Significance
A major problem in data science is representation of data so that the variables driving key functions can be uncovered and explored. Correlation analysis is widely used to simplify networks of feature variables by reducing redundancies, but makes limited use of the network topology, relying on comparison of direct neighbor variables. The proposed method incorporates relational or functional profiles of neighboring variables along multiple common neighbors, which are fitted with Gaussian mixture models and compared using a data metric based on a version of optimal mass transport tailored to Gaussian mixtures. Hierarchical interactive visualization of the result leads to effective unbiased hypothesis generation. In a cancer gene expression study, this method uncovered an unanticipated immunosuppressive mechanism resembling maternal–fetal immune tolerance.
Keywords: complex networks, Gaussian mixture models, community detection, optimal transport, immune tolerance
Abstract
We present a technique to construct a simplification of a feature network which can be used for interactive data exploration, biological hypothesis generation, and the detection of communities or modules of cofunctional features. These are modules of features that are not necessarily correlated, but nevertheless exhibit common function in their network context as measured by similarity of relationships with neighboring features. In the case of genetic networks, traditional pathway analyses tend to assume that, ideally, all genes in a module exhibit very similar function, independent of relationships with other genes. The proposed technique explicitly relaxes this assumption by employing the comparison of relational profiles. For example, two genes which always activate a third gene are grouped together even if they never do so concurrently. They have common, but not identical, function. The comparison is driven by an average of a certain computationally efficient comparison metric between Gaussian mixture models. The method has its basis in the local connection structure of the network and the collection of joint distributions of the data associated with nodal neighborhoods. It is benchmarked on networks with known community structures. As the main application, we analyzed the gene regulatory network in lung adenocarcinoma, finding a cofunctional module of genes including the pregnancy-specific glycoproteins (PSGs). About 20% of patients with lung, breast, uterus, and colon cancer in The Cancer Genome Atlas (TCGA) have an elevated PSG+ signature, with associated poor group prognosis. In conjunction with previous results relating PSGs to tolerance in the immune system, these findings implicate the PSGs in a potential immune tolerance mechanism of cancers.
Data analysis with a large number of variables always involves evaluating some kind of similarity between variables. This serves the purpose of finding mechanisms of action in the system, in which similar variables may indicate subsystems working together to accomplish some function. It also serves the purpose of dimensional reduction, reducing the complexity of the analysis by allowing a member of a group of related variables to serve as a proxy for the whole.
Practical unsupervised data analysis is often limited to similarity clustering based on standard sample-wise Pearson correlation, because of its ease of computation and straightforward interpretation. Correlation measures the goodness of fit of a linear relationship between two numerical variables. The disadvantage is that the comparison between variables is made without reference to other variables that could be essential for identifying related function.
For a higher-order approach, rather than directly comparing the values of two variables and , we compare only the function of the variables in the context of the whole system across a cohort. The idea is that we consider the bivariate distributions associated with each edge and then compare pairs of bivariate distributions associated with adjacent edges and . Accordingly, the functional profile of a given variable will mean the collection of joint distributions or scatter plots of against other variables (Figs. 1 and 2). To decide which variables to use in the functional profile of , we assume that the dataset is augmented with a network topology providing abstract connections between variables/features. A variable will be used in the profile of only if there is a connection or edge between and . In some contexts, such as a well-studied molecular pathway, this network may be known a priori and should be considered part of the dataset being analyzed (e.g., SI Appendix, Fig. S1). In other contexts, such as the large-scale transcriptomic analysis we performed on The Cancer Genome Atlas (TCGA) lung cancer samples, for an unbiased analysis this network should be data driven, inferred from the cohort data matrix itself. For network inference, we use liberal thresholds on the absolute value of the correlation between variables and to decide whether and will be connected by an edge.
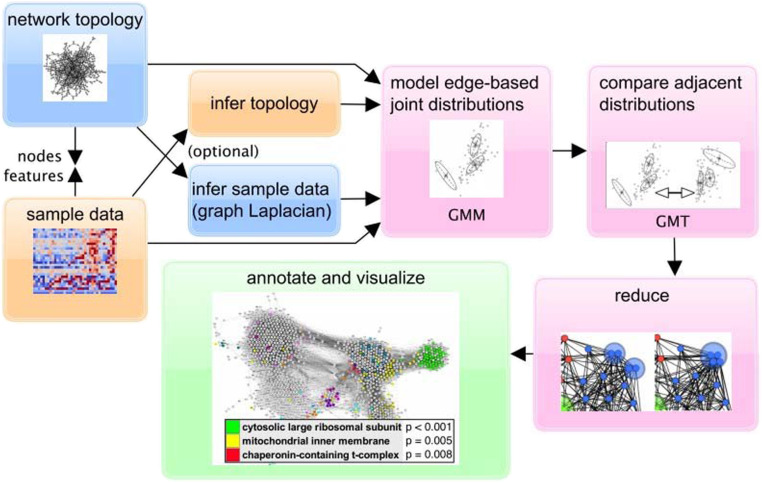
Fig. 1.
Flowchart summarizing the GMT analysis pipeline.
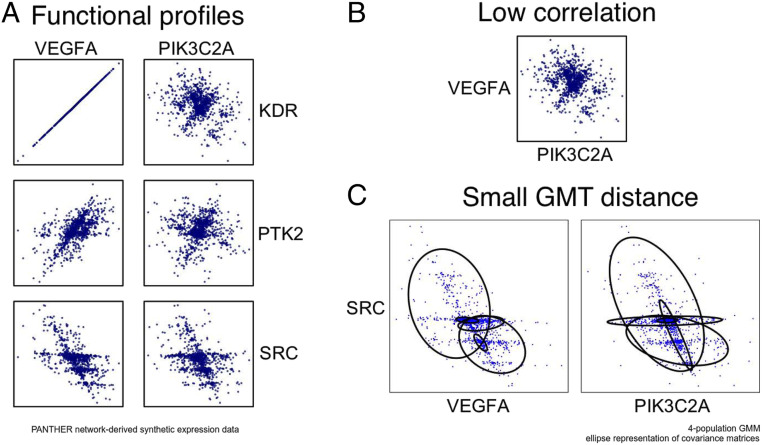
Fig. 2.
(A–C) An illustration of GMT comparison between two features. In this case the two features are not close because of correlation or a direct relationship between their values, but instead because of small GMT distance incorporating the relationships with other features. This comparison metric is an average of a Gaussian mixture model and optimal mass transport-based metric between the joint distributions (scatter plots) that are relevant according to the network topology. The gene network is the PANTHER curated database (http://www.pantherdb.org/), with numerical data synthesized from the network topology using the graph Laplacian described in Methods.
One often finds by informal investigation that some variables and have common functional profiles even when and are completely uncorrelated. To promote this type of informal investigation to objective analysis, one needs a comparison metric between joint distributions or scatter plots. To this end we first fit Gaussian mixture models (GMMs) to the distributions. This has a smoothing effect, filtering out noise, as well as making the distributions accessible to analytic formulas via the comparatively few fitted parameters. GMMs were selected because they are well studied and straightforward to fit. A computationally efficient version of optimal mass transport (OMT) adapted to GMMs (1) is used to measure the distance between the fitted models. It is important to note that distances based on OMT are (weakly) continuous as opposed to some other commonly used measures of distributions such as Kullback–Leibler divergence and total variation (2). Further, GMMs are natural models for representing probability distributions (3). Under very general conditions, probability density functions may be approximated (e.g., in ) by such weighted sums of Gaussians.
The final Gaussian mixture transport (GMT) distance between and is calculated as the average GMM/OMT distance between the functional profiles of and along variables that are common neighbors of and . The GMT metric may be used thereafter as the input to hierarchical clustering algorithms. We thus employ the GMT metric to create a force-directed graphical representation of the feature network in which close nodes are likely to share a common function with respect to other nearby nodes.
This analysis and visualization methodology is well suited to hypothesis generation in biological and medical applications with large gene-level datasets. In an investigation of mRNA expression data of lung adenocarcinoma samples, the graphical representation strongly grouped together a module of genes which were further singled out, among all modules found, for their comparatively high expression in a subsample belonging to a published unsupervised cluster with approximately 20% frequency (Fig. 3). This module turned out to include all the known 10 pregnancy-specific glycoproteins (PSGs) and several other genes known for expression in the placenta. This suggested the hypothesis that the PSG+ status is related to prognosis, which is confirmed by Kaplan–Meier analysis in the TCGA lung adenocarcinoma, breast, uterine corpus endometrial carcinoma, and colon adenocarcinoma cohorts. Approximately 20% of each cohort have PSG+ tumors, and these have an especially poor prognosis. Together with documented findings relating PSGs to regulation of the immune system, these results implicate the PSGs as a potential mechanism to mediate immune tolerance in cancers.
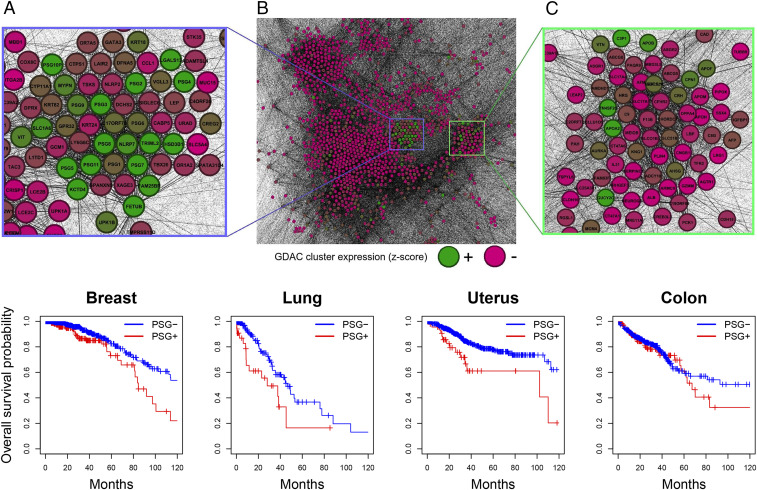
Fig. 3.
(Top, A–C) A special role for pregnancy-specific glycoproteins in cancer, uncovered with the proposed functional network analysis using the GMT metric. (B) A gene network inferred from the TCGA lung adenocarcinoma transcriptome using Pearson correlation cutoffs, analyzed with the GMT method and represented graphically using the GMT distances between genes. The gene coloring reflects the average -score profile for the third sample cluster identified in the consensus clustering published at the Broad GDAC Firehose (the profiles for the other four of the five GDAC sample clusters did not show clear patterns of expression with respect to the functional network representation). Green is high and red is low. The graph layout is force directed. Pearson correlation alone does not account for the grouping of the PSG genes: After removing 4 outliers out of 509, the mean of the absolute value of the Pearson correlation between the expression values for the 11 PSGs was only 0.177, with a SD of 0.201. (A) Highlight of the salient group containing highly expressed genes. The group contains all 10 of the PSG genes, as well as the pseudogene PSG10, and placental genes LGALS13 and HSD3B1. Several of the down-regulated genes in the group are closely related to certain cancer/testis antigens (CTAs): ADAM2 (ADAMTSL4), NLRP4 (NLRP2), SPATA19 (SPATA31D4), TMPRSS12 (TMPRSS11D), CRISP2 (CRISP1), XAGE3, and SPANXN5. See ref. 4 for the full list of 276 CTAs. (C) Highlight on the second group containing highly expressed genes, including genes similar to the CTAs: SSX4, DPPA2 (DPPA4), CT47A1, CT47A6, ARMC3 (ARMC9), TSPY1 (TSPYL6), and RGS22 (RGSL1). (Bottom) Kaplan–Meier survival analysis for the TCGA breast, lung adenocarcinoma, uterine corpus endometrial carcinoma, and colon cohorts stratified by the presence of at least one overexpressed PSG. The PSG+ phenotype confers a substantial survival disadvantage in these cancer types. In a few other cancer types, including ovarian cancer, a subset showing the PSG+ phenotype was present but did not confer a statistically significant advantage or disadvantage. No PSG+ cases were found in the TCGA pancreas cohort.
Methods
Overview.
A central problem in the field of network science is the representation of network data in a readily accessible format (5). Ideally the representation should be amenable to human-in-the-loop, interactive, exploratory data analysis. Compression methods have been used previously to reduce large networks to a desired level of resolution, mainly toward the goal of improving the computational performance of community detection algorithms (6). The key idea is to group nodes into modules and consider the new network composed of the connections between groups implied by the individual node connections. In its simplest form, the groups may be obtained by devising an edge weighting intended to measure similarity between neighboring nodes and successively “collapsing” edges, beginning with the greatest similarity, to create a hierarchical representation.
A natural improvement is to find approximations of a given network “from below” with gradually expanding small node subsets, an approach developed by Stanley et al. (7). Stanley et al. randomly distribute seed nodes which are then expanded into “supernodes” using direct neighborhoods. In a more global approach, Yang et al. (6) present a method of supernode network representation involving explicit consideration of known prior constraints on the set of network topologies of interest for a low-complexity approximation to the given network satisfying the constraints. For a more detailed survey, see Besta and Hoefler (8).
Unlike the method referred to in ref. 7, our proposed approach makes essential use of additional data beyond the network topology. Moreover, rather than qualitative constraints as in ref. 6, we assume that the nodes of the network are numerical feature variables, so the network is augmented with node weight data. For example, in genomics one can use RNA expression of genes across a tissue sample set. Such data can also be generated or synthesized from the underlying network topology if the topology is the primary structure of interest. Conversely the network topology may be inferred from the sample data if no prior network is known. The node weightings may be interpreted as defining samples from the joint distribution of random variables associated with the nodes.
A natural model for distributions is the Gaussian mixture, used in many data processing and analysis applications (3). In general, a mixture model is a weighted linear combination of distributions where each component represents a subpopulation. In particular, the GMM is a weighted average of Gaussians. GMMs are popular due to their versatility and overall simplicity in data representation. They are ubiquitous in statistics, hypothesis testing, decision theory, and machine learning. The idea is that real-world data may not be densely distributed on a high-dimensional space and instead are concentrated in a low-dimensional subspace. Further, in many cases of interest, the data are sparsely distributed into a number of subgroups, and so differences within a given subgroup are not as important as those among the subgroups. Mixture models capture these properties, and this motivated the work of Chen et al. (1) to modify OMT theory (9, 10) into a form suitable for Gaussian mixture models.
OMT provides the Gaussian mixture framework with a natural comparison metric between mixtures, and conversely mixtures provide a natural model with which to make the computation of OMT tractable. We use GMMs to model the functional role played by a node with respect to the data along its neighbors in the network. This role is quantified by the average GMM/OMT distance between two nodes, which we call the GMT distance. Hierarchical clustering then provides a simplified version of the network for each given level of complexity. The simplified or compressed network represents a projection of the prior network (rather than a subnetwork) which is most relevant according to the evidence observed in the data.
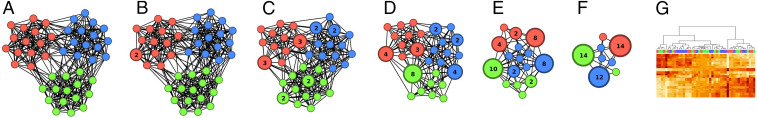
To illustrate the construction of GMT distance-based network reductions, we show the steps of the construction applied to a concrete example, a network synthesized to have a known community structure. The edge density is high within the communities and low between communities. Node weightings are randomly generated in terms of the network topology by neighbor averaging or the iterated graph Laplacian (11) of random weightings. An example of such a weighting is depicted in SI Appendix, Fig. S1. A preview of the series of network simplifications is shown in Fig. 4.
Fig. 4.
A synthetic network with K = 3 communities containing N = 45 nodes total. The network was randomly generated to have intercommunity edge connectivity 0.08 out of a possible maximum of 0.68 and intracommunity edge connectivity 0.28 out of a possible maximum of 0.32. Random node weightings are generated from the network by the iterated graph Laplacian (11) applied to an initial node weighting equal to 1 on a randomly selected node and 0 on every other node. Two hundred node weightings were generated in this manner. Hierarchical clustering was performed with respect to GMT-distance similarity edge scores. (A) The original network. (B–F) Five selected hierarchical levels from the series. Each node group is labeled by the number of nodes it contains. (G) Heatmap showing ordinary hierarchical clustering of the synthesized samples, with the usual rectilinear representation of the hierarchy tree.
Background on OMT for Gaussian Mixture Models.
A Gaussian mixture model is an important instance of the general mixture model structure, a structure that is commonly utilized to study properties of populations with several subgroups (3). Formally, a GMM is a probability density consisting of a weighted linear combination of several Gaussian components, namely
where each is a Gaussian distribution and is a probability vector. Here the finite number stands for the number of components of .
Let be two Gaussian mixture models of the form
The distribution is equivalent to a discrete measure with supports for each . The framework from ref. 1 is based on the discrete OMT problem
| [1] |
for these two discrete measures, where denotes the space of joint distributions with marginal distributions and . The cost is taken to be the 2-Wasserstein metric:
There is a closed formula for this metric (9, 10),
| [2] |
where and are Gaussian distributions with means and and covariances and , respectively.
The discrete OMT problem (1) always has at least one solution, and letting be a minimizer, we define
| [3] |
This formula, from ref. 1, is the key formula underlying our algorithm.
Gaussian Mixture Transport Distance.
A naive approach would group nodes together based on similar properties, for example, by making comparisons between the (univariate) distributions of the weight data associated with each node. Comparison in this case is a classic topic, addressed, for instance, by the Kolmogorov–Smirnov test. One step beyond this is to summarize the joint (bivariate) distribution associated with each edge–node pair by a Pearson correlation or related metric and then use this similarity metric for classic clustering.
As alluded to in the Introduction, in the present work we propose a higher-order method, in which we consider the bivariate distributions associated with each edge (i.e., the joint distributions of the variables associated with the two endpoint nodes) and then compare pairs of bivariate distributions associated with adjacent edges – and – (here the edges are adjacent along node ). If the distributions are similar, and will be considered to have a similar function in the network locally near . Ultimately, we will summarize this similarity over all intermediate between and . Fig. 2 shows an example of similar functional profiles. In this way we capture local parallelism, where some closely related nodes may provide alternative paths to similar effects.
How is the similarity between bivariate distributions quantified? The Bhattacharyya distance (12) is one similarity measure between distributions in higher dimensions. However, a direct calculation of this distance tends to require rasterization of the space involved and may be prohibitively costly to compute. The theory developed by Chen et al. (1) for Gaussian mixture models provides a nearly closed-form alternative which is well suited to the task, a metric we call Gaussian mixture transport similarity. First, the distributions are approximated by Gaussian mixtures with a given number of subpopulations. The mixture weights are interpreted as a probability distribution on the discrete set of subpopulations, which are themselves compared using the optimal mass transport metric or discrete earth mover’s distance (EMD). For this calculation of the EMD, the cost function corresponding to motion from the discrete point labeling a subpopulation of the first mixture to a discrete point labeling a subpopulation of the second mixture is taken to be the actual optimal mass transport distance between the corresponding Gaussian distributions. The GMT distance between two nodes (not necessarily connected by an edge) is this GMM/OMT distance averaged over all adjacent edges with the same free endpoints. For a detailed description, see SI Appendix, Table S2, summarizing the formulas in ref. 1 and in Background on OMT for Gaussian Mixture Models. The last step is classic hierarchical clustering using the GMT distance sparse similarity matrix. We use the average-distance–based hierarchical clustering method, although the standard alternatives single-linkage, complete-linkage, and Ward clustering, etc., may be used depending on the application.
Node Data Synthesis.
Although the GMT hierarchy is primarily designed for reducing the feature structure of a numerical dataset, it can also be applied to a pure network topology by synthesized node weights. For this we use the iterated graph Laplacian (11) applied to single-node weightings with randomly chosen support nodes. The resulting weightings can be understood as random linear combinations of the eigenfunctions, with emphasis on those eigenfunctions with large eigenvalues. The spectrum of is well studied and known to capture a lot of detailed information about the underlying graph.
Implementation and Runtime Complexity.
A naive version of our algorithm would iterate over all edge pairs, with complexity class where is the number of edges of the network. However, since only adjacent edge pairs are used, we instead iterate over the nodes and then over the pairs of its neighbors, with complexity class where is the number of nodes and is the maximum degree over all nodes.
The mixture modeling and GMT distance calculations are classically parallelizable: the mixture modeling, because it depends only on the variable pair distributions, and the GMT distances because they depend only on the resulting list of mixture models. This makes our algorithm feasible for rapid computation. The discrete earth mover’s distance is performed with the R package emdist (13). The mixture modeling itself is performed with the R package mclust (14). In practice, the number of mixture model populations has little effect on the overall output and performance as long as lies in the approximate range from 3 to 10. If the number of node-weighting samples is as low as a few hundred (), it is not meaningful to choose much greater than 10 anyway, since the number of data points per population () should not be too low. High accuracy of the mixture model as a representation of the joint distribution of two given node variables is not essential for the purpose of inferring distances between the distributions from distances between the models.
Visualization.
Once the hierarchy is computed, it is formatted for viewing in the Gephi graph visualization software using a custom plugin. Gephi is used to represent weight data with node size, color, and relative position in force-directed graph layouts. We use three different view types.
One view shows the compressed network at a user-selected level or scale, as depicted in Fig. 4.
For larger graphs, a second, static view is used to reduce the computational burden of real-time rendering. For this we use the hierarchy itself considered as a graph. We note that this visualization method applies to any hierarchical clustering and could serve as a general-purpose alternative to the usual rectilinear branch representation often used to decorate heatmaps. The graph is a tree or union of trees, with leaf nodes representing features and internal nodes representing feature groups. We choose leaf node sizes to reflect the linkage height, or hierarchical level, of the first internal node to which it is attached. Lower levels correspond to larger nodes, since nodes joining the tree at a lower level do so on the basis of stronger evidence of coordination with other nodes (smaller GMT distances), which we wish to highlight. Internal nodes are given negligible size. A planar representation with no edge crossings is possible since the graph consists of trees. A force-directed layout is used to arrange the nodes in a way guided by the tree structure and node sizes.
In the third view the original network topology, modified to include edges added between neighbors of neighbors, is visualized directly with edge weighting equal to the GMT distance. To emphasize a particular scale , we use a Gaussian transformation of GMT distance with mean and a chosen bandwidth . This representation has many of the computational advantages of a single preprocessed static view, but with some additional flexibility for interactive refinement via and .
Data Availability.
The gene expression data used in this study are publicly available from the TCGA (15) (https://www.cancer.gov/tcga) via cBioPortal (16). Cluster analysis published via the Gene Data Analysis Center (GDAC) Firehose (17) was used. Code for this paper is available on the public repository https://github.com/MSK-MedPhys-DeasyLab/functional-network-analysis.
Results
Benchmarking.
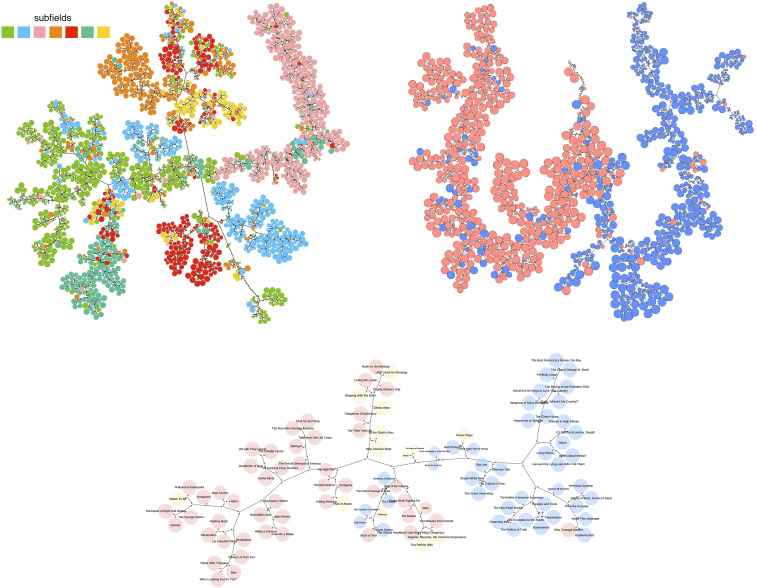
GMT-based hierarchical clustering can be completed to an unsupervised community detection algorithm using a numerical metric of modularity in the usual way, by selecting from among the level-cutoff clusterings the one with the best value of the metric. In the supervised setting, the level cutoff can be selected to give the best value of a cluster similarity metric, such as normalized mutual information (NMI) calculated against a known community structure. Fig. 5 compares this GMT community detection method with three established methods available in the R igraph library: greedy optimization, Louvain optimization, and label propagation.
Fig. 5.
Comparison of GMT community detection with greedy optimization of the modularity (18, 19), Louvain optimization (20), and label propagation (21), with respect to NMI. The hierarchy-tree graphical representations are shown. PolBlogs and PolBooks are respectively a network of political blog links and a copurchasing network for political books in the United States. Both are provided with manually annotated indications of political leaning for each node (red and blue). For these smaller networks with few, well-defined communities, the established methods outperform GMT. On the larger Cora academic paper citation network, presumably with greater real-world complexity, GMT outperforms the other methods by a factor of 3.7. The visualization substantially reveals the manually annotated subfields and also seems to suggest an improvement where some putative subfields are divided into multiple distinct groups and closely related to specific alternative subfields. For example, the subfield in red is divided into two tightly clustered groups, one group very close to the subfields in orange and yellow.
Analysis of Gene Regulatory Networks.
Gene and protein networks are now ubiquitous in medicine and biology. Although they are thought to be highly orchestrated, they are complex, and data-driven validation of known pathway mechanisms is still needed.
Functional network analysis of the lung adenocarcinoma RNA expression profiles in the TCGA database, using the GMT metric, produces a module of genes/nodes that are clustered together containing two sets of functionally related genes (Fig. 3). One set is expressed by trophoblasts, which reside only in the placenta (during pregnancy), and a second set belongs to the testis antigen family of genes. In Fig. 3, the green-colored genes contain high levels of transcription while the red-colored genes have low or no detectable levels of transcription. The quantification of these levels of mRNAs is from the average -score profile of one sample cluster from the data published by the Broad GDAC Firehose. The genes that are labeled in green (high levels) are largely from the placenta with all 11 genes of the PSG cluster of genes scoring as actively transcribing. The genes that are labeled largely red (little or no transcription) are enriched for the testis-specific antigens with only a few genes being expressed.
Thus, the GMT metric has identified a functional set of genes, the PSG genes, which are transcribed only in normal trophoblasts during pregnancy and whose transcripts were also detected in about 20% of lung adenocarcinomas in the TCGA. Furthermore, those cancers that expressed the PSG genes had the worst overall survivals as tested in Kaplan–Meier plots with statistical significance (Fig. 3). Similar results detecting the transcription of the PSG genes in about 20% of cancers of the breast, uterus, and colon were also observed. The PSG genes were not expressed in pancreatic cancers and in ovarian cancers statistical significance for lower overall survival was not observed. The PSG genes are in a cluster of genes expressed in the placenta and so any of these genes in the module shown in Fig. 3 might have some negative impact upon immune surveillance, resulting in a poor overall survival.
Discussion and Future Research.
We presented an unsupervised methodology for relational or functional analysis of networks based on a modification of optimal mass transport attuned to Gaussian mixture models, applicable to general feature network datasets with an intermediate-to-large number of variables. It identified known community structures in several benchmark datasets and suggested intriguing biological hypotheses in applications to cancer genomics, which we now discuss in more detail.
There are 10 PSG genes (numbered 1 to 9 and 11) and one pseudogene (PSG10) which are localized on a contiguous piece of DNA on chromosome 13.1–13.3. Their DNA sequences are related and so the transcription of one gene may cross-hybridize to other PSG genes. Because of this it has been difficult to know whether one or more of the PSG genes function in the same or similar ways. It will take better antibody reagents to distinguish between these gene products to elucidate their functions. The PSG proteins are produced in small amounts shortly after fertilization and as the placenta forms these glycoproteins are made and secreted by the trophoblasts whose origin is the embryo. The concentration of the PSGs in the blood stream increases to a maximum by the third trimester. The available evidence is that at least some of the PSGs are involved in immunosuppression of the mother’s CD-8 T cells, preventing a rejection of the fetus because of the allo-antigens expressed by the fetus (22). The PSGs act upon monocytes, resulting in the secretion of TGF-, IL-10, and IL-6. The TGF- and IL-10 induce FOX-P3 positive T-reg cells, which help to mediate immunosuppression during pregnancy and possibly some cancers (23, 24). In mice that are pregnant the administration of antibodies directed against two PSGs, alpha-2 and beta-1, results in spontaneous abortion of the embryos (25). Rousseaux et al. (26) have shown that adenocarcinomas of non–small-cell lung cancers that express PSGs are commonly very aggressive and metastatic. This is consistent with the observations in Fig. 3 demonstrating that non–small-cell lung cancer adenocarcinomas that express the PSGs have poorer overall survivals than those that do not express the PSGs. Very similar results are observed with breast, colon, uterus, and lung cancers.
Thus there is growing evidence to support the hypothesis that the PSGs initiate the production of CD-4 T-regs containing CTLA-4 and are utilized during pregnancy to effect an immunosuppressive state that prevents the fetus and embryo from being rejected and that this same mechanism is utilized to help prevent the immune system from rejecting tumors that also harbor foreign neoantigens by virtue of the mutations found in cancers. If these ideas are correct, the antibodies directed against the PSGs in some patients with cancer whose tumors secrete PSGs may well act like CTLA-4 or PD-1 and help to reject the tumor or at least extend the lifespan of the patient. If the mechanism by which the PSGs function to immunosuppress patients with cancer from attacking their tumors is solely through the production of CTLA-4 expressing T-regs (23, 24), then anti–CTLA-4 antibodies would be expected to function to reverse immunosuppression in PSG-expressing tumors. Unlike anti–CTLA-4 antibodies, one might anticipate that antibodies directed against the PSGs might not result in autoimmunity, because the PSGs are not expressed normally, so turning them off in a tumor should have little impact upon normal tissue. It is well known that the pregnant mother is immunosuppressed, and if she has an autoimmune disease (rheumatoid arthritis, lupus, multiple sclerosis, etc.), it is often less severe during pregnancy. If that is the case, perhaps one could treat the patient (these autoimmune diseases occur in females more often than males) with soluble PSGs, which could in turn moderate the autoimmune disease. Cancers may utilize normal physiological processes to escape immune surveillance and repurpose the PSGs to evade the immune system.
Supplementary Material
Acknowledgments
This study was supported by Air Force Office of Scientific Research Grant FA9550-17-1-0435, a grant from the National Institutes of Health (R01-AG048769), Memorial Sloan Kettering Cancer Center Support Grant/Core Grant P30 CA008748, and a grant from Breast Cancer Research Foundation (BCRF-17-193).
Footnotes
Competing interest statement: J.O.D. is a shareholder in PAIGE.AI. The authors have applied for patent protection related to this publication.
Data deposition: Code for this paper is available in GitHub at https://github.com/MSK-MedPhys-DeasyLab/functional-network-analysis.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002179117/-/DCSupplemental.
References
- 1.Chen Y., Georgiou T. T., Tannenbaum A., Optimal transport for Gaussian mixture models. IEEE Access 7, 6269–6278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arjovsky M., Chintala S., Bottou L., “Wasserstein generative adversarial networks” in International Conference on Machine Learning, Precup D., Teh Y. W., Eds. (Machinery, New York, NY, 2017), pp. 214–223. [Google Scholar]
- 3.McLachlan G., Peel D., Finite Mixture Models (John Wiley & Sons, 2004). [Google Scholar]
- 4.Almeida L. G., et al. , CTdatabase: A knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 37, D816–D819 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albert R., Barabási A., Statistical mechanics of complex networks. Rev. Mod. Phys. 74, 47–97 (2002). [Google Scholar]
- 6.Yang L., et al. , Improving the efficiency and effectiveness of community detection via prior-induced equivalent super-network. Sci. Rep. 7, 634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley N., Kwitt R., Niethammer M., Mucha P. J., Compressing networks with super nodes. Sci. Rep. 8, 10892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besta M., Hoefler T., Survey and taxonomy of lossless graph compression and space-efficient graph representations. arXiv:1806.01799 (5 June 2018).
- 9.Villani C., Topics in Optimal Transportation (American Mathematical Society, 2003), vol. 58. [Google Scholar]
- 10.Villani C., Optimal Transport: Old and New (Springer, 2008), vol. 338. [Google Scholar]
- 11.Chung F. R. K., Graham F. C., Spectral Graph Theory (CBMS Regional Conference Series, Conference Board of the Mathematical Sciences, 1997). [Google Scholar]
- 12.Georgiou T. T., Michailovich O., Rathi Y., Malcolm J., Tannenbaum A., Distribution metrics and image segmentation. Lin. Algebra Appl. 425, 663–672 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urbanek S., Rubner Y., emdist: Earth mover’s distance. https://cran.r-project.org/web/packages/emdist/. Accessed 12 December 2018.
- 14.Scrucca L., Fop M., Murphy T. B., Raftery A. E., Mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 8, 205–233 (2016). [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstein J. N., Collisson E. A., Mills E., The Cancer Genome Atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E., Gao J., Dogrusoz U., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broad Institute, TCGA genome data analysis center. https://gdac.broadinstitute.org/. Accessed 5 November 2018.
- 18.Adamic L. A., Glance N., “The political blogosphere and the 2004 U.S. election: Divided they blog” in Proceedings of the 3rd International Workshop on Link Discovery, LinkKDD ’05, Grossman R., Ed. (ACM, New York, NY, 2005), pp. 36–43. [Google Scholar]
- 19.Clauset A., Newman M. E. J., Moore C., Finding community structure in very large networks. arXiv:cond-mat/0408187 (30 August 2004). [DOI] [PubMed]
- 20.Lancichinetti A., Fortunato S., Community detection algorithms: A comparative analysis. Phys. Rev. 80, 056117 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Gregory S., Finding overlapping communities in networks by label propagation. New J. Physics 12, 103018 (2010). [Google Scholar]
- 22.Moore T., Dveksler G. S., Pregnancy-specific glycoproteins: Complex gene families regulating maternal-fetal interactions. Int. J. Dev. Biol. 58, 273–280 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Snyder S. K., et al. , Pregnancy-specific glycoproteins function as immunomodulators by inducing secretion of IL-10, IL-6 and TGF-beta1 by human monocytes. Am. J. Reprod. Immunol. 45, 205–216 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Jones K., et al. , PSG9 stimulates increase in FoxP3+ regulatory T-cells through the TGF-1 pathway. PloS One 11, e0158050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hau J., Gidley-Baird A. A., Westergaard J. G., Teisner B., The effect on pregnancy of intrauterine administration of antibodies against two pregnancy-associated murine proteins: Murine pregnancy-specific beta 1-glycoprotein and murine pregnancy-associated alpha 2-glycoprotein. Biomed. Biochim. Acta 44, 1255–1259 (1985). [PubMed] [Google Scholar]
- 26.Rousseaux S., Debernardi A., Jacquiau B., Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 5, 186ra66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression data used in this study are publicly available from the TCGA (15) (https://www.cancer.gov/tcga) via cBioPortal (16). Cluster analysis published via the Gene Data Analysis Center (GDAC) Firehose (17) was used. Code for this paper is available on the public repository https://github.com/MSK-MedPhys-DeasyLab/functional-network-analysis.