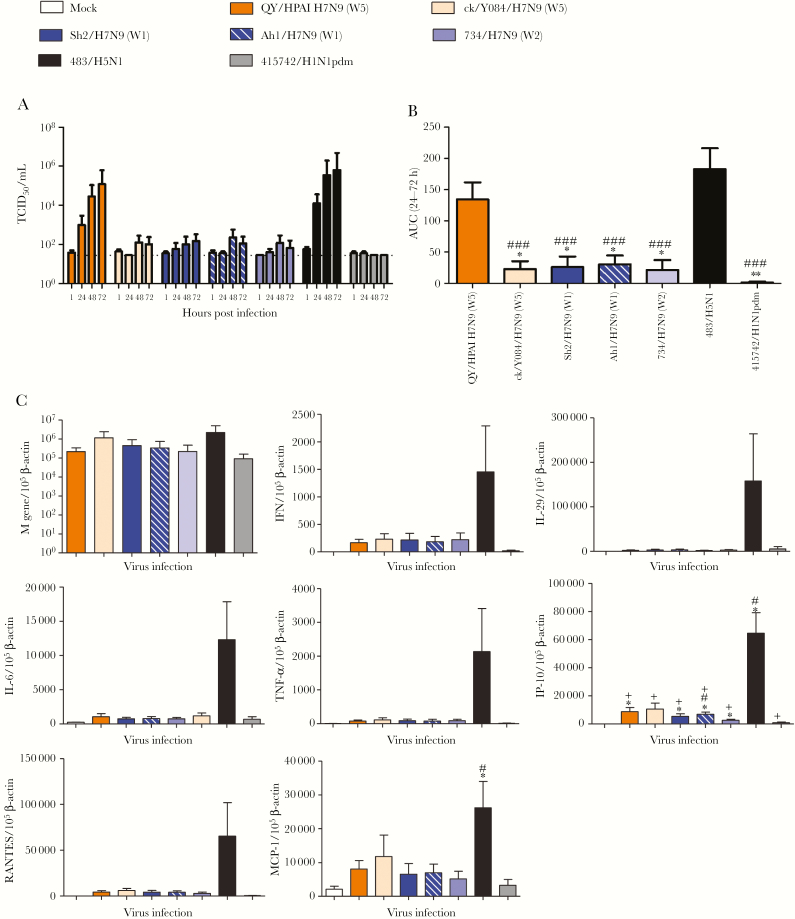
HPAI-H7N9 Infection of Polarized Human Alveolar Epithelial and Pulmonary Microvascular Endothelial Cells
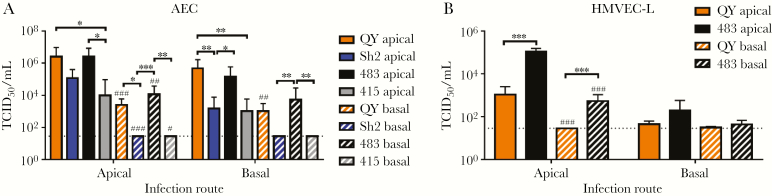
HPAI-H7N9 virus has been observed having tropism to brain tissues in ferrets and mice [8, 19]. To investigate whether HPAI-H7N9 might be at higher risk of extrapulmonary dissemination than LPAI-H7N9, the replication competence of QY/HPAI-H7N9, Sh2/H7N9, H5N1, and H1N1pdm in polarized AECs and HMVEC-L was determined. In AECs, all 4 viruses were able to release virus at the apical side regardless of the apical or basolateral infection routes (Figure 5A). QY/HPAI-H7N9 released significantly more virus than Sh2/H7N9 from basolateral infection and H1N1pdm from both infection routes. Likewise, basolateral virus release in AECs was restricted to QY/HPAI-H7N9 and H5N1 in both infection routes (Figure 5A). In HMVEC-L, only QY/HPAI-H7N9 and H5N1 could replicate to detectable titers (Figure 5B). Apical release of QY/HPAI-H7N9 was lower than that of H5N1, and titers from the apical infection route were statistical significant. Basolateral virus release was observed only in H5N1-infected HMVEC-L.
Figure 5.
Polarity of influenza A virus infection in human alveolar epithelial cells (AECs) and human lung microvascular endothelial cells (HMVEC-L). Virus titers of the tested viruses were determined after apical infection and basolateral infection of the AECs (A) and HMVEC-L (B) at 48 hours postinfection. Virus titers are pooled from at least 3 independent experiments and shown as mean ± standard error of the mean. *, P < .05; **, P < .01; ***, P < .001 (comparisons as indicated). #, P < .05; ##, P < .01; ###, P < .001 (compared with titers of apical release with the same infection route with corresponding virus). Statistical significances were calculated by one-way analysis of variance.
DISCUSSION
In recent studies, HPAI-H7N9 viruses have been detected in both poultry and humans causing public health concern. In our study, we evaluated the tissue tropism, replication kinetics, and innate immune induction of an HPAI-H7N9 strain isolated from a fatal human infection in comparison with its LPAI-H7N9 precursors. QY/HPAI-H7N9 replicated efficiently in ex vivo cultures of human bronchus and lung. Although it extensively infected the human lung tissue, only limited numbers of bronchial epithelial cells were infected. This differs from Sh2/H7N9, a human strain from the first epidemic, which infected both alveolar and bronchial epithelia extensively. This is probably related to the QY/HPAI-H7N9 HA substitutions of A160T and L226Q, which increased the binding affinity to avian-like α-2,3-linked SA receptors predominantly found in human alveolar epithelium and decreased binding to human-like α-2,6-linked SA receptors predominantly found in human bronchial epithelium [20]. This is confirmed by a slight reduction of hemagglutination observed with TRBCs after removal of α-2,3-linked glycans and modest binding affinity towards α-2,6-linked glycans. These data are consistent with others using a binding assay of synthetic sialyglycopolymers including 3’-SLN, 3’SLN-LN, 6’SLN, and 6’SLN-LN [21] where they confirmed dual receptor binding profile of the HPAI-H7N9 viruses. More detailed and precise receptor binding profiles of the viruses can be achieved by using glycan arrays in future study. Even though only limited number of cells was positive for viral antigen on the bronchial epithelium, QY/HPAI-H7N9 was still able to replicate efficiently in the human bronchus explants. The presence of polybasic HA cleavage site, mammalian adaptation markers PB2-E627K, and an additional substitution PB2-A588V may provide this virus with greater replication competence over LPAI-H7N9 viruses. Moreover, increased binding to α-2,3-linked SA receptors may have contributed to better replication of QY/H7N9 than the LPAI-H7N9 and H1N1pdm viruses in human AECs and HMVEC-L considering the higher abundance of α-2,3-linked SA receptors on these cell types [22, 23], although other factors may also contribute to this difference.
In a study comparing mild, severe, and fatal human infections with H7N9 virus, mutations of PB2-E627K, NA-R294K, and PA-V100A were found to be associated with increased fatality in the patients [24]. PB2-A588V substitution in H7N9, H10N8, and H9N2 viruses promotes polymerase activity and replication efficiency in mammalian and avian cells and increase virulence in mice [25]. In comparison with the H7N9 viruses from previous epidemics, the QY/HPAI-H7N9 virus in this study lost a mammalian adaptation marker of V100A in the PA gene but contained another adaptation marker of A588V substitution in PB2, which could be associated with the high replication competence in mammalian cell models. The presence of NA-R292K, which confers NA inhibitor resistance, in a human HPAI-H7N9 virus with PB2-A588V but not PB2-E627K has been associated with slightly reduced replication competence in human bronchial epithelial cells [8]. The HPAI-H7N9 virus used in this study also contains the same neuraminidase inhibitor (NAI) resistance gene, but there was no reduced fitness observed in comparison to LPAI-H7N9 viruses from previous waves. This may be due to the presence of both adaptation markers PB2-A588V and PB2-E627K. Furthermore, compared with LPAI-H7N9 viruses, HPAI-H7N9 virus has increased virulence and lethality in mice and ferrets [8, 19, 26]. Although the index strain was not lethal in mice or ferrets, the virus developed substitutions of 627K or 701N in the PB2 gene upon replication in ferrets, and hence it became highly lethal in mammals and even transmissible via respiratory droplets in ferrets [26].
Although there is no report on human-to-human transmission of HPAI-H7N9 infection so far, the HPAI-H7N9 virus transmitted efficiently via respiratory droplets among ferrets [8, 26]. High transmissibility between ferrets may be attributed to the stability of the viral HA protein. There is evidence showing that the HA protein of HPAI-H7N9 has greater thermal stability than H1N1pdm virus [26], and it fuses at a pH threshold lower than that of other LPAI-H7N9 viruses but comparable to that of seasonal influenza virus [19]. Therefore, the HPAI-H7N9 viruses have acquired a number of fitness markers for human-to-human transmission. The absence of human-to-human transmission may be explained by our findings on reduced bronchus tropism of HPAI-H7N9 because of its reduced binding affinity to human-like α-2,6-linked SA receptors, which are present predominantly on the human upper respiratory tract.
Endothelial cell tropism was reported to be a determinant of HPAI-H5N1 pathogenesis in ferrets and mice [16], and influenza viruses targeting endothelial cells might contribute to vascular leakage and severely progressing viral pneumonia [17, 18]. It may also contribute to dissemination of virus beyond the respiratory tract. In addition, endothelial cells have been recently found to be a source of proinflammatory cytokines in lung of mice infected with a mouse-adapted strain, A/WSN/1933 (H1N1) [27]. Influenza pathogenesis was partly contributed by the induction of proinflammatory cytokines from endothelial cells in the lungs, and the subsequent and excessive leukocytes infiltration increased vascular permeability resulting in viral pneumonia [28]. We found that QY/HPAI-H7N9 replicated efficiently in HMVEC-L, comparable with HPAI-H5N1 and significantly more efficient than LPAI-H7N9 or H1N1pdm viruses. Our data are consistent with others that these endothelial cells only support viral replication of HPAI viruses [23]. However, H5N1 elicited high mRNA levels encoding proinflammatory cytokines and chemokines, whereas QY/HPAI-H7N9 did not. This indicated that the role of lung endothelium in the pathogenesis of HPAI-H7N9 might not be the same as that of H5N1 infection. Further investigation should be performed using coculture models of both endothelial and epithelial cells for the infection of HPAI-H7N9.
Furthermore, H5N1 was reported to infect lung endothelial cells from the basolateral aspect, which facilitates virus dissemination via the blood stream [17]. We report here that HPAI-H7N9 is able to infect and egress from the apical compartment of HMVEC-L, but LPAI-H7N9 viruses did not. Although there is a lack of clinical evidence of extrapulmonary viral dissemination of HPAI-H7N9, the ability to replicate in polarized HMVEC-L suggested that HPAI-H7N9 has a higher chance than LPAI-H7N9 and H1N1pdm to disseminate from the lung parenchyma via the endothelial route. Imai et al [8] reported systemic spread of an HPAI-H7N9 virus to the brain of infected mice that was not observed with a LPAI-H7N9 virus. However, the minimal basolateral release of HPAI-H7N9 virus indicated probable less effective dissemination and thus lower pathogenicity compared with H5N1.
CONCLUSIONS
In summary, our findings demonstrated that an HPAI-H7N9 virus was capable of infecting and efficiently replicating in ex vivo cultures of human bronchus and lung. In addition, HPAI-H7N9 was more efficient at replicating in human AECs and HMVEC-L than its LPAI precursors. This replication capacity in endothelial cells may contribute to pathogenesis in the lung, and the HPAI-H7N9 virus might be at higher risk of extrapulmonary dissemination than the LPAI-H7N9 viruses. It is important that aggressive measures are taken to contain the spread of this virus within poultry to minimize zoonotic and pandemic risks.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. L. L. Y. C. and K. P. Y. H. planned and carried out the experiments with ex vivo cultures, analyzed the data, and contributed to drafting the manuscript. D. I. T. K., C. H. T. B., and C. K. P. M. carried out the in vitro culture and protein and molecular biology assays. Z. Y. and W. G. planned the experiments and isolated the H7N9 viruses. L. L. M. P. carried out the genetic sequencing and phylogenetic analysis. N. Z. planned the experiments and critical review the manuscript. J. M. N. carried out and interpreted the histology and immunohistochemistry studies, analyzed the data, and wrote the manuscript. J. S. M. P. planned the study, analyzed the data, and wrote the manuscript. M. C. W. C. obtained research funding, planned and coordinated the study, analyzed the data, and wrote the manuscript. All authors contributed to the data interpretation and analysis and provided critical comments on the draft manuscript.
Acknowledgments. We thank Kevin Fung (Department of Pathology, The University of Hong Kong) for help with immunohistochemical staining.
Financial support. This work was supported by the following: US National Institute of Allergy and Infectious Diseases under Centers of Excellence for Influenza Research and Surveillance contract (Grant HHSN27220140006C ); Theme Based Research Scheme (Grant T11-705/14-N ) by the Research Grants Council of the Hong Kong Special Administrative Region ; Health and Medical Research Fund by the Food and Health Bureau , Government of the Hong Kong Special Administrative Region (Grant 18170792 ); National Natural Science Foundation of China (Grant 81761128014 ); and the Science Research Project of the Guangdong Province (Grant 2016A050503047 ).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Shen Y, Lu H. Global concern regarding the fifth epidemic of human infection with avian influenza A (H7N9) virus in China. Biosci Trends 2017; 11:120–1. [DOI] [PubMed] [Google Scholar]
- 2. Wang X, Jiang H, Wu P, et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis 2017; 17:822–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huo X, Chen L, Qi X, et al. Significantly elevated number of human infections with H7N9 virus in Jiangsu in eastern China, October 2016 to January 2017. Euro Surveill 2017; 22:30496. doi: 10.2807/1560-7917.ES.2017.22.13.30496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. (OIE) WOfAH. OIE report of HPAI H7N9 virus infection in poultry, Guangdong Province, China. http://www.oie.int/wahis_2/temp/reports/en_imm_0000022933_20170221_163854.pdf. Accessed March 12, 2019. [Google Scholar]
- 5. FAO. H7N9 situation update. http://www.fao.org/ag/againfo/programmes/en/empres/h7n9/situation_update.html. Accessed March 12, 2019. [Google Scholar]
- 6. Zeng X, Tian G, Shi J, Deng G, Li C, Chen H. Vaccination of poultry successfully eliminated human infection with H7N9 virus in China. Sci China Life Sci 2018; 61:1465–73. [DOI] [PubMed] [Google Scholar]
- 7. Ke C, Mok CKP, Zhu W, et al. Human infection with highly pathogenic avian influenza A(H7N9) virus, China. Emerg Infect Dis 2017; 23:1332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imai M, Watanabe T, Kiso M, et al. A highly pathogenic Avian H7N9 influenza virus isolated from a human is lethal in some ferrets infected via respiratory droplets. Cell Host Microbe 2017; 22:615–26.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kang M, Lau EH, Guan W, et al. Epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus in Guangdong, 2016 to 2017. Euro Surveill 2017; 22 (27): 30568. doi: 10.2807/1560-7917.ES.2017.22.27.30568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou L, Tan Y, Kang M, et al. Preliminary epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus, China, 2017. Emerg Infect Dis 2017; 23:1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan MC, Chan RW, Chan LL, et al. Tropism and innate host responses of a novel avian influenza A H7N9 virus: an analysis of ex-vivo and in-vitro cultures of the human respiratory tract. Lancet Respir Med 2013; 1:534–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hui KP, Ching RH, Chan SK, et al. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir Med 2018; 6:846–54. [DOI] [PubMed] [Google Scholar]
- 13. Hui KP, Chan LL, Kuok DI, et al. Tropism and innate host responses of influenza A/H5N6 virus: an analysis of ex vivo and in vitro cultures of the human respiratory tract. Eur Respir J 2017; 49:1601710. doi: 10.1183/13993003.01710-2016. [DOI] [PubMed] [Google Scholar]
- 14. Alphonse RS, Vadivel A, Zhong S, et al. The isolation and culture of endothelial colony-forming cells from human and rat lungs. Nat Protoc 2015; 10:1697–708. [DOI] [PubMed] [Google Scholar]
- 15. van Beijnum JR, Rousch M, Castermans K, van der Linden E, Griffioen AW. Isolation of endothelial cells from fresh tissues. Nat Protoc 2008; 3:1085–91. [DOI] [PubMed] [Google Scholar]
- 16. Tundup S, Kandasamy M, Perez JT, et al. Endothelial cell tropism is a determinant of H5N1 pathogenesis in mammalian species. PLoS Pathog 2017; 13:e1006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan MC, Chan RW, Yu WC, et al. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir Res 2009; 10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ocaña-Macchi M, Bel M, Guzylack-Piriou L, et al. Hemagglutinin-dependent tropism of H5N1 avian influenza virus for human endothelial cells. J Virol 2009; 83:12947–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun X, Belser JA, Pappas C, et al. Risk assessment of fifth-wave H7N9 influenza A viruses in mammalian models. J Virol 2018 ; 93: e01740-18. doi: 10.1128/JVI.01740-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nicholls JM, Chan MC, Chan WY, et al. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med 2007; 13:147–9. [DOI] [PubMed] [Google Scholar]
- 21. Zhu W, Zhou J, Li Z, et al. Biological characterisation of the emerged highly pathogenic avian influenza (HPAI) A(H7N9) viruses in humans, in mainland China, 2016 to 2017. Euro Surveill 2017; 22:30533. doi: 10.2807/1560-7917.ES.2017.22.19.30533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hui CF, Chan RW, Fung K, et al. The regional distribution of different types of influenza receptors in cultured human alveolar epithelial cells and correlation with in vitro infection. Influenza Other Respir Viruses 2011; 5:436–7. [PMC free article] [PubMed] [Google Scholar]
- 23. Zeng H, Pappas C, Belser JA, et al. Human pulmonary microvascular endothelial cells support productive replication of highly pathogenic avian influenza viruses: possible involvement in the pathogenesis of human H5N1 virus infection. J Virol 2012; 86:667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sha J, Chen X, Ren Y, et al. Differences in the epidemiology and virology of mild, severe and fatal human infections with avian influenza A (H7N9) virus. Arch Virol 2016; 161:1239–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao C, Ma W, Sun N, et al. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci Rep 2016; 6:19474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi J, Deng G, Kong H, et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res 2017; 27:1409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teijaro JR, Walsh KB, Cahalan S, et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 2011; 146:980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Armstrong SM, Darwish I, Lee WL. Endothelial activation and dysfunction in the pathogenesis of influenza A virus infection. Virulence 2013; 4:537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.