Significance
Overnutrition leads to metabolic disorders including obesity and diabetes. Studies have shown that enhanced inflammation is an essential player in the progression of metabolic diseases. However, how immune cells sense nutritional status and contribute to whole-body metabolism are largely unknown. Protein O-linked β-N-acetylglucosamine (O-GlcNAc) modification is thought to be a metabolic sensor that modulates cell signaling. Here, we show that overnutrition stimulates O-GlcNAc signaling in macrophages. O-GlcNAc signaling suppresses macrophage proinflammatory activation and protects against diet-induced obesity and metabolic dysfunction. This study uncovers O-GlcNAc signaling as a homeostatic regulator at the interface of inflammation and metabolism and suggests that O-GlcNAc signaling may serve as a therapeutic target for obesity, diabetes, and other immune-related diseases.
Keywords: metabolic homeostasis, immunometabolism, knockout mice, RNA sequencing
Abstract
Enhanced inflammation is believed to contribute to overnutrition-induced metabolic disturbance. Nutrient flux has also been shown to be essential for immune cell activation. Here, we report an unexpected role of nutrient-sensing O-linked β-N-acetylglucosamine (O-GlcNAc) signaling in suppressing macrophage proinflammatory activation and preventing diet-induced metabolic dysfunction. Overnutrition stimulates an increase in O-GlcNAc signaling in macrophages. O-GlcNAc signaling is down-regulated during macrophage proinflammatory activation. Suppressing O-GlcNAc signaling by O-GlcNAc transferase (OGT) knockout enhances macrophage proinflammatory polarization, promotes adipose tissue inflammation and lipolysis, increases lipid accumulation in peripheral tissues, and exacerbates tissue-specific and whole-body insulin resistance in high-fat-diet-induced obese mice. OGT inhibits macrophage proinflammatory activation by catalyzing ribosomal protein S6 kinase beta-1 (S6K1) O-GlcNAcylation and suppressing S6K1 phosphorylation and mTORC1 signaling. These findings thus identify macrophage O-GlcNAc signaling as a homeostatic mechanism maintaining whole-body metabolism under overnutrition.
Modern global healthcare faces challenges that are different from past generations, as embodied by the prevalence of obesity worldwide. Obesity and associated insulin resistance play a pivotal role in the pathogenesis of type 2 diabetes, nonalcoholic fatty liver disease, atherosclerotic heart disease, and other disorders (1, 2). The association between obesity and the immune system has long been appreciated. The vicious cycle of inflammation between metabolic organs and immune cells in obese individuals is of major importance to the development of whole-body insulin resistance and metabolic dysfunction (3–6). For example, changes in adipose tissue (AT) microenvironment caused by chemokine release, adipocyte death, and so on during obesity lead to macrophage infiltration and proinflammatory activation, which further lead to detrimental effects on insulin action in metabolic tissues and contribute to whole-body insulin resistance (7–9). On the other hand, metabolic rewiring has been shown to be essential for immune cell maturation and acquisition of functional competency (10, 11). For example, classical (M1-like, proinflammatory) activation of macrophages is coupled to increased glycolysis, reduced mitochondrial oxidative phosphorylation, and fragmented tricarboxylic acid cycle, while alternative (M2-like, antiinflammatory) activation of macrophages is characterized by changes in iron metabolism, fatty acid oxidation, glutamine metabolism, and the uridine diphosphate β-N-acetylglucosamine (UDP-GlcNAc) pathway (12, 13). Despite the growing knowledge of the function of immune cells in tissue and whole-body metabolism and an emerging role of metabolic pathways in immune cell activation and immune responses, how immune cells sense and integrate nutritional cues and how the nutrient-sensing mechanisms control immune cell activation and whole-body metabolism remain overarching questions in the field.
O-GlcNAcylation is a nutrient-sensing posttranslational modification that involves the attachment of a single O-linked β-N-acetylglucosamine (O-GlcNAc) moiety to proteins (14–16). Nutrients including glucose, amino acid, fatty acid, and nucleotide flux into the hexosamine biosynthetic pathway (HBP) to generate the donor molecule for O-GlcNAcylation, UDP-GlcNAc. A single pair of enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), catalyze the addition and removal of O-GlcNAc moieties from serine and threonine residues of nuclear, cytoplasmic, and mitochondrial proteins, respectively. O-GlcNAc signaling, the dynamic and reversible modification in cells, has been shown to fluctuate with the availability of nutrients and regulate numerous signaling pathways (17–21). Dysregulated O-GlcNAc homeostasis has been linked to chronic human diseases associated with metabolic dysregulation, such as obesity, diabetes, cardiovascular diseases, Alzheimer’s disease, and cancer (22–24). Studies have shown that O-GlcNAcylation dampens insulin signaling both in vitro and in vivo (16, 17, 25). Key metabolic regulators such as protein kinase B (Akt), forkhead box protein O1 (FOXO1), carnitine palmitoyltransferase 2 (CPT2), and peroxisome proliferator-activated receptor-γ coactivator (PGC-1α) can be regulated by O-GlcNAcylation (26–29). O-GlcNAcylation was discovered in immune cells over 30 y ago and has been shown to regulate several immune signaling pathways in vitro, such as extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), p38 mitogen-activated protein kinase, and nuclear factor κB (NF-κB) signaling pathways (17–19, 30–32). However, the function of O-GlcNAc signaling in immune regulation, especially during metabolic perturbations, remains elusive.
In obesity and insulin resistance, chronic low-grade inflammation develops in multiple organs such as AT, liver, and skeletal muscle, which is reflected by immune cell infiltration and elevated proinflammatory cytokine production (3, 4, 6, 33–36). Macrophages are the primary cell type responsible for immune responses in metabolic tissues (8, 37). AT inflammation is characterized by macrophage infiltration and has been linked to impaired whole-body energy homeostasis (8, 9, 38). Approximately 10 to 15% of the stromal cells in AT are macrophages in lean mice, whereas it increases up to 60% in obese mice (33, 39). Furthermore, macrophages are capable of acquiring diverse functions in response to physiological and environmental signals. Classically activated (M1-like) macrophages produce proinflammatory cytokines and promote insulin resistance, while alternatively activated (M2-like) macrophages produce antiinflammatory cytokines and are associated with insulin sensitivity and lean phenotype (40, 41).
Here, we explored the role of O-GlcNAc signaling in macrophage activation and metabolic regulation in a mouse model of diet-induced obesity. We observed an increase in macrophage O-GlcNAc signaling during overnutrition and a transient decrease in O-GlcNAc signaling during macrophage proinflammatory polarization. Loss of OGT enhances macrophage proinflammatory activation in AT, which leads to enhanced adipocyte lipolysis, ectopic lipid accumulation in liver and muscle, and whole-body insulin resistance. These data elucidate an antiinflammatory role for O-GlcNAc signaling through inhibiting macrophage proinflammatory activation and provide a molecular link between overnutrition, inflammation, and metabolic dysfunction.
Results
O-GlcNAc Signaling Is Altered during Overnutrition and Macrophage Activation.
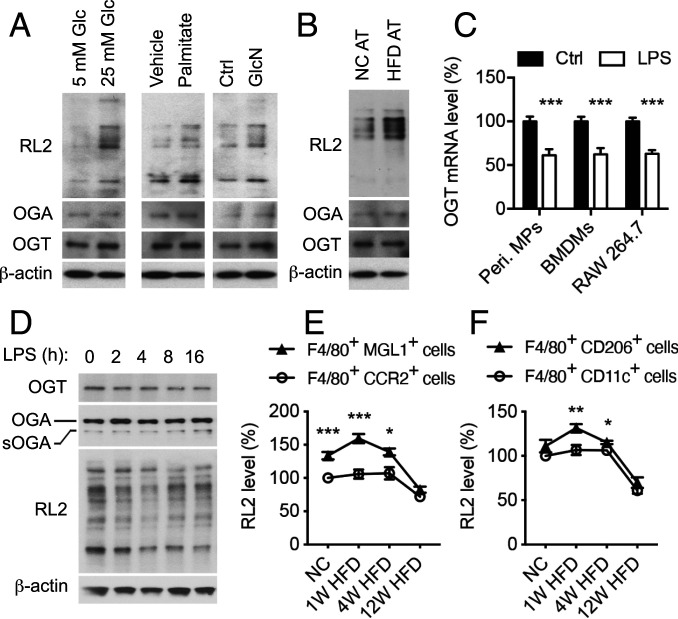
To study the nutritional regulation of macrophage function, we first examined how macrophages respond to different nutrient environments in vitro. Mouse peritoneal macrophages and bone marrow-derived macrophages (BMDMs) were first maintained in a culture medium containing physiological (5 mM) glucose and then challenged with 25 mM glucose or 0.3 mM palmitate to mimic glucolipotoxic conditions in type 2 diabetes. Glucosamine (GlcN) was used as a positive control to boost HBP and O-GlcNAcylation. Both conditions and GlcN led to enhanced overall O-GlcNAcylation levels, but not OGT and OGA levels, in cultured macrophages (Fig. 1A and SI Appendix, Fig. S1A). Moreover, peritoneal macrophages cocultured with epididymal white AT from the high fat diet (HFD)-fed mice have increased overall O-GlcNAcylation levels, but not OGT and OGA levels, as compared to macrophages cocultured with AT from normal chow (NC)-fed mice (Fig. 1B). These results suggest that overnutrition stimulates an increase of O-GlcNAc signaling in macrophages in vitro, possibly by boosting the HBP.
Fig. 1.
Nutrient-sensing O-GlcNAc signaling is regulated during overnutrition and macrophage activation. (A) Western blot analysis of OGT, OGA, and overall O-GlcNAcylation levels in mouse peritoneal macrophages after 2 h of treatment. “Vehicle” was 0.2% BSA and “Ctrl” was culture medium. GlcN was used as a positive control. RL2 recognizes O-GlcNAc modification on proteins. (B) Western blot analysis of OGT, OGA, and overall O-GlcNAcylation levels in mouse peritoneal macrophages cocultured with epididymal white AT of NC-fed and HFD-fed mice for 2 h. (C) Ogt mRNA level in mouse peritoneal macrophages (peri. MPs), mouse BMDMs, and RAW 264.7 macrophage cells (n = 4 to 8). LPS was used to stimulate M1 polarization. (D) Representative Western blots of OGT, OGA, and overall O-GlcNAcylation levels in mouse BMDMs. LPS was used to stimulate M1 polarization. (E and F) Flow cytometric analysis of average intensity of O-GlcNAc (RL2) staining of macrophage subpopulations including F4/80+ CCR2+ cells, F4/80+ MGL1+ cells (E), F4/80+ CD11c+ M1-like macrophages, and F4/80+ CD206+ M2-like macrophages (F) in the SVF of eWAT from NC-fed, 1-wk HFD-fed, 4-wk HFD-fed, and 12-wk HFD-fed WT mice (n = 4 to 6). Data are shown as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired Student’s t test.
We then sought to determine the activity of O-GlcNAc signaling during macrophage activation. Gene expression profiles retrieved from the Gene Expression Omnibus repository showed the level of OGT transcripts was decreased by ∼20% in lipopolysaccharides (LPS)/interferon-γ (IFN-γ) polarized M1-like human macrophages, while no change in OGA transcriptional level was found (42) (SI Appendix, Fig. S1B). To further determine the activation of O-GlcNAc signaling during macrophage activation, we induced proinflammatory M1 [M(LPS)] and antiinflammatory M2 [M(IL-4)] polarization by using LPS and interleukin-4 (IL-4), respectively, in mouse peritoneal macrophages, BMDMs, and macrophage cell line RAW 264.7. Consistent with the changes observed in human macrophages, qRT-PCR analysis showed that OGT transcriptional level was decreased during LPS-induced M1 polarization but not IL-4–induced M2 polarization, without any difference in OGA transcripts (Fig. 1C and SI Appendix, Fig. S1C). Next, we examined the activation of O-GlcNAc signaling at the protein level in BMDMs. The overall O-GlcNAc modification level was decreased by 25 to 40% during M1 polarization, accompanied by a ∼45% decrease in the OGT protein level (Fig. 1D and SI Appendix, Fig. S1D). A transient elevation of overall O-GlcNAc modification, but no significant changes in OGT and OGA protein levels, were observed during M2 polarization (SI Appendix, Fig. S1E). These results demonstrate that O-GlcNAc signaling is temporarily suppressed during M1 polarization and transiently enhanced at the onset of M2 polarization.
We then examined the dynamic changes of overall O-GlcNAcylation levels in various subpopulations of AT macrophages in the stromal vascular fraction (SVF) of epididymal white AT (eWAT) during HFD feeding by using flow cytometry (Fig. 1 E and F and SI Appendix, Figs. S1 F–I and S2). F4/80 served as a pan macrophage marker. CD11c and CD206 were used as markers for M1-like and M2-like macrophages, respectively. MGL1, highly expressed in resident population of AT macrophages, and CCR2, critical for macrophage recruitment to inflammation sites, were also used as markers (43–45). The results showed that 1) MGL1+ macrophages generally have higher overall O-GlcNAcylation levels, as compared to CCR2+ macrophages, 2) CD206+ M2-like macrophages generally have higher overall O-GlcNAcylation levels, as compared to CD11c+ M1-like macrophages, and 3) macrophage O-GlcNAc signaling is enhanced or maintained at a similar level at the early stage of obesity (1-wk and 4-wk HFD feeding) but decreased at the late stage of obesity (12-wk HFD feeding), as compared to the NC-fed mice. Based on these results, we hypothesized that macrophage O-GlcNAc signaling may have an antiinflammatory function and is involved in maintaining metabolic homeostasis, especially at the early stage of obesity.
Macrophage Ogt Knockout Mice Are Prone to Diet-Induced Metabolic Dysfunction.
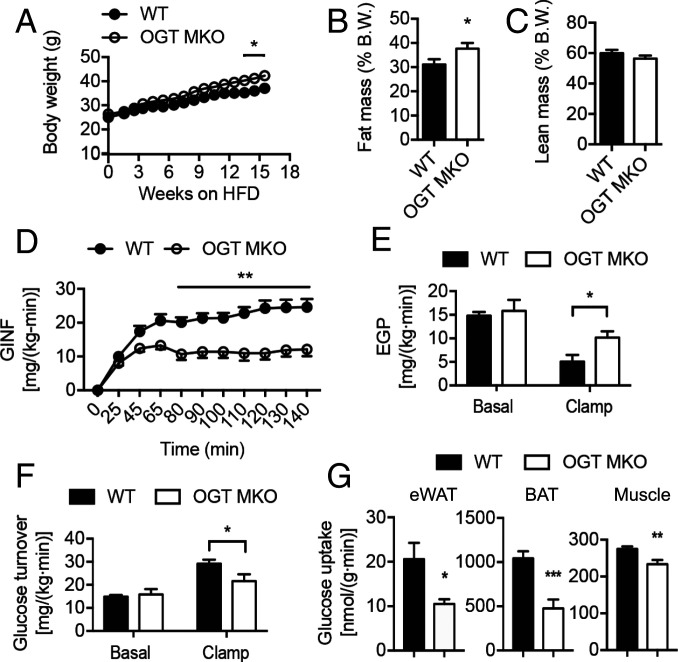
To test our hypothesis, we bred the Ogtflox strain with the LysM-Cre transgenic line to generate a mouse line with Ogt deletion in macrophages and other myeloid cells (hereafter termed OGT MKO mice) (SI Appendix, Fig. S3 A–C) and provided them with ad libitum access to NC and HFD. Flow cytometric analysis showed that OGT KO preferentially affect O-GlcNAc signaling in CCR2+ macrophages (SI Appendix, Fig. S3D). OGT MKO mice fed NC showed no difference in body weight, body composition, and whole-body insulin sensitivity compared to their wild-type (WT) littermates (SI Appendix, Fig. S3 E–K). We then challenged the mice with HFD. OGT MKO mice gained slightly more body weight and fat mass compared to their WT controls after 12 wk of HFD feeding, which could be attributed to increased food intake (Fig. 2 A and B and SI Appendix, Fig. S3L). In contrast, there is no difference in lean mass, oxygen consumption (VO2), respiratory exchange ratio, physical activity, and energy expenditure (Fig. 2C and SI Appendix, Fig. S3 M–P). Increases in tissue weights of subcutaneous inguinal white AT (iWAT) and interscapular brown AT (BAT), but not eWAT, were observed in OGT MKO mice (SI Appendix, Fig. S4 A–C). Despite an increase in BAT mass, the core body temperature of OGT MKO mice dropped more quickly than WT mice during an acute cold challenge (SI Appendix, Fig. S4D), which is likely due to BAT whitening (SI Appendix, Fig. S4E).
Fig. 2.
OGT MKO mice are prone to diet-induced metabolic dysfunction. (A) Body weight of HFD-fed WT and OGT MKO mice. (B and C) Fat mass (B) and lean mass (C) of 18-wk HFD-fed WT and OGT MKO mice (n = 12 to 19). (D) GINF rate to maintain euglycemia during hyperinsulinemic–euglycemic clamps in 8-wk HFD-fed WT and OGT MKO mice. (E and F) EGP rate (E) and whole-body glucose turnover rate (F) in both basal (without insulin stimulation) and clamps (with insulin stimulation) states in 8-wk HFD-fed WT and OGT MKO mice. (G) Glucose uptake in eWAT, BAT, and gastrocnemius muscle under insulin-stimulated condition in 8-wk HFD-fed WT and OGT MKO mice (n = 5 to 8). Data are shown as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired Student’s t test.
Whole-body glucose metabolism of HFD-fed WT and OGT MKO mice was determined by intraperitoneal glucose tolerance test (GTT), insulin tolerance test (ITT), and hyperinsulinemic–euglycemic clamps before their body weights diverged (Fig. 2A and SI Appendix, Fig. S4 F–H). OGT MKO mice showed a twofold increase of plasma insulin levels after overnight fasting without any significant difference in fasting plasma glucose, nonesterified fatty acid, or triacylglycerol (TAG) concentrations (SI Appendix, Fig. S4 I–L), reflecting increased whole-body insulin resistance. GTT and ITT showed that OGT MKO mice had impaired glucose metabolism and insulin resistance as compared to WT controls (SI Appendix, Fig. S4 M–Q), which is also illustrated by an increase in the homeostatic model assessment for insulin resistance (SI Appendix, Fig. S4R). During the hyperinsulinemic–euglycemic clamp, OGT MKO mice exhibited a marked impairment in whole-body insulin sensitivity as reflected by a ∼55% decline in the glucose infusion (GINF) rate required to maintain euglycemia (Fig. 2D and SI Appendix, Fig. S4 S and T). Despite no difference in the basal endogenous glucose production (EGP) rate, the EGP during the clamp was significantly higher in OGT MKO mice (Fig. 2E), demonstrating impaired hepatic insulin sensitivity. The increased EGP during the clamp may be attributable to increased acetyl-CoA (SI Appendix, Fig. S4U), an allosteric activator of pyruvate carboxylase (9). We also observed a reduced whole-body glucose turnover rate in OGT MKO mice (Fig. 2F), which could be at least partially attributed to reduced glucose uptake in eWAT, BAT, and gastrocnemius muscle (Fig. 2G). Together, these results demonstrate that loss of macrophage OGT exacerbates diet-induced whole-body insulin resistance independent of body weight changes.
Loss of Macrophage OGT Promotes Inflammation, AT Lipolysis, and Ectopic Lipid Accumulation and Insulin Resistance.
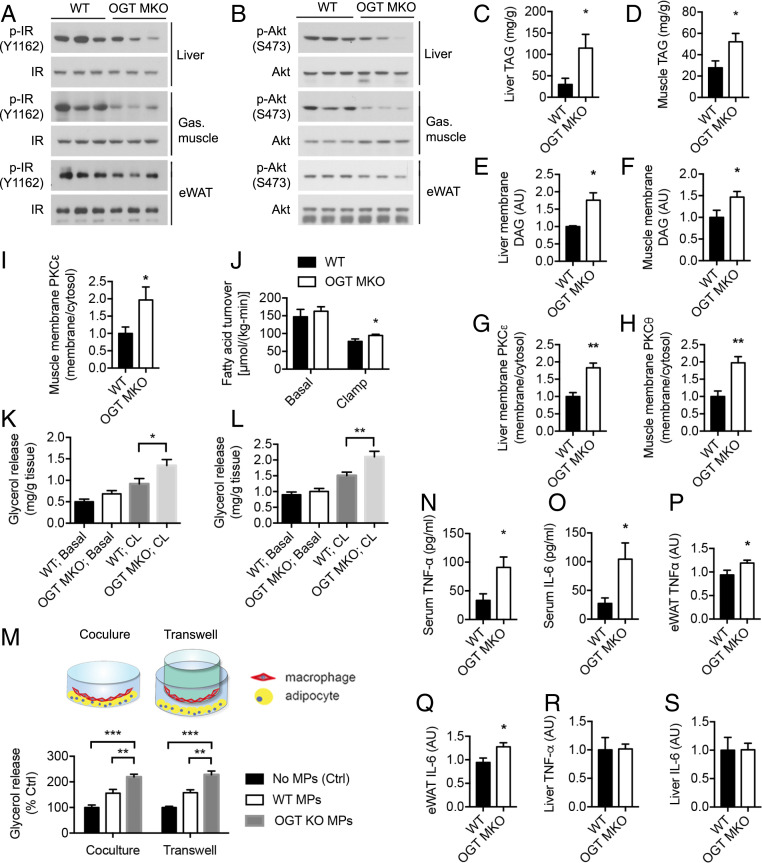
We then assessed the activation of insulin signaling in peripheral tissues of HFD-fed WT and OGT MKO mice. The phosphorylation of insulin receptor (IR) kinase and Akt were reduced under insulin stimulation in liver, gastrocnemius muscle, and eWAT of OGT MKO mice, indicating the impaired insulin signaling (Fig. 3 A and B and SI Appendix, Fig. S5A). Ectopic lipid accumulation is closely associated with insulin resistance (1, 46). To explore the underlying mechanism of insulin resistance in OGT MKO mice, levels of TAGs and diacylglycerols (DAGs) in liver and skeletal muscle were determined and the results showed that OGT MKO mice had a two- to threefold increase in TAG content in liver and muscle (Fig. 3 C and D). Consistently, a fatty liver phenotype including increased liver weight, pale-colored liver, and histopathologic features of steatosis was observed in OGT MKO mice (SI Appendix, Fig. S5 B–E). A 1.5- to 3-fold increase of DAG content was also found in all tested compartments (membrane, lipid droplets, and cytosol) in both liver and muscle of OGT MKO mice (Fig. 3 E and F and SI Appendix, Fig. S5 F–I). Previous studies demonstrated that membrane DAG induces peripheral insulin resistance by promoting the membrane translocation of protein kinase C (PKC)ε in liver and PKCθ in skeletal muscle (47, 48). Consistently, OGT MKO mice showed increased PKCε translocation from cytosol to the membrane in liver and increased PKCθ and PKCε translocation in muscle (Fig. 3 G–I and SI Appendix, Fig. S5 J and K). In contrast, no difference in hepatic and muscle ceramide content was observed (SI Appendix, Fig. S5 L and M). Taken together, these data suggest that loss of macrophage OGT promotes diet-induced whole-body insulin resistance by enhancing ectopic lipid accumulation and activating the DAG–PKC pathway in liver and skeletal muscle.
Fig. 3.
Loss of macrophage OGT promotes inflammation, AT lipolysis, and ectopic lipid accumulation and insulin resistance in liver and muscle. (A) Western blots of insulin-stimulated IR phosphorylation (Y1162) in liver, gastrocnemius muscle, and eWAT of 8-wk HFD-fed WT and OGT MKO mice after overnight fasting. (B) Western blots of insulin-stimulated Akt phosphorylation (S473) in liver, gastrocnemius muscle, and eWAT of 8-wk HFD-fed WT and OGT MKO mice after overnight fasting. (C and D) Triglyceride (TAG) levels in liver (n = 4) and gastrocnemius muscle (n = 9 to 11) of 12-wk HFD-fed WT and OGT MKO mice. (E and F) Membrane DAG levels in liver and gastrocnemius muscle of 12-wk HFD-fed WT and OGT MKO mice (n = 6 to 14). (G) Hepatic membrane/cytosolic PKCε ratio in 12-wk HFD-fed WT and OGT MKO mice. (H and I) Membrane/cytosolic PKCθ and PKCε ratio in gastrocnemius muscle of 12-wk HFD-fed WT and OGT MKO mice (n = 5). (J) Whole-body fatty acid turnover of 8-wk HFD-fed WT and OGT MKO mice (n = 6). (K and L) Basal and stimulated (10 μM CL-316,243) lipolysis measured by glycerol released from tissue trunks of iWAT (K) and eWAT (L) of 12-wk HFD-fed WT and OGT MKO mice (n = 10 to 12). (M) Scheme and glycerol release rate of in vitro cultured adipocytes when cocultured with WT and OGT KO peritoneal macrophages in contact (Coculture) and noncontact (Transwell) manners. (N–S) TNF-α and IL-6 levels in serum (N and O), eWATs (P and Q), and livers (R and S) of 8-wk HFD-fed WT and OGT MKO mice. Data are shown as mean ± SEM (n = 5 to 8). *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA with Dunnett multiple comparisons for K–M. *P < 0.05, **P < 0.01 by unpaired Student’s t test for other panels.
Next, we sought to understand the cause of ectopic lipid accumulation in peripheral tissues of OGT MKO mice. Studies have shown that excessive AT lipolysis plays an essential pathogenic role in diet-induced insulin resistance by enhancing fatty acid flux into metabolic tissues (9, 49, 50). Consistent with this idea, OGT MKO mice had increased fatty acid turnover rates during the clamp (Fig. 3J), suggesting that loss of macrophage OGT impaired insulin suppression of AT lipolysis. We then performed an ex vivo lipolysis assay with freshly prepared tissue trunks of WATs from HFD-fed WT and OGT MKO mice. The results showed that CL-316,243 (a β3-adrenergic receptor agonist)-stimulated lipolysis was significantly enhanced in sWAT and eWATs of OGT MKO mice (Fig. 3 K and L). To explore the underlying mechanism, we obtained cultured mature adipocytes differentiated from SVF of mouse eWAT. Then, we examined the effects of in vitro-cultured WT and OGT KO macrophages on adipocyte lipolysis in contact (coculture) and noncontact (transwell) manners. The results showed that both WT and OGT KO macrophages stimulated adipocyte lipolysis in a cell–cell contact-independent manner. More importantly, compared to WT macrophages, OGT KO macrophages were more potent to stimulate adipocyte lipolysis (Fig. 3M), demonstrating an essential role for macrophage OGT in suppressing adipocyte lipolysis through secreted factors. Previous studies have shown that inflammatory cytokines are important regulators of adipocyte lipolysis (9, 51). We then determined the levels of proinflammatory cytokines including tumor necrosis factor α (TNF-α) and IL-6 in the serum of 8-wk HFD-fed WT and OGT MKO mice and observed a three- to fourfold increase in OGT MKO mice (Fig. 3 N and O). Further analysis and histological analysis of hematoxylin–eosin staining showed that TNF-α and IL-6 levels were increased by ∼25% in eWAT of OGT MKO mice but no significant difference in hepatic inflammation (Fig. 3 P–S and SI Appendix, Fig. S5E), showing that AT is a primary target of macrophage-induced inflammation. Taken together, these data indicate that macrophage OGT depletion promotes AT inflammation and lipolysis, enhances fatty acid flux into liver and muscle, and finally leads to ectopic lipid accumulation and DAG–PKC-mediated insulin resistance in these metabolic tissues.
Loss of Macrophage OGT Promotes AT Inflammation.
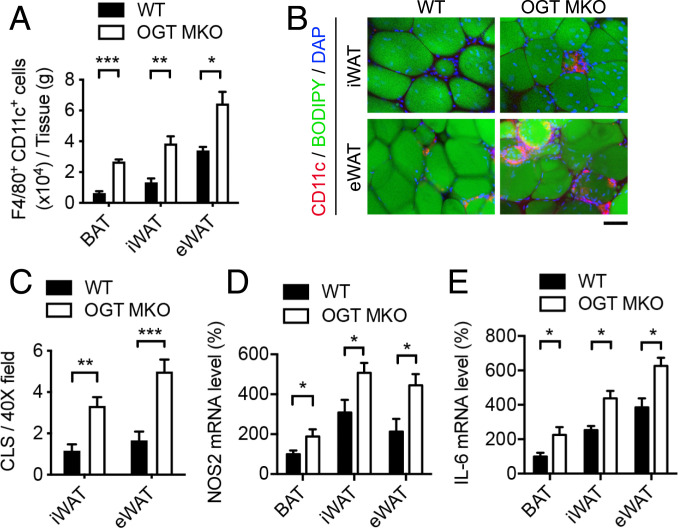
To determine the role of macrophage OGT in AT inflammation, flow cytometric analysis of AT macrophages and macrophage subpopulations was performed. In WT mice, 12-wk HFD feeding induced a 2.5-fold increase of CCR2+ macrophages in eWAT as compared to iWAT, while no difference in MGL1+ macrophages was found (SI Appendix, Fig. S6 A and B). The numbers of total macrophages (F4/80+ cells) were ∼10% in BAT and ∼30 to 40% in iWAT and eWAT (of SVF cells) (SI Appendix, Fig. S6C). In 12-wk HFD-fed OGT MKO mice, a significant increase in the number of total macrophages was found in BAT (∼2.5 fold), but not in iWAT and eWAT, as compared to WT mice (SI Appendix, Figs. S6D and S7A). Nevertheless, an approximately two- to fourfold increase of CD11c+ M1-like macrophages in all examined ATs and a ∼40 to 50% decrease of CD206+ M2-like macrophages in iWAT and eWAT (per gram of tissue) were observed (Fig. 4A and SI Appendix, Figs. S6 E and F and S7B).
Fig. 4.
Loss of macrophage OGT promotes AT inflammation. (A) Quantification of flow cytometric analysis of F4/80+ CD11c+ cells in BAT, iWAT, and eWAT from 12-wk HFD-fed WT and OGT MKO mice (n = 4). (B) Whole-mount staining of iWAT and eWAT of 12-wk HFD-fed WT and OGT MKO mice showing adipocytes (BODIPY FL, stains lipids) and macrophages (CD11c+ cells) in CLSs. (Scale bar, 80 μm.) (C) Quantitative results of CLSs in iWAT and eWAT of 12-wk HFD-fed WT and OGT MKO mice (n = 6). (D and E) Nos2 and Il-6 mRNA levels in BAT, iWAT, and eWAT of 12-wk HFD-fed WT and OGT MKO mice (n = 4 to 8). Data are shown as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired Student’s t test.
We identified a ∼2.5-fold increase in the number of crown-like structures (CLSs) formed by proinflammatory macrophages surrounding necrotic adipocytes in the whole-mount AT staining of iWAT and eWAT from OGT MKO mice (Fig. 4 B and C), which is consistent with enhanced accumulation of proinflammatory CD11c+ M1-like macrophages in WATs of OGT MKO mice. qRT-PCR analysis showed that the expression of macrophage proinflammatory polarization markers, Il-6 and Nos2, was increased in various ATs of OGT MKO mice compared with WT mice, without difference in Arg1 expression, an M2 polarization marker (Fig. 4 D and E and SI Appendix, Fig. S7C). These data demonstrate that loss of macrophage OGT aggravates AT inflammation by promoting proinflammatory macrophage activation and/or infiltration, which may contribute to increased WAT lipolysis.
O-GlcNAc Signaling Suppresses Macrophage Proinflammatory Activation.
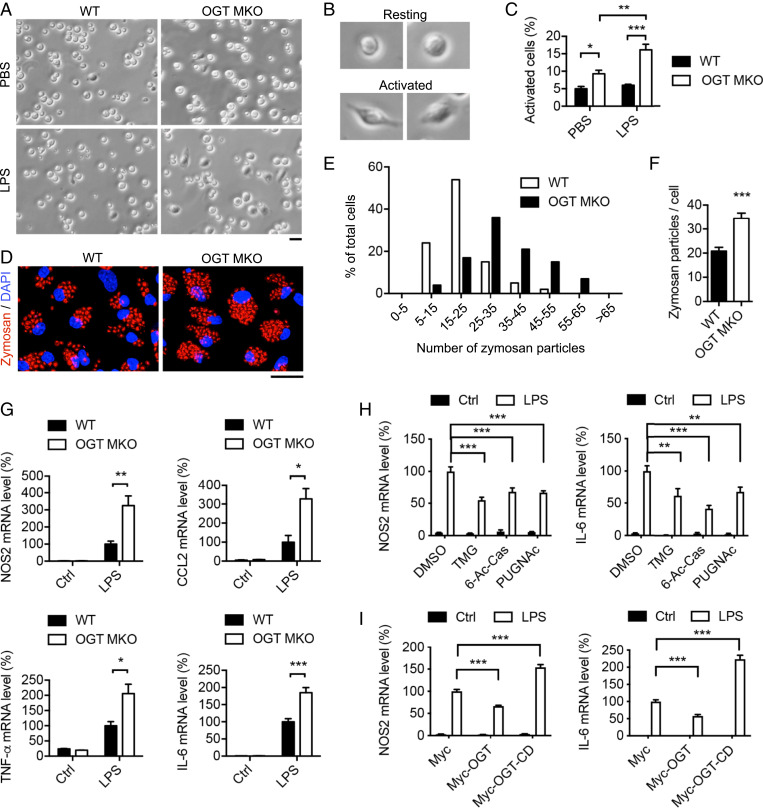
Based on above results, we hypothesized that O-GlcNAc signaling plays a suppressive role in macrophage proinflammatory activation. M1-like polarized macrophages are known to exhibit an elongated and spreading morphology with pseudopod-like protrusions (52). LPS-induced peritoneal macrophage activation was analyzed and we defined the spread cells as “activated cells” and the round-shaped cells as “resting cells” (Fig. 5B). Bright-field imaging showed that OGT KO group had a two- to threefold increase in the percentage of activated cells, as compared to the WT group, under both unstimulated and LPS-stimulated (30 min) conditions (Fig. 5 A–C), suggesting loss of OGT accelerates and promotes LPS-induced macrophage proinflammatory activation. We then treated WT and OGT KO BMDMs with fluorescein-labeled zymosan particles and found that OGT KO BMDMs internalized ∼75% more zymosan particles compared to WT BMDMs (Fig. 5 D–F), suggesting loss of OGT enhances macrophage phagocytic activity. Finally, qRT-PCR analysis showed that loss of OGT enhanced the expression of LPS-induced M1 markers including Nos2, Ccl2, TNF-α, and Il-6 or pro-Il-1b by 100 to 200% and IFN-γ–induced M1 markers by 50 to 100% (Fig. 5G and SI Appendix, Fig. S7D) but had no effect on IL-4–induced M2 markers expression including Arg1, Clec10a, Chi3l3, and Retnla (SI Appendix, Fig. S7E). Together, these data demonstrate a specific role of OGT in suppressing macrophage proinflammatory polarization.
Fig. 5.
O-GlcNAc signaling suppresses macrophage proinflammatory activation. (A–C) Bright-field imaging and statistical analysis of unstimulated and LPS-stimulated activation of peritoneal macrophages isolated from WT and OGT MKO mice (n = 4 to 8). (Scale bar, 20 μm.) (D–F) Fluorescence imaging and statistical analysis of phagocytosis of zymosan particles by WT and OGT KO BMDMs. (Scale bar, 20 μm.) (G) Macrophage M1 marker Nos2, Ccl2, TNF-α, and Il-6 mRNA levels in unstimulated and LPS-stimulated WT and OGT KO BMDMs (n = 3 to 6). (H) Nos2 and Il-6 mRNA levels in DMSO-, TMG-, 6-Ac-Cas-, and PUGNAc-treated BMDMs under unstimulated and LPS-stimulated conditions (n = 6). (I) Nos2 and Il-6 mRNA levels in Myc-, Myc-OGT–, and Myc-OGT-CD–overexpressing BMDMs under unstimulated and LPS-stimulated conditions (n = 4). Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA with Dunnett multiple comparisons for C, H, and I. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired Student’s t test for other panels.
To gain further insight into the role of O-GlcNAc signaling in macrophage proinflammatory polarization, O-GlcNAc signaling was interrogated in BMDMs and RAW 264.7 cells by treatment with various OGA inhibitors: thiamet-G (TMG), 6-acetamido-6-deoxy-castanospermine (6-Ac-Cas), and PUGNAc. The results showed that the pharmacological inhibition of OGA suppressed the expression of LPS-induced M1 markers, Nos2 and Il-6, by 40 to 60% (Fig. 5H and SI Appendix, Fig. S7F). Consistent with this finding, OGT overexpression suppressed LPS-induced Nos2 and Il-6 expression by 40 to 60%, while overexpression of the dominant negative OGT catalytic dead mutant (OGT-CD, D925N) significantly enhanced LPS-induced Nos2 and Il-6 expression (Fig. 5I and SI Appendix, Fig. S7 G and H). Taken together, these results demonstrate a suppressive role of O-GlcNAc signaling in macrophage proinflammatory activation.
RNA Sequencing Analysis Reveals a Preferential Regulation of Macrophage Proinflammatory Polarization by OGT.
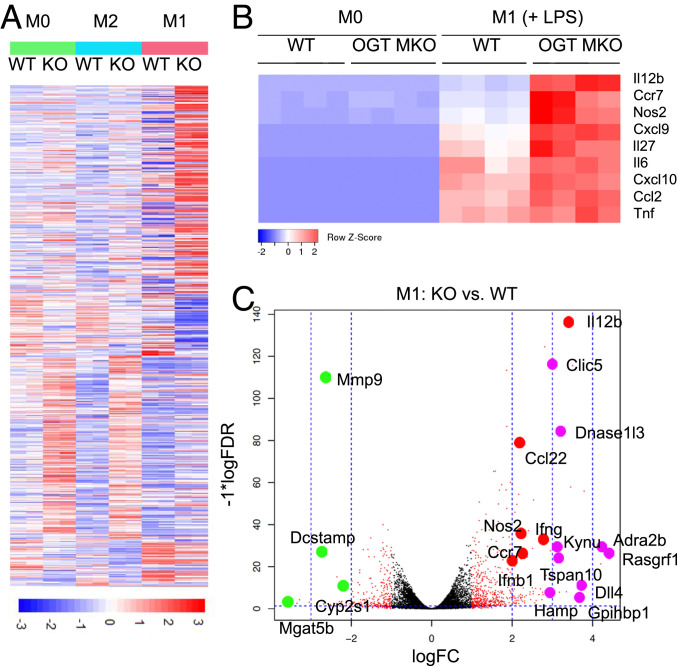
Macrophage activation involves extensive transcriptional rewiring (12). To assess the role of OGT in the systemic reprogramming during macrophage activation, we performed RNA sequencing-based transcriptional profiling for unstimulated (M0), LPS-stimulated (M1), and IL-4–stimulated (M2) WT and OGT KO BMDMs. The heat map of differentially expressed genes (DEGs) showed that loss of OGT led to a significantly increased expression of a large set of genes and a decreased expression of a small set of genes in M0, M1, and M2 polarized BMDMs (Fig. 6A). These results demonstrate a prevalent role of OGT in suppressing macrophage activation at the transcriptional level. Strikingly, DEGs between WT and OGT KO BMDMs in M0 and M2 groups shared a similar expression pattern, whereas DEGs in the M1 group showed a very unique pattern (Fig. 6A). A Venn diagram showing the overlap of DEGs between every two groups also supports the same conclusion (SI Appendix, Fig. S8A). Then, all three groups of DEGs were overlaid with a group of genes identified as macrophage-related genes (53, 54). Nine DEGs were found to be present in all groups, suggesting a robust OGT-mediated regulation of these genes. Again, a larger set of macrophage-related DEGs were found to be specifically present in the M1 group (57 in M1 vs. 19 in M2 and 14 in M0) (SI Appendix, Fig. S8B). Together, these results suggest that OGT in macrophages preferentially regulates macrophage M1 polarization, which is consistent with our previous results showing that loss of OGT affects LPS- and IFN-γ–induced M1 polarization but not IL-4–induced M2 polarization (Fig. 5G and SI Appendix, Fig. S7 D and E). The expression of selected M1 and M2 markers determined by RNA sequencing were also analyzed. Consistent with the previous results, loss of OGT enhanced the expression of M1 markers in LPS-treated BMDMs but did not affect the expression of M2 markers in IL-4–treated BMDMs (Fig. 6B and SI Appendix, Fig. S8C).
Fig. 6.
RNA sequencing analysis reveals a preferential regulation of macrophage M1 polarization by OGT. (A) Heat map showing the DEGs between WT and OGT KO BMDMs, where colors indicate counts per million values scaled by row (n = 4). (B) Heat maps of expression levels of M1 macrophage markers determined by RNA sequencing. (C) Volcano plot showing top DEGs between M1-polarized WT and OGT KO BMDMs. Red dot-labeled genes are up-regulated in OGT KO BMDMs and are closely related to M1 polarization. Purple and green dot-labeled genes have known functions related to M1 polarization and are up- and down-regulated in OGT KO BMDMs, respectively.
Gene set enrichment analysis (GSEA) for the Gene Ontology terms showed that loss of OGT in M1-polarized BMDMs enhanced the expression of genes that are highly enriched in the GO terms cytokine production, regulation of cytokine production, defense response, positive regulation of multicellular process, and immune response (SI Appendix, Fig. S8D), supporting a role of OGT in suppressing macrophage M1 activation and inflammation. Genes enriched in GO terms including myeloid cell differentiation and myeloid leukocyte differentiation were down-regulated in M1-polarized OGT KO BMDMs (SI Appendix, Fig. S8D). To determine whether macrophage maturation is affected by OGT, qRT-PCR analysis was performed and similar levels of F4/80, a macrophage marker, were observed in WT and OGT KO BMDMs (SI Appendix, Fig. S8E), demonstrating that LyzM-Cre–mediated OGT KO does not affect monocyte–macrophage differentiation. GSEA comparing M1- and M2-activated pathways between WT and OGT KO BMDMs showed that two pathways, inflammatory response and signaling receptor binding, were significantly enhanced in M1-polarized OGT KO BMDMs (SI Appendix, Fig. S8 F and G), suggesting that OGT may suppress macrophage M1 polarization by inhibiting signaling pathways involved in an inflammatory response.
To provide additional insights into the mechanism by which OGT regulates macrophage M1 polarization, top DEGs between WT and OGT KO BMDMs in the M1 group were showed in a Volcano plot (Fig. 6C). Among the top up-regulated DEGs in OGT KO BMDMs, genes closely related to M1 polarization including Il12b, Ccl22, Nos2, Ifng, Ccr7, and Ifnb1 were found. A large portion of the up-regulated DEGs including Clic5, Dnase1l3, Kynu, Adra2b, Rasgrf1, Tspan10, Dll4, Hamp, and Gpihbp1 have been known to play important roles in macrophage M1 polarization. Also, a small set of genes known to be involved in macrophage regulation including Mmp9, Dcstamp, Cyp2s1, and Mgat5b were identified in the top down-regulated DEGs (Fig. 6C). Together, these results demonstrate a profound role of OGT-mediated transcriptional rewiring in the suppression of macrophage proinflammatory polarization.
OGT Inhibits Macrophage Proinflammatory Activation by Suppressing mTORC1/S6K1 Signaling.
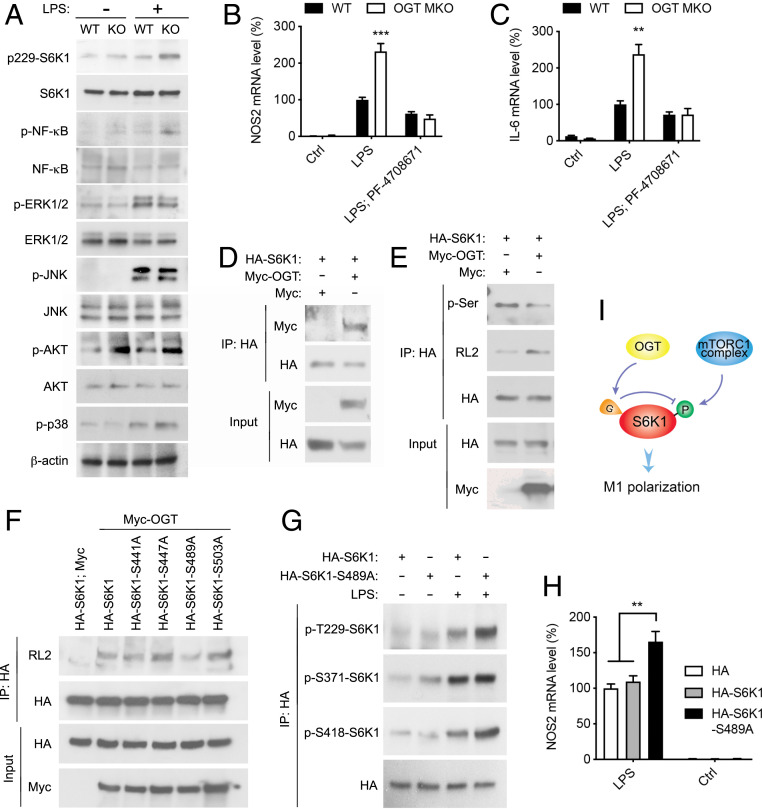
To understand the molecular mechanism by which O-GlcNAc signaling suppresses proinflammatory macrophage activation, we assessed the activation of several signaling pathways essential for macrophage proinflammatory polarization in WT and OGT KO BMDMs. We observed loss of OGT significantly enhanced LPS-induced activation of mTORC1/ribosomal protein S6 kinase beta-1 (S6K1) and NF-κB signaling but not the activation of ERK, JNK, Akt, and p38 signaling (Fig. 7A). Moreover, TMG treatment suppressed LPS-induced S6K1 activation in BMDMs (SI Appendix, Fig. S9A). To determine the role of NF-κB and mTORC1/S6K1 signaling in OGT-mediated macrophage proinflammatory polarization, we first treated WT and OGT KO BMDMs with PF-04708671 (S6K1 inhibitor) and rapamycin (mTORC1 inhibitor) and then with LPS to induce proinflammatory M1 polarization. qRT-PCR analysis showed that blocking mTORC1/S6K1 signaling, but not NF-κB signaling, completely abolished the enhancement of Nos2 and Il-6 expression in OGT KO macrophages upon LPS stimulation, suggesting that OGT suppresses macrophage M1 polarization by inhibiting the mTORC1/S6K1 pathway (Fig. 7 B and C and SI Appendix, Fig. S9 B–E).
Fig. 7.
OGT inhibits macrophage proinflammatory polarization by suppressing mTORC1/S6K1 signaling. (A) Western blot analysis showing the activation of S6K1, NF-κB, ERK, JNK, p38 MAPK, and Akt in unstimulated and LPS-stimulated (30 min) WT and OGT KO peritoneal macrophages. (B and C) Nos2 and Il-6 mRNA levels in unstimulated, LPS-stimulated, and PF-04708671-pretreated LPS-stimulated WT and OGT KO BMDMs (n = 4). (D) Immunoprecipitation (IP) and Western blot analysis showing the interaction between exogenously expressed HA-S6K1 and Myc-OGT in HeLa cells. (E) IP and Western blot analysis showing that OGT overexpression enhances S6K1 O-GlcNAcylation and decreases S6K1 serine phosphorylation in HeLa cells. (F) IP and Western blot analysis showing that serine 489 to alanine (S489A) mutation in S6K1 greatly abolished the overall O-GlcNAcylation on S6K1 in HeLa cells. (G) IP and Western blot analysis showing that S489A mutation in S6K1 enhanced LPS-induced S6K1 phosphorylation on threonine 229 (T229) and S418 in RAW 264.7 cells. (H) Nos2 mRNA levels in untreated and LPS-stimulated RAW 264.7 cells overexpressing HA, HA-S6K1, and HA-S6K1-S489A (n = 4 to 6). (I) Molecular model for OGT function in mTORC1/S6K1 signaling. Data are shown as mean ± SEM. **P < 0.01 by two-way ANOVA with Dunnett multiple comparisons for H. **P < 0.01, ***P < 0.001 by unpaired Student’s t test for other panels.
To determine how OGT regulates mTORC1 signaling, we first tested if tuberous sclerosis complex 2 (TSC2), a key regulator of mTORC1 signaling, can be modified by O-GlcNAcylation. Our previous proteome-wide analysis of OGT-binding proteins in HEK 293T cells showed that TSC2 is among the 853 putative OGT-binding proteins (27) (SI Appendix, Fig. S9F). However, we were not able to detect any notable O-GlcNAcylation on TSC2, even in OGT-overexpressing cells (SI Appendix, Fig. S9G). Moreover, the levels of TSC2 phosphorylation on multiple sites were not changed in OGT KO macrophages (SI Appendix, Fig. S9H), suggesting that OGT may not affect TSC2 activity. S6K1 is a major mTORC1 downstream signaling molecule. A previous study showed that ribosomal protein S6 kinase-like 1 (S6LK), which shares ∼40% homology with S6K1, can be modified by O-GlcNAcylation (55). We observed a robust interaction between exogenous OGT and S6K1 in HeLa cells and RAW 264.7 cells (Fig. 7D and SI Appendix, Fig. S10A). OGT overexpression in HeLa cells and RAW 264.7 cells led to enhanced O-GlcNAcylation and decreased serine phosphorylation on S6K1 (Fig. 7E and SI Appendix, Fig. S10B), indicating a competition between S6K1 O-GlcNAcylation and phosphorylation. We then sought to identify the glycosylation sites on S6K1. Based on a list of potential O-GlcNAcylation sites predicted by two independent online computational prediction analyses (56, 57), a series of S6K1 single-site mutants were generated and how these mutations affect overall S6K1 O-GlcNAcylation was determined. The results showed that the mutation of serine 489 to alanine (S489A) largely eliminated overall S6K1 O-GlcNAcylation in HeLa cells and RAW 264.7 cells (Fig. 7F and SI Appendix, Fig. S10 C–E), indicating that serine 489 is the primary site for S6K1 O-GlcNAcylation.
To establish the functional relevance of S6K1 O-GlcNAcylation in macrophage activation, we examined the effect of S6K1 O-GlcNAcylation on S6K1 phosphorylation in RAW 264.7 cells. The results showed that eliminating S6K1 serine 489 O-GlcNAcylation enhanced S6K1 phosphorylation at serine 418 and threonine 229 (Fig. 7G), supporting the notion that loss of S6K1 O-GlcNAcylation at serine 489 promotes S6K1 phosphorylation at the C terminus autoinhibitory domain, which further facilitates phosphorylation and activation of its N-terminal kinase domain (58–60). Consistently, LPS-induced proinflammatory M1 polarization was enhanced by 40 to 70% in RAW 264.7 cells overexpressing HA-S6K1-S489A mutation, as compared to HA- and HA-S6K1–overexpressing cells (Fig. 7H and SI Appendix, Fig. S10 F–H). These studies indicate that OGT suppresses macrophage proinflammatory polarization by modulating S6K1 O-GlcNAcylation/phosphorylation and S6K1 activation (Fig. 7I).
Discussion
In this study, we demonstrate that macrophage O-GlcNAc signaling plays a profound role in suppressing AT inflammation and insulin resistance. Recent studies have expanded our knowledge about the functional and molecular integration of metabolic and inflammatory pathways. Hyperglycemia and energy surplus associated with obesity and metabolic syndrome have been shown to promote inflammatory responses through various mechanisms (61, 62). Changes in glucose uptake, glycolysis, and UDP-GlcNAc biosynthesis were observed during macrophage polarization (12, 13). Additionally, free fatty acids have been shown to activate inflammatory pathways and impair insulin action in AT (63). These findings prompted us to postulate that nutrient-sensing O-GlcNAc signaling serves as a link between overnutrition and insulin resistance by regulating macrophage function in metabolic tissues. However, only a few studies have attempted to directly evaluate the role of O-GlcNAc signaling in macrophage activation, and contradictory results were reported (64–66). Here, using in vitro and in vivomodels, we demonstrate that O-GlcNAc signaling suppresses macrophage proinflammatory M1-like polarization and is dispensable for antiinflammatory M2-like polarization, which supports an immunosuppressive role of O-GlcNAc signaling in macrophage activation.
Increasing evidence has established a causative link between proinflammatory macrophage and insulin resistance in both rodents and humans (9, 41). Previous studies have shown that suppressing macrophage proinflammatory activation or entirely depleting the macrophage population is protective against diet-induced metabolic dysregulation (67, 68). Consistent with this, we found that genetic ablation of OGT in macrophages results in increased AT inflammation and excessive AT lipolysis, thus leading to increases in ectopic lipid accumulation and whole-body insulin resistance. Our study provides compelling evidence for the existence of a protective mechanism against inflammation through O-GlcNAc signaling in obesity. O-GlcNAc signaling enables macrophages to sense external nutrients and restrains macrophage proinflammatory activation, which contributes to AT and whole-body metabolic homeostasis. We speculate that macrophage O-GlcNAc signaling is part of a homeostatic mechanism maintaining metabolic homeostasis at the early stage of obesity. However, proinflammatory stimuli, such as adipocyte cell death and proinflammatory cytokines secreted by other cells, are aggravated by prolonged HFD feeding. Eventually, the protective mechanism is pushed to the limit and becomes dysfunctional in severely obese animals. Consistent with this idea, a study showed that proinflammatory macrophages in mouse epididymal fat appear 3 wk after the initiation of HFD feeding. In contrast, the accumulation of neutrophils peaks at day 3 after initiating HFD feeding (69). These results suggest that the proinflammatory activation of macrophages is inhibited at an early stage of exposure to excess amounts of nutrients.
mTOR signaling lies at the core of nutrient-sensing and inflammatory pathways (70). Previous studies implicate that TSC1, an upstream regulator of mTORC1, orchestrates macrophage M1 and M2 activation via separate pathways. Loss of TSC1 promotes M1-like activation by stimulating the ERK pathway, while inhibiting M2-like activation by suppressing the ERK and C/EBPβ pathways (71, 72). Several studies have suggested that obesity is associated with increased mTOR activity, and deficiency of the downstream mediator S6K1 can protect mice against diet- and age-induced insulin resistance (73, 74). The current study has identified OGT as a key regulator of the mTOR/S6K1 pathway. We have shown that O-GlcNAcylation suppresses mTORC1 signaling by directly regulating S6K1. S6K1 O-GlcNAcylation inhibits sequential phosphorylation required for kinase activation, resulting in suppression of proinflammatory gene transcription. Suppression of the mTORC1/S6K1 pathway abrogates the effect of OGT deletion on LPS-induced M1 activation, suggesting that S6K1 is integral to OGT-mediated macrophage regulation. Therefore, our study provides mechanistic insight into the regulation and the function of mTORC1/S6K1 pathway in immunometabolism.
Macrophages display considerable plasticity with M1 and M2 activation states at two extremes. How macrophages acquire diverse activation states in response to metabolic cues is still largely unknown. A previous study showed that mTOR/MYC signaling enhances O-GlcNAc signaling by promoting OGT protein expression (75). The AMP-activated protein kinase (AMPK) pathway is the other nutrient-sensing pathway involved in macrophage polarization, and AMPK can negatively regulate the mTOR pathway (76). Furthermore, there is evidence that O-GlcNAcylation of AMPK regulates its enzymatic activity (77). Taken together, it is likely that OGT, mTORC1 and AMPK pathways form a complex nutrient-sensing network that underpins a spectrum of macrophage activation states.
In summary, this study demonstrates O-GlcNAc signaling as a homeostatic regulator at the interface of inflammation and metabolism. The results have important implications for our understanding of the integration of metabolism and immunity, an evolutionary need for survival (78). Future studies to explore how macrophage O-GlcNAc signaling integrates nutritional signals to dictate the immune response would help develop new immunomodulatory strategies to treat chronic metabolic diseases. Glutamine and GlcN, the popular nutritional supplements implicated in immunometabolism, both are known to fuel the HBP and promote O-GlcNAc signaling. Studies have shown that human adipose glutamine levels correlate inversely with fat mass and white fat inflammation (79). Glutamine supplementation attenuates AT inflammation and reduces metabolic risk in rodents (79, 80). Supplementing glucosamine in HFD-fed rats improves glucose metabolism and reduces visceral fat mass (81). Further investigation is warranted to determine whether O-GlcNAc signaling in immune cells is involved in these processes. It is worth noting that GlcN also protects against sepsis-induced tissue injury and septic death in mouse and zebrafish models (66). Therefore, the immunosuppressive function of O-GlcNAc signaling may have broader implications in both obesity-associated morbidity and immune-related diseases.
Materials and Methods
Detailed descriptions of materials and methods are provided in SI Appendix, Materials and Methods. These describe animals used; biochemical and tissue analyses; GTTs and ITTs; basal and hyperinsulinemic–euglycemic clamp study and flux measurements; cold challenge; cell culture and treatments; macrophage polarization and phagocytosis assay; quantitative real-time PCR and RNA sequencing; Western blotting and immunoprecipitation; reagents and plasmids used; lipolysis assay; flow cytometry; histology; whole-mount AT staining; and statistical analysis.
All animal experiments were performed in accordance with protocols approved by Yale University’s Institutional Animal Care and Use Committee.
Data Availability.
All sequencing data generated for this study have been deposited in the GEO database (accession no. GSE145720).
Supplementary Material
Acknowledgments
We thank Dr. Sheng Ding and Dr. Tian Xu from Yale University for mouse phenotyping. This work was supported by US Public Health Service Grants R01DK089098, R01DK102648, P01DK057751, and American Diabetes Association (1-19-IBS-119) to X.Y.; K99CA215315 to R.J.P.; R01DK113984 and P30DK059635 to G.I.S.; and R01GM122078, R21CA209848, U01DA045300, and P30AR072582 to D.C. This work was also supported by a China Scholarship Council–Yale World Scholars Fellowship and American Heart Association Predoctoral Fellowship (19PRE34380268) to X.L. and American Heart Association Postdoctoral Fellowship 17POST33670873 to Y.Y. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.-H.L. is a guest editor invited by the Editorial Board.
Data deposition: RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE145720).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916121117/-/DCSupplemental.
References
- 1.Shulman G. I., Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N. Engl. J. Med. 371, 2237–2238 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Kahn S. E., Hull R. L., Utzschneider K. M., Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444, 840–846 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Gregor M. F., Hotamisligil G. S., Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 29, 415–445 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Saltiel A. R., Olefsky J. M., Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 127, 1–4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reilly S. M., Saltiel A. R., Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 13, 633–643 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Asghar A., Sheikh N., Role of immune cells in obesity induced low grade inflammation and insulin resistance. Cell. Immunol. 315, 18–26 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Chung K. J., Nati M., Chavakis T., Chatzigeorgiou A., Innate immune cells in the adipose tissue. Rev. Endocr. Metab. Disord. 19, 283–292 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Russo L., Lumeng C. N., Properties and functions of adipose tissue macrophages in obesity. Immunology 155, 407–417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry R. J. et al., Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell 160, 745–758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce E. L., Pearce E. J., Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olenchock B. A., Rathmell J. C., Vander Heiden M. G., Biochemical underpinnings of immune cell metabolic phenotypes. Immunity 46, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jha A. K. et al., Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Langston P. K., Shibata M., Horng T., Metabolism supports macrophage activation. Front. Immunol. 8, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond M. R., Hanover J. A., A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardivillé S., Hart G. W., Nutrient regulation of gene expression by O-GlcNAcylation of chromatin. Curr. Opin. Chem. Biol. 33, 88–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X., Qian K., Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X. et al., Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Golks A., Tran T. T., Goetschy J. F., Guerini D., Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 26, 4368–4379 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawauchi K., Araki K., Tobiume K., Tanaka N., Loss of p53 enhances catalytic activity of IKKbeta through O-linked beta-N-acetyl glucosamine modification. Proc. Natl. Acad. Sci. U.S.A. 106, 3431–3436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan H. B. et al., Calcium-dependent O-GlcNAc signaling drives liver autophagy in adaptation to starvation. Genes Dev. 31, 1655–1665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh J. P., Zhang K., Wu J., Yang X., O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivier-Van Stichelen S., Hanover J. A., You are what you eat: O-linked N-acetylglucosamine in disease, development and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care 18, 339–345 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson S. B., Hart G. W., New insights: A role for O-GlcNAcylation in diabetic complications. Crit. Rev. Biochem. Mol. Biol. 51, 150–161 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Wright J. N., Collins H. E., Wende A. R., Chatham J. C., O-GlcNAcylation and cardiovascular disease. Biochem. Soc. Trans. 45, 545–553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClain D. A. et al., Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc. Natl. Acad. Sci. U.S.A. 99, 10695–10699 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vosseller K., Wells L., Lane M. D., Hart G. W., Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 5313–5318 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan H. B. et al., O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dentin R., Hedrick S., Xie J., Yates J. 3rd, Montminy M., Hepatic glucose sensing via the CREB coactivator CRTC2. Science 319, 1402–1405 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Housley M. P. et al., O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283, 16283–16292 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres C. R., Hart G. W., Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 (1984). [PubMed] [Google Scholar]
- 31.Goldberg H., Whiteside C., Fantus I. G., O-linked β-N-acetylglucosamine supports p38 MAPK activation by high glucose in glomerular mesangial cells. Am. J. Physiol. Endocrinol. Metab. 301, E713–E726 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Jiang M. et al., Elevated O-GlcNAcylation promotes gastric cancer cells proliferation by modulating cell cycle related proteins and ERK 1/2 signaling. Oncotarget 7, 61390–61402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisberg S. P. et al., Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112, 1796–1808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park E. J. et al., Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140, 197–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saghizadeh M., Ong J. M., Garvey W. T., Henry R. R., Kern P. A., The expression of TNF alpha by human muscle. Relationship to insulin resistance. J. Clin. Invest. 97, 1111–1116 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y. S., Wollam J., Olefsky J. M., An integrated view of immunometabolism. Cell 172, 22–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chawla A., Nguyen K. D., Goh Y. P., Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 11, 738–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas D., Apovian C., Macrophage functions in lean and obese adipose tissue. Metabolism 72, 120–143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu X. Y. et al., Functional plasticity of adipose-derived stromal cells during development of obesity. Stem Cells Transl. Med. 5, 893–900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray P. J. et al., Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapouri-Moghaddam A. et al., Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Italiani P. et al., Transcriptomic profiling of the development of the inflammatory response in human monocytes in vitro. PLoS One 9, e87680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westcott D. J. et al., MGL1 promotes adipose tissue inflammation and insulin resistance by regulating 7/4hi monocytes in obesity. J. Exp. Med. 206, 3143–3156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serbina N. V., Pamer E. G., Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 7, 311–317 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Lumeng C. N., DelProposto J. B., Westcott D. J., Saltiel A. R., Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes 57, 3239–3246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuel V. T., Shulman G. I., The pathogenesis of insulin resistance: Integrating signaling pathways and substrate flux. J. Clin. Invest. 126, 12–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen M. C. et al., Insulin receptor Thr1160 phosphorylation mediates lipid-induced hepatic insulin resistance. J. Clin. Invest. 126, 4361–4371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roden M. et al., Mechanism of free fatty acid-induced insulin resistance in humans. J. Clin. Invest. 97, 2859–2865 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoiswohl G. et al., Impact of reduced ATGL-mediated adipocyte lipolysis on obesity-associated insulin resistance and inflammation in male mice. Endocrinology 156, 3610–3624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morigny P., Houssier M., Mouisel E., Langin D., Adipocyte lipolysis and insulin resistance. Biochimie 125, 259–266 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Grant R. W., Stephens J. M., Fat in flames: Influence of cytokines and pattern recognition receptors on adipocyte lipolysis. Am. J. Physiol. Endocrinol. Metab. 309, E205–E213 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Yan B. et al., HDAC6 deacetylase activity is critical for lipopolysaccharide-induced activation of macrophages. PLoS One 9, e110718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavin Y. et al., Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu Y. et al., Tet2 regulates osteoclast differentiation by interacting with Runx1 and maintaining genomic 5-hydroxymethylcytosine (5hmC). Genomics Proteomics Bioinformatics 16, 172–186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dias W. B., Cheung W. D., Hart G. W., O-GlcNAcylation of kinases. Biochem. Biophys. Res. Commun. 422, 224–228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kao H. J. et al., A two-layered machine learning method to identify protein O-GlcNAcylation sites with O-GlcNAc transferase substrate motifs. BMC Bioinformatics 16 (suppl. 18), S10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta R., Brunak S., Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput., 310–322 (2002). [PubMed] [Google Scholar]
- 58.Mahalingam M., Templeton D. J., Constitutive activation of S6 kinase by deletion of amino-terminal autoinhibitory and rapamycin sensitivity domains. Mol. Cell. Biol. 16, 405–413 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou Z., He L., Qi R. Z., Regulation of s6 kinase 1 activation by phosphorylation at ser-411. J. Biol. Chem. 282, 6922–6928 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Magnuson B., Ekim B., Fingar D. C., Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 441, 1–21 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Ptak W., Klimek M., Bryniarski K., Ptak M., Majcher P., Macrophage function in alloxan diabetic mice: Expression of adhesion molecules, generation of monokines and oxygen and NO radicals. Clin. Exp. Immunol. 114, 13–18 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wen Y. et al., Elevated glucose and diabetes promote interleukin-12 cytokine gene expression in mouse macrophages. Endocrinology 147, 2518–2525 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Sears B., Perry M., The role of fatty acids in insulin resistance. Lipids Health Dis. 14, 121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryu I. H., Do S. I., Denitrosylation of S-nitrosylated OGT is triggered in LPS-stimulated innate immune response. Biochem. Biophys. Res. Commun. 408, 52–57 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Zheng G. M., Yu C., Yang Z., Puerarin suppresses production of nitric oxide and inducible nitric oxide synthase in lipopolysaccharide-induced N9 microglial cells through regulating MAPK phosphorylation, O-GlcNAcylation and NF-κB translocation. Int. J. Oncol. 40, 1610–1618 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Hwang J. S. et al., Glucosamine improves survival in a mouse model of sepsis and attenuates sepsis-induced lung injury and inflammation. J. Biol. Chem. 294, 608–622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han M. S. et al., JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science 339, 218–222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patsouris D. et al., Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 8, 301–309 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elgazar-Carmon V., Rudich A., Hadad N., Levy R., Neutrophils transiently infiltrate intra-abdominal fat early in the course of high-fat feeding. J. Lipid Res. 49, 1894–1903 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Hotamisligil G. S., Erbay E., Nutrient sensing and inflammation in metabolic diseases. Nat. Rev. Immunol. 8, 923–934 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byles V. et al., The TSC-mTOR pathway regulates macrophage polarization. Nat. Commun. 4, 2834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu L. et al., TSC1 controls macrophage polarization to prevent inflammatory disease. Nat. Commun. 5, 4696 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Yoon M. S., The role of mammalian target of rapamycin (mTOR) in insulin signaling. Nutrients 9, E1176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Um S. H. et al., Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Sodi V. L. et al., mTOR/MYC Axis regulates O-GlcNAc transferase expression and O-GlcNAcylation in breast cancer. Mol. Cancer Res. 13, 923–933 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu J., Ji J., Yan X. H., Cross-talk between AMPK and mTOR in regulating energy balance. Crit. Rev. Food Sci. Nutr. 52, 373–381 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Gélinas R. et al., AMP-activated protein kinase and O-GlcNAcylation, two partners tightly connected to regulate key cellular processes. Front. Endocrinol. 9, 519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Man K., Kutyavin V. I., Chawla A., Tissue immunometabolism: Development, physiology, and pathobiology. Cell Metab. 25, 11–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petrus P. et al., Glutamine links obesity to inflammation in human white adipose tissue. Cell Metab. 31, 375–390 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Opara E. C., Petro A., Tevrizian A., Feinglos M. N., Surwit R. S., L-glutamine supplementation of a high fat diet reduces body weight and attenuates hyperglycemia and hyperinsulinemia in C57BL/6J mice. J. Nutr. 126, 273–279 (1996). [DOI] [PubMed] [Google Scholar]
- 81.Barrientos C., Racotta R., Quevedo L., Glucosamine attenuates increases of intraabdominal fat, serum leptin levels, and insulin resistance induced by a high-fat diet in rats. Nutr. Res. 30, 791–800 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated for this study have been deposited in the GEO database (accession no. GSE145720).