Significance
The assembly and function of membrane proteins depend on the lipid milieu. In recent years the protein components of the mitochondrial calcium uniporter have been identified, but its specific phospholipid requirements are not known. Utilizing yeast mutants defective in their ability to synthesize different phospholipids, we identify a specific requirement of cardiolipin (CL) in the stability and function of the mitochondrial uniporter. Our findings are translatable to higher organisms because endogenous uniporter abundance is decreased in patient-derived cells and cardiac tissue from Barth syndrome, an inherited deficiency in CL levels. This work shows that yeast phospholipid mutants can be leveraged to uncover specific lipid requirements of membrane proteins and suggests impaired mitochondrial calcium signaling in the pathogenesis of Barth syndrome.
Keywords: mitochondrial calcium uniporter (MCU), cardiolipin, Barth syndrome, EMRE, uniplex
Abstract
Calcium uptake by the mitochondrial calcium uniporter coordinates cytosolic signaling events with mitochondrial bioenergetics. During the past decade all protein components of the mitochondrial calcium uniporter have been identified, including MCU, the pore-forming subunit. However, the specific lipid requirements, if any, for the function and formation of this channel complex are currently not known. Here we utilize yeast, which lacks the mitochondrial calcium uniporter, as a model system to address this problem. We use heterologous expression to functionally reconstitute human uniporter machinery both in wild-type yeast as well as in mutants defective in the biosynthesis of phosphatidylethanolamine, phosphatidylcholine, or cardiolipin (CL). We uncover a specific requirement of CL for in vivo reconstituted MCU stability and activity. The CL requirement of MCU is evolutionarily conserved with loss of CL triggering rapid turnover of MCU homologs and impaired calcium transport. Furthermore, we observe reduced abundance and activity of endogenous MCU in mammalian cellular models of Barth syndrome, which is characterized by a partial loss of CL. MCU abundance is also decreased in the cardiac tissue of Barth syndrome patients. Our work raises the hypothesis that impaired mitochondrial calcium transport contributes to the pathogenesis of Barth syndrome, and more generally, showcases the utility of yeast phospholipid mutants in dissecting the phospholipid requirements of ion channel complexes.
The mitochondrial calcium uniporter is a highly selective calcium (Ca2+) channel complex present in the inner mitochondrial membrane (IMM) and is distributed widely across eukaryotic life with lineage specific losses (1–3). In response to physiological cues, the uniporter enables mitochondria to take up large amounts of Ca2+ from the cytoplasm, which contributes to the clearance of cytosolic Ca2+ transients and activates mitochondrial matrix dehydrogenases (4, 5). Although the physiological processes associated with mitochondrial Ca2+ uptake has been known for over six decades, the molecular identity of the uniporter has only been revealed in the past decade. The uniporter holocomplex is composed of the pore-forming subunit, MCU and small transmembrane subunit, EMRE, which are necessary and sufficient for Ca2+ transport, as well as the regulatory subunits MICU1, MICU2, and MCUb (6–9).
Localization of the uniporter holocomplex in the IMM predicts that its function is likely influenced by the phospholipid milieu. Previous studies have shown that the function and assembly of a number of IMM protein complexes, including the mitochondrial respiratory chain supercomplexes and the ADP/ATP translocase requires cardiolipin (CL), the signature phospholipid of mitochondria (10–12). More recent studies have identified the critical requirements of phosphatidylethanolamine (PE) for the catalytic activities of the respiratory complexes and phosphatidylcholine (PC) for the stabilization of the IMM protein translocase Tim23 (13–15). The structural evidence for the role of phospholipids in MCU function comes from a recent cryo-electron microscopy (EM) study that identified four lipid molecules on the membrane exposed surfaces of MCU, the pore-forming subunit (16). However, the molecular identities and the roles of these lipids in MCU function were not determined.
Deciphering the phospholipid requirements of the uniporter is challenging in part because we currently lack a robust in vitro biochemical reconstitution system in which to investigate the channel complex. Here, we have utilized yeast, Saccharomyces cerevisiae, as a facile, in vivo genetic system to address this problem. Our choice of yeast was guided by the following three considerations. First, the phospholipid composition of the IMM is highly conserved from yeast to mammals, with PC, PE, and CL being the three most abundant phospholipids (17). Second, the phospholipid composition of the IMM can be genetically and nutritionally manipulated in yeast without disrupting the gross structure of mitochondria (13). Third, S. cerevisiae do not contain any homologs of uniporter machinery (3), and previous studies have shown that it is possible to functionally reconstitute uniporter activity in yeast through heterologous expression of the uniporter machinery (18). Thus, yeast represents a suitable in vivo reconstitution system for dissecting the phospholipid requirements of the nonnative uniporter in a physiologically relevant mitochondrial membrane milieu (18, 19). In this study, we engineered isogenic yeast mutants with defined perturbations in the levels of PC, PE, and CL to uncover a CL-specific requirement for MCU stability and function. We validated the evolutionarily conserved requirement of CL for MCU in a mouse muscle cell line and in CL-depleted cardiac tissue of Barth syndrome (BTHS) patients.
Results
A Yeast System to Determine Specific Phospholipid Requirements of MCU.
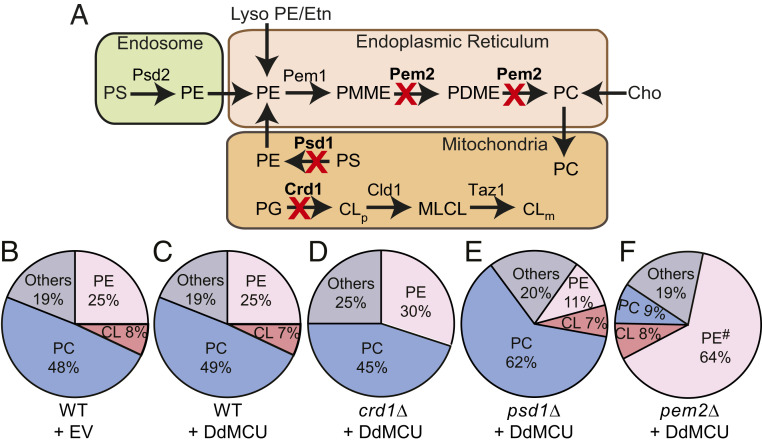
To test the specific phospholipid requirement of MCU, we utilized isogenic yeast mutants of CL, PE, and PC biosynthetic pathways with deletions in CRD1, PSD1, and PEM2, respectively (Fig. 1A). We focused on these three abundant mitochondrial phospholipids because together they form ∼80–90% of the total mitochondrial phospholipidome (Fig. 1B) (17) and are thus expected to influence membrane protein function. Previous work has shown that heterologous expression of membrane proteins in yeast can itself perturb the phospholipid composition (20), which can confound our study. Therefore, we first tested the effect of expressing MCU on the phospholipid composition of WT yeast cells. Heterologous expression of Dictyostelium discoideum MCU (DdMCU), an MCU homolog that can conduct Ca2+ independent of any other protein, did not alter the mitochondrial phospholipid composition (Fig. 1 B and C and SI Appendix, Fig. S1). We then expressed DdMCU in crd1Δ, psd1Δ, and pem2Δ cells that are characterized by a complete absence of CL, ∼2.5-fold decrease in PE, and a ∼5-fold decrease in PC, respectively (Fig. 1 D–F and SI Appendix, Fig. S1). These mutants are viable and exhibit normal growth in galactose-containing media, which allowed us to dissect the specific requirement of phospholipids for MCU function.
Fig. 1.
Yeast mutants with defined perturbations in mitochondrial phospholipid composition. (A) A schematic of yeast biosynthetic pathways of the most abundant mitochondrial phospholipids CL, PE, and PC. The enzymes for CL, PE, and PC biosynthesis that are deleted for phospholipid perturbation are highlighted in bold. Cho, choline; CL, cardiolipin; CLm, mature CL; CLp, premature CL; Etn, ethanolamine; lysoPE, lysophosphatidylethanolamine; MLCL, monolysocardiolipin; PC, phosphatidylcholine; PDME, phosphatidyldimethylethanolamine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PMME, phosphatidylmonomethylethanolamine; PS, phosphatidylserine. (B–F) Pie-charts depicting levels of major mitochondrial phospholipids, including CL, PE, and PC, are expressed as % of total phospholipid phosphorus in mitochondria isolated from WT cells expressing EV (empty vector) (B), and WT (C), crd1Δ (D), psd1Δ (E), and pem2Δ (F) expressing DdMCU, respectively. Data shown as mean from three independent experiments. Expanded data on levels of each mitochondrial phospholipid are shown in SI Appendix, Fig. S1. # represents sum of PE and PMME.
MCU Stability and Abundance Are Reduced in CL-Deficient Mitochondria.
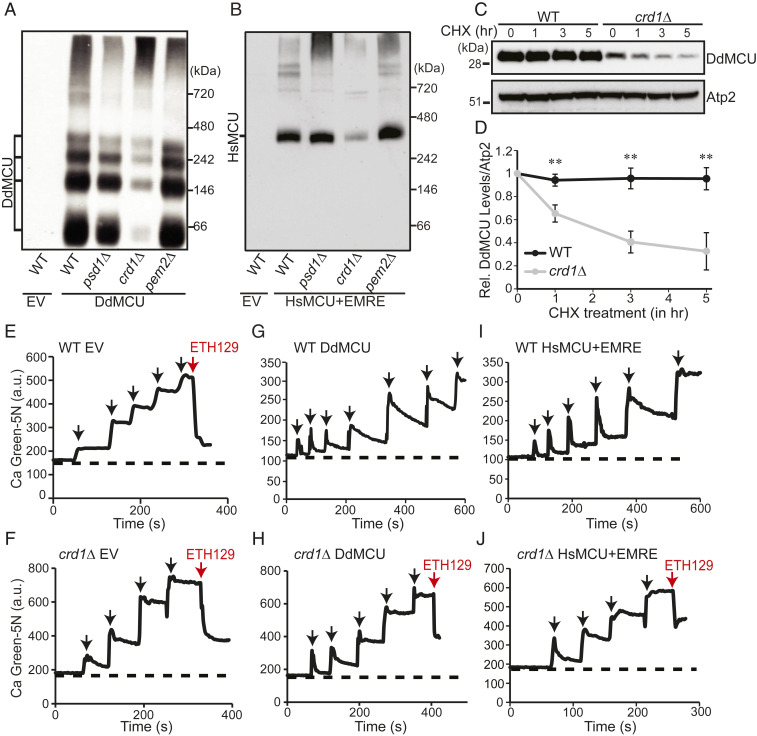
We next sought to determine how perturbations in these phospholipids impact the steady-state levels of DdMCU. Consistent with a previous report (18), we found that DdMCU is expressed as a mature ∼32-kDa protein in yeast mitochondria (SI Appendix, Fig. S2A). Notably, DdMCU levels in CL-deficient crd1Δ mitochondria were reduced by ∼50% (SI Appendix, Fig. S2 A and B), an effect not seen in PE- and PC-depleted psd1Δ and pem2Δ mitochondria, respectively (SI Appendix, Fig. S2 A and B). DdMCU monomers form higher-order oligomers when reconstituted in yeast (18). Consistent with the decrease in DdMCU monomers, Blue Native-polyacrylamide gel electrophoresis (BN-PAGE) immunoblot experiments showed a pronounced decrease in the abundance of DdMCU oligomeric complexes in crd1Δ mitochondria (Fig. 2A). In contrast, depletion of PE and PC did not impact the oligomeric assembly of DdMCU in psd1Δ and pem2Δ cells, respectively (Fig. 2A).
Fig. 2.
MCU stability and activity are reduced in CL-deficient yeast. (A) BN-PAGE immunoblot analysis of FLAG-tagged DdMCU in digitonin-solubilized mitochondria from the indicated yeast phospholipid mutants expressing empty vector (EV) or DdMCU. Blot is representative of three independent experiments. (B) BN-PAGE immunoblot analysis of FLAG-tagged HsMCU in digitonin-solubilized mitochondria from the indicated yeast phospholipid mutants expressing EV or coexpressing HsMCU and EMRE. Blot is representative of three independent experiments. (C) SDS/PAGE immunoblot analysis of FLAG-tagged DdMCU in cell lysates from WT and crd1Δ yeast treated with protein translation inhibitor CHX for the indicated time. Atp2 is used as loading control. (D) Quantification of relative DdMCU levels from C, normalized to 0 h of CHX treatment, by densitometry using ImageJ software. Data shown as mean ± SD, n = 3. **P < 0.005. (E and F) Representative traces monitoring extramitochondrial Ca2+ levels by Calcium Green-5N following repeated 10 μM CaCl2 additions (black arrows) to mitochondria isolated from WT (E) and crd1Δ cells (F) transformed with empty vector (EV). ETH129 is used as a Ca2+ ionophore (red arrow). (G and H) Representative traces monitoring extramitochondrial Ca2+ levels by Calcium Green-5N following repeated 10 μM CaCl2 additions (black arrows) to mitochondria isolated from DdMCU expressing WT (G) and crd1Δ cells (H). ETH129 is used as a Ca2+ ionophore (red arrow) in H. (I and J) Representative traces monitoring extramitochondrial Ca2+ levels by Calcium Green-5N following repeated 10 μM CaCl2 additions (black arrows) to mitochondria isolated from HsMCU and EMRE expressing WT (I) and crd1Δ cells (J). ETH129 is used in J (red arrow). All traces are representative of three independent experiments.
Next, we asked if CL requirement of MCU was evolutionarily conserved. To address this question, we reconstituted the human MCU (HsMCU) in our yeast phospholipid mutants. A previous study aimed at reconstitution of HsMCU in yeast showed that HsMCU alone is not sufficient for Ca2+ uptake and requires coexpression with EMRE—both of which are necessary and sufficient to reconstitute the uniporter activity (18). Therefore, we coexpressed HsMCU and EMRE in the yeast phospholipid mutants and found a specific reduction in monomeric (SI Appendix, Fig. S2 C and D) and oligomeric HsMCU (Fig. 2B) in CL-deficient mitochondria. Notably, the decrease in the oligomeric assemblies of MCU (both DdMCU and HsMCU) is more pronounced compared to its overall abundance, which suggests a critical role of CL for the assembly of higher-order MCU complexes in the IMM.
Given that loss of CL decreased the abundance of HsMCU, we asked if increasing CL would lead to an elevation in HsMCU. We manipulated CL levels by growing WT yeast in different carbon sources. Levels of CL tend to be much higher when yeast is grown in galactose or in lactate compared with glucose as the main carbon source. Under these conditions of increased CL levels, we also observed a concomitant increase in the abundance of HsMCU oligomers as assessed by BN-PAGE/immunoblotting (SI Appendix, Fig. S3). Collectively, our genetic “loss-of-function” and carbon source-based “gain-of-function” experiments perturbing CL levels show a consistent and concordant impact on MCU levels.
Reduced biosynthesis or an increased turnover of MCU could explain a decrease in the steady-state levels of MCU in CL-deficient cells. Because we observed decreased MCU protein levels in yeast where DdMCU was expressed under strong nonnative promoters, we argued that CL deficiency is unlikely to impact the expression of MCU but may affect its stability in the mitochondrial membrane. Therefore, we used a cycloheximide chase assay to test if MCU protein turnover in vivo is dependent on CL levels. DdMCU expressing WT and crd1Δ yeast cells were treated with cycloheximide to inhibit nascent protein translation from cytosolic ribosomes, and samples were collected over time to measure the turnover rate of DdMCU. We observed rapid turnover of DdMCU in CL-deficient crd1Δ cells compared to WT cells, where DdMCU was stable even after 5 h of cycloheximide treatment (Fig. 2 C and D). Thus, the decreased abundance of MCU in CL-deficient cells could be attributed to its reduced stability in the IMM.
Uniporter Activity Is Diminished in Yeast Cardiolipin Mutants.
A decrease in uniporter abundance is expected to cause a reduction in mitochondrial Ca2+ uptake in CL-deficient mitochondria. To test this, we used membrane impermeable Ca2+ indicator Calcium Green-5N (Ca Green-5N), which fluoresces upon binding with free Ca2+, to measure Ca2+ uptake in isolated mitochondria. When pulses of Ca2+ were added to WT yeast mitochondria, which lack endogenous Ca2+ uptake machinery, the Ca Green-5N fluorescence increased with every pulse because exogenously added Ca2+ remained outside of mitochondria and accessible to Ca Green-5N (Fig. 2E). Since mitochondrial integrity and membrane potential (ΔΨm) are crucial for Ca2+ uptake, we tested the intactness of these parameters by using the Ca2+ ionophore ETH129, which transports Ca2+ across the intact mitochondrial membrane in a ΔΨm-dependent manner (18). Experiments with ETH129 confirmed that these parameters were intact in our experimental system (Fig. 2 E and F), although, we did notice that the ETH129-mediated Ca2+ buffering capacity of crd1Δ mitochondria is slightly lower than WT mitochondria (Fig. 2 E and F). We further confirmed the requirement of ΔΨm for mitochondrial Ca2+ uptake by using an uncoupler (CCCP) or a respiratory chain inhibitor (antimycin), both of which abolished Ca2+ uptake (SI Appendix, Fig. S4 A and B). After validating our assay, we measured mitochondrial Ca2+ uptake in DdMCU expressing WT and crd1Δ mitochondria. WT yeast mitochondria expressing DdMCU were able to take up multiple pulses of 10 μM Ca2+, thereby rendering the added Ca2+ inaccessible to the membrane impermeable Ca Green-5N and lowering its fluorescence signal (Fig. 2G). In contrast, crd1Δ mitochondria expressing DdMCU could uptake only two pulses of 10 μM Ca2+, although it maintained sufficient ΔΨm to allow ETH129-mediated Ca2+ uptake (Fig. 2H). Consistent with these results, we observed a marked decrease in mitochondrial Ca2+ uptake in crd1Δ mitochondria when we coexpressed HsMCU and EMRE, despite the fact that ETH129-mediated Ca2+ uptake remained partially preserved (Fig. 2 I and J). Quantitative analysis showed reduced specific activity of HsMCU Ca2+ uptake kinetics in CL-deficient mitochondria (SI Appendix, Fig. S5), suggesting that lack of CL impairs mitochondrial Ca2+ uptake by decreasing both MCU abundance and activity. Taken together, these results demonstrate an evolutionarily conserved role of CL in MCU function.
MCU Function Is Impaired in a CL-Depleted Yeast Model of BTHS.
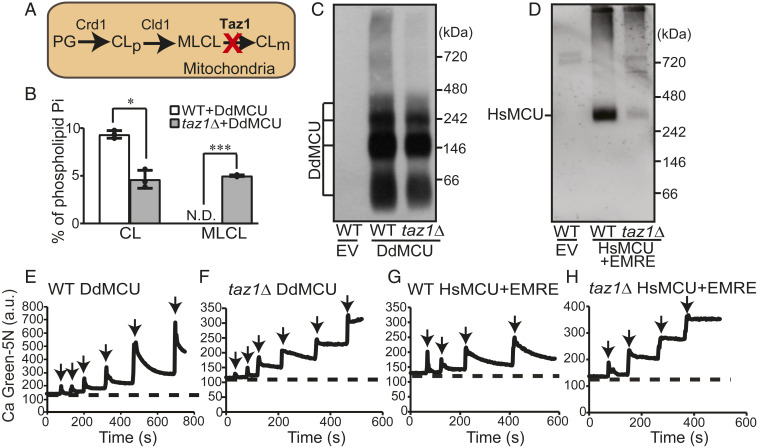
We next asked whether a partial loss of mitochondrial CL, as observed in BTHS patients and disease models, may perturb MCU abundance and activity. BTHS is a rare genetic disorder characterized by loss-of-function mutations in a conserved CL-remodeling enzyme Taz1, referred to as TAZ or Tafazzin in mammals. Loss of Taz1 leads to a reduction in the levels of CL and an accumulation of its precursor, monolysocardiolipin (MLCL) (Fig. 3A). We used yeast model of BTHS, taz1Δ cells, to determine if MCU function was compromised in yeast cells that are partially depleted in CL. We validated taz1Δ cells by measuring its mitochondrial phospholipid composition, which showed an expected ∼50% reduction in CL with a concomitant increase in MLCL (Fig. 3B). Consistent with our results in CL-deficient crd1Δ cells, we observed a modest decrease in the steady-state levels of DdMCU (SI Appendix, Fig. S6 A and B) and HsMCU, which was expressed along with EMRE, (SI Appendix, Fig. S6 C and D) in taz1Δ mitochondria. The oligomeric assemblies of both DdMCU and HsMCU/EMRE were also reduced in taz1Δ mitochondria, with the reduction being more pronounced for HsMCU (Fig. 3 C and D). As shown previously (21, 22), we find that a complete or partial loss of CL in crd1Δ and taz1Δ cells, respectively, does not lead to a universal decrease in the levels of IMM proteins or their supramolecular assemblies (SI Appendix, Fig. S7 A and B). In fact, none of the IMM protein levels were decreased in taz1Δ cells and levels of the respiratory chain subunits (Sdh1 and Atp2) and the protein import machinery (Tim44 and Tom70) were not reduced in crd1Δ cells either when these cells were cultured in galactose-containing media (SI Appendix, Fig. S7A). Thus, a decrease in the levels of DdMCU and HsMCU in crd1Δ and taz1Δ cells highlights their specific requirement of CL. In accordance with the reduced abundance of DdMCU and HsMCU in taz1Δ mitochondria, the mitochondrial Ca2+ uptake was also diminished in DdMCU (Fig. 3 E and F) and HsMCU/EMRE (Fig. 3 G and H) expressing taz1Δ mitochondria. Again, quantitative analysis of mitochondrial Ca2+ uptake kinetics showed reduced Ca2+ uptake rate by HsMCU in CL-depleted taz1Δ mitochondria (SI Appendix, Fig. S8). These results suggest that a partial loss of CL in taz1Δ mutant impairs MCU abundance, assembly, and activity.
Fig. 3.
MCU abundance and activity are reduced in taz1Δ cells, a yeast model of BTHS. (A) A schematic of CL biosynthetic and remodeling pathway in yeast. (B) CL and MLCL levels in mitochondria isolated from WT and taz1Δ cells expressing DdMCU. Data are shown as mean ± SD, n = 3. *P < 0.05; ***P < 0.0005. N.D., not detected. (C) BN-PAGE immunoblot analysis of FLAG-tagged DdMCU in digitonin-solubilized mitochondria from WT and taz1Δ cells expressing empty vector (EV) or DdMCU. Blot is representative of three independent experiments. (D) BN-PAGE immunoblot analysis of FLAG-tagged HsMCU in digitonin-solubilized mitochondria from WT and taz1Δ cells expressing EV or coexpressing HsMCU and EMRE. Blot is representative of three independent experiments. (E–H) Representative traces monitoring extramitochondrial Ca2+ levels with Calcium Green-5N following repeated 10 µM CaCl2 additions (black arrows) to mitochondria isolated from WT and taz1Δ cells expressing either DdMCU (E and F) or HsMCU and EMRE (G and H). All traces are representative of three independent experiments. CL, cardiolipin; CLm, mature CL; CLp, premature CL; MLCL, monolysocardiolipin; PG, phosphatidylglycerol.
MCU Function Is Disrupted in C2C12 Murine Myoblast Model of BTHS.
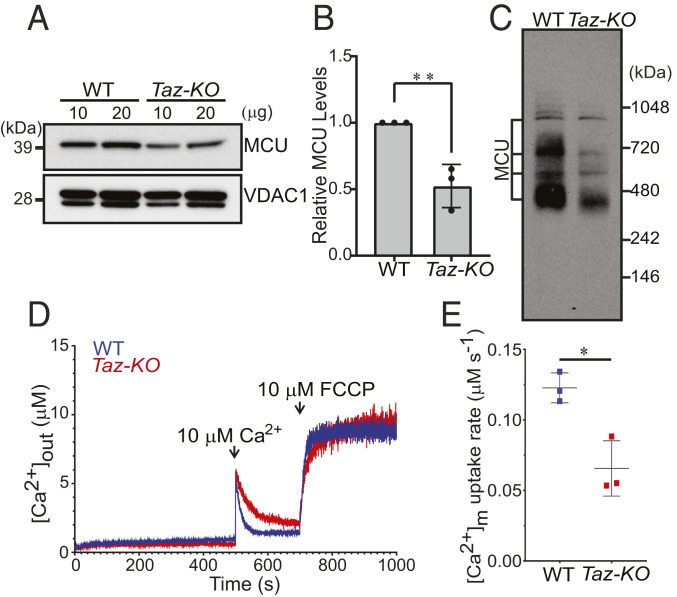
We wondered whether our findings with nonnative MCU expressed in yeast CL-deficient and CL-depleted mutants are applicable to endogenous MCU in mammalian cells. To this end, we utilized a Taz-KO C2C12 murine myoblast cell line, which has recently been developed as a murine skeletal muscle model of BTHS, a disease characterized by skeletal and cardiomyopathy (23). An expected decline in the levels of CL with a concomitant elevation in MLCL validated the Taz-KO C2C12 cells as a disease model of BTHS (SI Appendix, Fig. S9). Using this model of BTHS, we found that the steady-state levels of monomeric MCU and MCU-containing uniporter complexes were significantly reduced in Taz-KO cells when compared to WT (Fig. 4 A–C). A cycloheximide chase assay in Taz-KO C2C12 cells showed an increase in the turnover of endogenous MCU in Taz-KO cells compared to WT (SI Appendix, Fig. S10), suggesting that depletion of unsaturated CL or accumulation of MLCL reduces the stability of endogenous MCU.
Fig. 4.
Endogenous MCU abundance and activity are reduced in Taz-KO C2C12 myoblasts. (A) SDS/PAGE immunoblot analysis of MCU in mitochondria isolated from WT and Taz-KO C2C12 myoblasts. VDAC1 is used as a loading control. (B) Quantification of relative MCU levels from A by densitometry using ImageJ software. Data shown as mean ± SD n = 3. **P < 0.005. (C) BN-PAGE immunoblot analysis of MCU in digitonin-solubilized mitochondria isolated from WT and Taz-KO C2C12 myoblasts. Blot is representative of three independent experiments. (D) Permeabilized WT and Taz-KO cells were pulsed with 10 μM Ca2+ followed by 10 μM FCCP as indicated by arrows. Traces show bath [Ca2+] (μM). Traces are representative of three independent experiments. (E) MCU-mediated mitochondrial Ca2+ uptake rate calculated from D. Data shown as mean ± SEM, n = 3. *P < 0.05.
Next, we utilized permeabilized WT and Taz-KO C2C12 cells to test the role of CL on MCU-mediated mitochondrial Ca2+ uptake. After the permeabilized cells had reached a steady-state ΔΨm, we applied a 10 µM Ca2+ pulse and monitored the reduction in extramitochondrial Ca2+ ([Ca2+]out). Compared to WT, Taz-KO cells exhibited a delayed clearance of [Ca2+]out, indicating compromised mitochondrial Ca2+ uptake (Fig. 4 D and E). Collectively, these results reveal that optimal levels of CL are essential for maintaining endogenous MCU abundance and activity in the mouse muscle cell line model of BTHS.
MCU Abundance Is Reduced in BTHS Patient-Derived Cells and Cardiac Tissues.
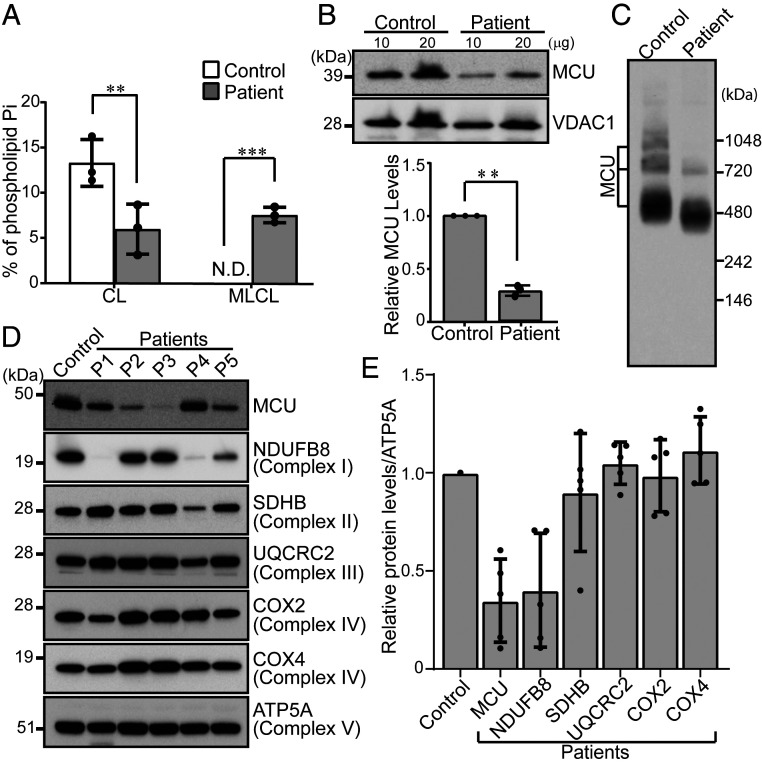
To test if our findings from yeast and C2C12 model systems of BTHS hold true in BTHS patient-derived cells and cardiac tissues, we procured a BTHS patient B-lymphocyte cell line and BTHS patient heart tissue samples. Phospholipid analysis of BTHS patient B-lymphocyte mitochondria showed reduced CL and elevated MLCL levels, validating the BTHS patient cell line (Fig. 5A). Immunoblot analysis revealed a ∼70% decrease in the steady-state levels of MCU in BTHS B-lymphocyte cells as compared to the control (Fig. 5B). We also observed a reduction in assembly of the higher-order, MCU-containing uniporter complexes in the patient cells (Fig. 5C). Consistent with our in vitro data, we observed a reduction in MCU abundance in cardiac tissues obtained from five different BTHS patients, with MCU being reduced to ∼40–90% as compared to the control sample (Fig. 5 D and E). Unlike MCU, other IMM proteins of the respiratory chain did not show reduced abundance, except for respiratory complex I subunit NDUFB8 (Fig. 5 D and E). These data are consistent with our yeast model of BTHS (SI Appendix, Fig. S7A) and further demonstrate a specific requirement of CL for MCU abundance.
Fig. 5.
MCU is reduced in BTHS patient-derived lymphocyte and cardiac tissues. (A) CL and MLCL levels in mitochondria isolated from control and BTHS patient B-lymphocyte. Data shown as mean ± SD, n = 3. **P < 0.005. ***P < 0.0005. N.D., not detected. (B, Upper) SDS/PAGE immunoblot analysis of MCU in mitochondria isolated from control and BTHS patient B-lymphocyte. VDAC1 is used as a loading control. (B, Lower) Quantification of relative MCU levels using ImageJ software. Data shown as mean ± SD, n = 3. **P < 0.005. (C) BN-PAGE immunoblot analysis of MCU in digitonin-solubilized mitochondria from control and BTHS patient B-lymphocyte. Blot is representative of three independent experiments. (D) SDS/PAGE immunoblot analysis of MCU and mitochondrial respiratory chain (MRC) complex subunits in cardiac tissue lysates obtained from control and five BTHS patients (P1–P5). ATP5A is used as a loading control. (E) Quantification of immunoblots from D using ImageJ software. Data shown as mean ± SD, n = 5. CL, cardiolipin; MLCL, monolysocardiolipin.
Discussion
The assembly and activity of integral membrane proteins often depend on the surrounding membrane phospholipid composition (24, 25). Recent studies using yeast phospholipid mutants have uncovered some unexpected findings regarding the roles of individual mitochondrial membrane phospholipids in the function and formation of IMM proteins. Studies from yeast phospholipid mutants show that most mitochondrial membrane proteins are able to tolerate a large perturbation in membrane phospholipid composition (13, 15). However, some IMM proteins specifically rely on a particular class of phospholipid for their protein:protein interactions, higher-order assembly, and activity (12, 13, 26). Here, we adapted a yeast heterologous expression system to uncover a CL-specific requirement for the stability and function of MCU. We demonstrate that the CL-specific requirement of MCU is conserved with a partial loss of CL reducing the stability of endogenous MCU in BTHS models and BTHS patient cardiac tissue, a finding that links MCU to BTHS disease pathology.
We attribute the decreased abundance of MCU in CL-deficient cells to its increased turnover (Fig. 2 C and D). We also considered the possibility that decreased MCU abundance could be due to the reduced mitochondrial membrane potential and protein import observed in CL-deficient yeast (27). However, this is unlikely because MCU levels were not decreased in PE-depleted psd1Δ cells (Fig. 2 A and B and SI Appendix, Fig. S2) that also exhibit similarly reduced mitochondrial membrane potential and protein import (26). Moreover, we have shown that crd1Δ mitochondria could uptake Ca2+ via the membrane potential-dependent Ca2+-ionophore ETH129 (Fig. 2 F, H, and J), indicating that these CL-deficient mitochondria maintain sufficient mitochondrial membrane potential to support Ca2+ uptake.
Prior studies have typically used in vitro patch-clamp approaches to identify regulators of ion channels. For example, phosphatidylinositides were shown to regulate the activity of Ca2+ ion channel, TRPML1, in an intracellular compartment (28). Similarly, M-type K+ channel activity was shown to be regulated by phosphatidylinositides as well as a wide range of phospholipids (29). Here, we provide a complementary in vivo approach to discover physiologically relevant phospholipid regulators of mitochondrial membrane localized ion channels. Our in vivo approach utilizing yeast phospholipid mutants provides information regarding the structural requirements of phospholipids in addition to their functional role, as seen in the case of MCU, where we found a specific requirement of CL for the stability of MCU (Fig. 2 A–D). Recently, it was shown that peripheral membrane proteins MICU1 and MICU2, which are two regulatory subunits of the uniporter, bind CL in vitro (30). This observation, combined with our finding, provides compelling biochemical and genetic evidence that CL plays a critical role in uniporter holocomplex formation and function. A recent cryo-EM study of a fungal MCU channel identified the presence of four acyl chains within the transmembrane helices of MCU that form lateral membrane openings, with basic residues on legs of the channel poised to interact with phospholipid head groups (16).
During the revision of this article, a new high-resolution cryo-EM structure of the human uniporter complex has been reported on a preprint server (31). The structure reports 8 CL and 16 PC molecules in the heterooctameric uniporter complex such that one CL and two PC molecules associate with each MCU subunit. The cavity formed by the two transmembrane helices of MCU is filled by these phospholipids, which the authors speculate may provide stability to the uniporter structure. However, in their report, functional studies were not included to establish the physiological roles of CL and PC. Our current in vivo work showing the requirement of CL for the stability of MCU (Figs. 2–5) strongly supports its structural role. Of note, our work also shows that MCU stability is not impacted by depletion of PC (Fig. 2 A and B), which may be replaceable with other phospholipids that accumulate in pem2Δ cells (SI Appendix, Fig. S1). Interestingly, the cryo-EM structure of the human uniporter complex also shows that acyl chain of CL interacts with EMRE, raising the possibility that the stabilizing effect of CL on MCU may be through its interactions with EMRE. However, since we find that CL is also required for the stability of DdMCU (Fig. 2A), which does not contain EMRE, it is very likely that it is the direct CL–MCU interactions that provide stability to MCU. Additionally, this study by Zhuo et al. (31) showed that Arg297 residue of MCU forms hydrogen bonds with the phosphate moiety of CL as well as with the N-terminal domain of EMRE, and its mutation to Asp abolished Ca2+ uptake. Our functional and biochemical studies perturbing CL are highly complementary to this structural observation and jointly support a specific role of CL in the stability and activity of MCU.
CL interacts strongly with many integral membrane proteins of the IMM and increases their stability (32). A recent study has revealed that by virtue of remodeling its acyl chain composition, CL relieves the elastic stress imposed on the mitochondrial membrane due to protein crowding. In doing so, CL allows the formation of stable lipid:protein complexes resulting in the normal functioning of mitochondrial membrane proteins (33). Our finding that loss of CL increases the turnover of MCU (Fig. 2 C and D and SI Appendix, Fig. S10) points to an underlying biochemical mechanism of how CL impacts the stability of mitochondrial membrane protein complexes and could potentially be extrapolated to other key membrane proteins of mitochondria known to be interacting with CL.
CL requirement of MCU is particularly relevant to BTHS, a rare genetic disorder caused by mutations in the CL-remodeling enzyme, TAZ (34, 35). The clinical presentations in BTHS patients and humans with loss-of-function mutations in MICU1 as well as the phenotypes observed in Mcu−/− or Micu2−/− mice show some overlapping features. For example, BTHS patients classically present with cardiomyopathy, skeletal muscle myopathy, proximal muscle myopathy, exercise intolerance, and fatigue (36). Loss-of-function mutations in MICU1 also result in similar clinical presentations in humans (37, 38). Mcu−/− and Micu2−/− mice exhibit exercise intolerance and cardiovascular phenotypes, respectively (39, 40). Together, these studies suggest that some aspects of BTHS pathology may be attributable to defective mitochondrial Ca2+ signaling via the uniporter.
Materials and Methods
Yeast Strains and Growth Conditions.
S. cerevisiae strains used in this study were obtained from Open Biosystems. All strains were isogenic to BY4741 WT and their authenticity was confirmed by PCR, replica plating on dropout plates, and phospholipid analyses. Yeast cells were precultured in standard growth media including YPD (1% yeast extract, 2% peptone and 2% dextrose) or YPGal (2% galactose). The final cultures were grown in synthetic complete (SC) medium, which contained 0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, 0.2% dropout mix containing amino acids, and 2% galactose (SC-galactose). Cells expressing either DdMCU or HsMCU and EMRE were grown in SC-galactose leucine drop-out or SC-galactose leucine/uracil double drop-out media, respectively, to resist plasmid curing. Growth in liquid media was measured spectrophotometrically at 600 nm. Final cultures were started at an optical density of 0.1 and were grown to late logarithmic phase at 30 °C.
Mammalian Cell Culture and BTHS Heart Tissue Samples.
The control (ND11500) and BTHS patient (GM22194) B-lymphocytes were obtained from the Coriell Institute for Medical Research. These cells were cultured in high glucose Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 15% fetal bovine serum (FBS) (Sigma). C2C12 WT and Taz-KO myoblasts were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. All cell lines were cultured in 5% CO2 at 37 °C. Deidentified BTHS patient heart samples were obtained from Barth Syndrome Registry and DNA Bank. Whole-cell protein was extracted in lysis buffer (BP-115, Boston BioProducts) supplemented with protease inhibitor mixture (cOmplete Mini EDTA-free; Roche Diagnostics), and the protein concentrations were determined by bicinchoninic acid assay (Pierce BCA Protein Assay).
Mitochondria Isolation.
Isolation of crude mitochondria from yeast cells was performed as previously described (41). Mitochondrial fractions were resuspended in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM Mops-KOH, pH 7.2) and stored at −80 °C for protein biochemistry experiments. Fresh mitochondrial isolates were used for calcium uptake assays. Mitochondria from mammalian cells were isolated using the Mitochondria Isolation Kit from Abcam (110170; Abcam). Protein concentrations were determined by BCA assay (Pierce BCA Protein Assay).
SDS/PAGE, BN-PAGE, and Immunoblotting.
Sodium dodecyl sulfate (SDS)/PAGE and BN-PAGE were performed to separate denatured and native protein complexes, respectively. For SDS/PAGE, mitochondrial lysate (10 μg, unless stated otherwise) was separated on NuPAGE 4–12% Bis-Tris gels (Thermo Fisher Scientific) and transferred onto polyvinylidene fluoride (PVDF) membranes using a Trans-Blot SD semidry transfer cell (Bio-Rad). For BN-PAGE, mitochondria were solubilized in buffer containing 1% digitonin (Thermo Fisher Scientific) at 6 g/g of yeast mitochondrial protein or 9 g/g of mammalian mitochondrial protein, followed by incubation for 15 min at 4 °C and centrifugation at 20,000 × g for 30 min. Clear supernatant was collected and 50× G-250 sample additive was added. Twenty micrograms of protein was loaded on 3–12% native PAGE Bis-Tris gel (Thermo Fisher Scientific) and transferred onto PVDF membranes using a wet transfer method. Following transfer, the membranes were probed with the following primary antibodies: For yeast proteins—Flag, 1:500,000 (F1804; Sigma); Pgk1, 1:50,000 (459250; Novex); Por1, 1:50,000 (ab110326; Abcam); Sdh1, 1:10,000; Sdh2, 1:5,000; Cor1, 1:5,000; Cor2, 1:80,000; Rip1, 1:50,000; Cox2, 1:5,000 (ab110271; Abcam); Cox4, 1:5,000 (ab110272; Abcam); Atp2, 1:40,000; Tim44, 1:5,000; Tom70, 1:5,000, and Aco1, 1:5,000. For mammalian proteins—MCU, 1:2,500 (14997S; Cell Signaling Technologies); VDAC1, 1:2,500 (ab14734; Abcam); NDUFB8, 1:2,500 (ab110242; Abcam); SDHB, 1:1,000 (ab14714; Abcam); UQCRC2, 1:2,500 (ab14745; Abcam); COX2, 1:2,500 (ab110258; Abcam); COX4, 1:2,500 (ab14744; Abcam); and ATP5A, 1:2,500 (ab14748; Abcam). Anti-mouse or anti-rabbit secondary antibodies (1:5,000) were incubated for 1 h at room temperature, and membranes were developed using Clarity Western ECL (Bio-Rad Laboratories).
Mitochondrial Phospholipid Measurement.
Phospholipids were extracted from mitochondrial pellets of yeast and mammalian cells (∼1.5 mg of protein) using the Folch method, as previously described (13, 42). Phospholipids were separated by two-dimensional TLC (2D-TLC), as previously described (13), using the following solvent systems: chloroform/methanol/ammonium hydroxide (32.5/17.5/2.5) in the first dimension, followed by chloroform/acetic acid/methanol/water (37.5/12.5/2.5/1.1) in the second dimension. Phospholipids were visualized with iodine vapor and scraped onto glass tubes to quantify phosphorus using the Bartlett assay (43).
Mitochondrial Ca2+ Uptake Measurement in Yeast.
This method was performed as previously described (18). For measuring mitochondrial Ca2+ uptake, freshly isolated yeast mitochondrial pellet was dissolved in resuspension buffer (0.6 M mannitol, 20 mM Hepes-KOH, pH 7.4, 1 mM EGTA, 0.2% [wt/vol] BSA supplemented with 1× protease inhibitor mixture). Mitochondrial protein concentration was measured by BCA assay. Five hundred micrograms of mitochondrial lysate was resuspended in assay buffer (0.6 M mannitol, 10 mM Hepes-KOH, pH 7.4, 10 mM potassium phosphate, 0.5 mg/mL BSA, 3 mM glutamate, 3 mM malate, 3 mM succinate, 10 μM EGTA, and 1 μM Calcium Green-5N, adjusted to pH 7.4 using KOH). Mitochondrial Ca2+ uptake was measured in the Spectramax M2 spectrofluorometer (Molecular Devices), by recording the fluorescence of Calcium Green-5N (Thermo Fisher Scientific) (excitation/emission at 506 nm/532 nm) every 2 s following repeated additions of 10 µM CaCl2 into the assay mix. For Ca2+ uptake rate measurements, 25 μM CaCl2 was added to the assay mix once and Calcium Green-5N fluorescence monitored every 2 s. Ten micromolar ETH129 (21193; Sigma), a Ca2+ ionophore, was used as a control. CCCP and antimycin (Sigma), 1 μM each, were used to uncouple and inhibit mitochondrial respiratory chain, respectively.
Protein Turnover Measurement.
The method was performed as previously described (44). Briefly, yeast cells expressing DdMCU were precultured in YPD at 30 °C overnight. Subsequently, cells were transferred into SC-galactose (leucine drop-out) media and allowed to grow for 6 h, after which cycloheximide (CHX) (C7698; Sigma) was added at a final concentration of 50 μg/mL. Cells (2 × 108) were subsequently collected at 0-, 1-, 3-, and 5-h time intervals, added to stop-solution (20 mM NaN3 and 0.5 mg/mL BSA) and pelleted down. Yeast cells were lysed using an alkaline lysis method. Briefly, cells were sequentially incubated in lysis buffer (0.2 M NaOH) at room temperature, followed by 0.1 M NaOH and LDS sample loading buffer at 70 °C to extract proteins. The lysates were centrifuged to pellet insoluble materials and the supernatant analyzed by gel electrophoresis and immunoblotting. Mammalian MCU turnover in C2C12 cells was measured by SDS PAGE/immunoblotting of protein lysate extracted following 10 μg/mL CHX treatment for 0, 24, 48, and 72 h, respectively.
Mitochondrial Ca2+ Uptake Measurement in Permeabilized Cells.
C2C12 cells (∼4 × 106 cells) were resuspended and permeabilized with digitonin (40 μg/mL) in 1.5 mL of medium composed of 120 mM KCl, 10 mM NaCl, 1 mM KH2PO4, 20 mM Hepes-Tris (pH 7.2), and 2 μM thapsigargin to block the SERCA pump. The experiments were performed in the presence of 5 mM succinate at 37 °C with constant stirring. For measuring mitochondrial Ca2+ uptake, the permeabilized cells were suspended in medium containing 1.0 μM Fura-2FF. Fluorescence was monitored in a multiwavelength excitation, dual-wavelength emission fluorimeter (DeltaRAM, Photon Technology International). Extramitochondrial Ca2+ was recorded at an excitation ratio (340 nm/380 nm) and emission at 510 nm of Fura-2FF fluorescence. Single 10 μM Ca2+ pulse was added at 500 s, and the changes in extramitochondrial Ca2+ fluorescence were monitored. Mitochondrial Ca2+ uptake rate was derived from the decay of bath [Ca2+] after Ca2+ pulses.
Statistical Analyses.
All statistical analyses were performed using two-tailed unpaired Student’s t test using data obtained from three independent experiments.
Data Availability.
All data are available in the main text or the supplementary materials.
Supplementary Material
Acknowledgments
We thank Dr. Miriam L. Greenberg for generously sharing BY4741 WT yeast strain and Taz-KO C2C12 cell lines and Drs. Sharon Ackerman, Chris Meisinger, Dennis R. Winge, Vincenzo Zara, and Jan Brix for their generous gift of antibodies. This work was supported by Welch Foundation Grant A-1810 (to V.M.G.) and NIH Grants R01GM111672 (to V.M.G.), R01AR071942 (to V.K.M.), and R01GM109882 (to M.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. V.K.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000640117/-/DCSupplemental.
References
- 1.Baughman J. M. et al., Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Stefani D., Raffaello A., Teardo E., Szabò I., Rizzuto R., A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bick A. G., Calvo S. E., Mootha V. K., Evolutionary diversity of the mitochondrial calcium uniporter. Science 336, 886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clapham D. E., Calcium signaling. Cell 131, 1047–1058 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Kamer K. J., Mootha V. K., The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 16, 545–553 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Sancak Y. et al., EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342, 1379–1382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perocchi F. et al., MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature 467, 291–296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plovanich M. et al., MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8, e55785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raffaello A. et al., The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 32, 2362–2376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M., Mileykovskaya E., Dowhan W., Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Pfeiffer K. et al., Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M., Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182, 937–950 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker C. D., Basu Ball W., Pryce E. N., Gohil V. M., Specific requirements of nonbilayer phospholipids in mitochondrial respiratory chain function and formation. Mol. Biol. Cell 27, 2161–2171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasseva G. et al., Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J. Biol. Chem. 288, 4158–4173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuler M. H., Di Bartolomeo F., Mårtensson C. U., Daum G., Becker T., Phosphatidylcholine affects inner membrane protein translocases of mitochondria. J. Biol. Chem. 291, 18718–18729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baradaran R., Wang C., Siliciano A. F., Long S. B., Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature 559, 580–584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu Ball W., Neff J. K., Gohil V. M., The role of nonbilayer phospholipids in mitochondrial structure and function. FEBS Lett. 592, 1273–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovács-Bogdán E. et al., Reconstitution of the mitochondrial calcium uniporter in yeast. Proc. Natl. Acad. Sci. U.S.A. 111, 8985–8990 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto T. et al., Analysis of the structure and function of EMRE in a yeast expression system. Biochim. Biophys. Acta 1857, 831–839 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Chen E. et al., Perturbation of the yeast mitochondrial lipidome and associated membrane proteins following heterologous expression of Artemia-ANT. Sci. Rep. 8, 5915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu Ball W. et al., Ethanolamine ameliorates mitochondrial dysfunction in cardiolipin-deficient yeast cells. J. Biol. Chem. 293, 10870–10883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baile M. G. et al., Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J. Biol. Chem. 289, 1768–1778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou W. et al., Loss of tafazzin results in decreased myoblast differentiation in C2C12 cells: A myoblast model of Barth syndrome and cardiolipin deficiency. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 857–865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowhan W., Mileykovskaya E., Bogdanov M., Diversity and versatility of lipid-protein interactions revealed by molecular genetic approaches. Biochim. Biophys. Acta 1666, 19–39 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee A. G., How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Böttinger L. et al., Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J. Mol. Biol. 423, 677–686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang F. et al., Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Li X., Xu H., Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc. Natl. Acad. Sci. U.S.A. 109, 11384–11389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telezhkin V., Reilly J. M., Thomas A. M., Tinker A., Brown D. A., Structural requirements of membrane phospholipids for M-type potassium channel activation and binding. J. Biol. Chem. 287, 10001–10012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamer K. J., Grabarek Z., Mootha V. K., High-affinity cooperative Ca2+ binding by MICU1-MICU2 serves as an on-off switch for the uniporter. EMBO Rep. 18, 1397–1411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhuo W., et al. , Structure of intact human MCU supercomplex with the auxiliary MICU subunits. bioRxiv:10.1101/2020.04.04.025205 (06 April 2020). [DOI] [PMC free article] [PubMed]
- 32.Musatov A., Sedlák E., Role of cardiolipin in stability of integral membrane proteins. Biochimie 142, 102–111 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Xu Y. et al., Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proc. Natl. Acad. Sci. U.S.A. 116, 11235–11240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bione S. et al., A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 12, 385–389 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Xu Y., Kelley R. I., Blanck T. J., Schlame M., Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 278, 51380–51385 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Clarke S. L. et al., Barth syndrome. Orphanet J. Rare Dis. 8, 23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logan C. V. et al.; UK10K Consortium , Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat. Genet. 46, 188–193 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Lewis-Smith D. et al., Homozygous deletion in MICU1 presenting with fatigue and lethargy in childhood. Neurol. Genet. 2, e59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan X. et al., The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat. Cell Biol. 15, 1464–1472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bick A. G. et al., Cardiovascular homeostasis dependence on MICU2, a regulatory subunit of the mitochondrial calcium uniporter. Proc. Natl. Acad. Sci. U.S.A. 114, E9096–E9104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meisinger C., Pfanner N., Truscott K. N., Isolation of yeast mitochondria. Methods Mol. Biol. 313, 33–39 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Folch J., Lees M., Sloane Stanley G. H., A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957). [PubMed] [Google Scholar]
- 43.Bartlett G. R., Phosphorus assay in column chromatography. J. Biol. Chem. 234, 466–468 (1959). [PubMed] [Google Scholar]
- 44.Veling M. T. et al., Multi-omic mitoprotease profiling defines a role for Oct1p in coenzyme Q production. Mol. Cell 68, 970–977.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.