Significance
We report high-resolution structures for the μ-opioid G protein-coupled receptor (μOR) complexed with β-arrestin2 while bound to either strong nonbiased agonists (morphine and DAMGO) or to biased agonist TRV130. We used these structures to identify a sequence of events leading to β-arrestin2 recruitment. We discovered that for nonbiased agonists, β-arrestin2 and Gi protein make strong anchors to the same intracellular loops of μOR, even though their recruitment leads to different outcomes. However, we find that for the biased ligand, β-arrestin2 binds much more weakly to μOR, possibly explaining the reduction in side effects. These structures provide the basis for structure-based design of biased drugs to mediate pain more effectively while reducing such side effects as respiratory depression.
Keywords: biased agonists, nonbiased agonists, molecular dynamics
Abstract
Agonists to the μ-opioid G protein-coupled receptor (μOR) can alleviate pain through activation of G protein signaling, but they can also induce β-arrestin activation, leading to such side effects as respiratory depression. Biased ligands to μOR that induce G protein signaling without inducing β-arrestin signaling can alleviate pain while reducing side effects. However, the mechanism for stimulating β-arrestin signaling is not known, making it difficult to design optimum biased ligands. We use extensive molecular dynamics simulations to determine three-dimensional (3D) structures of activated β-arrestin2 stabilized by phosphorylated μOR bound to the morphine and D-Ala2, N-MePhe4, Gly-ol]-enkephalin (DAMGO) nonbiased agonists and to the TRV130 biased agonist. For nonbiased agonists, we find that the β-arrestin2 couples to the phosphorylated μOR by forming strong polar interactions with intracellular loop 2 (ICL2) and either the ICL3 or cytoplasmic region of transmembrane (TM6). Strikingly, Gi protein makes identical strong bonds with these same ICLs. Thus, the Gi protein and β-arrestin2 compete for the same binding site even though their recruitment leads to much different outcomes. On the other hand, we find that TRV130 has a greater tendency to bind the extracellular portion of TM2 and TM3, which repositions TM6 in the cytoplasmic region of μOR, hindering β-arrestin2 from making polar anchors to the ICL3 or to the cytosolic end of TM6. This dramatically reduces the affinity between μOR and β-arrestin2.
During the past decade, both G protein-biased and β-arrestin (βarr)-biased ligands have been discovered and developed for ∼30 different G protein-coupled receptors (GPCRs). Such biased ligands provide functional selectivity to regulate more precisely biological functions of GPCRs, providing new drugs with superior efficacy but reduced side effects (1). Biased ligands are crucial for treatment of chronic neuropathic pain, a major challenge in clinical practice (2). Opioid analgesics, such as morphine, are prescribed to relieve severe pain by activating pain receptors in the central nervous system that induce G protein-mediated signaling to confer analgesia. However, these opioids are associated with such side effects as sedation, physical dependence, addiction, tolerance, and respiratory depression (3). These side effects, particularly respiratory depression, are thought to be mediated by activation of βarr signaling (3). To avoid such side effects, biased ligands that elicit Gi protein activation with minimal βarr recruitment have shown efficient pain treatment with minimal side effects, to replace traditional narcotic analgesics (2).
The μ-opioid GPCR (μOR) stimulates signaling via the adenylyl cyclase-inhibitory family of G proteins (Gi/o), leading to analgesic activity (4). Therefore, the detailed interplay between μOR, Gi protein, and ligands that induce Gi protein activation is crucial in the design of the active analgesics. The agonist modulates G protein signaling by triggering exchange of guanosine diphosphate (GDP) with guanosine triphosphate (GTP) bound to Gαi and decoupling of Gβγ from the GPCR to induce signaling (5). Afterward, the Gβγ helps recruit the G protein-coupled receptor kinase (6–8) that phosphorylates the activated GPCR (9, 10). In particular for μOR, phosphorylation takes place mainly at the serine and threonine resides in the long C-terminal tail, forming the phosphorylated-C (pp-C) tail (11–13). For nonbiased ligands the pp-C tail of the final agonist-GPCR complex reaches out to recruit βarr (11–13). Indeed, mutation of all serines and threonines to alanine on the μOR C tail cancels recruitment of βarr, thereby greatly diminishing such side effects as desensitization and internalization (11–13). In fact, a recent X-ray crystal structure of phosphorylated rhodopsin (pp-rhod)-arrestin-1 supports the significant role of the pp-C tail in recruiting arrestin since both phosphorylated residues on the C terminus form strong salt bridges with arrestin-1 (14). Therefore, the detailed interplay between μOR, βarr, and a nonbiased ligand orchestrates the critical steps toward recruiting and activating the βarr. Understanding these interactions should be useful for in silico design of biased ligands that might prevent a critical step of βarr activation.
Unfortunately, neither crystal nor cryo-electron microscopy (EM) structures are available for the μOR-βarr complex, making it difficult to carry out structure-based design and development of new biased agonists. To this end, we report here three-dimensional (3D) structures of the final activated state of βarr2 coupled to the active state of μOR bound to a full agonist D-Ala2, N-MePhe4, Gly-ol]-enkephalin (DAMGO), a partial agonist morphine, and a biased agonist TRV130. These 3D structures provide the basis for obtaining a deep understanding of βarr2 interaction with both biased and nonbiased ligands. This should enable in silico design of biased agonists with low activity for βarr2 activation.
To build the agonist-pp-μOR-βarr2 complex, we started with the recent ∼3.0-Å-resolution crystal structure of pp-rhod-arrestin-1 complex. Although this structure provides the overall shape and conformation of the complex, including most hydrophobic interactions, the resolution does not identify the important polar interactions (salt bridges and hydrogen bonds). Therefore, we refined the pp-rhod-arrestin-1 complex to discover several important polar interactions not identified in the crystal structure (SI Appendix, Fig. S1). To do this, we first carried out 4 ns of simulated annealing while imposing strong restraints on the backbone atoms to retain the main features of the crystal structure. Here, all side chains were flexible so that they could form new favorable polar interactions. Subsequently, we performed a long (∼450-ns) molecular dynamics (MD) simulation to relax the complex while imposing heavy restraints on the backbone atoms to retain the main features of the crystal structure. We then used this refined structure of the pp-rhod-arrestin-1 as the basis for predicting the high-affinity βarr2-pp-μOR-DAMGO, βarr2-pp-μOR-morphine, and βarr2-pp-μOR-Oliceridine (known as TRV130) complexes. We discovered that the active conformation of βarr2 couples to the core of pp-μOR by forming polar anchors to the intracellular loop 2 (ICL2) and either the ICL3 or the cytosolic end of transmembrane (TM6) domain, which further stabilizes the fully engaged complex of βarr2-pp-μOR-agonist. In this regard, the biased ligand differs substantially from the nonbiased ligands by TRV130 showing much lower binding affinity between pp-μOR and βarr2 because βarr2 does not form polar anchors with the ICL3 or the cytosolic end of TM6.
For the nonbiased ligands, we find extensive interactions between the μOR pp-C tail and βarr2 that stabilize the active conformation of the βarr2, allowing the βarr2 to fully engage with the core of pp-μOR to eventually form the polar anchors. Indeed, we find that the mobility of the finger loop is the main driving force during recruitment, contributing to the full engagement of βarr2 with pp-μOR.
Strikingly, we showed (15) that the Gi protein couples to the opioid receptors by forming polar anchors to each of three ICLs of the μOR. Thus, for nonbiased ligands βarr2 binds to μOR in the same fashion as Gi, competing for the same binding sites, while their recruitment leads to very different outcomes.
To understand how a biased agonist, such as TRV130, selectively stimulates Gi protein while disfavoring βarr2 signaling, we followed the same strategy as above, modeling and optimizing the activated state of the human μOR-Gi-TRV130 complex. The TRV130 binding site to human μOR is distinctly different from the nonbiased agonists, with TRV130 binding more strongly with TM2 and TM3 in the extracellular portion of μOR, which repositions TM6 that dramatically reduces βarr2 binding to pp-μOR. However, the Gi protein forms the same polar anchors to ICL2 and the cytosolic end of TM6 as the nonbiased ligands.
For the active βarr2-pp-μOR-TRV130 complex, we find that the active conformation of βarr2 forms a polar anchor with the ICL2 of the pp-μOR, just as for nonbiased ligands. However, due to the repositioned TM6, we find that βarr2 is unable to make polar anchors with either ICL3 or the cytosolic end of TM6, in contrast to the nonbiased ligands. This lack of polar anchors prevents the active conformation of βarr2 from binding properly to the pp-μOR core, significantly lowering the binding affinity between the βarr2 and μOR.
This insight provides the basis for designing biased ligands acting on the μOR.
Result and Discussion
Modeling the βarr2-pp-μOR Interface.
To build the βarr2-pp-μOR-agonist complex, we first optimized the recent crystal structure of pp-rhod bound to arrestin-1 (Protein Data Bank [PDB] ID code 5W0P) (14) and then used this structure as the template for building our models. Prior to optimization, we removed the artificial N-acetylglucosamine and T4 lysozyme protein from the pp-rhod. In addition, we built in residues 324 to 330 missing in the C tail of the 3.0-Å X-ray structure (SI Appendix, SI Methods). Then, we immersed the complex in the lipid bilayer including water and salt and performed 450 ns of MD simulation with positional restraints on the protein backbone atoms to ensure that the crystal structure is not disturbed while the residue side chains are refined to find the optimum polar interactions between the arrestin-1 and pp-rhod. Our final structure (SI Appendix, Fig. S1) is in perfect agreement with the crystal structure, with rmsd = 0.3 Å.
We used the GEnSeMBLE (16) method to predict the active conformation of human-μOR starting from the mouse-μOR (PDB ID code 5C1M) (17). We then docked morphine into the predicted human-μOR structures using DarwinDock (18). Next, we separately matched DAMGO and TRV130 into the human morphine binding pocket. Subsequently, we added and modeled all residues in the C terminus of μOR using the MODELER program (19). It has been shown that the main phosphorylation sites on the μOR for recruiting the βarr2 are serine and threonine residues on the C tail (12). However, the degree of phosphorylation can depend on the type of agonist (11, 20, 21). Interestingly, a recent study (12) showed that mutation of all serines and threonines to alanine on the C tail of μOR in the presence of DAMGO and morphine blocks the βarr2 recruitment. Therefore, we used the full degree of phosphorylation for this study. To find the conformation of the pp-C tail that has the maximum number of interactions with βarr2, we started with a complex between the inactive state of βarr2 (22) (PDB ID code 3P2D) and the activated state of μOR (SI Appendix, SI Methods). To optimize the interactions between the pp-C tail and the βarr2, we performed several 25- to 90-ns meta-MD simulations to identify the maximum number of salt bridges between the N domain of βarr2 and the pp-C tail of μOR while heavy restraints were placed on the backbone atoms of the protein except for the pp-C tail. Next, to model the final activated state of βarr2-μOR-DAMGO, βarr2-μOR-morphine, and βarr2-μOR-TRV130, we replaced the inactive βarr2 with the active one (23) (PDB ID code 5TV1) by superimposing the refined arrestin-1 and pp-rhod complex on both active βarr2 and μOR. This structure was subjected to several minimization and optimization steps using meta-MD and MD simulations.
Active-State Complex of βarr2-pp-μOR Bound to DAMGO, a Full Agonist.
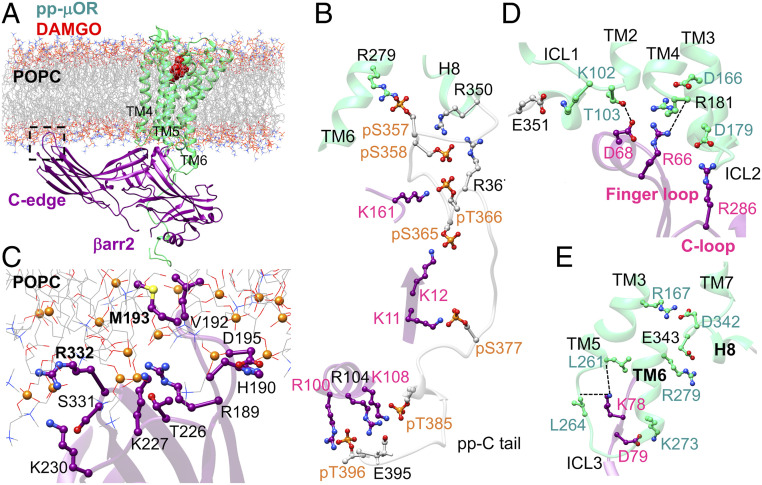
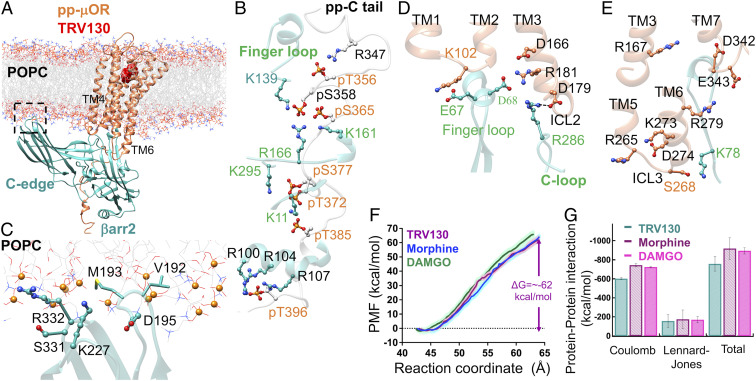
DAMGO is a full agonist in the βarr2 assay, with 8.8-fold higher efficacy than morphine for recruiting βarr2 (24). Thus, we first characterized the fully engaged pp-μOR-βarr2-DAMGO complex by performing a 500-ns MD simulation. An overview of the optimized complex (25) immersed in the lipid bilayer is shown in Fig. 1A. Since mutation of all serine and threonine in the carboxyl-terminal receptor to alanine was shown to inhibit recruitment of βarr2 (12), we optimized the structure with all 10 serine (pS) and threonine (pT) residues on the C tail phosphorylated (Fig. 1B). This allows the pp-C tail to engage tightly the N domain of βarr2, leading finally to emergence of persistent salt bridges between pS365-K12, pS366-K161, pS377-K11, pT385-R108, E395-R108, and pT396-R100. These extensive interactions play a pivotal role in stabilizing the complex.
Fig. 1.
(A) The high-affinity βarr2-pp-μOR-DAMGO complex immersed in the membrane bilayer. (B) The strong polar interactions between the N domain of βarr2 and the pp-C tail of μOR, mostly involving pS and pT residues on the pp-C tail interacting with positively charged residues on the N domain of βarr2. (C) The membrane anchoring from the C edge of βarr2, which is dominated by hydrophobic contacts. Here, P atoms are shown as orange spheres. (D) The polar anchor from βarr2 to ICL2 of the μOR, which creates a polar network of interactions from the finger loop to ICL2 and the cytosolic end of TM2. (E) Polar anchors from the βarr2 to both ICL3 and the bottom end of TM6, fully engaging the body of the βarr2 to the core of the μOR.
Our optimized complex shows that the C edge of the βarr2 makes extensive contacts to the lipid bilayer (Fig. 1C), mostly dominated by hydrophobic interactions. Remarkably, one of the C-edge loops, 189RHFLMSDRS197, penetrates into the lipid bilayer, allowing H190, L192, M193, and D195 to establish hydrophobic contacts to the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane, while the R189 involves a polar interaction with a POPC phosphate group. In contrast, the other C-edge loops, 224NSTKTVKKI232 and 330VSRGG334, reside on the membrane surface with K227, K230, and R332 making polar contacts to phosphate groups of POPC. A previous computational study showed that lipid interactions with the C edge of arrestin function as a membrane anchor that is essential for stabilizing the high-affinity arrestin-GPCR complex (26). Another recent computational study (27) indicates that the lack of such lipid anchoring transforms the active βarr1 to its inactive conformation, suggesting that such contacts between the arrestin and lipid bilayer stabilize the active conformation of βarr1. Indeed, our MD studies show that βarr2 retains its active conformation with an averaged interdomain twist angle of ∼18° ± 3° (SI Appendix, Figs. S2 A–C and S3), similar to the 17° twist reported for the βarr2 crystal structure of βarr2 (23), confirming that lipid anchoring by the C edge of βarr2 is pivotal for the stability of the high-affinity pp-μOR-βarr2 complex.
We discovered that βarr2 forms strong anchors to ICL2, ICL3, and the cytosolic end of TM6 in our high-affinity pp-μOR-βarr2-DAMGO complex. The high affinity between βarr2 and ICL2 creates a network of polar interactions that stabilize the active-state complex (Fig. 1D). In this network, the salt bridge between D179ICL2 and R286 on the C loop serves as an anchor that aligns the finger loop to establish polar interactions with ICL2 and the cytosolic end of TM2. Here, R181ICL2 plays a crucial role in regulating βarr2 binding. R181ICL2 exhibits a charge–charge interaction with D1663.49 [the superscript is Ballesteros–Weinstein numbering for GPCRs (28) taken from ref. 29], while its carbonyl oxygen forms a hydrogen bond to R66 on the finger loop (Fig. 1D). The coordinated R66 makes a salt bridge with D68Finger loop, orienting the D68Finger loop to involve a hydrogen bond with T1032.37. Moreover, we find that D79 in the N domain of βarr2 makes an ionic contact to the K2736.26 at the end of TM6 (Fig. 1E). We consider this salt bridge as the second ionic anchor that induces K78 in the N domain to engage two hydrogen bonds with the carbonyl oxygen of L2615.65 and L264ICL3. Our MD simulation indicates that the ionic anchors from the pp-μOR to βarr2 play vital roles in βarr2 recruitment.
Active-State Complex of βarr2-pp-μOR Bound to Morphine, a Partial Agonist.
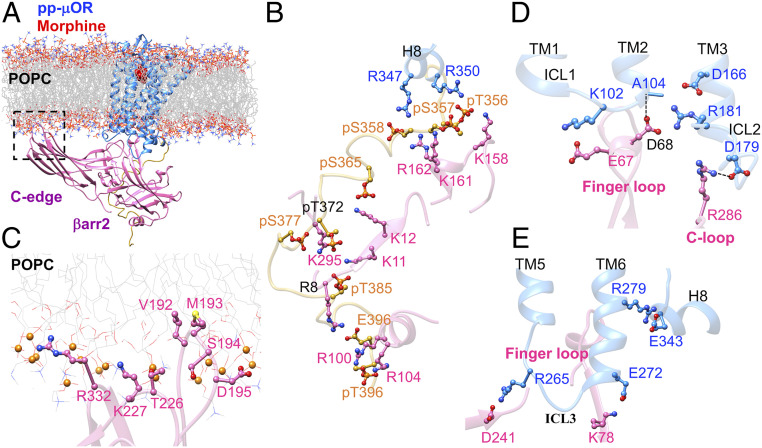
To find if emergence of the ionic anchors is statistically significant, we performed an independent 500-ns MD simulation to characterize the fully engaged βarr2-pp-μOR complex using morphine, a clinical drug considered to be a partial agonist (24) for βarr2 recruitment. An overview of the optimized complex immersed in the lipid bilayer is shown in Fig. 2A. Our MD simulation shows that the N domain of βarr2 couples tightly to the phosphorylated C tail with strong electrostatic attractions (Fig. 2B). We find that numerous salt bridges: K161-pS357, R162-pS358, K12-pS365, K11-pT372, K11-pT385, R8-E396, R100-pT396, and R104-pT396 participate in this tight coupling. Of these interactions, pS377 makes a persistent salt bridge with K295, which is known to play an important role in βarr2 recruitment (20). A previous experiment on the human embryo kidney (HEK) cells showed that blocking the residue equivalent to S377 in human-μOR from phosphorylation greatly diminishes association of μOR with βarr2 (20).
Fig. 2.
(A) The high-affinity βarr2-pp-μOR-morphine complex immersed in the membrane bilayer. (B) Strong interactions between the N-terminal domain of βarr2 and the pp-C tail of the μOR, mostly involving pS and pT residues on the pp-C tail interacting with positively charged residues on the N-terminal domain of βarr2. (C) The membrane anchoring from the C edge of βarr2, which is dominated by hydrophobic contacts. Here, the P atoms are shown by orange spheres. (D) The polar anchor from βarr2 to ICL2 of the μOR, which creates a polar network of interactions from the finger loop to ICL2 and the cytosolic end of TM2. (E) Polar anchors from the βarr2 to both ICL3 and the bottom end of TM6, fully engaging the body of the βarr2 to the core of the μOR.
We find that the C edge of βarr2 anchors to the membrane bilayer (Fig. 2C), allowing the βarr2 to maintain its active conformation with an averaged interdomain twist angle of ∼20° ± 4° (SI Appendix, Figs. S2 D–F and J and S4A). However, when we eliminate the lipid anchors (SI Appendix, Fig. S5), βarr2 shifts from the active conformation to an inactive conformation with an averaged interdomain twist angle of ∼5°. This is consistent with a recent computational study (27) showing that without a lipid anchor, the βarr1 is not able to remain its active conformation, with the interdomain twist angle changing from ∼17° to between ∼0° and 7°. This indicates that interactions of the membrane bilayer with the C edge of βarr2 are essential to stabilize the active conformation of βarr2.
For morphine, the C-edge loop, 189RHFLMSDRS197, also penetrates the lipid bilayer, with L192 and M193 forming hydrophobic anchors to the membrane. However, the other C-edge loops, 223NSTKTVKKI232 and 329VSRGG334, stay on the membrane surface, coordinating K227 and R332 to form polar interactions with the phosphate groups of POPC. Indeed, this second independent simulation confirms that lipid anchors are essential for stimulating the βarr2 signaling, aligning the βarr2 to bind effectively to the cytoplasmic part of the pp-μOR.
We find that the high-affinity complex of βarr2-pp-μOR-morphine features polar anchors from the βarr2 to the ICL2 and ICL3 of the pp-μOR, similar to DAMGO. In this structure, R286 on the C loop forms a hydrogen bond with D179ICL2 (Fig. 2D). This anchor coordinates the ICL2 to tightly engage the finger loop. Here, R181ICL2 establishes persistent salt bridges with D1663.49 and D68Finger loop. This induces D68Finger loop to form a hydrogen bond with carbonyl oxygen of A1042.38 at the cytosolic end of TM2. Indeed, this network of polar interactions is similar to the one created by DAMGO. In addition, βarr2 forms a second anchor from D241 to R265 on ICL3 (Fig. 2E). Also, our analysis shows that E2726.25 at the bottom of TM6 frequently forms a weak salt bridge with K78. To eliminate the possibility that emergence of these ionic anchors is not statistically significant, we repeated the 500-ns MD simulation with the velocities reassigned (SI Appendix, Fig. S6). Importantly, our second optimized structure identifies similar anchors between D179ICL2-R286C-loop and R265ICL3-E314Back loop. Overall, the emergence of polar anchors between βarr2 and pp-μOR in the presence of both the full and partial agonists indicates that these anchors effectively coordinate the βarr2 to have strong interactions with the core of the receptor.
Activated State of Mouse μOR-DAMGO-Gi Complex.
Strikingly, we found recently that ionic anchors are also essential for activation of Gi protein mediated by opioid receptors (15). Thus, in the presence of nonbiased agonists, we find a similar pattern in binding of Gi protein and βarr2 to the μOR.
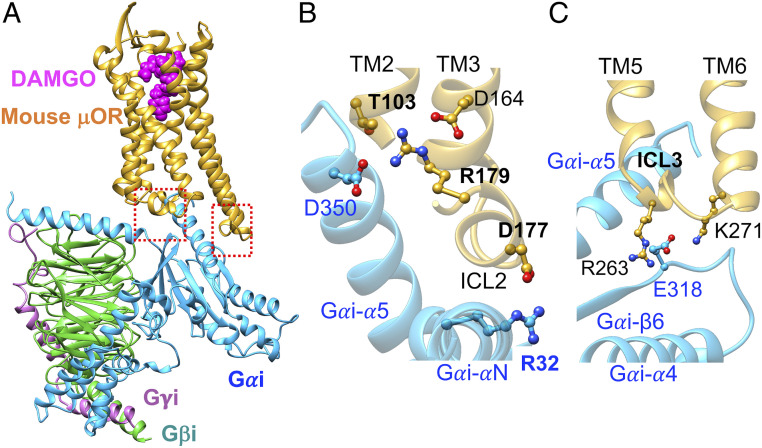
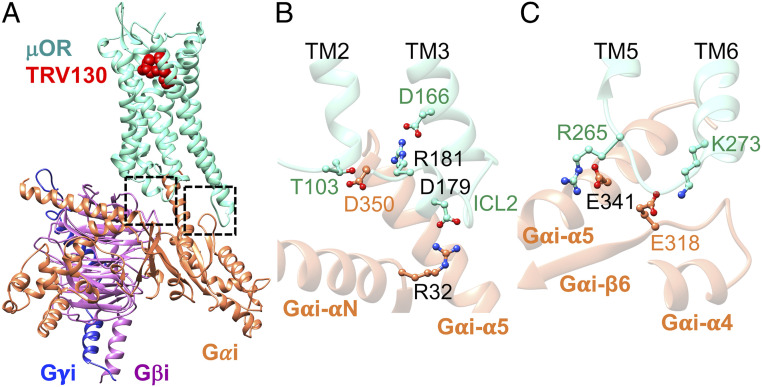
Optimizing the cryo-EM structure (Fig. 3) of the active state of mouse-μOR bound to DAMGO stabilized by Gi protein (15, 30) shows that ICL2 of μOR also involves extensive polar interactions with the Gi protein in which the salt bridge from D177ICL2 (equivalent to the human D179ICL2) to R32 on the Gαi subunit (Fig. 3B) serves as an ionic anchor that engages Gi protein recruitment. Moreover, we find that R179ICL2 (equivalent to the human R181ICL2) forms a charge–charge interaction with D350 in the Gαi-α5 helix (Fig. 3B), which allows D350 to establish a hydrogen bond with T1032.39 at the cytosolic end of TM2. Indeed, this complex network of polar interactions induced by the Gi protein is analogous to that created by the βarr2. Here, the D177ICL2 and R179ICL2 play similar crucial roles to coordinate this network. The similar recruitment of Gi protein and βarr2 by μOR indicates that these two effectors compete for the same binding site even though their recruitment leads to opposite outcomes.
Fig. 3.
Activated Gi protein binds to activated mouse μOR in the same fashion as βarr2 couples to pp-μOR. (A) Well-optimized Gi-mouse-μOR-DAMGO complex obtained from MD simulation (15), starting from the recent cryo-EM (30). (B) The polar anchor from R32Gαi to D177ICL2 (equivalent to D179 in the human μOR). This salt bridge creates a network of polar interactions from the Gαi-α5 helix to the ICL2 and the cytosolic end of TM2, which is similar to the network that emerges from βarr2 coupling to the pp-μOR. (C) The polar anchors between Gi protein and μOR: from E318 to R263ICL3 (equivalent to R265 in the human μOR) and K2716.26 (equivalent to K273 in the human μOR). These anchors show that Gi protein and βarr2 compete for the same binding site in the μOR, even though their recruitment results to totally opposite outcomes.
We also found that ICL3 of opioid receptors serves an important role in recruiting the Gi protein (15). Our optimized μOR-Gi protein shows that both R263 and K2716.26 in mouse μOR (equivalent to R265ICL3 and K2736.26, respectively, in the human μOR) form salt bridges with E318 in the RAS-like domain of Gαi (Fig. 3C). As we showed above, these positively charged residues also play crucial roles in recruiting the βarr2. Indeed, this finding confirms that both the Gi protein and βarr2 compete to bind to the same sites in μOR.
Recruitment of Activated βarr2 by pp-μOR-Morphine.
The primary role of the phosphorylated carboxyl terminus of μOR is to recruit and subsequently to activate arrestin, which displaces the C-terminal tail of arrestin (31–34). Upon activation, arrestin undergoes a remarkable ∼20° twist of the C domain relative to the N domain, which is widely considered as a primary metric to assess arrestin activation (14, 23, 35, 36). Recent structures of arrestins bound to phosphorylated GPCRs (14, 27, 37) indicate that the activated arrestins bind tightly to the pp-C tail of receptors while also interacting strongly with the 7TM core of the GPCR. In fact, a computational study showed that the 7TM core and phosphorylated C tail independently stimulate the arrestin activation but both together take part in formation of high-affinity GPCR-arrestin complex (36).
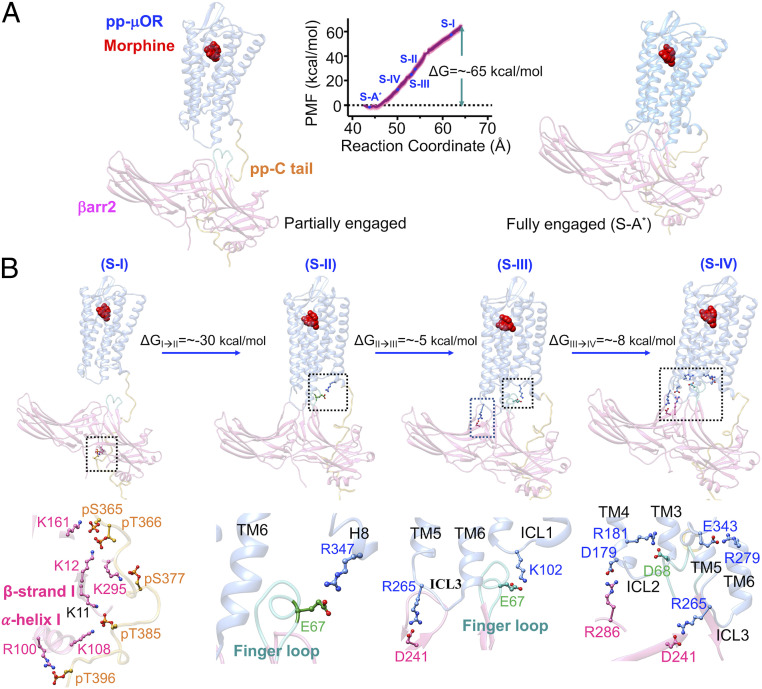
To understand how βarr2 bound to the pp-C tail migrates from water to interact strongly with the core of the pp-μOR, we performed a free energy calculation using umbrella sampling (38–41). In this calculation, we gradually disengage the βarr2 from the high-affinity complex to form a partially engaged βarr2-pp-μOR structure, where the βarr2 binds solely to the pp-C tail (Fig. 4A). Importantly, βarr2 remains in its active conformation with an averaged interdomain twist angle of ∼18° ± 5°, showing that the pp-C tail alone is sufficient to activate βarr2. Indeed, this finding is consistent with multiple studies revealing that just the pp-C tail is sufficient to stimulate arrestin activation and signaling (35, 41–45). On the other hand, βarr2 is characterized to have an averaged interdomain twist angle of ∼20° ± 5° in the high-affinity complex (denoted as fully engaged complex) (Figs. 2A and 4A), where the βarr2 is fully engaged in the 7TM core of μOR. This finding confirms that a strong coupling between βarr2 and pp-μOR stabilizes the active conformation of βarr2, showing the significant role of the 7TM core in βarr2 recruitment. Our findings are in excellent agreement with a recent study on pp-rhod-arrestin-1, indicating that both the pp-C tail and the core of receptor together further stabilize the active conformation of arrestin 1 (36).
Fig. 4.
Process of forming the fully engaged complex between the pp-μOR and the βarr2 in the presence of morphine. (A) The averaged potential of mean force (PMF) indicates that the recruited βarr2 by the pp-C tail spontaneously couples the core of pp-μOR. The free energy was obtained by umbrella sampling MD, where the reaction coordinate is the distance along the z component between the center of mass of Cαs in the pp-μOR for residues 54 to 340 and the center of mass of Cαs in βarr2. The errors (shaded as pink) were assessed by the bootstrap method (38). (B) Sequence of important events in the recruitment of the βarr2 by the pp-μOR bound to morphine. Our free energy calculation suggests the following pathway. 1) S-I: the βarr2 couples to the pp-C tail of μOR, involving mainly salt bridges from pS and pT residues to positively charged residues on the N domain of the βarr2. 2) S-II: the flexible finger loop extends to the receptor core to engage the H8 helix. 3) S-III: the extended finger loop moves toward ICL1 to form an anchor from E67 to K102ICL2. Anchoring to the ICL1 allows the rest of βarr2 to ascend to form another anchor from D241 to R265 on the ICL3. 4) S-IV: finally, the βarr2 forms a polar anchor from R286 on the C loop to D179 on the ICL2 that transforms the finger loop to the final position.
We find that after βarr2 is activated by the pp-C tail, it spontaneously couples to the 7TM core of pp-μOR (Movie S1). Our free energy calculation shows high affinity between the activated βarr2 and pp-μOR (a partially engaged βarr2 with the pp-μOR, denoted as S-I), leading to forming the fully engaged complex, while reducing the energy substantially, by ∼−65 kcal/mol (Fig. 4A). This high binding affinity between pp-μOR and βarr2 is expected since the pp-μOR carries negative net charges, while the βarr2 has positive net charges, facilitating spontaneous electrostatic attraction. During the free energy calculation, we identified a sequence of important events that lead to recruitment of βarr2 by the pp-C tail and the 7TM core (Fig. 4B).
The βarr2 recruitment is triggered by strong interactions from the negatively charged residues on the pp-C tail, mostly from pS and pT residues, to positively charged amino acids on the N domain of βarr2 (S-I in Fig. 4B). The N domain binds the pp-C tail with its α-helix I and β-strand I. Interestingly, we find that the pp-C tail primarily forms a salt bridge from the pS377 to K295βarr2, indicating that S377 is crucial for recruiting the βarr2. Indeed, it is well known from experiments on HEK293 cells that morphine is a full agonist (24) for phosphorylation at S375 (equivalent to S377 in human μOR), whereas inhibiting this serine from phosphorylation greatly reduces βarr2 recruitment (12).
Our MD simulations show that the finger loop penetrates to the core of μOR to eventually promote βarr2 to fully engage with the 7TM core. The finger loop has a flexible conformation when it is not yet engaged in interactions with the pp-μOR (Movie S2). This flexibility enables the finger loop to extend from water to the core of the receptor to establish a loose salt bridge from E67 to R3478.52 in the H8 helix (S-II in Fig. 4B). This loose charge–charge interaction between E67-R3478.52 is not able to stabilize the finger loop conformation and mobility. Consequently, the finger loop fluctuates inside the receptor to finally make another ionic contact from E67 to K102ICL1 (S-III in Fig. 4B). This salt bridge serves as an anchor that induces the C domain of the βarr2 to ascend and interact with both the ICL3 and ICL2. Our free energy calculations suggest that the finger loop is a major driver of the βarr2 coupling to the 7TM core pp-μOR.
The anchor from the finger loop to the ICL1 induces the tilted C domain of βarr2 to move toward the pp-μOR to make an ionic anchor from D241 to R265ICL3(S-III in Fig. 4B; Movie S3), which reduces the energy by ∼8 kcal/mol compared with S-II. Subsequently, this ionic anchor facilitates pushing up the rest of the C domain of the βarr2, leading to emergence of an ionic anchor between R286 in the C loop and D179 in ICL2 (S-IV in Fig. 4B; Movie S4), which substantially lowers the energy by ∼8 kcal/mol compared with S-III. Remarkably, emergence of the anchor between ICL2 and βarr2 coincides with breaking the salt bridge between E67 and K102, which displaces the finger loop to its final position, in which D68 establishes a salt bridge with R181ICL2 (S-IV in Fig. 4B; Movie S4). Eventually, the system energy decreases by ∼18 kcal/mol to reach the high-affinity complex (S-A*) described earlier, where βarr2 is fully engaged with the core of pp-μOR. Indeed, our free energy calculations indicate that the ICLs of pp-μOR play a crucial role in stimulating βarr2 recruitment, with polar anchors to all three ICLs significantly mediating the binding. In fact, a recent computational study revealed that binding of the visual arrestin-1 to the core of rhodopsin is primarily mediated by interactions of ICLs with the body of the arrestin (36). Interestingly, our optimized structure of pp-rhod-arrestin-1 complex (SI Appendix, Fig. S1) features strong ionic anchors, K141ICL2-D253C-loop and E239ICL3-Arg318Back loop, confirming that the ionic anchors from ICL2 and ICL3 play key roles in coupling of arrestin-1 to the pp-rhod.
To determine whether the penetration of finger loop into the cytoplasmic region of the pp-μOR is the main driver of the βarr2 coupling to the 7TM core of pp-μOR, we repeated our free energy calculation to follow the recruitment of βarr2 by the pp-μOR in the presence of DAMGO (SI Appendix, Fig. S7). Our free energy calculation reveals that the finger loop penetrates to the 7TM core to make an ionic contact from R66 to D179ICL2, which eventually induces the βarr2 to form ionic anchors with ICL2 and the cytosolic end of TM6, leading to formation of a high-affinity complex between the proteins. This calculation indicates the finger loop plays a crucial role in recruitment of the βarr2 by pp-μOR.
Active-State Complex of βarr2-pp-μOR Bound to TRV130, a Biased Agonist.
To understand how TRV130, a biased clinical agonist, selectively stimulates Gi protein while disfavoring βarr2 signaling, we started with modeling and optimizing the activated state of μOR-Gi-TRV130 (Fig. 5A). We find that Gi binds to the human μOR by forming ionic anchors to ICL2 and the cytosolic end of TM6, which is consistent with our previous computational study showing that ionic anchors to the ICLs are essential for Gi protein signaling mediated by opioid receptors (15). Here, R179ICL2 forms a salt bridge to R32 on the Gαi subunit (Fig. 5B), while K2736.26 forms a salt bridge with E318 in the RAS-like domain of Gαi (Fig. 5C). Moreover, R265ICL3 makes a salt bridge contact to D341 on the Gαi-α5 helix. Our MD simulations show that the Gi protein couples to the μOR bound to TRV130 in a fashion similar to the coupling of Gi to μOR in the presence of DAMGO. This behavior is expected since the main role of the biased ligand is to favor only Gi signaling. In this regard, measurements (24) in HEK293 cells reveal that the TRV130, morphine, and DAMGO exhibit a comparable efficacy for G protein recruitment.
Fig. 5.
Activated Gi protein binds to activated human μOR. (A) Well-optimized Gi-human-μOR-TRV130 complex obtained from an ∼250-ns MD simulation. (B) The polar anchor from R32Gαi to D179ICL2. This salt bridge creates a network of polar interactions from the Gαi-α5 helix to ICL2 and the cytosolic end of TM2, which is similar to the network that emerges in the presence of DAMGO. (C) The polar anchor between Gi protein and μOR: E318 to K2736.26.
TRV130 selectively disfavors the pathway of βarr2 signaling. Thus, it exhibits 14% of the morphine efficacy for βarr2 recruitment (24) in the HEK293 cells. To determine why TRV130 deviates from morphine and DAMGO regarding βarr2 signaling, we predicted the fully activated βarr2-pp-μOR-TRV130 complex by performing a 500-ns MD simulation. To have a fair comparison between DAMGO, morphine, and TRV130, we considered the full phosphorylation at all 10 serine and threonine residues on the C tail, even though this phosphorylation is a βarr2 agonist property. Indeed, TRV130 is known to lead to less phosphorylation at S375 in the HEK293 experiments (equivalent to S377 in human μOR) compared with morphine, which is a full agonist for phosphorylation of S375 (24). Thus, the lack of full phosphorylation at S375 may lower the efficacy of TRV130 for βarr2 coupling (24). Even so, our model with full phosphorylation at the C tail already shows that TRV130 is less able to recruit βarr2, which is consistent with TRV130 leading to only the 14% of the morphine efficacy for βarr2 coupling (24).
An overview of the optimized complex immersed in the lipid bilayer is shown in Fig. 6A. Our βarr2-pp-μOR-TRV130 complex features two important sites for interactions: 1) strong interactions between the N domain of βarr2 and the pp-C tail (Fig. 6B), which are dominated mostly by ionic contacts (Fig. 6C), and 2) hydrophobic contacts between the C edge of βarr2 and lipid membrane. These interactions, which are similar to those we found for complexes to nonbiased ligands, stabilize the active conformation of the βarr2 with an averaged interdomain twist angle of ∼19° ± 6° (SI Appendix, Figs. S2 G–J and S4B).
Fig. 6.
(A) The high-affinity βarr2-pp-μOR-TRV130 immersed in the membrane bilayer. (B) Strong interactions between the N domain of βarr2 and the pp-C tail of the μOR, mostly involving pS and pT residues on the pp-C tail with positively charged residues on the N domain of βarr2. (C) The membrane anchoring from the C edge of βarr2, which is mostly dominated by hydrophobic contacts. Here, the P atoms are shown by orange spheres. (D) The polar anchor from βarr2 to ICL2 of μOR, which creates a polar network of interactions from the finger loop to ICL2 and the cytosolic end of TM2. (E) The lack of polar anchoring from βarr2 to ICL3 or the bottom end of TM6 hinders the proper coupling of βarr2 to the core of μOR. (F) The averaged potential of mean force (PMF) for βarr2 coupling to the 7TM core μOR. The free energy was obtained by umbrella sampling where the reaction of coordinate is the distance along the z component between the center of mass of Cαs in the pp-μOR for residues 54 to 340 and the center of mass of Cαs in βarr2. The errors (shaded as pink for TRV130, light green for DAMGO, and light blue for morphine) were assessed by the bootstrap method (38). (G) The nonbonded interactions within 12 Å between βarr2 and the core of pp-μOR for DAMGO (full), morphine (partial), and TRV130 (biased) agonist in the fully engaged complexes, where we excluded water, and ions from these calculations.
We find that in the presence of TRV130 the active conformation of βarr2 forms a polar anchor with the ICL2 of the pp-μOR. In this structure, R286 on the C loop makes a hydrogen bond with D179ICL2 (Fig. 6D), which is identical to the one we identified in the presence of morphine (Fig. 2D). This anchor coordinates ICL2 to interact strongly with the finger loop. R181ICL2 establishes a persistent salt bridge with D68Finger loop. Indeed, this network of polar interactions is similar to the one created by DAMGO and morphine.
Strikingly, for the TRV130 case, the βarr2 is unable to make polar anchors with either ICL3 or the cytosolic end of TM6, in stark contrast to the complexes obtained by the nonbiased ligands. Here, R265ICL3 does not interact with any polar residues on βarr2, while K2736.26 forms an internal salt bridge with D2746.25, making K2736.26 inaccessible for forming anchors with βarr2 (Fig. 6E). In the presence of nonbiased ligands, R265ICL3 and K2736.26 play crucial roles in mediating βarr2 coupling to the pp-μOR. These results suggest that the ability of pp-μOR to make polar anchors to the ICL3 or to the cytosolic end of TM6 to βarr2 may distinguish biased from nonbiased ligands. We propose that this lack of polar anchors between βarr2 and pp-μOR has significant consequences on the binding affinity between the βarr2 and the pp-μOR. To test this idea, we examined the binding free energy of the βarr2 to the pp-μOR in the presence of DAMGO, morphine, and TRV130 (Fig. 6F and SI Appendix, Figs. S7 and S8). We find that the free energy differences as the system evolves from the partially engaged to the fully engaged state are ∼−68, −65, and −62 kcal/mol for the DAMGO, morphine, and TRV130, respectively. Thus, all three ligands lead to a comparable affinity between the βarr2 and the pp-μOR. Even so, these values do indicate that the highest affinity between the βarr2 and the pp-μOR is from binding of DAMGO to the pp-μOR, while the lowest affinity is from binding of TRV130, which correlates with experimental recruitment data (24).
To determine whether the modest difference in the binding free energies is really significant, we further analyzed the nonbonded interactions (enthalpy contribution) in the fully engaged complexes. We evaluated the nonbonded interactions between the pair proteins within 12 Å excluding the effects of water and ions (Fig. 6G). We find that pp-μOR exhibits a comparable nonbonded interaction (∼−900 kcal/mol) with βarr2 when bound to morphine or DAMGO, but the nonbonded interactions decrease dramatically (by 17%) for TRV130 bound to pp-μOR (∼−750 kcal/mol). Thus, our free energy calculations and our analysis of nonbonded interactions both suggest that the affinity between the βarr2 and the pp-μOR decreases remarkably when biased TRV130 binds to the pp-μOR. This is consistent with biased activity for TRV130.
The comparable binding free energies between the pp-μOR and the βarr2 in the presence of DAMGO, morphine, and TRV130 may arise because we assume the same degree of phosphorylation for all three. Thus, the actual level of phosphorylation for each case, which is not yet available from experiment, might further differentiate the ligands in terms of their activity for the βarr2 coupling.
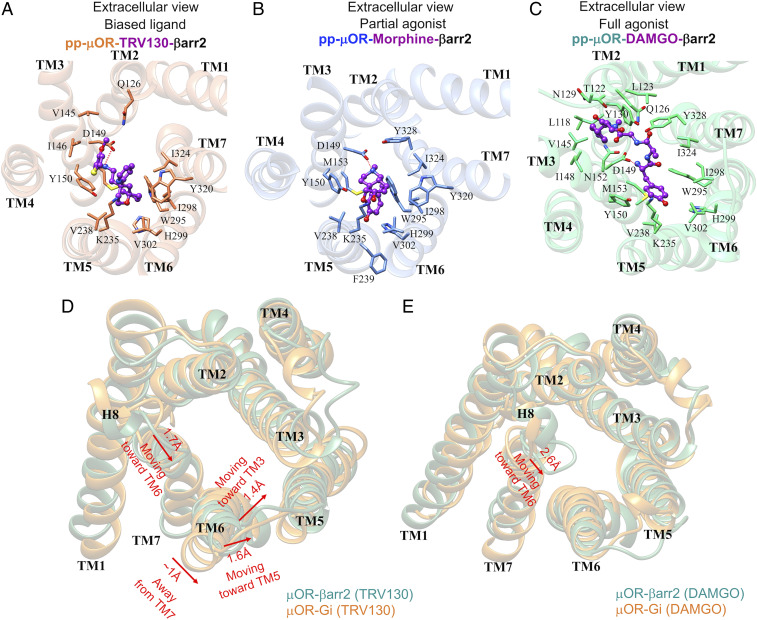
Analysis of the extracellular portion of pp-μOR reveals that all three agonists make a persistent salt bridge with D1493.32 (Fig. 7 A–C), which is a well-known anchoring point for binding of various agonists and antagonists to μOR (17, 30, 31, 46). In addition, these agonists are locked into the human μOR through extensive hydrophobic plus a few polar interactions (Fig. 7 A–C). DAMGO as a full agonist for the βarr2 coupling makes the greatest number of contacts with the orthosteric binding pocket, including TM2, TM3, TM5, TM6, and TM7. This behavior is anticipated as DAMGO is a relatively bulky ligand. Compared with DAMGO, morphine makes significantly fewer interactions with the μOR binding pocket, and it does not make any contact to TM2, explaining why morphine has 8.8-fold lower efficacy than DAMGO (24) for βarr2 recruitment.
Fig. 7.
The pp-μOR binding pocket after ∼500 ns of MD simulation stabilized by the βarr2 in the presence of (A) TRV130, a biased agonist; (B) morphine, a partial agonist; and (C) DAMGO, a full agonist for the βarr2 coupling. The dotted lines represent the hydrogen binding. (D) The structural differences between the cytoplasmic region of the μOR after recruiting the Gi protein (orange) and the βarr2 (green) in the presence of TRV130. The red arrows represent movements of the μOR induced by the βarr2. (E) The structural differences between the cytoplasmic region of the μOR after recruiting the Gi protein (orange) and the βarr2 (green) in the presence of DAMGO. The red arrow represents the only significant movement of the μOR induced by the βarr2.
On the other hand, our MD simulations show that biased ligand TRV130 binds to μOR differently even though TRV130 has a size and structure comparable with morphine. We find that TRV130 has a stronger interaction with TM2 and TM3, with a hydrophobic interaction to Q1262.60 in contrast to morphine, which has no contact to TM2. Also, TRV130 forms two hydrophobic interactions with V1453.28 and I1463.29, whereas these two residues do not contribute to morphine binding. While V1453.28 has a hydrophobic interaction with the DAMGO, I1463.29 is specific for binding of TRV130. Moreover, TRV130 also fails to make contacts to several residues involved in the binding pockets of DAMGO and morphine. Thus, M1533.36 is not involved in binding of TRV130. In contrast M1533.36 is a well-known hydrophobic residue that defines the morphinan hydrophobic pocket for antagonists and agonists (17, 46). In addition, TRV130 does not interact with Y3287.42, which contrasts dramatically with the nonbiased morphine and DAMGO. In fact, Y3287.42 plays an important role in mediating the binding pocket of morphine since mutation of Y3287.42 to phenylalanine dramatically reduces morphine binding affinity (47). We attribute the lack of interaction between TRV130 and Y3287.42 to the tendency of TRV130 to tightly engage the TM2 and TM3, which pulls TRV130 away from TM7 so that it cannot properly interact with Y3287.42. Overall, we find that the binding pocket of TRV130 differs significantly from that of the nonbiased agonist, which modifies the structure of the ICLs of the TRV130-μOR complex sufficiently that the affinity of the pp-μOR to the βarr2 is diminished substantially (Fig. 6F).
To better understand how TRV130 selectively discourages coupling of βarr2 to the pp-μOR, we compare the cytoplasmic region of μOR stabilized by both Gi protein and βarr2 (Fig. 7D). We find that for TRV130 the cytoplasmic region of μOR undergoes remarkable structural changes when βarr2 is recruited. Notably, TM6 moves ∼1 Å away from TM7 [measuring the distance between L2776.30 (Cα)-N3347.49(Cα)], while it approaches TM5 by ∼1.6 Å [measuring the distance between K2716.24(Cα)-K2625.66(Cα)] and TM3 by ∼1.1 Å [measuring the distance between K2716.24(Cα)-V1713.54(Cα)]. This remarkable repositioning of TM6 does not allow βarr2 to bind properly to ICL3 or the cytosolic end of TM6, which impedes the proper coupling of βarr2 to μOR (Fig. 7D).
Interestingly, analysis of the intracellular (IC) portion of the μOR stabilized by Gi protein compared with βarr2 for DAMGO shows that the TM6 hardly repositions, encouraging βarr2 to form an ionic anchor with the bottom of TM6 (Fig. 7E). Moreover, for DAMGO, the H8 helix on μOR has a similar movement of ∼2.6 Å toward the TM6, allowing E3438.48 to make a persistent salt bridge with R2796.32 (Fig. 1D). In addition, this movement coordinates D3428.47 to establish a charge–charge interaction with R1673.50. In contrast, TRV130 is not able to promote the creation of these two salt bridges, while morphine favors only the formation of the salt bridge between the E3438.48 and R2796.32 (Fig. 2D). Summarizing, our MD simulations indicate that the reconfiguration of the cytoplasmic region of the μOR induced by TRV130 is the main cause preventing the βarr2 from binding to μOR with high affinity.
Conclusions
We report 3D structures of the final activated state of βarr2 stabilized by the active state of pp-μOR bound to a full agonist DAMGO, a partial agonist morphine, and TRV130, which is biased against for the βarr2 coupling. We found that in the presence of the nonbiased agonists, βarr2 couples to the pp-μOR by forming strong polar interactions with ICL2 and either the ICL3 or cytoplasmic region of TM6. Interestingly, we found that Gi protein couples with μOR in a similar fashion, with Gi making similar polar contacts to the identical residues on the ICL2 and either of the ICL3 or the cytoplasmic region of TM6. These results indicate that Gi protein and βarr2 compete for the same binding site even though their recruitment leads to opposite outcomes.
On the other hand, we found that biased TRV130 has a greater tendency to bind to the extracellular portion of TM2 and TM3, inducing a repositioning of TM6 in the cytoplasmic region of the μOR, which hinders βarr2 from properly binding to pp-μOR. We found that for TRV130, βarr2 is unable to form any polar anchors to the ICL3 or the cytosolic end of TM6 (although it does make an anchor to ICL2 similar to nonbiased agonists), which causes a remarkable reduction in the affinity between the pp-μOR and βarr2. This dramatic difference in the pharmacophore for biased and nonbiased agonists suggests that ligands could be designed to have much greater biased activity.
Methods
As described in detail in SI Appendix, we built active-state complexes of βarr2-pp-μOR bound to DAMGO (full agonist), morphine (partial agonist), and TRV130 (biased agonist). Subsequently, we performed long MD simulations (∼500 ns) to optimize these complexes with lipid (POPC), water, and ions, which resulted in a simulation box of 123 × 104 × 138 Å3 with ∼180,000 atoms.
All molecules were described using AMBER force fields. The proteins were described using AMBER14 (48), while parameters for the POPC were borrowed from LIPID17, which is incorporated in Ambertools 16 (49). The phosphorylated serine and threonine residues with a net charge of −2 were parameterized using Ambertools 16 (49) with phosaa10 (50) parameters. The morphine, DAMGO, and TRV130 ligands were described using parameters obtained from the Generalized Amber force field (51) using ACPYPE (52) and Antechamber16 (53). The partial charges for the ligands were assigned with the semiempirical AM1-BCC model (54), which is incorporated in USCF chimera (55). The transferable intermolecular potential 3P (TIP3P) (56) model was used to treat the water.
We used the following simulation algorithms for the final equilibration. The temperature was maintained at 310 K using a Nose–Hoover (57, 58) thermostat with a damping constant of 1.0 ps, and the pressure was controlled at 1 bar using a Parrinello–Rahman barostat algorithm (59) with a damping constant of 5.0 ps. Semi-isotropic pressure coupling was applied during this calculation. The Lennard–Jones cutoff radius was 1.2 nm, where the interaction was smoothly shifted to zero after 1.0 nm. Unlike-atom interactions were computed using the standard Lorentz–Berthelot combination rules. Periodic boundary conditions were applied to all three directions. The short-range columbic interaction was treated within a cutoff radius of 1.2 nm, while the particle mesh Ewald (PME) algorithm (60) with a grid spacing of 0.16 nm was used to calculate the long-range electrostatic interactions. The compressibility of 4.5 × 10−5 bar−1 was used in the xy plane and z axis to relax the box volume. All simulations were performed using GROMACS (61) graphics processing unit computing algorithm with an autotuning PME. Water OH bonds were constrained by the SETTLE algorithm (62), and the remaining H bonds were constrained using the P-LINCS algorithm (63).
All data and procedures are included in the manuscript, Movies S1–S4, and the GitHub depository.
Supplementary Material
Acknowledgments
This work was partially supported through a Cargill Incorporated–Caltech Research Collaboration Project. It was also funded by gifts to the Materials and Process Simulation Center. We thank Brian Guthrie for helpful comments and suggestions.
Footnotes
The authors declare no competing interest.
Data deposition: Our optimized structure has been deposited in GitHub, https://github.com/amafi-gpcr/Beta-arrestin2-mu-opioid-receptor-agonist-complex-PNAS-2020.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918264117/-/DCSupplemental.
References
- 1.Tan L., Yan W., McCorvy J. D., Cheng J., Biased ligands of G protein-coupled receptors (GPCRs): Structure–functional selectivity relationships (SFSRs) and therapeutic potential. J. Med. Chem. 61, 9841–9878 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Singla N. et al., A randomized, Phase IIb study investigating oliceridine (TRV130), a novel µ-receptor G-protein pathway selective (μ-GPS) modulator, for the management of moderate to severe acute pain following abdominoplasty. J. Pain Res. 10, 2413–2424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manglik A. et al., Structure-based discovery of opioid analgesics with reduced side effects. Nature 537, 185–190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Hasani R., Bruchas M. R., Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology 115, 1363–1381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traut T. W., Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Pitcher J. A. et al., Role of beta gamma subunits of G proteins in targeting the beta-adrenergic receptor kinase to membrane-bound receptors. Science 257, 1264–1267 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Pitcher J. A., Touhara K., Payne E. S., Lefkowitz R. J., Pleckstrin homology domain-mediated membrane association and activation of the beta-adrenergic receptor kinase requires coordinate interaction with G beta gamma subunits and lipid. J. Biol. Chem. 270, 11707–11710 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Li J. et al., Agonist-induced formation of opioid receptor-G protein-coupled receptor kinase (GRK)-G β γ complex on membrane is required for GRK2 function in vivo. J. Biol. Chem. 278, 30219–30226 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Pitcher J. A., Freedman N. J., Lefkowitz R. J., G protein–coupled receptor kinases. Annu. Rev. Biochem. 67, 653–692 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Kahsai A. W., Pani B., Lefkowitz R. J., GPCR signaling: Conformational activation of arrestins. Cell Res. 28, 783–784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams J. T. et al., Regulation of μ-opioid receptors: Desensitization, phosphorylation, internalization, and tolerance. Pharmacol. Rev. 65, 223–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kliewer A. et al., Phosphorylation-deficient G-protein-biased μ-opioid receptors improve analgesia and diminish tolerance but worsen opioid side effects. Nat. Commun. 10, 367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arttamangkul S., Heinz D. A., Bunzow J. R., Song X., Williams J. T., Cellular tolerance at the µ-opioid receptor is phosphorylation dependent. eLife 7, e34989 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X. E. et al., Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell 170, 457–469.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mafi A., Kim S.-K., Goddard W. A. 3rd, The atomistic level structure for the activated human κ-opioid receptor bound to the full Gi protein and the MP1104 agonist. Proc. Natl. Acad. Sci. U.S.A. 117, 5836–5843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bray J. K., Abrol R., Goddard W. A. 3rd, Trzaskowski B., Scott C. E., SuperBiHelix method for predicting the pleiotropic ensemble of G-protein-coupled receptor conformations. Proc. Natl. Acad. Sci. U.S.A. 111, E72–E78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W. et al., Structural insights into µ-opioid receptor activation. Nature 524, 315–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith A. R., “Darwindock & GAG-dock: methods and applications for small molecule docking,” PhD thesis, California Institute of Technology, Pasadena, CA (2017).
- 19.Webb B., Sali A., Functional Genomics, (Springer, 2017), pp. 39–54. [Google Scholar]
- 20.Lau E. K. et al., Quantitative encoding of the effect of a partial agonist on individual opioid receptors by multisite phosphorylation and threshold detection. Sci. Signal. 4, ra52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doll C. et al., Agonist-selective patterns of µ-opioid receptor phosphorylation revealed by phosphosite-specific antibodies. Br. J. Pharmacol. 164, 298–307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan X., Gimenez L. E., Gurevich V. V., Spiller B. W., Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J. Mol. Biol. 406, 467–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Q. et al., Structural basis of arrestin-3 activation and signaling. Nat. Commun. 8, 1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeWire S. M. et al., A G protein-biased ligand at the μ-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphine. J. Pharmacol. Exp. Ther. 344, 708–717 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Mafi A., Kim S.-K., Goddard W. A., Beta-arrestin2-mu-opioid-receptor-agonist-complex-PNAS-2020. GitHub. https://github.com/amafi-gpcr/Beta-arrestin2-mu-opioid-receptor-agonist-complex-PNAS-2020. Deposited 19 May 2020.
- 26.Lally C. C. M., Bauer B., Selent J., Sommer M. E., C-edge loops of arrestin function as a membrane anchor. Nat. Commun. 8, 14258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staus D. P. et al., Structure of the M2 muscarinic receptor-β-arrestin complex in a lipid nanodisc. Nature 579, 297–302 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballesteros J. A., Weinstein H., Methods in Neurosciences, (Elsevier, 1995), Vol. 25, pp. 366–428. [Google Scholar]
- 29.Pándy-Szekeres G. et al., GPCRdb in 2018: Adding GPCR structure models and ligands. Nucleic Acids Res. 46, D440–D446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koehl A. et al., Structure of the µ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tohgo A. et al., The stability of the G protein-coupled receptor-β-arrestin interaction determines the mechanism and functional consequence of ERK activation. J. Biol. Chem. 278, 6258–6267 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Palczewski K., Buczyłko J., Imami N. R., McDowell J. H., Hargrave P. A., Role of the carboxyl-terminal region of arrestin in binding to phosphorylated rhodopsin. J. Biol. Chem. 266, 15334–15339 (1991). [PubMed] [Google Scholar]
- 33.Gurevich V. V., Benovic J. L., Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J. Biol. Chem. 268, 11628–11638 (1993). [PubMed] [Google Scholar]
- 34.Shukla A. K. et al., Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla A. K. et al., Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Latorraca N. R. et al., Molecular mechanism of GPCR-mediated arrestin activation. Nature 557, 452–456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W. et al., Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 579, 303–308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hub J. S., De Groot B. L., Van Der Spoel D., g_wham– A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 6, 3713–3720 (2010). [Google Scholar]
- 39.Patey G. N., Valleau J. P., The free energy of spheres with dipoles: Monte Carlo with multistage sampling. Chem. Phys. Lett. 21, 297–300 (1973). [Google Scholar]
- 40.Torrie G. M., Valleau J. P., Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 23, 187–199 (1977). [Google Scholar]
- 41.Kumar S., Rosenberg J. M., Bouzida D., Swendsen R. H., Kollman P. A., The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992). [Google Scholar]
- 42.Nobles K. N., Guan Z., Xiao K., Oas T. G., Lefkowitz R. J., The active conformation of β-Arrestin1 direct evidence for the phosphate sensor IN the N-domain and conformational differences IN the active states OF β-ARRESTINS1 AND-2. J. Biol. Chem. 282, 21370–21381 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Kumari P. et al., Functional competence of a partially engaged GPCR-β-arrestin complex. Nat. Commun. 7, 13416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomsen A. R. B. et al., GPCR-G protein-β-arrestin super-complex mediates sustained G protein signaling. Cell 166, 907–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cahill T. J. 3rd et al., Distinct conformations of GPCR-β-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc. Natl. Acad. Sci. U.S.A. 114, 2562–2567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manglik A. et al., Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raynor K. et al., Pharmacological characterization of the cloned kappa-, delta-, and mu-opioid receptors. Mol. Pharmacol. 45, 330–334 (1994). [PubMed] [Google Scholar]
- 48.Dickson C. J. et al., Lipid14: The amber lipid force field. J. Chem. Theory Comput. 10, 865–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Case D. A., et al. , AMBER16 (University of California, San Francisco, CA 2016).
- 50.Homeyer N., Horn A. H., Lanig H., Sticht H., AMBER force-field parameters for phosphorylated amino acids in different protonation states: Phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J. Mol. Model. 12, 281–289 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A., Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Sousa da Silva A. W., Vranken W. F., ACPYPE-Antechamber python parser interface. BMC Res. Notes 5, 367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J., Wang W., Kollman P. A., Case D. A., Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Jakalian A., Jack D. B., Bayly C. I., Fast, efficient generation of high-quality atomic charges. AM1-BCC model. II. Parameterization and validation. J. Comput. Chem. 23, 1623–1641 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Pettersen E. F. et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 57.Nosé S., A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268 (1984). [Google Scholar]
- 58.Hoover W. G., Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 59.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 60.Essmann U. et al., A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995). [Google Scholar]
- 61.Abraham M. J. et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015). [Google Scholar]
- 62.Miyamoto S., Kollman P. A., Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992). [Google Scholar]
- 63.Hess B., P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.