Significance
Low availability of nitrogen (N), mainly nitrate in aerobic soils, is a primary limiting factor for crop production. Most terrestrial plants live in symbiosis with arbuscular mycorrhizal (AM) fungi to increase nutrient uptake, including N, from soil. Research on the AM symbiosis field has focused almost exclusively on ammonium as the form of N transferred to the plants, and there has been no direct evidence of N transfer as nitrate thus far. Here, we report that mycorrhizal rice could receive more than 40% of its N via the mycorrhizal pathway and that the AM-specific nitrate transporter OsNPF4.5 accounted for approximately 45% of the mycorrhizal nitrate uptake. Our work suggests the presence of a mycorrhizal route for nitrate uptake in plants.
Keywords: arbuscular mycorrhiza, RNA sequencing, nitrate transporter, nitrogen uptake, OsNPF4.5
Abstract
Low availability of nitrogen (N) is often a major limiting factor to crop yield in most nutrient-poor soils. Arbuscular mycorrhizal (AM) fungi are beneficial symbionts of most land plants that enhance plant nutrient uptake, particularly of phosphate. A growing number of reports point to the substantially increased N accumulation in many mycorrhizal plants; however, the contribution of AM symbiosis to plant N nutrition and the mechanisms underlying the AM-mediated N acquisition are still in the early stages of being understood. Here, we report that inoculation with AM fungus Rhizophagus irregularis remarkably promoted rice (Oryza sativa) growth and N acquisition, and about 42% of the overall N acquired by rice roots could be delivered via the symbiotic route under N-NO3− supply condition. Mycorrhizal colonization strongly induced expression of the putative nitrate transporter gene OsNPF4.5 in rice roots, and its orthologs ZmNPF4.5 in Zea mays and SbNPF4.5 in Sorghum bicolor. OsNPF4.5 is exclusively expressed in the cells containing arbuscules and displayed a low-affinity NO3− transport activity when expressed in Xenopus laevis oocytes. Moreover, knockout of OsNPF4.5 resulted in a 45% decrease in symbiotic N uptake and a significant reduction in arbuscule incidence when NO3− was supplied as an N source. Based on our results, we propose that the NPF4.5 plays a key role in mycorrhizal NO3− acquisition, a symbiotic N uptake route that might be highly conserved in gramineous species.
In a natural soil ecosystem, the majority of land plants can form mutualistic symbiosis with arbuscular mycorrhizal (AM) fungi of Glomeromycotina to better adapt to limited nutrient supplies (1). AM association is an endosymbiotic process that requires the differentiation of both symbionts to create novel contact interfaces within the cells of plant roots. In the AM symbiosis, the fungal hyphae penetrate the root epidermis, grow through the intercellular spaces of the root, and subsequently invade cortical cells, developing highly branched tree-like structures called arbuscules (2). Cortical cells develop a specialized membrane, the periarbuscular membrane (PAM), to envelop each branching hypha to separate the fungus from the plant cell cytoplasm, resulting in an extensive plant–fungal interface specialized for nutrient exchange (3). Upon the formation of AM symbiosis, mycorrhizal plants have two pathways for nutrient uptake, either direct uptake from the soil via root hairs and root epidermis or indirectly through the AM fungal hyphae at the plant–fungus interface. It has been demonstrated that AM fungi dominates Pi uptake in symbiotic plants (4, 5).
Nitrogen (N) is the most important nutrient for plant growth and development. The primary forms of N absorbed by plant roots are nitrate (NO3−) in aerobic upland soil and ammonium (NH4+) in flooding soil. An increasing number of reports suggest that AM fungi can take up both NO3− and NH4+, as well as organic N forms, from the surrounding soils (6–13). Although N transfer in the AM symbiosis has been receiving increasing attention (10), the mechanism underlying the AM-mediated N acquisition pathway remains largely unknown. Current data propose that, once N has been transported into the fungal cytoplasm, it is assimilated into arginine (10), translocated probably together with Poly-P through the intraradical hyphae, and, after hydrolysis in the arbuscule, NH4+ is exported from the AM fungus to the periarbuscular space (12). The import of NH4+ across the PAM, probably in the form of NH3, into the root cell is then mediated by plant NH4+ transporters (AMTs). In some mycorrhizal plants living in aerobic environments examined so far, such as Medicago truncatula, Lotus japonicus, Glycine max, and Sorghum bicolor, two to five AMT transporters were found to be specifically expressed or strongly up-regulated in mycorrhizal roots (11, 14–16). Immunolocalization evidence showed that two mycorrhiza-induced AMTs, GmAMT4.1 (14) and SbAMT3.1 (11), from G. max and S. bicolor, respectively, localize exclusively on the PAM, strongly suggesting the existence of a symbiotic NH4+ uptake pathway at least in these plant species. Nonetheless, AM association occurs preferably in aerobic soil condition, in which NO3− is the major form of inorganic N, due to rapidly nitrification of NH4+ (17). Therefore, it is possible that a symbiotic pathway for NO3− uptake that could be more important and/or prevalent than the mycorrhizal NH4+ uptake route exists, at least in some plant species. Consistent with this notion, previous studies, through transcriptome hunting, have showed the presence of putative NO3− transporter genes with AM-induced expression in several plant species (18, 19). However, it is still unclear whether NO3− could be directly translocated from the extraradical hyphae to the fungal structures within roots and whether NO3− could be directly transferred across the intraradical symbiotic interface into the root cells. This lack of knowledge restricts our understanding regarding both the global N underground movement and the nutrient exchange capacity of what is arguably the world’s most ancient, widespread, and important symbiosis.
Rice (Oryza sativa), a semiaquatic crop plant that can grow in both flooding paddy and upland conditions, is one of the most important food crops worldwide. As with most vascular flowering plants, rice has also inherited the capacity to be well colonized by AM fungi under aerobic growth conditions. Moreover, evidence from different research groups showed enhanced biomass production of rice plants inoculated with AM fungi (12, 20, 21). Because of the availability of technology to produce gene knockouts and overexpressing lines of specific genes, rice is a good model system to study the role of mycorrhizal N uptake routes on plant growth and the symbiotic interaction. Here we report that about 42% of the overall N acquired by rice roots could be delivered via the symbiotic route under N-NO3− supply conditions, in which the mycorrhizal root-specific OsNPF4.5 nitrate transporter plays a crucial role. We also report that repressing NO3− transport across the intraradical symbiotic interface in loss-of-function osnpf4.5 mutants decreases AM colonization efficiency and reduces arbuscule incidence.
Results and Discussion
RNA Sequencing Uncovered the Up-Regulation of Multiple Genes Involved in Nitrate Transport and Metabolism in Mycorrhizal Rice Plants.
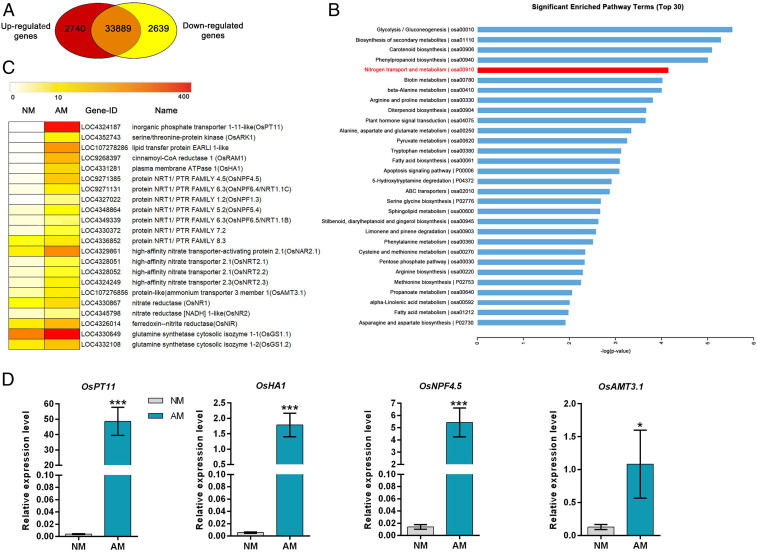
To gain an overview of rice transcriptional responses to AM fungal colonization, an Illumina HiSeq 2500 sequencing platform was used to conduct high-throughput RNA-seq analysis of both mycorrhizal and nonmycorrhizal roots collected from wild-type rice plants (O. sativa cv. Nipponbare) inoculated or mock-inoculated with Rhizophagus irregularis for 6 wk (22). Differentially expressed genes (DEGs) between the two treatments were identified applying a P value < 0.05 and a twofold change threshold. RNA-seq analysis revealed a total of 5,379 DEGs, of which 2,740 genes were up-regulated (Dataset S1) and 2,639 genes were down-regulated in the rice mycorrhizal roots, whereas 33,889 genes did not show significant alteration in transcript levels (Fig. 1A). To better understand the potential functions of these DEGs and their related biological processes, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed for up-regulated genes (Fig. 1B). The Glycolysis/Gluconeogenesis pathway was found to be the most significantly enriched pathway, followed by pathways for biosynthesis of secondary metabolites, carotenoid biosynthesis, and phenylpropanoid biosynthesis. Interestingly, the N metabolism pathway was identified as the fifth most predominant enriched pathway in the KEGG analysis (Fig. 1B), with a ranking higher than the pathway of fatty acid biosynthesis. Several components involved in fatty acid biosynthesis and transport have been shown to be highly up-regulated in the AM fungal-colonized roots, and play an essential role in maintaining AM symbiosis through modulating lipid export from the host plant to AM fungi (3, 23–27).
Fig. 1.
RNA sequencing analysis of the rice mycorrhizal and nonmycorrhizal roots. (A) Venn diagram showing the relationships between genes that show statistically significant differential expression in response to AM symbiosis in roots. The up-regulated genes are shown in red color, while the down-regulated genes are indicated in yellow color. The genes with no significant alteration in transcripts are shown in the intersection. (B) The 30 most significantly enriched pathways analyzed by KEGG algorithm. (C) A heat map of the up-regulated genes involved in nitrogen transport and metabolism, as well as several previously described AM–up-regulated genes that were shown as marker genes. (D) Quantitative RT-PCR analysis showed a more than 500-fold up-regulation of OsNPF4.5 and an 11-fold up-regulation of OsAMT3.1 in response to AM symbiosis. The AM-specific Pi transporter gene OsPT11 and H+-ATPase gene OsHA1 were used as control genes. The relative expression level of the assayed genes was normalized to a constitutive Actin gene. Values are the means ± SE of three biological replicates (n = 3). The asterisks indicate significant differences (*P < 0.05; **P < 0.01, ***P < 0.001).
Careful scrutiny of the DEGs uncovered the substantial up-regulation (2 to 500 fold) of 14 genes involved in NO3− transport and metabolism in rice mycorrhizal roots. Ten of these genes encode putative nitrate transporters from the NRT1/NPF and NRT2 families, of which OsNPF4.5 was the strongest up-regulated gene from a barely detectable expression level in nonmycorrhizal roots. We found that OsNPF4.5, as a substantially AM-induced gene, could also be traced in a previously released microarray data of rice mycorrhizal roots (28), in which only 256 genes showing more than a threefold change were detected. The AM–up-regulated expression nature of some rice NPF genes, including OsNPF4.5, was confirmed in a recent study (29). Among the other four genes related to NO3− transport or metabolism, one encodes the high-affinity nitrate transporter-activating protein OsNAR2.1 (30, 31), and the remaining three encode two putative nitrate reductases (NR) and a nitrite reductase (NiR), respectively (Fig. 1C). Comparing our data (22) with the previously released microarray data of rice mycorrhizal roots (28), we found only one common DEG encoding a putative ammonium transporter OsAMT3.1 (32), with an 11-fold up-regulation in rice mycorrhizal roots. The previously described mycorrhiza-specific phosphate transporter gene OsPT11 (5, 33) and plasma membrane H+-ATPase gene OsHA1 (34), which were used as positive controls for the mycorrhiza-specific accumulation of transcripts, were strongly up-regulated in our transcriptome of rice mycorrhizal roots (Dataset S1). Quantitative RT-PCR (qRT-PCR) analysis of one of the two RNA preparations used for RNA-seq validated the transcriptome results regarding the mycorrhiza-inducible nature of all N transport- and metabolism-related DEGs (SI Appendix, Fig. S1) and confirmed that OsNPF4.5 was the strongest up-regulated putative nitrate transport gene, with the transcripts increased by over 500 fold in mycorrhizal roots relative to the mock control (Fig. 1D). These findings suggest the presence of a symbiotic pathway for nitrate uptake in the mycorrhizal rice plants.
AM Fungal Colonization Promotes Rice Growth and Nitrate Uptake.
To investigate the potential role of AM symbiosis in plant nitrate acquisition, rice plants were grown in a sand/soil mixture-based substrate inoculated or mock-inoculated with AM fungus (R. irregularis) and supplemented with 0.25, 1.0, 2.5, and 5.0 mM of NO3− as N sources. After 8 wk of growth, all mycorrhizal rice plants supplied with NO3− showed a statistically significant increase in root and shoot biomass and N and P accumulation in both shoots and roots compared with nonmycorrhizal plants except those supplied with 0.25 mM NO3−, which did not differ significantly in biomass with the mock-inoculated control plants (SI Appendix, Fig. S2). Our findings highlight that AM fungal colonization could promote rice plant growth and nitrate uptake. The lack of growth promotion observed in mycorrhizal rice plants supplemented with 0.25 mM NO3− might be partially ascribed to a relatively smaller shoot N increment and lower colonization efficiency and arbuscule incidence compared with those grown under high NO3− (SI Appendix, Fig. S3 A and B). It has been documented that the mycelium of AM fungi constitutes considerable N sink, and competition for N would potentially reduce N delivery and mycorrhizal benefits to the host plant under N-limited conditions, which may in turn lead to a negative effect on AM fungal colonization (12, 35). The reduced mycorrhization in low-NO3−–treated plants was confirmed by a decreased expression of the AM-specific marker gene OsPT11 (SI Appendix, Fig. S3C). The reduced colonization efficiency caused by low NO3− application was also observed in mycorrhizal sorghum plants (SI Appendix, Fig. S3 D–F). In contrast to high phosphate that is well known to inhibit the symbiotic process (5, 36), we found that high NO3− concentrations (5 mM) did not inhibit mycorrhization. These results suggest that phosphate but not nitrogen availability is the major signal that the plants perceive to activate or repress the AM symbiosis.
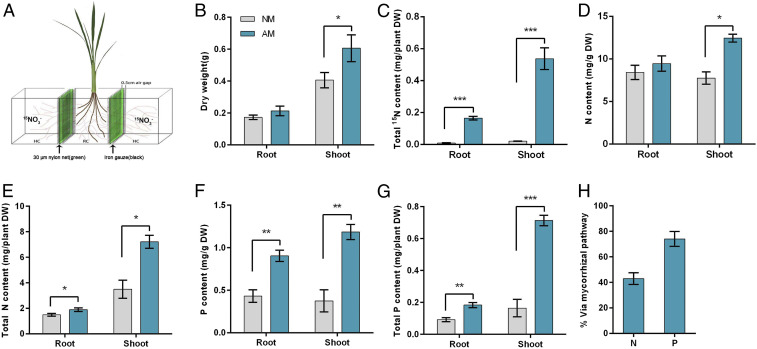
To further evaluate the contribution of symbiotic NO3− uptake to the overall N nutrition of the mycorrhizal rice plants, 15NO3−-labeled uptake measurement was performed using a compartmented growth system (Fig. 2A) containing a middle root/fungal compartment (RFC) that was separated by two 30-mm nylon meshes from two hyphal compartments (HCs) with a 0.5-cm air gap between the RFC and HC compartments to prevent 15NO3− diffusion (diagram in Fig. 2A) (37). Control and R. irregularis-inoculated rice seedlings were grown in the RFC compartment supplemented with 2.5 mM NO3− as sole N source, and an equal amount of 15NO3− was provided to the two HC compartments. The 15N, total N, and total P contents were determined in both roots and shoots of mock and mycorrhizal rice plants at 6 wk post inoculation (wpi). Mycorrhizal plants showed an increase of 49 ± 15% in shoot biomass (dry weight) compared with the nonmycorrhizal controls (Fig. 2B). High 15N accumulation was readily detectable in the roots and shoots of inoculated plants, but barely detectable in all of the mock-inoculated plants (Fig. 2C), indicating that fungal hyphae could reach and take up nutrients from HCs and that no NO3− diffusion across the nylon meshes occurred. Mycorrhizal plants also showed increases of 60 ± 8% in shoot N content and 106 ± 15% in total shoot N content per plant as compared to the controls (Fig. 2 D and E). We also found that mycorrhizal plants had a threefold increase in shoot P content and a fivefold increase in total shoot P content per plant over the control (Fig. 2 F and G). In contrast to P content that was significantly increased in the root of mycorrhizal plants, N content in the root did not differ significantly between mycorrhizal plants and mock-inoculated plants (Fig. 2 D–G), suggesting a more rapid transport of N than P from root to shoot in mycorrhizal plants. A determination of the percentage of N and P transferred via the mycorrhizal pathway showed that 42 ± 4% N and 74 ± 7% P was taken up via the mycorrhizal pathway (Fig. 2H). Our results on P uptake via the symbiotic pathway are similar to that of a previous report demonstrating that mycorrhizal rice received over 70% of its Pi via the symbiotic uptake pathway (5), suggesting that our experimental setup is adequate to measure the contribution of the mycorrhizal route on nutrient uptake. These findings highlight that, in addition to the mycorrhizal P uptake pathway, rice also activates an efficient route for symbiotic N acquisition upon the formation of AM symbiosis.
Fig. 2.
AM fungal colonization promotes rice growth and nitrate uptake. (A) A diagrammatic representation (not to scale) of the compartmented culture system used in the experiment. Two inoculated or mock-inoculated seedlings of WT or mutant plants were grown in the middle root/fungal compartment (RFC) and watered weekly with nutrient solution containing 2.5 mM NO3−. The hyphal compartments (HCs) aside were watered with nutrient solution containing an equal amount of 15NO3−. (B) Biomass of inoculated and mock-inoculated plants. (C) Assay of 15N content in both roots and shoots of inoculated and mock-inoculated plants. (D–G) N (D and E) and P (F and G) contents of inoculated and mock-inoculated plants. (H) The percentage of N and P transferred via the mycorrhizal pathway. Values are the means ± SE of five independent biological replicates (n = 5). The asterisks indicate significant differences (*P < 0.05; **P < 0.01, ***P < 0.001).
Identification and Characterization of the AM-Induced OsNPF4.5 in Rice.
The increased nitrate uptake of mycorrhizal rice plants prompts us to speculate that AM-induced nitrate transporter(s) might be required for nitrate uptake at the symbiotic interface. Since OsNPF4.5 is the gene encoding a putative nitrate transporter of the NRT1/NPF family with the highest up-regulated expression in mycorrhizal roots, we decided to further investigate its expression pattern and possible function. An attempt to clone the full-length open reading frame (ORF) of OsNPF4.5 based on the predicted online information (Os01g0748950/LOC_Os01g54515.1) was unsuccessful. Thus, RNA-based RACE-PCR (First Choice RLM-RACE Kit; Ambion) was employed to obtain a full-length cDNA of OsNPF4.5. By comparing the cDNA and its genomic DNA sequences, OsNPF4.5 was found to contain a 1,830-bp-length ORF separated by six introns (SI Appendix, Fig. S4A). As with most known plant NPF transporters (17, 38), OsNPF4.5 putatively harbors 12 transmembrane domains with an intracellular central loop (SI Appendix, Fig. S4B). Phylogenetic analysis grouped OsNPF4.5 and its orthologs together with several NPF homologs that have been evidenced to possess nitrate transport capacity, such as the rice OsNPF6.3 (39) and OsNPF6.5 (40) (SI Appendix, Fig. S4C). Overall comparison of the crystal structure of the well-known nitrate transporter AtNRT1.1/CHL1 (41) and the model structure of OsNPF4.5 revealed a high level of superposition between the two protein structures. The model structure of OsNPF4.5 suggests the presence of 12 transmembrane helices disposed in a similar orientation as those of AtNRT1.1 forming the NO3− transport tunnel, in which some important residues such as L49, V53, and K164, and the phosphorylation site T101 in AtNRT1.1 are also conserved in OsNPF4.5 (SI Appendix, Fig. S5). A sequence alignment of AtNRT1.1, OsNRT1.1, OsNPF4.5, and multiple OsNPF4.5 orthologs from diverse monocot and dicot plant species, and secondary structure assignment according to the OsNPF4.5 model and the AtNRT1.1 reported structure, showed that the 12 putatively transmembrane helices and the residues mentioned earlier are also highly conserved in OsNRT1.1, the rice ortholog of AtNRT1.1, and in the different OsNPF4.5 orthologs (SI Appendix, Fig. S6). However, some other residues forming part of the transport tunnel and the binding pocket in OsNPF4.5 are different from those present in equivalent positions in AtNRT1.1 and OsNRT1.1, such as L373, Q377, D499, and Y534 (in reference to OsNPF4.5 residue position), but highly conserved among NPF4.5 orthologs (SI Appendix, Figs. S5 and S6). Further experimentation is needed to determine if these residues are functional and biologically relevant as for the case of AtNRT1.1.
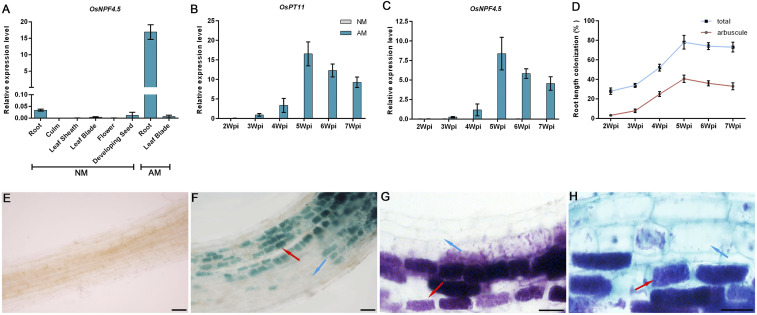
Besides mycorrhizal roots, OsNPF4.5 transcripts were barely detectable in other tissues, including culm, leaf sheath and blade, flower, and developing seeds (Fig. 3A). Unlike the known nitrate transporters, such as OsNPF6.3/NRT1.1A (39) and OsNPF6.5/NRT1.1B (40), having an inducible expression in response to NO3−, or even NH4+, supply, OsNPF4.5 showed no conspicuous response to external NO3− or NH4+ application or deprivation (SI Appendix, Fig. S7). A time-course expression analysis further revealed a similar kinetics of transcript accumulation between OsNPF4.5 and OsPT11 in rice mycorrhizal roots, with expression starting to be detected 3 wpi and reaching a maximum at 5 wpi in both cases (Fig. 3 B and C). The kinetic of expression of OsNPF4.5 and OsPT11 also correlated well with mycorrhizal colonization intensity (Fig. 3 B–D). To explore in more detail the expression pattern of OsNPF4.5, we constructed a transcriptional fusion between the promoter of this nitrate transporter and the coding sequence of the GUS reporter gene. Histochemical GUS assays confirmed that OsNPF4.5 expression was practically undetectable in nonmycorrhizal roots (Fig. 3E), whereas intense GUS staining was detected in mycorrhizal roots (Fig. 3 F and G). Colocalization of GUS expression and AM fungal structure by overlay of magenta-GUS with Trypan blue staining showed that the GUS activity driven by the OsNPF4.5 promoter was exclusively confined to cells containing arbuscules (Fig. 3H). Subcellular localization analysis showed that the eGFP-OsNPF4.5 fusion protein expressed under control of the 35S cauliflower mosaic virus promoter in Nicotiana benthamiana epidermal cells was exclusively localized to the plasma membrane (SI Appendix, Fig. S8A). Expression of the OsNPF4.5-eGFP fusion protein from its own promoter in mycorrhizal rice showed a distinct localization signal, likely the PAM, in arbuscule-containing cells (SI Appendix, Fig. S8B). These results confirm that the expression of OsNPF4.5 is specific in arbuscule-containing cells and that OsNPF4.5 is a membrane-localized protein probably present in the PAM upon AM symbiosis. Further studies are required to determine the precise subcellular localization of OsNPF4.5.
Fig. 3.
Tissue-specific expression assay of OsNPF4.5 in response to AM symbiosis. (A) Transcripts of OsNPF4.5 in different tissues of mycorrhizal (AM) and nonmycorrhizal (NM) plants. (B–D) Time-course expression of OsNPF4.5 and OsPT11 (used as a control) in rice mycorrhizal roots. (D) Quantification of AM fungal colonization at different sampling time points. (E and F) Histochemical GUS staining of rice roots expressing pOsNPF4.5::GUS in the absence (E) and presence (F) of inoculation. (G) Magenta-GUS staining of the mycorrhizal roots. (H) Colocalization of GUS activity (indicated by the purple color from the overlay of the trypan blue and magenta-GUS stains). Red arrows indicate arbuscules. Blue arrows denote noncolonized cells in mycorrhizal roots. (Scale bars: 50 μm.)
To determine whether the NPF4.5 orthologs in other mycorrhizal plant species were also inducible in response to AM symbiosis, we quantitatively assayed the expression of the NPF4.5 orthologs in Medicago (MtNPF4.5), maize (ZmNPF4.5), and sorghum (SbNPF4.5; SI Appendix, Fig. S4). Our results showed that expression of all these three NPF4.5 orthologs was barely detectable in roots of non–AMF-inoculated roots (SI Appendix, Fig. S9). By contrast, AMF inoculation strongly induced expression of ZmNPF4.5 in maize and SbNPF4.5 in sorghum, while MtNPF4.5 was slightly induced in Medicago (SI Appendix, Fig. S9). The strong inducibility of SbNPF4.5 transcripts in response to AM symbiosis was confirmed in the RNA-seq data from a recent report on the global transcriptional changes induced by arbuscular mycorrhizal fungi on several S. bicolor accessions (42). These results suggest the likely presence of a conserved symbiotic NO3− uptake route, at least in gramineous species. It was previously reported that symbiosis with R. irregularis strongly induced the expression of the OsNPF4.5 orthologs in Populus trichocarpa (POPTR_004g064100) (43) and Helianthus annuus (HanXRQChr15g0472261) (44), suggesting that NPF4.5 could play an important role in symbiotic NO3− nutrition in plants outside gramineae.
OsNPF4.5 Possesses Nitrate Transport Capacity In Vitro and In Vivo.
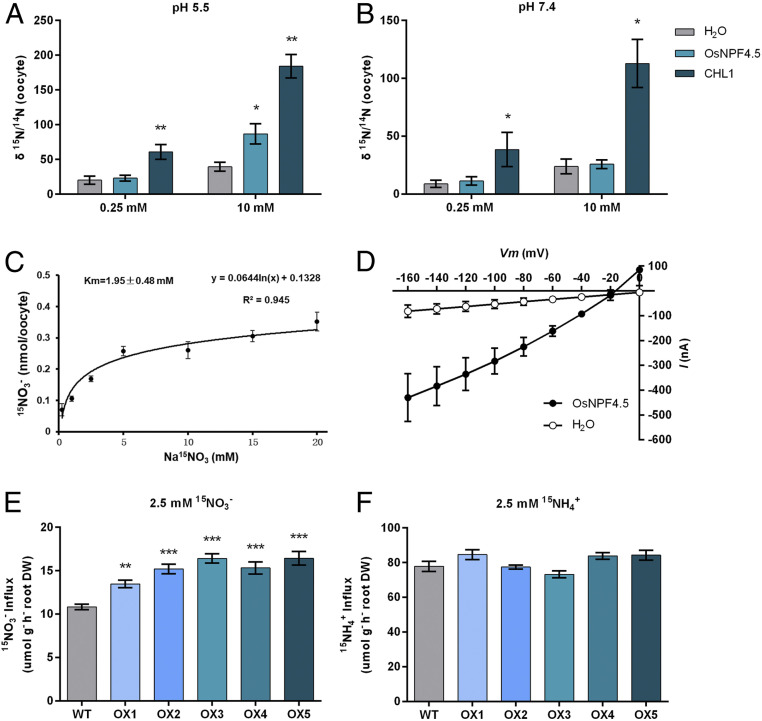
The NO3− transport capacity of OsNPF4.5 was initially evaluated by heterologous expression in Xenopus oocytes. CHL1/AtNRT1.1, the well-established dual-affinity NO3− transporter (45), was used as a positive control. Assays of 15N-nitrate uptake showed that the NO3− uptake was much higher in oocytes injected with CHL1 cRNA (complementary RNA) than those in water-injected controls under both low (0.25 mM) and high (10 mM) NO3− concentrations. Oocytes injected with OsNPF4.5 cRNA and incubated in 0.25 mM NO3− showed no significant difference in nitrate uptake activity versus the water-injected controls, while those incubated in 10 mM NO3− showed a twofold increase in NO3− uptake when compared with the water-injected oocytes at pH 5.5 (Fig. 4A), but not at pH 7.4 (Fig. 4B). The Km of OsNPF4.5 affinity for NO3− uptake was calculated from the net NO3− accumulation of the oocytes incubated in a series of concentrations (0.25, 1, 2.5, 5, 10, 15, and 20 mM) of 15N-NO3−, and was estimated as 1.95 ± 0.48 mM (Fig. 4C). Inward currents responding to alterations in membrane potential could also be evoked by 10 mM NO3− supply for OsNPF4.5-injected oocytes (Fig. 4D). These results demonstrate that OsNPF4.5 functions as a low-affinity, pH-dependent NO3− transporter when expressed in Xenopus oocytes.
Fig. 4.
Functional characterization of OsNPF4.5 in vitro and in vivo. (A and B) Results of nitrate-uptake assay in Xenopus oocytes injected with OsNPF4.5 and CHL1 cRNAs using 15N-nitrate at pH 5.5 (A) and pH 7.4 (B). CHL1 was used as a positive control. (C) Nitrate uptake kinetics of OsNPF4.5 in oocytes. OsNPF4.5 cRNA was injected into oocytes, which were incubated in the ND96 solution containing 0.25, 1, 2.5, 5, 10, 15, and 20 mM Na15NO3, respectively, for 2 h at pH 5.5. (D) Current–voltage curves of oocytes expressing OsNPF4.5. The I–V curves shown were recorded from OsNPF4.5- and H2O-injected oocytes, which were treated with 10 mM nitrate at pH 5.5. Values are means ± SE (n = 10 oocytes). (E and F) The 15N accumulation in roots of WT and OsNPF4.5-overexpressing plants under 15NO3− (E) or 15NH4+ (F) supply hydroponic conditions. In the uptake experiment, WT and OsNPF4.5-overexpressing transgenic lines, referred as OX lines, suffered from N deprivation for 4 d and then were resupplied with 15N-labled 2.5 mM NO3− or 2.5 mM NH4+ for 10 min. Values are means ± SE of five biological replicates (n = 5). The asterisks indicate significant differences (*P < 0.05; **P < 0.01, ***P < 0.001).
To assess whether overexpression of OsNPF4.5 can facilitate NO3− uptake in vivo, we generated transgenic rice plants constitutively overexpressing OsNPF4.5 under the control of a maize ubiquitin promoter and performed both short-term and long-term hydroponic uptake experiments. In the short-term uptake experiment, wild-type (WT) control plants and five independent OsNPF4.5-overexpressing transgenic lines, referred as OX lines (SI Appendix, Fig. S10), were subjected to N deprivation for 4 d and then resupplied with 2.5 mM 15N-labeled NO3− or NH4+ for 10 min. When supplied with 2.5 mM 15NO3−, all of the OX lines showed a 24 to 50% higher 15N uptake than WT plants (Fig. 4E). By contrast, no difference in 15N accumulation could be observed between the WT and OX plants when supplied with 15NH4+ supply (Fig. 4F). For the long-term uptake experiment, seedlings of WT plants and three OX lines were subjected to N deprivation for 4 d and then resupplied with 2.5 mM NO3− or NH4+ for 3 wk. When supplemented with 2.5 mM NO3−, OX transgenic lines showed a 25 to 46% increase in shoot biomass, a six- to eightfold increase in NO3− content in roots, a two- to threefold increase in NO3− content in shoots, and an increase of 80 to 110% in total N content in both shoot and root when compared to WT plants (SI Appendix, Fig. S11 A–C). The high NO3− and total N content phenotype of OX plants seems to be due to the high level of OsNPF4.5 transcripts in OX transgenic rice lines, as it was increased thousands of folds compared to that in WT plants (SI Appendix, Fig. S10). In the long-term uptake experiment, no significant difference in either plant biomass or total N content could be observed between the WT and OX transgenic plants supplied with NH4+ (SI Appendix, Fig. S11 D and E). In OsNPF4.5-overexpressing rice plants supplied with NO3−, increased expression of some N assimilation-related genes such as OsNR1/2 and OsGS1 was observed (SI Appendix, Fig. S11 F–H). All these results lend solid evidence to support that OsNPF4.5 has NO3− but not NH4+ transport capacity. The significantly superior capacity of OX plants in NO3− uptake opens the possibility of using OsNPF4.5 in breeding programs to improve rice N use efficiency, as had been proposed for several other NO3− transporter genes (38–40).
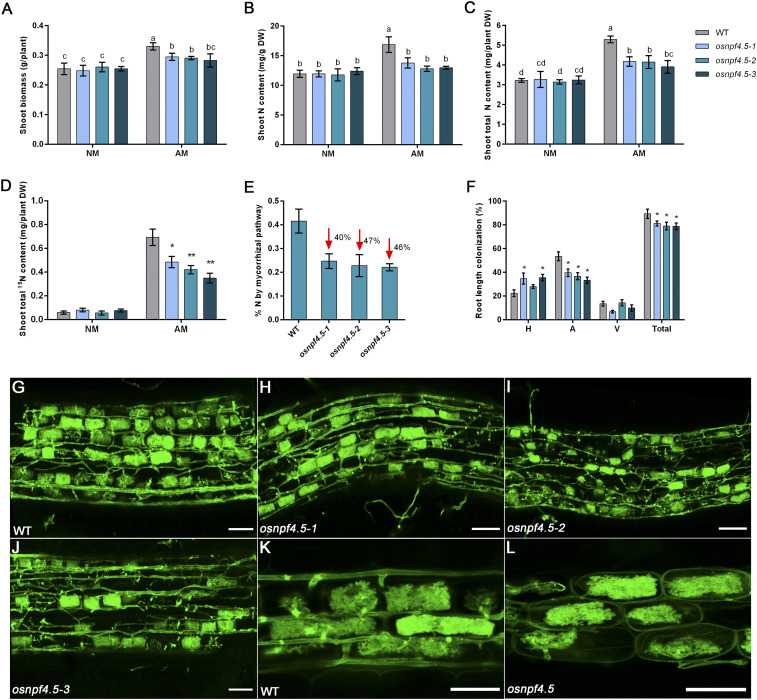
Loss of OsNPF4.5 Function Decreases Symbiotic Nitrate Transport and Arbuscule Incidence.
The mycorrhiza-specific property of OsNPF4.5 inspired us to investigate whether OsNPF4.5 contributes to the symbiotic NO3− uptake and/or AM formation. To test this, osnpf4.5 knockout mutants were generated with the CRISPR-Cas9 system using three different spacers targeting the coding sequence of OsNPF4.5 (Materials and Methods). Two of the three spacers worked effectively in the editing system, resulting in the generation of nine mutant lines, which were screened by PCR sequencing, and three independent homozygous lines were used for further study (SI Appendix, Fig. S12). Osnpf4.5–1 contains a “T” insertion at nucleotide 483 of the ORF that causes a shift in reading frame, and osnpf4.5–2 harbor a “G” deletion at position 482 and osnpf4.5–3 an “A” deletion at position 708 (SI Appendix, Fig. S12). In all cases, CRISPR-Cas9 mutations resulted in frame shifts and premature termination in the first half of OsNPF4.5 (SI Appendix, Fig. S13). No significant difference in N accumulation could be observed between the three osnpf4.5 mutants and WT plants grown under hydroponic conditions supplied with either 2.5 mM NO3− or NH4+ as an N source (SI Appendix, Fig. S14) or a sand-based pot culture supplied with 2.5 mM NO3− in the absence of AM fungal inoculation (SI Appendix, Fig. S15). When inoculated with R. irregularis, the mycorrhizal WT plants increased shoot biomass and shoot N content by 31 ± 6% and 39 ± 7%, respectively, relative to noninoculated plants, whereas osnpf4.5 plants showed only a 10 ± 4% increase in shoot biomass and no significant increase in shoot N content as compared to noninoculated WT or mutant lines (SI Appendix, Fig. S15). When total N and P content was quantified, we found that inoculated WT plants increased 65 ± 6% and 275 ± 19% in total N and P content, respectively, compared to noninoculated WT plants. By contrast, osnpf4.5 plants displayed an increase of 28 to 34% and 234 to 247% in total shoot N and P content relative to that determined in mock-inoculated WT and mutant lines. These results strongly suggest that OsNPF4.5 plays an important role in the mycorrhizal NO3− uptake pathway, but not in the direct uptake pathway. Moreover, the reduction in the growth promotion of inoculated osnpf4.5 mutants is most probably due to a reduction in N supply because of the lack of a functional OsNPF4.5 transporter. However, we could not rule out that the reduction in growth promotion in inoculated osnpf4.5 plants might be partially caused by a colonization difference between the WT and osnpf4.5 plants.
To quantify the potential contribution of OsNPF4.5 to mycorrhizal NO3− uptake, seedlings of WT and osnpf4.5 plants were cultivated in the compartmented growth system, and 2.5 mM NO3− and 15NO3− were supplied to the RFC compartment and HC compartments, respectively (Fig. 2A). Consistent with the results obtained from the pot culture, inoculated WT plants increased shoot biomass by about 30 ± 4%, shoot N content by about 42 ± 5%, and total N content by 64 ± 5% relative to mock-inoculated WT (Fig. 5 A–C). By contrast, mycorrhizal osnpf4.5 mutant plants showed an increase of only 15 ± 4% in shoot biomass and no difference in shoot N content relative to mock-inoculated WT and the respective mutant lines (Fig. 5 A–C). Both the WT and osnpf4.5 mycorrhizal plants contained higher 15N than the corresponding mock-inoculated control plants (Fig. 5D), indicating that both the WT and osnpf4.5 can take up NO3− from hyphal compartments via the fungal hyphae. However, the significant decrease in 15N accumulation observed in the shoots of osnpf4.5 mycorrhizal plants compared with that in the mycorrhizal WT plants highlights the important role of OsNPF4.5 in mycorrhizal NO3− uptake. Mutation of OsNPF4.5 led to a decrease of the percentage of mycorrhizal N uptake contribution from 42% in WT plants to less than 25% in osnpf4.5 mutant lines (Fig. 5E), indicating that OsNPF4.5 may account for ∼45% of the mycorrhizal N uptake when supplied with NO3− as N sources. Since we have solid evidence showing that OsNPF4.5 has NO3− transporter activity, we propose that NO3− is the molecule that is released into the periarbuscular space and imported by root cells using NPF4.5 and other nitrate transporters. However, since some NO3− transporters have also been shown to be able to transport amino acids and small peptides (12), we cannot exclude the possibility that at least a fraction of the symbiotic N is supplied to the plant in the form of organic N molecules.
Fig. 5.
Physiological analysis of the OsNPF4.5 loss function mutants. WT and three osnpf4.5 mutant lines generated by CRISPR-Cas9 were cultivated in a compartmented growth system containing a middle root/hyphal compartment (RHC) that was separated by 30-mm nylon meshes from two hyphal compartments (HCs). The RHC and HC were irrigated with 2.5 mM NO3− and 15NO3− weekly, respectively. The inoculated and mock-inoculated WT and osnpf4.5 plants were harvested for physiological analysis at 6 wpi. (A) Shoot biomass (dry weight), shoot N content (B and C), and 15N accumulation (D) of the WT and osnpf4.5 mutant plants inoculated with R. irregularis (AM) or mock-inoculated controls (NM). (E) The contribution of the symbiotic NO3− acquisition pathway to overall N uptake of WT and osnpf4.5 mutants. (F–L) The mycorrhizal colonization level (F) determined in hypha (labeled “H”), arbuscule (“A”), and vesicle (“V”) and arbuscule incidence and morphology in WT (G and K) and osnpf4.5 mutants (H–J and L). Values are means ± SE of five independent biological replicates (n = 5). Different letters and asterisks indicate significant differences (*P < 0.05; **P < 0.01). (Scale bars: G–J, 50 μm; K and L, 25 μm.)
The bidirectional nutrient exchange between host plants and AM fungi is thought to follow a “free-market” model, in which both symbionts can exert control over their partners (46, 47). A mutually stimulating mechanism has been repeatedly proposed during the simultaneous exchange of C and Pi between the two partners (47). Blocking mycorrhizal P transport via silencing the Pi transporters or H+-ATPases located in the PAM caused a remarkable defect in mycorrhization and arbuscule development (5, 34, 48, 49). To determine whether alteration in symbiotic nitrate transport caused by mutation of OsNPF4.5 affects AM symbiosis, the degree of AM colonization, as well as the arbuscule morphology and populations in the mycorrhizal roots of WT and osnpf4.5 mutant lines, were assessed 6 wpi (Fig. 5 F–L and SI Appendix, Fig. S16). Compared to WT plants, a small but statistically significant decrease of ∼10% in total root length colonization and nearly 20% in arbuscule colonization rate was observed in osnpf4.5 mutant lines (Fig. 5 F–J). It is worth noting that, although reduced in arbuscule colonization rate, well-developed arbuscules were clearly observed in osnpf4.5 plants (Fig. 5 K and L and SI Appendix, Fig. S16), suggesting that symbiotic NO3− transport might not be an essential requirement for arbuscule development. Considering that in osnpf4.5 plants there is still significant NO3− transport through the mycorrhizal route, it would be of great significance to further explore whether the arbuscule development would be impaired when mycorrhizal NO3− transport route is entirely or severely blocked by simultaneously silencing other AM-induced NO3− transporters.
Conclusion
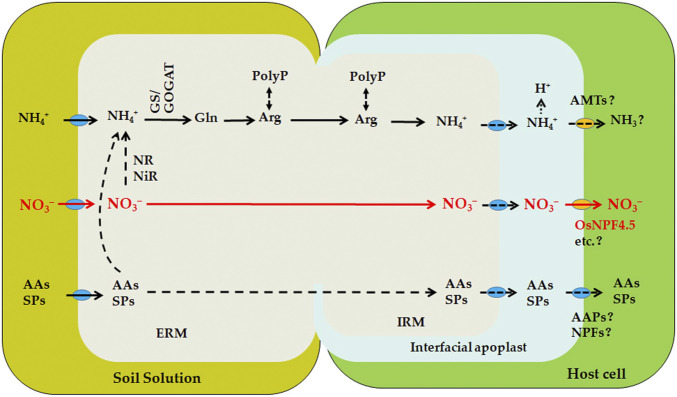
NH4+ and NO3− are the two major inorganic forms of N taken up by plants. Previous studies in several plant species have suggested the presence of a symbiotic NH4+/NH3 transport route via the interfacial apoplast into plant root cells, probably mediated by the AM-induced plant NH4+ transporters located on the PAM (16, 50, 51). Rice is thought to have evolved a high-efficiency NH4+ transport system, as, in paddy fields, NH4+ is the major N source. RNA sequencing analysis in this study (22), however, allowed us to identify multiple genes involved in nitrate transport and metabolism, but only one NH4+ transporter gene that was significantly up-regulated in rice mycorrhizal roots (Fig. 1C and SI Appendix, Fig. S1). Our findings obtained from the compartmented culture system enrich the previously proposed mycorrhizal N uptake model by clearly indicating the presence of a symbiotic NO3− acquisition route (Fig. 6), from NO3− uptake by extraradical mycelium to NO3− translocation at the fungus–root interface mediated by plant NO3− transporters (Fig. 6). We show that the mycorrhizal NO3− uptake route could contribute up to 42% of the overall rice N uptake when NO3− was supplied as N source. Moreover, our results demonstrate that about 45% of the mycorrhizal NO3− was delivered via OsNPF4.5, the strongest AM-induced NO3− transporter. Given that several NPF homologs in diverse plant species have been shown to have the ability to transport dipeptides and amino acids, as well as other substrates, we cannot completely exclude the possibility that, in addition to NO3−, OsNPF4.5 might also have the ability to transport other organic N substrates, such as small peptides and amino acids. Our findings generate an interesting evolutionary question, that is, as a typical NH4+-preferred crop plant, why rice prefers to up-regulate much more genes involved in NO3− transport rather than NH4+ transport upon AM symbiosis, or, in other words, why rice retained a mycorrhizal NO3− acquisition route during the process of domestication. A possible explanation might be that rice has evolved well-developed aerenchyma and strong root oxygen-released capacity, which can generate rapid nitrification in rhizosphere, in particular in drained soils. It has been estimated that nitrate contributes up to 40% of total N acquired by rice roots even under wetland conditions (52). Repeated evidence has also shown that manipulation of NO3− transporters, but not NH4+ transporters, substantially increases rice N use efficiency (NUE) (38–40). The induction of mycorrhizal NO3− acquisition route thus might be an evolutionary strategy of rice to adapt to the NH4+-NO3− shift environment between the flood and drained soils. Our results suggest that AM symbiosis not only activates the transport of NO3− but also N assimilation in general because genes encoding nitrate reductase, nitrite reductase, glutamine synthetase, and glutamate synthase are also up-regulated during mycorrhization with R. irregularis. In this study, we also revealed a high conservation in both the secondary structure and residues potentially involved in NO3− binding and transport among rice OsNPF4.5 and its orthologs from other dicot and monocot plant species. The strong induction of the orthologs ZmNPF4.5 and SbNPF4.5 observed in maize and sorghum, respectively, in response to AM symbiosis suggests that the NPF4.5-mediated symbiotic NO3− uptake route as an important pathway for mycorrhizal N acquisition might be highly conserved, at least in gramineous species.
Fig. 6.
A model for N uptake, assimilation, and translocation in AM symbiosis. AM fungi can take up both NH4+ and NO3−, as well as organic N forms, such as amino acids (AAs) and small peptides (SPs), from soil solution via their extraradical mycelium (ERM). The NH4+ in fungal cytoplasm can be rapidly assimilated into amino acids, mainly arginine, via the glutamine synthetase-glutamate synthase (GS-GOGAT) pathway and translocated, probably coupled with Poly-P through the intraradical hyphae. After hydrolysis in the arbuscule, NH4+ is exported from the AM fungus to the periarbuscular space (PAS) and subsequently imported, probably in the form of NH3, into the root cell by the putative plant NH4+ transporters located on the periarbuscular membrane (PAM). The NO3− absorbed by extraradical mycelium can be directly translocated into intraradical hyphae and released into the interfacial apoplast. The import of NO3− into root cell is mediated by the PAM-localized NO3− transporters, such as OsNPF4.5. NR, nitrate reductase; NiR, nitrite reductase; GS, glutamine synthetase; GOGAT, glutamate synthase; AMT, ammonium transporter, AAP, amino acid permease. Question marks and dotted lines indicate that the putative transporters or transport/metabolic processes have not yet been established.
Materials and Methods
Plant Materials and Growth Conditions.
The rice (O. sativa ssp. japonica) wild-type and transgenic plants used in this study were in the cv Nipponbare background. Rice seeds were surface-sterilized and germinated in a growth chamber programmed for 14 h light at 28 °C and 10 h dark at 22 °C and maintained to grow in one-half IRRI nutrient solution (SI Appendix, Table S1, for rice) for 1 wk. Seedlings produced as mentioned earlier were then transferred to pot or compartmented culture inoculation with AM fungus. For pot culture, eight seedlings of WT or each line of individual mutants were transplanted to four holes (two seedlings as a replicate were placed into each hole) in a pot (35-cm diameter × 24-cm height) filled with a 4:1 mixture of sterilized sand and low-N soil (the soil contains 2.2 mg kg−1 NH4+, 3.7 mg kg−1 NO3−, and 1.4 mg kg−1 available P). The seedlings in each hole were inoculated with ∼200 R. irregularis spores around the roots. The nonmycorrhizal control plants were obtained by inoculation with autoclaved inoculum. The plants in each pot were regularly watered and fertilized weekly with 500 mL nutrient solution containing 2.5 mM NO3− (or other concentrations for different treatments) and 30 μM Pi, as well as the other essential nutrients from the modified IRRI nutrient solution recipe (SI Appendix, Table S1, for rice).
Determination of Mycorrhizal Nitrate Uptake Contribution.
A compartmented culture system was employed to investigate the contribution of symbiotic NO3− uptake to the overall N nutrition of mycorrhizal rice WT and osnpf4.5 mutant plants (Fig. 2A). The culture system contains a middle root/fungal compartment (RFC) and two hyphal compartments (HCs; each compartment is 10 × 10 × 12 cm in length, width, and height), as described by Liu et al. (37). All three compartments were filled with ∼1 L sand/low-N soil mixture. Two seedlings of WT or mutant plants were grown in the RFC inoculated with R. irregularis or autoclaved inoculum (as control) for 6 wk. Each treatment comprised five compartmented boxes as independent biological replicates. The plants in RFC were regularly watered and fertilized weekly with 250 mL nutrient solution containing 2.5 mM NO3− as the N source, and simultaneously the two HCs were supplied with equal amounts of nutrient solution containing 2.5 mM 15NO3−. To monitor whether fungal hyphae could reach and take up NO3− from HCs, the 15N content in the inoculated and mock-inoculated plants was determined. To assay 15N content, harvested plants were rinsed for 1 min in 0.1 mM CaSO4 solution and then roots and shoots were separated. The collected root and shoot samples were dried at 70 °C and weighed before being ground. One milligram of the finely ground powder for each sample was used to determine the 15N content by an isotope ratio mass spectrometer with an elemental analyzer (DELTA V ADVANTAGE isotope Ratio MS; Thermo Fisher) (40). Total shoot N or root N or 15N content (in milligrams per plant) was equal to shoot N or root N or 15N content (in milligrams per gram) multiplied by shoot or root biomass (in grams, dry weight). Total N content in the plant equals total shoot N content plus total root N content. The percentage of contribution of the mycorrhizal pathway to total N uptake in WT or osnpf4.5 mutants was calculated with the formula [(total N content in AM plant − total N content in NM plant)/total N content in AM plant] × 100%. The contribution of OsNPF4.5 to mycorrhizal pathway of NO3− uptake was calculated with the formula [mycorrhizal N uptake contribution in WT plants − mycorrhizal N uptake contribution in osnpf4.5 mutants/mycorrhizal N uptake contribution in WT plants] × 100%.
RNA Sequencing.
The inoculated and mock-inoculated seedlings were irrigated with IRRI nutrient solution containing 1.25 mM NH4NO3 and 30 μM Pi weekly. The roots of the mycorrhizal and nonmycorrhizal plants were collected 6 wk post inoculation. Total RNA was isolated using the RNEasy Plant Maxi kit (Qiagen). Three biological replicates for each treatment were used for RNA sequencing reaction performed on an Illumina HiSeq 2500. After trimming and eliminating low-quality reads, 39,463,820 and 38,621,548 clean reads were obtained for inoculated and control plants, respectively, which accounted for over 95% of the total sequences (22). The transcriptome data analysis was commercially conducted by CapitalBio (Beijing, China).
RNA-Based RACE PCR.
The full-length cDNA of OsNPF4.5 was obtained by rapid amplification of cDNA ends (RACE; First Choice RLM-RACE Kit; Ambion). One and 10 µg of total RNA were used for the 3′ and 5′ RLM-RACE protocols, respectively, following the manufacturer’s instructions strictly. The specific primers used for amplifying the 5′ and 3′ ends of OsNPF4.5 cDNA are: 5′ outer primer, ggccaatgaaagtgtccgcgaag; 5′ inner primer, acggctagagacaacgaggcaagg; 3′ outer primer, gccgcagttcaccgtgtt; and 3′ inner primer, tcatcgggctcctcgagtt.
Phylogenetic Analysis.
The unrooted phylogenetic tree of the plant NPF homologs was constructed using their protein sequences by the neighbor-joining algorithm within the MEGA 6 program with bootstrapping value (range 0 to 100). For tree construction, we used the OsNPF4.5 orthologs in Medicago (MtNPF4.5), maize (ZmNPF4.5), and sorghum (SbNPF4.5) as previously identified by other authors (42) and confirmed by bidirectional BLAST analysis, and other nitrate transporters. The reference numbers of the protein sequences used for constructing the tree are the following: OsNPF1.3, XP_015636060.1; OsNPF5.4, XP_015612792.1; OsNPF8.3, XP_015634046.1; LjNPF8.6, IPR000109; MtNPF1.7, XP_003588616.1; MtNPF6.8, XP_003616931.1; OsNPF6.3 (OsNRT1.1A), XP_015650127.1; OsNPF6.5 (OsNRT1.1B), XP_015614015.1; OsNPF6.4 (OsNRT1.1C), XP_015632236.1; OsNPF2.4, XP_015630690.1; OsNPF2.2 (OsPTR2), XP_015620477.1; OsNPF7.2, XP_015627752.1; ZmNPF6.6, XP_008658424.1; ZmNPF6.4, NP_001145735.1; AtNPF6.4 (AtNRT1.1), NP_563899.1; AtNPF4.6 (AtNRT1.2), NP_564978.1; AtNPF5.12 (AtTOB1), NP_177359.1; AtNPF6.2 (AtNRT1.4), NP_850084.1; AtNPF5.5, NP_181345.1; AtNPF1.1 (AtNRT1.12), NP_188239.1; AtNPF6.4 (AtNRT1.3), NP_188804.1; AtNPF4.1 (AtNIT3), NP_189163.1; AtNPF4.2 (AtNIT4), NP_189165.1; AtNPF2.7 (AtNAXT1), NP_190151.1; AtNPF2.3, NP_190154.1; AtNPF2.10 (AtGTR1), NP_566896.2; AtNPF7.2 (AtNRT1.8), NP_193899.2; AtNPF2.9 (AtNRT1.9), NP_173322.1; AtNPF5.10, NP_173670.2; AtNPF4.5 (AtAIT2), NP_973919.1; AtNPF2.12 (AtNRT1.6), NP_174028.2; AtNPF7.3 (AtNRT1.5), NP_174523.2; AtNPF1.2 (AtNRT1.11), NP_175630.1; AtNPF8.5 (AtPTR6), NP_176411.2; AtNPF3.1 (AtNitr1), NP_177024.1; OsNPF7.3 (OsPTR6), XP_015633790.1; ZmNPF4.5, XP_020406064.1; SbNPF4.5, XP_021311980.1; SbNPF1.2, XP_002458530.1; GmNPF4.5, XP_003532772.2; and MtNPF4.5, XP_024627880.1.
Vectors, Strains, and Rice Gene Transformation.
For promoter-GUS assays, a 2,030-bp promoter fragment of OsNPF4.5 immediately upstream of the translation start ATG was amplified and inserted into the pCAMBIA1300 binary vector to replace the CaMV35S promoter in front of the GUS reporter gene. To construct the OsNPF4.5 overexpression vector, the coding sequence of OsNPF4.5 was amplified and cloned into the binary vector pTCK303 under the control of a maize ubiquitin promoter using the ClonExpress II One Step Cloning Kit (Vazyme Biotech). The CRISPR-Cas9 gene knockout constructs were generated using the pH-Ubi-cas9-7 vector. Three different spacers (spacer1, ggggaagacctgcaataaga; spacer2, gttcgaccccaagtgcgaga; and spacer3, gtgtggatccagagctacaa) targeting the coding sequence of OsNPF4.5 were selected from the rice-gene–specific spacers library provided by Miao et al. (53). These spacers were first cloned into the intermediate vector pOs-sgRNA via BsaI and then introduced into the expression vector pH-Ubi-cas9-7 using the Gateway recombination technology (Invitrogen). All of the resulting constructs were transformed into the Agrobacterium tumefaciens EHA105 strain. The transformation of rice plants was carried out as described previously (54). The screening of mutant lines was performed by PCR sequencing. spacer1 did not work effectively in the editing system, but the other two spacers successfully resulted in the generation of several homozygous mutant rice lines.
Subcellular Localization Analysis.
The CDS of OsNPF4.5 was fused in frame with eGFP via cloning into the binary vector pRCS2-ocs-nptII. The resulting vector, named 35S::eGFP-OsNPF4.5, was transformed into the EHA105 strain. The agroinfiltration of tobacco leaves and the imaging of eGFP fluorescence were performed as described by Liu et al. (37). For assaying the subcellular localization of OsNPF4.5 in mycorrhizal roots, the native promoter of OsNPF4.5 was amplified and inserted into the pCAMBIA1300 vector to replace the CaMV35S promoter, and then the OsNPF4.5-eGFP chimeric gene was cloned and inserted into the vector under the control of the OsNPF4.5 promoter. The resulting vector, named NPF4.5pro::OsNPF4.5-eGFP, was introduced into the EHA105 strain and used for rice transformation. The transgenic plants were then transferred to sand-based pot culture for inoculation with the AM fungus R. irregularis. The eGFP image was observed with a confocal microscope (Leica Confocal TCS-SP8) 6 wk post inoculation.
Mycorrhizal Colonization Quantification.
Histochemical staining of the GUS activity in transgenic plants was performed as described previously (55). Mycorrhizal colonization was quantified based on the grid line intersect method (56) using a binocular microscope (Leica). The measurement of arbuscule sizes in the arbuscule populations was performed according to the procedures described by Breuillin-Sessoms et al. (15). To visualize the fungus, roots were stained in 0.2 mg/mL WGA Alexa Fluor 488 solution as described by Javot et al. (48). For assessment of arbuscule populations, the stained root segments were observed using the confocal microscope, arbuscules were grouped into three size classes (0 to 30 μm, 30 to 50 μm, and >50 μm) based on their lengths, and the percentage of arbuscules in each size class was counted. Arbuscule size was determined by measuring the length of all of the visible arbuscules (at least 200 arbuscules) in 5 to 10 independent infection units for each root sample, and the average and the SE of each arbuscule size are graphed from three independent biological replicates.
Determination of N and P Contents.
The digestion of dried plant material with 98% H2SO4 and 30% H2O2 and the assay of total P content with the molybdate blue method were performed as described previously (57). The assay of total N and nitrate was performed as described previously by Tang et al. (58).
Analysis of Gene Expression.
RNAs were extracted by using TRIzol reagent (Invitrogen). Two micrograms of total RNA was used for RT-PCR reactions using an MLV reverse transcription kit (TaKaRa). Quantitative RT-PCR was performed based on the instructions of the SYBER premix ExTaq kit (TaKaRa) on an Applied Biosystems Plus Real-Time PCR System by using gene-specific primers (SI Appendix, Table S2). The expression of Os-Actin (Os03g50885) was used for normalization. Four biological replications were performed.
The 15N-Nitrate Uptake Assay in Xenopus laevis Oocytes.
The CDS of OsNPF4.5 was amplified and cloned into the X. laevis oocyte expression vector pT7Ts between the restriction sites Bgl II and SpeI and then linearized with XbaI. Capped mRNA (cRNA) was synthesized in vitro using the Ambion mMessage mMachine kit (Ambion; AM1340) according to the manufacturer’s protocol. X. laevis oocytes were injected with 50 ng of OsNPF4.5 cRNA or 50 nL nuclease-free water. After injection, oocytes were cultured in ND-96 medium for 48 h and used for 15NO3− uptake assays. High- and low-affinity uptake assays in oocytes were conducted using 250 µM and 10 mM 15N-NaNO3, respectively, as described previously by Xia et al. (59). Two-electrode voltage clamp assay was performed as described previously (60).
The 15N-Nitrate Uptake Activity In Vivo.
Nitrate-uptake activity was determined using a 15N-labeling assay under hydroponic condition. Two-week-old seedlings of WT and transgenic plants were grown in IRRI nutrient solution containing 1 mM NH4+ for 3 wk and then deprived of N supply for 4 d. The N-starved plants were transferred to 0.1 mM CaSO4 solution for 1 min and then resupplied with the nutrient solution containing either 2.5 mM 15NO3− or 2.5 mM 15NH4+ for 10 min. The treated plants were transferred to 0.1 mM CaSO4 solution for 1 min before sampling. The 15N content in roots was determined with a DELTA V ADVANTAGE isotope ratio MS as described earlier, and the uptake activity was calculated as the amount of 15N taken up per unit weight of roots per unit time.
Structural Alignment of Nitrate Transporters and Structure Modeling of OsNPF4.5.
Multiple sequence alignment of NRT1.1 transporters and NPF4.5 orthologs was performed using MAFFT, and secondary structures were assigned using ESPript 3.0 (http://espript.ibcp.fr) (61). NCBI accession numbers used in the analyses are as follows: Brachypodium distachyon (Bd), XP_014754374.1; Zea mays (Zm), XP_020406064.1; M. truncatula (Mt), XP_024627880.1; G. max (Gc), XP_003532772.2; Vitis vinifera (Vv), XP_019078273.1; Populus euphratica (Pe), XP_011009674.1; P. trichocarpa (Pt), XP_002305708.2; H. annuus (Ha), XP_022013935.1; Solanum tuberosum (St), XP_006356126.1; Cannabis sativa (Cs), XP_030479547.1; and Amborella trichopoda (At), XP_011624609.1. OsNPF4.5 structure was modeled using Rosetta (62) and visualized with PyMOL (Schrödinger, version 2.3.2). Structure alignment between crystal structure of AtNRT1.1 and the model structure of OsNPF4.5 was analyzed using PyMOL and SuperPose server, version 1.0 (63).
Statistical Analysis.
The data were analyzed by ANOVA (SPSS 16.0; SPSS) and Student’s t test. Significance of differences was defined at P < 0.05, P < 0.01, and P < 0.001.
Accession Numbers.
The sequence data from this article can be found in NCBI with the following accession numbers: OsNPF4.5 (LOC9271385), OsNPF6.4 (LOC9271131), OsPT11 (LOC4324187), OsHA1 (LOC4331281), OsNAR2.1 (LOC4329861), OsNRT2.1 (LOC4328051), OsNRT2.2 (LOC4328052), OsNPF1.3 (LOC4327022), OsNPF5.4 (LOC4348864), OsNPF7.2 (LOC4330372), OsNPF8.3 (LOC4336852), OsAMT3.1 (LOC107276856), OsNR1 (LOC4330867), OsNR2 (LOC4345798), OsGS1.1 (LOC4330649), MtNPF4.5 (LOC11406786), ZmNPF4.5 (LOC103652484), and SbNPF4.5 (LOC8062188).
Supplementary Material
Acknowledgments
This work was supported by National Key Research and Development Program of China (2016YFD0100700), National Natural Science Foundation of China (31572188, 31372121), the Basic Research Program of Jiangsu province in China (BK20181324), the Innovative Research Team Development Plan of the Ministry of Education of China (Grant no. IRT_17R56; KYT201802), and Jiangsu Collaborative Innovation Center for Solid Organic Waste Resource Utilization. We thank Prof. Lijia Qu (College of Life Sciences, Peking University, Beijing) for providing the vectors for the CRISPR-Cas9 system in rice, Ms. Hongye Qu for technical assistance, and Ms. Xiaoli Dai and Kaiyun Qian from MOA Key Laboratory of Plant Nutrition and Fertilization in Lower-Middle Reaches of the Yangtze River, Nanjing Agriculture University, for technical support in 15N assay and membrane localization of OsNPF4.5.
Footnotes
The authors declare no competing interest.
Data deposition: All the RNA-seq data presented in this paper, including the raw data, are available in the NCBI with the accession number PRJNA635697.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000926117/-/DCSupplemental.
References
- 1.Smith S. E., Smith F. A., Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62, 227–250 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Gutjahr C., Parniske M., Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29, 593–617 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Jiang Y. et al., Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during Mycorrhizal Symbiosis. Mol. Plant 11, 1344–1359 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Smith S. E., Smith F. A., Jakobsen I., Functional diversity in arbuscular mycorrhizal (AM) symbioses: The contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol. 162, 511–524 (2003). [Google Scholar]
- 5.Yang S. Y. et al., Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 24, 4236–4251 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ames R. N., Reid C. P., Porter L. K., Cambardella C., Hyphal uptake and transport of nitrogen from two 15N-labeled sources by Glomus mosseae, a vesicular-arbuscular mycorrhizal fungus. New Phytol. 95, 381–396 (1983). [Google Scholar]
- 7.Johansen A., Jakobsen I., Jensen E. S., Hyphal transport by a vesicular arbuscular mycorrhizal fungus of N applied to the soil as ammonium or nitrate. Biol. Fertil. Soils 16, 66–70 (1993). [Google Scholar]
- 8.Bago B., Vierheilig H., Piché Y., Azcón-Aguilar C., Nitrate depletion and pH changes induced by the extraradical mycelium of the arbuscular mycorrhizal fungus Glomus intraradices grown in monoxenic culture. New Phytol. 133, 273–280 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Hodge A., Campbell C. D., Fitter A. H., An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413, 297–299 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Govindarajulu M. et al., Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435, 819–823 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Koegel S. et al., The family of ammonium transporters (AMT) in Sorghum bicolor: Two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198, 853–865 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Chen A., Gu M., Wang S., Chen J., Xu G., Transport properties and regulatory roles of nitrogen in arbuscular mycorrhizal symbiosis. Semin. Cell Dev. Biol. 74, 80–88 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Hestrin R., Hammer E. C., Mueller C. W., Lehmann J., Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Commun. Biol. 2, 233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobae Y., Tamura Y., Takai S., Banba M., Hata S., Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell Physiol. 51, 1411–1415 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Breuillin-Sessoms F. et al., Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 27, 1352–1366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guether M. et al., A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol. 150, 73–83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G., Fan X., Miller A. J., Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Guether M. et al., Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 182, 200–212 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Gomez S. K. et al., Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 9, 10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solaiman M., Hirata H., Effect of arbuscular mycorrhizal fungi inoculation of rice seedlings at the nursery stage upon performance in the paddy field and greenhouse. Plant Soil 191, 1–12 (1997). [Google Scholar]
- 21.Maiti D., Toppo N. N., Variar M., Integration of crop rotation and arbuscular mycorrhiza (AM) inoculum application for enhancing AM activity to improve phosphorus nutrition and yield of upland rice (Oryza sativa L.). Mycorrhiza 21, 659–667 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Wang S., Chen A., Xu G., Sequences of mycorrhizal rice root. National Center for Biotechnology Information (NCBI). https://www.ncbi.nlm.nih.gov/sra/PRJNA635697. Deposited 28 May 2020.
- 23.Bravo A., Brands M., Wewer V., Dörmann P., Harrison M. J., Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 214, 1631–1645 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y. et al., Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Keymer A. et al., Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6, 29107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luginbuehl L. H. et al., Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Xue L. et al., AP2 transcription factor CBX1 with a specific function in symbiotic exchange of nutrients in mycorrhizal Lotus japonicus. Proc. Natl. Acad. Sci. U.S.A. 115, E9239–E9246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Güimil S. et al., Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. U.S.A. 102, 8066–8070 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drechsler N., Courty P. E., Brulé D., Kunze R., Identification of arbuscular mycorrhiza-inducible Nitrate Transporter 1/Peptide Transporter Family (NPF) genes in rice. Mycorrhiza 28, 93–100 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Chen J. et al., Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14, 1705–1715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J. et al., OsNAR2.1 positively regulates drought tolerance and grain yield under drought stress conditions in rice. Front. Plant Sci. 10, 197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pérez-Tienda J., Corrêa A., Azcón-Aguilar C., Ferrol N., Transcriptional regulation of host NH4+ transporters and GS/GOGAT pathway in arbuscular mycorrhizal rice roots. Plant Physiol. Biochem. 75, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Paszkowski U., Kroken S., Roux C., Briggs S. P., Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 13324–13329 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang E. et al., A H+-ATPase that energizes nutrient uptake during mycorrhizal symbioses in rice and Medicago truncatula. Plant Cell 26, 1818–1830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cliquet J. B., Murray P. J., Boucaud J., Effect of the arbuscular mycorrhizal fungus Glomus fasciculatum on the uptake of amino nitrogen by Lolium perenne. New Phytol. 137, 345–349 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Nagy R., Drissner D., Amrhein N., Jakobsen I., Bucher M., Mycorrhizal phosphate uptake pathway in tomato is phosphorus-repressible and transcriptionally regulated. New Phytol. 181, 950–959 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Liu J. et al., The potassium transporter SlHAK10 is involved in mycorrhizal potassium uptake. Plant Physiol. 180, 465–479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan X. et al., Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. U.S.A. 113, 7118–7123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W. et al., Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30, 638–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu B. et al., Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Sun J. et al., Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watts-Williams S. J. et al., Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ. 42, 1758–1774 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Calabrese S. et al., Transcriptome analysis of the Populus trichocarpa-Rhizophagus irregularis mycorrhizal symbiosis: Regulation of plant and fungal transportomes under nitrogen starvation. Plant Cell Physiol. 58, 1003–1017 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Vangelisti A. et al., Transcriptome changes induced by arbuscular mycorrhizal fungi in sunflower (Helianthus annuus L.) roots. Sci. Rep. 8, 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu K. H., Huang C. Y., Tsay Y. F., CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiers E. T. et al., Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333, 880–882 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Wang W. et al., Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol. Plant 10, 1147–1158 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Javot H., Penmetsa R. V., Terzaghi N., Cook D. R., Harrison M. J., A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 1720–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krajinski F. et al., The H+-ATPase HA1 of Medicago truncatula is essential for phosphate transport and plant growth during arbuscular mycorrhizal symbiosis. Plant Cell 26, 1808–1817 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian C. et al., Regulation of the nitrogen transfer pathway in the arbuscular mycorrhizal symbiosis: Gene characterization and the coordination of expression with nitrogen flux. Plant Physiol. 153, 1175–1187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tisserant E., et al. , Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. U.S.A. 110, 20117–20122 (2013). Erratum in: Proc. Natl. Acad. Sci. U.S.A.111, 563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirk G. J. D., Kronzucker H. J., The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: A modelling study. Ann. Bot. 96, 639–646 (2005). (Lond). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao J. et al., Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Upadhyaya N. M. et al., Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Funct. Plant Biol. 27, 201–210 (2000). [Google Scholar]
- 55.Chen A. et al., Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol. 189, 1157–1169 (2011). [DOI] [PubMed] [Google Scholar]
- 56.McGonigle T. P., Miller M. H., Evans D. G., Fairchild G. L., Swan J. A., A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115, 495–501 (1990). [DOI] [PubMed] [Google Scholar]
- 57.Chen A., Hu J., Sun S., Xu G., Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol. 173, 817–831 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Tang Z. et al., Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 160, 2052–2063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia X. et al., Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 66, 317–331 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Huang N. C., Liu K. H., Lo H. J., Tsay Y. F., Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song Y. et al., High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maiti R., Van Domselaar G. H., Zhang H., Wishart D. S., SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 32, W590–W594 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.