Significance
Most of the known viruses of amoeba have been seen to share some features that eventually prompted authors to classify them into common evolutionary groups. Here we describe Yaravirus, an entity that could represent either the first isolated virus of Acanthamoeba spp. out of the group of NCLDVs or, in an alternative scenario, a distant and extremely reduced virus of this group. Contrary to what is observed in other isolated viruses of amoeba, Yaravirus does not have a large/giant particle or a complex genome, but at the same time carries a number of previously undescribed genes, including one encoding a divergent major capsid protein. Metagenomic approaches also testified for the rarity of Yaravirus in the environment.
Keywords: Yaravirus, ORFan, NCLDV, metagenomics, capsid
Abstract
Here we report the discovery of Yaravirus, a lineage of amoebal virus with a puzzling origin and evolution. Yaravirus presents 80-nm-sized particles and a 44,924-bp dsDNA genome encoding for 74 predicted proteins. Yaravirus genome annotation showed that none of its genes matched with sequences of known organisms at the nucleotide level; at the amino acid level, six predicted proteins had distant matches in the nr database. Complimentary prediction of three-dimensional structures indicated possible function of 17 proteins in total. Furthermore, we were not able to retrieve viral genomes closely related to Yaravirus in 8,535 publicly available metagenomes spanning diverse habitats around the globe. The Yaravirus genome also contained six types of tRNAs that did not match commonly used codons. Proteomics revealed that Yaravirus particles contain 26 viral proteins, one of which potentially representing a divergent major capsid protein (MCP) with a predicted double jelly-roll domain. Structure-guided phylogeny of MCP suggests that Yaravirus groups together with the MCPs of Pleurochrysis endemic viruses. Yaravirus expands our knowledge of the diversity of DNA viruses. The phylogenetic distance between Yaravirus and all other viruses highlights our still preliminary assessment of the genomic diversity of eukaryotic viruses, reinforcing the need for the isolation of new viruses of protists.
Viral evolution and classification have been subjects of an intense debate, especially after the discovery of giant viruses that infect protists (1–4). These viruses are predominantly characterized by the large size of their virions and genomes encoding hundreds to thousands of proteins, of which a large proportion currently remain without homologs in public sequence databases (5–10). These coding sequences are commonly referred as ORFans, and due to the lack of phylogenetic information, their origin and function still represent a mystery (11–14). Strikingly, the increasing number of available viral genomes demonstrates that there is a huge set and great diversity of genes without homologs in current databases, which needs to be further explored (11). Importantly, many amoebal virus ORFan genes have already been proven to be functional, being expressed and encoding for components of the viral particles (6, 15). However, the large set of ORFans makes it difficult to predict the biology of viruses discovered through cultivation-independent methods, such as metagenomics analysis, reinforcing the need for the complementary isolation and experimental characterization of new viruses. All currently known isolated amoebal viruses are related to nucleocytoplasmic large DNA viruses (NCLDVs) (16). This group comprises families of eukaryotic viruses (Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycodnaviridae, Marseilleviridae, and Mimiviridae) as well as other amoebal virus lineages including pithoviruses, pandoraviruses, molliviruses, medusaviruses, pacmanviruses, faustoviruses, klosneuviruses, and others. NCDLVs have dsDNA genomes and were proposed to share a monophyletic origin based on criteria that include the sharing of a set of ancestral vertically inherited genes (17, 18). From this handful set of genes, a core gene cluster is found to be present in almost all members of the NCLDVs, being composed by five distinct genes, namely a DNA polymerase family B, a primase-helicase, a packaging ATPase, a transcription factor, and a major capsid protein (MCP) for which the double jelly-roll (DJR) fold constitutes the main protein architectural class (19, 20). Recently, the International Committee on Taxonomy of Viruses (ICTV) brought forward a proposal for megataxonomy of viruses (21). The DJR major capsid protein (MCP) supermodule of DNA viruses includes NCLDVs and other icosahedral viruses that infect prokaryotes and eukaryotes. In addition to the signature DJR-MCPs, the majority of these viruses also encode for additional single jelly-roll minor capsid proteins (e.g., penton proteins) and genome packaging ATPases of the FtsK-HerA superfamily.
According to this proposal, the evolutionary conservation of the three genes of the morphogenetic module in the DJR-MCP supermodule, to the exclusion of all other viruses, justifies the establishment of a realm named Varidnaviria (21). Although some NCLDVs, as pandoraviruses, seem to have lost the DJR-MCP gene, their genome harbors a large set of genes that support their classification into NCLDVs (and Varidnaviria, consequently). Here we describe the discovery of Yaravirus, an amoeba virus with a puzzling origin and evolution. This virus has a genome that mainly consists of a near full set of genes that are ORFans. Yaravirus could represent either the first isolated virus of Acanthamoeba spp. out of the group of NCLDVs, inaugurating a group belonging to Varidnaviria, or, in an alternative evolutive scenario, a distant and extremely reduced virus of this group. Viral particles have a size of 80 nm, escaping the concept of large and giant viruses. Thus, Yaravirus expands our knowledge about viral diversity and evolution.
Results
Yaravirus Isolation and Replication Cycle.
A prospecting study was conducted by collecting samples of muddy water from creeks of an artificial urban lake called Pampulha, located at the city of Belo Horizonte, Brazil. Here, by using a protocol of direct inoculation of water samples on cultures of Acanthamoeba castellanii (Neff strain, ATCC 30010), we have managed to isolate an amoebal virus that we named Yaravirus brasiliensis, as a tribute to an important character (Yara, the mother of waters) of the mythological stories of the Tupi-Guarani indigenous tribes (22). Negative staining revealed the presence of small icosahedral particles on the supernatant of infected amoebal cells, measuring about 80 nm in diameter (Fig. 1A). Cryoelectron microscopy images of purified particles suggest that Yaravirus particles present two capsid shells, as previously described for Faustovirus, although future studies are needed to confirm this (23) (SI Appendix, Fig. S1).
Fig. 1.
Yaravirus particle and the beginning of the viral cycle. (A) Negative staining of an isolated Yaravirus virion. (Scale bar: 100 nm.) (B) Transmission electron microscopy (TEM) representing the beginning of the viral cycle, in which one particle is associated to the host cell membrane and the second one was already incorporated by the amoeba inside an endocytic vesicle. (Scale bar: 200 nm.) (C) Detailed image of an incorporated Yaravirus particle in the interior of an endocytic vesicle. (Scale bar: 100 nm.) (D) Viral uptake by the amoeba may occur individually but also in groups of particles, as observed in the micrograph. (Scale bar: 250 nm.) (E) The viral factory completely develops, occupying the nuclear region and recruiting mitochondria around it. Two different regions can be distinct: an electron-lucent region where the virions are assembled as empty shells and a second region formed by several electron-dense points where the genome is packaged inside the particles. (Scale bar: 500 nm.)
At the beginning of infection in A. castellanii, Yaravirus particles are found attached to the outside part of the amoebal plasma membrane, suggesting the participation of a host receptor in order to internalize the virions (Fig. 1B, SI Appendix, Fig. S2, blue arrows, and Movie S1). The replication cycle is then followed by the incorporation of individual or grouped Yaravirus particles inside endocytic vesicles, which, in a later stage of infection, are found next to a region occupied by the nucleus (Fig. 1 B–D and SI Appendix, Fig. S2, red arrows). The viral factory then takes place and completely develops into its mature form, replacing the region formerly occupied by the cell nucleus and recruiting mitochondria around its boundaries, likely to optimize the availability of energy to construct the virions (Fig. 1E). The step corresponding to viral morphogenesis happens similarly as how it is observed for other viruses of amoeba. First, it starts by the appearance of small crescents in the electron-lucent region of the factory (Fig. 1E and 2A). Next, step by step, the virions gain an icosahedral symmetry by the sequential addition of more than one layer of protein or membranous components around its structure (Fig. 2A). The constructed virions, with a capsid still empty, start then to migrate to the periphery of the viral factory, where there is the accumulation of corpuscular electron-dense material (Figs. 1E and 2 B and C, SI Appendix, Fig. S3 A–C, and Movie S2). These enucleations are scattered throughout the periphery of the infected cell and seem to represent different regions or morphogenesis points where the final step for Yaravirus maturation occurs. In these regions, the capsid of Yaravirus is filled with electron-dense material and the virus is finally ready to be released (Fig. 2C, red arrow, and SI Appendix, Fig. S4). Sometimes it is also possible to observe several particles of Yaravirus being packed in the interior of vesicle-like structures, suggesting a potential release by exocytosis, as observed for other viruses of amoeba (24, 25) (Fig. 2D and Movie S3). Most of the viral shedding, however, still occurs by lysis of the amoebal cell, followed by the release of Yaravirus particles, which later reach the supernatant of the infected culture, or sometimes might get attached to the debris of the cellular membranes (Fig. 2E). We have also evaluated Yaravirus replication by concomitantly investigating the decrease of the host cell numbers together with the increase of viral genome during infection. Interestingly, during the first hours of infection, the A. castellanii cultures seem to progressively grow until 24 h.p.i., showing a fastidious character for Yaravirus replication (Fig. 2F). The cells then start to suffer lysis induced by the virus only after 72 h.p.i. (Fig. 2F). On the same level, from 96 h.p.i. to 7 d post infection, there is no change of the detection levels of Yaravirus genome and the lysis seems to stop, and the remaining trophozoites turn to cysts.
Fig. 2.
Yaravirus morphogenesis and release. (A) The virions are assembled by the addition of more than one layer of protein or membranous components around its structure. (Scale bar: 70 nm.) (B) The particles then start to migrate to the periphery of the cell, where there is the presence of several electron-dense points that function as morphogenetic structures to package the DNA inside the Yaravirus particles (regions inside dashed lines). (Scale bar: 1,000 nm.) (C) Detailed image of the morphogenetic regions where the DNA (red arrow) is incorporated inside the Yaravirus virion (black arrow). (Scale bar: 150 nm.) (D) Sometimes, the final step of viral replication is marked by the particles being packaged inside vesicle-like structures, suggesting a potential release by exocytosis. (Scale bar: 500 nm.) (E) Most of the particles, however, are released by cellular lysis and have a high affinity to the membranes of cellular debris. (Scale bar: 150 nm.) (F) Graph comparing concomitantly the decrease of host cell numbers (red bars) with the increase of Yaravirus genome during the infection (black line). Replication of viral genome was measured by qPCR and calculated by delta-delta Ct.
Genome.
Sequencing of the Yaravirus genome has shown the presence of a double-stranded DNA molecule with a length of 44,924 bp and harboring a total of 74 predicted genes (Fig. 3A). Although two of these genes (73 and 74) seemed to be truncated and do not start with codons belonging to the methionine amino acid, both were detected in the proteomic analysis (Yaravirus Proteomics). Despite a smaller genome than other viruses of amoeba, Yaravirus encodes for six tRNA genes: tRNA-Ser (gct), tRNA-Ser (tga), tRNA-Cys (gca), tRNA-Asn (gtt), tRNA-His (gtg), and tRNA-Ile (aat) (Fig. 3 A and D and SI Appendix, Tables S1 and S2 and Dataset S1). All of them are colocated on an intergenic region between genes 29 and 30 (Fig. 3 A and D). In contrast to tRNA genes in Tupanviruses, we did not observe a correlation between the Yaravirus tRNA isoacceptors and the codons most frequently used by the virus or its A. castellanii host (SI Appendix, Fig. S5). The genome has a GC content of 57.9%, which is one of the highest found in any amoebal virus discovered to date (SI Appendix, Fig. S6A). When analyzed gene by gene, Yaravirus has a spectrum of GC content that varies between 46% and 65% (SI Appendix, Table S3). The analysis of the intergenic regions of the genome (46% GC content) did not reveal any enriched sequence motifs that might indicate a conserved promoter, as opposed to what is observed in many other NCLDV members (26).
Fig. 3.
Yaravirus genome features. (A) Circular representation of Yaravirus genome highlighting the predicted ORFs (arrows). Red arrows represent ORFs predicted by analyses of similarity of amino acid sequences with information regarding their best hits. Black dots indicate ORFs encoding predicted proteins whose functions were suggested by HHPred (structural analyses). Yellow stars indicate proteins found in virion proteomics. (B) The percentage of ORFan genes among the complete genome of different viruses of amoeba is represented by the graph with red scale bars. (C) The graph with greenish scale bars represents the absolute number of genes with homologs in databases (non-ORFan genes) for each of the same amoebal viruses previously analyzed. (D) All of the six Yaravirus predicted tRNAs, as well as their corresponding sequences, are pictured with information about their anticodon (in parentheses), their nucleotide length, the percentage of GC content, and the position in the intergenic regions of genes 29 and 30. Regarding the asterisk, note that the number of ORFan/non-ORFan genes represented in C and D do not take into account the structural annotation made by the HHpred servers, as for most of the amoebal viruses represented in the graphs.
By considering only the portions of genome that are part of coding regions, Yaravirus also has a similar coding capacity as observed in other viruses when their genome was first annotated, ∼90% (SI Appendix, Fig. S6B). Surprisingly, Yaravirus genome annotation showed that none of its genes matched with sequences of known organisms when we compared them at the nucleotide level. When we looked for homology at the amino acid levels, we found that only two predicted proteins had hits in the Pfam-A database and, in total, six had distant matches in the nr database. Therefore, considering the same criteria that have been used to analyze other giant viruses’ genomes, about 90% (n = 68) of the Yaravirus predicted genes are ORFans. The six genes whose product has some homology with known protein sequences (Fig. 3A and Table 1) are homologous to fragments of proteins predicted to have different functions, such as an exonuclease/recombinase bacterial protein (gene 2; best hit, Timonella senegalensis), a hypothetical protein (gene 3; best hit, A. castellanii), a hypothetical protein (gene 28; best hit, Acytostelium subglobosum LB1, a dictyostelid), a packaging ATPase (gene 40; best hit, Pleurochrysis endemic virus), a conserved hypothetical protein (gene 46; best hit, Melbournevirus, a marseillevirus strain), and a bifunctional DNA primase/polymerase (gene 69; best hit, Marinobacter sp., Alphaproteobacteria; Table 1). Complimentary prediction of three-dimensional structures of these proteins indicated a potential function of 11 more genes (Table 2 and Yaravirus Proteomics). If we consider an additional 11 genes whose functions were predicted by structural analyses, the Yaravirus proportion of ORFans would be 80% (n = 57; still a high number, considering that, usually, in other studies involving giant viruses, methodologies based on protein structures are not used for this accounting) (27, 28). Phylogenetic analyses were then performed for genes 02, 40, 46, and 69 after aligning them with protein sequences of similar function belonging to different members of the virosphere and to organisms of the three cellular domains of life. Other identified genes (Table 2) did not have enough genetic information to be included in a phylogenetic analysis. For analysis corresponding to the putative exonuclease/recombinase (gene 02), three major groups were observed to construct the morphology of the tree. Yaravirus was placed in one of those clades, clustering with some members of Eukarya, specifically with stony coral and insects (SI Appendix, Fig. S7). Analyses of gene 40 (virion packing ATPase) revealed that Yaravirus clustered in a polyphyletic branch, with members belonging to Mimiviridae family, bacteria (although many of these sequences seem to represent misclassified NCLDVs from metagenome-assembled genomes), and Pleurochrysis sp. endemic virus 1a and 2 (SI Appendix, Fig. S8). For the phylogenetic analysis corresponding to gene 46 (hypothetical protein conserved in Marseillevirus), Yaravirus was clustered with Marseillevirus strains (SI Appendix, Fig. S9). For the last tree, representing analysis for gene 69 (bifunctional DNA primase/polymerase), we have observed that Yaravirus was clustered with members of eukaryotes corresponding to the Streblomastix and Phytophthora groups (SI Appendix, Fig. S10). However, it should be noted that, in a previous study, the authors detected sequences of mimivirus genes among the Phytophthora parasitic strain INRA-310 genome (29). After all those analyses, it is important to note that, although Yaravirus has some genes with representatives in the genome of other organisms, their homology with orthologs is very low (25.24 to 44.12%), highlighting that Yaravirus genome content is essentially divergent among the other members of the virosphere (Table 1).
Table 1.
Yaravirus genes with similarity on current databases and their best-hits (BLASTp)
| Yaravirus gene ID | Best hit | Total score | Query cover, % | E-value | Identity, % | Annotation of best BLASTp hit |
| 2 | Timonella senegalensis WP_019148817.1 | 67.0 | 87 | 1e-09 | 27.88 | Exonuclease/recombinase |
| 3 | A. castellanii XP_004339080.1 | 51.6 | 80 | 1e-06 | 44.12 | Hypothetical protein |
| 28 | Acytostelium subglobosum LB1 XP_012747655.1 | 57.8 | 85 | 4e-07 | 27.59 | Hypothetical protein |
| 40 | Pleurochrysis sp. endemic virus 1a AUD57256.1 | 121 | 66 | 4e-28 | 33.05 | Virion packaging ATPase |
| 46 | Melbournevirus YP_009094634.1 | 181 | 88 | 5e-47 | 32.63 | Hypothetical protein conserved in marseilleviruses |
| 69 | Marinobacter sp. MAB50943.1 | 106 | 35 | 8e-20 | 25.24 | Bifunctional DNA primase/polymerase |
Table 2.
Annotation of Yaravirus proteins based on the predicted tridimensional structure of the proteins coded by the virus
| Gene | Best hit | Functional prediction | Probability | E-value |
| 2 | Laribacter hongkongensis | Exonuclease | 99.98 | 2.5e-30 |
| 3 | Eubacterium eligens | Uncharacterized protein | 94.49 | 0.88 |
| 11 | Homo sapiens | Adiponetctin | 95.76 | 0.096 |
| 12 | Rattus norvegicus | Cerebellin-1; synapse protein | 97.54 | 5,00e-03 |
| 17 | Rattus norvegicus | prkc apoptosis wt1 regulator protein | 95.86 | 0.17 |
| 26 | Homo sapiens | Retinoblastoma-binding protein 6 | 94.71 | 0.026 |
| 28 | Faba Bean Necrotic Yellows | Replication-associated protein; endonuclease | 95.07 | 0.11 |
| 40 | Sulfolobus turreted icosahedral virus 2 | Genome packaging NTPase B204; FtsK-HerA superfamily | 99.67 | 7.9e-15 |
| 41 | Singapore grouper iridovirus | Major capsid protein | 98.89 | 3.1e-8 |
| 43 | Arabidopsis thaliana | Transcription factor HY5 | 96.6 | 0.016 |
| 46 | Enterobacteria phage HK620 | DNA stabilization protein; tail needle, viral genome-ejection | 86.58 | 0.033 |
| 54 | Haloarcula marismortui | Ribosome 50S | 90.84 | 0.81 |
| 57 | H. sapiens | BEN domain-containing protein 3 | 86.72 | 0.43 |
| 58 | Oryctolagus cuniculus | Potential copper-transporting atpase | 91.25 | 0.62 |
| 67 | Xenopus laevis | DNA-(apurinic or apyrimidinic site) lyase | 92.31 | 0.45 |
| 69 | Bovine papillomavirus type 1 | Replication protein E1/DNA Complex; DNA helicase, AAA+, ATPase | 99.18 | 4.5e-10 |
| 70 | Escherichia coli | Holliday junction resolvase | 99.85 | 9.2e-20 |
Note: The bold genes represent proteins observed on the viral proteomics.
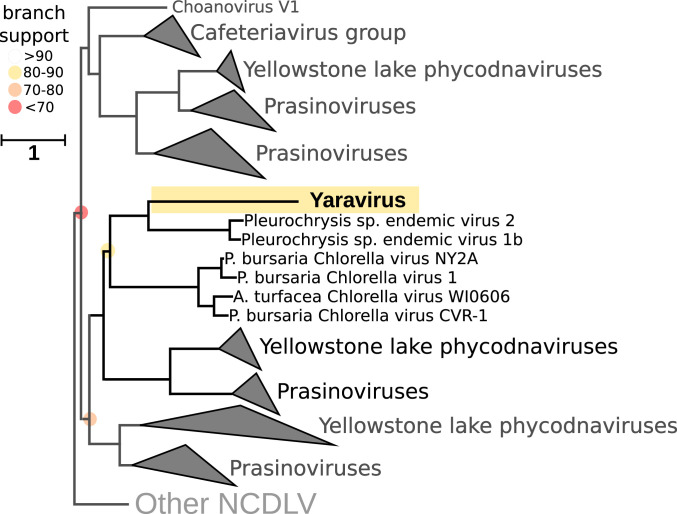
In order to detect sequences related to Yaravirus, we surveyed 8,535 publicly available metagenomes in the IMG/M database that have been generated from samples from diverse habitats across our planet (30). We discovered distant homologs of the Yaravirus ATPase (NCVOG0249) with an amino acid homology of up to 33.9% in the metagenomic data, while the closest homolog in the NCBI nr database was that of Pleurochrysis sp. endemic virus 1a, with 33.1%. In a phylogenetic tree of the viral ATPase, the Yaravrus branched within the Mimiviridae as part of a highly supported clade made up by its distant metagenomic relatives and Pleurochrysis sp. endemic viruses (Fig. 4 and SI Appendix, Fig. S11). In contrast to known members of the Mimiviridae, viral contigs and viral genomes in this clade featured a high GC content, with up to 62%. Adding sequences of polinton and virophage ATPases to this dataset resulted in a slightly altered topology, but the position of Yaravirus remained stable, not grouping with these additional elements (SI Appendix, Fig. S12). We also searched for proteins similar to the Yaravirus putative MCP but were not able to retrieve closely related sequences in the metagenomic data. In parallel, we used 18 hidden Markov models to detect MCPs in 235 NCLDV reference genomes (plus contigs of three Pleurochrysis endemic viruses). After dereplicating the hits by clustering at 90% sequence similarity using accurate mode in cd-hit, we prepared a structure-guided alignment with Expresso in T-Coffee (using PDB structures), and also a conventional alignment with mafft-ginsi (–unalignlevel 0.8–allowshift). Both alignments returned surprisingly good results for Yaravirus MCP, and then phylogenetic trees were calculated with iq-tree LG+F+R8. Yaravirus MCP was shown to group together with the MCPs of Pleurochrysis endemic viruses in a well-supported clade; however, in contrast to the ATPase tree, they were affiliated with Phycodnaviridae (chlorella viruses in particular) and not with the Mimiviridae (Fig. 5 and SI Appendix, Figs. S13 and S14 and Supplementary Files).
Fig. 4.
Phylogenetic position of Yaravirus and related viral sequences in the Mimiviridae based on the viral ATPase (NCVOG0249). The Yaravirus ATPase is highlighted in yellow. Branch support is indicated as colored circles for support values of 90 or below. The tree is rooted at the Poxviruses. (Scale bar: substitutions per site.) GC content of viral genomes and contigs containing NCVOG0249 is shown together with the average GC content of collapsed clades. In addition, environmental origin and assembly sizes of Yaravirus and related viral contigs and genomes are shown.
Fig. 5.
Structure-guided phylogeny based on hmmsearch employed to identify the Yaravirus MCP in 235 NCDLV reference genomes using specific hidden Markov models. The alignments were made with Expresso in the software T-Coffee, using PDB structures. After, the phylogenetic trees were built using IQ-tree (v1.6.12; ref. 61) with LG+F+R8 based on the built-in model select feature (62) and 1,000 ultrafast bootstrap replicates (63). The Yaravirus MCP is highlighted in yellow. Branch support is indicated as colored circles for support values of 90 or below. The tree is unrooted. (Scale bar: substitutions per site.)
Finally, trying to investigate a potential relationship between Yaravirus and different members of the Pleurochrysis sp. endemic virus group, we have looked for protein similarities between these organisms. Comparisons were made using the BLASTp database and have suggested some ortholog candidates, but with a low percentage of protein identity (around 24 to 33%). For Pleurochrysis sp. endemic virus 1a and Pleurochrysis sp. endemic virus 1b (Genbank accession codes KY131436 and KY203336, respectively), there is some similarity with genes 28, 40, and 41 of Yaravirus. For Pleurochrysis sp. endemic virus 2 (Genbank accession code KY346835), two genes are related to gene 28 in Yaravirus and two others are similar to genes 40 and 02. Finally, for Pleurochrysis sp. endemic virus unk (accession code KY203337), we have found similarity with gene 69 of Yaravirus.
Yaravirus Proteomics.
As aforementioned, most Yaravirus proteins had no detectable homologs in public databases. This peculiarity prompted us to have a closer look at the proteins responsible to form the mature particles of Yaravirus. Proteomics revealed a total of 26 viral proteins present in purified particles (SI Appendix, Table S4). We then analyzed the predicted three-dimensional structures of those 26 proteins by using three platforms for domain comparison, the HHpred, the Phyre2, and the Swiss-model tools (30–37). Only five sequences (genes 11, 12, 28, 41, and 46) were observed to have structural features similar to known proteins (Table 2 and Dataset 2). That means that about 80% of its virion proteome consists of ORFans. It is important to mention that the same approach (in silico structure prediction) has been used in parallel to evaluate all of the 74 predicted genes on the genome of Yaravirus, resulting in total in the discovery of 17 gene-encoded products with structural resemblance to other proteins in public databases (Table 2). Proteomics data revealed that the most abundant proteins in the viral particles corresponded to genes 41, 46, and 51 (from most to least abundant; SI Appendix, Table S4). While, for the third highest expressed protein, we were not able to find any structural candidates with known biological function, for sequences represented by genes 41 and 46, we observed fragments of protein resembling the three-dimensional structure of the capsid of other viruses (Dataset S2 and Table 2). With a confidence of 97%, a relevant portion (65%) of gene 41 was found to have a structural similarity with the double jelly-roll domain of the MCP of the Paramecium bursaria Chlorella virus type 1 (Dataset S2). Gene 46-encoded predicted protein was found to have structural similarity with bacterial secreted protein pcsB and tail needle protein (Table 2 and Dataset S2), a portion composed of a long alpha-helix. The function of protein encoded by gene 46 remains to be investigated. Therefore, we were not able to convincingly find any minor capsid protein. It is also important to note that sequences represented by gene 46 are the same described earlier to be highly conserved in marseilleviruses and in medusaviruses (Table 1). Finally, the last two proteins observed in the proteome that had matches with structural deposits in public databases are represented by genes 11 and 12, the first one predicted by HHpred to encode for an adiponectin and the second one for a protein called cerebellin-1 (Table 2).
Discussion
In recent years, amoebal large and giant viruses have frequently been found around the world (5–10, 22, 24, 27, 38–41). Here, we describe Yaravirus brasiliensis, an 80-nm-sized virus with a genome containing a notable proportion of genes that have never been observed before. Using standard protocols, our very first genetic analysis was unable to find any recognizable sequences of capsid or other classical viral genes in Yaravirus. This is a relevant feature to highlight the importance of studies related to the isolation of new viral samples, as, by following the current metagenomic protocols for viral detection, Yaravirus would not even be recognized as a viral agent (42, 43).
According to our knowledge, Yaravirus represents the first virus isolated in Acanthamoeba spp. that is potentially not part of the complex group of NCLDVs. Several characteristics unite previously discovered amoebal viruses: large-sized virions, genomes coding for hundreds to thousands of genes, and presumably a monophyletic origin that is reflected in the presence of a set of about 20 most likely vertically inherited genes (17, 18). None of these features are present in Yaravirus, and that makes it potentially the first isolate of a novel bona fide group of amoebal virus. Of course, we cannot exclude the possibility that Yaravirus may represent a reduced NCLDV, presenting highly divergent or even absent NCLDV hallmark proteins. Recently, a similar case was described for three small crustacean viruses (44). However, despite their reduced genome when compared to other members of the NCLDV, an important number of hallmark genes were shared with this group, differently as observed for Yaravirus (44). In this not less exciting scenario, supported by the analysis regarding the ATPase and MCP phylogenetic trees, Yaravirus would represent the to-date smallest member of the NCLDVs, both in particle and genome size. The presence of six copies of tRNAs in Yaravirus also impresses when analyzed by the perspective of a selective pressure forcing to maintain these genes in such a small genome when compared to larger viruses of amoeba. Even more interestingly, none of the isoacceptors related to the Yaravirus tRNAs correspond to codons of amino acids abundantly used by the virus or the amoeba. Considering the fastidious infection cycle of Yaravirus in Acanthamoeba, it is conceivable that, in nature, a different organism might act as the preferred host of Yaravirus. Some genes were found to be shared between Yaravirus and members of the Pleurochrysis endemic virus group; however, the low coverage supporting their putative orthologs make difficult a close relationship between these organisms.
Most members of the to-date isolated giant viruses of amoeba show a capsid specially composed by copies of an MCP related to the D13L of Vaccinia virus (15, 45). Pandoraviruses are an exception, as they seem to lack a protein shell to protect their genomes (28). Interestingly, even some of the amoeba hosts of these viruses may carry copies of MCP genes, suggesting possible horizontal gene transfer between virus and protist host (46, 47). By the structure-guided alignment and analysis of the constructed phylogenetic trees, Yaravirus does seem to share a common origin of MCP capsid with other NCLDVs, even though their sequences show an incredibly low similarity (48). Taken together, we can conclude that Yaravirus represents a divergent lineage of viruses isolated from A. castellanii. The large amount of unknown proteins encoded by Yaravirus reflects the variability existing in the viral world and the astonishing coding potential of new viral genomes yet to be discovered.
Methods
Origin of Samples and Viral Isolation.
In 2017, searching to isolate novel variants of viruses infecting amoebas, we collected samples of muddy water from a creek of Lake Pampulha, an artificial lagoon located at the city of Belo Horizonte, Brazil (19 51 0.60S and 43 58 18.90W). As soon as they were collected, the samples were quickly taken to our lab and stored at 4°C until they were further processed. Following the protocol, 4 × 104 amoebas of the A. castellanii Neff strain (ATCC 30010) were seeded in each well of a 96-well plate, inoculating to each one a volume of around 100 µL of the collected samples, originally diluted 1:10 in PBS buffer. The plates were then incubated for 7 d at 32°C and observed daily for the appearance of cytopathic effect, which may indicate a probable viral infection. All of the content from the wells was then collected and submitted to three processes of freezing and thawing and analysis of the possible isolates by negative staining technique. By the end, the collected content was submitted to another two blind passages in fresh cultures of amoeba, but this time in 25-cm2 Nunc Cell Culture Treated Flasks with Filter Caps (Thermo Fisher Scientific) containing around 1 million amoebal cells. After viral isolation, all of the following experiments were made by infecting A. castellanii cells in a low multiplicity of infection (MOI), given the Yaravirus’s fastidious replication cycle.
Transmission Electron Microscopy (TEM), TEM Tomography, Cryo-Electron Microscopy.
For resin embedding and transmission electron microscopy (TEM), A. castellanii cells infected with Yaravirus were fixed at 20 h postinfection with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer. Cells were washed three times with a solution of 0.2 M saccharose in 0.1 M sodium cacodylate. Cells were postfixed for 1 h with 1% OsO4 diluted in 0.2 M potassium hexa-cyanoferrate (III)/0.1 M sodium cacodylate. After washes with distilled water, cells were gradually dehydrated with ethanol by successive 10-min baths in 30, 50, 70, 96, 100, and 100% ethanol.
Substitution was achieved by successively placing the cells in 25, 50, and 75% Epon solutions for 15 min. Cells were placed for 1 h in 100% Epon solution and in fresh Epon 100% overnight at room temperature. Polymerization took place with cells in fresh 100% Epon for 48 h at 60°. Ultrathin 70- or 300-nm-thick sections were cut with a UC7 ultramicrotome (Leica) and placed on HR25 300 Mesh Copper/Rhodium grids (TAAB). Ultrathin sections were contrasted according to Reynolds (49). Electron micrographs were obtained on a Tecnai G20 TEM operated at 200 keV equipped with a 4,096 × 4,096-pixel resolution Eagle camera (FEI). For tomography, gold nanoparticles 10 nm in diameter (ref. 741957; Sigma-Aldrich) were deposited on both faces of the sections prior to contrasting. Tomography tilt series were acquired on the G20 Cryo TEM (FEI) with the Explore 3D software (FEI) for tilt ranges of 110° with 1° increments. The mean applied defocus was −2 μm. The magnification ranged between 3,500 and 29,000 with pixel sizes between 3.13 and 0.37 nm, respectively. The image size was 40,962 pixels. The tilt series were aligned using ETomo from the IMOD software package (University of Colorado) by cross-correlation (50). The tomograms were reconstructed using the weighted back-projection algorithm in ETomo from IMOD. The average thickness of the obtained tomograms was 268.40 ± 64 nm (n = 16). Fiji/ImageJ (NIH) was used for making tomography movies (51). For cryoelectron microscopy assays, the supernatant of infected cultures of A. castellanii was collected after 7 d postinfection and submitted to a first round of centrifugation at 1,500 × g for 10 min, looking to pellet the cell debris from the virus present on the supernatant. Next, the portion containing the Yaravirus was then submitted to a second round of centrifugation, and the virus was concentrated by ultracentrifugation at 100,000 × g for 2 h. The following steps were previously described by Klose et al. (23). Briefly, 3 µL of virus solution was placed on glow-discharged C-Flat 2/2 grids (EMS) and plunge-frozen into liquid ethane using a Gatan Cryoplunge 3. Samples were then imaged on a Talos F200C (ThermoFisher Scientific) equipped with a Ceta camera (ThermoFisher Scientific).
Genome Sequence and Analysis.
The Yaravirus genome was sequenced two times by using the Illumina MiSeq platform (Illumina) with the paired-end application. The generated reads were then assembled de novo by using the software ABYSS and SPADES, with the resulting contigs ordered by the Python-based CONTIGuator.py software. After, gene predictions were made by using the GeneMarkS tool (52). The functional annotation for the Yaravirus predicted proteins was made through searches against the GenBank NCBI nonredundant protein sequence database (nr), considering as homologous proteins only the sequences that presented an e-value < 1 × 10−3. The annotation was refined by comparing the in silico-predicted protein structures of Yaravirus with domains of proteins present in different databases using three platforms: the HHpred, the Phyre2, and the Swiss-model tools (30–37). It is important to note that, for the HHpred, we considered true-positive protein structures only matches that had a probability >80% and e-value ∼1, as suggested by Söding and colleagues (37). For the qPCR assays, the increase in genome replication was assessed in cultures of A. castellanii cells infected by Yaravirus in different time points (H4, H6, H8, H12, H24, H72, H96, and H168), using primers which were constructed based on the sequence of the gene 69 of Yaravirus (primers, 5′TGCAGCAAGTCGGTCAAGAT3′ and 5′AACTTCCACATGCGAAACGC3′). Conditions used in the assay were previously described (53).
The amino acid and codon usage data were compared to those presented by A. castellanii and by different strains of amoebal viruses. For this, the sequences were downloaded from the NCBI database and analyzed by using the software Artemis 18.0.3. The percentage of GC content and GC skew have also been analyzed by using the same software. Transfer RNA (tRNA) sequences were identified using the ARAGORN tool. Phylogenetic analyses were performed for the six proteins of Yaravirus holding similarities with other organisms on the NCBI database (Table 1). By using the ClustalW tool in the Mega 10.0.5 software program, amino acid sequences of these Yaravirus proteins were previously aligned with the corresponding sequences of representatives of the virosphere and from other cellular organisms belonging to the three domains of life. The analysis involved 55 amino acid sequences for gene 40, 49 amino acid sequences for gene 02, 13 amino acid sequences for gene 46, and 34 amino acid sequences for gene 69. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All of the trees were constructed by using the maximum likelihood evolution method, with the JTT matrix-based model and a bootstrap of 1,000 replicates (54).
Yaravirus Proteomics.
In order to identify the proteins that make up Yaravirus particles, thirty 75-cm2 cell culture flasks (Nunc), containing 7 × 106 A. castellanii cells per flask, were infected with the isolated virus, and the cytopathic effect was followed up to 7 d.p.i. After severe amoebal lysis, the content was collected and submitted to a first round of centrifugation, at 1,500 × g for 10 min, looking to pellet the cell debris from the virus present on the supernatant. Then, this viral portion was submitted to a second round of centrifugation, and the virus was concentrated by ultracentrifugation at 100,000 × g for 2 h. To finish, viral pellet was then prepared for a two-dimensional gel electrophoresis and analysis by matrix-assisted laser desorption/ionization and liquid chromatography-tandem mass spectrometry as described before by Reteno and colleagues (55).
Metagenomic Survey.
The Yaravirus ATPase (NCVOG0249) and the putative major capsid protein (MCP) were used to query 8,535 publicly available metagenomes in the IMG/M database (30) using diamond BLASTp (v0.9.25.126; ref. 56). Resulting protein hits with more than 30% query and subject coverage and an e-value of at least 1e-5 were extracted from the metagenomic data. In parallel, hmmsearch (version 3.1b2; hmmer.org) was employed to identify and extract ATPases (NCVOG0249) and MCPs (multiMCP) from 235 NCDLV reference genomes using specific hidden Markov models (https://bitbucket.org/berkeleylab/mtg-gv-exp/). Extracted proteins were then combined with the Yaravirus queries. To remove most redundant sequences, the MCP data set was clustered in cd-hit (57) at an amino acid similarity level of 90% using the accurate mode. ATPases were then aligned with MAFFT-linsi (v7.294b; ref. 58) and MCPs in Expresso (59) (most accurate mode in t-coffee v_13.41.0.28, alignment guided by PDB structures), and the resulting alignments trimmed with trimal (v1.4, -gt 0.1; ref. 60). Phylogenetic trees were built using IQ-tree (v1.6.12; ref. 61) with LG+F+R5 (ATPase) and LG+F+R8 (MCP) based on the built-in model select feature (62) and 1,000 ultrafast bootstrap replicates (63). Phylogenetic trees were visualized with iTol [v5.8 (61)] and ete3 (64).
Data Availability.
Data for the Yaravirus genome have been deposited to GenBank under accession number MT293574.
Supplementary Material
Acknowledgments
We thank our colleagues from IHU (Aix Marseille University) and from Laboratório de Vírus (Universidade Federal de Minas Gerais) for their assistance, especially Said Mougari, Issam Hasni, Lina Barrassi, Priscilla Jardot, Erna Kroon, Claudio Bonjardim, Paulo Ferreira, Giliane Trindade, and Betania Drumond. In addition, we thank the Méditerranée Infection Foundation, Centro de Microscopia da Universidade Federal de Minas Gerais (UFMG), Pro Reitorias de Pesquisa e Pós-graduação da UFMG, Programa de Pós-graduação em Microbiologia da UFMG, CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), and FAPEMIG (Fundação de Amparo à Pesquisa do estado de Minas Gerais) for their financial support. J.S.A. is a CNPq researcher. B.L.S., J.S.A., P.C., P.V.M.B. and G.P.O. are members of a CAPES-Comitê Francês de Avaliação da Cooperação Universitária project. This work was supported by the Agence Nationale de la Recherche (ANR), including the ‘Programme d'investissement d'avenir' under the reference Méditerranée Infection 10-1AHU-03 and European funding FEDER PRIMI. The work that was conducted by F.S. at the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract DE-AC02–05CH11231. It made use of resources of the National Energy Research Scientific Computing Center, which is also supported by the DOE Office of Science under Contract DE-AC02–05CH11231.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Yaravirus genome data have been deposited at GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession number MT293574.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001637117/-/DCSupplemental.
References
- 1.Guglielmini J., Woo A. C., Krupovic M., Forterre P., Gaia M., Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 116, 19585–19592 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koonin E. V., Yutin N., Multiple evolutionary origins of giant viruses. F1000 Res. 7, 1840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colson P. et al., Ancestrality and mosaicism of giant viruses supporting the definition of the fourth TRUC of microbes. Front. Microbiol. 9, 2668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colson P., Ominami Y., Hisada A., La Scola B., Raoult D., Giant mimiviruses escape many canonical criteria of the virus definition. Clin. Microbiol. Infect. 25, 147–154 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Abrahão J. et al., Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 9, 749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legendre M. et al., In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. U.S.A. 112, E5327–E5335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreani J. et al., Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfarviridae and Faustoviruses. J. Virol. 91, e00212-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreani J. et al., Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses 8, 300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajrai L. H. et al., Kaumoebavirus, a new virus that clusters with faustoviruses and Asfarviridae. Viruses 8, 278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulz F. et al., Giant virus diversity and host interactions through global metagenomics. Nature 578, 432–436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer M., Gimenez G., Suzan-Monti M., Raoult D., Classification and determination of possible origins of ORFans through analysis of nucleocytoplasmic large DNA viruses. Intervirology 53, 310–320 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Siew N., Fischer D., Twenty thousand ORFan microbial protein families for the biologist? Structure 11, 7–9 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Yin Y., Fischer D., Identification and investigation of ORFans in the viral world. BMC Genomics 9, 24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siew N., Fischer D., Unravelling the ORFan puzzle. Comp. Funct. Genomics 4, 432–441 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renesto P. et al., Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J. Virol. 80, 11678–11685 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan N. A., Acanthamoeba: Biology and Pathogenesis (Caister Academic Press, ed. 2, 2015). [Google Scholar]
- 17.Koonin E. V., Yutin N., Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv. Virus Res. 103, 167–202 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Iyer L. M., Balaji S., Koonin E. V., Aravind L., Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117, 156–184 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Krupovic M., Koonin E. V., Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U.S.A. 114, E2401–E2410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutin N., Wolf Y. I., Raoult D., Koonin E. V., Eukaryotic large nucleo-cytoplasmic DNA viruses: Clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 6, 223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin E. V. et al., Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 84, e00061-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrade A. C. D. S. P. et al., Ubiquitous giants: A plethora of giant viruses found in Brazil and Antarctica. Virol. J. 15, 22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klose T. et al., Structure of faustovirus, a large dsDNA virus. Proc. Natl. Acad. Sci. U.S.A. 113, 6206–6211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira Andrade A. C. D. S. et al., New isolates of pandoraviruses: Contribution to the study of replication cycle steps. J. Virol. 93, e01942-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legendre M. et al., Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 9, 2285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira G. P. et al., Promoter motifs in NCLDVs: An evolutionary perspective. Viruses 9, 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raoult D. et al., The 1.2-megabase genome sequence of Mimivirus. Science 306, 1344–1350 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Philippe N. et al., Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Sharma V., Colson P., Giorgi R., Pontarotti P., Raoult D., DNA-dependent RNA polymerase detects hidden giant viruses in published databanks. Genome Biol. Evol. 6, 1603–1610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen I. A. et al., IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterhouse A. et al., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bienert S. et al., The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 45, D313–D319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guex N., Peitsch M. C., Schwede T., Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30 (suppl. 1), S162–S173 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Benkert P., Biasini M., Schwede T., Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27, 343–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T., Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7, 10480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer M. et al., Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. U.S.A. 106, 21848–21853 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boratto P. V. et al., Niemeyer virus: A new mimivirus group A isolate harboring a set of duplicated aminoacyl-tRNA synthetase genes. Front. Microbiol. 6, 1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodrigues R. A. L. et al., Morphologic and genomic analyses of new isolates reveal a second lineage of cedratviruses. J. Virol. 92, e00372-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva L. K. D. S. et al., Cedratvirus getuliensis replication cycle: An in-depth morphological analysis. Sci. Rep. 8, 4000 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang Y. et al., Metagenomic analysis of the diversity of DNA viruses in the surface and deep sea of the South China sea. Front. Microbiol. 10, 1951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Corte D. et al., Viral communities in the global deep ocean conveyor belt assessed by targeted viromics. Front. Microbiol. 10, 1801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramaniam K. et al., A new family of DNA viruses causing disease in Crustaceans from diverse aquatic biomes. MBio 11, e02938-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilhelm S. W. et al., A student’s guide to giant viruses infecting small eukaryotes: From Acanthamoeba to Zooxanthellae. Viruses 9, 46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chelkha N. et al., A phylogenomic study of Acanthamoeba polyphaga draft genome sequences suggests genetic exchanges with giant viruses. Front. Microbiol. 9, 2098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maumus F., Blanc G., Study of gene trafficking between Acanthamoeba and giant viruses suggests an undiscovered family of amoeba-infecting viruses. Genome Biol. Evol. 8, 3351–3363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krupovic M., Bamford D. H., Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr. Opin. Virol. 1, 118–124 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Reynolds E. S., The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J. et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Besemer J., Lomsadze A., Borodovsky M., GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bajrai L. H. et al., Isolation of yasminevirus, the first member of klosneuvirinae isolated in coculture with vermamoeba vermiformis, demonstrates an extended arsenal of translational apparatus components. J. Virol. 94, e01534-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones D. T., Taylor W. R., Thornton J. M., The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992). [DOI] [PubMed] [Google Scholar]
- 55.Reteno D. G. et al., Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J. Virol. 89, 6585–6594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K., Standley D. M., A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32, 1933–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tommaso P. et al., T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for the Yaravirus genome have been deposited to GenBank under accession number MT293574.