Significance
Facioscapulohumeral dystrophy (FSHD) is an inherited disabling muscular disorder caused by misexpression of DUX4 in skeletal muscles. FSHD has variable onset; its infantile form has a more severe disease course. There is no cure for FSHD. Here, we show the potential of antisense oligonucleotides called locked nucleic acid (LNA) gapmers for treating FSHD. We designed LNA gapmers to knock down DUX4 messenger RNA and found that its expression was effectively reduced in patient-derived cells and an FSHD mouse model. Functional benefits and minimal off-targeting were observed in vitro. Our study facilitates progress toward finding new candidates for treating FSHD. The screening protocol used here for antisense oligonucleotides targeting DUX4 can also be adapted by other efforts developing similar treatments for FSHD.
Keywords: facioscapulohumeral muscular dystrophy, DUX4, LNA gapmers, antisense therapy, FLExDUX4 mouse model
Abstract
Facioscapulohumeral muscular dystrophy (FSHD), characterized by progressive muscle weakness and deterioration, is genetically linked to aberrant expression of DUX4 in muscle. DUX4, in its full-length form, is cytotoxic in nongermline tissues. Here, we designed locked nucleic acid (LNA) gapmer antisense oligonucleotides (AOs) to knock down DUX4 in immortalized FSHD myoblasts and the FLExDUX4 FSHD mouse model. Using a screening method capable of reliably evaluating the knockdown efficiency of LNA gapmers against endogenous DUX4 messenger RNA in vitro, we demonstrate that several designed LNA gapmers selectively and effectively reduced DUX4 expression with nearly complete knockdown. We also found potential functional benefits of AOs on muscle fusion and structure in vitro. Finally, we show that one of the LNA gapmers was taken up and induced effective silencing of DUX4 upon local treatment in vivo. The LNA gapmers developed here will help facilitate the development of FSHD therapies.
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal-dominant disorder characterized by progressive, asymmetric muscle weakness beginning at the face, shoulders, and upper limbs, which spreads to the lower regions of the body with age (1). It is the third most common muscular dystrophy, with ∼1:8,000–1:22,000 people affected worldwide (1). Age of onset is variable, ranging from birth to adulthood. Patients with the rare infantile form of FSHD, presenting symptoms before 5 y of age, follow a more severe and rapid course of the disease (2). At present, FSHD is incurable.
The majority of FSHD patients (∼95%, FSHD1) have a contraction of the D4Z4 repeat array in chromosome 4q35, which is typically 11–100 D4Z4 units long in healthy individuals (1, 3). FSHD1 was thought to be caused by having 10 or fewer D4Z4 repeats in the array; however, recent evidence indicates that factors other than contraction size determine whether one will manifest the disorder. For instance, individuals with 7–10 D4Z4 units exhibit wide clinical variability, with some remaining asymptomatic (4). In this case, factors influencing D4Z4 array methylation play a larger role in disease penetrance. This is unlike in those having <7 D4Z4 units, for whom the degree of contraction primarily determines array demethylation and disease severity (4). Each D4Z4 repeat contains the first two exons of the double homeobox protein 4 (DUX4) gene, with its third (and final) exon located immediately downstream of the array (5–7). There are several DUX4 isoforms (8); we refer to the full-length pathogenic one here unless otherwise stated. The D4Z4 array is normally hypermethylated in the course of development. Studies show that the contraction relaxes the chromatin and demethylates DNA in this region, resulting in aberrant DUX4 expression in skeletal muscle (9). In other patients (∼5%, FSHD2), the D4Z4 array is shorter in comparison to the average array size in the general population, with an average of 12–16 D4Z4 units observed (10, 11). The more important determinant in FSHD2, however, is the presence of mutations in the genes of epigenetic regulators that similarly lead to D4Z4 array demethylation (1, 9). In both cases, FSHD only develops when chromosome 4 is of certain permissive A haplotypes, in which exon 3 has a polyadenylation signal (PAS) required for DUX4 messenger RNA (mRNA) stability (6).
The aberrant expression of DUX4 in skeletal muscle is thought to cause FSHD and thus serves as an attractive therapeutic target. DUX4 encodes a transcription factor that activates pathways involved in muscle degeneration and apoptosis, events observed in patient muscles (12, 13). DUX4 also inhibits myogenic differentiation and increases the sensitivity of muscle cells to oxidative stress (14, 15). Several studies have demonstrated the feasibility of using antisense oligonucleotides (AOs) to reduce DUX4 expression. Phosphorodiamidate morpholino oligomer (PMO) AOs against the PAS, interfering with its recognition, knocked down the expression of DUX4 and its downstream targets in immortalized and primary FSHD muscle cells (16, 17). Approximately 25–50% DUX4 knockdown was observed by RT-PCR in one study, where 50 nM of each of these PMOs were transfected (16). Electroporation of one PAS-targeting PMO into mice with FSHD patient biopsy xenografts led to ∼100% DUX4 knockdown and a reduction in downstream gene expression in the transplanted muscle (17).
Another strategy uses AOs to disrupt proper DUX4 mRNA splicing. Phosphorothioated 2′-O-methyl RNA AOs against splice sites in DUX4 exons 2 and 3 reduced DUX4 expression by ∼30–50% and significantly prevented atrophy in primary FSHD myotubes (7, 12). The in vivo efficacy of an exon 3 splice site-targeting vivo-PMO was also tested in mice with recombinant adeno-associated virus-mediated DUX4 expression. Intramuscular (i.m.) injection of the AO into the tibialis anterior (TA) reduced DUX4 expression by ∼30-fold compared to control AO-injected mice (7).
While the above studies are promising, there remains a need to more effectively knock down DUX4 expression and screen for AOs against DUX4. AO chemistries are available that directly degrade target mRNA, and not passively act via mRNA processing interference. One example would be locked nucleic acid (LNA) gapmers, which consist of a central segment of DNA flanked by short LNA stretches. LNA gapmers bind targets by sequence complementarity, producing a DNA/RNA hybrid that is cleaved by RNase H, leading to gene knockdown (18). Having enhanced stability, specificity, and potency at low doses, LNA gapmers are expected to more effectively reduce DUX4 expression than previously used chemistries.
In this work, we sought to develop LNA gapmer AOs for the treatment of FSHD. We designed LNA gapmers targeting the DUX4 transcript and screened them for efficacy using immortalized FSHD patient-derived muscle cells. We use a method capable of reliably detecting changes in endogenous DUX4 expression posttreatment that should be useful for future AO and drug-screening efforts, given that endogenous DUX4 detection in vitro has proven difficult due to its low expression (19). We then proceeded to determine the in vivo uptake and efficacy of a selected LNA gapmer from this screen using the recently developed FLExDUX4 FSHD mouse model (20). Ours is a unique study reporting the use of this model for DUX4 AO testing. FLExDUX4 mice naturally express low amounts of DUX4 without suffering from severe disease phenotypes. This makes them a useful model for in vivo screening, especially since they develop no embryonic lethality despite the toxicity of DUX4, an issue that has immensely hampered the use of FSHD animal models in the past (21).
Results
LNA Gapmers Effectively Knock Down DUX4 Expression In Vitro.
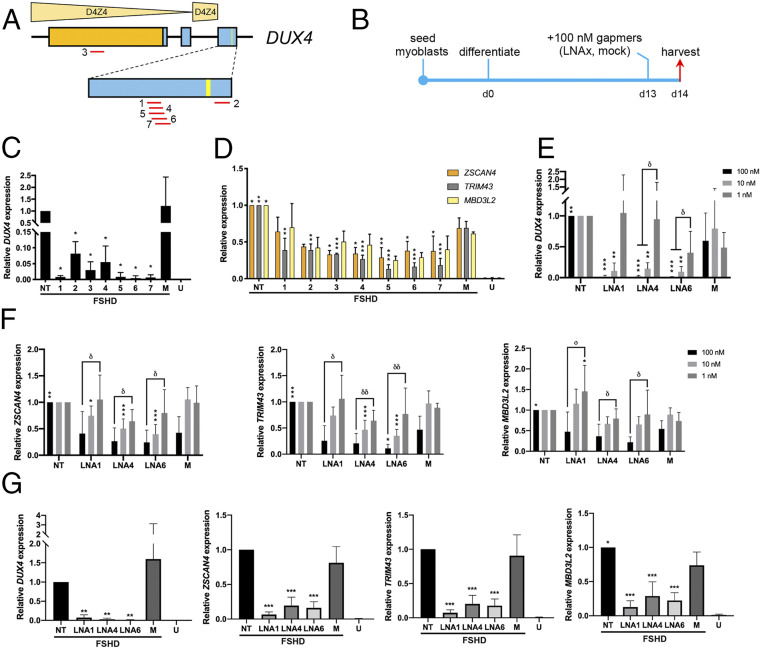
To induce DUX4 mRNA expression to detectable levels in vitro in immortalized patient-derived muscle fibers, KOSR-supplemented medium was used for differentiation (22). DUX4 expression significantly increased with differentiation (SI Appendix, Fig. S1). At 13 d postdifferentiation, DUX4 expression was on average ∼20 times greater than at 4 d postdifferentiation. Since no further increase in DUX4 expression was detected past this 13-d mark, we screened our LNA gapmers (Table 1 and Fig. 1A) by transfecting them at 100 nM into myotubes during this time (Fig. 1B). We chose to mainly target DUX4 exon 3, which is specifically associated with the pathogenic DUX4 transcript. We also avoided the PAS, given its similarity to other such sequences in the genome. All gapmers significantly reduced DUX4 expression in vitro (n = 3, P < 0.05), demonstrating ∼100% knockdown of the transcript on average (Fig. 1C). Corresponding decreases in expression of ZSCAN4, TRIM43, and MBD3L2—known activated downstream targets of DUX4—were also observed (Fig. 1D). Considering gapmers against exon 3, efficacy appeared to correlate with the openness of the target site secondary structure (SI Appendix, Fig. S2). Based on these results, three gapmers were selected for further analysis: LNA1, LNA4, and LNA6.
Table 1.
Characteristics of the LNA gapmers used in this study
| ID | Sequence* (5′ to 3′) | Length (nt) | Target DUX4 exon | GC content (%) |
| LNA1 | AGCGTCGGAAGGTGG | 15 | 3 | 66.7 |
| LNA2 | ATAGGATCCACAGGGA | 16 | 3 | 50.0 |
| LNA3 | AGATCCCCTCTGCC | 14 | 1 | 64.3 |
| LNA4 | CAGCGTCGGAAGGTG | 15 | 3 | 66.7 |
| LNA5 | ACAGCGTCGGAAGGTG | 16 | 3 | 62.5 |
| LNA6 | GACAGCGTCGGAAGGT | 16 | 3 | 62.5 |
| LNA7 | AGACAGCGTCGGAAGG | 16 | 3 | 62.5 |
All sequences are fully phosphorothioated; bold indicates LNA, nonbold indicates DNA.
Fig. 1.
LNA gapmer screen and evaluation of knockdown efficacy by qPCR. (A) LNA gapmers (red lines) were designed across DUX4. The relative positions of the distal D4Z4 units are shown. Exons, boxes; ORF, orange; exon 3 PAS, yellow line. (B) Culture schedule for the LNA gapmer screen. Days postdifferentiation are indicated. (C) DUX4 expression posttreatment with 100 nM of the various gapmers, indicated by numbers. (D) ZSCAN4, TRIM43, and MBD3L2 expression posttreatment with 100 nM gapmers. For C and D, U = unaffected/healthy control. (E and F) LNA1, LNA4, LNA6, and the mock gapmer were transfected at 100, 10, or 1 nM following B. Expression of (E) DUX4 and (F) ZSCAN4, TRIM43, and MBD3L2 after these treatments is shown. (G) DUX4 and the expression of its downstream targets 10 d after treatment with 100 nM gapmers at 4 d postdifferentiation. Error bars: SD (n = 3 for C, D, and healthy controls in G; n = 5–6 for E and F, and treatment groups in G). *P < 0.05, **P < 0.005, ***P < 0.0005, one-way ANOVA with Dunnett’s test versus mock (M), δP < 0.05, δδP < 0.005, one-way ANOVA with Tukey’s test.
Dose-dependent activity was displayed by these gapmers in vitro, with significant DUX4 knockdown at ∼90% achievable at a 10-fold lower dose of 10 nM (n = 6, P < 0.005) (Fig. 1E). No significant reduction in DUX4 expression was observed at the 1 nM dose. Corresponding dose-dependent decreases in DUX4 target gene expression were similarly observed (Fig. 1F). When cells were transfected with the gapmers at 4 d postdifferentiation, we observed significant knockdown of DUX4 (n = 5, P < 0.005), ZSCAN4, TRIM43, and MBD3L2 (n = 6, P < 0.0005) by LNA1, LNA4, and LNA6 at 14 d postdifferentiation (Fig. 1G). The downstream genes, in particular, had lower levels of expression than observed previously (Fig. 1D), indicating that DUX4 protein levels were most likely reduced by treatment.
LNA4 Decreases the Expression of FSHD-Associated Genes.
RNA sequencing was conducted to evaluate the in vitro therapeutic efficacy of 100 nM LNA4 treatment on a more global scale. Approximately 30 million reads per sample were obtained, mapping to ∼93–96% of the genome (23). With the healthy control as reference, we identified 877 genes whose expression was significantly affected (i.e., at least a twofold increase/decrease in expression, P < 0.05) in immortalized FSHD patient-derived cells. To extract a subset of these genes that are associated with FSHD, we compared our dataset with that of Rickard et al. (24). Rickard et al. used FSHD patient-derived primary myoblasts carrying a DUX4-responsive fluorescent reporter that were differentiated and sorted by flow cytometry to obtain a pool of myocytes currently or recently expressing DUX4, which was then used as a sample for RNA sequencing. We decided to use this dataset as it had the most similar cell culture conditions to ours, in particular with the use of KOSR for differentiation. The comparison revealed 91 overlapping genes between the lists of significantly affected genes of the two studies (Fig. 2A). These were considered our FSHD signature genes: 86 significantly up-regulated and 5 significantly down-regulated genes (SI Appendix, Table S1). When the fold-change values obtained by the two different RNA sequencing experiments were plotted for each gene, we found that the majority of genes were regulated in the same direction (Fig. 2B), validating use of the signature.
Fig. 2.
RNA sequencing analysis of LNA gapmer therapeutic efficacy. (A) FSHD signature genes were determined via comparison with the Rickard et al. dataset (24). For the current study, clonal immortalized healthy control and FSHD patient-derived myotube populations were compared. (B) Two-dimensional plot of log2(fold-change) values between the present study and Rickard et al., with a log2(fold-change) cutoff of +2/−2 for the significantly affected genes of both datasets. (C) Volcano plots of RNA sequencing data from nontreated and LNA4-treated myotubes, with the FSHD-unaffected control as reference. The 91 FSHD signature genes are marked (red: up-regulated, blue: down-regulated in FSHD); gray vertical lines indicate twofold log2(fold-change) values in either direction. (D) Heat map displaying expression changes for the 91 FSHD signature genes after LNA4 treatment, with the FSHD-unaffected control as reference. High expression is indicated by more purple shades, and genes whose expression values were significantly restored to healthy levels (P < 0.05) are marked by asterisks on the right-hand side (n = 3 independent experiments per condition).
The 91 FSHD signature genes were marked on volcano plots of the RNA sequencing data from nontreated FSHD and LNA4-treated FSHD muscle cells, with the unaffected control as reference (Fig. 2C). Upon treatment with LNA4, there is an overall observable shift in the position of the marked genes toward the center of the plot, indicating restoration of expression to levels found in FSHD-unaffected muscle cells. This is similarly depicted using a heat map showing changes in expression for each of the 91 FSHD signature genes posttreatment (Fig. 2D). We found that the expression of the downstream DUX4 target gene ZSCAN4 was significantly decreased posttreatment compared to nontreated controls (P < 0.05). Of the 86 up-regulated FSHD signature genes, 27 had significantly reduced expression after treatment with LNA4 (P < 0.05) compared to the nontreated FSHD muscle cells. Expression levels of the five down-regulated FSHD signature genes were not significantly affected by the treatment. To further validate our results, we performed a similar analysis using the dataset of Geng et al., who performed microarray analysis on DUX4-transduced healthy primary myoblasts (25). Our comparison revealed 50 overlapping significantly affected genes (P < 0.05), 47 up-regulated and 3 down-regulated, the majority of which were similarly regulated between the two studies (SI Appendix, Fig. S3 A and B). Of the 47 up-regulated genes, 16 had significantly decreased expression posttreatment while of the 3 down-regulated genes, 2 had significantly increased expression posttreatment (P < 0.05) (SI Appendix, Fig. S3 C and D). Overall, 39 common FSHD signature genes were identified from both comparisons (our study versus Rickard et al. and Geng et al.), consisting of 1 down- and 38 up-regulated genes (SI Appendix, Fig. S3E). The common down-regulated gene, FGFR3, was not significantly affected by LNA4 treatment. However, 15 of the common up-regulated signature genes had significantly decreased expression posttreatment (P < 0.05) (SI Appendix, Table S2).
Effects of LNA Gapmer Treatment on Myogenic Fusion and Apoptosis.
DUX4 negatively regulates MYOD, which is involved in establishing and ensuring proper muscle differentiation (26). A manifestation of this DUX4-induced differentiation defect is the impaired fusion of myoblasts into muscle fibers in FSHD. We observed a significant decrease in myogenic fusion in immortalized FSHD patient-derived muscle cells compared to FSHD-unaffected controls (Fig. 3 A and B). Treatment with LNA6 significantly increased the myogenic fusion index of FSHD myotubes compared to the mock-treated group by ∼56% (n = 6, P < 0.005), whereas no significant improvement was observed with LNA1 and LNA4. As FSHD patient-derived myotubes have been found to display hypotrophy in vitro (27), we decided to quantify myotube diameters in our different treatment groups. LNA-treated muscle fibers displayed an observable shift toward larger diameters that were characteristic of healthy myotubes (Fig. 3C). FSHD patient-derived myotubes treated with LNA1, LNA4, or LNA6 had significantly increased myotube diameters at 16–20 μm on average compared to the mock-treated control, which had an average myotube diameter of 14.8 μm (n = 3, P < 0.05 for LNA4 and P < 0.0005 for LNA1, LNA6) (Fig. 3D).
Fig. 3.
Effect of LNA gapmer treatment on in vitro muscle cell fusion and apoptosis. (A) Representative images of immunostained immortalized healthy control and FSHD patient-derived cells with and without treatment using various LNA gapmers at 10 nM. Nuclei, blue; desmin, green. (Scale bar, 100 μm.) (B) Quantification of myogenic fusion indices (MFIs) from immunostaining images. MFI values were divided by the average MFI of the nontreated (NT) groups to eliminate batch effect. Error bars: SEM (n = 6). (C) Frequency distribution of muscle fiber diameters in the different conditions. (D) Quantification of the diameters in C. The central line marks the median, the box covers the 25th to 75th percentiles, and the whiskers represent the range. (n = 3) (E) Flow cytometry-based (Annexin V/propidium iodide) quantification of early and late apoptotic cells from immortalized healthy FSHD-unaffected control and FSHD patient-derived cells before and after 10 nM LNA gapmer treatment. For B and E, error bars: SEM (n = 6). *P < 0.05, **P < 0.005, ***P < 0.0005, one-way ANOVA with Dunnett’s test versus the mock (M), δP < 0.05, δδP < 0.005, δδδP < 0.0005, unpaired two-tailed t test. U = unaffected/healthy control.
Transfection of DUX4 constructs has also been reported to induce apoptosis in vitro (13, 26). We found that treatment with LNA1, LNA4, and LNA6 at 13 d postdifferentiation did not significantly affect early apoptotic cell numbers, either upon evaluation at 1 or 5 d after transfection using an Annexin V/propidium iodide-based flow cytometry assay (Fig. 3E and SI Appendix, Fig. S4). No significant differences in late apoptotic cell populations were observed across treatments and between nontreated and FSHD-unaffected controls at 14 d postdifferentiation (Fig. 3E), whereas late apoptosis was interestingly significantly increased in FSHD-unaffected controls at 18 d postdifferentiation (SI Appendix, Fig. S4B).
Minimal Potential Off-Target Toxicity with LNA Gapmer Treatment In Silico and In Vitro.
To assess off-target effects resulting from LNA gapmer treatment in vitro, a list of potential targets with sequences having at least one base mismatch to the DUX4 target sequence was compiled (SI Appendix, Table S3). Of the listed genes, three have detectable expression in muscle: RASA4, PLEKHH3, and MGAT4B. At both the 100 and 10 nM doses, PLEKHH3 and MGAT4B expression levels were not affected by LNA gapmer treatment (SI Appendix, Fig. S5). On the contrary, RASA4 expression was significantly increased at both doses upon treatment by LNA1 (n = 5, P < 0.05 at 100 nM and P < 0.005 at 10 nM), which does not constitute a direct off-target knockdown effect by the LNA gapmer.
In Vivo Uptake and Efficacy of LNA4.
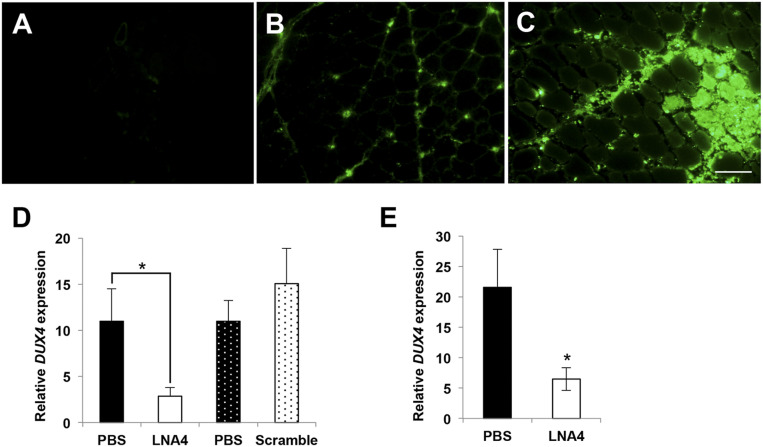
FLExDUX4 mice leak DUX4 at a very low level, similar to amounts found in human FSHD myoblasts. At this level, no DUX4 downstream genes are induced and no overt pathologies are reported (20). To visualize LNA gapmers after in vivo delivery, we injected 20 µg of fluorescein-tagged LNA4 into the TA muscles of FLExDUX4 mice. Two FLExDUX4 mice received i.m. injections of phosphate-buffered saline (PBS) instead. Fluorescence imaging showed no positive signal in the PBS-injected TA (Fig. 4A), whereas TAs that received LNA4 showed fluorescent signals in the muscles. The signals can be clearly seen in the interstitial space (Fig. 4B) and within muscle fibers (Fig. 4C).
Fig. 4.
In vivo uptake and efficacy of LNA gapmers. (A–C) FLExDUX4 mice were injected with single 20-µL injection volumes of PBS in the left TA (A) and 20 µg fluorescently tagged LNA4 (green) in the right TA (B and C). Muscles were collected 1 d later. The image in B depicts muscle tissue away from the injection site, while C shows the injection site. (n = 2). (Scale bar, 100 µm.) (D) Intramuscular injections of 20 µg LNA4 in TA muscles (one leg with LNA, contralateral leg with PBS; solid and hashed bars indicate leg pairs) of FLExDUX4 mice every other day for a total of three injections showed knockdown of DUX4 mRNA by qPCR 1 d after the last injection. No knockdown was observed when a scrambled LNA gapmer control was injected. (n = 5 each) (E) DUX4 transcript expression was evaluated 7 d after intramuscular injections of LNA4 in the TA. Significant DUX4 knockdown was observed. Error bars: SEM (n = 5 each). *P < 0.05, paired t test.
To determine if the LNA4 that significantly suppressed DUX4 expression in FSHD patient myoblasts was also effective in vivo, FLExDUX4 mice were given either three i.m. injections of 20 µg LNA4 or three injections of LNA gapmer control with a nonspecific, scrambled sequence every other day. Both groups received i.m. injections of LNA gapmers in one TA and of PBS in the contralateral TA. DUX4 mRNA expression was reduced by 84% upon treatment with LNA4 compared to the PBS-injected contralateral TA 1 d after the last injection (Fig. 4D). Injections of the scrambled control did not have a significant effect on DUX4 mRNA expression (Fig. 4D). We then repeated the experiment, but this time instead of collecting muscles 1 d after the last injection, we collected them 7 d postinjection. Our results showed a significant 70% knockdown of DUX4 mRNA (n = 5, P < 0.05) in the treated muscles (Fig. 4E). Thus, LNA4 significantly reduced DUX4 expression in the skeletal muscle of FLExDUX4 mice, and the effect lasted for at least 7 d postinjection.
Discussion
Our study shows the potential of using LNA gapmers for FSHD therapy through DUX4 transcript knockdown. Gapmers are gaining traction in therapeutic development, with a number approved by the US Food and Drug Administration (FDA) or in clinical trials (28). One of the earliest successes is mipomersen (Kynamro; Ionis, Genzyme), a 2′-O-methoxyethyl (2′-MOE) gapmer targeting apolipoprotein B-100 that was approved by the FDA for familial hypercholesterolemia treatment in 2013 (28, 29). In 2018 the FDA approved another 2′-MOE gapmer, inotersen (Tegsedi; Ionis, Akcea), for treating hereditary transthyretin amyloidosis-associated polyneuropathy (28, 30). Gapmers have been tested for Huntington’s disease, familial chylomicronemia, familial partial lipodystrophy, and various cancers (28, 31). Available results from these, particularly for Huntington’s disease, are encouraging. To our knowledge, only three LNA gapmers (Enzon) have been evaluated in clinical trials thus far, for treating certain cancers. However, studies on these have all been discontinued, likely due to concerns related to safety and/or efficacy. The potential of LNA gapmers for therapy remains to be seen, particularly for neuromuscular disorders.
Aside from showing nearly complete reduction of DUX4 transcript levels (Fig. 1C) and likely a decrease of DUX4 protein levels (Fig. 1 D, F, and G) in immortalized FSHD patient-derived myotubes as a result of LNA gapmer treatment, we demonstrated significant knockdown of DUX4 mRNA in the TA muscle of an FSHD mouse model with local injections (Fig. 4 D and E). We also provided evidence of safety against potential off-target effects identified by a short input sequence-optimized search engine (SI Appendix, Fig. S5).
Previous studies have mostly used PMOs to interfere with PAS recognition or DUX4 pre-mRNA splicing (7, 12, 16, 17). While these are viable strategies to knock down DUX4 expression, certain limitations to efficacy are imposed by the PMO chemistry. PMOs are charge-neutral AOs that have been proven safe and stable in vivo (32). Despite their promising efficacy in vitro, PMOs display poor uptake into target tissues in vivo and are rapidly cleared from circulation, with an elimination half-life of less than 4 h (33). In contrast, LNA gapmers have a longer half-life, reaching up to 15 h (34), and their negatively charged backbone likely allows better recognition by cell surface receptors, leading to more effective internalization (35). This is supported by our study in which we observed good LNA gapmer uptake into injected muscles (Fig. 4C). In addition, LNA gapmers are stable, have a stronger affinity to RNA, and have higher RNase H-mediated cleavage activity than 2′-O-methyl or phosphorothioated DNA AOs (34). This allows for LNA gapmers to be administered at lower doses and yet retain appreciable potency. Whereas transfection of LNA gapmers at 100 nM knocked down DUX4 to nearly undetectable levels (Fig. 1C), transfection at a 10-fold lower dose still led to significant DUX4 knockdown, strikingly by ∼90% lower than nontreated controls on average (Fig. 1E). This is more efficient than in previous studies, which used PMOs at higher concentrations (50–150 nM) and achieving at most 50% knockdown (12, 16). Finally, knocking down DUX4 through RNase H-mediated degradation offers more assurance of efficacy rather than blocking PAS or splice sites. For instance, alternative PAS sequences are potentially available to DUX4 (36), and the introduction of splice-switching AOs can trigger the use of alternative splice sites.
We also introduce a pipeline for reliably screening DUX4-targeting AOs for FSHD treatment. One challenge working with DUX4 is its low expression in FSHD muscle, with only 1/1,000 myoblasts and 1/200 myotube nuclei expressing DUX4 in primary cultures from FSHD muscle (8, 19). Conventional techniques such as RT-PCR and Western blot used previously (12, 16) are limited in their ability to detect DUX4 and may not provide consistent, sufficiently quantitative results. Here, we use a cell culture procedure to induce endogenous DUX4 mRNA expression in immortalized patient-derived myotubes for AO screening (22). This allowed for qPCR-based detection of DUX4 at high sensitivity (SI Appendix, Fig. S1) and helped distinguish among our LNA gapmers based on efficacy (Fig. 1C). Such an in vitro system would be simpler than one involving transfection of DUX4-containing vectors, which requires lengthy optimization and dealing with the inherent variability in transfection efficiencies across cells. Furthermore, we introduce the use of the FLExDUX4 FSHD mouse model (20) for the in vivo testing of AOs (Fig. 4). This model expresses DUX4 transcripts at levels sufficient for qPCR detection even without induction of the inserted DUX4 transgene (20). Together, the in vitro and in vivo methods used here provide a valuable system for screening AOs and other drugs for FSHD therapy.
LNA gapmers targeting sites upstream of the PAS in exon 3 were determined most efficacious (Fig. 1 A and C). In the context of exon 3, DUX4 knockdown efficacy appears to correlate with the predicted RNA folding conformation at the target site (SI Appendix, Fig. S2). LNA2, whose target site adopts an open conformation at only one end, exhibited lower efficacy than LNA1 and LNA4-7, whose target sites were open in the middle (Fig. 1C). This is despite both groups of LNA gapmers being complementary to similar numbers of target bases in open regions. We believe this is because recruitment of RNase H occurs in the central DNA/RNA hybrid region of the LNA gapmer-target RNA complex; an open conformation in that region likely facilitates RNase H binding and activity. This stresses the need for the proper in silico design and screening of AOs prior to testing in cellular and animal models. In our previous work on PMOs for exon skipping in Duchenne muscular dystrophy (37), we showed that certain features between AOs and their target sequences (e.g., binding energy, GC content) can be used to create a predictive tool to determine which sites would lead to the highest levels of exon skipping when targeted. Perhaps the same logic can be applied for LNA gapmers in the selection of target sites for future studies.
We observed significant improvements in muscle cell fusion and fiber diameters upon treatment compared to nontreated controls (Fig. 3 A–D). Impaired fusion as a result of DUX4 has been demonstrated using transfection studies in vitro (38) and in some patient muscle samples (14). Our result is encouraging, as it shows that LNA gapmer treatment is beneficial both in reversing the molecular effects of DUX4 misexpression (Fig. 2) and in ameliorating pathological in vitro phenotypes. Unlike for fusion, we did not find a significant effect of treatment on apoptosis (Fig. 3E and SI Appendix, Fig. S4B). It is likely that more time is required posttreatment to observe therapeutic effects on this phenotype. Furthermore, our data suggest that, using our culture system, at 14 d postdifferentiation, immortalized FSHD patient-derived myotubes are only still beginning to undergo the early phases of apoptosis and are yet to exhibit differences in the number of late apoptotic cells observed (Fig. 3E). However, it is encouraging to note that LNA gapmer treatment did not induce apoptosis, adding favorably to the safety of this treatment.
In conclusion, using immortalized FSHD patient-derived muscle cells and the FLExDUX4 FSHD mouse model, we were able to show that our designed LNA gapmers could significantly and selectively knock down DUX4 expression in skeletal muscle. Effects on muscle structure and function, as well as an evaluation of the pharmacokinetic properties of LNA gapmers in vivo, remain to be determined with a systemic treatment study. This will complement our findings in vitro, which show a limited albeit promising view of the potential of our designed LNA gapmers for therapy. Additionally, we outline the use of a consistent, reliable method for screening DUX4-targeting AOs in vitro and in vivo. Taken together, we expect the promising therapeutic we have developed and the AO screening method used in this study to facilitate progress in the field toward the production of viable treatments for FSHD.
Methods
FLExDUX4 mice were used for the study (20). Animal care and use were approved by the Institutional Animal Care and Use Committee of the Children’s Research Institute of Children’s National Health System, Washington, DC. All animal procedures were carried out in accordance with approved guidelines. The mice were maintained in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Detailed information on animal procedures, AO design, cell culture and transfections, qPCR, RNA sequencing, in vitro functional tests, off-target evaluation, and statistical analysis are provided in SI Appendix.
Data Availability.
All data and relevant materials are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank Aja Rieger and Juan Jovel (University of Alberta) for their technical assistance and support in the study. This study is supported by the FSH Society, Muscular Dystrophy Canada, University of Alberta, Women and Children’s Health Research Institute Graduate Studentship award and Innovation Grant, Friends of FSH Research, FSHD Global Research Foundation, The Friends of Garrett Cumming Research Fund, HM Toupin Neurological Science Research Fund, Canadian Institutes of Health Research, Canada Foundation for Innovation, and Alberta Enterprise and Advanced Education.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. G.S.B.-B. is a guest editor invited by the Editorial Board.
Data deposition: RNA sequencing data have been deposited in the National Center for Biotechnology Information Sequence Read Archive portal (BioProject PRJNA606474).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909649117/-/DCSupplemental.
References
- 1.Wang L. H., Tawil R., Facioscapulohumeral dystrophy. Curr. Neurol. Neurosci. Rep. 16, 66 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Klinge L. et al., Severe phenotype in infantile facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 16, 553–558 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Wijmenga C. et al., Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 2, 26–30 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Lemmers R. J. L. F. et al., Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 24, 659–669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabriëls J. et al., Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236, 25–32 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Lemmers R. J. L. F. et al., A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansseau E. et al., Antisense oligonucleotides used to target the DUX4 mRNA as therapeutic approaches in FaciosScapuloHumeral muscular dystrophy (FSHD). Genes (Basel) 8, 93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snider L. et al., Facioscapulohumeral dystrophy: Incomplete suppression of a retrotransposed gene. PLoS Genet. 6, e1001181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hewitt J. E., Loss of epigenetic silencing of the DUX4 transcription factor gene in facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 24, R17–R23 (2015). [DOI] [PubMed] [Google Scholar]
- 10.de Greef J. C. et al., Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology 75, 1548–1554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himeda C. L., Jones P. L., The genetics and epigenetics of facioscapulohumeral muscular dystrophy. Annu. Rev. Genomics Hum. Genet. 20, 265–291 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Vanderplanck C. et al., The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One 6, e26820 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowaljow V. et al., The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 17, 611–623 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Barro M. et al., Myoblasts from affected and non-affected FSHD muscles exhibit morphological differentiation defects. J. Cell. Mol. Med. 14, 275–289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosnakovski D. et al., An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 27, 2766–2779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsollier A.-C. et al., Antisense targeting of 3′ end elements involved in DUX4 mRNA processing is an efficient therapeutic strategy for facioscapulohumeral dystrophy: A new gene-silencing approach. Hum. Mol. Genet. 25, 1468–1478 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Chen J. C. et al., Morpholino-mediated knockdown of DUX4 toward facioscapulohumeral muscular dystrophy therapeutics. Mol. Ther. 24, 1405–1411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J. J. A., Yokota T., Antisense therapy in neurology. J. Pers. Med. 3, 144–176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tassin A. et al., DUX4 expression in FSHD muscle cells: How could such a rare protein cause a myopathy? J. Cell. Mol. Med. 17, 76–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones T., Jones P. L., A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS One 13, e0192657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dandapat A. et al., Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep. 8, 1484–1496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey S. N., Khawaja H., Chen Y.-W., Culture conditions affect expression of DUX4 in FSHD myoblasts. Molecules 20, 8304–8315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim K. R. Q., et al. , RNA sequencing of control, non-treated FSHD, and LNA gapmer-treated FSHD myotubes. National Center for Biotechnology Information Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA606474. Deposited 13 February 2020.
- 24.Rickard A. M., Petek L. M., Miller D. G., Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 24, 5901–5914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng L. N. et al., DUX4 activates germline genes, retroelements, and immune mediators: Implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knopp P. et al., DUX4 induces a transcriptome more characteristic of a less-differentiated cell state and inhibits myogenesis. J. Cell Sci. 129, 3816–3831 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerji C. R. S. et al., Dynamic transcriptomic analysis reveals suppression of PGC1α/ERRα drives perturbed myogenesis in facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 28, 1244–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen X., Corey D. R., Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 46, 1584–1600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hair P., Cameron F., McKeage K., Mipomersen sodium: First global approval. Drugs 73, 487–493 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Keam S. J., Inotersen: First global approval. Drugs 78, 1371–1376 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Moreno P. M. D., Pêgo A. P., Therapeutic antisense oligonucleotides against cancer: Hurdling to the clinic. Front Chem. 2, 87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim K. R. Q., Yokota T., “Invention and early history of exon skipping and splice modulation” in Exon Skipping and Inclusion Therapies: Methods and Protocols, Yokota T., Maruyama R., Eds. (Springer, New York, 2018), pp. 3–30. [DOI] [PubMed] [Google Scholar]
- 33.Amantana A. et al., Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate. Bioconjug. Chem. 18, 1325–1331 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Kurreck J., Wyszko E., Gillen C., Erdmann V. A., Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 30, 1911–1918 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooke S. T., Wang S., Vickers T. A., Shen W., Liang X. H., Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 35, 230–237 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Ozsolak F. et al., Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143, 1018–1029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Echigoya Y., Mouly V., Garcia L., Yokota T., Duddy W., In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS One 10, e0120058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yip D. J., Picketts D. J., Increasing D4Z4 repeat copy number compromises C2C12 myoblast differentiation. FEBS Lett. 537, 133–138 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and relevant materials are included in the manuscript and SI Appendix.