Significance
Obesity is associated with hepatic steatosis and activation of the cJun NH2-terminal kinase (JNK) stress-signaling pathway. Studies in mice demonstrate that JNK deficiency in the liver prevents the development of hepatic steatosis. This observation suggests that inhibition of JNK signaling may represent a possible treatment for hepatic steatosis. However, the long-term consequences of JNK inhibition are poorly understood. Here we demonstrate that loss of JNK causes changes in cholesterol and bile acid metabolism that promote cholestasis, bile duct proliferation, and intrahepatic cholangiocarcinoma. We identify PPARα activation as the molecular mechanism that accounts for this phenotype. Our analysis has important implications for the long-term use of JNK inhibitors for the treatment of obesity.
Keywords: JNK, PPARa, bile acid, cholangiocarcinoma
Abstract
Metabolic stress causes activation of the cJun NH2-terminal kinase (JNK) signal transduction pathway. It is established that one consequence of JNK activation is the development of insulin resistance and hepatic steatosis through inhibition of the transcription factor PPARα. Indeed, JNK1/2 deficiency in hepatocytes protects against the development of steatosis, suggesting that JNK inhibition represents a possible treatment for this disease. However, the long-term consequences of JNK inhibition have not been evaluated. Here we demonstrate that hepatic JNK controls bile acid production. We found that hepatic JNK deficiency alters cholesterol metabolism and bile acid synthesis, conjugation, and transport, resulting in cholestasis, increased cholangiocyte proliferation, and intrahepatic cholangiocarcinoma. Gene ablation studies confirmed that PPARα mediated these effects of JNK in hepatocytes. This analysis highlights potential consequences of long-term use of JNK inhibitors for the treatment of metabolic syndrome.
Liver cancer is the fifth most common cancer and the second leading cause of cancer deaths worldwide (1, 2). Cholangiocarcinoma the second most common liver cancer, is a malignancy of bile duct epithelia with a clinically silent development and an increasing global incidence (3). Due to the absence of early markers for its diagnosis, most cholangiocarcinoma patients are identified at an advanced stage and die of metastasis (4).
Hepatic cJun NH2-terminal kinase (JNK) has been identified as a signal transduction pathway that is critically required for obesity-induced insulin resistance and hepatic steatosis (5, 6). Indeed, mice with hepatocyte-specific JNK deficiency are resistant to high-fat diet–induced insulin resistance and steatosis (7); therefore, this signaling pathway represents a potential target for therapeutic intervention. Biochemical studies demonstrate that JNK suppresses Peroxisome Proliferator-activated Receptor α (PPARα) activation in hepatocytes, affecting lipid metabolism and steatosis through the hepatokine FGF21 (encoded by a PPARα target gene) (7, 8).
The transcription factor PPARα plays a pivotal role in intracellular free fatty acid and triglyceride metabolism by regulating genes involved in fatty acid transport and degradation in mitochondria and peroxisomes (9–11). PPARα is expressed primarily in liver, heart, and muscle and is a major regulator of fatty acid transport, catabolism, and energy homeostasis (12). PPARα activation in the liver is increased in metabolic diseases and obesity (12), and PPARα agonists appear to be therapeutically beneficial for the treatment of metabolic syndrome. Indeed, PPARα activity protects against steatosis in the mouse (13) and suppresses hepatic inflammation (14). However, long-term studies in rodents showed an association of PPARα agonists with hepatic carcinogenesis (15). These findings conflict with the growth inhibitory effects reported for PPARα agonists in cancer cell lines, including liver cancer cell lines (16–18). PPARα may therefore cause context-specific actions on liver cancer development.
The activation of PPARα modifies bile acid (BA) synthesis, conjugation, and transport (19). This change in BA metabolism can protect against steatosis, but may also promote liver cancer. An example is represented by fibroblast growth factor 15/19 (FGF15/19) that is expressed in the liver during the development of hepatocellular carcinoma and intrahepatic cholangiocarcinoma (20, 21). Notably, FGF15/19 improves glycemic responses and reduces hepatic steatosis, but also promotes liver cancer (22). These functions of BAs prompted us to examine whether long-term blockade of JNK signaling in hepatocytes, and the resulting activation of PPARα, could alter BA homeostasis and promote liver disease.
Results
Hepatic JNK Deficiency Alters Bile Acid Homeostasis.
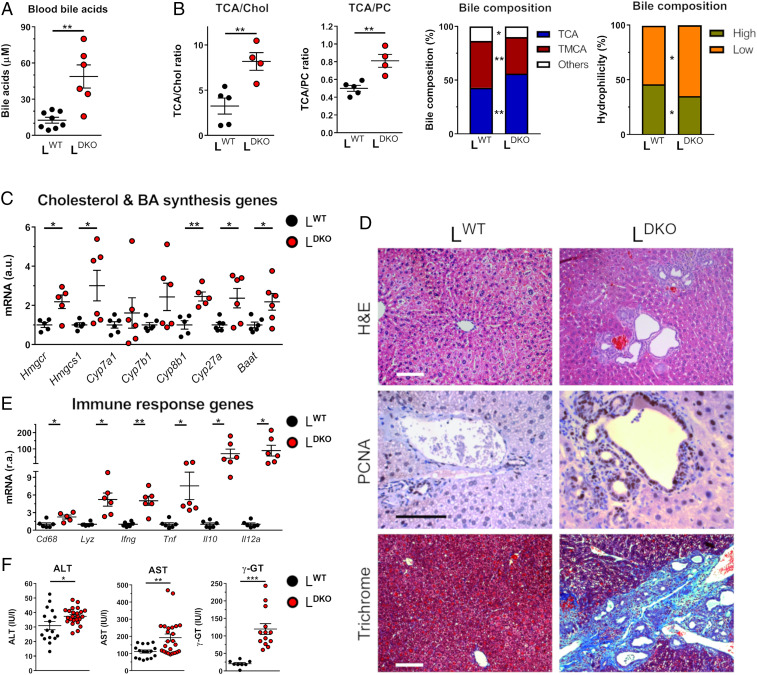
We have previously shown that hepatic JNK deficiency results in the activation of the transcription factor PPARα and causes protection against diet-induced insulin resistance and hepatic steatosis (7). The activation of PPARα may cause altered BA metabolism (19). We therefore examined BA in hepatic JNK1 plus JNK2-deficient mice (LDKO) and control mice (LWT) at 6 mo of age. We found that the total BA concentration in the blood of LDKO mice was significantly increased compared with LWT mice (Fig. 1A). The increase in circulating BA concentration is consistent with the possibility that LDKO mice exhibit cholestasis.
Fig. 1.
Hepatic JNK deficiency alters bile acid production and causes cholestasis. (A) LWT and LDKO mice (age 6 mo) were fasted overnight, and blood was collected for measuring bile acids (mean ± SEM; n = 6–11). Student’s t test differences between LDKO and LWT are indicated (**P < 0.01). (B) The composition of bile fluid collected from the gall bladder was examined by measurement of the ratio of BAs to cholesterol (Chol) or PC and the different type of BA. TCA, taurocholic acid; TMCA, tauromuricholic acids. The data presented are the mean ± SEM (n = 4–5). Student’s t test differences between LDKO and LWT are indicated (*P < 0.05; **P < 0.01). (C) LKO and LWT mice (age 6 mo) were fasted overnight prior to removal of the liver. The expression of genes related to cholesterol synthesis (Hmgcr and Hmgcs1) and BA synthesis (Cyp7a1, Cyp7b1, Cyp27a1, Baat, Cyp27a, Cyp8b1) was measured by quantitative RT-PCR (mean ± SEM; n = 5–6) and normalized to the amount of 18S RNA in each sample. Student’s t test differences between LDKO and LWT are indicated (*P < 0.05; **P < 0.01). (D) Representative liver sections prepared from mice (age 10 mo) stained with hematoxylin and eosin (H&E), an antibody to PCNA, and Masson Trichrome (Trichrome) are presented. (Scale bar, 100 µm.) (E) The expression of genes related to inflammation was evaluated by RT-PCR. (mean ± SEM; n = 5–6) and normalized to the amount of 18S RNA in each sample. Student’s t test differences between LDKO and LWT are indicated (*P < 0.05). (F) Liver damage assessed from serum measurements of ALT, AST, and γ-GT. (mean ± SEM; n = 11–24). Student’s t test differences between LDKO and LWT are indicated (*P < 0.05; **P < 0.01, ***P < 0.001).
Analysis of bile collected from the gall bladder of LDKO and LWT mice revealed significantly increased amounts of BA relative to the amount of cholesterol and phosphatidylcholine (PC) (Fig. 1B). Hepatic expression of genes related to hepatic PC synthesis (Scd2, Chpt1, and Chkb) or hepatocyte-mediated transport of PC (Abcb4 and Atp8b1) and BA (Abc11 and Slc10a1) was markedly increased in LDKO mice (SI Appendix, Fig. S1 A and B). Similarly, increased expression of genes related to cholesterol synthesis (Hmgcs1, Hmgcr) and BA synthesis (Baat, Cyp8b1 and Cyp27a) was detected in LDKO mice (Fig. 1C). These data are consistent with altered biosynthesis and secretion of both cholesterol and BA through PPARα activation.
Hepatic JNK Deficiency Causes Cholestasis and Liver Damage.
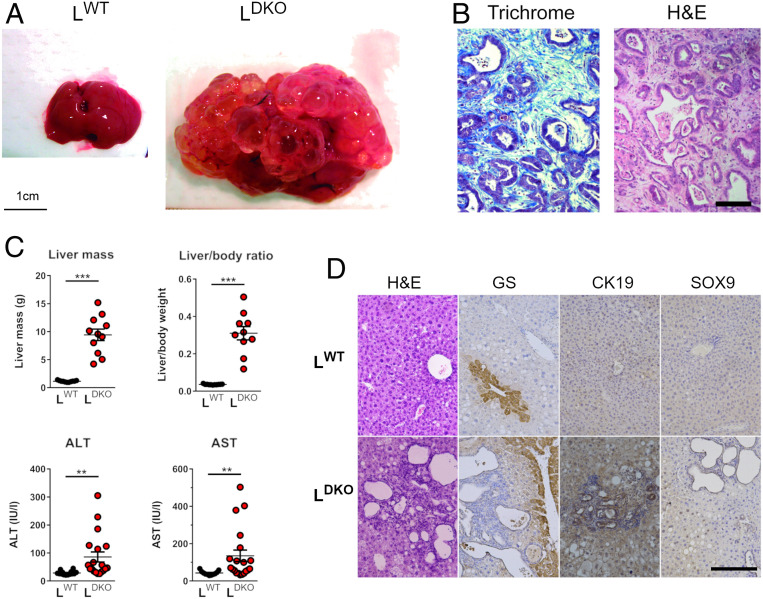
It is established that cholangitis is a major risk factor for the development of cholangiocarcinoma (23). We therefore examined the liver of mature adult LDKO and LWT mice. No evidence of hepatic disease was found in LWT mice. Similarly, analysis of hepatic sections prepared from young adult LDKO mice (age 4 mo) did not indicate the presence of liver pathology (SI Appendix, Fig. S1C). However, at age 10 mo, 82% of LDKO mice displayed multifocal bile duct hyperplasia together with fibrosis and inflammatory cell infiltrates (Fig. 1D). Cholangiocytes stained positively with PCNA (proliferating cell nuclear antigen), a marker for proliferation (Fig. 1D). These changes were associated with increased expression of myeloid genes (Cd68 and Lyz) and inflammatory cytokines (Ifng, Tnf, Il10, and Il12a) in the liver (Fig. 1E) and increased liver damage, as monitored by the high levels of liver enzymes (ALT, AST, and γ-GT) in the blood of LDKO mice (Fig. 1F). The remaining LDKO mice exhibited intrahepatic cholangiocarcinoma (6%) or appeared to be healthy (12%). At age 14 mo, 95% of LDKO mice displayed intrahepatic cholangiocarcinoma (Fig. 2A) associated with fibrosis (Fig. 2B) and a large increase in liver mass together with significant increases in serum levels of ALT and AST (Fig. 2C). The remaining LDKO mice (6%) exhibited cystic livers with bile duct hyperplasia. Histological analysis indicated increased staining for glutamine synthetase (GS) in liver tumor lesions, together with neoplastic nodules with positive staining of the ductular markers CK19 and Sox9 (Fig. 2D). Together, these data confirm that the majority of mature mice with compound deficiency of JNK1 and JNK2 progressively develop intrahepatic cholangiocarcinoma.
Fig. 2.
Hepatic JNK deficiency progress to cholangiocarcinoma. (A) Representative livers of LWT and LDKO mice at age 14 mo are shown. (B) Representative sections of the liver of chow-fed LDKO mice stained with H&E and Masson Trichrome (Trichrome). (Scale bar, 100 µm.) (C) The liver mass and liver damage measured by levels of ALT and AST (mean ± SEM; n = 11–20) are presented. Student’s t test differences between LDKO and LWT are indicated (**P < 0.01, ***P < 0.001). (D) Representative liver sections of 10-mo-old LDKO and LWT mice stained with GS, Cytokeratin 19 (CK19), and Sox9. (Scale bar, 100 µm.)
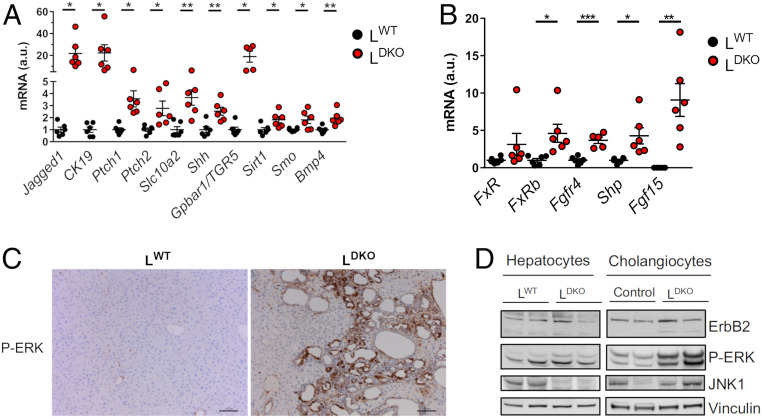
The development of bile duct hyperplasia and intrahepatic cholangiocarcinoma in LDKO mice was associated with increased hepatic expression of Cytokeratin 19 (Krt19), a cholangiocyte-specific epithelial marker, together with the G protein-coupled BA receptor 1 (Gpbar1) and the apical sodium-dependent BA transporter (Slc10a2) that are expressed in cholangiocytes (24, 25) (Fig. 3A). The Notch receptor ligand Jagged-1 promotes the formation of intrahepatic bile ducts (26) and was overexpressed in the liver of LDKO mice (Fig. 3A). Moreover, bone morphogenetic protein 4 (Bmp4) mediates cholestasis-induced fibrosis (27) and cooperates with FGF to promote the development of cholangiocytes from hepatoblasts (28); expression of hepatic Bmp4 was increased in LDKO mice compared with LWT mice (Fig. 3A). Hepatoblasts can differentiate to cholangiocytes through activation of the ERK pathway (29), and BA can increase proliferation by ERK activation through the FXR/FGF15/FGFR4 pathway (30). We evaluated this pathway in mice, before cancer has developed. In concordance with elevated BA production, we found increased Farnesoid X receptor (FXR) activity, as monitored by high levels of FXR target gene (Shp and Fgfr4) expression, in LDKO mice (Fig. 3B). Moreover, while control mice did not express Fgf15, we could detect Fgf15 mRNA (messenger RNA) in LDKO livers (Fig. 3B). Moreover, histological analysis indicated increased staining of phospho-ERK in cholangiocytes from LDKO mice compared with LWT mice (Fig. 3C). The increased phospho-ERK in cholangiocytes, but not hepatocytes, was confirmed by immunoblot analysis (Fig. 3D). Together, these changes in FXR/FGF15/FGFR4/ERK pathway activity may contribute to cholangiocyte proliferation and maturation from hepatoblasts, resulting in bile duct hyperplasia and the development of cholangiocarcinoma detected in LDKO mice.
Fig. 3.
ERK pathway activation accompanies the development of cholangiocarcinoma induced by hepatic JNK deficiency. (A) The expression of genes related to cholangiocytes proliferation was evaluated by RT-PCR in 14-mo-old LWT and LDKO mice and normalized to the amount of 18S RNA in each sample (mean ± SEM; n = 5–6). Student’s t test differences between LDKO and LWT are indicated (*P < 0.05; **P < 0.01). (B) The expression of genes related to the nuclear factor FXR (Fxr, Fxrb, Shp, Fgfr4) and Fgf15 was measured by quantitative RT-PCR in LWT and LDKO liver from mice at age 6 mo (mean ± SEM; n = 5–6) normalized to the amount of Actb mRNA in each sample. Student’s t tests between LDKO and LWT are indicated (*P < 0.05; **P < 0.01, ***P < 0.001). (C) Representative liver sections of LDKO and LWT mice (age 10 mo) stained with phospho-ERK. (Scale bar, 100 µm.) (D) Comparative protein expression of ErbB2, phospho-ERK, and JNK1 in hepatocytes and cholangiocytes in LWT and LDKO mice was evaluated by immunoblot analysis. Vinculin was used as a loading control.
PPARα Deficiency Reduces Liver Cancer in JNK1/2 Deficient Liver.
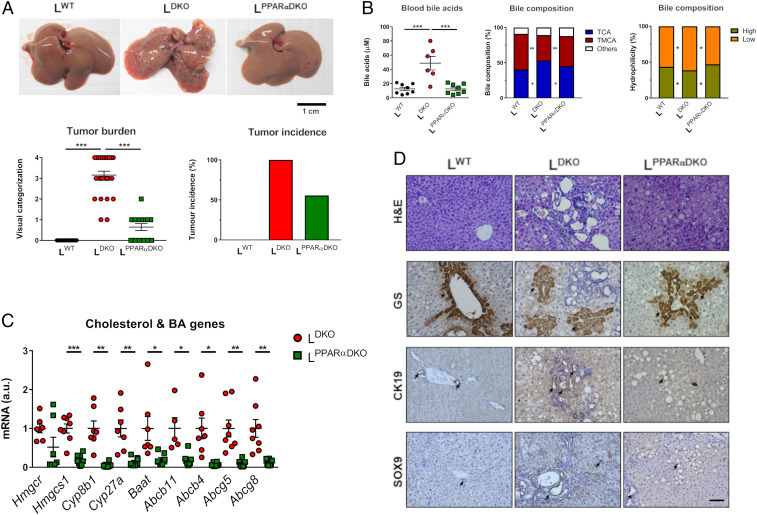
We examined hepatic gene expression in chow-fed WT and LDKO mice using RNA-seq data (7) and found evidence for activation of PPARα and FXR transcription factors in the liver of LDKO mice compared to LWT mice (SI Appendix, Fig. S2). To test the role of PPARα, we ablated the Ppara gene in LDKO mice to examine whether PPARα is required for the development of intrahepatic cholangiocarcinoma. PPARα plus JNK1/2 liver-deficient mice (LPPARαDKO) exhibited reduced tumor burden and tumor incidence compared with LDKO mice (Fig. 4A). The major changes in BA were also reversed (Fig. 4B). This is consistent with reduced hepatic expression in LPPARαDKO mice of genes involved in cholesterol and BA synthesis (Hmgcr, Baat, Cyp8b1, and Cyp27a) and hepatocyte-mediated BA transport (Abc11, Abc4, Abcg5, Abcg8) (Fig. 4C). Histological analyses indicated that PPARα deficiency caused increased liver steatosis, but reduced hallmarks of carcinogenesis (anisokaryosis, apoptosis, ductogenesis, dysplasia, and mitosis) in LPPARαDKO compared with LDKO mice (Table 1 and Fig. 4D). Moreover, CK19 and SOX9 staining were increased in LDKO mice compared with LPPARαDKO, consistent with cholangiocyte proliferation. Furthermore, the prominent staining for GS by liver tumor lesions in LDKO mice was suppressed in the liver of LPPARαDKO mice (Fig. 4D).
Fig. 4.
PPARα deficiency reduces liver cancer induced by hepatic JNK deficiency. (A) Representative livers, tumor burden, and incidence in 11-mo-old LWT, LDKO, and LPPARαDKO mice (mean ± SEM; n = 14 ∼ 25). (B) The amount of bile acid in the blood was measured (mean ± SE; n = 7–11). The composition of bile fluid collected from the gall bladder was examined. Two-way ANOVA differences between LPPARαDKO, LDKO, and LWT are indicated (*P < 0.05; **P < 0.01). (C) The expression of genes related to cholesterol synthesis (Hmgcr and Hmgcs1), BA synthesis, and transporters (Cyp27a1, Baat, Cyp27a, Cyp8b1, Abcb11, Abcb4, Abcg5, Abcg8) was measured by quantitative RT-PCR (mean ± SEM; n = 5–8) normalized to the amount of Actb mRNA in each sample. Two-way ANOVA differences between LDKO and LPPARαDKO are indicated (*P < 0.05; **P < 0.01, ***P < 0.001). (D) Representative liver sections of 10-mo-old LWT, LDKO, and LPPARαDKO mice stained with GS, Cytokeratin 19 (CK19), and Sox9. (Scale bar, 100 µm.)
Table 1.
PPARα deficiency reduces carcinogenesis markers in JNK1/2-deficient livers
| LWT | LDKO | LPPARαDKO | |
| Anisokaryosis | − | ++ | + |
| Apoptosis | − | +++ | + |
| Necrotic foci | − | +++ | ++ |
| Cellular hypertrophy | − | +++ | ++ |
| Ductogenesis | − | +++ | + |
| Cystogenesis | − | +++ | ++ |
| Microsteatosis | + | + | +++ |
| Macrosteatosis | − | ++ | +++ |
| Lymphocytic inflammation | − | +++ | + |
| Dysplasia | − | +++ | + |
| Mitosis | + | +++ | + |
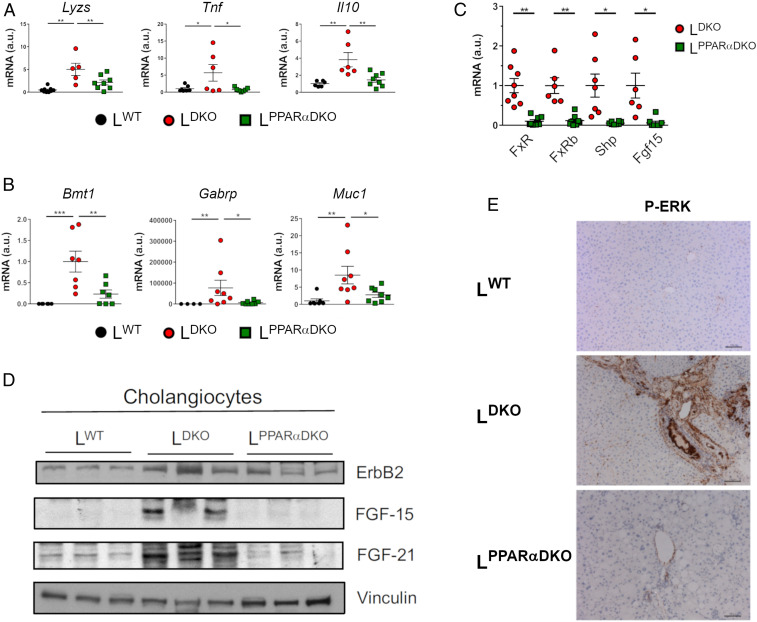
Gene expression analysis demonstrated that both inflammation and cholangiocarcinoma markers were reduced in LPPARαDKO mice compared with LDKO mice (Fig. 5 A and B). This evidence suggests that PPARα deficiency protected against the promotion of cholangiocyte proliferation in mice lacking hepatocyte JNK1/2. To evaluate whether PPARα deficiency and subsequent normalization of BA production blunted the FXR/FGF15/FGFR4/ERK pathway, we evaluated FXR target gene expression. We found that hepatic expression of Fgf15 and Shp was reduced in LPPARαDKO mice compared with LDKO mice (Fig. 5C). Indeed, immunoblot analysis confirmed that PPARα deficiency suppressed the increased FGF15/21 expression detected in JNK-deficient cholangiocytes (Fig. 5D). This is consistent with the observation of lower levels of ERK activation, detected by immunohistochemistry, in LPPARαDKO cholangiocytes (Fig. 5E).
Fig. 5.
PPARα deficiency reduces liver cholangiocarcinoma induced by hepatic JNK deficiency. (A and B) The expression of genes related to inflammation and cholangiocarcinoma was evaluated by quantitative RT-PCR (mean ± SEM; n = 4–8) normalizing to the amount of Actb mRNA in each sample. Two-way ANOVA differences between LDKO and LPPARαDKO are indicated (*P < 0.05; **P < 0.01, ***P < 0.001). (C) The expression of genes related to nuclear factor FXR pathway (Fxr, Fxrb, Shp, Fgfr4, and Fgf15) was evaluated in LKO and LPPARαDKO livers by quantitative RT-PCR (mean ± SEM; n = 6–8) normalizing to the amount of Actb mRNA in each sample. Student’s t test differences between LDKO and LPPARαDKO are indicated (*P < 0.05; **P < 0.01). (D) Representative Western blots of ErbB2, FGF15, and FGF21 in cholangiocytes in LDKO, LPPARαDKO, and LWT mice (n = 3). Vinculin was used as a loading control. (E) Representative sections of the liver of 10-mo-old LDKO, LPPARαDKO, and LWT mice stained with Phospho-ERK. (Scale bar, 100 µm.)
Discussion
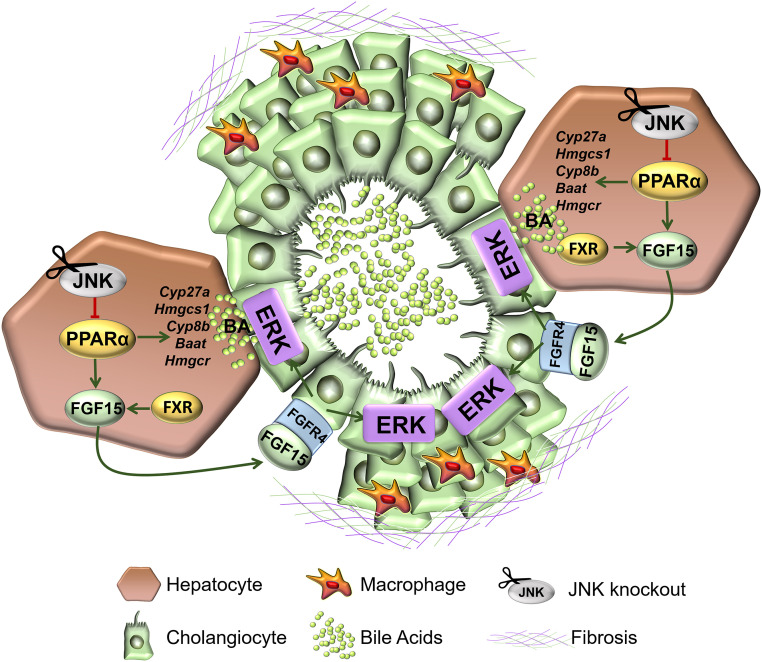
Since increased BA can lead to inflammation, apoptosis, and necrosis of hepatocytes (31, 32), long-term elevated BA levels in patients are considered a risk factor for liver cancer development (33). Indeed, serum BA might be useful for the diagnosis of cholangiocarcinoma (34). Our findings provide an animal model in which defects in BA homeostasis are linked to cholangiocarcinoma (Fig. 6).
Fig. 6.
Schematic illustration of JNK-regulated cholangiocarcinoma development in the liver.
Previous studies have established that hepatic JNK deficiency can suppress cholangiocyte proliferation and oncogenic transformation in a p53/Kras-induced model of cholangiosarcoma (35) and promotes cholangiocarcinoma in dethylnitrosamine and NEMO deficiency models of liver cancer (36). The results of the present study demonstrate that hepatic JNK deficiency is sufficient for the development of cholangiocyte malignancy (Fig. 2).
PPARα is an important modulator of liver metabolism controlling lipid and BA homeostasis, and its activation has been shown to decrease fatty liver disease (19, 37, 38). We report that JNK-mediated repression of PPARα causes changes in BA homeostasis which suppress cholangiocyte proliferation. Consequently, JNK deficiency stimulates cholangiocyte proliferation and promotes the development of cholangiocarcinoma. This increased proliferation is mediated by the altered BA metabolism and the elevated hepatic expression of FXR/FGF15/FGFR4 that triggers ERK activation in cholangiocytes (Fig. 6). Our results have strong translational implications for obesity treatment. Activation of FXR by BA triggers the secretion of FGF15/FGF19 in humans (39), and the beneficial effects of FGF expression on the obese metabolic profile has been well characterized (40). However, the clinical use of FGF has been debated due to the potential for increasing liver tumor development (41, 42).
Hepatic PPARα is an important mediator of this regulatory cascade. Indeed, PPARα deficiency dramatically suppresses the phenotypes caused by JNK deficiency. Nevertheless, the role of PPARα in liver cancer remains unclear. While some studies have demonstrated that PPARα activation might promote liver cancer (43–45), others indicate that PPARα activation may be neutral or suppress liver cancer development (46–49). This could be due to different experimental conditions used in these studies. In our system, the protumorigenic effect of PPARα activation is due, in part, to an alteration in BA metabolism that drives ERK activation, suggesting that PPARα activation is a critical factor in cholangiocarcinoma development and progression.
The role of JNK/PPARα/FGF signaling in lipid metabolism suggests that this pathway could represent a target for the treatment for steatosis and obesity. However, the possible contribution of this pathway to carcinogenic progression represents a serious problem for long-term treatment. Our analysis suggests that treatment strategies using long-term JNK inhibition should consider the potential risk of cholangiocarcinoma development among the possible secondary effects of this treatment.
Methods
Animals.
PPARα knockout mice (B6;129S4-Pparatm1Gonz/J; RRID:IMSR_JAX:008154) and Albumin-Cre mice (B6.Cg-Speer6-ps1Tg(Alb-cre)21Mng/J; RRID:IMSR_JAX:003574) were purchased from the Jackson Laboratory and backcrossed for 10 generations to the C57BL/6J background (Jackson Laboratory; RRID:IMSR_JAX:000664). Mice with compound JNK1/2 deficiency in hepatocytes (LDKO) have been described (50, 51). Genotypes were identified by PCR analysis of genomic DNA isolated from mouse tails. All experiments were performed using male mice. Mice were housed in a pathogen-free animal facility and kept on a 12-h light/dark cycle at constant temperature and humidity.
All animal experiments conformed to European Union (EU) Directive 2010/63EU and Recommendation 2007/526/EC, enforced in Spanish law under Real Decreto 53/2013 and the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
Serum Analysis.
Plasma transaminase activity was assessed with the ALT and AST Reagent Kit (Biosystems Reagents) using a Benchmark Plus microplate spectrophotometer (Bio-Rad). Plasma concentration of nonsulfated bile acids was measured with the Bile Acid Assay Kit (Sigma-Aldrich) using a Fluoroskan Ascent fluorescence multiwell plate reader (Thermo Labsystems).
Histochemistry.
Histology was performed using tissue fixed in 10% formalin for 24 h, dehydrated, and embedded in paraffin. Sections (7 μm) were cut and stained using hematoxylin and eosin (American Master Tech Scientific). Sections were also incubated with Bouin´s fluid overnight, counterstain with hematoxylin (Sigma), and then stained with Masson-Trichrome stain (American Master Tech Scientific). Immunohistochemistry was performed by staining tissue sections with antibodies against PCNA (biotinylated from Thermofisher MS-106-B; RRID:AB_64272), SOX9 (Abcam ab3697; RRID:AB_304012), glutamine synthetase (Abcam ab73593; RRID:AB_2247588), cytokeratin 19 (Abcam ab15463; RRID:AB_2281021), or phospho-p44/42 MAPK (Thr202/Tyr204) (Cell Signaling Technology #9101). Streptavidin-conjugated horseradish peroxidase (Biogenex) and the substrate 3,3′-diaminobenzidene (Vector Laboratories) were used followed by brief counterstaining with Mayer’s hematoxylin (Sigma).
Analysis of Biliary Lipids.
Bile was collected from the gall bladder following cholecystectomy. We determined cholesterol and phospholipids using an enzymatic assay (Wako). Total bile acids were measured using Hall's Bile Stain Kit (American MasterTech). Bile acid species were examined by a modification of the method described by ref. 52 using an HPLC-MS/MS (6410 Triple Quad LC/MS, Agilent Technologies). Chromatographic separation was achieved with gradient elution using a Zorbax Eclipse XDB-C18 column (150 × 4.6 mm, 5 µm) kept at 35 °C and a flow rate of 500 µL/min. Initial mobile phase was 80:20 methanol/water, both containing 5 mM ammonium acetate and 0.01% formic acid, pH 4.6, and it was changed to 97:3 methanol/water over 9 min and then returned to 80:20 in 1 min. Electrospray ionization in negative mode was used, with the following conditions: gas temperature 350 °C, gas flow 8 L/min, nebulizer 10 psi, capillary voltage 2,500 V. MS/MS acquisition was performed in multiple reaction monitoring mode using the specific m/z transitions: [M-H]− ion to 80.2 for taurine-conjugated bile acids and [M-H]− ion to 74 for glycine-conjugated bile acids. Free bile acids did not generate characteristic ion fragments, as reported by others (52), and transition from unfragmented precursor molecular ions 407.1–407.1, 391.3–391.3, and 375.3–375.3 were selected for trihydroxylated, dehydroxylated, and monohydroxylated free bile acids, respectively.
Isolation of Hepatocytes and Cholangiocytes.
Mice livers were perfused using a peristaltic pump with 40 mL of Hanks Balanced Salt Solution (with 10 mM Hepes [pH 7.4] and 1 mM EGTA; without MgCl2 and CaCl2) and then with 1 mg/mL of collagenase type I (Worthington). Hepatocytes were collected as a cell suspension in Dulbecco’s Modified Eagle’s Medium/F12 Medium (10% FBS, 0.2 mg/mL BSA, 5% sodium pyruvate, 10 mM Hepes [pH 7.4], 1% l-glutamine, 1% Penicillin/Streptomycin, 0.51 mg/mL NaHCO3). Biliary trees were collected in Williams´medium supplemented with 5 mM Hepes (pH 7.4) and 1 mg/mL collagenase type I prior to incubation (1 h) at 37 °C to isolate cholangiocytes. The cells were washed with PBS before protein extraction as previously described (53).
Western Blot Analysis.
Hepatocytes and cholangiocytes proteins were extracted in lysis buffer (50 mM Tris⋅HCl pH 7.5, 1 mM EGTA, 1 mM EDTA pH 8.0, 50 mM sodium fluoride, 1 mM sodium β-glycerophosphate, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 0.27 M sucrose, 1% Triton X-100, 0.1 mM PMSF, 0.1% 2-mercaptoethanol, 1 µg/mL leupeptin, and 1 µg/mL aprotinin). Extracts were separated by SDS–PAGE and transferred to 0.2-µm pore-size nitrocellulose membranes (Bio-Rad). Blots were probed with primary antibodies to ErbB2 (Cat# ab16901, Abcam; RRID: AB_443537), phospho-ERK (Cat#9101, Cell Signaling Biotechnology; RRID: AB_330744), JNK1 (Cat# sc-1648, Santa Cruz Biotechnology; RRID:vAB_675868), FGF-15 (Cat# sc-514647, Santa Cruz Biotechnology), FGF-21 (Cat# RD281108100, BioVendor Laboratory Medicine; RRID: AB_2034054), and Vinculin (Cat#V4505, Sigma; RRID:AB_477617). All antibodies were used at 1:1,000 dilution. The membranes were washed and incubated with an appropriate horseradish peroxidase-conjugated secondary antibody (GE Healthcare), and immune complexes were detected using an enhanced chemiluminescent substrate (Clarity Western ECL substrate; Bio-Rad).
RNA-seq Analysis.
Liver RNA-seq data from chow-fed LWT and LDKO (GEO GSE55190) were examined (7). Sequencing reads were preprocessed by means of a pipeline that used FastQC, to assess read quality, and Cutadapt to trim sequencing reads, eliminating Illumina adaptor remains, and to discard reads that were shorter than 30 bp. Resulting reads were mapped against reference transcriptome GRCm38.91 and quantified using RSEM. Percentages of reads participating in at least one reported alignment were around 80%. Expected expression counts calculated with RSEM were then processed with an analysis pipeline that used the Bioconductor package Limma for normalization (using TMM method) and differential expression testing, taking only into account those genes expressed with at least 1 count per million in a number of samples equal to the number of replicate samples of the condition with less replicates. Significant expression changes between wild type (WT) and JNK1/2 double-KO conditions, with Benjamini and Hochberg adjusted P value < 0.05, were detected for 44 genes. Given that the collection of differentially expressed genes was relatively small, an adjusted P value threshold of 0.2 was applied for further analyses. The resulting collection of 739 genes was then used for functional enrichment analyses with IPA (Ingenuity Pathway Analysis), to discover overrepresented gene lists derived from Ingenuity's proprietary knowledge-base (IPAKB). IPAKB-derived gene lists consisted of collections of genes belonging to the same signaling or metabolic pathway (Canonical Pathway analyses), or regulated by the same molecule (Upstream Regulator analyses). In general, enrichments associated to Benjamini-Hochberg adjusted P value < 0.05 are considered significant. Importantly, IPA may issue predictions on the activation state of pathways or regulators in the form of a parameter called z score; activation or inhibition is indicated by positive or negative values, respectively. Other data manipulations and graphical representations (heatmaps, bar plots, and scatter plots) were produced with statistical package R.
Real-Time qPCR.
Total RNA was isolated from liver and tumor tissue using the RNeasy Mini Kit (Qiagen) with on-column DNase I-digestion. cDNA (complementary DNA) was synthesized with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Taqman© assays were performed using the probes listed in SI Appendix, Table S1 (Applied Biosystems). Sequences of primers used for quantitative real-time PCR (RT-PCR) are provided in SI Appendix, Table S2. Expression levels were normalized to 18S using Taqman© assays (430449011032, Applied Biosystems) (Figs. 1 and 3A and SI Appendix, Fig. S1) or Actb (Figs. 3B, 4, and 5) mRNA. qRT-PCR was performed using the Fast SYBR Green system (Applied Biosystems) in a 7900HT Fast Real-Time PCR thermal cycler (Applied Biosystems). A dissociation curve program was employed after each reaction to verify purity of the PCR products. The expression of mRNA was examined by quantitative PCR analysis using a 7500 Fast Real-Time PCR machine.
Statistics.
Differences between groups were examined for statistical significance using two-tailed unpaired Student’s t test (with Welch’s correction when variances were different) or ANOVA coupled to Bonferroni’s posttest. Kaplan-Meier analysis was performed using the log-rank test. Statistical details and experimental n are specified in figure legends. Statistical analyses were performed with the GraphPad Prism 7 software (RRID:SCR_002798).
Material and Data Availability.
Sources for materials used in this study are described in Methods. The raw data obtained for this study are presented in SI Appendix, Fig. 3D and Dataset S1.
Supplementary Material
Acknowledgments
We thank S. Bartlett for English editing and David Garlick (University of Massachusetts Medical School) for pathological examination of tissue sections. We are grateful to the CNIC Advanced Imaging and Animal facility for technical support. G.S. (RYC-2009-04972) and F.J.C. (RYC-2014-15242) are investigators of the Ramón y Cajal Program. E.M. was awarded a La Caixa fellowship. C.F. was awarded a Sara Borrell contract (CD19/00078). This work was funded by grants supported in part by funds from the European Regional Development Fund: EU’s Seventh Framework Programme (FP7/2007-2013) ERC 260464, EFSD/Lilly European Diabetes Research Programme Dr. Sabio, 2017 Leonardo Grant for Researchers and Cultural Creators, BBVA Foundation (Investigadores-BBVA-2017) IN[17]_BBM_BAS_0066, MINECO-FEDER SAF2016-79126-R, and Comunidad de Madrid IMMUNOTHERCAN-CM S2010/BMD-2326 and B2017/BMD-3733 and La Asociación Española contra el Cáncer (to G.S.); EXOHEP-CM S2017/BMD-3727 and the European Cooperation in Science & Technology (COST) Action CA17112 (to F.J.C.); MINECO Retos SAF2016-78711, the AMMF Cholangiocarcinoma Charity 2018/117, NanoLiver-CM Y2018/NMT-4949, UCM-25-2019, ERAB EA/18-14 (to F.J.C.). F.J.C. is a Gilead Liver Research Scholar. Grant DK R01 DK107220 from the NIH (to R.J.D.); and PI16/00598 from Carlos III Institute of Health, Spain (to J.J.G.M.). The CNIC is supported by the Ministerio de Ciencia, Innovación y Universidades, and the Pro-CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
Footnotes
The authors declare no competing interest.
Data deposition: The RNA-seq analysis (Fig. S2) was performed using a dataset that we have previously published (7) that was deposited in the GEO database (GSE55190).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002672117/-/DCSupplemental.
References
- 1.Parkin D. M., Bray F., Ferlay J., Pisani P., Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 (2005). [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H. B., Davila J. A., Petersen N. J., McGlynn K. A., The continuing increase in the incidence of hepatocellular carcinoma in the United States: An update. Ann. Intern. Med. 139, 817–823 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Khan S. A., Tavolari S., Brandi G., Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 39 (suppl. 1), 19–31 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Khan S. A. et al.; British Society of Gastroenterology , Guidelines for the diagnosis and treatment of cholangiocarcinoma: An update. Gut 61, 1657–1669 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Manieri E., Sabio G., Stress kinases in the modulation of metabolism and energy balance. J. Mol. Endocrinol. 55, R11–R22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabio G., Davis R. J., cJun NH2-terminal kinase 1 (JNK1): Roles in metabolic regulation of insulin resistance. Trends Biochem. Sci. 35, 490–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vernia S. et al., The PPARα-FGF21 hormone axis contributes to metabolic regulation by the hepatic JNK signaling pathway. Cell Metab. 20, 512–525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vernia S., Cavanagh-Kyros J., Barrett T., Tournier C., Davis R. J., Fibroblast growth factor 21 mediates glycemic regulation by hepatic JNK. Cell Rep. 14, 2273–2280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulick T., Cresci S., Caira T., Moore D. D., Kelly D. P., The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl. Acad. Sci. U.S.A. 91, 11012–11016 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unger R. H., Zhou Y. T., Lipotoxicity of beta-cells in obesity and in other causes of fatty acid spillover. Diabetes 50 (suppl. 1), S118–S121 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Evans R. M., Barish G. D., Wang Y. X., PPARs and the complex journey to obesity. Nat. Med. 10, 355–361 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Memon R. A. et al., Up-regulation of peroxisome proliferator-activated receptors (PPAR-alpha) and PPAR-gamma messenger ribonucleic acid expression in the liver in murine obesity: Troglitazone induces expression of PPAR-gamma-responsive adipose tissue-specific genes in the liver of obese diabetic mice. Endocrinology 141, 4021–4031 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Ip E. et al., Central role of PPARalpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology 38, 123–132 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Teoh N. C. et al., Short-term therapy with peroxisome proliferation-activator receptor-alpha agonist Wy-14,643 protects murine fatty liver against ischemia-reperfusion injury. Hepatology 51, 996–1006 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Holden P. R., Tugwood J. D., Peroxisome proliferator-activated receptor alpha: Role in rodent liver cancer and species differences. J. Mol. Endocrinol. 22, 1–8 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Panigrahy D. et al., PPARalpha agonist fenofibrate suppresses tumor growth through direct and indirect angiogenesis inhibition. Proc. Natl. Acad. Sci. U.S.A. 105, 985–990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggiora M., Oraldi M., Muzio G., Canuto R. A., Involvement of PPARα and PPARγ in apoptosis and proliferation of human hepatocarcinoma HepG2 cells. Cell Biochem. Funct. 28, 571–577 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Yamasaki D. et al., Fenofibrate suppresses growth of the human hepatocellular carcinoma cell via PPARα-independent mechanisms. Eur. J. Cell Biol. 90, 657–664 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Li T., Chiang J. Y., Regulation of bile acid and cholesterol metabolism by PPARs. PPAR Res. 2009, 501739 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo C. et al., Multiplexed gene expression profiling identifies the FGFR4 pathway as a novel biomarker in intrahepatic cholangiocarcinoma. Oncotarget 8, 38592–38601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miura S. et al., Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer 12, 56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro H., Kolodziejczyk A. A., Halstuch D., Elinav E., Bile acids in glucose metabolism in health and disease. J. Exp. Med. 215, 383–396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Groen P. C., Gores G. J., LaRusso N. F., Gunderson L. L., Nagorney D. M., Biliary tract cancers. N. Engl. J. Med. 341, 1368–1378 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Keitel V. et al., The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology 45, 695–704 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Dawson P. A., Lan T., Rao A., Bile acid transporters. J. Lipid Res. 50, 2340–2357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piccoli D. A., Spinner N. B., Alagille syndrome and the Jagged1 gene. Semin. Liver Dis. 21, 525–534 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Fan J. et al., Bone morphogenetic protein 4 mediates bile duct ligation induced liver fibrosis through activation of Smad1 and ERK1/2 in rat hepatic stellate cells. J. Cell. Physiol. 207, 499–505 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Yanai M. et al., FGF signaling segregates biliary cell-lineage from chick hepatoblasts cooperatively with BMP4 and ECM components in vitro. Dev. Dyn. 237, 1268–1283 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Yang L. et al., A single-cell transcriptomic analysis reveals precise pathways and regulatory mechanisms underlying hepatoblast differentiation. Hepatology 66, 1387–1401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Chiang J. Y., Bile acids as metabolic regulators. Curr. Opin. Gastroenterol. 31, 159–165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen P. L. et al., The ascending pathophysiology of cholestatic liver disease. Hepatology 65, 722–738 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Allen K., Jaeschke H., Copple B. L., Bile acids induce inflammatory genes in hepatocytes: A novel mechanism of inflammation during obstructive cholestasis. Am. J. Pathol. 178, 175–186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W. et al., A weighted relative difference accumulation algorithm for dynamic metabolomics data: Long-term elevated bile acids are risk factors for hepatocellular carcinoma. Sci. Rep. 5, 8984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sombattheera S. et al., Total serum bile acid as a potential marker for the diagnosis of cholangiocarcinoma without jaundice. Asian Pac. J. Cancer Prev. 16, 1367–1370 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Yuan D. et al., Kupffer cell-derived Tnf triggers cholangiocellular tumorigenesis through JNK due to chronic mitochondrial dysfunction and ROS. Cancer Cell 31, 771–789.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cubero F. J. et al., Loss of c-Jun N-terminal kinase 1 and 2 function in liver epithelial cells triggers biliary hyperproliferation resembling cholangiocarcinoma. Hepatol. Commun 4, 834–851 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelmegeed M. A. et al., PPARalpha expression protects male mice from high fat-induced nonalcoholic fatty liver. J. Nutr. 141, 603–610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeon J. E. et al., Reduced expression of peroxisome proliferator-activated receptor-alpha may have an important role in the development of non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 19, 799–804 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Inagaki T. et al., Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Nies V. J. et al., Fibroblast growth factor signaling in metabolic regulation. Front. Endocrinol. (Lausanne) 6, 193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M. et al., Mouse species-specific control of hepatocarcinogenesis and metabolism by FGF19/FGF15. J. Hepatol. 66, 1182–1192 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Cui G. et al., Up-regulation of FGF15/19 signaling promotes hepatocellular carcinoma in the background of fatty liver. J. Exp. Clin. Cancer Res. 37, 136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters J. M., Cattley R. C., Gonzalez F. J., Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis 18, 2029–2033 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Hays T. et al., Role of peroxisome proliferator-activated receptor-alpha (PPARalpha) in bezafibrate-induced hepatocarcinogenesis and cholestasis. Carcinogenesis 26, 219–227 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Nishimura J. et al., Effect of fenofibrate on oxidative DNA damage and on gene expression related to cell proliferation and apoptosis in rats. Toxicol. Sci. 97, 44–54 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Takashima K., Ito Y., Gonzalez F. J., Nakajima T., Different mechanisms of DEHP-induced hepatocellular adenoma tumorigenesis in wild-type and Ppar alpha-null mice. J. Occup. Health 50, 169–180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heindryckx F., Colle I., Van Vlierberghe H., Experimental mouse models for hepatocellular carcinoma research. Int. J. Exp. Pathol. 90, 367–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung C. et al., Diminished hepatocellular proliferation in mice humanized for the nuclear receptor peroxisome proliferator-activated receptor alpha. Cancer Res. 64, 3849–3854 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Morimura K., Cheung C., Ward J. M., Reddy J. K., Gonzalez F. J., Differential susceptibility of mice humanized for peroxisome proliferator-activated receptor alpha to Wy-14,643-induced liver tumorigenesis. Carcinogenesis 27, 1074–1080 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das M., Garlick D. S., Greiner D. L., Davis R. J., The role of JNK in the development of hepatocellular carcinoma. Genes Dev. 25, 634–645 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das M. et al., Induction of hepatitis by JNK-mediated expression of TNF-alpha. Cell 136, 249–260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye L., Liu S., Wang M., Shao Y., Ding M., High-performance liquid chromatography-tandem mass spectrometry for the analysis of bile acid profiles in serum of women with intrahepatic cholestasis of pregnancy. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 860, 10–17 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Barbier-Torres L. et al., Histone deacetylase 4 promotes cholestatic liver injury in the absence of prohibitin-1. Hepatology 62, 1237–1248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.