Significance
Peroxiredoxins are most central to the cellular adaptation against oxidative stress. They act as oxidant scavengers, stress sensors, transmitters of signals, and chaperones, and they possess a unique quaternary switch that is intimately related to these functions. However, so far it has not been possible to monitor peroxiredoxin structural changes in the intact cellular environment. This study presents genetically encoded probes, based on homo-FRET (Förster resonance energy transfer between identical fluorophores) fluorescence polarization, that allow following these quaternary changes in real time, in living cells. We envisage that these probes can be used to address a broad range of questions related to the function of peroxiredoxins.
Keywords: peroxiredoxins, protein oligomerization, dimer-decamer equilibrium, homo-FRET, fluorescence polarization
Abstract
Peroxiredoxins are central to cellular redox homeostasis and signaling. They serve as peroxide scavengers, sensors, signal transducers, and chaperones, depending on conditions and context. Typical 2-Cys peroxiredoxins are known to switch between different oligomeric states, depending on redox state, pH, posttranslational modifications, and other factors. Quaternary states and their changes are closely connected to peroxiredoxin activity and function but so far have been studied, almost exclusively, outside the context of the living cell. Here we introduce the use of homo-FRET (Förster resonance energy transfer between identical fluorophores) fluorescence polarization to monitor dynamic changes in peroxiredoxin quaternary structure inside the crowded environment of living cells. Using the approach, we confirm peroxide- and thioredoxin-related quaternary transitions to take place in cellulo and observe that the relationship between dimer–decamer transitions and intersubunit disulfide bond formation is more complex than previously thought. Furthermore, we demonstrate the use of the approach to compare different peroxiredoxin isoforms and to identify mutations and small molecules affecting the oligomeric state inside cells. Mutagenesis experiments reveal that the dimer–decamer equilibrium is delicately balanced and can be shifted by single-atom structural changes. We show how to use this insight to improve the design of peroxiredoxin-based redox biosensors.
Peroxiredoxins are highly versatile proteins. They have long been recognized for their enzymatic role as thiol peroxidases, acting as highly efficient scavengers of H2O2 (and other peroxides) and protecting the cell against oxidative damage (1–3). It is now clear that they can also serve as highly sensitive peroxide sensors and as mediators of protein redox regulation (4). As such, they form part of protein complexes involved in signal transduction and, by either preventing or facilitating the oxidation of redox-regulated proteins, initiate or modulate a multitude of adaptive cellular processes (5–9). They also seem to fulfill additional functions, less well characterized, in particular as chaperones (10). Consistent with their regulatory role, peroxiredoxins appear to be themselves regulated by numerous posttranslational modifications (PTMs) (11). Among the different peroxiredoxins, the typical 2-Cys peroxiredoxins (Prxs), members of the Prx1 class (12), are of particular interest, as they are the major Prxs in the cytosol, nucleus, and mitochondrial matrix of mammalian cells, centrally involved in both redox homeostasis and cell signaling.
Almost all peroxiredoxins, including the typical 2-Cys peroxiredoxins, are homooligomeric proteins. They exist in different oligomeric states and undergo dynamic oligomeric transitions. The basic units are stable homodimers. These have been observed to form toroidal complexes of 8, 10, or 12 subunits. The typical 2-Cys peroxiredoxins usually form doughnut-shaped decamers (i.e., pentamers of dimers) (12). Electron micrographs showing toroidal rings provided the first description of “torin,” now known as Prx2 (13, 14). Prx toroids have been seen to interlock or stack, to form nanotubes or even dodecahedra (10). It should, however, be stressed that the in vivo relevance of Prx oligomeric forms and transitions remains uncertain. The reason is that Prx oligomeric states are usually prepared using recombinant proteins in vitro and under special conditions (e.g., for electron microscopy or crystallography) or are analyzed outside their natural environment (size-exclusion chromatography [SEC] or native gel electrophoresis). It seems that in particular the existence of the very-high-molecular-weight complexes (i.e., stacked toroids) in an intact intracellular environment remains to be established. In contrast, the switching between dimers and decamers/dodecamers is well studied and likely to be relevant inside living cells (15). However, little is known about the dynamics of the dimer–toroid transition in the crowded intracellular environment. What is still missing is a method that allows the quantitative real-time monitoring of Prx oligomeric states in their native cellular setting.
Prx oligomeric states and their transitions are connected to Prx function (16). Concerning the peroxide scavenging function of typical 2-Cys Prxs, the connection between redox and oligomeric state has been described (based on in vitro experiments) as follows: Prx dimers in the reduced state prefer to assemble into decamers, which are observed to be more reactive toward H2O2 than dimers (17). The conformational change associated with Prx oxidation (i.e., sulfenic acid formation, followed by intersubunit disulfide bond formation), the so-called local unfolding (LU) transition, then destabilizes dimer–dimer interfaces, leading to dissociation of decamers into disulfide-linked dimers. Reduction of the intersubunit disulfide bonds returns the Prx dimer to the so-called fully folded (FF) conformation, which allows reassembly into decamers and restarting of the cycle (15). Concerning other Prx functions, it has been suggested that stacking of at least two toroids is required to enable Prx chaperone activity (10). The oligomeric state relevant to Prx signaling functions remains to be defined. The situation is further complicated by the fact that the Prx oligomeric state is influenced by additional factors beyond the redox state; these include pH (18), oxidative thiol modifications that are not part of the canonical dithiol–disulfide cycle (i.e., glutathionylation and hyperoxidation) (19–21), and various non-thiol-based PTMs, including acetylation, phosphorylation, and nitration (11, 22, 23). Most PTMs have been shown to alter the dimer–decamer equilibrium, suggesting that cells modulate Prx function through regulation of their oligomeric state.
Given that oligomeric state is central to the understanding of Prx function and regulation, and given the many unknowns about Prx oligomer dynamics inside living cells, we aimed to establish a noninvasive technique that allows following Prx–Prx homotypic interactions inside living cells. We therefore employed homo-FRET (Förster resonance energy transfer), which reflects homotypic crowding by altering fluorescence polarization (anisotropy), depending on the proximity relationships between two or more identical fluorophores. Homo-FRET has been used previously to measure homotypic protein clustering by confocal and two-photon microscopy (24–27). Here we establish the use of a microplate reader to quantitatively monitor Prx oligomeric state by fluorescence polarization in real time and in living cells. We characterize the behavior of Prx-based homo-FRET probes in cellulo and in vitro, demonstrate how fluorophore density and oligomeric state relate to polarization, and clarify how probe responses are influenced by heterooligomerization with endogenous Prx. We then show that intracellular decamer–dimer transitions are only partially mirrored by the formation of Prx intersubunit disulfide bonds. By comparing Prx point mutants we also show that the dimer–decamer equilibrium is in a delicate balance, as it can be almost completely shifted by a single-atom sulfur-to-oxygen replacement (Cys-to-Ser) within the C-terminal tail. Based on the knowledge of how individual mutations influence the dimer–decamer equilibrium in cellulo, we designed a roGPF2-based probe for the redox state of human Prx2 and show that it can resolve subtle differences in peroxide levels between different types of tumor cells. Furthermore, we give examples of how homo-FRET can be used to compare the behavior of different Prx isoforms and to assess the influence of nutrient conditions and small molecules on oligomer dynamics inside cells.
Results
The Fluorescence Polarization Associated with Prx2-Fused Monomeric Cerulean Reflects the Relationship Between Intracellular H2O2 Levels and Reducing Capacity.
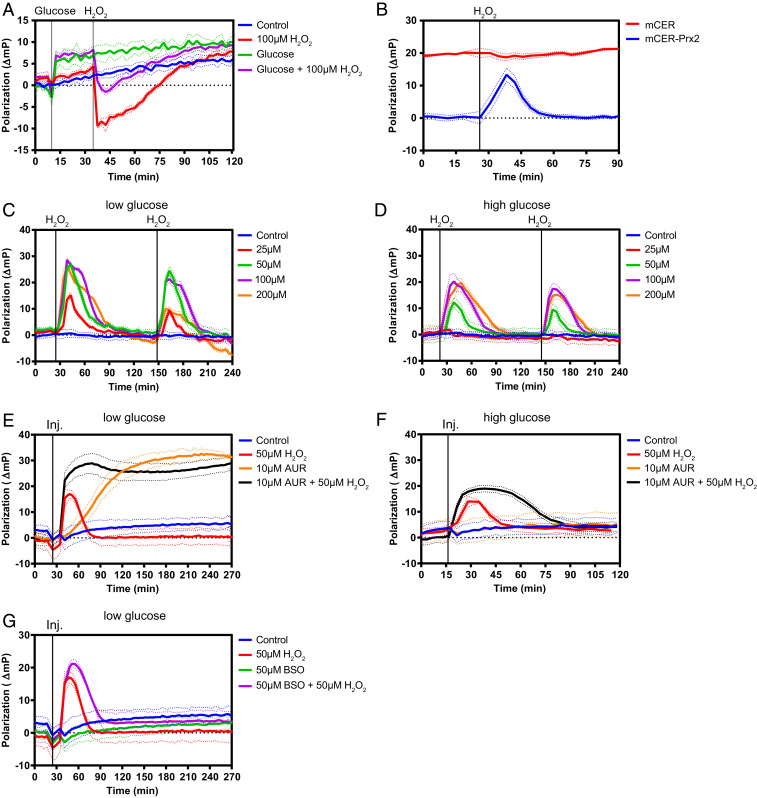
To evaluate the potential of microplate reader-based homo-FRET measurements we initially tested the previously described Apollo-NADP+ probe, which is a fusion between monomeric cerulean (mCER) and truncated glucose-6-phosphate dehydrogenase (G6PDH). This probe measures NADP+-induced homodimerization of G6PDH in living cells, as previously demonstrated by two-photon microscopy (26). Polarization changes observed in the microplate reader were consistent with results previously obtained by microscopy: glucose supply (leading to NADP+ reduction) increased polarization (indicating stabilization of the monomeric state) and peroxide treatment (leading to NADPH oxidation) decreased polarization (indicating NADP+-induced dimerization) (Fig. 1A). This result suggested that a microplate reader-based approach may also be suitable to monitor the dynamics of higher-order homooligomeric protein complexes, which are currently difficult to assess by microscopy. We then asked if homo-FRET fluorescence polarization is capable of measuring the oligomeric state of a typical 2-Cys peroxiredoxin, a class of protein known to reversibly transition between dimeric and decameric states. To this end we translationally fused mCER to the N terminus of human peroxiredoxin-2 (Prx2). The mCER-Prx2 fusion protein was stably expressed in HEK293 cells, at levels similar to or slightly higher than endogenous Prx2. Decamers incorporating mCER-Prx2 are expected to exhibit higher homo-FRET efficiency and therefore lower fluorescence polarization, relative to dimers. Indeed, exposure of cells to a bolus of exogenous H2O2 (expected to induce dissociation of decamers into dimers) led to a transient and fully reversible increase in fluorescence polarization, while cells expressing unfused mCER did not show a response (Fig. 1B). In vitro measurements comparing untagged and mCER-tagged Prx2 did not reveal kinetic differences in H2O2 reactivity (SI Appendix, Fig. S1A), suggesting that the mCER tag does not compromise Prx2 responsiveness. Mutagenesis of Cys-48, the single solvent-exposed cysteine of mCER, did not influence the probe response, suggesting that this cysteine does not interfere with probe function (SI Appendix, Fig. S1B). Titration of H2O2 demonstrated the dose dependency of the polarization change and confirmed the reversibility and repeatability of the response (Fig. 1 C and D). Under conditions of low glucose availability (limiting the regeneration of NADPH) the maximal polarization change was ∼30 millipolarization (mP) units (Fig. 1C), while under high glucose (supporting NADPH regeneration) the polarization change was more limited (Fig. 1D). We then investigated the influence of the intracellular reducing systems. Under conditions of low glucose availability, inhibition of thioredoxin reductase (TrxR), alone or in combination with H2O2 treatment, strongly induced the decamer-to-dimer transition (Fig. 1E). When the same experiment was conducted in glucose-rich medium the degree of Prx decamer dissociation induced by TrxR inhibition was substantially decreased (Fig. 1F). In comparison, the inhibition of glutathione biosynthesis had relatively minor effects (Fig. 1G). In conclusion, the fluorescence emitted by intracellular mCER-Prx2 polarizes in response to H2O2 and depolarizes in response to the NADPH-dependent Trx/TrxR system. Therefore, the photophysical behavior of mCER-Prx2 matches the well-accepted notion of a Prx decamer–dimer–decamer oscillation that is driven by H2O2 and the Trx system.
Fig. 1.
The fluorescence polarization associated with Prx2-fused mCER reflects the relationship between intracellular H2O2 levels and cellular reducing capacity. (A) Polarization changes facilitated by mCER-tagged G6PDH (Apollo-NADP+) in response to glucose and/or H2O2. (B) mCER-Prx2, but not unfused mCER, facilitates a reversible polarization change in response to a single bolus of H2O2 (50 µM). (C) Polarization changes in response to repeated H2O2 boli applied in low-glucose medium (5.5 mM). (D) Polarization changes in response to repeated H2O2 boli applied in high-glucose medium (25 mM). (E) Polarization changes in response to the TrxR inhibitor auranofin (AUR) and/or H2O2 in low-glucose medium (5.5 mM). (F) Polarization changes in response to the TrxR inhibitor AUR and/or H2O2 in high-glucose medium (25 mM). (G) Polarization changes in response to the glutathione biosynthesis inhibitor buthionine sulphoximine (BSO) and/or H2O2 in low-glucose medium (5.5 mM). All graphs in this figure are based on n = 3 biological replicates with six technical replicates each.
Heterooligomerization with Endogenous Prx2 Alters the Amplitude but Not the Onset or Duration of Probe Responses.
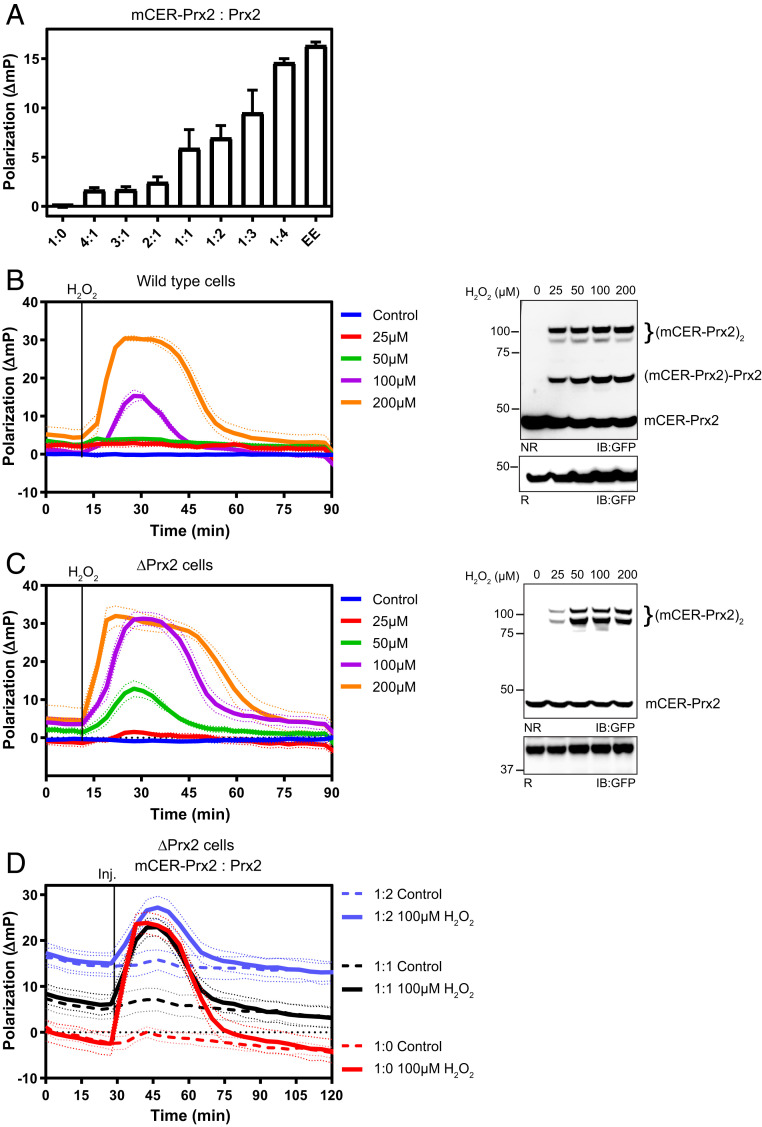
In the preceding experiments we ectopically expressed mCER-Prx2 in cells already expressing endogenous Prx2, at similar levels. We therefore expected mCER-Prx2 to form homodimers (mCER-Prx2:mCER-Prx2) as well as heterodimers with endogenous Prx2 (mCER-Prx2:Prx2). Accordingly, we expected decamers to be composed of a mixture of three kinds of dimers: Prx2:Prx2, mCER-Prx2:mCER-Prx2, and mCER-Prx2:Prx2. Assuming equimolar expression of mCER-Prx2 and Prx2, the average dimer is expected to contain one fluorophore and the average decamer to contain five fluorophores (SI Appendix, Fig. S2A). While fluorophore crowding will be higher in decamers than in dimers at any stoichiometry, the polarization difference between dimers and decamers (i.e., the dynamic range) is predicted to increase with the mCER-Prx2-to-Prx2 ratio. To test these basic assumptions about stoichiometry-dependent heterocomplex formation and fluorophore crowding, we mixed purified mCER-Prx2 and Prx2 under decamer-stabilizing conditions in vitro and measured the resulting polarization. An increasing proportion of Prx2 led to increased polarization (Fig. 2A), confirming that average fluorophore density determines the degree of decamer-specific polarization (corresponding to the “baseline” polarization in Fig. 1 B–G). To test these notions in the cellular context, we expressed mCER-Prx2 in Prx2-proficient (wild-type, WT) and -deficient (ΔPrx2) cells (SI Appendix, Fig. S2B) and monitored the response to H2O2 side by side (Fig. 2 B and C, Left). The responses were of similar shape but more pronounced in ΔPrx2 cells, in agreement with an enhanced dynamic range in polarization. The corresponding immunoblots confirmed that mCER-Prx2 forms covalent heterodimers with endogenous Prx2 in WT cells (Fig. 2 B, Right), which are naturally absent in ΔPrx2 cells (Fig. 2 C, Right). To achieve a direct comparison of different stoichiometries within the same cell line, we then coexpressed different ratios of untagged Prx2 and mCER-Prx2 in ΔPrx2 cells. The lower the mCER-Prx2-to-Prx2 ratio, the higher the decamer polarization (i.e., the “baseline”) and the lower the decamer-to-dimer dynamic range (Fig. 2D). Importantly, the responses were identical, except for the amplitude of the polarization change, demonstrating that the ratio of mCER-Prx2 to Prx2 determines sensitivity but not the temporal aspects of the probe response.
Fig. 2.
Heterooligomerization of the probe with endogenous Prx2 determines the amplitude of the polarization change but does not alter the onset or duration of the response. (A) Polarization associated with recombinant mCER-Prx2, mixed with Prx2 at different ratios, under decamer stabilizing conditions. EE denotes the mCER-Prx2(T82E,A85E) mutant, which does not form decamers and therefore only supports minimal depolarization. The bars indicate the mean ± SEM. (B, Left) H2O2-dependent polarization responses of WT HEK293 cells transiently transfected with mCER-Prx2. (B, Right) Corresponding anti-GFP immunoblot, 5 min after H2O2 exposure. NR/R: non-reducing/reducing gel electrophoresis. (C, Left) H2O2-dependent polarization responses of ΔPrx2 cells transiently transfected with mCER-Prx2. (C, Right) Corresponding anti-GFP immunoblot, 5 min after H2O2 exposure. (D) H2O2-dependent polarization responses of ΔPrx2 cells transiently cotransfected with mCER-Prx2 and Prx2 at different ratios. All graphs in this figure are based on n = 3 biological replicates with six technical replicates each. All immunoblots in this figure are representative of n ≥ 3 experiments.
The fact that mCER-Prx2 forms two kinds of covalent dimers (Fig. 2 B, Right) raised the question of whether the mCER tag influences the disulfide reduction step and if there are kinetic differences in the reduction of the three types of covalent dimers (Prx2:Prx2, mCER-Prx2:mCER-Prx2, and mCER-Prx2:Prx2). While some experiments did not show any significant difference (SI Appendix, Fig. S2C), others indicated that probe homodimers are more slowly reduced than probe heterodimers (SI Appendix, Fig. S2D). In vitro reduction experiments using a recombinant Trx/TrxR/NADPH system confirmed that Prx2:Prx2 homodimers are slightly better reduced than mCER-Prx2:mCER-Prx2 homodimers (SI Appendix, Fig. S2E), while mCER-Prx2:Prx2 heterodimers showed reduction kinetics closely matching Prx2 homodimers (SI Appendix, Fig. S2F). These results suggest that the presence of two mCER tags within one Prx dimer slightly retards Trx-mediated reduction, most likely due to steric hindrance.
The formation of probe heterodimers furthermore raised the question of whether endogenous Prx1 may also be forming heterodimers with mCER-Prx2. We were able to detect covalent mCER-Prx2:Prx1 heterodimers in both WT and ΔPrx2 cells (SI Appendix, Fig. S2 G and H). However, their relative abundance appears to be minor in relation to mCER-Prx2:Prx2 heterodimers (SI Appendix, Fig. S2I) or Prx1:Prx1 homodimers (SI Appendix, Fig. S2 G and H). This indicates that Prx1 strongly prefers to form homodimers over heterodimers with Prx2 and/or that mCER-Prx2 strongly prefers pairing with Prx2 over Prx1. We therefore conclude that the presence of Prx1 is not a major complicating factor for the interpretation of mCER-Prx2-based measurements.
H2O2 Triggers Dissociation of Decamers into a Mixture of Covalent and Noncovalent Dimers.
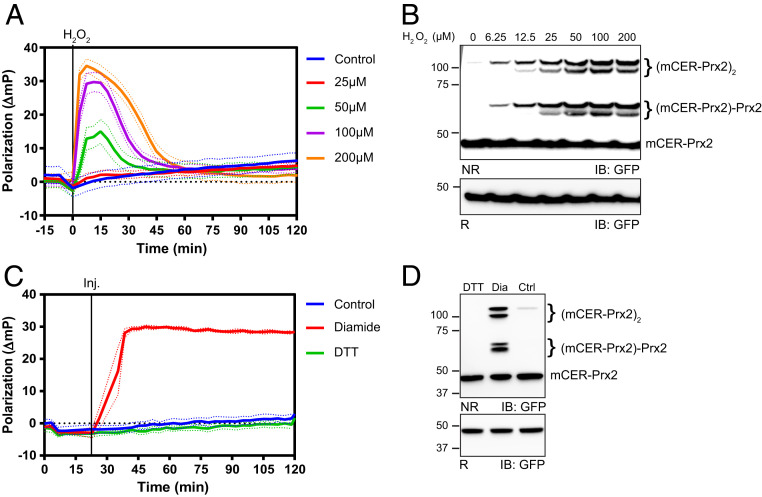
Having established a photophysical readout of redox-related dimer–decamer transitions (Fig. 1) and having clarified the influence of heterocomplex formation (Fig. 2), we then asked about the relationship between fluorescence polarization changes and intersubunit disulfide bond formation. To this end we monitored the decamer–dimer–decamer oscillation as induced by increasing concentrations of H2O2 and analyzed samples (at time t = 2 min, representing maximal polarization) from an identical side-by-side-experiment by non-reducing gel electrophoresis. At a bolus concentration of 200 µM (corresponding to 60 fmoles of H2O2 per cell under the conditions of the experiment) the polarization increase approached 30 mP units, suggesting that the whole intracellular pool of Prx2:mCER-Prx2 heterodecamers was dissociated into dimers (Fig. 3A). The peak polarization change induced by 200 µM H2O2 was confirmed to be close to maximal, as higher H2O2 concentrations did not lead to further increases (SI Appendix, Fig. S3 A and B). Complete reassociation of dimers into decamers occurred over the next ∼45 min, which is also the time required by HEK293 cells to clear an H2O2 bolus from the supernatant (28). The corresponding non-reducing gel electrophoresis (for t = 2 min) confirmed that H2O2 triggered the formation of covalent homo- and heterodimers (each appearing as a double band representing one and two intersubunit disulfide bonds) (Fig. 3B). Interestingly, complete (or near-complete) dissociation of decamers into dimers (as indicated by maximal polarization) was not accompanied by complete (or near-complete) covalent dimerization. Probed side by side under identical conditions, endogenous Prx2 in cells not expressing mCER-Prx2 also showed incomplete covalent dimerization (SI Appendix, Fig. S3C). Likewise, when the decamer-to-dimer transition was irreversibly driven to the maximum by diamide (which oxidizes glutathione and thereby depletes NADPH) (Fig. 3C), only part of the fully dissociated mCER-Prx2 pool was converted into covalent dimers (Fig. 3D). In conclusion, these findings suggest that partial oxidation of the Prx2 decamer, that is, intermolecular disulfide formation in just one or two dimers, is sufficient to trigger complete decamer dissociation. They also show that the ratio of covalent dimers to monomers (as visualized on nonreducing gels) cannot be interpreted to represent the intracellular dimer–decamer ratio.
Fig. 3.
H2O2 triggers dissociation of decamers into a mixture of covalent and noncovalent dimers. (A) Polarization response of mCER-Prx2–expressing cells treated with different concentrations of H2O2 in low-glucose medium (5.5 mM). (B) Non-reducing immunoblot analysis from the same cells as in A, 2 min after H2O2 exposure. NR/R: non-reducing/reducing gel electrophoresis. (C) Polarization response of mCER-Prx2 expressing cells treated with 2 mM diamide or 10 mM DTT. (D) Non-reducing immunoblot analysis from the same cells as in C, 15 min after diamide exposure. All graphs in this figure are based on n = 3 biological replicates with six technical replicates each. All immunoblots in this figure are representative of n ≥ 3 experiments.
Mutational Perturbation of the mCER-Prx2 Dimer–Decamer Equilibrium.
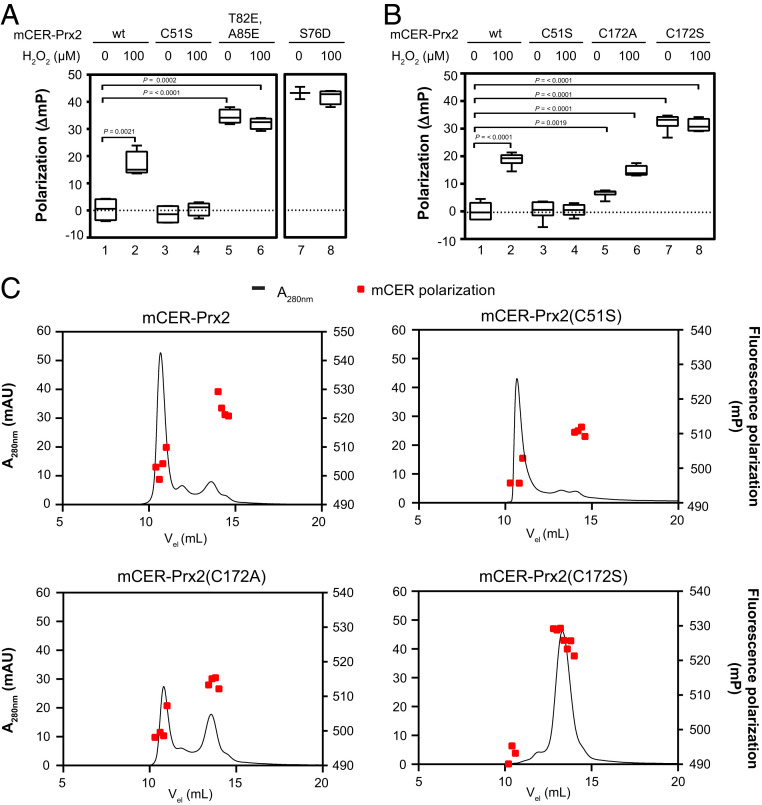
To further test the notion that mCER-Prx2 fluorescence polarization reflects the Prx2 oligomeric state, we introduced point mutations expected to affect the dimer–decamer equilibrium. In a first step, we aimed to lock mCER-Prx2 in the decameric state. To this end, we rendered mCER-Prx2 oxidation-insensitive by removing the peroxidatic cysteine (C51S). As expected, the C51S mutant remained decameric in the presence of H2O2 (Fig. 4A, columns 3 and 4 and SI Appendix, Fig. S4A). In the next step, we aimed to lock mCER-Prx2 in the dimeric state. The introduction of charged residues at the A-type dimer–dimer interface has previously been described to prevent decamer formation (29, 30). As expected, the double mutation T82E/A85E completely prevented decamer formation (Fig. 4A, columns 5 and 6 and SI Appendix, Fig. S4A). Based on structural considerations, we predicted that replacement of Ser-76 (a potential phosphorylation site at the dimer–dimer interface) by aspartate (mimicking phosphorylation) should also prevent decamer formation. This was indeed the case (Fig. 4A, columns 7 and 8), showing that the approach allows evaluating the influence of previously untested Prx mutations on the intracellular oligomeric state. We then asked how mutations of the resolving cysteine (CR) would influence the oligomeric state. We mutated CR to either alanine (C172A) or serine (C172S). Under nonoxidizing conditions, the Cys-to-Ala mutant showed slightly higher polarization relative to WT Prx2 (Fig. 4B, compare columns 1 and 5, and SI Appendix, Fig. S4B), suggesting that the loss of the thiol group mildly destabilizes the decameric state. Yet, the mutant responded to H2O2 by further dissociating into dimers (Fig. 4B, column 6 and SI Appendix, Fig. S4B). Interestingly, the Cys-to-Ser mutant exhibited maximal polarization already in the absence of oxidizing conditions and showed no further response to H2O2, suggesting that the presence of the hydroxyl group in the CR position strongly destabilizes the decameric state (Fig. 4B, columns 7 and 8 and SI Appendix, Fig. S4B). Non-reducing gel electrophoresis showed that all tested mutants, except T82E/A85E, lost their ability to form intersubunit disulfide bonds (SI Appendix, Fig. S4C) and confirmed that the intensity of the monomer band is uninformative about the decameric state. In vitro measurements with recombinant proteins furthermore confirmed that the C51S mutant is completely unreactive toward H2O2, the S76D mutant is partially reactive, and all other mutants (C172A, C172S, and T82E/A85E) are almost fully reactive (SI Appendix, Fig. S4D). To further validate the interpretation of the polarization measurements, we analyzed the recombinant proteins by conventional SEC (Fig. 4C). A serial dilution experiment demonstrated that a minimum concentration of 2 μM is required to maintain recombinant mCER-Prx2 in the decameric state (SI Appendix, Fig. S4E), in close agreement with the value previously reported for untagged Prx2 (31). At this concentration and in agreement with cell-based polarization measurements, mCER-Prx2 was predominantly decameric (Fig. 4 C, Upper Left), the C51S mutant almost exclusively decameric (Fig. 4 C, Upper Right), the C172A mutant a mixture of dimers and decamers (Fig. 4 C, Lower Left), and the C172S mutant exclusively dimeric (Fig. 4 C, Lower Right). When SEC fractions were collected and promptly measured in the microplate reader, the correlation between elution volume and fluorescence polarization was largely maintained in that dimer peak fractions showed higher polarization than decamer peak fractions (Fig. 4C, red squares). In conclusion, the comparison between polarization and SEC, across a range of mutants known to affect the oligomeric state, strongly supports the notion that mCER-Prx2 polarization is a trustworthy readout of the Prx oligomeric state.
Fig. 4.
Mutational perturbation of the mCER-Prx2 dimer–decamer equilibrium. (A) Polarization recorded from cells expressing mCER-Prx2 or the mutants C51S, T82E/A85E, and S76D, before and after H2O2 exposure. (B) Polarization recorded from cells expressing mCER-Prx2 or the mutants C51S, C172A, and C172S, before and after H2O2 exposure. (C) Oligomeric state of recombinant mCER-Prx2 proteins (WT, C51S, C172A, C172S) as analyzed by SEC. The black line indicates protein absorbance at 280 nm; red squares indicate fluorescence polarization values. The box plots in this figure are based on n = 3 biological replicates with six technical replicates each. The chromatograms in this figure are representative of n ≥ 3 experiments.
Applications for mCER-Prx2 and Related Probes.
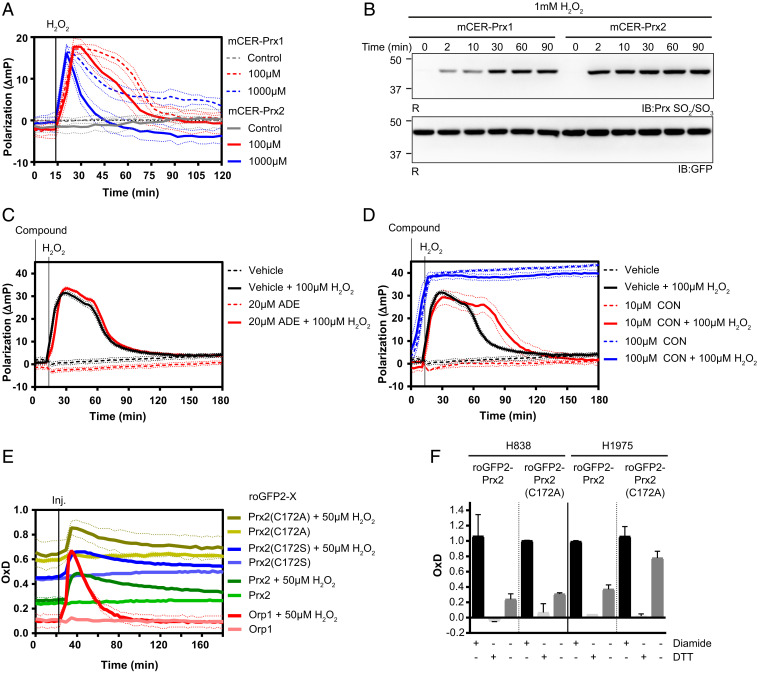
Several applications can be envisaged for mCER-Prx fusion proteins. For example, mCER fusion proteins can be used to compare the intracellular behavior of different Prx family members, for example the closely related human cytosolic typical 2-Cys peroxiredoxin isoforms Prx1 and Prx2. For proof of concept, we monitored the intracellular response of mCER-Prx2 and mCER-Prx1 side by side (Fig. 5A). While the kinetics of H2O2-induced dissociation of decamers into dimers was similar, reassociation into decamers was slower for mCER-Prx1. Interestingly, very high H2O2 concentrations induced a drop of mCER-Prx2 probe polarization below the “baseline” level corresponding to the decamer state (the same phenomenon is also seen in SI Appendix, Fig. S3A at 400 μM H2O2), suggesting that mCER-Prx2 (but not mCER-Prx1) assembles into higher-than-decamer oligomeric states, most likely stacked decamers. Formation of such higher-order aggregates may be triggered by overoxidation, as previously suggested (32, 33). Indeed, mCER-Prx2 was found to be rapidly hyperoxidized, while mCER-Prx1 was more resistant (Fig. 5B).
Fig. 5.
Applications for mCER-Prx2 and related probes. (A) Comparison of the polarization response associated with mCER-Prx1 and mCER-Prx2 in response to H2O2. (B) Hyperoxidation of mCER-Prx1 and mCER-Prx2 in response to H2O2 (1 mM), as analyzed by reducing (R) gel electrophoresis and immunoblotting. (C) mCER-Prx2 polarization response to adenanthin (ADE) in the presence or absence of H2O2. (D) mCER-Prx2 polarization response to conoidin A (CON) in the presence or absence of H2O2. (E) Degree of oxidation (OxD) measurements for various roGFP2 fusion (roGFP2-X) proteins [roGFP2-Prx2, roGFP2-Prx2(C172A), roGFP2-Prx2(C172S) and roGFP2-Orp1] expressed in HEK293 cells, at steady state and in response to H2O2. (F) Steady-state OxD measurements for roGFP2-Prx2 and roGFP2-Prx2(C172A), as expressed in two non-small-cell lung cancer cell lines (H838 and H1975), together with reference treatments (diamide for full oxidation, DTT for full reduction). The bars indicate the mean ± SEM. All graphs in this figure are based on n = 3 biological replicates with six technical replicates each. All immunoblots in this figure are representative of n ≥ 3 experiments.
Another potential application for mCER-Prx probes is to identify small-molecule modulators of the Prx oligomeric state. For proof of concept, mCER-Prx2–expressing cells were separately exposed to two small molecules, adenanthin and conoidin A, previously proposed to act as Prx2 inhibitors (34, 35). Adenanthin did not cause any shift in polarization, alone or in combination with H2O2 (Fig. 5C). A high concentration of conoidin A directly induced decamer-to-dimer dissociation, and a lower concentration delayed the reassociation of dimers into decamers following exposure to H2O2 (Fig. 5D).
The insights obtained through polarization measurements can also be exploited to develop or optimize complementary probes and detection systems. We recently developed the roGFP2-Tsa2ΔCR redox probe which detects peroxide-mediated oxidation of the yeast 2-Cys peroxiredoxin Tsa2. In this construct, removal of the resolving cysteine (ΔCR) by mutagenesis (C171A) slows down the reduction of oxidized Tsa2, thus promoting transfer of oxidation to roGFP2 and prolonging the half-life of the probe’s oxidized state. The probe has been confirmed to form mixed decamers with endogenous Tsa2 (36). The results shown in Fig. 4B suggested that a corresponding probe based on human Prx2 (for use in human cells) would form decamers if the resolving cysteine was replaced by alanine, but not if it was replaced by serine. Decamers are supposed to react more sensitively to H2O2 than dimers (37), and hence we predicted that the alanine mutant would be a more sensitive probe than the serine mutant. Based on these considerations, we generated, expressed, and compared roGFP2-Prx2(WT), roGFP2-Prx2(C172S), and roGFP2-Prx2(C172A) (Fig. 5E). Indeed, the Prx2(C172A) construct exhibited the highest steady-state degree of oxidation (OxD) relative to the other constructs. Steady-state OxD reflects the balance between probe oxidation and reduction (38). From this we concluded that the roGFP2-Prx2(C172A) probe may be especially useful to detect small differences in H2O2 homeostasis between different human (tumor) cell lines. To test this notion, we stably expressed the roGFP2-Prx2(WT) and roGFP2-Prx2(C172A) probes in nonsmall lung cancer cell lines H838 and H1975 and compared their steady-state OxD (Fig. 5F). H838 cells are expected to exert a stronger reducing activity on the Prx system, due to constitutive Nrf2 activation (39). Indeed, the C172A probe shows a larger difference in steady-state oxidation between H838 and H1975 cells than the WT probe does, demonstrating the greater resolution afforded by the roGFP2-Prx2(C172A) probe.
Discussion
Acting as peroxide scavengers, sensors, and signal transmitters, peroxiredoxins are most central to redox biology. Their manifold functions are closely connected to oligomeric states and transitions. The fact that little is known about the dynamics of Prx oligomer assembly and disassembly inside intact living cells prompted us to look for new experimental tools.
Already a few years back, the use of hetero-FRET to monitor the oligomeric state of Prxs was investigated by the Dietz laboratory. Working on plant peroxiredoxins, they explored three different strategies. One study fused cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) to the N and C terminus, respectively, of chloroplast 2-Cys Prx (40). Another study investigated so-called two-step hetero-FRET, which requires three separate fusion constructs (Prx-CFP, Prx-YFP, and Prx-mCherry) to be coexpressed (41). Yet another study fused a photoconvertible fluorescent protein (Kaede) to Prx (42). While all three hetero-FRET approaches were considered blueprints for further development, it seems that they were not advanced or applied beyond those initial studies. More recently, a hetero-FRET construct for human Prx2 (Clover-Prx2-mRuby2) was presented (43). Although the construct was shown to respond to H2O2, it has not been clarified which conformational and/or oligomeric change is actually reflected by the measured change in hetero-FRET.
In this study, we made use of energy transfer between identical fluorophores (i.e., homo-FRET) to monitor the assembly of fluorophore-tagged typical 2-Cys peroxiredoxins into different kinds of proximity relationships (i.e., types of oligomeric complexes). Although donor and acceptor fluorophores are spectrally identical, energy transfer depolarizes incident polarized light upon excitation–emission and can therefore be detected by measuring fluorescence polarization (25). An absolute photophysical parameter such as polarization is especially suitable for quantitative measurements. Homo-FRET is already an established tool for the quantification of protein clustering by imaging (44). In principle, it can be employed to quantify oligomer sizes as well as oligomer size distributions (45). While previous fluorescence polarization measurements were based on microscopy, we here demonstrate the use of a microplate reader to monitor real-time changes in fluorescence polarization from genetically encoded fluorophores expressed in living cells.
Considering the design of a Prx oligomerization probe, an obvious advantage of homo- over hetero-FRET is the need for just one type of fluorophore. Hence, only one protein terminus needs to be modified, reducing the risk of altering normal Prx behavior. As fluorophore we chose the CFP variant mCER. The design of the mCER-Prx conformational probes paralleled that of the previously developed roGFP2-Prx redox probes, in that they share the same domain order, the fluorescent protein domain being fused to the N terminus of the Prx domain. We previously showed for the roGFP2-Tsa2ΔCR fusion protein that the N-terminal fluorescent protein tag does not interfere with Prx oligomerization (36).
Measuring mCER-Prx2 both in cells and in vitro, we demonstrate that the resolution provided by fluorescence polarization easily resolves dimeric and decameric states. We confirm that H2O2-induced decamer–dimer oscillations actually take place in the crowded environment of intact cells. This result has been expected, but nevertheless seems significant, as previous studies on oligomeric transitions were almost exclusively based on in vitro studies. We further confirm that the peroxide-induced mCER-Prx decamer-to-dimer transition is reversed by action of the thioredoxin system, with only little contribution of the glutathione system, as expected. Point mutations known to disrupt the dimer–dimer interface prevented formation of mCER-Prx decamers, both in vitro and in cellulo.
When working with fluorescent protein tags, a general concern is the possibility of perturbing the protein of interest. While the mCER tag did not exert detectable influence on the oxidation kinetics of Prx2, it slightly slowed down the reduction of covalent mCER-Prx homodimers (but not of mCER-Prx2:Prx2 heterodimers), presumably because the presence of two mCER tags on one dimer makes it sterically more difficult for Trx to approach the intersubunit disulfide bond. Nevertheless, peroxide-induced mCER-Prx2 polarization changes closely paralleled oxidation of unmodified Prx2, suggesting that mCER-Prx2 is a dependable proxy readout for the behavior of unmodified Prx2.
The expression of mCER-Prx2 in cells which also express endogenous Prx2 raised the question if and how heterooligomerization affects the measured polarization changes. As expected, mCER-Prx2 was confirmed to form heterodimers with endogenous Prx2, and the average stoichiometry of the resulting heterooligomers generally reflected the relative expression levels. We found that increasing the mCER-Prx2-to-Prx2 ratio increases the polarization difference between dimers and decamers, thus making the probe response more sensitive. Accordingly, expression of mCER-Prx2 in ΔPrx2 cells allowed the most sensitive measurements of small shifts in the dimer–decamer equilibrium. Importantly, while the amplitude of the polarization response was a function of the mCER-Prx2-to-Prx2 ratio, the temporal aspects of the polarization response were not affected (Fig. 2D). Thus, heterodimer formation is not found to cause complications, as long as the mCER-Prx2-to-Prx2 ratio is high enough to provide the necessary dynamic range. The approach presented herein is likely to be further refined by using genome engineering to integrate mCER (or other homo-FRET tags) directly into Prx genes, thus maintaining the expression level and endogenous regulation of the respective Prx while maximizing the polarization range.
One insight obtained from comparing in cellulo polarization measurements with corresponding non-reducing gels is that the complete dissociation of Prx2 decamers into dimers, as triggered by H2O2, is not paralleled by complete conversion of noncovalent into covalent dimers. On the one hand, this finding emphasizes the fact that non-reducing gels cannot be used to draw conclusions about the presence or absence of decamers, as the monomer band can represent any noncovalent oligomer, including noncovalent dimers. On the other hand, it raises questions about how exactly the oxidation–reduction cycle is related to oligomeric transitions. One possible interpretation is that partial oxidation of a Prx2 decamer, that is, disulfide formation in just one or two of the five dimeric units, leads to complete decamer dissociation, thus releasing both covalent and noncovalent dimers. A recent study has shown that the decamer-disrupting effect of Prx oxidation can differ between closely related peroxiredoxins (e.g., Tsa1 vs. Tsa2) depending on small structural differences (Thr-44 vs. Ser-45) (46). Hence, it is conceivable that different Prxs have different “oxidation thresholds” at which their decamers fall apart. This may also imply that both free and decamer-embedded dimers run through their own redox cycles, with distinct oxidation and reduction kinetics, perhaps serving different functions.
We found preliminary evidence suggesting that polarization can detect and resolve oligomeric states higher than decamers. Under a very high oxidative load, mCER-Prx2, but not mCER-Prx1, decreased polarization to a value lower than the baseline corresponding to decamers (SI Appendix, Fig. S3A and Fig. 5A). This high degree of depolarization may represent stacked decamers, their formation being caused by hyperoxidation (47), and potentially associated with chaperone-like activity. This observation is in line with previous findings that hyperoxidized Prx2 forms oligomeric stacks in vitro (47). Prx2 is found to be more sensitive to hyperoxidation than Prx1, as documented previously (48), probably explaining the difference in the polarization response.
Mutagenesis of the CR in mCER-Prx2 provided an interesting insight: In the absence of any peroxide treatment, the substitution of the CR to serine completely shifted the intracellular steady-state equilibrium from the decamer to the dimer while the corresponding alanine mutation almost completely maintained the decameric state. Apparently, very small structural changes close to the C terminus, here the presence or absence of a hydroxyl group, can have a major influence on the oligomeric state, probably reflecting a major shift in the LU:FF equilibrium. In agreement with this observation, a study of the typical 2-Cys peroxiredoxin from Salmonella typhimurium, AhpC, has concluded that the FF:LU free energy difference is small and delicate, so even a minor structural perturbance can substantially impact the enzyme’s oligomeric state (49). Indeed, the FF form of the S. typhimurium enzyme was also shown to be destabilized by replacement of CR with serine (17). These observations appear to be relevant for the interpretation of past and future experiments. The resolving cysteine has been mutated in various kinds of peroxiredoxins for various purposes in a range of studies, sometimes to serine and sometimes to alanine. It now seems important to consider the possibility that the choice of Ala vs. Ser as a replacement for CR influences the oligomeric state and potentially also the outcome and interpretation of the experiment.
We then investigated the implications of this insight for the rational design of novel Prx-based redox sensors. Previously described Prx-based H2O2 sensors were developed using fungal peroxiredoxins, first roGFP2-Tsa2ΔCR [= roGFP2-Tsa2(C171A)] for use in budding yeast (36) and, more recently, roGFP2-Tpx1(C169S) (= roGFP2-Tpx1ΔCR) for use in fission yeast (50). In these probes the CR is mutated to promote transfer of oxidation to the fused roGFP2 moiety and to lessen competition with the Trx system (36). Considering that Prxs are more sensitively oxidized as decamers than as dimers (37), we reasoned that for a probe based on human Prx2 (roGFP2-Prx2ΔCR) it should make a difference if CR is replaced by Ser or Ala. Comparing roGFP2-Prx2(C172S) and roGFP2-Prx2(C172A), we confirmed that the alanine variant is more oxidized under basal steady-state conditions. The expression of this probe in two different lung carcinoma cell lines (one with and one without a mutation that confers constitutive Nrf2 activation) revealed differences in the basal oxidation state of the probe, reflecting endogenous differences in redox homeostasis between the two cancer cell lines that cannot be resolved with roGFP2-Prx2(WT).
Finally, we also showed that mCER-Prx2 can be used to assess the influence of small-molecule drugs on peroxiredoxin oligomeric states and transitions, which opens the possibility to systematically screen compound libraries in a cell-based assay. For proof of concept we tested two thiol-reactive compounds previously claimed to selectively inhibit Prxs. While adenanthin (35) did not show an effect, in line with another study showing that the compound is slow-reacting and nonspecific (51), conoidin A promoted dimerization of Prx2, perhaps indicating an inhibition of Prx reducing systems rather than the suggested covalent irreversible inhibition of Prx2 (52). Another perspective for the homo-FRET approach is the study of Prx PTMs, either by introducing mutations that prevent or mimic such modifications (as exemplified here with the S76D mutant), or by comparing isogenic cell lines differing in the expression of Prx-modifying enzymes, for example kinase PIN1, proposed to regulate Prx1 activity (53).
In conclusion, mCER-Prx probes are valuable tools for the study of Prx structure–function relationships, both in vitro and in living cells. Beyond peroxiredoxins, we expect the plate reader-based homo-FRET assay to be a valuable tool for the study of many other proteins.
Methods
DNA Constructs.
Plasmids encoding mCER and mCER-tagged G6PDH (Apollo-NADP+) were obtained from Addgene. A gateway donor vector for human peroxiredoxin-2 (Prx2) was obtained from the DKFZ Genomics and Proteomics Core Facility. Open reading frames for Prx2 and mCER were amplified by PCR and ligated into pcDNA3.1(−), pLPCX (Clontech) or pET-SUMO (Life Technologies) vectors using the Gibson Assembly Cloning Kit (New England BioLabs). Mutations were introduced by using the QuikChange site-directed mutagenesis kit (Agilent). The list of primers is provided in SI Appendix, Table S1. Plasmids used in this study were pcDNA3.1(−), pcDNA3.1(−) Prx2, pcDNA3.1(−) mCER, pcDNA3.1(−) mCER-Prx2, pcDNA mCER(C48S)-Prx2, pcDNA3.1(−) mCER-Prx2(C51S), pcDNA3.1(−) mCER-Prx2(C172S), pcDNA3.1(−) mCER-Prx2(C172A), pcDNA3.1(−) mCER-Prx2(T82E/A85E), pcDNA3.1(−) mCER-Prx2(S76D), pLPCX mCER-Prx2, pLPCX mCER-Prx2(C51S), pLPCX mCER-Prx2(C172S), pLPCX mCER-Prx2(C172A), pLPCX mCER-Prx1, pCMV-VSV-G, pET-SUMO mCER-Prx2, pET-SUMO mCER-Prx2(C51S), pET-SUMO mCER-Prx2(C172A), and pET-SUMO mCER-Prx2(C172S).
Cell Lines, Antibodies, and Reagents.
GripTite HEK293 cells (Thermo Fisher Scientific), and human lung adenocarcinoma cell lines H838 (ATCC) and H1976 (ATCC) were used for stable cell line generation. Phoenix Ampho cells (ATCC) were used for virus production. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (Life Technologies), supplemented with 10% (vol/vol) fetal bovine serum (FBS; Life Technologies), 2 mM L-glutamine (Life Technologies), and 50 units/mL of penicillin and streptomycin (Pen-Strep; Life Technologies). In addition, 50 units/mL of Geneticin (Life Technologies) was used for GripTite HEK293 cells. Primary antibodies used in this study were rabbit anti-GFP (green fluorescent protein) (8334; Santa Cruz), rabbit anti-Prx2 (109367; Abcam), rabbit anti-Prx1 (8499; Cell Signaling), and rabbit anti-Prx-SO3 (LF-PA0004; AbFrontier). All antibodies were used at a dilution of 1:1,000. Secondary antibodies used in this study were peroxidase-conjugated AffiniPure goat anti-rabbit IgG (111-035-144; Jackson ImmunoResearch). All chemicals were from Sigma-Aldrich, unless stated otherwise. Adenanthin was from Aobious and conoidin A from Cayman Chemicals. Human Prx2 purified from red blood cells was from Abfrontier (YIF-LF-P0007).
Generation of Stably Expressing Cell Lines.
pLPCX retroviral expression vectors were transfected into the packaging cell line Phoenix Ampho. After 2 d, viral supernatant was collected, filtered through a 0.45-μm cellulose acetate filter and used to infect a freshly thawed culture of the respective target cell line. Transduced cells were FACS-sorted for mCER or GFP fluorescence, expanded, and frozen for later use.
Generation of Transiently Transfected Cell Lines.
Cells (2.5 × 105) were seeded in a six-well plate. Lipofectamine 2000 (Life Technologies) was used according to the manufacturer’s protocol. In the specific case of cotransfection of mCER-Prx2 and Prx2 (Fig. 2D), the amount of mCER-Prx2 encoding plasmid was 2 µg and the amount of Prx2 encoding plasmid was either 4 µg, 2 µg, or 0 µg (corresponding to 1:2, 1:1, and 1:0 ratios). The total amount of DNA for transfection was adjusted to 6 µg using a noncoding plasmid. The next day, 5 × 104 transfected cells were seeded in black/clear 96-well plates and cultured for 24 h in 100 µL FluoroBrite, 2% FBS, 2 mM L-glutamine, and 50 units/mL Pen-Strep at 37 °C and 5% CO2.
Generation of Prx2 Knockout Cell Lines.
A single-guide RNA (sgRNA) targeting the coding region of the human PRDX2 gene was selected from the GeCKO library. An oligonucleotide encoding the sgRNA (AGGGGCCTCTTTATCATCGA) was cloned into the lentiCRISPRv1 vector containing the SpCas9 gene and a puromycin resistance cassette (54, 55). GripTite HEK293 cells were transfected with the sgRNA-encoding plasmid using Lipofectamine 2000 and grown for 48 h. Cells were then selected with puromycin (750 ng/mL) for 6 d. Subsequently, single-cell clones were grown, expanded, and analyzed by immunoblotting. The selected Prx2 knockout clones were further verified by PCR and sequencing of the PCR product.
Fluorescence Polarization Measurements.
For the cell polarization measurements, 5 × 104 stably or transiently transfected GripTite HEK293 cells were seeded into black/clear 96-well plates (Falcon; Thermo Fischer Scientific) and cultured for 24 h in 100 µL FluoroBrite (Thermo Fisher Scientific), 2% FBS, 2 mM L-glutamine, and 50 units/mL Pen-Strep at 37 °C and 5% CO2. For in vitro polarization measurements, individual recombinant proteins, or mixtures thereof, at a final concentration of 10 µM, were diluted into 20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, and 10 mM dithiothreitol (DTT) in a final volume of 20 µL. The mixture was incubated for 10 min and measured in a black/clear 384-well plate (BD Falcon), using a PHERAstar FS microplate reader (BMG Labtech) equipped with a fluorescence polarization filter, excitation at 430 nm and emission at 480 nm. Polarization was calculated using MARS data analysis software (BMG Labtech) according to the following equation:
The G factor (compensation factor for the plate reader) was set automatically for each measurement based on gain adjustment settings.
Immunoblot Analysis.
HEK293 cells (5 × 104) cultivated in flat-bottom 96-well plates were incubated for 5 min with 80 mM methanethiosulfonate (MMTS) in Dulbecco’s phosphate-buffered saline (DPBS; Life Technologies). Cells were washed once with DPBS and lysed in 40 µL 1% Triton X-100 in TBS (50 mM Tris, 150 mM NaCl, pH 7.4) supplemented with Complete Protease Inhibitor Mixture tablets (Roche). Post nuclear supernatants were mixed with SDS sample buffer and equally divided into non-reduced and reduced (25 mM DTT) fractions. Protein samples of 25 µg were run on SDS/PAGE and transferred to polyvinyl difluoride membranes (Immobilon-P; Millipore) using a tank transfer unit (TE22; Hoefer). Membranes were probed with appropriate antibodies and analyzed using SuperSignal West Femto chemiluminescent substrate (Thermo Fischer Scientific).
Protein Expression and Purification.
The bacterial expression vector pET-SUMO mCER-Prx2 (or variants thereof) was transformed into Escherichia coli BL21 (DE3). Single colonies were inoculated into 10 mL terrific broth (TB) medium (Roth) supplemented with 30 mg/L kanamycin and grown at 37 °C overnight. On the next day, the overnight culture was amplified into 1 L of TB medium with 30 mg/L kanamycin. The culture was grown at 37 °C until the optical density at 600 nm (A600) reached 0.7 to 0.8, induced with 0.5 mM isopropyl-beta-d-thiogalactoside to induce expression, and grown at 16 °C overnight. The cells were harvested by centrifugation at 4,000 × g for 15 min. The pellet was resuspended in lysis buffer (50 mM Tris⋅HCl [pH 8.0], 500 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine, and 10% glycerol) supplemented with 1 µg/mL leupeptin (Sigma) and 0.1 mg/mL AEBSF hydrochloride (Applichem) and 0.1 mg/mL DNase I from bovine pancreas (Sigma) and lysed by sonication for 5 min, in 10-s pulse–10-s break intervals. The lysate was cleared by centrifugation at 14,500 × g for 1 h and the clarified supernatant was added to Ni2+-Sepharose (GE Healthcare) affinity beads equilibrated in lysis buffer. The mixture was incubated for 1 h at 4 °C and packed into a column. The beads were washed with lysis buffer plus 5 mM imidazole and eluted with the linear gradient to 300 mM imidazole in the same buffer. The protein was concentrated using an Amicon Ultra filter (EMD Millipore) and dialyzed against 20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT. Protein was quantified by absorbance at 280 nm (mCER-Prx2: ε280nm = 47,330 M−1⋅cm−1; Prx2: ε280nm = 21,430 M−1⋅cm−1).
Size Exclusion Chromatography.
A defined amount (0.5 nmol) of reduced purified mCER-Prx2 (WT, C51S, C172A, C172S) was injected on a Superdex 200 Increase 10/300 GL column (GE Healthcare), equilibrated with 20 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 1 mM DTT. Protein elution was monitored by the recording of A280nm on an ÄKTA pure fast-liquid protein chromatography device (GE Healthcare). Protein-containing eluates were collected in 0.2-mL fractions.
Measurement of roGFP2 Probe Oxidation.
The roGFP2 redox state was measured using either a CLARIOstar or a PHERAstar FS plate reader (BMG Labtech), as described previously (56). Cells were grown to 80% confluency in FluoroBrite, 2% FBS, 2 mM l-glutamine, and 50 units/mL Pen-Strep, in a flat-bottom black/clear 96-well imaging plate. To determine the degree of probe oxidation (OxD), tetramethylazodicarboxamide (diamide) was added to a final concentration of 2 mM to generate a fully oxidized control sample. DTT was added to a final concentration of 10 mM to generate a fully reduced control sample. OxD was calculated as described previously (56).
Statistics.
All data are presented as mean ± SEM. Statistical significance was determined using the unpaired Student´s two-tailed t test. Data points were fitted using GraphPad Prism.
Data Availability.
The data supporting the findings of this study are available within the paper or in SI Appendix.
Supplementary Material
Acknowledgments
We thank Virginie Malak and Gabriele Kuntz for technical assistance, Dr. Chintan Shah (Division of Quantum Dynamics and Control, Max-Planck-Institute for Nuclear Physics, Heidelberg), and Dr. Felix Bestvater (Core Facility Unit Light Microscopy, German Cancer Research Center, Heidelberg) for valuable discussions. We acknowledge support by the Deutsche Forschungsgemeinschaft (SFB1036 and SPP1710 to T.P.D.) and by the European Research Council (742039 to T.P.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915275117/-/DCSupplemental.
References
- 1.Wood Z. A., Schröder E., Robin Harris J., Poole L. B., Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Zeida A. et al., Catalysis of peroxide reduction by fast reacting protein thiols. Chem. Rev. 119, 10829–10855 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Perkins A., Nelson K. J., Parsonage D., Poole L. B., Karplus P. A., Peroxiredoxins: Guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 40, 435–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee S. G., Kil I. S., Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 86, 749–775 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Rhee S. G., Woo H. A., Multiple functions of peroxiredoxins: Peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid. Redox Signal. 15, 781–794 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Stöcker S., Van Laer K., Mijuskovic A., Dick T. P., The conundrum of hydrogen peroxide signaling and the emerging role of peroxiredoxins as redox relay hubs. Antioxid. Redox Signal. 28, 558–573 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Rhee S. G., Woo H. A., Kang D., The role of peroxiredoxins in the transduction of H2O2 signals. Antioxid. Redox Signal. 28, 537–557 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Park J., Lee S., Lee S., Kang S. W., 2-cys peroxiredoxins: Emerging hubs determining redox dependency of mammalian signaling networks. Int. J. Cell Biol. 2014, 715867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stöcker S., Maurer M., Ruppert T., Dick T. P., A role for 2-Cys peroxiredoxins in facilitating cytosolic protein thiol oxidation. Nat. Chem. Biol. 14, 148–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Z., Lindsay J. G., The peroxiredoxin family: An unfolding story. Subcell. Biochem. 83, 127–147 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Skoko J. J., Attaran S., Neumann C. A., Signals getting crossed in the entanglement of redox and phosphorylation pathways: Phosphorylation of peroxiredoxin proteins sparks cell signaling. Antioxidants 8, E29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole L. B., Nelson K. J., Distribution and features of the six classes of peroxiredoxins. Mol. Cells 39, 53–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris J. R. et al., Comparison of the decameric structure of peroxiredoxin-II by transmission electron microscopy and X-ray crystallography. Biochim. Biophys. Acta 1547, 221–234 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Harris J. R., The isolation and purification of a macromolecular protein component from the human erythrocyte ghost. Biochim. Biophys. Acta 188, 31–42 (1969). [DOI] [PubMed] [Google Scholar]
- 15.Wood Z. A., Poole L. B., Hantgan R. R., Karplus P. A., Dimers to doughnuts: Redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry 41, 5493–5504 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Barranco-Medina S., Lázaro J. J., Dietz K. J., The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 583, 1809–1816 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Nelson K. J. et al., Experimentally dissecting the origins of peroxiredoxin catalysis. Antioxid. Redox Signal. 28, 521–536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morais M. A. et al., How pH modulates the dimer-decamer interconversion of 2-Cys peroxiredoxins from the Prx1 subfamily. J. Biol. Chem. 290, 8582–8590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chae H. Z., Oubrahim H., Park J. W., Rhee S. G., Chock P. B., Protein glutathionylation in the regulation of peroxiredoxins: A family of thiol-specific peroxidases that function as antioxidants, molecular chaperones, and signal modulators. Antioxid. Redox Signal. 16, 506–523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J. W., Piszczek G., Rhee S. G., Chock P. B., Glutathionylation of peroxiredoxin I induces decamer to dimers dissociation with concomitant loss of chaperone activity. Biochemistry 50, 3204–3210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veal E. A., Underwood Z. E., Tomalin L. E., Morgan B. A., Pillay C. S., Hyperoxidation of peroxiredoxins: Gain or loss of function? Antioxid. Redox Signal. 28, 574–590 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Pan Y., Jin J. H., Yu Y., Wang J., Significant enhancement of hPrx1 chaperone activity through lysine acetylation. ChemBioChem 15, 1773–1776 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Randall L. M. et al., Nitration transforms a sensitive peroxiredoxin 2 into a more active and robust peroxidase. J. Biol. Chem. 289, 15536–15543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Ham T. J. et al., Towards multiparametric fluorescent imaging of amyloid formation: Studies of a YFP model of alpha-synuclein aggregation. J. Mol. Biol. 395, 627–642 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Warren S. C., Margineanu A., Katan M., Dunsby C., French P. M., Homo-FRET based biosensors and their application to multiplexed imaging of signalling events in live cells. Int. J. Mol. Sci. 16, 14695–14716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron W. D. et al., Apollo-NADP(+): A spectrally tunable family of genetically encoded sensors for NADP(+). Nat. Methods 13, 352–358 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Ross B. L. et al., Single-color, ratiometric biosensors for detecting signaling activities in live cells. eLife 7, e35458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobotta M. C. et al., Exposing cells to H2O2: A quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Radic. Biol. Med. 60, 325–335 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Loberg M. A. et al., Aromatic residues at the dimer-dimer interface in the peroxiredoxin Tsa1 facilitate decamer formation and biological function. Chem. Res. Toxicol. 32, 474–483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunniff B. et al., Disabling mitochondrial peroxide metabolism via combinatorial targeting of peroxiredoxin 3 as an effective therapeutic approach for malignant mesothelioma. PLoS One 10, e0127310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manta B. et al., The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch. Biochem. Biophys. 484, 146–154 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Jang H. H. et al., Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117, 625–635 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Saccoccia F. et al., Moonlighting by different stressors: Crystal structure of the chaperone species of a 2-Cys peroxiredoxin. Structure 20, 429–439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haraldsen J. D. et al., Identification of conoidin a as a covalent inhibitor of peroxiredoxin Ii. Org. Biomol. Chem. 7, 3040–3048 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C. X. et al., Adenanthin targets peroxiredoxin I and II to induce differentiation of leukemic cells. Nat. Chem. Biol. 8, 486–493 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Morgan B. et al., Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat. Chem. Biol. 12, 437–443 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Parsonage D. et al., Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 44, 10583–10592 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastor-Flores D., Becker K., Dick T. P., Monitoring yeast mitochondria with peroxiredoxin-based redox probes: The influence of oxygen and glucose availability. Interface Focus 7, 20160143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itoh K. et al., An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236, 313–322 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Muthuramalingam M. et al., Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol. Plant 2, 1273–1288 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Seidel T., Seefeldt B., Sauer M., Dietz K. J., In vivo analysis of the 2-Cys peroxiredoxin oligomeric state by two-step FRET. J. Biotechnol. 149, 272–279 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Wolf H., Barisas B. G., Dietz K. J., Seidel T., Kaede for detection of protein oligomerization. Mol. Plant 6, 1453–1462 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Langford T. F., Huang B. K., Lim J. B., Moon S. J., Sikes H. D., Monitoring the action of redox-directed cancer therapeutics using a human peroxiredoxin-2-based probe. Nat. Commun. 9, 3145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan F. T., Kaminski C. F., Kaminski Schierle G. S., HomoFRET fluorescence anisotropy imaging as a tool to study molecular self-assembly in live cells. ChemPhysChem 12, 500–509 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Bader A. N., Hofman E. G., Voortman J., en Henegouwen P. M., Gerritsen H. C., Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys. J. 97, 2613–2622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tairum C. A. et al., Catalytic thr or ser residue modulates structural switches in 2-Cys peroxiredoxin by distinct mechanisms. Sci. Rep. 6, 33133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon J. C. et al., Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J. Biol. Chem. 280, 28775–28784 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Dalla Rizza J., Randall L. M., Santos J., Ferrer-Sueta G., Denicola A., Differential parameters between cytosolic 2-Cys peroxiredoxins, PRDX1 and PRDX2. Protein Sci. 28, 191–201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perkins A. et al., The sensitive balance between the fully folded and locally unfolded conformations of a model peroxiredoxin. Biochemistry 52, 8708–8721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carmona M. et al., Monitoring cytosolic H2O2 fluctuations arising from altered plasma membrane gradients or from mitochondrial activity. Nat. Commun. 10, 4526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soethoudt M. et al., Interaction of adenanthin with glutathione and thiol enzymes: Selectivity for thioredoxin reductase and inhibition of peroxiredoxin recycling. Free Radic. Biol. Med. 77, 331–339 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Nguyen J. B. et al., Peroxiredoxin-1 from the human hookworm Ancylostoma ceylanicum forms a stable oxidized decamer and is covalently inhibited by conoidin A. Chem. Biol. 20, 991–1001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu K. L. et al., Regulation of PRDX1 peroxidase activity by Pin1. Cell Cycle 12, 944–952 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shalem O. et al., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morgan B., Sobotta M. C., Dick T. P., Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic. Biol. Med. 51, 1943–1951 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper or in SI Appendix.