Significance
In photosynthetic reaction centers from purple bacteria (PbRC) and the water-splitting enzyme, photosystem II (PSII), light-induced electron transfer occurs only in one of the two branches irrespective of the apparent symmetry in the structures. In PSII, the protein components that are involved in the Mn4CaO5 cluster or the proceeding proton-transfer pathway facilitate electron transfer along the active branch. In PbRC, most of these components are not conserved and polar residues on the active side facilitate electron transfer. The energy profile suggests that the initial electron donor differs between PbRC and PSII. It seems likely that the acquirement of oxygen-evolving ability alters the protein environment near the accessory chlorophyll and makes it the initial electron donor in PSII.
Keywords: unidirectional electron transfer, oxygen evolution, P680, excitation energy transfer, artificial photosynthesis
Abstract
In photosynthetic reaction centers from purple bacteria (PbRC) and the water-oxidizing enzyme, photosystem II (PSII), charge separation occurs along one of the two symmetrical electron-transfer branches. Here we report the microscopic origin of the unidirectional charge separation, fully considering electron–hole interaction, electronic coupling of the pigments, and electrostatic interaction with the polarizable entire protein environments. The electronic coupling between the pair of bacteriochlorophylls is large in PbRC, forming a delocalized excited state with the lowest excitation energy (i.e., the special pair). The charge-separated state in the active branch is stabilized by uncharged polar residues in the transmembrane region and charged residues on the cytochrome c2 binding surface. In contrast, the accessory chlorophyll in the D1 protein (ChlD1) has the lowest excitation energy in PSII. The charge-separated state involves ChlD1•+ and is stabilized predominantly by charged residues near the Mn4CaO5 cluster and the proceeding proton-transfer pathway. It seems likely that the acquirement of water-splitting ability makes ChlD1 the initial electron donor in PSII.
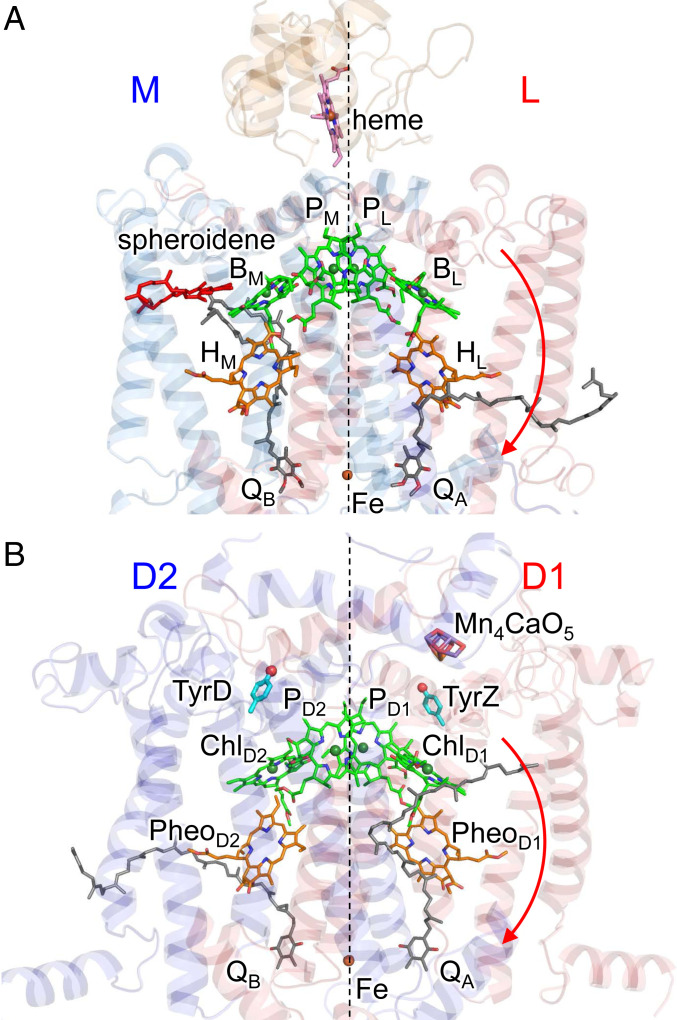
In photosystem II (PSII), the driving force of water oxidation is provided by light-induced charge separation in the reaction center. In PSII, the reaction center has a pair of chlorophylls, PD1/PD2, accessory chlorophylls, ChlD1/ChlD2, pheophytins, PheoD1/PheoD2, and plastoquinones, QA/QB, in the heterodimeric D1/D2 protein subunit pairs (Fig. 1) (1). Notably, the crystal structures of PSII and purple bacterial photosynthetic reaction centers (PbRC) show a large structural similarity (2). In PbRC, the reaction center has a pair of bacteriochlorophylls, PL/PM, accessory bacteriochlorophylls, BL/BM, bacteriopheophytins, HL/HM, and ubiquinones (or menaquinones), QA/QB, in the heterodimeric L/M protein subunit pairs (Fig. 1). PL and PM form the electronically coupled special pair [PLPM]. Electronic excitation of [PLPM] leads to the formation of the charge-separated state, [PLPM]•+BL•–, and subsequent electron transfer occurs to QB via HL and QA (3). The cationic state [PLPM]•+ is reduced by electron transfer from an outer protein subunit, cytochrome c2 (in PbRC from Rhodobacter sphaeroides or tetraheme cytochrome in PbRC from Blastochloris viridis). In PSII, transfer of excitation energy from the surrounding antenna chlorophylls to the reaction-center chlorophylls (4, 5) leads to the formation of the cationic state [PD1PD2]•+, which has a significantly high redox potential (Em) for one-electron oxidation [>1,100 mV (6–9)] with respect to [PLPM]•+ [500 mV (10)]. The high Em makes [PD1PD2]•+ abstract electrons from the substrate water molecules at the Mn4CaO5 moiety [700 to 800 mV (11)] via redox-active D1-Tyr161 (TyrZ).
Fig. 1.
Electron transfer chains in photosynthetic reaction centers of (A) PSII (PDB ID code 3ARC) and (B) PbRC from R. sphaeroides (PDB ID codes 3I4D and 1L9B). Red arrows indicate electron transfer. Dotted lines indicate pseudo-C2 axes. Electron-transfer active branches are labeled in red and inactive branches in blue.
In both PbRC and PSII, electron transfer predominantly occurs along the L- and D1-branches, not the M- and D2-branches, irrespective of the pseudo-C2 symmetry between the two branches (12–14). Because exciton is stabilized by electron–hole Coulomb interaction, charge separation necessitates a sufficient potential offset between electron donor and acceptor cofactors. In PbRC, the energy levels of the charge-separated states were investigated in mutant proteins (15–18). Recent theoretical studies suggested that the redox potential of BL for one-electron reduction, Em(BL)0/•–, is significantly higher than Em(BM)0/•–, which suggests that the BL•– formation is more stabilized than the BM•– formation (19). The factors that increase Em(BL)0/•– with respect to Em(BM) 0/•– are 1) the difference in the polarity of the uncharged L/M residue pairs in the hydrophobic transmembrane region [e.g., Phe-L181/Tyr-M210 (19, 20)], 2) acidic residues that provide the binding interface of cytochrome c2 on the protein surface of subunit M [e.g., Glu-M95 and Asp-M184 (19, 21)], and 3) a cluster of hydrophobic residues that form the carotenoid binding site (19).
In contrast to PbRC, Em(ChlD1)0/•– is lower than Em(ChlD2)0/•– in PSII, which suggests that ChlD1•– is unstable and [PD1/PD2]•+ChlD1•– is not relevant as a charge-separated intermediate (19). This is in line with the proposal that ChlD1•+PheoD1•– is the charge-separated state in PSII (22–24). The D1/D2 residue pairs that (destabilize ChlD1•– with respect to ChlD2•– and) stabilize ChlD1•+ with respect to ChlD2•+ are involved in the Mn4CaO5 cluster or the proceeding proton transfer pathway (e.g., D1-Asp61/D2-His61, D1-Asp170/D2-Phe169, D1-Asn181/D2-Arg180, and D1-Glu189/D2-Phe188) (19). The absence of the corresponding residues in PbRC implies that the energetic difference between BL/BM in PbRC and ChlD1/ChlD2 in PSII is associated with the presence of the water-splitting components in PSII.
The reported Em values of cofactors show the energetic difference between the two symmetrical electron transfer branches before light-induced charge separation occurs, that is, the ground state (19). However, the Em profile also shows that both electron-transfer branches are energetically downhill in PSII (19). This suggests that the unidirectional charge separation cannot be explained solely from the Em profile, because the charge-separated intermediate states are involved in the electron transfer process. As far as we are aware, the energy levels of the electronically excited states and the subsequent charge-separated states in the two branches and the protein components that facilitate unidirectional electron transfer are not reported for the two reaction centers. It remains thus far unclear why light-induced electron transfer occurs only in one of the two electron transfer branches irrespective of the apparent symmetry in the protein structure. It is also an open question why the charge-separation mechanism is likely to differ between PbRC and PSII, irrespective of the apparent structural similarity (14).
Here we report the energetics of electronically excited states and charge-separated states in PbRC and PSII, using a combined quantum mechanical/molecular mechanical/polarizable continuum model (QM/MM/PCM) approach with density functional theory (DFT) and time-dependent DFT (TDDFT) and considering electron–hole interaction (i.e., exciton binding energy), electronic coupling, and electrostatic interactions with the polarizable, entire protein environments.
Results
Initial Electron Donor [PLPM] in PbRC.
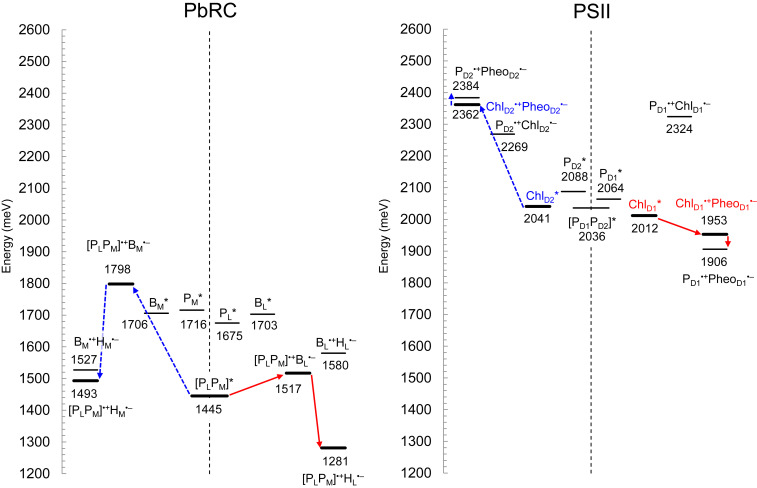
The excitonic coupling of 27 meV between PL* and PM* in the PbRC protein environment (Table 1) is sufficiently larger than interaction between the excited state and the protein electrostatic environment [e.g., 10 to 15 meV (25)]. The TDDFT-QM/MM calculations show that the [PLPM] pair has the lowest excitation energy among the bacteriochlorophylls (Fig. 2). The formation of the [PLPM] pair leads to a decrease of 230 to 270 meV in the excitation energy with respect to the monomeric PL and PM bacteriochlorophylls (Fig. 2). The large couplings between other excited states (e.g., 136 meV between PL* and PL•+PM•–; SI Appendix, Table S1) also contributes to the decrease in the [PLPM] pair excitation energy.
Table 1.
Electronic and excitonic coupling for the [PLPM] bacteriochlorophyll pair in PbRC and the [PD1PD2] chlorophyll pair in PSII in millielectron volts (centimeters–1)
| PbRC | PSII | |
| Excitonic coupling | 27 (218) | 10 (81) |
| Electronic coupling | 114 (919) | 13 (105) |
Fig. 2.
Energy values for electronic excitation and charge-separated states of (bacterio)chlorophylls and (bacterio)pheophytins in PbRC (Left) and PSII (Right), calculated using a QM/MM approach, where the interaction between electron and hole was considered quantum-chemically. Thick solid bars indicate the major intermediate states. Red solid arrows indicate major electron transfer in the active branch, and blue dotted arrows indicate the corresponding electron transfer in the inactive branch.
For other factors, electrostatic interactions with the protein environment contribute to a decrease of 70 to 110 meV in the excitation energy of the monomeric PL and PM bacteriochlorophylls (Table 2). The larger contribution of the protein electrostatics to PL* (109 meV) than PM* (70 meV) is due to the presence of the H-bond between PL and His-L168. Deformations of the chlorin rings induced by interactions with the protein environment (26, 27) contribute to a decrease of 50 meV in the excitation energy of the monomer PL and PM bacteriochlorophylls. It seems likely that [PLPM] is an electronically coupled special pair, serving as the initial electron donor in PbRC (Fig. 2).
Table 2.
Factors that decrease the excitation energy of (bacterio)chlorophyll [(B)Chl] in the reaction center in millielectron volts
| BM* | PM* | PL* | BL* | ChlD2* | PD2* | PD1* | ChlD1* | |
| (B)Chl* in vacuum | 1,834 | 1,834 | 1,834 | 1,834 | 2,129 | 2,129 | 2,129 | 2,129 |
| +Ring deformation† | −45 | −48 | −50 | −67 | −32 | −3 | −26 | −37 |
| +Protein electrostatics‡ | −83 | −70 | −109 | −64 | −62 | −38 | −39 | −85 |
| (B)Chl* in protein without special pair formation | 1,706 | 1,716 | 1,675 | 1,703 | 2,041 | 2,088 | 2,064 | 2,012 |
| +Special pair formation§ | 0 | −271 | −230 | 0 | 0 | 0 | 0 | 0 |
| (B)Chl* in protein | 1,706 | 1,445 | 1,445 | 1,703 | 2,041 | 2,088 | 2,064 | 2,012 |
Influence of deformation of the chlorin ring due to interactions with the protein environment (e.g., van der Waals contact and H-bond interactions).
Influence of electrostatic interactions with the protein environment.
Influence of the formation of the special pair.
Charge-Separated State [PLPM]•+BL•– in PbRC.
When the interaction between electron and hole is considered quantum-chemically in TDDFT-QM/MM/PCM calculations (28), the charge-separated state in the L-branch, [PLPM]•+BL•–, is 281 meV more energetically stable than the corresponding charge-separated state in the M-branch, [PLPM]•+BM•– (Fig. 2). The energy difference remains unchanged even when the intramolecular reorganization energy is considered (285 meV) (SI Appendix, Fig. S2). In the presence of the intramolecular reorganization energy, charge separation from [PLPM]* to [PLPM]•+BL•– is 55 meV energetically downhill, whereas charge separation from [PLPM]* to [PLPM]•+BM•– is 230 meV uphill.
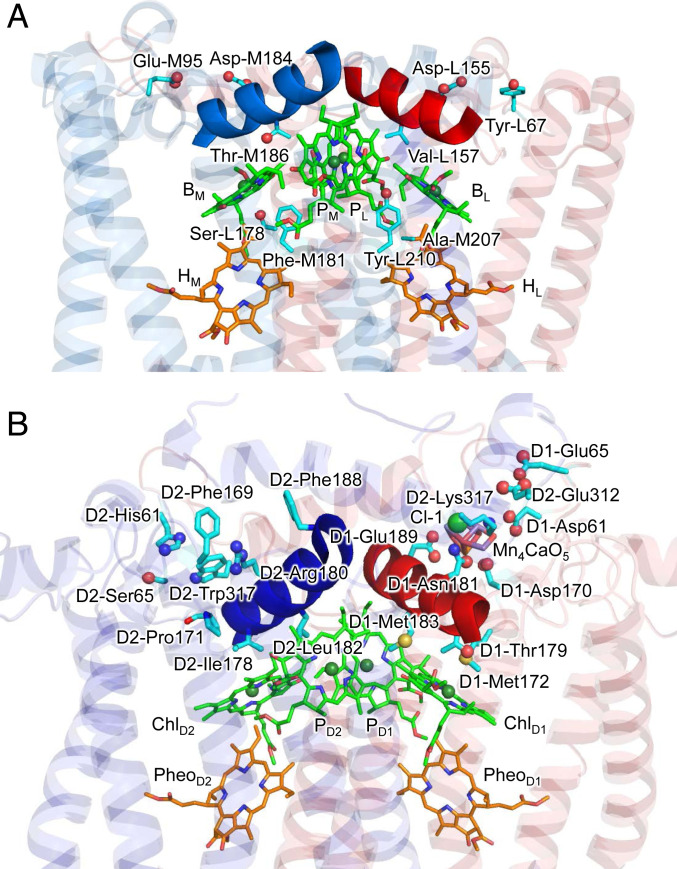
The energy difference between [PLPM]•+BL•– and [PLPM]•+BM•– is predominantly due to the energy difference between BL•– and BM•– (i.e., the difference in the lowest unoccupied molecular orbital [LUMO] energy, which corresponds to the difference in Em for one-electron reduction) because [PLPM]•+ can be considered to be equidistant from BL and BM (even if the PL•+/PM•+ population alters slightly, depending on BL•– and BM•–). The key L/M residue pairs that stabilize BL•– with respect to BM•– are summarized in Table 3. In the Tyr-L67/Glu-M95 and Asp-L155/Asp-M184 pairs, Glu-M95 and Asp-M184 provide the binding interface of the electron-donor cytochrome c2 (21), destabilize the BM•– formation, and thus decrease Em(BM) (Fig. 3A) (19). It is characteristic to PbRC that polar, uncharged residues stabilize BL•– with respect to BM•–. The Phe-L181/Tyr-M210 pair is located near BM/BL, and the difference has been suggested to be crucial to the initial electron transfer (29): Mutations of Tyr-M210 to phenylalanine decreased the initial electron transfer with a time constant from 3.5 ps to 16 ps (20). The polar –OH group of Tyr-M210, which is oriented toward BL, stabilizes the BL•– formation and increases Em(BL) (19). The absence of the residue that corresponds to Tyr-M210 in PSII (i.e., D2-Leu205) (2) implies that there is a difference in the charge-separation mechanism between PbRC and PSII (discussed below). Thr-M186 is in van der Waals contact with the BM chlorin ring. Because Thr-M186 forms an H-bond with the backbone carbonyl group of the ligand residue of BM, His-M182, the hydroxyl O is oriented toward BM, destabilizing BM•–. Ser-L178 is located at a weak H-bond distance from the ester keto O of BM (O…O = 3.5 Å). However, it forms an H-bond with the backbone carbonyl O of Met-L174 (O…O = 2.8 Å), which destabilizes BM•–. Electrostatic interactions between these residues and BM are pronounced in the hydrophobic protein environment near BM, that is, ∼30 hydrophobic residues from subunit M that form the carotenoid binding site (19). All these residue pairs listed in Table 3 have already been suggested to stabilize [PLPM]•+BL•– with respect to [PLPM]•+BM•– in electrostatic calculations for PbRC (19).
Table 3.
L/M residue pairs that stabilize BL•– with respect to BM•– (>40 meV) in the LUMO energy level in millielectron volts, which corresponds to Em for one-electron reduction
| Em(BL) | Em(BM) | Em(BL) | Em(BM) | Stabilizing BL•– | ||
| Tyr-L67 | 5 | 0 | Glu-M95* | −14 | −141 | 132 |
| Phe-L181 | 3 | 71 | Tyr-M210 | 161 | −5 | 98 |
| Val-L157 | 30 | 0 | Thr-M186 | −3 | −49 | 76 |
| Ser-L178 | 5 | −54 | Ala-M207 | −11 | 0 | 48 |
| Asp-L155 | −150 | −35 | Asp-M184*,† | −35 | −193 | 43 |
Cytochrome c2 binding site (21).
Corresponding to D1-Arg180 in PSII.
Fig. 3.
(A) L/M residue pairs that stabilize BL•– with respect to BM•– in PbRC. Most of the residue pairs are located in the hydrophobic transmembrane region. Helix cd in L and M are colored red and blue, respectively. (B) D1/D2 residue pairs that stabilize ChlD1•+PheoD1•– with respect to ChlD2•+PheoD2•– or that stabilize ChlD1* with respect to ChlD2* in PSII. In contrast to PbRC, most of the residue pairs are located in the membrane-extrinsic region. Helix cd in D1 and D2 are colored red and blue, respectively.
The Absence of the Special Pair Chlorophylls in PSII.
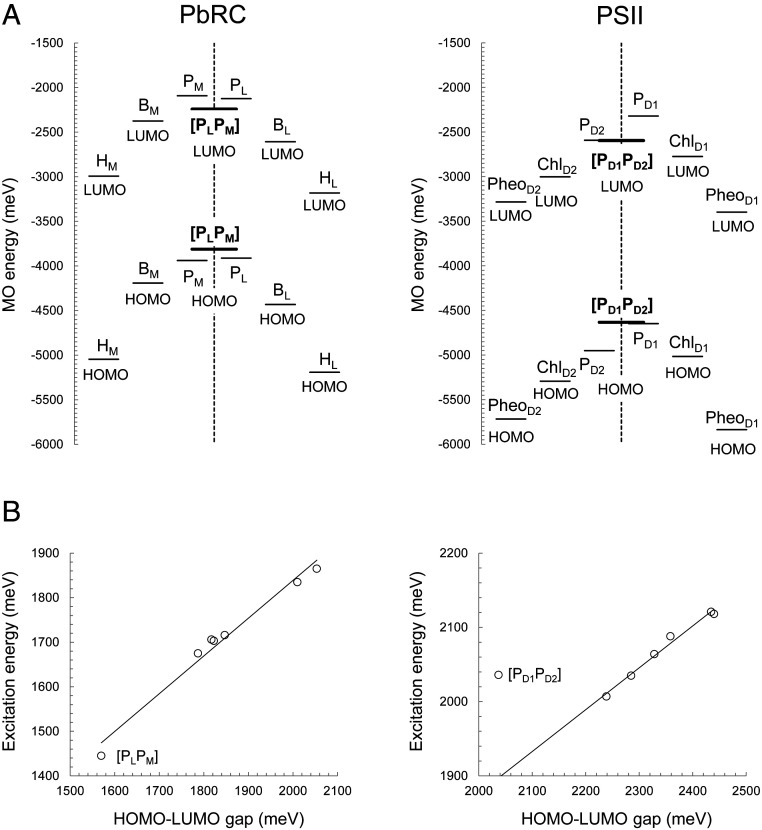
In contrast to [PLPM] in PbRC, the calculated value of the excitonic coupling between PD1 and PD2 for the chlorophyll pair in PSII is small, 10 meV (Table 1). The electronic coupling between PD1 and PD2 is also very small, 13 meV, with respect to 114 meV between PL and PM in PbRC (Table 1). In addition, the highest occupied molecular orbital (HOMO) energy level of [PD1PD2] is essentially the same as that of PD1, whereas the LUMO energy level of [PD1PD2] is the same as that of PD2 (Fig. 4A), suggesting that PD1 and PD2 do not form a special pair. Consistently, the HOMO–LUMO energy gap is correlated with the excitation energy in [PLPM], but not in [PD1PD2] (Fig. 4B). The absence of the [PD1PD2] special pair is largely due to the low overlap of π-orbital between PD1 and PD2. See SI Appendix, Table S2 for couplings of other excited states.
Fig. 4.
(A) LUMO and HOMO energy levels in PbRC (Left) and PSII (Right) in millielectron volts, calculated including the four (bacterio)chlorophylls and two (bacterio)pheophytins in the QM regions. Thick bars indicate [PLPM] and [PD1PD2]. (B) Correlation between calculated HOMO–LUMO gap and excitation energy in PbRC (Left) (coefficient of determination R2 = 0.98) and PSII (Right) (R2 = 0.98, excluding [PD1PD2]). [PD1PD2] does not fit to the correlation, because excitation of [PD1PD2] is excitation from HOMO of PD1 to LUMO of PD2 (A), which corresponds to charge transfer process, that is, the PD1•+PD2•– formation.
The Lowest Excitation-Energy Site, ChlD1.
Among PD1, PD2, [PD1PD2], ChlD1, and ChlD2, the excitation energy of ChlD1 is the lowest (Fig. 2), as also indicated by the lowest HOMO–LUMO energy gap (Fig. 4A). The stretch in the chlorin ring along the Qy transition dipole moment may stretch in the π-conjugation system and decrease the excitation energy. The chlorin-ring deformation differs among PD1, PD2, ChlD1, and ChlD2 (27). The N–N distance along the Qy transition dipole moment in the chlorin ring is longer in ChlD1 and ChlD2 than PD1 and PD2 (SI Appendix, Table S3 and Fig. S1). Accordingly, the N–N distance along the Qx transition dipole moment is shortest in ChlD1. It seems likely that the N–N distance along the Qy transition dipole moment is partially associated with the excitation energy (SI Appendix, Fig. S1). However, it should also be noted that it does not explain the detailed difference, for example, between PD1 and PD2 (SI Appendix, Supplementary Information Text). The following factors contribute to the low excitation energy of ChlD1 with respect to other chlorophylls.
Low excitation energy at the accessory positions.
PD1 and PD2 have histidine ligands (D1-His198 and D2-His197, respectively) and ChlD1 and ChlD2 have water ligands (W424D and W1009A, respectively). The difference in the ligand group, histidine or water, is not the primary reason for the lower excitation energy at the accessory chlorophyll sites than the pair chlorophyll sites, because two ligands decrease the chlorophyll excitation energy equally, by ∼10 meV (SI Appendix, Table S4).
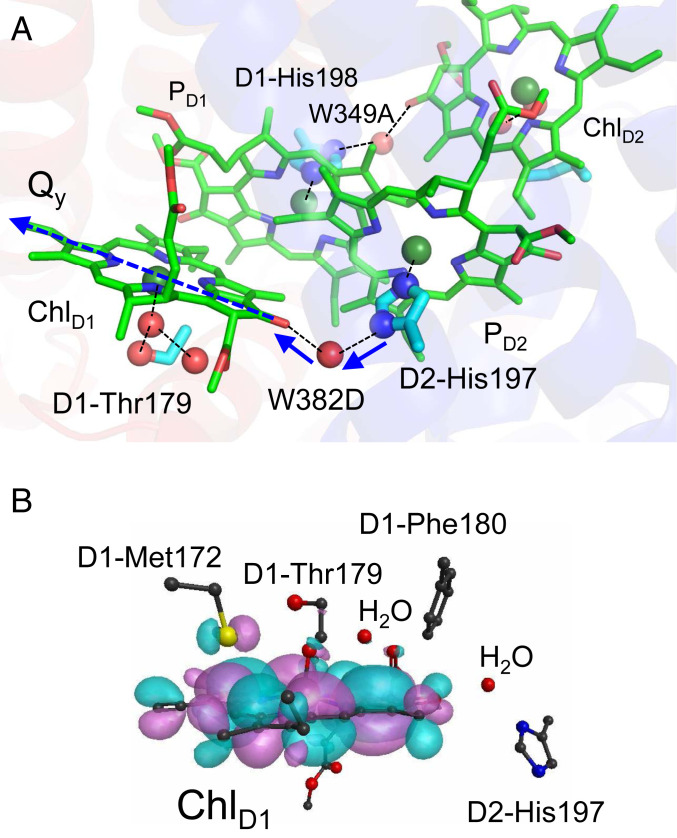
Remarkably, the excitation energy of ChlD1 is decreased by the PD2 ligand, D2-His197, and a water molecule (W382D) that bridges between the Nδ site of D2-His197 and the keto O site of ChlD1 (SI Appendix, Table S4) because both -Nδ-H of D2-His197 and –OH of the bridging water molecule orient toward the keto O site of ChlD1 along the Qy transition dipole (Fig. 5A). On the ChlD2 site, the corresponding components, the PD1 ligand (D1-His198) and the bridging water molecule (W349A), also decrease the excitation energy (SI Appendix, Table S4). It seems that the H-bond network, [PD2…D2-His197…H2O…ChlD1] and [PD1…D1-His198…H2O…ChlD2], decrease the excitation energy on the accessory chlorophyll sites with respect to the pair chlorophyll sites.
Fig. 5.
(A) The bridging water molecule (W382D) that connects between the ligand Nδ site of PD2 and the keto O site of ChlD1. H-bonds and ligand interactions are indicated by dotted lines. The donor to acceptor orientations of the H-bonds are indicated by blue solid arrows. The Qy transition dipole is indicated by the dotted blue arrow (see SI Appendix, Fig. S1 for the orientations of the Qx and Qy transition dipoles). (B) Distribution of HOMO (pink and cyan spaces) at the ChlD1 moiety, which were obtained based on QM/MM/PCM (r = 3.0) with the CAM-B3LYP functional (μ = 0.14). The QM region was defined as ChlD1, the ligand (W424D), second sphere ligand (W1003A), and bridging (W382D) water molecules, and the side chains of D1-Met172 and D1-Phe180 in van der Waals contact with ChlD1, and the ligand (or ligand-associated) side chains of D1-Thr179 and D2-His197.
The bridging water molecule is also conserved in PbRC. It may contribute to a decrease in the excitation energy at the accessory BL and BM sites. However, the influence can practically be ignored because the special pair [PLPM] formation decreases the excitation energy at PL and PM more significantly in PbRC (>200 meV; Table 2).
Low excitation energy of ChlD1 with respect to ChlD2.
QM/MM calculations show that the difference in the protein electrostatic environment between D1 and D2 is the major factor that decreases the excitation energy of ChlD1 with respect to ChlD2 (by 23 meV; Table 2). Most of the D1/D2 residue pairs that stabilize ChlD1* with respect to ChlD2* are located at the van der Waals distance from ChlD1 and ChlD2. D1-Met172, which is 3.7 Å away from ChlD1 in the loop region near helix cd (D1-176 to 190 and D2-176 to 188 in PSII, Fig. 3B), stabilizes ChlD1* by 10 meV (Table 4) by hybridizing the sulfur lone-pair molecular orbital with the HOMO of ChlD1 (Fig. 5B) and interacting electrostatically. The corresponding stabilization is absent in ChlD2* because D1-Met172 is replaced with D2-Pro171 near ChlD2. As far as we are aware, the role of D1-Met172 in the PSII function is not reported, but the absence of the corresponding methionine near BL in PbRC implies that D1-Met172 plays a role in stabilizing ChlD1* in PSII, and that the charge separation mechanism differs between the two type-II reaction centers. D1-Met172 is replaced with leucine in the PsbA2 protein, which can be expressed under microaerobic conditions in cyanobacterial PSII. In PsbA2, in turn, the next residue D1-Pro173 is replaced with methionine (30), which might partially substitute a role of D1-Met172 of the PsbA protein. The difference in the H-bond network of the ligand water molecule between ChlD1 and ChlD2, i.e., the presence of an H-bond donor (D1-Thr179) on the ChlD1 side and the absence of the H-bond donor (D2-Ile178) on the ChlD2 side also decreases the excitation energy of specifically ChlD1 (4 meV; Table 4).
Table 4.
D1/D2 residue pairs that decreases the excitation energy of ChlD1 with respect to ChlD2 in millielectron volts
| ChlD1* | ChlD2* | ChlD1* | ChlD2* | Stabilizing ChlD1* | ||
| D1-Met172 | −10 | 0 | D2-Pro171 | 0 | 1 | −11 |
| D1-Thr179 | −4 | 0 | D2-Ile178 | 0 | 0 | −4 |
| Cl-1 | −2 | 0 | −2† | |||
| +D1-Asn181 | 0 | 0 | +D2-Arg180 | 0 | 2 | |
| +D1-Trp317 | 0 | 0 | +D2-Lys317 | 2 | 0 |
As a Cl-1 binding site with Cl-1, D1-Asn181, and D2-Lys317.
The only electrostatic component that decreases the excitation energy of ChlD1 with respect to ChlD2 is Cl-1 and the binding sites D1-Asn181 and D2-Lys317 (2 meV; Table 4 and Fig. 3B). Molecular dynamics simulations by Rivalta et al. (31) suggested that the removal of Cl-1 from the binding site lead to the formation of a salt-bridge between D1-Asp61 and D2-Lys317; this would inhibit the release of a proton from W1. The counterpart of polar D1-Asn181 is basic D2-Arg180, which provides a driving force for proton transfer from TyrD toward the bulk surface (32, 33). Mutations of D2-Arg180 resulted in a loss and/or serious modifications of the electron paramagnetic resonance signal from TyrD and perturbations in the PSII photochemistry (34). The D1-Asn181/D2-Arg180 pair also contributes to PD1•+ > PD2•+ (8) and Em(ChlD1) < Em(ChlD2) (i.e., ChlD1•+ > ChlD2•+) (19). See below for further details of the D1-Asn181/D2-Arg180 pair.
Charge-Separated State ChlD1•+PheoD1•– in PSII.
As described, ChlD1 has the lowest excitation energy among the four chlorophylls and can serve as the initial electron donor. In this case, ChlD1•+PheoD1•– is the initial charge-separated state. In the TDDFT-QM/MM/PCM calculations, where the interaction between electron and hole is considered quantum-chemically, ChlD1•+PheoD1•– in the D1-branch is 409 meV is more stable than the corresponding ChlD2•+PheoD2•– in the D2-branch, which suggests that the charge separation occurs predominantly along the D1-branch via ChlD1•+PheoD1•– (Fig. 2). The HOMO energy level is highest at PD1 among the four chlorophylls (Fig. 4A). This indicates that the hole on ChlD1•+PheoD1•– is transferred to and stabilized predominantly at PD1, which is consistent with the larger population of PD1•+ than PD2•+ in the [PD1PD2]•+ pair (8, 35–37). The significant energy difference between ChlD1•+PheoD1•– and ChlD2•+PheoD2•– is mainly due to the energy difference in the oxidized accessory chlorophylls, ChlD1•+ and ChlD2•+ (272 meV), rather than in the reduced pheophytins, PheoD1•– a and PheoD2•– (114 meV, SI Appendix, Fig. S3).
Larger stability of PheoD1•– than PheoD2•–.
The D1/D2 residue pairs that stabilize PheoD1•– with respect to PheoD2•– were D1-Met214/D2-Ile213 (106 meV), D1-Arg27/D2-Phe27 (73 meV), D1-Tyr126/D2-Phe125 (67 meV), and D1-Arg136/D2-Leu135 (52 meV) (Table 5). D1-Met214 provides the hydrophobic binding pocket to the isoprenoid side-chain of QB. D1-Met214 destabilizes PheoD2•– (SD1-Met214… OPheoD2 = 3.1 Å) in the hydrophobic environment, whereas the Cγ of D2-Ile213 stabilizes PheoD1•– (CγD2-Ile213… OPheoD1 = 3.1 Å; Table 5). D1-Arg27 stabilizes the binding of the anionic head group of sulfoquinovosyl diacylglycerol. Although D1-Arg27 and D1-Arg136 are located near the stromal surface, their electrostatic influence is not completely shielded at PheoD1 due to the presence of the hydrophobic environment around QA. D1-Tyr126 donates an H-bond to the ester carbonyl O of PheoD1 and stabilizes PheoD1•– (38).
Table 5.
D1/D2 residue pairs that stabilize PheoD1•– with respect to PheoD2•– (>40 meV) in the LUMO energy level in millielectron volts (corresponding to Em for one-electron reduction)
| Em(PheoD1) | Em(PheoD2) | Em(PheoD1) | Em(PheoD2) | Stabilizing PheoD1•– | ||
| D1-Met214 | 8 | −41 | D2-Ile213 | 65 | 8 | 106 |
| D1-Arg27 | 84 | 19 | D2-Phe27 | 0 | −8 | 73 |
| D1-Tyr126 | 84 | 3 | D2-Phe125 | 0 | 14 | 67 |
| D1-Arg136 | 71 | 14 | D2-Leu135 | 0 | 5 | 52 |
In PbRC, HL•– is also more stable than HM•– (Figs. 2 and 4). However, the L/M residues that stabilize HL•– with respect to HM•– (e.g., Glu-L104 (39), SI Appendix, Table S5) do not correspond to those that stabilize PheoD1•– with respect to PheoD2•– (Table 5), which implies the difference in the protein electrostatic environment and the charge-separation mechanism between the two reaction centers.
Larger stability of ChlD1•+ than ChlD2•+.
The D1-Asp61/D2-His61 and D1-Asp170/D2-Phe169 pairs stabilize ChlD1•+ with respect to ChlD2•+ (Table 6 and Fig. 3B); the two residue pairs are reported to be responsible for the lower Em(ChlD1) than Em(ChlD2) (19). The D1-Met172/D2-Pro171 and D1-Thr179/D2-Ile178 pairs stabilize not only ChlD1•+ with respect to ChlD2•+ (Table 6 and Fig. 3B) but also ChlD1* with respect to ChlD2* (Table 4), facilitating the ChlD1* formation and the subsequent ChlD1•+ formation. Remarkably, many of the residues listed in Table 6 play a key role in the release of electrons or protons from the substrate water molecules. D1-Asp61/D2-His61, D1-Asn181/D2-Arg180, and D1-Asp170/D2-Phe169 contribute to PD1•+ > PD2•+ (8), which is advantageous for electron transfer from the substrate water molecules to PD1•+. D1-Asp61 (32, 40–43), D2-Lys317 (31, 44), and Cl− (32, 45, 46) are involved in the proton transfer pathway that proceeds from the oxygen-evolving complex toward the protein bulk surface. D1-Asp170 and D1-Glu189 listed in Table 6 are the ligand residues of the Mn4CaO5 cluster.
Table 6.
D1/D2 residue pairs that stabilize ChlD1•+ with respect to ChlD2•+ (>40 meV) in the HOMO energy level in millielectron volts (corresponding to Em for one-electron oxidation)
| Em(ChlD1) | Em(ChlD2) | Em(ChlD1) | Em(ChlD2) | Stabilizing ChlD1•+ | ||
| D1-Asp61* | −117 | −35 | D2-His61† | 35 | 141 | −188 |
| Cl-1 | −152 | −57 | −141‡ | |||
| +D1-Asn181† | 14 | 0 | +D2-Arg180† | 68 | 234 | |
| +D1-Trp317 | −5 | −5 | +D2-Lys317§ | 152 | 46 | |
| D1-Thr179¶ | −35 | −5 | D2-Ile178 | −8 | 52 | −90 |
| D1-Asp170# | −122 | −44 | D2-Phe169 | −5 | −11 | −72 |
| D1-Met183 | −19 | 3 | D2-Leu182 | −5 | 35 | −62 |
| D1-Arg323 | 38 | 65 | D2-Glu323 | −82 | −52 | −57 |
| D1-Asp59 | −73 | −22 | D2-Tyr59 | 0 | 3 | −54 |
| D1-Met172 | −22 | 0 | D2-Pro171 | 5 | 35 | −52 |
| D1-Glu65‖ | −79 | −19 | D2-Ser65 | −3 | −8 | −55 |
| +D1-Asn315 | +D2-Glu312‖ | |||||
| D1-Glu189# | −79 | −41 | D2-Phe188 | 3 | 14 | −49 |
| D1-Val306 | 0 | 0 | D2-Glu302 | −87 | −38 | −49 |
Proton transfer pathway proceeding from the Mn4CaO5 cluster.
As a Cl-1 binding site with Cl-1, D1-Asn181, and D2-Lys317.
Cl-1 binding site.
H-bond partner of the second sphere water ligand of ChlD1.
Ligand of the Mn4CaO5 cluster.
PD1•+PheoD1•– in PSII.
Fig. 2 shows that PD1•+PheoD1•– is more stable than ChlD1•+PheoD1•–, which suggests that the hole on ChlD1•+ is transferred to PD1•+ during the charge-separation process. Because PD1 is closer to TyrZ than ChlD1, the PD1•+ formation is more advantageous to accept an electron from the substrate water molecules via TyrZ in water oxidation. The corresponding state on the D2 side, PD2•+PheoD2•–, is 478 meV less stable than PD1•+PheoD1•–, which suggests that the charge separation predominantly occurs along the D1-branch and the charge separation via ChlD1•+PheoD1•– is further stabilized by the PD1•+PheoD1•– formation (Fig. 2).
Discussion
The present result indicates that the absence of the special pair formation between PD1 and PD2 prevents [PD1PD2] from serving the initial electron donor, but it increases Em for one-electron oxidation and makes PSII capable of oxidizing the substrate water molecules (11, 47). The absence of the special pair formation along the pseudo-C2 axis also requires PSII to localize the initial charge separation site either on the D1 or D2 side. The excitation energy is lowest at ChlD1, although the difference in the energy between ChlD1* and ChlD2* is not particularly large, 29 meV (Fig. 2). The initial charge-separated state, ChlD1•+PheoD1•–, is 409 meV more stable than ChlD2•+PheoD2•– (Fig. 2) due to the more positively charged protein environment on the D2 side than on the D1 side (e.g., D1-Asp61/D2-His61, D1-Asn181/D2-Arg180, and D1-Trp317/D2-Lys317 in Table 6 (19)). In the presence of the intramolecular reorganization energy, the corresponding energy difference is 386 meV; charge separation from [ChlD1]* to ChD1•+PheoD1•– is 264 meV energetically downhill, whereas charge separation from [ChlD2]* to ChD2•+PheoD2•– is 95 meV uphill (SI Appendix, Fig. S2), in agreement with electron transfer predominantly occurring along the D1 branch.
Remarkably, many of these residues that play a key role in stabilizing ChlD1* (Table 4) and ChlD1•+ (Table 6) are located in the lumenal helix cd (D1-176 to 190 and D2-176 to 189) and adjacent loop (D1-166 to 175 and D2-164 to 175) region (Fig. 3B). It has been reported that the lumenal helix cd and adjacent loop region characterizes PSII with respect to PbRC most significantly, making Em(ChlD1) < Em(ChlD2) in PSII, whereas Em(BL) > Em(BM) in PbRC (19). Two Mn4CaO5 ligand residues, D1-Asp170 and D1-Glu189, are also located in this region and stabilize ChlD1•+ (Table 6). Intriguingly, in the earliest-evolving D1 proteins (e.g., Gloeobacter kilaueensis, Chroococcidiopsis thermalis PCC7203, and Fischerella sp. JSC-11), the two ligand residues are not conserved (48). D1-Asp170 is replaced with alanine or serine, which would fail to bind Ca2+ and the dangling Mn (Mn4). D1-Glu189 is often replaced with aspartate (asparagine, arginine, glutamine, or alanine in some D1 proteins) in the earliest-evolving D1 proteins. The side chain of D1-Glu189 accepts an H-bond from a water molecule (W7), which is the H-bond donor to TyrZ and is required for TyrZ and D1-His190 to form a remarkably short, low-barrier H-bond (OTyrZ…ND1-His190 = 2.5 Å) (49). Shortening the side-chain length from glutamate to aspartate would alter the W7 position and fail to form the low-barrier H-bond. Based on these, the earliest evolving D1 proteins are unlikely to proceed water oxidation (48). Indeed, the earliest evolving D1 proteins also lack D1-Glu65 in the proton transfer pathway (40, 41, 50), which suggests that not only the Mn4CaO5 cluster but also the proton transfer pathway is incomplete.
In contrast, the protein environment near PheoD1 is highly conserved even in the earliest evolving D1 proteins, as D1-residues listed in Table 5 are mostly conserved (note that D1-Met214 is replaced with alanine in G. kilaueensis) (48). In addition, the D1/D2 residue pairs that facilitate the PheoD1•– formation (Table 5) are neither in common with the residue pairs that facilitate the ChlD1* formation (Table 4) nor the ChlD1•+ formation (Table 6). It seems thus far likely that the acquirement of the oxygen-evolving ability predominantly alters the protein environment that is crucial specifically for the ChlD1, without affecting the PheoD1 energetics.
The difference in the electrostatic properties of the residues that stabilize the charge-separated intermediate states between PbRC and PSII is also remarkable. In PSII, charged residues are predominantly involved in the stabilization of ChlD1•+PheoD1•– (Table 6), whereas in PbRC, polar, uncharged residues are involved in the stabilization of [PLPM]•+BL•– (Table 3). Because the binding sites of ChlD1 and BL are located in the uncharged transmembrane regions, the stabilization of [PLPM]•+BL•– by polar, uncharged residues is energetically favored. The less-charged protein environment of PbRC is also indicated as Glu-M95 on the protein surface of subunit M destabilizes not only BM•– (Table 3) but also HM•– (SI Appendix, Table S5). These polar, uncharged residues are not conserved in PSII. It is highly likely that the membrane-extrinsic charged region that provides the cationic Mn4CaO5 binding site and the proceeding proton transfer pathway can stabilize the charge-separated state more effectively (e.g., PSII) than the polar protein environment in the uncharged transmembrane region (e.g., PbRC). This could also explain why PSII does not require Tyr-M210, the residue that is most crucial to the stabilization of [PLPM]•+BL•– (19, 20, 29).
Conclusions
Based on the energetics of the electronically excited states and charge-separated states presented here, we are able to explain why electron transfer predominantly occurs along the L-branch in PbRC and the D1-branch in PSII, irrespective of the pseudo-C2 symmetry of the two electron transfer branches (Fig. 2). Notably, these findings cannot be obtained solely from the Em profiles, as the LUMO energy levels (i.e., Em for one-electron reduction) are downhill along the two branches in both PbRC and PSII (Fig. 4A).
In PbRC, PL and PM form the special pair [PLPM], as indicated by a decrease of 230 to 270 meV in the excitation energy (Table 2 and Fig. 2). [PLPM]•+BL•– is 281 meV more stable than [PLPM]•+BM•– (Fig. 2). Glu-M95 and Asp-M184, which provide the cytochrome c2 binding interface (21), are the charged components that destabilize BM•–. In contrast to PSII, it is characteristic to PbRC that polar residues in der van der Waals contact with BL and BM also contribute to the significant energy difference between [PLPM]•+BL•– and [PLPM]•+BM•–. Tyr-M210 stabilizes BL•–, and Thr-M186 and Ser-L178 destabilize BM•– (Table 3). The values of electronic coupling and excitonic coupling between PD1 and PD2 in PSII are significantly smaller than those between PL and PM in PbRC (Table 1), which indicates that PD1 and PD2 do not form the special pair. The absence of the special pair in PSII displaces the excitation site from the pseudo-C2 axis to the D1 site. The H-bond network [PD2…D2-His197…H2O…ChlD1] and [PD1…D1-His198…H2O…ChlD2], which orients toward the Qy transition dipole of ChlD1 and ChlD2, decrease the excitation energy of ChlD1 and ChlD2, respectively (SI Appendix, Table S4 and Fig. 5A). The presence of D1-Met172 that hybridizes the sulfur lone-pair molecular orbital with the HOMO of ChlD1 (Fig. 5B) and D1-Thr179 that forms the H-bond network with the ChlD1 ligand water molecule contribute to a decrease in the excitation energy of ChlD1 (Table 4). The excitation energy of ChlD1 is thus far slightly (29 meV) lower than that of ChlD2 (Fig. 2), whereas ChlD1•+PheoD1•– is 409 meV more stable than ChlD2•+PheoD2•– (SI Appendix, Fig. S3). This suggests that the large stability of ChlD1•+PheoD1•– is the main factor that facilitates the D1-branch electron transfer. The large stability of PD1•+PheoD1•– with respect to ChlD1•+PheoD1•– (Fig. 2) makes PD1•+ serve as an electron acceptor for the substrate water molecules via TyrZ.
In PSII, the key components that play a role in stabilizing the intermediate ChlD1•+ state and facilitating the D1-branch electron transfer (Table 6) also play a role in 1) decreasing the excitation energy of ChlD1 (D1-Met172, D1-Thr179, and Cl-1; Table 4), 2) constructing the Mn4CaO5 cluster (D1-Asp170, D1-Glu189, and the second sphere ligand D1-Asp61; Table 6), 3) mediating proton transfer from the substrate water molecules [D1-Asp61, D1-Glu65, D2-Glu312, and Cl-1 (41, 51)], and 4) facilitating electron transfer from the substrate water molecule by pushing the cationic state toward PD1•+ [D1-Asp61/D2-His61, D1-Asp170/D2-Phe169, D1-Asn181/D1-Arg180, and D1-Glu189/D2-Phe188 (8)]. Thus, the charge-separation mechanism, the ChlD1* formation and the subsequent electron transfer via the ChlD1•+ intermediate, is largely associated with the water-splitting ability.
As viewed, the localization of the acidic environment for hosting the Mn4CaO5 cluster (e.g., the Mn4CaO5 ligands and the proceeding proton transfer pathway) contributes to the ChlD1•+PheoD1•– stabilization in the charge separation process (Table 6). This can explain why both the water-splitting site and the charge-separation site are located on the same D1 protein. The localization of the acidic environment for hosting the Mn4CaO5 cluster also contributes to the PD1•+ stabilization in the [PD1PD2]•+ pair (8). This also indicates that the localization of the Mn4CaO5 cluster on the D1 protein is ultimately the basis of restricting photodamage to the D1 protein (47). It should be noted that the absence of large π-coupling between PD1 and PD2 can also contribute to confining the subsequent cationic state to the D1 side (as PD1•+).
Most of the D1/D2 residue pairs that facilitate the D1-branch electron transfer (Tables 4 and 6) are not conserved in PbRC (2) and some of these residues are even not conserved in the earliest evolving D1 proteins (48). Most of the L/M residue pairs that facilitate the L-branch electron transfer in PbRC (Table 3) are also not conserved in PSII (2). The independent components (i.e., uncharged polar groups near BL and BM in the transmembrane region in PbRC and charged groups near the water-splitting/proton-conducting site in the membrane-extrinsic region in PSII) facilitate the similar, unidirectional electron transfer in completely different mechanisms. It seems likely that the unique charge separation via ChlD1•+PheoD1•– is pronounced after PSII obtains the complete, functional Mn4CaO5 cluster and the proceeding electron and proton transfer pathways.
Methods
Coordinates and Atomic Partial Charges.
The atomic coordinates were taken from the X-ray structures, PbRC from R. sphaeroides at 2.01-Å resolution (Protein Data Bank [PDB] ID code 3I4D), and PSII monomer unit (designated monomer A) of the PSII complexes from Thermosynechococcus vulcanus at 1.9-Å resolution (PDB ID code 3ARC) (52). The polarizable AMBER-02 force field (53) was used to consider the induced dipoles of the MM atoms. The positions of all heavy atoms were fixed and all titratable groups (e.g., acidic and basic groups) were ionized in the MM region. The residue protonation states were consistent with those used for Em calculations in PbRC and PSII (19). The protonation states of the titratable residues were considered to be ionized for acidic and basic residues and charge-neutral for histidine residues unless otherwise specified (listed in ref. 50). Because D1/D2 residue pairs that affect the excitation energy (Table 4) or the charge-separated state (Table 6) do not contain the doubly protonated histidine residues [e.g., D1-His92 (50)] except for D2-His61, the present results are unlikely to depend on the protonation states. The large stability of ChlD1•+ with respect to ChlD2•+ will be less pronounced if D2-His61 is not doubly protonated. However, this does not affect the conclusions, because D1-Asp61 is ionized (41). D1-His337, which forms an H-bond with the Mn4CaO5 cluster, was considered to be protonated (54). Water molecules in the crystal structures were represented explicitly. The atomic charges of the other cofactors [(bacterio)chlorophyll, (bacterio)pheophytin, ubiquinone, plastoquinone, spheroidene, sulfoquinovosyl diacylglycerol, heptyl 1-thiohexopyranoside, and the Fe complex] were taken from previous studies (19), which were determined by fitting the electrostatic potential in the neighborhood of these molecules using the RESP (Restrained Electrostatic Potential) procedure (55). To obtain the atomic charges of the Mn4CaO5 cluster or the Fe complex, backbone atoms are not included in the RESP procedure (except for D1-Ala344). The resulting atomic partial charges of the cofactors, including the nonheme Fe complex in PbRC and PSII and the Mn4CaO5 cluster (S1), where (Mn1, Mn2, Mn3, Mn4) = (III, IV, IV, III), are listed in ref (19). For the atomic charges of the nonpolar CHn groups in cofactors (e.g., the phytol chains of (bacterio)chlorophyll and (bacterio)pheophytin and the isoprene side chains of quinones), the value of +0.09 was assigned for nonpolar H atoms.
QM/MM Calculations.
We employed the electrostatic embedding QM/MM/PCM scheme for all of the electronic structure calculations. We used the QuanPol method (56) implemented in the GAMESS code (57), in which Lennard-Jones parameters describe interactions between QM and MM atoms.
Geometry Optimization.
For geometry optimization, the DFT method was employed with the B3LYP functional plus the Grimme’s dispersion correction (58) and 6-31G(d) basis sets. The PCM method was not used for geometry optimization (i.e., QM/MM). All of the atomic coordinates in the QM region were fully relaxed (i.e., not fixed) during the QM/MM-geometry optimization. The coordinates of the heavy atoms in the surrounding MM region were fixed at their original X-ray coordinates. The polarizable AMBER-02 force field (53) was used to consider the induced dipoles of the MM atoms and to reproduce the dielectric screening. For geometry optimization of (bacterio)chlorophyll, the QM region was defined as the PL/PM/BL/BM bacteriochlorophyll tetramer for PbRC and the PD1/PD2/ChlD1/ChlD2 chlorophyll tetramer for PSII, with the ligand groups and the water molecules as an H-bond partner of (bacterio)chlorophyll. The H-bond partners of (bacterio)chlorophylls and (bacterio)pheophytins (i.e., His-L168 for PL, protonated Glu-L104 for HL, D1-Tyr126 and D1-Gln130 for PheoD1, and D2-Gln129 and D2-Asn142 for PheoD2) were also included in the QM region. See Datasets S1 and S2 for the QM/MM-optimized atomic coordinates.
Contribution of Residues to the HOMO and LUMO Energy Levels.
To analyze contributions of residues to the HOMO and LUMO levels (which correspond to Em for one-electron oxidation and reduction, respectively), we used the QM/MM/PCM scheme with the B3LYP functional (i.e., polarizable QM/MM/PCM).
Electronic Coupling, Excitonic Coupling, Excited States, and Charge-Separated States.
To calculate electronic coupling of the (bacterio)chlorophyll pair, we employed a QM/MM approach with PCM method with the dielectric constant of 80, in which electrostatic and steric effects created by a protein environment were explicitly considered in the presence of bulk water. Here, the polarizable amber-02 force field (53) was applied for the MM region, where induced dipoles of the MM atoms were considered to reproduce the dielectric screening (i.e., polarizable QM/MM/PCM). In the PCM method, the polarization points were put on the spheres with the radius of 3.0 Å from each atom center. The intramolecular reorganization energies of (bacterio)chlorophylls were calculated through QM/MM-geometry optimization.
To analyze excitonic coupling of the (bacterio)chlorophyll pair and the energetics of electronically excited states and charge separated states, the TDDFT was employed with the CAMB3LYP functional (59), where the range-separation parameters μ of 0.14 (25), α of 0.19, and β of 0.46 were used (i.e., polarizable TDDFT-QM/MM/PCM). The electronic couplings between excited states are calculated with the diabatization scheme of adiabatic electronic states (28). Here, the exciton coupling includes contributions from both long-range Coulomb (Förster) and electron exchange (Dexter) mechanisms. When excited states were not involved in the electronic coupling between the pair (bacterio)chlorophylls, charge transfer integrals were calculated based on (bacterio)chlorophyll monomer orbitals and the Fock matrix of (bacterio)chlorophyll dimer (60). The calculation scheme for the electronic coupling (28) is implemented in the modified version of the GAMESS program.
Data Availability.
All of the data supporting the findings of this study are available within the paper, SI Appendix, and Datasets S1 and S2.
Supplementary Material
Acknowledgments
This research was supported by Japan Science and Technology Agency Core Research for Evolutional Science and Technology (JPMJCR1656 to H.I.), Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (JP18H01937 to H.T. and H.I., JP18H05155, JP20H03217, and JP20H05090 to H.I., JP18H01186 to K.S., and JP16H06560 to K.S.), and the Interdisciplinary Computational Science Program in the Center for Computational Sciences, University of Tsukuba.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000895117/-/DCSupplemental.
References
- 1.Shen J. R., The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Michel H., Deisenhofer J., Relevance of the photosynthetic reaction center from purple bacteria to the structure of photosystem II. Biochemistry 27, 1–7 (1988). [Google Scholar]
- 3.Zinth W., Wachtveitl J., The first picoseconds in bacterial photosynthesis—ultrafast electron transfer for the efficient conversion of light energy. ChemPhysChem 6, 871–880 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Vasil’ev S., Orth P., Zouni A., Owens T. G., Bruce D., Excited-state dynamics in photosystem II: Insights from the x-ray crystal structure. Proc. Natl. Acad. Sci. U.S.A. 98, 8602–8607 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasil’ev S., Bruce D., A protein dynamics study of photosystem II: The effects of protein conformation on reaction center function. Biophys. J. 90, 3062–3073 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klimov V. V., Allakhverdiev S. I., Demeter S., Krasnovskii A. A., Photoreduction of pheophytin in the photosystem 2 of chloroplasts with respect to the redox potential of the medium. Dokl. Akad. Nauk SSSR 249, 227–230 (1979). [Google Scholar]
- 7.Rutherford A. W., Mullet J. E., Crofts A. R., Measurement of the midpoint potential of the pheophytin acceptor of photosystem II. FEBS Lett. 123, 235–237 (1981). [Google Scholar]
- 8.Saito K. et al., Distribution of the cationic state over the chlorophyll pair of the photosystem II reaction center. J. Am. Chem. Soc. 133, 14379–14388 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Rappaport F., Guergova-Kuras M., Nixon P. J., Diner B. A., Lavergne J., Kinetics and pathways of charge recombination in photosystem II. Biochemistry 41, 8518–8527 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Williams J. C. et al., Effects of mutations near the bacteriochlorophylls in reaction centers from Rhodobacter sphaeroides. Biochemistry 31, 11029–11037 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Mandal M., Kawashima K., Saito K., Ishikita H., Redox potential of the oxygen-evolving complex in the electron transfer cascade of photosystem II. J. Phys. Chem. Lett. 11, 249–255 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Gunner M. R., Nicholls A., Honig B., Electrostatic potentials in Rhodopseudomonas viridis reaction centers: Implications for the driving force and directionality of electron transfer. J. Phys. Chem. 100, 4277–4291 (1996). [Google Scholar]
- 13.Lockhart D. J., Boxer S. G., Stark effect spectroscopy of Rhodobacter sphaeroides and Rhodopseudomonas viridis reaction centers. Proc. Natl. Acad. Sci. U.S.A. 85, 107–111 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardona T., Sedoud A., Cox N., Rutherford A. W., Charge separation in photosystem II: A comparative and evolutionary overview. Biochim. Biophys. Acta 1817, 26–43 (2012). [DOI] [PubMed] [Google Scholar]
- 15.McDowell L. M., Kirmaier C., Holten D., Charge transfer and charge resonance states of the primary electron donor in wild-type and mutant bacterial reaction centers. Biochim. Biophys. Acta 1020, 239–246 (1990). [Google Scholar]
- 16.McDowell L. M., Gaul D., Kirmaier C., Holten D., Schenck C. C., Investigation into the source of electron transfer asymmetry in bacterial reaction centers. Biochemistry 30, 8315–8322 (1991). [DOI] [PubMed] [Google Scholar]
- 17.Heller B. A., Holten D., Kirmaier C., Effects of Asp residues near the L-side pigments in bacterial reaction centers. Biochemistry 35, 15418–15427 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Harris M. A. et al., Protein influence on charge-asymmetry of the primary donor in photosynthetic bacterial reaction centers containing a heterodimer: Effects on photophysical properties and electron transfer. J. Phys. Chem. B 117, 4028–4041 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Kawashima K., Ishikita H., Energetic insights into two electron transfer pathways in light-driven energy-converting enzymes. Chem. Sci. 9, 4083–4092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkele U., Lauterwasser C., Zinth W., Gray K. A., Oesterhelt D., Role of tyrosine M210 in the initial charge separation of reaction centers of Rhodobacter sphaeroides. Biochemistry 29, 8517–8521 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Axelrod H. L. et al., X-ray structure determination of the cytochrome c2: Reaction center electron transfer complex from Rhodobacter sphaeroides. J. Mol. Biol. 319, 501–515 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Rutherford A. W., . “Photosystem II, the oxygen evolving photosystem” in Light-Energy Transduction in Photosynthesis: Higher Plant and Bacterial Models, Stevens S., Bryant D. A., Eds. (American Society of Plant Physiology, Rockville, MD, 1988), pp. 163–177. [Google Scholar]
- 23.Prokhorenko V. I., Holzwarth A. R., Primary process and structure of the photosystem II reaction center: A photon echo study. J. Phys. Chem. B 104, 11563–11578 (2000). [Google Scholar]
- 24.Dekker J. P., van Grondelle R., Primary charge separation in Photosystem II. Photosynth. Res. 63, 195–208 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Saito K., Suzuki T., Ishikita H., Absorption-energy calculations of chlorophyll a and b with an explicit solvent model. J. Photochem. Photobiol. Chem. 358, 422–431 (2018). [Google Scholar]
- 26.Barkigia K. M., Chantranupong L., Smith K. M., Fajer J., Structural and theoretical models of photosynthetic chromophores. Implications for redox, light-absorption properties and vectorial electron flow. J. Am. Chem. Soc. 110, 7566–7567 (1988). [Google Scholar]
- 27.Saito K. et al., Deformation of chlorin rings in the Photosystem II crystal structure. Biochemistry 51, 4290–4299 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Tamura H., Diabatization for time-dependent density functional theory: Exciton transfers and related conical intersections. J. Phys. Chem. A 120, 9341–9347 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Parson W. W., Chu Z.-T., Warshel A., Electrostatic control of charge separation in bacterial photosynthesis. Biochim. Biophys. Acta 1017, 251–272 (1990). [DOI] [PubMed] [Google Scholar]
- 30.Sugiura M. et al., The D1-173 amino acid is a structural determinant of the critical interaction between D1-Tyr161 (TyrZ) and D1-His190 in Photosystem II. Biochim. Biophys. Acta 1837, 1922–1931 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Rivalta I. et al., Structural-functional role of chloride in photosystem II. Biochemistry 50, 6312–6315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito K., Rutherford A. W., Ishikita H., Mechanism of tyrosine D oxidation in photosystem II. Proc. Natl. Acad. Sci. U.S.A. 110, 7690–7695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito K., Sakashita N., Ishikita H., Energetics of the proton transfer pathway for tyrosine D in photosystem II. Aust. J. Chem. 69, 991–998 (2016). [Google Scholar]
- 34.Manna P., LoBrutto R., Eijckelhoff C., Dekker J. P., Vermaas W., Role of Arg180 of the D2 protein in photosystem II structure and function. Eur. J. Biochem. 251, 142–154 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Rigby S. E. J., Nugent J. H. A., O’Malley P. J., ENDOR and special triple resonance studies of chlorophyll cation radicals in photosystem 2. Biochemistry 33, 10043–10050 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Diner B. A. et al., Site-directed mutations at D1-His198 and D2-His197 of photosystem II in Synechocystis PCC 6803: Sites of primary charge separation and cation and triplet stabilization. Biochemistry 40, 9265–9281 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Sugiura M. et al., Site-directed mutagenesis of Thermosynechococcus elongatus photosystem II: The O2-evolving enzyme lacking the redox-active tyrosine D. Biochemistry 43, 13549–13563 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Kato Y., Nagao R., Noguchi T., Redox potential of the terminal quinone electron acceptor QB in photosystem II reveals the mechanism of electron transfer regulation. Proc. Natl. Acad. Sci. U.S.A. 113, 620–625 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saggu M., Fried S. D., Boxer S. G., Local and global electric field asymmetry in photosynthetic reaction centers. J. Phys. Chem. B 123, 1527–1536 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S., Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Ishikita H., Saenger W., Loll B., Biesiadka J., Knapp E.-W., Energetics of a possible proton exit pathway for water oxidation in photosystem II. Biochemistry 45, 2063–2071 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Debus R. J., Evidence from FTIR difference spectroscopy that D1-Asp61 influences the water reactions of the oxygen-evolving Mn4CaO5 cluster of photosystem II. Biochemistry 53, 2941–2955 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Kawashima K., Takaoka T., Kimura H., Saito K., Ishikita H., O2 evolution and recovery of the water-oxidizing enzyme. Nat. Commun. 9, 1247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pokhrel R., Service R. J., Debus R. J., Brudvig G. W., Mutation of lysine 317 in the D2 subunit of photosystem II alters chloride binding and proton transport. Biochemistry 52, 4758–4773 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Boussac A., Rutherford A. W., The origin of the split S3 EPR signal in Ca2+-depleted photosystem II: Histidine versus tyrosine. Biochemistry 31, 7441–7445 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Wincencjusz H., van Gorkom H. J., Yocum C. F., The photosynthetic oxygen evolving complex requires chloride for its redox state S2→S3 and S3→S0 transitions but not for S0→S1 or S1→S2 transitions. Biochemistry 36, 3663–3670 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Rutherford A. W., Faller P., Photosystem II: Evolutionary perspectives. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 245–253 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cardona T., Murray J. W., Rutherford A. W., Origin and evolution of water oxidation before the last common ancestor of the cyanobacteria. Mol. Biol. Evol. 32, 1310–1328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito K., Shen J.-R., Ishida T., Ishikita H., Short hydrogen bond between redox-active tyrosine YZ and D1-His190 in the photosystem II crystal structure. Biochemistry 50, 9836–9844 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Sakashita N., Watanabe H. C., Ikeda T., Saito K., Ishikita H., Origins of water molecules in the photosystem II crystal structure. Biochemistry 56, 3049–3057 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Debus R. J., FTIR studies of metal ligands, networks of hydrogen bonds, and water molecules near the active site Mn4CaO5 cluster in Photosystem II. Biochim. Biophys. Acta 1847, 19–34 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Umena Y., Kawakami K., Shen J.-R., Kamiya N., Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Cieplak P., Caldwell J., Kollman P., Molecular mechanical models for organic and biological systems going beyond the atom centered two body additive approximation: Aqueous solution free energies of methanol and N-methyl acetamide, nucleic acid base, and amide hydrogen bonding and chloroform/water partition coefficients of the nucleic acid bases. J. Comput. Chem. 22, 1048–1057 (2001). [Google Scholar]
- 54.Nakamura S., Noguchi T., Infrared determination of the protonation state of a key histidine residue in the photosynthetic water oxidizing center. J. Am. Chem. Soc. 139, 9364–9375 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Bayly C. I., Cieplak P., Cornell W. D., Kollman P. A., A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 97, 10269–10280 (1993). [Google Scholar]
- 56.Thellamurege N. M. et al., QuanPol: A full spectrum and seamless QM/MM program. J. Comput. Chem. 34, 2816–2833 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Schmidt M. W. et al., General atomic and molecular electronic-structure system. J. Comput. Chem. 14, 1347–1363 (1993). [Google Scholar]
- 58.Grimme S., Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 27, 1787–1799 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Yanai T., Tew D. P., Handy N. C., A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 393, 51–57 (2004). [Google Scholar]
- 60.Tamura H. et al., Theoretical analysis on the optoelectronic properties of single crystals of thiophene-furan-phenylene co-oligomers: Efficient photoluminescence due to molecular bending. J. Phys. Chem. C 117, 8072–8078 (2013). [Google Scholar]
- 61.Nakamura S., Noguchi T., Infrared detection of a proton released from tyrosine YD to the bulk upon its photo-oxidation in photosystem II. Biochemistry 54, 5045–5053 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Ho F. M., “Substrate and product channels in photosystem II” in Molecular Solar Fuels, Wydrzynski T. J., Hillier W., Eds. (RSC Energy and Environment Series, RSC Publishing, 2012), pp. 208–248. [Google Scholar]
- 63.Iwata S., Barber J., Structure of photosystem II and molecular architecture of the oxygen-evolving centre. Curr. Opin. Struct. Biol. 14, 447–453 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data supporting the findings of this study are available within the paper, SI Appendix, and Datasets S1 and S2.