Significance
Plant mitochondrial and plastid genomes maintain unusually low mutation rates, but the mechanisms responsible for their accurate transmission of DNA sequences have remained mysterious. Application of high-fidelity DNA sequencing techniques enabled detection of new mutations still present at low frequencies within tissue samples from the flowering plant Arabidopsis and showed that disrupting a gene in the mutS mismatch repair family (MSH1) increases the frequency of mitochondrial and plastid sequence variants approximately 10-fold to 1,000-fold. This gene has an unusually disjunct distribution across the tree of life, implying a history of horizontal gene transfer among eukaryotes, prokaryotes, and viruses. Its presence in plants and absence in lineages such as animals may contribute to radical differences in organelle mutation rates across eukaryotes.
Keywords: duplex sequencing, mutation rate, plant mitochondria, chloroplast, organelle genomes
Abstract
Mitochondrial and plastid genomes in land plants exhibit some of the slowest rates of sequence evolution observed in any eukaryotic genome, suggesting an exceptional ability to prevent or correct mutations. However, the mechanisms responsible for this extreme fidelity remain unclear. We tested seven candidate genes involved in cytoplasmic DNA replication, recombination, and repair (POLIA, POLIB, MSH1, RECA3, UNG, FPG, and OGG1) for effects on mutation rates in the model angiosperm Arabidopsis thaliana by applying a highly accurate DNA sequencing technique (duplex sequencing) that can detect newly arisen mitochondrial and plastid mutations even at low heteroplasmic frequencies. We find that disrupting MSH1 (but not the other candidate genes) leads to massive increases in the frequency of point mutations and small indels and changes to the mutation spectrum in mitochondrial and plastid DNA. We also used droplet digital PCR to show transmission of de novo heteroplasmies across generations in msh1 mutants, confirming a contribution to heritable mutation rates. This dual-targeted gene is part of an enigmatic lineage within the mutS mismatch repair family that we find is also present outside of green plants in multiple eukaryotic groups (stramenopiles, alveolates, haptophytes, and cryptomonads), as well as certain bacteria and viruses. MSH1 has previously been shown to limit ectopic recombination in plant cytoplasmic genomes. Our results point to a broader role in recognition and correction of errors in plant mitochondrial and plastid DNA sequence, leading to greatly suppressed mutation rates perhaps via initiation of double-stranded breaks and repair pathways based on faithful homologous recombination.
It has been apparent for more than 30 y that rates of nucleotide substitution in plant mitochondrial and plastid genomes are unusually low (1, 2). In angiosperms, mitochondrial and plastid genomes have synonymous substitution rates that are, on average, ∼16-fold and 5-fold slower than the nucleus, respectively (3). The fact that these low rates are evident even at sites that are subject to relatively small amounts of purifying selection (4, 5) suggests that they are the result of very low underlying mutation rates: a surprising observation especially when contrasted with the rapid accumulation of mitochondrial mutations in many eukaryotic lineages (6, 7).
Although the genetic mechanisms that enable plants to achieve such faithful replication and transmission of cytoplasmic DNA sequences have not been determined, a number of hypotheses can be envisioned. One possibility is that the DNA polymerases responsible for replicating mitochondrial and plastid DNA (8) might have unusually high fidelity. However, in vitro assays with the two partially redundant bacterial-like organellar DNA polymerases in Arabidopsis thaliana, PolIA (At1g50840) and PolIB (At3g20540), have indicated that they are highly error-prone (9), with misincorporation rates that exceed those of Pol γ, the enzyme responsible for replicating DNA in the rapidly mutating mitochondrial genomes of humans (10). These observations make it unlikely that the low mutation rates in plant cytoplasmic genomes are attributable to exceptional polymerase accuracy, although it is unclear whether these in vitro assays conducted in the absence of associated proteins fully reflect the function of the polymerases in vivo. This work also found that PolIB had a measured error rate (5.45 × 10−4 per base pair) that is 7.5-fold higher than that of PolIA (7.26 × 10−5 per base pair). Therefore, although knocking out both of these genes results in lethality (8), disrupting one of the two polymerases to make the cell rely on the other may provide an opportunity to investigate the effects of polymerase misincorporation on the overall mutation rate.
It is also possible that plant mitochondria and plastids are unusually effective at preventing or repairing DNA damage resulting from common mechanisms such as guanine oxidation (e.g., 8-oxo-G) and cytosine deamination (uracil). Like most organisms, plants encode dedicated enzymes to recognize these forms of damage and initiate base-excision repair. In Arabidopsis, a pair of enzymes (FPG [At1g52500] and OGG1 [At1g21710]) both appear to function in repair of 8-oxo-G in mitochondrial and nuclear DNA, as evidenced by increased oxidative damage in the double mutant background (11). Another DNA repair enzyme, uracil N-glycosylase (UNG [At3g18630]), recognizes and removes uracil in all three genomic compartments (12, 13). A recent study investigated the effects of knocking out UNG on mitochondrial sequence variation in Arabidopsis but did not find any nucleotide substitutions that rose to high frequency, nor any difference in variant frequencies relative to wild-type in 10-generation mutation accumulation lines (14). The apparent low fidelity of plant organellar DNA polymerases and tolerance of disruptions to the UNG base-excision repair pathway suggest that other mechanisms are at play in dealing with mismatches and DNA damage.
Mitochondrial and plastid genomes are present in numerous copies per cell, and it is often hypothesized that recombination and homology-directed repair (HDR) may eliminate mutations and damaged bases in plant cytoplasmic genomes (9, 14–18). In both mitochondria and plastids, there is extensive recombination and gene conversion between homologous DNA sequences (19–21), and the large inverted repeats in plastid genomes have slower sequence evolution than single-copy regions (1, 22), suggesting that increased availability of homologous templates may improve the accuracy of error correction. The extensive recombinational dynamics in plant mitochondrial genomes often extend to short repeat sequences, resulting in structural rearrangements. As such, the slow rate of sequence evolution in these genomes is juxtaposed with rapid structural change (21).
MutS Homolog 1 (MSH1 [At3g24320]) is involved in recombination in both mitochondria and plastids and represents a natural candidate for maintaining low mutation rates in plant cytoplasmic genomes. Plants homozygous for mutated copies of MSH1 or with RNAi-based suppression of this gene exhibit extensive phenotypic variation (23–25) and often develop variegated leaf phenotypes that subsequently follow a pattern of maternal inheritance, indicating alterations in cytoplasmic genomes (26–29). MSH1 is distinguished from other members of the larger mutS mismatch repair (MMR) gene family by an unusual C-terminal domain predicted to be a GIY-YIG endonuclease (30). This observation led Christensen (17) to hypothesize that MSH1 recognizes mismatches or DNA damage and introduces double-stranded breaks (DSBs) at those sites as a means to initiate accurate repair via HDR. However, analysis and sequencing of mitochondrial and plastid genomes in msh1 mutants has not detected de novo base substitutions or small indels (16, 28, 29, 31). Instead, characterization of cytoplasmic genomes in these mutants has revealed structural rearrangements resulting from ectopic recombination between small repeats (16, 28, 29, 32). These findings have led to the prevailing view that the primary role of MSH1 is in recombination surveillance rather than in correction of mismatches or damaged bases (28, 30, 33–36), as is the case for some other members of the mutS gene family (37). Numerous other genes have also been identified as playing a role in mitochondrial and plastid recombination (21). One example is the mitochondrial-targeted RECA3 gene (At3g10140), with recA3 and msh1 mutants exhibiting similar but nonidentical effects in terms of repeat-mediated rearrangements and aberrant growth phenotypes (33, 38).
It is striking that so many genes have been identified in controlling the structural stability of plant mitochondrial and plastid genomes (21) and yet researchers have not been able to identify any gene knockouts in plants that lead to increased cytoplasmic mutation rates despite the many promising hypotheses and candidates. This gap may reflect the inherent challenges in studying rare mutational events in long-lived multicellular organisms. The advent of high-throughput DNA sequencing has raised the possibility of using deep sequencing coverage to catch de novo mutations essentially as they arise and are still present at extremely low frequencies among the many cytoplasmic genome copies that are found within cells and tissue samples (heteroplasmy). However, the error rate of standard sequencing technologies such as Illumina is relatively high—often above 10−3 errors per base pair and much worse in certain sequence contexts (39)—setting a problematic noise threshold for accurate detection of rare variants. Fortunately, numerous specialized methods have been introduced to improve these error rates (40). The most accurate technique is known as duplex sequencing (41), which entails tagging both ends of each original DNA fragment with adapters containing random barcodes such that it is possible to obtain a consensus from multiple reads originating from the same biological molecule, including those from each of the two complementary strands. Duplex sequencing has been found to reduce error rates ∼10,000-fold to levels below 10−7 errors per base pair (41), opening the door for accurate detection of extremely rare variants.
Here, we have applied duplex sequencing to detect de novo mitochondrial and plastid mutations in wild-type Arabidopsis and a number of mutant backgrounds carrying disrupted copies of key nuclear candidate genes involved in cytoplasmic DNA recombination, replication, and repair (RRR). We find that, of these candidates, only msh1 mutants show massive increases in rates of point mutations and small indels in cytoplasmic genomes, identifying this gene as a key player in maintaining the remarkably low mutation rates in plant mitochondria and plastids.
Results
Detection of Mitochondrial and Plastid Mutations in Wild-Type Arabidopsis.
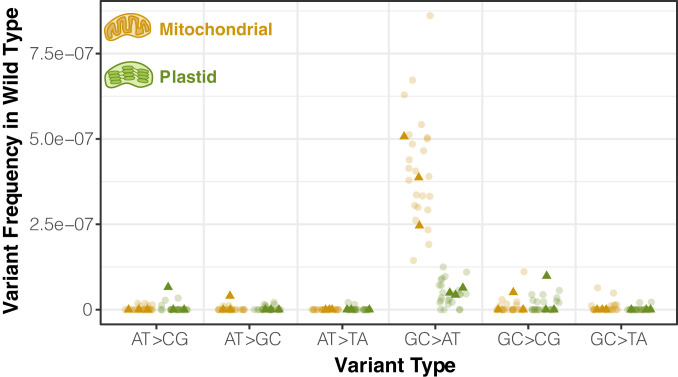
We modified the standard duplex sequencing protocol to include treatment with repair enzymes that correct single-stranded DNA damage and established a noise threshold of ∼2 × 10−8 sequencing errors per base pair using Escherichia coli samples derived from single colonies (SI Appendix, Tables S1 and S2). We then applied this sequencing method to purified mitochondrial and plastid DNA from A. thaliana Col-0 to provide a baseline characterization of the variant spectrum in wild-type plants. To obtain sufficient quantities of purified mitochondrial and plastid DNA, we pooled rosette tissue from multiple individuals from full-sib families (∼35 g per replicate). Following the removal of spurious variants that resulted from contaminating nuclear copies of mitochondrial- or plastid-derived sequences [known as NUMTs and NUPTs (42), respectively] or from chimeric molecules produced by recombination between nonidentical repeats, we found that almost all single-nucleotide variant (SNV) types were present at a frequency of less than 10−7, suggesting that levels of standing variation were generally at or near the noise threshold despite the extreme sensitivity of this method. The one obvious exception was GC → AT transitions in mtDNA, which were detected at a mean frequency of 3.8 × 10−7 across three biological replicates. The dominance of GC → AT transitions in the mitochondrial mutation spectrum was further supported by subsequent sequencing of 24 additional wild-type control replicates (Fig. 1) that were part of later experiments investigating individual candidate genes. GC → AT transitions also tended to be the most common type of SNV in plastid DNA samples, but at a level that was almost an order of magnitude lower than that observed in mitochondrial samples (mean frequency of 4.6 × 10−8).
Fig. 1.
Observed frequency of mitochondrial and plastid SNVs in wild-type Arabidopsis tissue based on duplex sequencing. Dark triangles represent three biological replicate families of wild-type A. thaliana Col-0. Lighter circles are F3 families derived from homozygous wild-type plants that segregated out from a heterozygous parent containing one mutant copy of an RRR candidate gene. Variant frequencies are calculated as the total number of observed mismatches in mapped duplex consensus sequences divided by the total base pairs of sequence coverage.
Screen of Candidate Genes Reveals Greatly Increased Frequency of Mitochondrial and Plastid Mutations in msh1 Knockout.
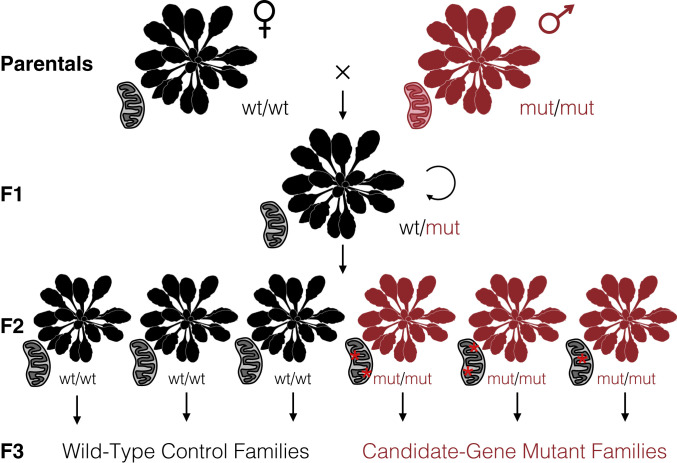
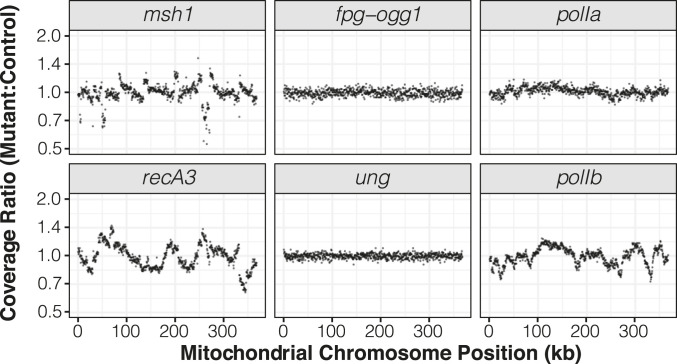
To test the effects of disrupting key RRR genes (SI Appendix, Table S3) on cytoplasmic mutation rates, we applied crossing designs that enabled direct comparisons between families of homozygous mutants and matched wild-type controls that all inherited their cytoplasmic genomes from the same grandparent (Fig. 2). We performed duplex sequencing with purified mitochondrial and plastid DNA from the resulting samples, generating a total of 1.2 Tbp of raw Illumina reads that were collapsed down into 10 Gbp of processed and mapped duplex consensus sequence (DCS) data (SI Appendix, Table S4). Many of the candidate genes have been previously found to affect structural stability in the mitochondrial genome (8, 33, 38). Consistent with these expected structural effects, we found that msh1, recA3, and polIb mutants all showed their own distinct patterns of large and repeatable shifts in coverage in regions of the mitochondrial genome (Fig. 3). Shifts in coverage were weaker in polIa mutants and not detected in ung mutants or fpg/ogg1 double mutants. Such coverage variation was not found in the plastid genome for any of the mutants (SI Appendix, Fig. S1).
Fig. 2.
Crossing design to test candidate nuclear genes involved in RRR of cytoplasmic genomes. Using a wild-type maternal plant (black) and either a homozygous mutant (red) or heterozygous pollen donor, we generated a heterozygous F1 individual that carried cytoplasmic genomes inherited from a wild-type lineage (as indicated by the black mitochondrion). After selfing the F1, we genotyped the resulting F2 progeny to identify three homozygous mutants and three homozygous wild-type individuals. Given that the mutations in candidate RRR genes are expected to be recessive, the F2 generation would be the first in which the sampled cytoplasmic genomes were exposed to the effects (red asterisks) of these mutants. The identified F2 individuals were each allowed to self-fertilize and set seed to produce multiple F3 families that all inherited their cytoplasmic genomes from the same F1 grandparent. The F3 families were used for purification of mitochondrial and plastid DNA for duplex sequencing. Sequencing was performed on three replicate families for each genotype. Arabidopsis silhouette image is from PhyloPic (Mason McNair).
Fig. 3.
Sequencing coverage variation across the mitochondrial genome in mutants relative to their matched wild-type controls. Each panel represents an average of three biological replicates. The reported ratios are based on counts per million mapped reads in 500-bp windows. The msh1 mutant line reported in this figure is CS3246.
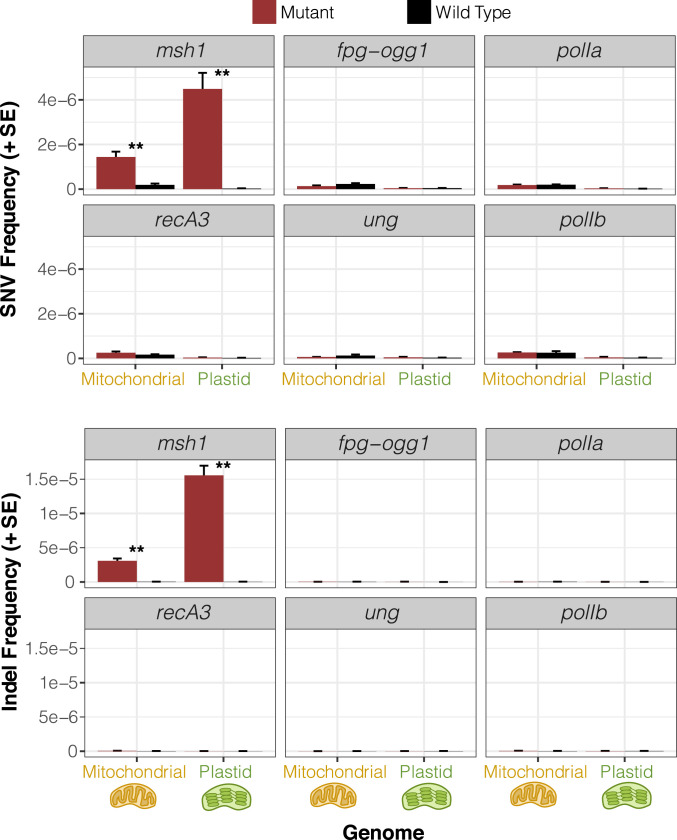
Most of the analyzed candidate genes did not have detectable effects on cytoplasmic mutation rates. Despite the difference in measured misincorporation rates for PolIA and PolIB in vitro (9), we did not find that disrupting either of these genes had an effect on the frequencies of SNVs or small indels in vivo (Fig. 4 and SI Appendix, Fig. S2). Likewise, ung mutants and fpg/ogg1 double mutants did not exhibit any detectable increase in sequence variants. In recA3 mutants, there was a weak trend toward higher rates of mitochondrial SNVs and small indels compared to wild-type controls (SI Appendix, Fig. S2), but neither of these effects were statistically significant.
Fig. 4.
Observed frequency of mitochondrial and plastid SNVs (Top) and indels (Bottom) based on duplex sequencing in Arabidopsis mutant backgrounds for various RRR genes compared to matched wild-type controls. Variant frequencies are calculated as the total number of observed mismatches or indels in mapped duplex consensus sequences divided by the total base pairs of sequence coverage. Means and SEs are based on three replicate F3 families for each genotype (Fig. 2). The msh1 mutant line reported in this figure is CS3246. **Significant differences between mutant and wild-type genotypes at a level of P < 0.01 (t tests on log-transformed values). All other comparisons were nonsignificant (P > 0.05).
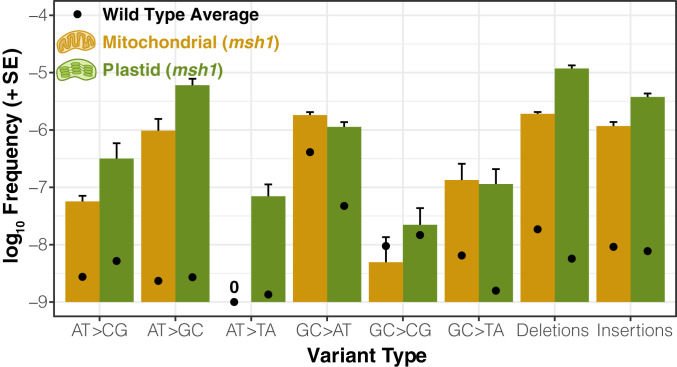
Unlike the rest of the candidate genes, the msh1 mutant line (CS3246) exhibited a striking increase in SNVs compared to wild type controls: close to 10-fold in the mitochondrial genome and more than 100-fold in the plastid genome (Fig. 4 and SI Appendix, Fig. S2). The msh1 mutation spectrum in both mitochondrial and plastid DNA was dominated by transitions. GC → AT substitutions remained the most common mitochondrial SNV in msh1 mutants, but there was a disproportionate increase in AT → GC variants such that both transition types reached comparable levels (Fig. 5). The increased frequency of AT → GC transitions in msh1 mutants was even more dramatic for plastid DNA, making them by far the most abundant type of SNV. Disruption of MSH1 also affected transversion rates, with substantial increases in GC → TA and AT → CG SNVs in both genomes (Fig. 5).
Fig. 5.
Spectrum of point mutations and indels in Arabidopsis msh1 mutants (CS3246 allele). Variant frequencies are calculated as the total number of observed mismatches or indels in mapped duplex consensus sequences divided by the total base pairs of sequence coverage. Means and SEs are based on three replicate F3 families. Black points represent the mean variant frequency calculated from all wild-type libraries (Fig. 1). Note that the “0” indicates that no mitochondrial AT → TA transversions were observed in any of the msh1-CS3246 or wild-type libraries.
The rate of small indel mutations also increased dramatically in the msh1 mutant line, with indel frequencies jumping approximately two or three orders of magnitude in the mitochondrial and plastid genomes, respectively (Fig. 4 and SI Appendix, Fig. S2). The indels in msh1 mutants overwhelmingly occurred in homopolymer regions (i.e., single-nucleotide repeats). On average, 92.1% of indels from mapped DCS data in the mitochondrial genome and 98.0% in the plastid genome were in homopolymers at least 5 bp in length, and many of the remaining indels represented expansions or contractions of short tandem repeats (Dataset S1). There was a clear bias toward deletions in the msh1 mutants, with deletions 1.6-fold and 3.1-fold more abundant than insertions on average in mitochondrial and plastid genomes, respectively (Fig. 5), which is a common feature of indel spectra across the diversity of life (43). Just as in the initial wild-type analysis described above, all SNVs and indels were filtered to exclude variants resulting from chimeric recombination products. Therefore, the observed increases in variant frequency in msh1 mutants are not simply the result of the well described role of MSH1 in recombination surveillance (16, 28, 29, 32).
Confirmation of msh1 Mutator Effects in Additional Mutant Backgrounds.
To verify that disruption of msh1 was indeed responsible for the observed elevation in mitochondrial and plastid mutation rates, we repeated our crossing design and duplex sequencing analysis with two additional independently derived mutant alleles in this gene (SI Appendix, Table S3). All three msh1 mutant backgrounds showed the same qualitative pattern of increased SNVs, small indels, and structural variation (SI Appendix, Figs. S3 and S4). The magnitude of these effects was equivalent in the initial mutant line (CS3246) and a second mutant (CS3372), which both harbor point mutations that appear to generate null msh1 alleles (28). In contrast, the increases in sequence and structural variation were much smaller for a third msh1 allele (SALK_046763). The SALK_046763 mutants also exhibited weaker phenotypic effects, with lower rates of visible leaf variegation (SI Appendix, Fig. S5). The 3,357-bp MSH1 coding sequence is distributed across 22 exons, and this mutant allele carries a T-DNA insertion in the eighth intron (28), which we reasoned might reduce but not eliminate expression of functional MSH1 protein, resulting in weaker effects on phenotype and mutation rates. In support of this prediction, complementary DNA (cDNA) sequencing across the boundary between exons 8 and 9 confirmed the presence of properly spliced transcripts in homozygous SALK_046763 mutants despite the large T-DNA insertion in the intron (SI Appendix, Fig. S6A. Furthermore, quantitative reverse-transcriptase PCR (qRT-PCR) showed that expression levels in leaf tissue were roughly fivefold lower than in wild-type individuals (SI Appendix, Fig. S6B). Therefore, reducing the expression level of MSH1 also appears to increase cytoplasmic mutation rates, though to a lesser degree than effective knockouts.
Inheritance of msh1-Induced Heteroplasmies.
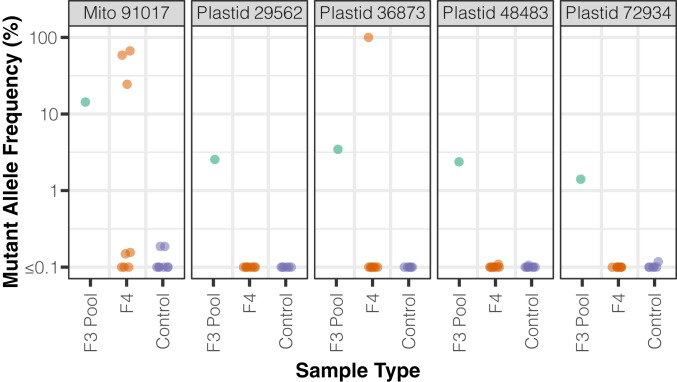
Because we performed our duplex sequencing analysis on whole-rosette tissue, it was not immediately clear whether the increase in observed mutations included changes that could be transmitted across generations or only variants that accumulated in vegetative tissue and would not be inherited. The majority of SNVs (∼80%) in msh1 mutants and all SNVs in their matched wild-type controls were detected in only a single DCS read family (Dataset S1), implying very low heteroplasmic frequency in the pooled F3 tissue as would be expected for new mutations. However, we did identify a total of 433 SNVs across the msh1 mutant samples that were each supported by multiple DCS read families (i.e., distinct biological molecules in the original DNA samples), in some cases reaching frequencies of >2%. We reasoned that, to be found at such frequencies in a pool of tissue from dozens of F3 individuals, a variant likely had to have occurred in the F2 parent and been inherited in a heteroplasmic state by multiple F3s. Although the individuals used for duplex sequencing were sacrificed in the process of extracting mitochondrial and plastid DNA from whole rosettes, we had collected F4 seed from siblings of the F3 plants that were grown up in parallel. Therefore, we developed droplet digital PCR (ddPCR) markers to test for the inheritance of some identified high-frequency SNVs in the F4 generation.
We assayed five SNVs with ddPCR markers, each of which was found at substantial frequencies in the corresponding F3 cytoplasmic DNA sample (1.4 to 14.3%), confirming the variant identification from our duplex sequencing. As controls, we sampled F4 msh1 mutants derived from other F3 families that did not show evidence of the variant in duplex sequencing data. All of these controls exhibited a frequency of below 0.2%, which we considered the noise threshold for the assay. For two of the five markers (one mitochondrial and one plastid), we were also able to detect the SNV in DNA samples from individual F4 plants. The frequency of these heteroplasmic mutations varied dramatically across F4 individuals: anywhere from below the noise threshold to as high as apparent homoplasmy (>99.9%) in one case (Fig. 6). These high frequencies indicate the potential for de novo mutations to spread to majority status remarkably fast. To further assess whether these SNVs represented heritable mutations, we performed follow-up assays on siblings of three different F4 individuals that showed detectable mutations in the initial round of ddPCR analysis. In each case, we identified siblings sharing the same variant (SI Appendix, Table S5). Therefore, these SNVs provide clear evidence that de novo cytoplasmic mutations can occur in meristematic tissue in an msh1 mutant background and be transmitted across generations, thereby increasing the heritable mutation rate.
Fig. 6.
Estimates of heteroplasmic frequency of select SNVs using ddPCR. The F3 pool is the same msh1-CS3246 mutant mitochondrial or plastid DNA sample in which the SNV was discovered by duplex sequencing. F4 indicates plants descended from the F3 family used in duplex sequencing. Controls are F4 plants from msh1-CS3246 mutant lines other than the one in which the SNV was originally discovered.
For the other three SNVs, we did not detect the heteroplasmic mutation in a sample of eight F4 individuals, which could indicate that the variant was restricted to vegetative tissue in a single individual within the F3 pool. However, we suspect that it is more likely that the negative F4 individuals lost the corresponding variant via a heteroplasmic sorting process or descended from a subset of F3 parents that did not carry it.
Plant MSH1 Is Part of a Widely Distributed Gene Family in Diverse Eukaryotic Lineages, as Well as Some Bacteria and Viruses.
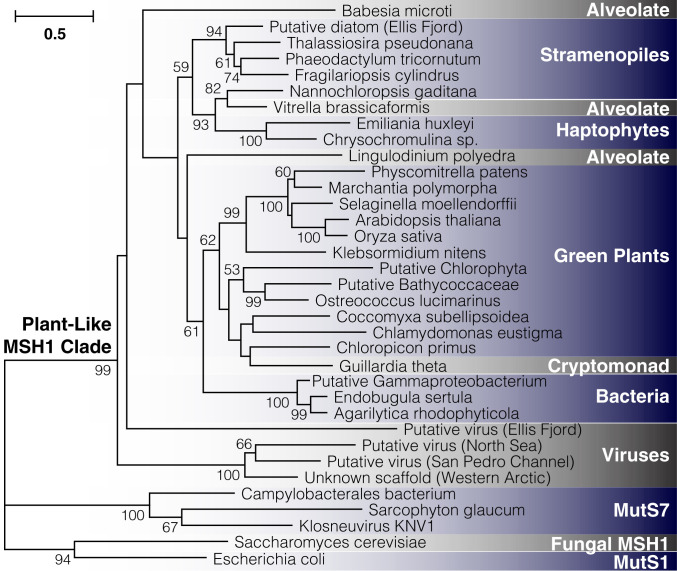
MSH1 is divergent in sequence and domain architecture relative to all other members of the mutS MMR gene family (44). Although named after the MSH1 gene in yeast, which also functions in mitochondrial DNA repair (45), plant MSH1 is from an entirely different part of the large mutS family (44). It is known to be widely present across green plants (46), but its evolutionary history beyond that is unclear. Taxon-specific searches of public genomic and metagenomic repositories failed to detect copies of MSH1 in red algae and glaucophytes, the other two major lineages of Archaeplastida. Likewise, we did not find evidence of this gene in Amorphea (which includes Amoebozoa, animals, fungi, and related protists). Although these initial results implied a distribution that might be truly restricted to green plants (Viridiplantae), searches of other major eukaryotic lineages found that plant-like MSH1 homologs carrying the characteristic GIY-YIG endonuclease domain were present in numerous groups, specifically stramenopiles, alveolates, haptophytes, and cryptomonads (Fig. 7). More surprisingly, we found that it was present in the genomes of two closely related bacterial species within the Cellvibrionaceae (Gammaproteobacteria) and another gammaproteobacterium of uncertain classification, as well as some unclassified viruses curated from environmental and metagenomic datasets (47, 48). Phylogenetic analysis confirmed that these sequences represented a well-resolved clade within the mutS family (Fig. 7). Therefore, the plant-like MSH1 gene appears to have an unusually disjunct distribution across diverse lineages in the tree of life.
Fig. 7.
Detection of plant-like MSH1 genes across diverse evolutionary lineages. The maximum-likelihood tree is constructed based on aligned protein sequences, with branch lengths indicating the number of amino acid substitutions per site. Support values are percentages based on 1,000 bootstrap pseudoreplicates (only values >50% are shown). Metagenomic samples are putatively classified based on other genes present on the same assembled contig (SI Appendix, Table S6, provides information on sequence sources). The sample labeled “unknown scaffold” lacks additional genes for classification purposes, but it clustered strongly with two sequences from the IMG/VR repository of viral genomic sequences.
Discussion
The Role of MSH1 in Maintaining the Low Mutation Rates.
The striking differences in mutation rates between cytoplasmic genomes in land plants vs. those in many other eukaryotes, including mammals, have posed a longstanding mystery because reactive oxygen species (ROS) are expected to be a potent source of DNA damage in all of these compartments. The presence of MSH1 in plants and its dual targeting to the mitochondria and plastids may provide an explanation for their unusually low rates. Although previous efforts to analyze cytoplasmic genomes in msh1 mutants with conventional sequencing technologies did not detect de novo SNV or indel mutations (16, 29, 31), our application of the higher-sensitivity duplex sequencing method found that msh1 mutants exhibit major increases in the frequency of these variants. These findings align with a growing theme that the distinctive mutational properties of cytoplasmic genomes relative to the nucleus may be driven more by differences in RRR machinery than by ROS or the biochemical environment associated with cellular respiration and photosynthesis (49–51).
How does MSH1 suppress cytoplasmic mutation rates? As a mutS homolog, it could conceivably be part of a conventional MMR pathway that has yet to be described in plant organelles. Postreplicative mismatch repair typically relies on the heuristic that mismatches in double-stranded DNA are more likely to reflect errors in the newly synthesized strand, with various mechanisms being used to specifically identify and repair that strand (52). However, the presence of a conventional MMR pathway would not explain how plant mitochondria and plastids maintain mutation rates substantially lower than in most eukaryotic genomes. An alternative, nonconventional pathway could involve use of the GIY-YIG endonuclease domain to introduce a DSB near sites identified by the mismatch recognition domain, followed by HDR of the DSB (9, 14, 17). This previously proposed model could lead to unusually high repair accuracy because it does not require use of a heuristic to determine which strand carries the error at mismatched sites, instead employing homologous recombination with an unaffected genome copy to “break the tie.” As such, incipient mutations would be detected and accurately corrected when they are still in the mismatched state before they become true double-stranded base pair substitutions.
A model based on DSBs and HDR might also explain some surprising features in our data. We found that the frequency of SNVs in wild-type plants was much higher in the mitochondrial genome than the plastid genome, which is opposite the rates of evolution observed in these genomes on phylogenetic scales (1, 3). The mutation spectrum in wild-type mitochondrial DNA was also dominated by GC → AT transitions (Fig. 1), which is inconsistent with the relatively neutral transition:transversion ratio observed in natural sequence variation both within and among species (53, 54). We speculate that these apparent contrasts can be explained by the different copy numbers of mitochondrial and plastid genomes in vegetative tissues. Whereas individual plastids each contain numerous genome copies, it has been estimated that there is less than one genome copy per mitochondrion in Arabidopsis leaf tissue (55). Therefore, even when MSH1 is intact, HDR pathways may be less available for repair of mitochondrial DNA in vegetative tissues due to a paucity of homologous template copies (14), which would imply that the abundant GC → AT SNVs in wild-type mitochondrial DNA are generally restricted to vegetative tissue and not transmitted to future generations. The role of HDR may also be limited by the lower expression of MSH1 in certain vegetative tissues and cell types (24, 33, 56). In contrast, the fusion of mitochondria into a large network within meristematic cells could provide an opportunity for mitochondrial genome copies to cooccur and utilize HDR (57). Because of the high genome copy number in plastids, they may rely more heavily on HDR even in some vegetative tissues, which would explain why knocking out MSH1 has a much larger proportional effect on observed variant frequencies in the plastid genome (Fig. 4 and SI Appendix, Fig. S2).
We find growing support for the model in which MSH1 is the link between mismatches, DSBs, and HDR. However, much remains to be done to validate this model, as researchers have yet to successfully express and purify full-length MSH1 for in vitro biochemical studies, and a recent analysis of the purified GIY-YIG domain was unable to detect endonuclease activity (34). Moreover, our use of pools of whole rosettes from F3 families means that the observed variants in msh1 mutant lines are a combination of those that arose in meristematic tissue in F2 parents and throughout F3 development and vegetative tissue differentiation. As such, our analysis does not provide a direct quantification of the mutation rate per generation or per round of DNA replication. However, given the large effect of disrupting MSH1 on variant frequencies, we anticipate that propagation and resequencing of msh1 mutation accumulation lines may be an effective means to quantify the per-generation rate of heritable mitochondrial and plastid mutations, which has been unmeasurably low in other Arabidopsis mutation accumulation lines (14, 54). Future analysis and technological advances may also help map the dynamics of organelle mutations at a finer scale, including cell- and organelle-level resolution. Given the evidence that differential expression and subcellular localization leads to variation in the abundance of MSH1 across tissue, cell, and plastid types (24, 33, 56), such progress would improve our understanding of how mutation accumulation aligns with the activity of MSH1, DNA replication, and other organelle functions throughout development.
Which DNA aberrations does MSH1 recognize? The ability to bind to multiple types of disruptions in Watson–Crick pairing is a common feature of many MutS homologs (52). The fact that we observed increased frequencies of indels in msh1 mutants implies that MSH1 can recognize indel loops, including in homopolymer regions, which are likely to be one of the most prevalent sources of polymerase errors, especially in the AT-rich genomes of plastids (58). The increased frequency of SNVs in msh1 mutants also implies recognition of the bulges in DNA caused by mismatches and/or damaged bases. There is some evidence to suggest that MSH1 is capable of repairing both of these sources of mutation. The most prominent feature of the msh1 mutation spectrum is the enormous increase in AT → GC transitions, which does not correspond to a major class of damage like cytosine deamination (GC → AT) or guanine oxidation (GC → TA). This aspect of the mutation spectrum is more likely explained by polymerase misincorporations during DNA replication, as steady-state kinetic analysis has indicated that PolIA and PolIB are highly prone to misincorporate Gs opposite Ts in the template strand (9). We reasoned that disrupting the POLIA gene would increase mutation rates because of higher misincorporation rates for PolIB (9). The failure to find such an effect suggests a general insensitivity to polymerase errors when MSH1 is intact, presumably because of its ability to recognize and repair these errors. This proposed role of MSH1 would similarly explain why sequence evolution is so slow in these genomes despite polymerases with unusually high misincorporation rates (9). Disrupting genes involved in the repair of uracil (UNG) and 8-oxo-G (FPG and OGG1) failed to measurably affect the frequency of mitochondrial or plastid variants, which could indicate that MSH1 is capable of recognizing and correcting such damage. Alternatively, these sources of damage may be too minor under the tested growth conditions to contribute meaningfully to variant frequencies, or there may be additional uncharacterized plant genes with overlapping base-excision repair functions. For example, some animals have a second enzyme with uracil N-glycosylase activity (59). However, the fact that MSH1 was recently shown to exhibit higher expression in ung mutants (14) points toward a capability to recognize damaged bases in addition to conventional mismatches.
Although land plants generally exhibit low rates of sequence evolution in cytoplasmic genomes, major exceptions have been documented in certain angiosperm lineages (60). These observations raise the question of whether changes in MSH1 contributed to such accelerations. Previous studies have confirmed that MSH1 is present and transcribed in accelerated lineages such as Geraniaceae and Silene (61, 62), suggesting that these accelerations cannot be explained by the outright loss of MSH1. However, functional investigations would be needed to assess the possibility that there have been changes in MSH1 expression patterns, localization, or activity in these taxa.
Disrupting MSH1 has been shown to affect numerous traits and result in extensive phenotypic variation in descendent lineages (23, 24). However, we caution against assuming that specific phenotypes are attributable to the observed accumulation of point mutations and small indels in cytoplasmic genomes. In addition to these effects on mutation rates, msh1 mutants also exhibit dramatic structural reorganization of organelle genomes via repeat-mediated recombination (27–29, 32), as well as widespread changes to patterns of epigenetic modifications in the nucleus (23, 24, 31), both of which have been associated with large phenotypic consequences. Moreover, despite the massive proportional increases in the frequency of SNVs and small indels that we observed in msh1 mutants, the abundance of mutations is not necessarily high in an absolute sense (e.g., less than one SNV per cytoplasmic genome copy on average; Fig. 4). Hemicomplementation assays, in which MSH1 is targeted to either mitochondria or plastids but not both, have been an effective way to distinguish phenotypic effects mediated by mitochondrial vs. plastid functions of MSH1 (24, 29, 31). Likewise, the use of segregation and backcrossing designs with msh1 mutants or lines that suppress MSH1 expression via RNAi have helped partition effects of cytoplasmic genetics vs. heritable nuclear epigenetic modifications (23, 27, 31). To distinguish among the phenotypic effects of structural vs. sequence changes in organelle genomes, it may be helpful that structural rearrangements and copy-number changes show a high degree of repeatability among msh1 replicates and lines (SI Appendix, Fig. S3), in contrast to the random nature of de novo point mutations.
The Evolutionary History of MSH1 and Parallels with Other mutS Lineages.
To date, MSH1 has been identified and studied only in green plants. Researchers have previously noted the similarities in domain architecture between MSH1 and MutS7 (30), a lineage within the MutS family that independently acquired a C-terminal fusion of an endonuclease domain (44). MutS7 is encoded in the mitochondrial genome itself of octocorals, another eukaryotic lineage with unusually slow rates of mitochondrial genome evolution (63), as well as in the genomes of a small number of bacterial lineages and some giant viruses (64). In this sense, our results extend the parallels between MSH1 and MutS7 to include features of their phylogenetic distribution, as each is scattered across disparate lineages of eukaryotes, bacteria, and viruses. The distribution of MSH1 (Fig. 7) clearly implies some history of horizontal gene transfer. However, the ancient divergences, sparse representation outside of eukaryotes, and poor phylogenetic resolution at deep splits within the gene tree make the timing of such events or specific donors and recipients unclear. Another open question is the functional role of MSH1 outside of land plants. The similarities in its effects on organelle genome stability between angiosperms and mosses (28, 32) suggest that much of the role of MSH1 in cytoplasmic genome maintenance is likely ancestral, at least in land plants. Notably, all of the eukaryotes that we identified as having MSH1 outside of green plants harbor a plastid derived from secondary endosymbiosis. It will therefore be interesting to assess whether it has mitochondrial and/or plastid functions in these eukaryotes (some show in silico targeting predictions to the organelles; SI Appendix, Table S6). This pattern also raises the question as to whether it was ancestrally present deep in the eukaryotic tree and subsequently lost in many lineages or transferred among major eukaryotic lineages perhaps in conjunction with secondary endosymbiosis.
Because the apparent viral copies of MSH1 were curated from metagenomic assemblies and bulk environmental virus sampling (SI Appendix, Table S6), we were not able to assign these sequences to a specific type of virus. Interestingly, however, one of these cases was found on a viral-like metagenomic contig in the IMG/VR database that is >100 kb in size, and another cooccurs on a contig with a gene that has a top BLAST hit to the Mimiviridae, a clade of giant viruses. Therefore, similar to mutS7, it appears that MSH1 may reside in giant viruses. We speculate that such viruses, which are also known as nucleocytoplasmic large DNA viruses or NCLDVs (65), have acted as a repository for distinctive RRR machinery and a repeated source of horizontal acquisition by eukaryotic lineages, reshaping the mechanisms of cytoplasmic mutation rate and genome maintenance.
Materials and Methods
A complete description of the methods is available in the SI Appendix. In brief, mitochondrial and plastid DNA isolations were performed on rosette tissue harvested after 7 to 9 wk of growth from either A. thaliana Col-0 or F3 families derived from crossing mutant lines (SI Appendix, Table S3) against A. thaliana Col-0 (Fig. 2). Duplex sequencing followed a modified version of the protocol of Kennedy et al. (41) that was first optimized in our lab by testing on single-colony E. coli samples. Sequencing was performed on an Illumina NovaSeq 6000 at the University of Colorado Cancer Center. Data processing was performed with a custom pipeline available at https://github.com/dbsloan/duplexseq. Inheritance of selected high-frequency SNVs in the F4 generation was assessed with droplet digital PCR on a Bio-Rad QX200 system using fluorescently labeled allele-specific probes. To assess the phylogenetic distribution of plant-like MSH1 genes, searches were performed against the NCBI nr protein database, as well as metagenomic and viral repositories hosted by JGI (47, 48). Identified sequences were used for maximum-likelihood phylogenetic analysis with PhyML v3.3.20190321.
Data Availability.
All raw sequencing data are available via the NCBI Sequence Read Archive (PRJNA604834 and PRJNA604956), and the code used for data analysis is available at https://github.com/dbsloan/duplexseq.
Supplementary Material
Acknowledgments
We thank Dolores Córdoba‐Cañero for providing fpg and ogg1 mutant Arabidopsis lines and Claudia Gentry-Weeks for providing the E. coli K12 MG1655 strain. We also thank Mychaela Hodous, Jocelyn Lapham, Holly Harroun, Amber Torres, and Mariella Rivera for assistance with plant growth, seed collection, and PCR genotyping. Luis Brieba, Jeff Palmer, members of the Sloan laboratory, and three anonymous reviewers also provided insightful comments and discussion. This work was supported by a grant from the NIH (R01 GM118046) and an NSF graduate fellowship (DGE-1450032).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All raw sequencing data are available via the NCBI Sequence Read Archive (PRJNA604834 and PRJNA604956), https://www.ncbi.nlm.nih.gov/sra (SRR11018557–SRR11018568 and SRR11025077–SRR11025178) and the code used for data analysis is available in GitHub at https://github.com/dbsloan/duplexseq.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001998117/-/DCSupplemental.
References
- 1.Wolfe K. H., Li W. H., Sharp P. M., Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer J. D., Herbon L. A., Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J. Mol. Evol. 28, 87–97 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Drouin G., Daoud H., Xia J., Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 827–831 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Sloan D. B., Taylor D. R., Testing for selection on synonymous sites in plant mitochondrial DNA: The role of codon bias and RNA editing. J. Mol. Evol. 70, 479–491 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Wynn E. L., Christensen A. C., Are synonymous substitutions in flowering plant mitochondria neutral? J. Mol. Evol. 81, 131–135 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Brown W. M., George M. Jr., Wilson A. C., Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 76, 1967–1971 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havird J. C., Sloan D. B., The roles of mutation, selection, and expression in determining relative rates of evolution in mitochondrial versus nuclear genomes. Mol. Biol. Evol. 33, 3042–3053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parent J. S., Lepage E., Brisson N., Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol. 156, 254–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayala-García V. M., Baruch-Torres N., García-Medel P. L., Brieba L. G., Plant organellar DNA polymerases paralogs exhibit dissimilar nucleotide incorporation fidelity. FEBS J. 285, 4005–4018 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Longley M. J., Nguyen D., Kunkel T. A., Copeland W. C., The fidelity of human DNA polymerase γ with and without exonucleolytic proofreading and the p55 accessory subunit. J. Biol. Chem. 276, 38555–38562 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Córdoba-Cañero D., Roldán-Arjona T., Ariza R. R., Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 79, 824–834 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Boesch P. et al., Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res. 37, 5690–5700 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Córdoba-Cañero D., Dubois E., Ariza R. R., Doutriaux M. P., Roldán-Arjona T., Arabidopsis uracil DNA glycosylase (UNG) is required for base excision repair of uracil and increases plant sensitivity to 5-fluorouracil. J. Biol. Chem. 285, 7475–7483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynn E., Purfeerst E., Christensen A., Mitochondrial DNA repair in an Arabidopsis thaliana uracil N-glycosylase mutant. Plants (Basel) 9, 261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khakhlova O., Bock R., Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46, 85–94 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Davila J. I. et al., Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9, 64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen A. C., Genes and junk in plant mitochondria-repair mechanisms and selection. Genome Biol. Evol. 6, 1448–1453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevigny N., Schatz-Daas D., Lotfi F., Gualberto J. M., DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 21, 328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maréchal A., Brisson N., Recombination and the maintenance of plant organelle genome stability. New Phytol. 186, 299–317 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Arrieta-Montiel M. P., Mackenzie S. A., “Plant mitochondrial genomes and recombination” in Plant Mitochondria, Kempken F., Ed. (Springer Verlag, New York, 2011), Vol. 1, pp. 65–82. [Google Scholar]
- 21.Gualberto J. M., Newton K. J., Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 68, 225–252 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Zhu A., Guo W., Gupta S., Fan W., Mower J. P., Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 209, 1747–1756 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Virdi K. S. et al., Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth. Nat. Commun. 6, 6386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virdi K. S. et al., MSH1 is a plant organellar DNA binding and thylakoid protein under precise spatial regulation to alter development. Mol. Plant 9, 245–260 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Yang X. et al., Segregation of an MSH1 RNAi transgene produces heritable non-genetic memory in association with methylome reprogramming. Nat. Commun. 11, 2214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redei G. P., Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat. Res. 18, 149–162 (1973). [Google Scholar]
- 27.Martínez-Zapater J. M., Gil P., Capel J., Somerville C. R., Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell 4, 889–899 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdelnoor R. V. et al., Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. U.S.A. 100, 5968–5973 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y. Z. et al., MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23, 3428–3441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelnoor R. V. et al., Mitochondrial genome dynamics in plants and animals: Convergent gene fusions of a MutS homologue. J. Mol. Evol. 63, 165–173 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Xu Y.-Z. et al., The chloroplast triggers developmental reprogramming when mutS HOMOLOG1 is suppressed in plants. Plant Physiol. 159, 710–720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odahara M., Kishita Y., Sekine Y., MSH1 maintains organelle genome stability and genetically interacts with RECA and RECG in the moss Physcomitrella patens. Plant J. 91, 455–465 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Shedge V., Arrieta-Montiel M., Christensen A. C., Mackenzie S. A., Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19, 1251–1264 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukui K. et al., The GIY-YIG endonuclease domain of Arabidopsis MutS homolog 1 specifically binds to branched DNA structures. FEBS Lett. 592, 4066–4077 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Morley S. A., Ahmad N., Nielsen B. L., Plant organelle genome replication. Plants (Basel) 8, 358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odahara M., Factors affecting organelle genome stability in Physcomitrella patens. Plants (Basel) 9, 145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Z., Nei M., Ma H., The origins and early evolution of DNA mismatch repair genes–Multiple horizontal gene transfers and co-evolution. Nucleic Acids Res. 35, 7591–7603 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller-Messmer M. et al., RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 159, 211–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schirmer M., D’Amore R., Ijaz U. Z., Hall N., Quince C., Illumina error profiles: Resolving fine-scale variation in metagenomic sequencing data. BMC Bioinformatics 17, 125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sloan D. B., Broz A. K., Sharbrough J., Wu Z., Detecting rare mutations and DNA damage with sequencing-based methods. Trends Biotechnol. 36, 729–740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy S. R. et al., Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 9, 2586–2606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazkani-Covo E., Zeller R. M., Martin W., Molecular poltergeists: Mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 6, e1000834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo C. H., Ochman H., Deletional bias across the three domains of life. Genome Biol. Evol. 1, 145–152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogata H. et al., Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment. ISME J. 5, 1143–1151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pogorzala L., Mookerjee S., Sia E. A., Evidence that msh1p plays multiple roles in mitochondrial base excision repair. Genetics 182, 699–709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackenzie S. A., Kundariya H., Organellar protein multi-functionality and phenotypic plasticity in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen I. A. et al., IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paez-Espino D. et al., IMG/VR v.2.0: An integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 47, D678–D686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy S. R., Salk J. J., Schmitt M. W., Loeb L. A., Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 9, e1003794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itsara L. S. et al., Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 10, e1003974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kauppila J. H. K. et al., Base-excision repair deficiency alone or combined with increased oxidative stress does not increase mtDNA point mutations in mice. Nucleic Acids Res. 46, 6642–6669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiricny J., Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 5, a012633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christensen A. C., Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 5, 1079–1086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Z., Waneka G., Sloan D. B., The tempo and mode of angiosperm mitochondrial genome divergence inferred from intraspecific variation in Arabidopsis thaliana. G3 (Bethesda) 10, 1077–1086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preuten T. et al., Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 64, 948–959 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Beltrán J. et al., Specialized plastids trigger tissue-specific signaling for systemic stress response in plants. Plant Physiol. 178, 672–683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seguí-Simarro J. M., Staehelin L. A., Mitochondrial reticulation in shoot apical meristem cells of Arabidopsis provides a mechanism for homogenization of mtDNA prior to gamete formation. Plant Signal. Behav. 4, 168–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massouh A. et al., Spontaneous chloroplast mutants mostly occur by replication slippage and show a biased pattern in the plastome of Oenothera. Plant Cell 28, 911–929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nilsen H. et al., Excision of deaminated cytosine from the vertebrate genome: Role of the SMUG1 uracil-DNA glycosylase. EMBO J. 20, 4278–4286 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mower J. P., Touzet P., Gummow J. S., Delph L. F., Palmer J. D., Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol. Biol. 7, 135 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J. et al., Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity. Genome Biol. Evol. 8, 622–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Havird J. C., Trapp P., Miller C. M., Bazos I., Sloan D. B., Causes and consequences of rapidly evolving mtDNA in a plant lineage. Genome Biol. Evol. 9, 323–336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pont-Kingdon G. et al., Mitochondrial DNA of the coral Sarcophyton glaucum contains a gene for a homologue of bacterial MutS: A possible case of gene transfer from the nucleus to the mitochondrion. J. Mol. Evol. 46, 419–431 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Bilewitch J. P., Degnan S. M., A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evol. Biol. 11, 228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koonin E. V., Yutin N., Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv. Virus Res. 103, 167–202 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data are available via the NCBI Sequence Read Archive (PRJNA604834 and PRJNA604956), and the code used for data analysis is available at https://github.com/dbsloan/duplexseq.