Significance
Epigenetic regulation of gene expression is associated with switching between chromatin states characterized by distinct histone modifications. Polycomb/Trithorax regulation involves the mutually exclusive H3K27me3/H3K36me3 modifications, but how these states are faithfully inherited through DNA replication, yet can switch from one to another, is still poorly understood. One mechanism that would aid switching is the association of histone methyltransferases with factors demethylating the opposing histone modification. Here, we show that an Arabidopsis 2-oxoglutarate–dependent dioxygenase, an activity associated with histone demethylation in other organisms, physically associates with the Polycomb Repressive Complex 2. We propose that physical association of histone methylation/demethylation activities will be generally important to coordinate switching between chromatin states.
Keywords: epigenetic, ICU11, Polycomb, chromatin
Abstract
Molecular mechanisms enabling the switching and maintenance of epigenetic states are not fully understood. Distinct histone modifications are often associated with ON/OFF epigenetic states, but how these states are stably maintained through DNA replication, yet in certain situations switch from one to another remains unclear. Here, we address this problem through identification of Arabidopsis INCURVATA11 (ICU11) as a Polycomb Repressive Complex 2 accessory protein. ICU11 robustly immunoprecipitated in vivo with PRC2 core components and the accessory proteins, EMBRYONIC FLOWER 1 (EMF1), LIKE HETEROCHROMATIN PROTEIN1 (LHP1), and TELOMERE_REPEAT_BINDING FACTORS (TRBs). ICU11 encodes a 2-oxoglutarate–dependent dioxygenase, an activity associated with histone demethylation in other organisms, and mutant plants show defects in multiple aspects of the Arabidopsis epigenome. To investigate its primary molecular function we identified the Arabidopsis FLOWERING LOCUS C (FLC) as a direct target and found icu11 disrupted the cold-induced, Polycomb-mediated silencing underlying vernalization. icu11 prevented reduction in H3K36me3 levels normally seen during the early cold phase, supporting a role for ICU11 in H3K36me3 demethylation. This was coincident with an attenuation of H3K27me3 at the internal nucleation site in FLC, and reduction in H3K27me3 levels across the body of the gene after plants were returned to the warm. Thus, ICU11 is required for the cold-induced epigenetic switching between the mutually exclusive chromatin states at FLC, from the active H3K36me3 state to the silenced H3K27me3 state. These data support the importance of physical coupling of histone modification activities to promote epigenetic switching between opposing chromatin states.
Epigenetic silencing is mediated by conserved histone-based mechanisms in many organisms. Polycomb Repressive Complex 2 (PRC2) delivers a H3K27me3-based silencing that plays major roles in developmental and environmental epigenetic regulation (1–3). The PRC2 core components are conserved between organisms and associate with more diverse PRC2 accessory proteins that recognize sequence-specific or chromatin features such as CpG islands (4–13). A well-studied PRC2 target in Arabidopsis is the floral repressor locus, FLOWERING LOCUS C (FLC) (14). The prolonged cold of winter epigenetically silences the locus enabling expression in the spring of the genes required for the meristem to adopt a floral fate (15–18). FLC silencing occurs in a two-step cis-mediated PRC2 switching mechanism mediated by distinct complexes. During the cold, a PRC2 containing SWINGER (SWN) and associated with the accessory proteins VERNALIZATION5 (VRN5) and VERNALIZATION INSENSITIVE3 (VIN3) nucleates H3K27me3 silencing at a small intragenic site and this confers a metastable silencing (19). After return to warm, a PRC2 containing CURLY LEAF (CLF) and LHP1 mediate spreading of H3K27me3 silencing across the gene body, and this confers long-term stable silencing (19). Resetting FLC expression then occurs as the embryos develop to ensure vernalization is required each generation, a process requiring the H3K27me3 demethylase, EARLY FLOWERING 6 (ELF6) (20).
Modeling of the mechanism at FLC has provided a generic view of how opposing chromatin states, marked by a transcriptionally active H3K36me3 state and a silenced H3K27me3 state, can provide epigenetic stability, yet switch from one to another (1, 15, 21). This mechanism involves positive feedbacks to maintain each state and nonlinearity in the interaction mechanisms (22). Physical coupling of methylase and demethylase activities has been proposed to contribute to cooperativity in the system, a prediction partly validated for the Arabidopsis system by the finding that the H3K27me3 demethylase ELF6 physically interacted in vivo with the H3K36me3 methyltransferase SET DOMAIN GROUP 8 (SDG8) (23). Whether there was a similar physical interaction linking H3K36me3 demethylation with the Polycomb methyltransferases was not known.
Here, through analysis of a Ds transposon-tagged Arabidopsis mutation showing pleiotropic developmental phenotypes we identify an activity associated with H3K36me3 demethylation. The mutation was initially called wavy leaves and cotyledons furled back (wlc-1) and was identified as an early flowering, deformed leaf mutant in the Landsberg erecta genotype (24). Cloning revealed the gene (At1g22950) corresponds to the recently described ICU11 locus (25), so the mutant is hereafter referred to as icu11-3. ICU11 is part of a small family of genes in the Arabidopsis genome with partially redundant functions. icu11 mutants misexpress many developmental regulators, share many phenotypes with embryonic flower mutants, and genetic analysis linked ICU11 activity with PRC2 function (25). Here, we extend this understanding and show that ICU11 robustly associates with PRC2 components in plants, and when defective, PRC2-mediated repression is compromised. Through analysis of a direct target, the floral repressor locus FLC, we find that ICU11 facilitates H3K36me3 demethylation and promotes the switch to the Polycomb H3K27me3 silenced state. icu11 also shows other subtle epigenetic changes, so we propose that perturbed histone demethylation causes increased transcriptional activity of ICU11 targets with direct effects on PRC2 silencing and indirect effects on the wider Arabidopsis epigenome.
Results
Ds Insertion into At1g22950 Results in Weak PcG Mutant Phenotypes.
The wlc-1 mutant, hereafter icu11-3, was identified in an Ac/Ds transposon-tagging mutagenesis screen (24) through its phenotypes, which include small size, pronounced cotyledon and leaf curling and early flowering in short days (Fig. 1 A and B). Subsequent cloning revealed it to be ICU11, a 397-amino acid protein of unknown function (At1G22950, TAIR10), with the InterPro EMBL-EBI protein domain database predicting an Fe2+/2-oxoglutarate–dependent dioxygenase (2OG) domain with homology to Arabidopsis prolyl-4-hydroxylases (P4Hs) and alpha ketoglutarate-dependent dioxygenase Bs (AlkBs) (26). In icu11-3, the insertion of the transposed Ds into the fifth exon introduces a premature stop codon, truncating ICU11 before the predicted enzymatic domain (SI Appendix, Fig. S1A). In addition, the icu11-3 mutant expresses approximately fourfold less ICU11 at the mRNA level (SI Appendix, Fig. S1B). Transformation of the icu11-3 mutant with genomic ICU11 alone, or genomic ICU11 with C-terminal enhanced green fluorescent protein (eGFP) or 3xHA fusion constructs complemented icu11-3, rescuing the observed morphological defects (SI Appendix, Figs. S1C and S2 A and B) and the misexpression of target genes in young seedlings (SI Appendix, Figs. S1D and S2 C and D). In addition, revertants were generated through Ac-induced remobilization of Ds to generate wild-type (WT) plants and fully stable mutant plants; in the latter the excision had led to a frame shift in the coding sequence. ICU11 is widely expressed, appearing elevated during later plant development (SI Appendix, Fig. S1E), while pICU11::ICU11-eGFP constructs showed broad expression of the protein in roots, nuclear localization at the subcellular level and association with metaphase chromosomes (SI Appendix, Fig. S1F).
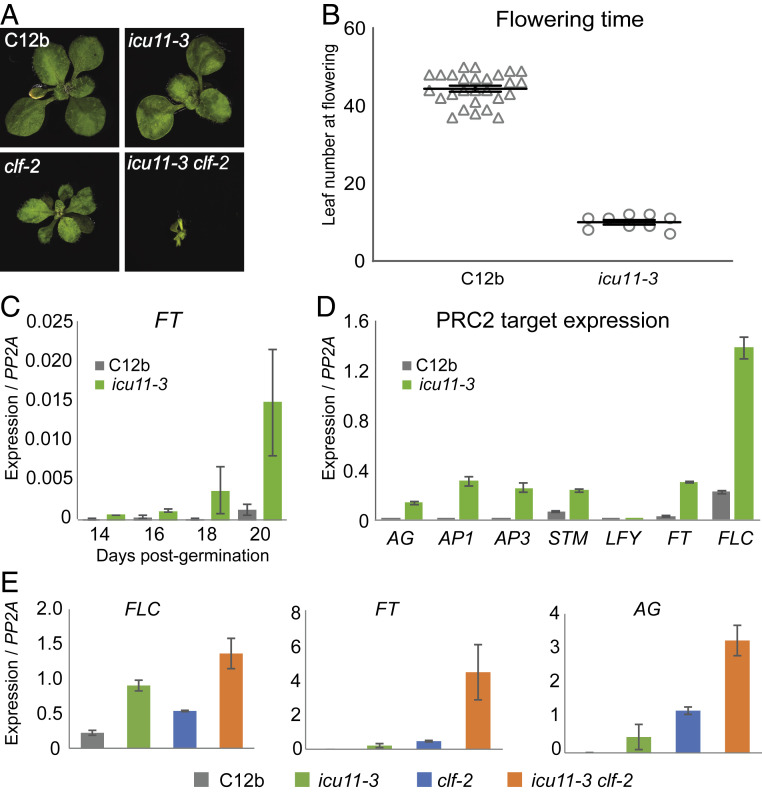
Fig. 1.
icu11-3 phenotypes are reminiscent of mutants defective in PRC2. (A) Morphological phenotypes of WT line C12b, the icu11-3 and clf-2 mutants, and synergistic interaction in icu11-3 clf-2 double mutant. (B) Flowering is significantly accelerated under short-day conditions in icu11-3 (P < 0.0001, unpaired Student’s t test). Error bars represent SEM (n = 9 [icu11-3]; n = 27 [WT]). (C–E) Mean gene expression in C12b WT, icu11-3, clf-2, and icu11-3 clf-2 double-mutant lines. Target gene expression is normalized to the housekeeping gene PP2A; error bars represent SEM for three biological replicates. (C) The floral activator FT is overexpressed in early seedling development in icu11-3 compared with WT under short-day conditions. (D) The icu11-3 mutant overexpresses PRC2 target genes during early seedling development. (E) The icu11-3 clf-2 double mutant synergistically overexpresses PRC2 targets compared with either single mutant.
To investigate the early flowering of icu11-3 in short days, we compared the expression of the floral activator FT, a known PRC2 target, during early seedling development under short-day growth conditions. FT was up-regulated in icu11-3 compared with the progenitor line (C12b), with the overexpression phenotype becoming more pronounced with seedling age (Fig. 1C). To investigate the potential misregulation of PRC2 targets more generally, we performed quantitative RT-PCR (qRT-PCR) on RNA extracted from 14-d-old seedlings of both icu11-3 and the progenitor line (Fig. 1D) and found the icu11-3 mutant overexpresses a range of PRC2 target genes compared with WT, including the reproductive transition regulators FT and FLC, floral genes AG, AP1, and AP3, and the shoot meristem maintenance gene STM. We crossed icu11-3 with clf-2 and compared morphological and gene expression phenotypes in the single and double mutants. As found previously, double mutants exhibit severe morphological defects (Fig. 1A) and higher up-regulation of the flowering targets FLC, FT, and AG compared with either single mutant alone (Fig. 1E), similar to combination of clf with mutations in other PRC2 accessory proteins such as lhp1 (27).
ICU11 Is a PRC2 Accessory Protein.
To investigate ICU11 function further, we performed coimmunoprecipitation/mass spectrometry (coIP-MS) experiments using 3xHA- and GFP-tagged ICU11 proteins as bait. Our IP-MS revealed core PRC2 complex components CLF, SWN, FERTILIZATION INDEPENDENT ENDOSPERM (FIE), MULTICOPY SUPPRESSOR OF IRA1 (MSI), and EMBRYONIC FLOWER (EMF)2 and accessory proteins EMF1, LHP1, and TELOMERE-REPEAT-BINDING (TRB) 1–3, as ICU11 interactors. EMF1 and LHP1 have been found in several studies as direct interactors of PRC2 (28, 29), and TRBs have been found associated with CLF and SWN (11). JMJ14, a H3K4me3 demethylase (30), was also detected in the ICU11-enriched peptides. Reciprocal coIP-MS with CLF-GFP, SWN-GFP, and EMF1-FLAG confirmed the interaction between ICU11 and PRC2 (Table 1 and Dataset S1). Interestingly, we did not find ICU11 interacting with VIN3 or its homologs VRN5 and VEL1, reinforcing the view of distinct Polycomb complexes operating over different spatial and temporal timescales.
Table 1.
ICU11 associates with the PRC2
| Protein | 35S::GFP | gICU11-GFP icu11-3 | icu11-3 | gICU11-HA icu11-3 | Col-0 | EMF1-3XFLAG emf1 | 35S::GFP | 35S:GFP-CLF clf-50 | gSWN-GFP | |
| Core PRC2 | ICU11 | 0–0 | 13–19 | 0–0 | 12–11 | 0–0 | 11–20 | 0–0 | 13–2 | 5–5 |
| EMF2 | 0–0 | 11–4 | 0–0 | 29–0 | 0–0 | 2–22 | 0–0 | 33–18 | 4–36 | |
| MSI1 | 0–0 | 12–6 | 0–0 | 25–2 | 3–2 | 6–18 | 2–0 | 22–18 | 6–26 | |
| FIE | 0–0 | 10–1 | 0–0 | 18–0 | 0–0 | 5–17 | 2–0 | 23–22 | 6–25 | |
| SWN | 0–0 | 10–0 | 0–0 | 27–0 | 0–0 | 0–26 | 0–0 | 11–0 | 21–68 | |
| CLF | 0–0 | 7–0 | 0–0 | 26–0 | 0–0 | 3–17 | 9–0 | 75–46 | 8–4 | |
| Accessory | EMF1 | 0–0 | 5–3 | 0–0 | 26–2 | 0–0 | 6–40 | 0–0 | 46–17 | 2–37 |
| LHP1 | 0–0 | 2–1 | 0–0 | 11–0 | 0–0 | 2–14 | 0–0 | 22–8 | 3–18 | |
| TRB1 | 0–0 | 5–9 | 0–0 | 12–2 | 0–0 | 0–1 | 0–0 | 0–0 | 0–0 | |
| TRB2 | 0–0 | 9–5 | 0–0 | 15–1 | 0–0 | 1–11 | 0–0 | 11–3 | 2–10 | |
| TRB3 | 0–0 | 9–6 | 0–0 | 15–3 | 0–0 | 3–12 | 0–0 | 9–4 | 2–7 | |
| VRN5 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 24–13 | 0–24 | |
| VEL1 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 41–28 | 0–46 | |
| ALL PEPTIDES | 787–1817 | 1286–7239 | 2944–3524 | 5481–6267 | 11483–16737 | 11170–13095 | 6705–2489 | 7376–7714 | 5127–6855 |
The numbers indicate uniquely identified peptides from each protein found by mass spectrometry in two independent experiments. The total number of peptides identified in each experiment is also indicated at the bottom of the columns. PRC2 core components are shown at the top, with accessory components below. Columns are paired with control on left and immunoprecipitated to the right. For example, 35S:GFP samples in column 1 were the control for the ICU11-GFP samples in column2 in the gICU11-GFP analysis. 35S:GFP-CLF clf50 and gSWN-GFP share the 35S::GFP control. The full list of proteins identified is presented as an excel sheet in Dataset S1.
ICU11 as a Putative Histone Demethylase.
To further investigate the role of ICU11 in epigenetic regulation we used Western blots to determine levels of histone modifications associated with active chromatin (H3K4me1, H3K4me2, H3K4me3; H3K36me3) and those associated with silenced chromatin (H3K27me3) in WT and icu11-3 14-d-old seedlings. We observed a small increase in H3K4me2/me3, and H3K36me3 in icu11-3 relative to WT (Fig. 2A and SI Appendix, Figs. S3 and S4). These data, together with the analysis of FLC derepression in the icu11-3 mutant, raised the possibility that a primary activity of ICU11 was demethylation of histone modifications associated with active chromatin states, for example, H3K4me2/3 and H3K36me3. Loss of one protein may destabilize the whole-protein complex that contains putative demethylases specific for different histone modifications. A combination of activities, e.g., ICU11 and JMJ14 in the whole complex would then link demethylation of active histone modifications at multiple sites on the histone tail. This linking of activities in vivo means in vitro analyses are the best way to define the specific activity of different demethylases. The 2OGD domain of ICU11 falls within the same enzymatic superfamily as Jumonji C-domain histone demethylases. Despite considerable effort using in vitro assays using recombinant protein and commercial histones, or by colocalization assays of demethylation and expressed ICU11 protein in Nicotiana benthamiana transient assays, we could not generate reproducible data to clearly support a direct H3K4 or K36me3 demethylase role for ICU11. This is not uncommon for this family of proteins and may indicate that ICU11 requires accessory proteins or specific posttranslational modification for histone demethylase activity. It is also possible that ICU11 modifies another component of the complex, which then allosterically influences histone demethylation.
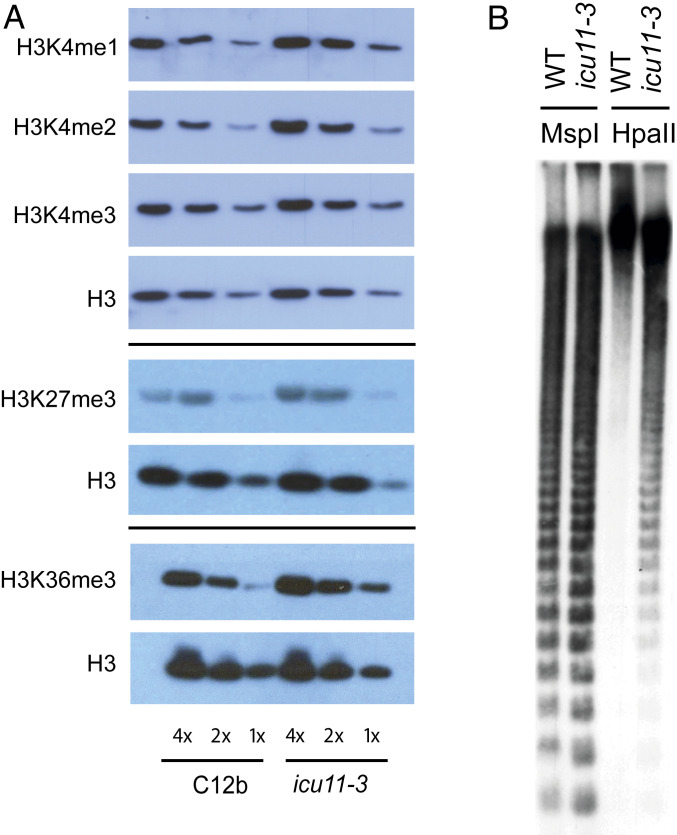
Fig. 2.
icu11-3 affects global histone methylation and DNA methylation at the pericentromeric cen180 repeats. (A) The icu11-3 mutant shows elevation of H3K36me3 and H3K4 methylation, but no change in H3K27me3 compared with WT C12b. Histones were prepared from a nuclear extract from 14-d-old seedlings and serially diluted; Western blots are shown with total histone H3 from the same extraction as loading controls. (B) Southern blot analysis of methylation at the pericentromeric cen180 repeat region. Genomic DNA digested with the methylation-insensitive restriction enzyme MspI (control) and methylation-sensitive enzyme HpaII indicates slightly reduced DNA methylation over centromeric repeats in icu11-3.
DNA Methylation Is Subtly Changed in icu11-3.
The wide range of phenotypes in icu11 mutants prompted us to look for changes in other epigenetic marks. In plants, DNA methylation occurs in CG, CHG, and CHH contexts (H = A, T, or C). CG methylation is broadly distributed, present in both gene bodies and repeat-rich regions; while CHG and CHH methylation are primarily found in transposable elements (31, 32). Southern blot analysis of genomic DNA digested with the differentially methylation-sensitive enzymes MspI and HpaII revealed slightly reduced CG methylation of centromeric 180-bp repeat regions in icu11-3 (Fig. 2B), that was reverted to WT CG methylation in a line where Ds had retransposed restoring the WT ICU11 allele (SI Appendix, Fig. S5A). To further investigate the consequence of the observed changes in DNA methylation, we compared expression of a subset of transposons in early-stage seedlings (SI Appendix, Fig. S5 B–D). We observed some slight up-regulation in icu11-3 compared to WT and clf-2, though to a lesser extent than in DNA methylation mutants ddm1-1 and cmt3-7.
To further examine the effect of icu11-3 on the cytosine methylome, we performed bisulfite sequencing on seedlings of icu11-3 mutant and the WT progenitor (SI Appendix, Table S2). Consistent with the previous report (25), we found a marginal decrease in CG methylation over genes (SI Appendix, Fig. S6A) and no substantial changes of CG methylation levels over transposons (SI Appendix, Fig. S6B). The repetitive nature of the pericentromeric regions complicates their analysis, but we observed a slight reduction in CG methylation at cen180 repeats in icu11-3 (SI Appendix, Table S3). From the combination of bisulfite sequencing and Southern analysis we can conclude that a small proportion of the centromeric repeats lose their CG DNA methylation in icu11-3. CHG and CHH (H = A, C or T) methylation over transposon sequences were slightly increased in the icu11-3 mutant (SI Appendix, Fig. S6B), but these slight changes in the non-CG contexts are consistent with the variations seen among plant tissues and plants grown in different environmental conditions due to the dynamics of non-CG methylation (33, 34). Overall, therefore these data agree with previous observations that ICU11 function does not have a large impact on DNA methylation (25). The observed effects may be due to indirect effects of increased transcription at ICU11 targets on the wider Arabidopsis epigenome.
icu11 Perturbs the Vernalization Response in Arabidopsis.
To clarify the functional implications of the physical and genetic interaction of ICU11 with PRC2, we asked how the icu11 mutation affected vernalization, a well understood PRC2-regulated developmental transition. To set a high FLC expression state and therefore a vernalization requirement, we crossed icu11-3 to fca-1. icu11-3 fca-1 plants showed an increased variability in flowering compare to fca-1 (Fig. 3 A and B and SI Appendix, Table S4). Cold treatment of different durations revealed a clear vernalization defect in icu11-3 fca-1 plants; flowering was significantly delayed after 2 wk cold treatment (SI Appendix, Table S4) and was only marginally accelerated with longer cold. No difference in flowering was observed in the icu11-3 single mutant following vernalization (SI Appendix, Fig. S7).
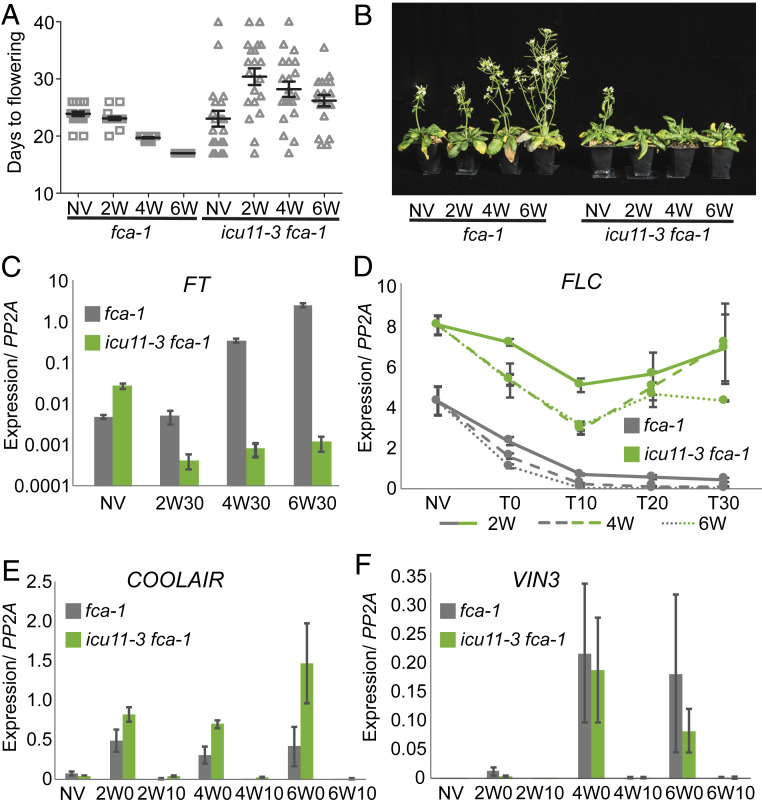
Fig. 3.
icu11-3 is defective in vernalization. (A and B) Flowering is significantly more asynchronous at all time points and significantly delayed in the icu11-3 fca-1 genotype after 2 wk vernalization (SI Appendix, Table S4). Days to flowering counted from the end of a pregrowth period of 14 d in warm conditions (NV), or from the end of pregrowth followed by a 2-, 4-, or 6-wk vernalization treatment. Bars represent mean and SEM (n = 20). (B) Flowering plants imaged at 25 d postvernalization. (C–F) Gene expression in the vernalization-requiring fca1 and icu11-3 fca-1 backgrounds. Gene expression is normalized to PP2A; error bars represent SEM for three biological replicates. (C) FT expression at 30 d posttreatment is higher in fca1 than icu11-3 fca-1. (D) Expression of the flowering repressor FLC is higher in icu11-3 fca1 than fca-1 before (NV), during (T0), and 10, 20, or 30 d after vernalization (T10–30), reactivating strongly in icu11-3 fca1 compared with the stable postvernalization repression observed in fca-1. Increasing vernalization time (2, 4, and 6 wk) leads to lower and more stably silenced FLC expression. (E) The antisense RNA COOLAIR is expressed at higher levels in icu11-3 fca1 than fca-1. (F) Expression of VIN3, a cold-induced PRC2 target, is not affected by the icu11-3 mutation.
We collected tissue before vernalization (NV), on the final cold day after 2, 4, or 6 wk cold (T0), and at 10-d intervals (T10, 20, 30) upon return of plants to warm, long-day conditions, allowing us to examine the expression of key regulatory genes in the vernalization-induced flowering pathway. Expression of FT in fca-1 and icu11-3 fca-1 mirrored the differences observed in flowering time. Without cold, FT was expressed at ∼10-fold higher levels in icu11 fca-1, even though expression of FLC (its repressor) was also high. This presumably reflects misregulation of all Polycomb target genes in an icu11 background (Fig. 3C). Following increasing periods of cold exposure, FT expression levels increased in WT plants, but less so in icu11 fca-1. In fca-1, FLC expression decreased quantitatively by prolonged cold and was maintained in the transcriptionally silent state after return to warm conditions. In contrast, in icu11-3 fca-1, FLC expression was approximately twofold higher prior to vernalization (Fig. 3D), and although expression was reduced by cold, this was not maintained following vernalization. We interpret this as reflecting a reduced ability for the locus to digitally switch to an epigenetically silenced state. Consistent with this, the icu11-3 mutant showed higher expression of the FLC antisense transcript COOLAIR during the cold (Fig. 3E), but expression of the cold-induced FLC repressor VIN3, another PRC2 target, was unaffected (Fig. 3F).
ICU11 Functions Directly at FLC to Affect the Balance of Histone Modifications.
Vernalization-induced silencing of FLC is dependent on PRC2-mediated switching between bistable opposing epigenetic states: an active state, marked by high levels of H3K36me3 at the nucleation region of FLC; and a silent state, marked by a peak of H3K27me3 in the nucleation region that accumulates during cold exposure and spreads to cover the entire FLC locus upon return to the warm. We had previously shown that H3K4me3 at FLC follows transcription rather than functioning as the opposing chromatin state to H3K27me3 (1).
Comparison of icu11-3 fca-1 compared to fca-1 over the vernalization time series was used to define the role of ICU11 in FLC epigenetic switching. As expected, H3K36me3 was lost from the nucleation region in fca-1 after 4 wk of cold exposure and replaced by a peak of H3K27me3 that spread to cover FLC after 10 d subsequent growth in warm long-day conditions (Fig. 4). In contrast, in icu11-3 fca-1 the H3K36me3 peak was higher before vernalization and was not lost from the nucleation region following cold exposure or subsequent warm growth conditions. This was coincident with an attenuated accumulation of H3K27me3 accumulation at the locus following cold (Fig. 4). Comparison of H3K27me3 and H3K36me3 levels at ACT7 and STM indicated that histone methylation was relatively stable across time points and between genotypes at control genes (SI Appendix, Fig. S8). Thus, ICU11 is required for the cold-induced epigenetic switch between the active H3K36me3 state to the silenced H3K27me3 state at FLC.
Fig. 4.
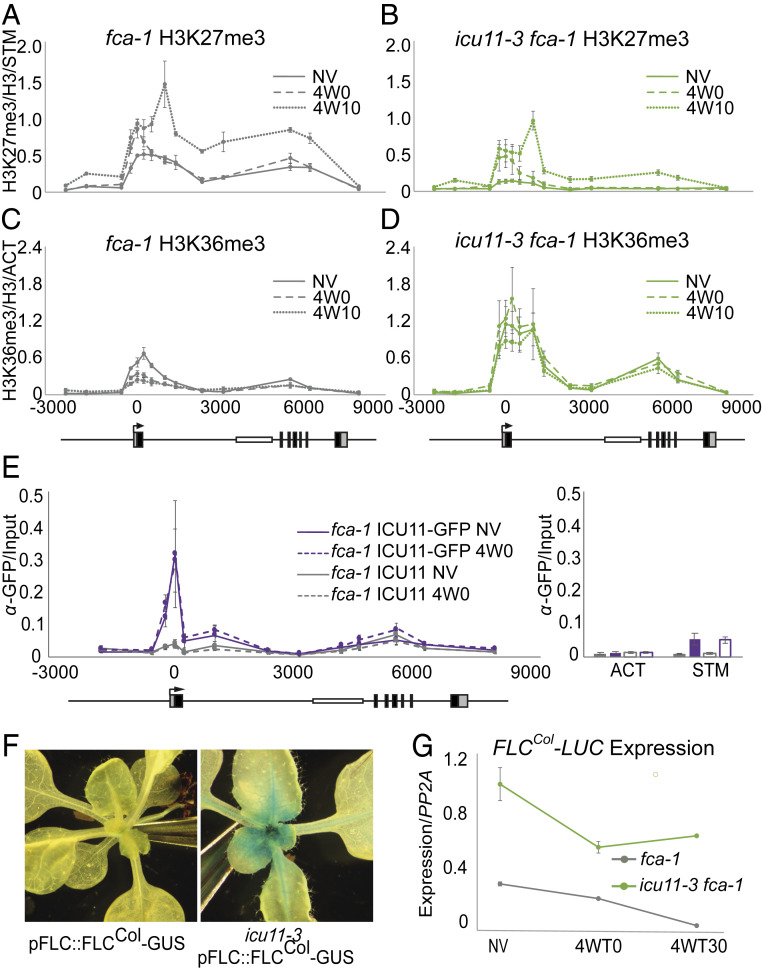
ICU11 influences H3K27me3 and H3K36me3 dynamics at FLC through a vernalization timecourse. (A–E) ChIP analysis of H3K27me3 (A and C) and H3K36me3 (B and D) histone modifications and ICU11 localization (E) at FLC; error bars are SEM of three biological replicates. Abundance, relative to total histone H3 and normalized to STM (H3K27me3) or ACT7 (H3K36me3) was measured in nonvernalized plants, after 4 wk vernalization and vernalization followed by 10 d growth in warm conditions. The icu11-3 fca-1 mutant (C and D) has reduced H3K27me3 and increased H3K36me3 relative to the fca-1 control (A and B). (E) ChIP analysis of ICU11 localization at FLC before and after 4 wk vernalization treatment, normalized to input. Purple lines represent plants containing the GFP-tagged ICU11 construct, while gray lines represent control plants with untagged ICU11. Localization of ICU11-GFP at ACT and STM before (solid bar) and after (empty bar) vernalization shown as positive and negative controls. Underlying A–E, an FLC locus schematic showing UTRs (gray boxes), exons (black boxes), and the Ler background MULE insertion in intron one (white box). (F) Gus staining of FLC expression in FLCCol-0-GUS, which lacks the MULE in intron one found in Ler background. Expression of this allele is higher in icu11-3, indicating misregulation of the MULE insertion is not responsible for the FLC overexpression phenotype. (G) Expression of FLC-LUC from FLCCol-0-LUC (no MULE in intron one) is higher in the icu11-3 background than WT before and after 4 wk vernalization and shows reactivation after vernalization followed by postcold growth. Error bars are SEM of three biological replicates.
These analyses had been undertaken using icu11-3, a mutation in the Landsberg erecta (Ler) background. In Ler, a Mutator-like transposable element (MULE) in the first intron of FLC leads to low expression before vernalization compared with other alleles. We wanted to confirm the effect of ICU11 at FLC was on Polycomb switching rather than an influence FLC through an effect of the MULE insertion. First, we performed chromatin immunoprecipitation to define sites of association of ICU11 at FLC, using a GFP-tagged ICU11 construct transformed into the icu11-3 fca-1 background. ICU11 was localized at the FLC 5′ end coincident with the H3K36me3 enrichment, both before vernalization and at the end of a 4-wk cold treatment (Fig. 4E). We also observed a small increase in ICU11 occupancy over the gene body after 4 wk cold, similar to that seen for other PRC2 components (1). Thus, FLC is a direct target of ICU11, colocalizing with PRC2 components before cold and at distinct phases of the cold silencing process. Notably, our ICU11-GFP chromatin immunoprecipitation (ChIP) experiment did not indicate enrichment of ICU11 signal over the MULE.
To confirm our hypothesis that ICU11 does not influence FLC via an effect of the MULE, we utilized a pFLC::FLC-GUS reporter construct generated using the Col-0 FLC allele (pFLC::FLCCol-0-GUS), which lacks the MULE. This construct was crossed into the icu11-3 (Ler) background, and WT and icu11-3 siblings from an F2 population examined in nonvernalizing conditions. In icu11-3, FLC expression was strongly up-regulated before cold even in the absence of the MULE (Fig. 4F). We used a second Col-0 allele construct, pFLC::FLCCol-0-LUCIFERASE, to examine whether the FLC reactivation seen following vernalization was also independent of transposon derepression in icu11-3. The pFLC::FLCCol-0-LUCIFERASE construct was backcrossed into the icu11-3 fca-1 background. Paralleling our observation of the native FLCLer allele in icu11-3 fca-1 (Fig. 3D), we found that FLC-LUC expression was higher in the icu11-3 background than in WT prior to vernalization, was reduced by cold, but that transcription reactivated on return to the warm (Fig. 4G). Thus, defective silencing of FLC in icu11-3 is PRC2-dependent and independent of the MULE insertion in FLCLer.
Discussion
Here, through analysis of a mutant displaying a range of PcG-impaired phenotypes we identify the ICU11 Fe2+/2 oxoglutarate-dependent oxygenase activity that influences Arabidopsis PRC2 regulation. The large 2OGD superfamily in Arabidopsis, of which ICU11 is a member, contains prolyl-4-hydroxylases, AlkB DNA repair enzymes and enzymes integral to an array of biosynthetic processes (26). However, ICU11 and its most closely related sequences form a separate clade within the Arabidopsis genome (25), suggesting distinct functionality. Our analysis of global changes and specific defects in the vernalization silencing process suggests that impairment of ICU11 primarily influences active histone marks—especially H3K36me3. A 2-OGD is the catalytically active demethylation domain of Jumonji histone demethylases; however, we could not reproducibly show using in vitro assays that ICU11 has this biochemical activity. Notably, previous studies have similarly struggled to demonstrate recombinant activity in vitro despite clear functional evidence for their function in vivo (35, 36).
The subtle alterations in DNA methylation in the icu11-3 mutant were unexpected for a mutation predominantly associated with Polycomb silencing. Such pleiotropic changes have not previously been reported for PRC2 mutants in Arabidopsis; indeed, a comparable Southern blot analysis indicates that even the severe PRC2 clf swn double mutant maintains effective cen180 repeat methylation (37). Thus, we speculate that any DNA methylation icu11 phenotypes are linked to altered transcriptional activity. ICU11 may work specifically as an H3K36me3 demethylase, influencing directly PRC2 targets and then indirectly a larger set of targets. Alternatively, ICU11 may act as a promiscuous histone demethylase targeting distinct methylation types according to the specificity of its binding partners. However, the robust immunoprecipitation with PRC2 components, but not other chromatin regulators, argues against this. ICU11 may also influence methylation levels of trans factors. For example, the histone demethylase LSD1 demethylates both H3K4 methylation and the DNA methyltransferase Dnmt1 in mouse embryonic stem cells, directly influencing H3K4 and indirectly reducing H3K9me2 (38). Further analysis on defining demethylase activities is clearly important if we are to fully understand epigenetic homeostasis within the Arabidopsis genome.
Materials and Methods
The transposon-tagged lines are in the Landsberg erecta genotype. Other material is in the Col-0 genotype. Detailed descriptions of the growth conditions and the experimental procedures used—cloning, qRT-PCR, proteomics, Southern blots, whole-genome methylation analysis, Western blots, chromatin immunoprecipitation, and the in vitro histone demethylation assay—are in SI Appendix, Materials and Methods.
Materials and Data Availability.
The full list of mass spectrometry results is provided as Dataset S1. The DNA methylation analysis data have been deposited at Gene Expression Omnibus (accession no. GSE151449). All of the other raw data and materials that support the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We would like to thank Scott Berry for help with ICU-GFP nuclear localization microscopy, Hao Yu and Lisha Shen for assistance with 6mA DNA methylation analysis, Donna Gibson for graphic design assistance, and members of the C.D. and Howard laboratories for helpful discussions. This work was funded by the European Research Council grants to “MEXTIM” (to C.D.) and “SexMeth” (to X. Feng), by the Biotechnological and Biological Sciences Research Council (BBSRC) Institute Strategic Programmes GRO (BB/J004588/1), GEN (BB/P013511/1), BBSRC grant (to X. Feng) (BB/S009620/1), and the Marie Sklodowska–Curie Postdoctoral Fellowships “UNRAVEL” (to R.H.B.) and "WISDOM" (to X. Fang). Additional funding via the Wellcome Trust through a Senior Research Fellowship (to J.R.) (103139) and a multiuser equipment grant (108504). The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149).
Footnotes
The authors declare no competing interest.
Data deposition: DNA methylation analysis data have been deposited in the Gene Expression Omnibus (GEO) repository (accession no. GSE151449).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920621117/-/DCSupplemental.
References
- 1.Yang H., Howard M., Dean C., Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 24, 1793–1797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan W. et al., H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R., Strome S., Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2, 1169–1177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossniklaus U., Vielle-Calzada J. P., Hoeppner M. A., Gagliano W. B., Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Mylne J. S. et al., LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl. Acad. Sci. U.S.A. 103, 5012–5017 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Lucia F., Crevillen P., Jones A. M. E., Greb T., Dean C., A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 105, 16831–16836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greb T. et al., The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 17, 73–78 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Sung S., Amasino R. M., Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Calonje M., Sanchez R., Chen L., Sung Z. R., EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20, 277–291 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y., Hartwig B., James G. V., Schneeberger K., Turck F., Complementary activities of TELOMERE REPEAT BINDING proteins and polycomb group complexes in transcriptional regulation of target genes. Plant Cell 28, 87–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y. et al., Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat. Genet. 50, 638–644 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Derkacheva M. et al., H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat. Plants 2, 16126 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Qüesta J. I., Song J., Geraldo N., An H., Dean C., Arabidopsis transcriptional repressor VAL1 triggers polycomb silencing at FLC during vernalization. Science 353, 485–488 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Bloomer R. H., Dean C., Fine-tuning timing: Natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 68, 5439–5452 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Angel A. et al., Vernalizing cold is registered digitally at FLC. Proc. Natl. Acad. Sci. U.S.A. 112, 4146–4151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry S., Hartley M., Olsson T. S. G., Dean C., Howard M., Local chromatin environment of a polycomb target gene instructs its own epigenetic inheritance. eLife 4, e07205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helliwell C. A., Wood C. C., Robertson M., James Peacock W., Dennis E. S., The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Searle I. et al., The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H. et al., Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357, 1142–1145 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Crevillén P. et al., Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry S., Dean C., Howard M., Slow chromatin dynamics allow polycomb target genes to filter fluctuations in transcription factor activity. Cell Syst. 4, 445–457.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodd I. B., Micheelsen M. A., Sneppen K., Thon G., Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell 129, 813–822 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Yang H., Howard M., Dean C., Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. U.S.A. 113, 9369–9374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bancroft I., Jones J. D. G., Dean C., Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell 5, 631–638 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateo-Bonmatí E. et al., INCURVATA11 and CUPULIFORMIS2 are redundant genes that encode epigenetic machinery components in Arabidopsis. Plant Cell 30, 1596–1616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai Y., Ono E., Mizutani M., Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 78, 328–343 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y. et al., Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, 4833–4838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derkacheva M. et al., Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang S. C. et al., Kicking against the PRCs–A domesticated transposase antagonises silencing mediated by polycomb group proteins and is an accessory component of polycomb repressive complex 2. PLoS Genet. 11, e1005660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W., Jiang D., Jiang J., He Y., A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 62, 663–673 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Lister R. et al., Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cokus S. J. et al., Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouyer D. et al., DNA methylation dynamics during early plant life. Genome Biol. 18, 179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartels A. et al., Dynamic DNA methylation in plant growth and development. Int. J. Mol. Sci. 19, 2144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audergon P. N. C. B. et al., Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348, 132–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba A. et al., PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 13, 668–675 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Lindroth A. M. et al., Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23, 4286–4296 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J. et al., The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.