Significance
LIN28B is well known as a RNA-binding protein and a suppressor of microRNA biogenesis by selectively blocking the processing of let-7 precursors. However, little is known about let-7–independent roles of LIN28B. Here, we show that LIN28B is recruited to active promoters by binding to the zinc-finger transcription factor ZNF143. LIN28B acts as a cofactor to upregulate expression of a subset of downstream target genes that are essential for neuroblastoma cell survival and migration. Our paper reveals an unexpected role of LIN28B in transcriptional regulation that is independent of let-7 during neuroblastoma pathogenesis.
Keywords: neuroblastoma, zebrafish model, transcriptional regulation, LIN28B, ZNF143
Abstract
LIN28B is highly expressed in neuroblastoma and promotes tumorigenesis, at least, in part, through inhibition of let-7 microRNA biogenesis. Here, we report that overexpression of either wild-type (WT) LIN28B or a LIN28B mutant that is unable to inhibit let-7 processing increases the penetrance of MYCN-induced neuroblastoma, potentiates the invasion and migration of transformed sympathetic neuroblasts, and drives distant metastases in vivo. Genome-wide chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIP-seq) and coimmunoprecipitation experiments show that LIN28B binds active gene promoters in neuroblastoma cells through protein–protein interaction with the sequence-specific zinc-finger transcription factor ZNF143 and activates the expression of downstream targets, including transcription factors forming the adrenergic core regulatory circuitry that controls the malignant cell state in neuroblastoma as well as GSK3B and L1CAM that are involved in neuronal cell adhesion and migration. These findings reveal an unexpected let-7–independent function of LIN28B in transcriptional regulation during neuroblastoma pathogenesis.
Neuroblastoma is an intractable solid tumor of the peripheral sympathetic nervous system (PSNS) that arises in children and accounts for 15% of childhood cancer deaths (1). High-risk neuroblastoma patients have a 5-y event-free survival rate below 50%, and patients who relapse have a dismal prognosis (2). Neuroblastoma derives from the sympathoadrenal lineage of neural crest cells and typically arises in the adrenal medulla or paraspinal ganglia (1). Over half of neuroblastomas exhibit metastases at diagnosis with frequent spread to bone marrow, cortical bone, lymph nodes, liver, brain, orbital sites, and lung (1–5). It is, therefore, critical to devise new strategies for the treatment of this high-risk pediatric malignancy.
MYCN amplification is present in ∼20–25% of neuroblastomas and is the most common genetic aberration associated with poor disease outcome (2). A genome-wide association study identified an association between single-nucleotide polymorphisms in the LIN28B locus and the development of neuroblastoma, suggesting that LIN28B may function as a predisposition gene or oncogenic driver during neuroblastoma pathogenesis (6). Furthermore, genome-wide CRISPR analysis has implicated LIN28B as a selective genetic dependency in MYCN-amplified neuroblastoma (7), indicating that therapies designed to inactivate the neoplastic functions of LIN28B may be particularly beneficial for patients with MYCN-amplified neuroblastomas.
LIN28B and its homolog LIN28A are important regulators of mammalian stem cell self-renewal, and LIN28A is one of four factors in a mixture capable of reprogramming differentiated somatic cells into pluripotent stem cells (8). LIN28B has been implicated in malignant transformation and is highly expressed in a variety of solid tumors and hematological malignancies including neuroblastoma (6, 9–11), Wilms’ tumor (12), hepatocellular carcinoma (13), colon cancer (14, 15), peripheral T-cell lymphoma (16), pediatric leukemia (17), and malignant germ-cell tumors (18). Both LIN28A and LIN28B are known to selectively block the processing of let-7 microRNA (miRNA) precursors into mature miRNAs by directly binding primary let-7 transcripts (19, 20). LIN28B may promote neuroblastoma, at least, in part, through suppression of let-7 because LIN28B overexpression has been shown to enhance MYCN expression in the sympathoadrenal lineage of cells in mice (10). LIN28B also promotes neuroblastoma tumorigenesis through a LIN28B-RAN-AURKA signaling network by mechanisms that are both let-7–dependent and let-7–independent (21). We found that LIN28B overexpression is one of multiple mechanisms that disrupt the let-7 miRNA family to promote neuroblastoma (11). Although let-7–independent functions of LIN28B have been implicated in neurogenesis and tumorigenesis of the PSNS (22–24), the nature of the let-7–independent functions of LIN28B during neuroblastoma tumorigenesis remains elusive.
In this study, we demonstrate that stable transgenic zebrafish lines coexpressing MYCN and either WT or let-7-suppression–deficient mutant LIN28B in the PSNS enhance the penetrance and promote metastasis of MYCN-induced neuroblastoma. Genome-wide chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIP-seq) experiments reveal a novel let-7–independent function for LIN28B. It binds active gene promoters in neuroblastoma cells through an interaction with a zinc-finger transcription factor ZNF143 and regulates the transcription of genes involved in neuroblastoma cell migration and survival. The LIN28B-associated transcriptional network may, therefore, provide a source of “druggable” targets for neuroblastoma tumors that require LIN28B for malignancy.
Results
Overexpression of WT or Mutant LIN28B Accelerates MYCN-Induced Neuroblastoma.
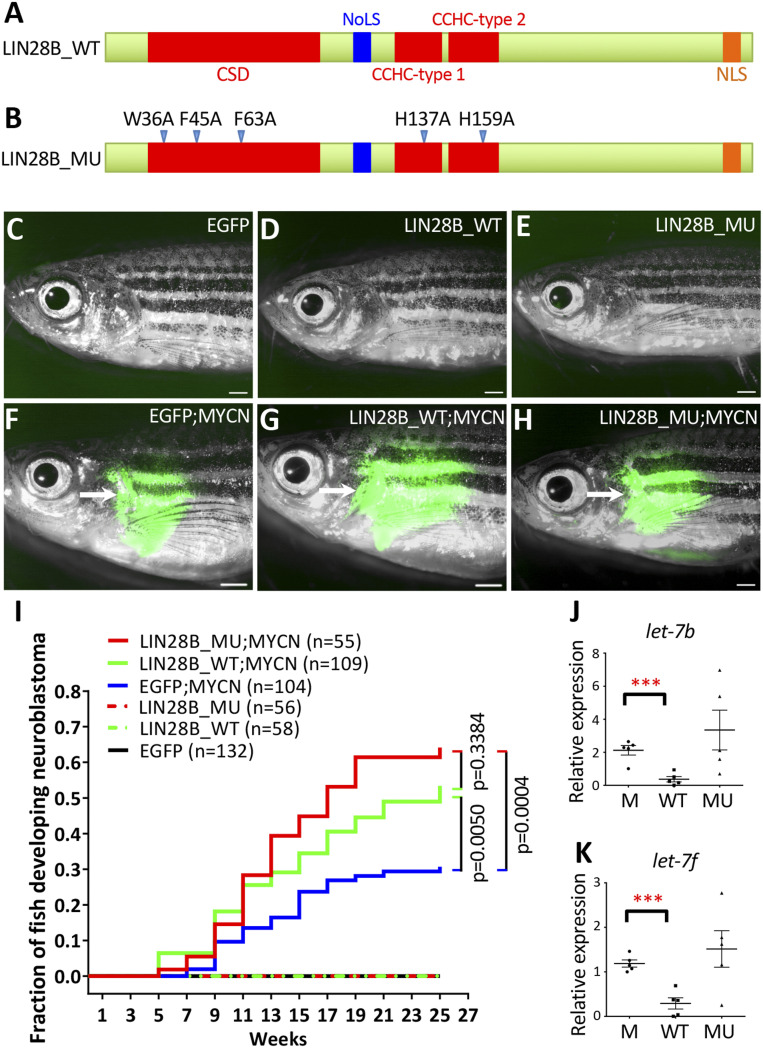
Large scale experiments have shown that LIN28B is highly expressed in neuroblastoma (SI Appendix, Fig. S1A), and elevated LIN28B expression is strongly associated with a lower probability of overall survival in neuroblastoma patients (SI Appendix, Fig. S1B). To investigate the let-7–dependent and let-7–independent functions of LIN28B during neuroblastoma pathogenesis, we generated stable transgenic zebrafish lines that coexpress enhanced green fluorescent protein (EGFP) with human WT LIN28B (LIN28B_WT) or let-7-suppression–deficient mutant LIN28B (LIN28B_MU) in the PSNS by the zebrafish dopamine-β-hydroxylase gene (dβh) promoter. The mutant LIN28B contains five point mutations spread across the cold shock domain (CSD) and CysCysHisCys (CCHC) zinc-finger RNA-binding motifs (Fig. 1 A and B) that render the protein incapable of let-7 suppression in the closely related LIN28B paralog LIN28A (22, 25). The transgenes were cointegrated into the genome such that EGFP marks expression of the transgene and facilitates visualization of tumor development (26). Two stable transgenic zebrafish lines were identified, Tg(dβh:EGFP;dβh:LIN28B_WT) and Tg(dβh:EGFP;dβh:LIN28B_MU) and designated LIN28B_WT and LIN28B_MU hereafter. Fish transgenic for EGFP [Tg(dβh:EGFP)], LIN28B_WT or LIN28B_MU alone did not develop neuroblastoma in this assay (Fig. 1 C–E and I).
Fig. 1.
Overexpression of WT or mutant LIN28B increases the penetrance of MYCN-induced neuroblastoma. (A and B) Schematic of the WT (A, LIN28B_WT) and mutant (B, LIN28B_MU) human LIN28B proteins. CSD; NoLS, nucleolar localization signal; CCHC zinc-finger domain; NLS, nuclear localization signal. Five point mutations (arrowheads) were created in the LIN28B_MU. (C–H) Lack of detectable EGFP expression in the IRG of a 17-wpf EGFP transgenic fish (C) and 39-wpf LIN28B_WT (D) and LIN28B_MU (E) fish; and EGFP expression in the tumors (arrows) arising from the IRG of 17-wpf EGFP;MYCN (F), LIN28B_WT;MYCN (G) and LIN28B_MU;MYCN (H) fish. (Scale bar, 1 mm.) (I) Kaplan–Meier analysis of cumulative frequency of neuroblastoma in transgenic lines. Statistical analysis was performed using the logrank test. (J and K) The relative expression of zebrafish let-7b (J) and let-7f (K) miRNAs in EGFP;MYCN (M), LIN28B_WT;MYCN (WT), and LIN28B_MU;MYCN (MU) tumors by qRT-PCR. Five independent tumors from 6-mo-old fish were analyzed for each group. Each dot represents the expression levels of the indicated gene normalized to U6 snRNA. Horizontal bars indicate means ± SD. Statistical analysis was performed using the two-tailed unpaired t test. ***P < 0.001.
To determine whether LIN28B collaborates with MYCN during neuroblastoma development, we first analyzed available databases for coexpression of LIN28B and MYCN in patient tumors. Indeed, we found a positive correlation between LIN28B and MYCN expressions in human primary neuroblastomas (SI Appendix, Fig. S1 C and D) and neuroblastoma cell lines (SI Appendix, Fig. S1E). We then bred a zebrafish neuroblastoma model Tg (dβh:EGFP-MYCN) (designated MYCN) (27) with both LIN28B_WT and LIN28B_MU lines as well as the EGFP control line. Both LIN28B_WT;MYCN and LIN28B_MU;MYCN compound transgenic lines developed tumors in the interrenal gland (IRG), the zebrafish counterpart to the human adrenal medulla (Fig. 1 F–H). These tumors arose with an earlier onset at 5-wk postfertilization (wpf) and increased penetrance of 50–60% by 25 wpf, compared to ∼30% in EGFP;MYCN control fish (P = 0.0050 and P = 0.0004 for LIN28B_WT;MYCN and LIN28B_MU;MYCN lines, respectively) (Fig. 1I). In addition, tumors arising in the LIN28B_WT;MYCN line but not the LIN28B_MU;MYCN line had dramatically reduced levels of the mature let-7 miRNA family members let-7b and let-7f compared to those arising in the EGFP;MYCN line (Fig. 1 J and K). LIN28B can, therefore, collaborate with MYCN to induce neuroblastoma through a mechanism that does not involve processing of the let-7 miRNA.
Nontransformed cells of the IRG predominantly exist as chromaffin cells that express tyrosine hydroxylase (TH) (SI Appendix, Fig. S2 A–C, E–G, and I–K) and do not have detectable LIN28B unless transgenically expressed in the LIN28B_WT and LIN28B_MU stable lines (SI Appendix, Fig. S2 D, H, and L). By contrast, all MYCN-induced tumor cells, with or without LIN28B_WT/MU overexpression, were composed of TH-expressing undifferentiated small and round neuroblasts with hyperchromatic nuclei and scant cytoplasm (SI Appendix, Fig. S2 M–O, Q–S, and U–W). Human LIN28B was highly expressed in the tumor cells of LIN28B_WT;MYCN and LIN28B_MU;MYCN transgenic fish but not of EGFP;MYCN fish (SI Appendix, Fig. S2 P, T, and X). Notably, while WT LIN28B was found in both the cytoplasm and the nucleus, mutant LIN28B appeared to be specifically enriched in the nucleus (SI Appendix, Fig. S2 H, L, T, and X), which is in agreement with previously reported findings (25). These results suggest that both WT and mutant versions of LIN28B accelerate the development of MYCN-induced neuroblastoma but do not cause overt changes affecting the tumor-cell histology.
Both WT and Mutant LIN28B Promote Distant Metastases of MYCN-Induced Neuroblastoma.
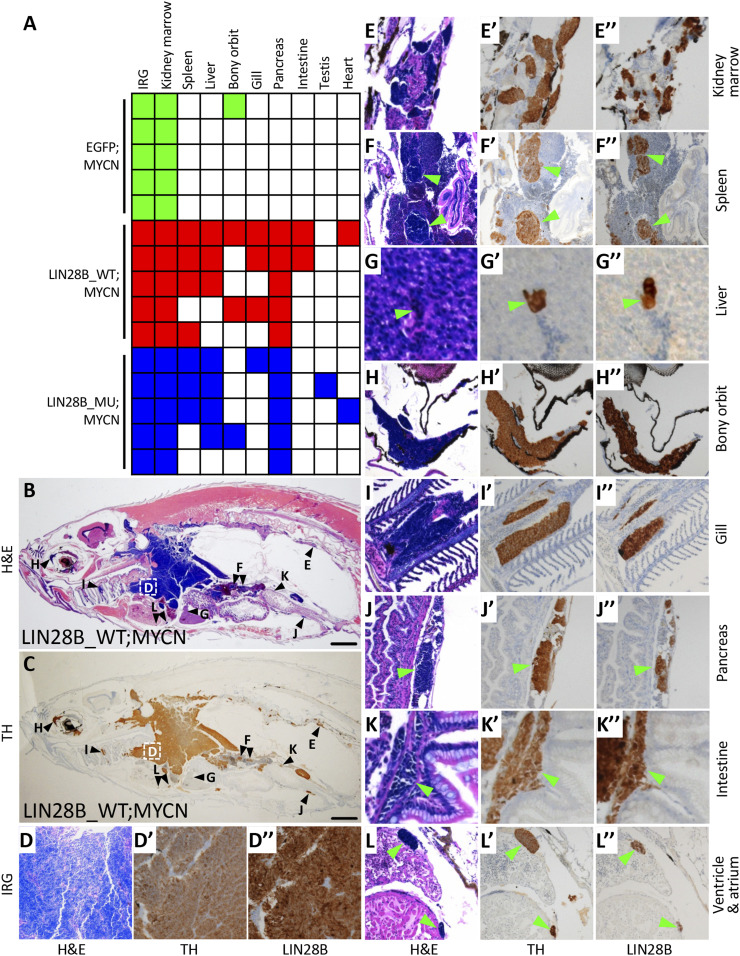
Overexpression of LIN28B has been shown to enhance cellular proliferation, migration, invasion, and metastasis in colorectal cancer xenograft mouse models (14, 15). To investigate whether LIN28B also promotes the metastasis of neuroblastoma, five tumor-bearing fish at 6 mo of age were randomly selected from each of the experimental groups: i) EGFP;MYCN, ii) LIN28B_WT;MYCN, and iii) LIN28B_MU;MYCN. Hematoxylin and eosin (H&E) staining and immunohistochemical staining for the neuronal marker TH (characteristic of neuroblastoma) and LIN28B revealed tumor-cell masses in the IRG and the adjacent kidney marrow of all 15 fish (Fig. 2 A–E″ and SI Appendix, Fig. S3). Disseminated tumor metastases were also present in multiple additional organs and sites in LIN28B_WT;MYCN and LIN28B_MU;MYCN lines but only very rarely in the EGFP;MYCN line (P = 0.0476, Fig. 2A and SI Appendix, Fig. S3).
Fig. 2.
LIN28B promotes the metastasis of MYCN-induced neuroblastoma. (A) Summary of neuroblastoma metastases in EGFP;MYCN, LIN28B_WT;MYCN, and LIN28B_MU;MYCN transgenic lines at 6 mo of age, five fish per group. Colored boxes indicate that metastasis was observed in the indicated organ while white boxes indicate no apparent metastasis. The difference in metastases between EGFP;MYCN and either LIN28B_WT;MYCN or LIN28B_MU;MYCN fish (P = 0.0476) was compared using the two-tailed Fisher’s exact test. (B and C) Representative images of neuroblastoma metastases in the sagittal sections of LIN28B_WT;MYCN fish by H&E staining (B) and immunohistochemical staining of TH (C). The organs harboring metastatic tumor cells are marked in B and C and are correspondingly magnified in D–L″. (Scale bar, 1 mm.) (D–L″) Magnified views of the organs shown in B and C showing H&E staining as well as immunostaining of TH and LIN28B in LIN28B_WT;MYCN fish. Green arrowheads indicate metastatic cells.
The most common metastatic site for neuroblastoma in humans is the bone marrow (3) where hematopoietic stem and progenitor cells normally reside. Hematopoiesis in zebrafish takes place in the kidney marrow (28), accounting for the fact that all of the fish had involvement of kidney marrow due to local invasion extending from the IRG. LIN28B_WT;MYCN and LIN28B_MU;MYCN fish harbored metastases in the spleen (the zebrafish equivalent of human lymph nodes, Fig. 2 F–F″), liver (Fig. 2 G–G″), bony orbit (Fig. 2 H–H″), and gill (the zebrafish equivalent of human lung, Fig. 2 I–I″), corresponding to the most prevalent sites of neuroblastoma metastases in humans. Of note, metastases in bone marrow, bony orbit, and lung are associated with poor prognosis and decreased survival in neuroblastoma patients (3–5). Tumor cells from both LIN28B_WT;MYCN and LIN28B_MU;MYCN lines were also identified in sites that are less commonly associated with human neuroblastoma metastasis, such as the pancreas (Fig. 2 J–J″), intestine (Fig. 2 K–K″), and testis (Fig. 2A). The identification of tumor cells in the cardiac ventricle and atrium in both LIN28B_WT;MYCN and LIN28B_MU;MYCN zebrafish lines (Fig. 2 A and L–L″) suggests that metastases developed through the hematogenous spread of tumor cells. These data indicate that LIN28B promotes the colonization of MYCN-induced neuroblastoma in multiple tissues in a manner that is independent of let-7 regulation.
Both WT and Mutant LIN28B Promote Human Neuroblastoma Cell Invasion and Migration.
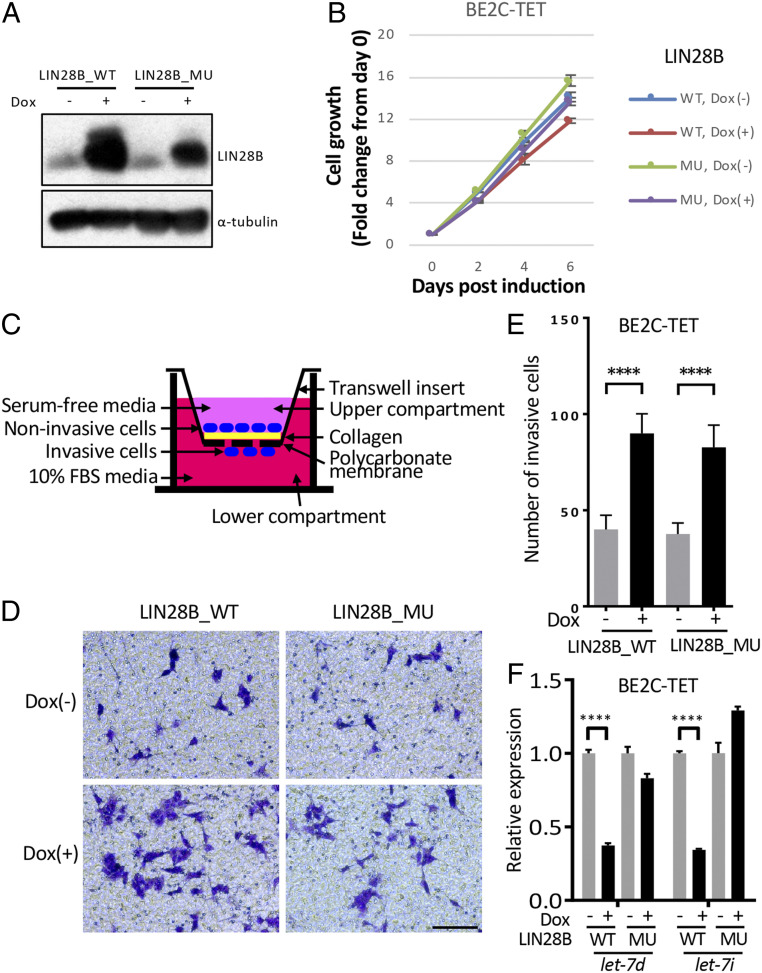
To determine whether WT or mutant LIN28B promotes the invasion and migration of human neuroblastoma cells, we engineered a doxycycline-inducible Flag-tagged LIN28B_WT or LIN28B_MU overexpression system in the human MYCN-amplified neuroblastoma cell lines BE2C and CHP134 (designated BE2C-TET and CHP134-TET lines, respectively). Both LIN28B_WT and LIN28B_MU were strongly induced by doxycycline treatment (Fig. 3A and SI Appendix, Fig. S4A) but did not affect the proliferation of BE2C-TET or CHP134-TET cells (Fig. 3B and SI Appendix, Fig. S4B). However, transwell migration and invasion assays (Fig. 3C) showed that doxycycline-treated BE2C-TET or CHP134-TET cells expressing either LIN28B_WT or LIN28B_MU exhibited ∼1.6–2 fold more cells that pass through collagen-coated permeable membranes (Fig. 3 D and E and SI Appendix, Fig. S4 C and D) compared to control cells with inducible expression of red fluorescent protein (SI Appendix, Fig. S4 C and D). Importantly, the expression of let-7d and let-7i were both robustly inhibited in BE2C-TET cells by overexpression of WT but not mutant LIN28B (Fig. 3F). These data suggest that overexpression of LIN28B_WT and LIN28B_MU promotes the migration and invasion of human neuroblastoma cells.
Fig. 3.
Both WT and mutant LIN28B promote cell invasion and migration in BE2C cells. (A) Western blotting of LIN28B_WT/MU using the BE2C-TET cells untreated or treated with 50 ng/mL doxycycline for 3 d. α-Tubulin was used as a loading control. (B) Relative cell growth of BE2C-TET cells untreated or treated with 50-ng/mL doxycycline. Values represent means ± SEM of triplicate experiments. (C) Schematic of transwell migration and invasion assay. (D and E) Transwell migration and invasion assay of the BE2C-TET cells untreated or treated with 50-ng/mL doxycycline. Cells were stained with 0.1% crystal violet (D), and the number of migrated cells through the membrane per field was compared with the two-tailed unpaired t test (E). Values represent means ± SD of triplicate experiments. ****P < 0.0001. (Scale bar, 100 µm.) (F) qRT-PCR to detect let-7d and let-7i expressions in BE2C-TET cells that were either untreated or treated with 50-ng/mL doxycycline for 3 d. Values were normalized to U6, U47, and RNU44 small nuclear RNAs and represent the means ± SD of triplicate experiments. Statistical analysis was performed using the two-tailed unpaired t test. ****P < 0.0001.
LIN28B Binds Active Promoters in Human Neuroblastoma Cells.
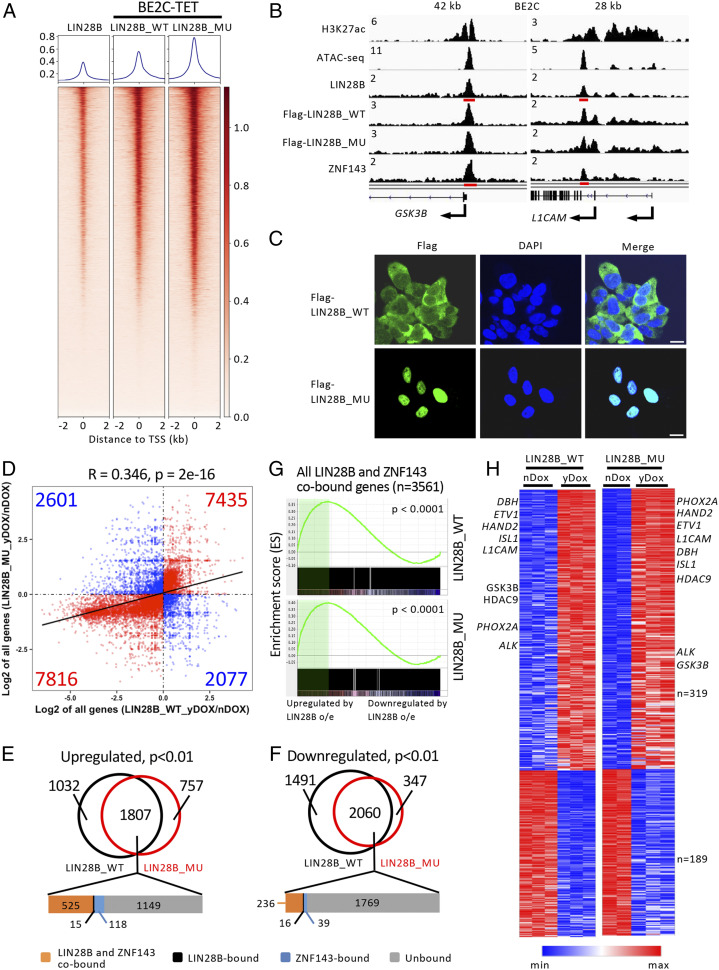
Superenhancer-regulated and cell-requisite transcription factors may compose elements of a core regulatory circuitry (CRC), which is required for both the survival and the establishment of the unique transcriptional profile of a particular cell type (29). We have shown that LIN28B represents a selective gene dependency in MYCN-amplified neuroblastoma and is associated with a superenhancer (7). Furthermore, endogenous LIN28B protein is localized to both the cytoplasmic and the nuclear fractions of BE2C, CHP134, and Kelly neuroblastoma cells (SI Appendix, Fig. S5A). Therefore, we tested the hypothesis that LIN28B functions as a transcriptional cofactor associated with the CRC in the adrenergic subtype of neuroblastoma cells (7, 30, 31). ChIP-seq experiments revealed that many high-confidence genomic regions of enrichment for LIIN28B (P value cutoff of 1e-9) were shared among BE2C, CHP134, and Kelly cells (Fig. 4A and SI Appendix, Table S1), which are neuroblastomas of the adrenergic subtype that have MYCN gene amplification, indicating that LIN28B is associated with particular DNA sequences. However, we quickly ascertained that LIN28B is not consistently recruited to the adrenergic CRC as a cofactor by one of the DNA-binding members of the CRC. CRC members typically bind very closely to each other on DNA, forming dense clusters of transcription factors within their own enhancer regions, collectively regulating the expression of each gene, and creating an interconnected feed-forward autoregulatory loop (7, 32). We found that LIN28B co-occupies a subset of genomic regions with the CRC transcription factors (HAND2, ISL1, PHOX2B, GATA3, TBX2, and MYCN) and may regulate the expression of a number of genes (SI Appendix, Fig. S5 B and C). However, LIN28B is not consistently present at either its own enhancer or the known CRC enhancers (marked by H3K27ac; SI Appendix, Fig. S5B), effectively ruling out involvement of LIN28B as a bona fide cofactor of the neuroblastoma CRC. Rather, LIN28B is a member of the extended regulatory network (ERN) (32) of the adrenergic CRC because each of the other CRC transcription factors is localized to the LIN28B superenhancer (SI Appendix, Fig. S5B).
Fig. 4.
LIN28B binds active promoters in human neuroblastoma cells. (A) Heatmaps of genome-wide occupancy for LIN28B as determined by ChIP-seq in BE2C, CHP134, and Kelly cells. Regions (rows) were defined as those enriched in ChIP-seq reads for, at least, one cell line. Color keys indicating signals normalized to reads per million. The percentage overlap of ChIP-seq peaks in CHP134 and Kelly cells compared with that in BE2C cells are indicated underneath the heatmaps. (B) Annotation of LIN28B ChIP-seq peaks identified in BE2C cells. (C) Representative ChIP-seq tracks for LIN28B at two different promoter regions in BE2C, CHP134, and Kelly cells. ChIP-seq read densities are plotted as the number of reads per million reads sequenced for each sample. Regions with significant LIN28B binding are denoted by red bars underneath each of the ChIP-seq tracks. (D) Heatmaps and metagenes of genome-wide co-occupancy for LIN28B, POLR2A, H3K4me3, and H3K27ac as determined by ChIP-seq in BE2C cells. Regions (rows) spanning 2 kb from each side of the TSS are ranked by the strength of the LIN28B signal therein. Color keys indicating signals normalized to reads per million are displayed below each heatmap. (E) The MEME-ChIP program was used to create a position weight matrix depicting the top predicted LIN28B DNA-binding motif in BE2C cells. The distribution of the motif around the ChIP-seq peaks is shown. (F) Heatmaps (using same parameters as in D) of genome-wide co-occupancy for LIN28B and ZNF143 as determined by ChIP-seq in BE2C cells. (G) The MEME-ChIP program was used to create a position weight matrix depicting the top predicted ZNF143 DNA-binding motif in BE2C cells. The distribution of the motif around the ChIP-seq peaks is shown. (H) Reciprocal co-IP experiments were performed using BE2C nuclear lysates to test for interactions between LIN28B and ZNF143. IgG, isotype control IgG antibody. (I) BE2C-TET cells were untreated or treated with 50-ng/mL doxycycline for 3 d. Co-IP experiments were performed using nuclear lysates to test for interactions between ZNF143 and either Flag-LIN28B_WT or Flag-LIN28B_MU. (J) BE2C cells expressing doxycycline-inducible short hairpin RNAs (shRNAs), including control luciferase (shLuc), LIN28B (shLIN28B no. 1 and no. 4) and ZNF143 (shZNF143 no. 1 and no. 3), were treated with 1.5-µg/mL doxycycline for 6 d. The expression of LIN28B and ZNF143 was then analyzed by Western blot using chromatin-bound protein extracts. Histone H3 was used as a loading control. The positions of molecular weight markers are indicated for H–J.
Further analysis showed that more than 70% of the genomic regions with specific enrichment of LIN28B are located at promoter-proximal regions within 1 kilobase (kb) from the transcription start site (TSS) in all three cell lines, indicating that LIN28B could be involved in the transcriptional regulation of the associated genes (Fig. 4 B and C and SI Appendix, Fig. S5 D–F). In addition, RNA polymerase II/POLR2A and histone marks that are associated with active transcription, such as H3K4me3 and H3K27ac, are highly enriched at LIN28B-binding sites (Fig. 4D and SI Appendix, Fig. S5G). Motif analysis using the MEME-ChIP and HOMER programs predicted a major bipartite consensus sequence 5′-TGGGA-RW-TGTAGTYC-3′ with a distribution that is centered near the summits of peak enrichment in all three cell lines (Fig. 4E and SI Appendix, Table S2). Gene ontology term analysis using Enrichr (33) revealed that LIN28B-bound genes in BE2C, CHP134, and Kelly cells are significantly enriched for genes that function in the regulation of transcription, RNA binding, and mRNA splicing (SI Appendix, Fig. S5 H and I), indicating that LIN28B may regulate multiple aspects of RNA metabolism through transcriptional activation in addition to its well-known roles in suppression of miRNA processing (19) and direct binding to messenger RNAs with effects on splicing, stability, and translation (34).
To determine how LIN28B is localized to specific genomic regions, we performed a motif enrichment analysis (35) and found that a ZNF143-binding motif (36) is significantly enriched in the top 500 LIN28B ChIP-seq peaks (AUROC = 0.97 and P < 0.001), present in 94% compared to 2% of a genomically matched reference set and centered around the peak summits in BE2C cells. We, therefore, performed ChIP-seq with an anti-ZNF143 antibody in BE2C cells and found that LIN28B binds coordinately with ZNF143 to promoter regions (Fig. 4F). The ZNF143-binding motif predicted by the MEME-ChIP program revealed the consensus sequence 5′-RKGCMTKC-TGGGA-RW-TGTAGT-3′ (Fig. 4G), which is highly similar to the consensus sequence for LIN28B (Fig. 4E). We also demonstrated a protein–protein interaction between endogenous LIN28B and ZNF143 proteins in BE2C cells by reciprocal coimmunoprecipitation (co-IP) experiments (Fig. 4H and SI Appendix, Fig. S6A). ZNF143 also interacts with exogenously expressed WT or mutant LIN28B in BE2C cells (Fig. 4I and SI Appendix, Fig. S6B). Furthermore, Western blotting of chromatin-bound protein extracts demonstrated that while a doxycycline-inducible knockdown of LIN28B did not affect ZNF143 binding to chromatin, a doxycycline-inducible knockdown of ZNF143 decreased LIN28B binding to chromatin (Fig. 4J and SI Appendix, Fig. S6C). These data indicate that LIN28B is predominantly enriched in genomic regions located near gene promoters due to its specific association with the zinc-finger transcription factor ZNF143.
WT and Mutant LIN28B Activate Transcriptional Programs that Promote Neuroblastoma Pathogenesis.
To understand how LIN28B promotes the development of MYCN-induced neuroblastoma, we sought to identify direct or indirect target genes with an expression that is significantly altered due to LIN28B binding to ZNF143 within gene-specific promoter regions. BE2C-TET cell lines engineered to express Flag-tagged LIN28B_WT or LIN28B_MU were treated with doxycycline for 3 d, and samples were analyzed by ChIP-seq and RNA sequencing (RNA-seq). ChIP-seq with an anti-Flag antibody revealed that both LIN28B_WT and LIN28B_MU share similar genome-wide occupancy at the promoter regions as the endogenous LIN28B (Fig. 5 A and B). Immunofluorescence staining showed that LIN28B_WT is distributed to both the cytoplasm and the nucleus, while LIN28B_MU is predominantly found in the nucleus (Fig. 5C). These observations are consistent with our findings in zebrafish (SI Appendix, Fig. S2 T and X). RNA-seq demonstrated that 76.5% of the differentially expressed genes (15,251 of 19,929) were either co-up-regulated or co-down-regulated by LIN28B_WT and LIN28B_MU compared to untreated controls (Fig. 5D). Moreover, there was a large overlap between genes that were differentially expressed (P < 0.01) in LIN28B_WT- and LIN28B_MU-expressing cells (Fig. 5 E and F). For the genes that were co-up-regulated or co-down-regulated by LIN28B_WT and LIN28B_MU, most of the LIN28B-bound genes were also bound by ZNF143 (Fig. 5 E and F), consistent with a role for ZNF143 in localizing LIN28B within the genome. Furthermore, genes up-regulated by LIN28B_WT only but not those up-regulated by LIN28B_MU only were enriched for let-7 targets (SI Appendix, Fig. S7A), indicating that the LIN28B mutant is unable to inhibit let-7 processing. Gene set enrichment analysis (GSEA) revealed that a subset of common gene signatures related to tumor metastasis and the epithelial-to-mesenchymal transition were enriched in LIN28B WT- and mutant-expressing BE2C-TET cells (SI Appendix, Fig. S7B), while gene signatures involved in cell adhesion were negatively enriched (SI Appendix, Fig. S7C). These data suggest that LIN28B activates transcriptional programs that drive tumor metastasis in a let-7–independent manner.
Fig. 5.
WT and mutant LIN28B show similar genome-wide binding profiles and share a subset of common downstream targets. (A) Metagenes and heatmaps of genome-wide co-occupancy for endogenous LIN28B in BE2C cells as well as LIN28B_WT and LIN28B_MU in BE2C-TET cells treated with 50-ng/mL doxycycline for 3 d as determined by ChIP-seq. See Fig. 4D legend for detail. (B) Representative ChIP-seq tracks for H3K27ac, endogenous LIN28B and ZNF143, ATAC-seq tracks in BE2C cells, as well as ChIP-seq tracks for Flag-LIN28B_WT and Flag-LIN28B_MU in BE2C-TET cells treated with 50-ng/mL doxycycline for 3 d. See Fig. 4C legend for details. (C) Immunostaining images of Flag-LIN28B_WT/MU (green) in BE2C-TET cells treated with 50-ng/mL doxycycline for 3 d. Nuclei were stained with DAPI (blue). (Scale bar, 10 μm.) (D) Fold change (log2 of yDox/nDox) in gene expression following overexpression of LIN28B_WT or LIN28B_MU in BE2C-TET cells treated with doxycycline (yDox) versus untreated cells (nDox). Gene numbers within each group are indicated. (E and F) Venn diagrams of genes that were significantly (P < 0.01) up-regulated (E) or down-regulated (F) in doxycycline-treated LIN28B_WT or LIN28B_MU expressing BE2C-TET cells compared to untreated controls. (G) GSEA to determine the correlation of DNA binding with gene expression changes upon overexpression of LIN28B_WT or LIN28B_MU in BE2C-TET cells. The GSEA plot indicates the degree to which LIN28B and ZNF143 cobound genes are overrepresented at the extreme left (up-regulated by overexpression) or right (down-regulated by overexpression) of the entire ranked list. Solid bars represent LIN28B-bound genes. The green shading indicates the leading edge subset of genes. o/e, overexpression. (H) Heatmaps depicting the relative expression levels of high-confidence let-7–independent LIN28B target genes in BE2C-TET cells either untreated (nDox) or treated (yDox) with doxycycline. Each row corresponds to a gene, and signal intensity is normalized across the row. Genes are ranked by P value with the lowest at the top.
To further analyze the individual genes that are regulated by LIN28B, we performed GSEA using all of the LIN28B and ZNF143 cobound genes in BE2C cells as a gene set and found that they were more likely to be up-regulated upon LIN28B_WT or LIN28B_MU overexpression (P = 1.04e-21 for LIN28B_WT and P = 4.58e-14 for LIN28B_MU by the Kolmogorov–Smirnov test) (Fig. 5G). This finding suggests that LIN28B acts more frequently as a transcriptional activator than a repressor. To identify high-confidence let-7–independent LIN28B target genes (n = 508; Fig. 5H and SI Appendix, Table S3), we selected LIN28B and ZNF143 cobound genes that show significant changes in expression in response to overexpression of WT and mutant LIN28B (P < 0.01 with an absolute log2-fold-change ≥ 0.2 for both). Interestingly, many of these LIN28B-regulated genes are selective neuroblastoma gene dependencies and transcription factors that comprise the adrenergic core regulatory circuitry, including ISL1, PHOX2A, and HAND2 (7). Other directly up-regulated genes have been shown to regulate neural crest development and cell lineage specification, such as GSK3B, DBH, and HDAC9 (37–39), and several are associated with tumor metastasis, including GSK3B, L1CAM, and ETV1 (37, 40–43). To validate these findings, we analyzed both human and zebrafish neuroblastoma cells by qRT-PCR. We detected increased expression of GSK3B, L1CAM (zebrafish homolog l1.1) and ETV1 in BE2C-TET cells overexpressing LIN28B_WT or LIN28B_MU (Fig. 6A) as well as in tumors derived from the LIN28B_WT;MYCN or LIN28B_MU;MYCN fish compared to EGFP;MYCN-derived tumors (Fig. 6 B–D). These data suggest that aberrant overexpression of LIN28B induces the expression of key mediators of neuroblastoma metastasis.
Fig. 6.
Expression of WT or mutant LIN28B reverses the cell invasion and migration defects induced by GSK3B inhibition. (A) qRT-PCR to detect LIN28B, GSK3B, L1CAM, and ETV1 expression in BE2C-TET cells that were either untreated or treated with 50-ng/mL doxycycline for 3 d. Values were normalized to EEF1A1, ACTB, and B2M and represent the means ± SD of triplicate experiments. (B–D) The expression of zebrafish gsk3b (B), l1.1 (C) and etv1 (D) mRNAs in EGFP;MYCN (M), LIN28B_WT;MYCN (WT), and LIN28B_MU;MYCN (MU) tumors was analyzed by qRT-PCR. Five independent tumors from 6-mo-old fish were analyzed for each group. Each dot represents the expression levels of the indicated gene normalized to EEF1A1. Horizontal bars indicate means ± SD. (E) The LIN28B- and ZNF143-binding sequences within the GSK3B promoter region were cloned upstream of the firefly luciferase reporter gene and transfected into BE2C or BE2C-TET cells expressing Flag-tagged LIN28B_WT (WT) or LIN28B_MU (MU) that were untreated or treated with doxycycline. A reporter construct encoding renilla luciferase under control of the cytomegalovirus promoter was cotransfected to normalize firefly luciferase values. Luciferase activities were measured 48-h posttransfection and expressed as a ratio of the cells treated with doxycycline to untreated controls. Values represent means ± SEM of triplicate experiments. (F and G) Transwell migration and invasion assay of the BE2C-TET cells untreated or treated with 50-ng/mL doxycycline or 25-μM GSK3B inhibitor SB415286. Cells were stained with 0.1% crystal violet (F), and the number of migrated cells through the membrane per field was plotted as means ± SD of triplicate experiments (G). (Scale bar, 100 µm.) All of the statistical analyses were performed using the two-tailed unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Previous studies have shown that GSK3B promotes cell migration during neural crest development and cancer cell progression and invasion (37, 40, 41). An analysis of publicly available datasets revealed that GSK3B is highly expressed in neuroblastoma (SI Appendix, Fig. S8A) and is positively correlated with the expression of LIN28B (SI Appendix, Fig. S8 B and C). We also found that the 50 most significantly down-regulated genes upon treatment of IMR32 neuroblastoma cells with the GSK3B inhibitor BIO-acetoxime (44) were enriched in LIN28B WT- or mutant-expressing BE2C-TET cells compared to controls (SI Appendix, Fig. S8D), suggesting that GSK3B is an important target of LIN28B. To test the possibility that LIN28B interacts via protein–protein binding with ZNF143 at the GSK3B promoter, we cloned the putative consensus sequence upstream of a firefly luciferase reporter gene and transfected the construct into BE2C-TET cells. We observed an ∼1.4-fold increase in reporter activity in response to overexpression of WT or mutant Flag-LIN28B compared to the BE2C parent cells (Fig. 6E), suggesting that GSK3B expression is activated by LIN28B through its interaction with ZNF143. To evaluate whether GSK3B functions in neuroblastoma-cell invasion and migration, we treated BE2C-TET cells with a small molecule inhibitor of GSK3B known as SB415286 (45). In the absence of doxycycline, SB415286 treatment robustly inhibited invasion and migration. The addition of doxycycline in cells engineered to overexpress WT or mutant LIN28B was able to partially rescue this phenotype, presumably through up-regulation of GSK3B (Fig. 6 F and G). Our results, therefore, indicate that GSK3B is an important let-7–independent transcriptional target of LIN28B that contributes to neuroblastoma invasion and migration.
Discussion
LIN28s have previously been shown to influence development, metabolism, and disease processes in a manner that can be either dependent or independent of their effects on let-7 processing. LIN28A is normally substantially down-regulated upon the differentiation of mouse P19 neuronal cells into glial cells, but constitutive expression of either WT or a mutated version of LIN28A that fails to suppress let-7 maturation completely blocks neurogliogenesis (22). Furthermore, let-7–dependent and let-7–independent functions of LIN28 synergize to regulate glucose metabolism and tissue repair (46, 47). In neuroblastomas, LIN28B coordinates expression of the oncogenes RAN and AURKA through both let-7–dependent and let-7–independent mechanisms (21). High LIN28B expression is one of multiple mechanisms that disrupt the let-7 and, consequently, results in elevated MYCN protein expression to promote neuroblastoma pathogenesis in mice, at least, in part, through suppression of let-7 (10, 11). In our zebrafish model, MYCN cDNA lacking the 3′ untranslated region is overexpressed in the PSNS under the dβh promoter so that direct effects of LIN28B on let-7 processing would not affect MYCN protein levels. We find that both WT and let-7-suppression–deficient LIN28B promote MYCN-induced neuroblastoma by a mechanism that does not require suppression of let-7 but, instead, is due to LIN28B-mediated up-regulated expression of genes that are essential for neuroblastoma cell survival and migration. LIN28B is recruited to the promoters of these genes not through an intrinsic DNA binding but rather through protein–protein interactions with the zinc-finger transcription factor ZNF143. In the future, it will be important to determine how the loss of function of LIN28B affects both global gene expression and the expression of ZNF143 target genes in MYCN-amplified neuroblastoma.
The adrenergic neuroblastoma CRC contributes to the formation of hundreds of superenhancers at additional transcriptional regulators and genes critical for neuroblastoma cell identity (7). The ERN is composed of the downstream genes regulated by superenhancers and the CRC transcriptional factors (32). The proteins encoded by these genes are not necessarily transcription factors, do not bind to the superenhancers of the CRC genes, or are not likely autoregulatory. Thus, genes regulated by superenhancers that are bound by the CRC proteins can be either CRC or ERN genes, depending on whether their products bind to the CRC gene enhancers and autoregulate themselves and the other CRC members. In this context, while the LIN28B superenhancer is driven by the CRC, LIN28B itself is not part of the CRC as it does not bind through ZNF143 to the enhancers of all of the CRC genes and, thus, does not reinforce their expression. Rather, LIN28B facilitates the expression of its own targets (e.g., GSK3B and L1CAM). While LIN28B is not a CRC gene, its status as a superenhancer-driven downstream target gene of the CRC proteins qualifies it as a member of the ERN.
The CSD found in LIN28A and LIN28B is thought to be one of the most ancient nucleic acid-binding domains in existence, and all CSD-containing proteins bind to RNA (48). The CCHC zinc-finger domain, a prominent feature of both LIN28A and LIN28B, is also found in many RNA-binding proteins (48). Here, we show that point mutations of the CSD and CCHC domains in LIN28B abrogate its ability to block let-7 processing but do not dramatically affect the transcriptional program induced by LIN28B, indicating that these mutations are, therefore, likely to be dispensable for LIN28B’s ability to regulate transcription. Instead, we found that both WT and mutant LIN28B bind to chromatin through binding to the zinc-finger transcription factor ZNF143 (Fig. 4 H–J). ZNF143 has been shown to be a regulator of chromatin loops and to secure chromatin interactions at gene promoters (36, 49), suggesting that LIN28B acts as a cofactor with ZNF143 to regulate chromatin looping and gene expression.
LIN28B overexpression has been shown to promote colon cancer and pancreatic ductal adenocarcinoma cell migration and invasion and is correlated with reduced patient survival and increased tumor recurrence (14, 15, 50). A recent study in zebrafish showed that overexpression of LIN28B increases neural crest cell migration and supports neuroblastoma onset (23). Indeed, our model showed neuroblastomas arising in both LIN28B_WT;MYCN and LIN28B_MU;MYCN transgenic fish exhibited a clear increase in tissue invasion and distant metastases compared to tumors arising from the MYCN-only fish (Fig. 2). Our paper has also revealed that LIN28B activates a subset of genes that have previously been shown to promote tumor metastasis, including GSK3B, L1CAM, and ETV1 (37, 40–43). Consistent with previous studies implicating a role for GSK3 in controlling migration of the neural crest lineage (37, 51) and cancer cell migration and invasion (41), our data suggest that GSK3B serves as an important target of ZNF143 and its cofactor LIN28B, whose up-regulated activity promotes the migration and invasion of neuroblastoma cells. However, many questions remain unanswered. For example, what are the critical residues that mediate LIN28B binding to ZNF143? Does LIN28B form transcriptional complexes with other factors? How does the cell coordinate both the let-7–dependent and let-7–independent functions of LIN28B? The resolution of these questions will undoubtedly advance our understanding of the important and diverse roles of LIN28B in normal development and malignant transformation.
Materials and Methods
A detailed description of zebrafish lines and maintenance, tumor watch, nucleic acid, and protein analysis methods, tissue processing and staining, cell culture and analysis methods, ChIP-seq and analysis methods, and RNA-seq and analysis methods are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Cicely Jette for an editorial review of the paper and critical comments, Grace Thurston, Daniel Debiasi, Michael T. Coute, and Monica Alves for their excellent care of the zebrafish, and Z. Herbert of the Dana-Farber Molecular Biology Core Facility for genomics support. This work was supported by a NIH Grant R01-CA180692 (A.T.L.), an Innovation Award from Alex’s Lemonade Stand Foundation (A.T.L.), a NIH Grant R21-HG009268 (M.L.B.), the Hope Funds for Cancer Research (B.J.A.), the American Lebanese Syrian Associated Charities (B.J.A.), and a research grant from the Rally Foundation for Childhood Cancer Research and Open Hands Overflowing Hearts (T.T.). A.D.D. was supported by funding from the Damon-Runyon Cancer Research Foundation (Grant DRSG-24-18), the Alex’s Lemonade Stand Foundation, and the CureSearch for Children’s Cancer Foundation. B.J.A. is the Hope Funds for Cancer Research Grillo-Marxuach Family Fellow.
Footnotes
Competing interest statement: B.J.A. is a shareholder in Syros Pharmaceuticals, Inc. The other authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/(accession no. GSE138743).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922692117/-/DCSupplemental.
References
- 1.Cheung N. K., Dyer M. A., Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 13, 397–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang M., Weiss W. A., Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 3, a014415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DuBois S. G. et al., Metastatic sites in stage IV and IVS neuroblastoma correlate with age, tumor biology, and survival. J. Pediatr. Hematol. Oncol. 21, 181–189 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Harreld J. H. et al., Orbital metastasis is associated with decreased survival in stage M neuroblastoma. Pediatr. Blood Cancer 63, 627–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith S. J., Diehl N. N., Smith B. D., Mohney B. G., Incidence, ocular manifestations, and survival in children with neuroblastoma: A population-based study. Am. J. Ophthalmol. 149, 677–682.e2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diskin S. J. et al., Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 44, 1126–1130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin A. D. et al., Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat. Genet. 50, 1240–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J. et al., Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Beckers A. et al., MYCN-driven regulatory mechanisms controlling LIN28B in neuroblastoma. Cancer Lett. 366, 123–132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molenaar J. J. et al., LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 44, 1199–1206 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Powers J. T. et al., Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 535, 246–251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urbach A. et al., Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 28, 971–982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen L. H. et al., Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 26, 248–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King C. E. et al., LIN28B promotes colon cancer progression and metastasis. Cancer Res. 71, 4260–4268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King C. E. et al., LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 30, 4185–4193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beachy S. H. et al., Enforced expression of Lin28b leads to impaired T-cell development, release of inflammatory cytokines, and peripheral T-cell lymphoma. Blood 120, 1048–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helsmoortel H. H. et al., LIN28B is over-expressed in specific subtypes of pediatric leukemia and regulates lncRNA H19. Haematologica 101, e240–e244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray M. J. et al.; CCLG , LIN28 Expression in malignant germ cell tumors downregulates let-7 and increases oncogene levels. Cancer Res. 73, 4872–4884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piskounova E. et al., Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147, 1066–1079 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viswanathan S. R., Daley G. Q., Gregory R. I., Selective blockade of microRNA processing by Lin28. Science 320, 97–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnepp R. W. et al., A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell 28, 599–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balzer E., Heine C., Jiang Q., Lee V. M., Moss E. G., LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 137, 891–900 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Corallo D. et al., LIN28B increases neural crest cell migration and leads to transformation of trunk sympathoadrenal precursors. Cell Death Differ. 27, 1225–1242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hennchen M. et al., Lin28B and let-7 in the control of sympathetic neurogenesis and neuroblastoma development. J. Neurosci. 35, 16531–16544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balzer E., Moss E. G., Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 4, 16–25 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Langenau D. M. et al., Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene 27, 4242–4248 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu S. et al., Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 21, 362–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menke A. L., Spitsbergen J. M., Wolterbeek A. P., Woutersen R. A., Normal anatomy and histology of the adult zebrafish. Toxicol. Pathol. 39, 759–775 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Bradner J. E., Hnisz D., Young R. A., Transcriptional addiction in cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boeva V. et al., Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 49, 1408–1413 (2017). [DOI] [PubMed] [Google Scholar]
- 31.van Groningen T. et al., Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 49, 1261–1266 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Saint-André V. et al., Models of human core transcriptional regulatory circuitries. Genome Res. 26, 385–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuleshov M. V. et al., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilbert M. L. et al., LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell 48, 195–206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mariani L., Weinand K., Vedenko A., Barrera L. A., Bulyk M. L., Identification of human lineage-specific transcriptional coregulators enabled by a glossary of binding modules and tunable genomic backgrounds. Cell Syst. 5, 187–201.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey S. D. et al., ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat. Commun. 2, 6186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez Malagon S. G. et al., Glycogen synthase kinase 3 controls migration of the neural crest lineage in mouse and Xenopus. Nat. Commun. 9, 1126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu N., Strobl-Mazzulla P. H., Bronner M. E., Epigenetic regulation in neural crest development. Dev. Biol. 396, 159–168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huber K., The sympathoadrenal cell lineage: Specification, diversification, and new perspectives. Dev. Biol. 298, 335–343 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Chikano Y. et al., Glycogen synthase kinase 3β sustains invasion of glioblastoma via the focal adhesion kinase, Rac1, and c-Jun N-terminal kinase-mediated pathway. Mol. Cancer Ther. 14, 564–574 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Domoto T. et al., Glycogen synthase kinase-3β is a pivotal mediator of cancer invasion and resistance to therapy. Cancer Sci. 107, 1363–1372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh S., Shin S., Janknecht R., ETV1, 4 and 5: An oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta 1826, 1–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raveh S., Gavert N., Ben-Ze’ev A., L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett. 282, 137–145 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Duffy D. J., Krstic A., Schwarzl T., Higgins D. G., Kolch W., GSK3 inhibitors regulate MYCN mRNA levels and reduce neuroblastoma cell viability through multiple mechanisms, including p53 and Wnt signaling. Mol. Cancer Ther. 13, 454–467 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Forde J. E., Dale T. C., Glycogen synthase kinase 3: A key regulator of cellular fate. Cell. Mol. Life Sci. 64, 1930–1944 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu H. et al.; DIAGRAM Consortium; MAGIC Investigators , The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shyh-Chang N. et al., Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155, 778–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudson W. H., Ortlund E. A., The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 15, 749–760 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen Z., Huang Z. T., Zhang R., Peng C., ZNF143 is a regulator of chromatin loop. Cell Biol. Toxicol. 34, 471–478 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Wang Y. et al., Lin28B facilitates the progression and metastasis of pancreatic ductal adenocarcinoma. Oncotarget 8, 60414–60428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hur E. M., Zhou F. Q., GSK3 signalling in neural development. Nat. Rev. Neurosci. 11, 539–551 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.