Abstract
Lasso peptides are a class of ribosomally synthesized natural product which possess a unique lariat knot conformation. The low entropy “threaded” conformation endows lasso peptides with considerable resistance to heat and proteolytic degradation, which are attractive properties for the development of peptide-based therapeutics. Despite their discovery nearly 30 years ago, the molecular mechanism underlying lasso peptide biosynthesis remain poorly characterized due to low stability of the purified biosynthetic enzymes. Here, we report the biosynthetic reconstitution of a lasso peptide derived from Thermobifida fusca, termed fusilassin. Beyond robust catalytic activity, the fusilassin enzymes demonstrate extraordinary substrate tolerance during heterologous expression in E. coli and upon purification in cell-free biosynthetic reconstitution reactions. We provide evidence that the fusilassin biosynthetic enzymes are not capable of forming branched-cyclic products but can produce entirely unrelated lasso peptides. Finally, we leveraged our bioinformatic survey of all lasso peptides identified in GenBank to perform coevolutionary analysis of two requisite biosynthetic proteins. This effort correctly identified an important protein-protein interaction site, illustrating how genomics insight can accelerate the characterization of natural product biosynthetic pathways. The fusilassin enzymes described within represent a model system for both designing future lasso peptides of biomedical importance and also for elucidating the molecular mechanisms that govern lasso peptide biosynthesis.
Graphical Abstract
Introduction.
The ribosomally synthesized and post-translationally modified peptides (RiPPs) are a natural product class comprised of no fewer than two dozen structural subclasses encoded by all three domains of life.1 RiPP biosynthesis begins with the translation of a ribosomally synthesized precursor peptide which, for the majority of RiPPs, is composed of an N-terminal “leader peptide” and a C-terminal “core peptide.” A specific motif within the leader peptide, the recognition sequence, is bound by the RiPP recognition element (RRE)2 which acts to recruit the cognate biosynthetic enzymes to modify the core peptide. During lasso peptide biosynthesis, the leader peptidase (a homolog of the enzyme transglutaminase) site-specifically removes the leader peptide in an RRE-dependent fashion.3 Subsequently, a lasso cyclase enzyme (homologous to asparagine synthetase) presumably adenylates an Asp or Glu residue of the core peptide while folding the substrate into a pre-lasso conformation.4 The liberated N-terminus then attacks the adenylated residue, forming a macrolactam linkage and a sterically locked lasso peptide. Some lasso peptides display additional modification that include C-terminal methyl ester formation (e.g. lassomycin5), arginine deimination (e.g. citrulassin3), or epimerization (e.g. MS-2716). Steric constraints within the three-dimensional structure endow lasso peptides with extraordinary resistance to heat denaturation and proteolytic degradation. On rare occasions, some lasso peptides are known to unthread after prolonged periods at elevated temperatures.7,8
Nearly 50 lasso peptides have been isolated to date although less than half report a biological activity. Known bioactivities include cell-surface receptor antagonism, HIV fusion inhibition, RNA polymerase inhibition, and mycobacterial Clp protease inhibition.9 Beyond activities possessed by the native lasso peptides, the scaffold is highly amenable to alteration, as previously characterized lasso peptides have demonstrated tolerance to core peptide variation. A major exception to this flexibility is an intolerance to variation for the two residues involved in macrolactam formation. Introduction of non-native epitopes has conferred new functions to lasso peptides. Specifically, epitope grafting has been used to endow lasso peptides with activity towards the human integrin receptor.10,11 The rational engineering of lasso peptides is especially relevant when considering their translational potential. For instance, lasso peptides have demonstrated good stability under simulated gastrointestinal conditions12 and possess efficacy in mouse models of Helicobacter pylori13 and Salmonella enterica14 infections.
Given the potential of lasso peptides for future therapeutic development, there have been significant efforts from a number of research groups to isolate and characterize novel lasso peptides. Given their disparate biological activities, lasso peptides discovered by bioassay-guided isolation are serendipitous and low throughput. By leveraging recent advances in bioinformatics, rapid progress has been made in the identification and isolation of novel natural products. Frustratingly, the compactness of lasso peptide biosynthetic gene clusters (BGCs, sometimes less than 3 kb total) and the high sequence similarity of these enzymes to counterparts in primary metabolism (i.e. transglutaminase and asparagine synthetase) complicate genome-mining efforts. Additionally, the short length and hypervariability of the lasso precursor peptides typically means the coding sequences are not found by automated gene-finders and only nearly identical sequences can be found by sequence similarity search tools such as BLAST.
To address these longstanding issues, we developed the bioinformatic algorithm RODEO (Rapid ORF Description & Evaluation Online: www.ripprodeo.org), which automates the identification of lasso peptide3 and other RiPP BGCs15 from genomes available in GenBank. RODEO uses profile Hidden Markov Models and supervised machine learning to locate RiPP BGCs and predict the most likely precursor peptide, respectively. Previously, we leveraged RODEO to define the extent of lasso peptides encoded by all sequenced bacteria and archaea, which expanded the number of putative lasso peptide BGCs by two orders of magnitude and revealed significant new insights into the natural product family as a whole. Using the data set generated during this study, we selected several candidate BGCs from thermophilic bacteria for enzymatic reconstitution. We surmised that thermophilic proteins would exhibit higher general stability in vitro, a strategy that we have successfully applied to the study of other RiPP biosynthetic pathways.16,17
Results and Discussion.
Heterologous expression of fusilassin in E. coli.
Thermobifida fusca, a mesothermophilic actinomycete, was found to encode a lasso peptide BGC with a putative precursor peptide (FusA, coding sequence not annotated as a gene on NC_007333.1) adjacent to a lasso cyclase (FusC, WP_011291592.1), RRE (FusE, WP_011291591.1), leader peptidase (FusB, WP_011291590.1), and an ABC transporter (FusD, WP_011291589.1) (Fig. 1A). Attempts to detect “fusilassin” from the native producer by mass spectrometry (MS) were not successful. We thus cloned FusA into a modified pET28 vector that provides an N-terminal fusion to maltose-binding protein (MBP, see Supporting Methods, Table S1). FusB, FusC, and FusE were cloned into a separate pACYC expression vector, and the two plasmids were co-transformed into E. coli. After a 3 h expression, fusilassin was readily detected by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS in the crude methanolic extracts of the induced cells (Fig. 1B).
Figure 1. E. coli-based expression of fusilassin.
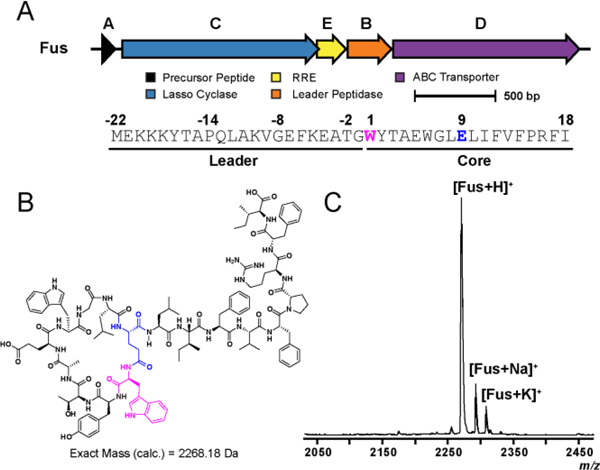
(A) Biosynthetic gene cluster diagram and primary sequence of FusA. (B) Predicted structure of lasso peptide fusilassin shown in an unthreaded conformation for clarity. The residues involved in macrolactam formation (Trp1 and Glu9) are highlighted pink and blue, respectively. (C) MALDI-TOF-MS spectrum demonstrating the expression of fusilassin (Fus) in E. coli.
Fusilassin represents a new lasso peptide of novel composition and is the first observed lasso peptide containing tryptophan at position one of the core peptide. It was previously believed that only small residues would be biosynthetically tolerated at this position.9 Given that Trp is the largest proteinogenic residue, we hypothesized that fusilassin would possess intrinsic tolerance to substitution at position 1, a site that has been historically recalcitrant to substitution.23–28 Fusilassin analogs were successfully produced via E. coli expression for all classes of amino acid (Fig. S1), an ability not demonstrated by any previously characterized lasso peptide. Only the Pro1 variant failed to generate a detectable lasso peptide. To discriminate if FusB, FusC, or both, were incapable of tolerating Pro1, we first sought to reconstitute fusilassin biosynthesis in vitro using purified proteins.
Enzymatic reconstitution and mutational analysis.
The genes encoding FusA, B, C, and E were individually cloned into pET28-MBP vectors, expressed in E. coli, and purified by amylose-affinity chromatography (Supporting Methods, Fig. S2). Production of a dehydrated product (m/z 2269 Da) was readily observed in reactions that included ATP and all four proteins (Fig. 2A). The 2269 Da species was further analyzed by high-resolution and tandem MS (HRMS-MS/MS), which confirmed a macrolactam linkage between Trp1 and Glu9 of the FusA core peptide (Fig. S3). Omission of ATP or FusC resulted in the formation of a mass corresponding to the linear FusA core peptide (m/z 2287). Generation of the core peptide was dependent upon the presence of both FusE and FusB, as omission of either protein led to no observed mass changes.
Figure 2. Enzymatic reconstitution of fusilassin.
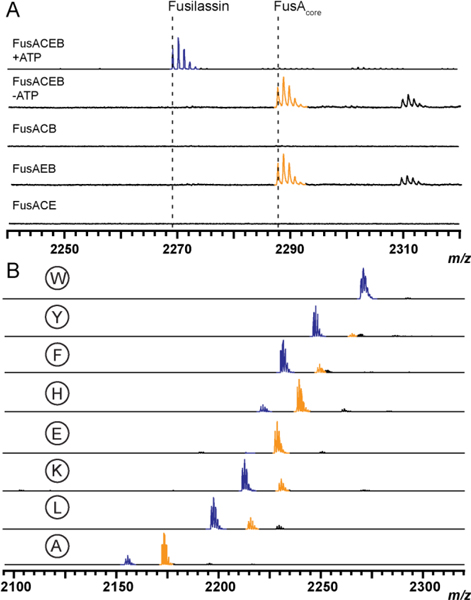
(A) MALDI-TOF-MS of all requisite biosynthetic components (top) shown alongside parallel reactions with individual components omitted from the reaction. (B) Relative processing of core position 1 variants from purified enzyme assays with wild-type Trp1 as the top spectrum. W1E product formation was at the limit of detection (Fig. S8). Spectra represent end-point assays after 3 h reactions. Macrolactam-containing products are blue and linear core (uncyclized) peptides are orange. Low-intensity peaks at +22 Da represent sodiated ions of either the linear core or mature lasso peptide.
To probe the importance of highly conserved residues of the leader peptidase and lasso cyclase, we generated separate multiple sequence alignments for FusB and FusC using 11 homologous proteins from previously characterized lasso peptide BGCs. Six residues of FusB were targeted for replacement with Ala, including the predicted catalytic Cys79 and His112 residues, a nearby highly conserved Trp, and three Glu residues (Fig. S4). The catalytic activity of the FusB variants was then assessed using the purified enzymes. Ala-substitution of Cys79 and His112 abolished catalytic activity while the E116A variant was severely impaired (Fig. S5). FusB-W114A exhibited extensive proteolytic degradation upon expression in E. coli, leading us to conclude this residue is of structural significance (Fig. S2). Ala-replacement of FusB-E123 and -E124 did not significantly perturb proteolytic efficiency under the reaction conditions employed. Four residues of FusC predicted to be involved in ATP-binding were targeted for replacement with Ala, based on the known structure of E. coli asparagine synthetase B (Protein DataBank entry 1CT9, Fig. S6).21 The catalytic activity of FusC-D238A and -D340A was abolished, even when assaying at high concentration (5 mM) of ATP. The effects were less severe for FusC-S239A and G336A, which displayed catalytic impairment only when supplied with 500 μM ATP (Fig. S7). The concentration-dependent activity of these variants likely arises from an increased KM for ATP, which corroborate the predicted ATP-binding residues of FusC.
To address enzymatic processing of FusA variants at core position 1, each variant was prepared as above and subjected to identical reconstitution reactions using wild-type FusB, FusC, and FusE. In good agreement with E. coli expression, all variants, except Pro1, were tolerated by the Fus biosynthetic enzymes although the ion intensity for the Glu1 variant was at the limit of detection (Figs. 2, S8). We next evaluated if the leader peptidase, the lasso cyclase, or both were intolerant of Pro1. While leader proteolysis was efficient on wild-type FusA treated with FusB and FusE (Fig. 2A), no reaction was observed with FusA-W1P as the substrate (Fig. S8). The intolerance of Pro at the P1’ position of many proteases, including cysteine hydrolases, is well known.22 To evaluate if FusC could tolerate Pro1, we first determined if the Fus biosynthetic proteins were capable of processing the linear core of wild-type FusA in a leader peptide-independent manner. While FusC was incapable of forming product on its own, the presence of FusB and FusE in the reaction led to a low but detectable level of product formation (Fig. S9) in the absence of leader peptide. Similar results were obtained when repeating the reaction with the linear core of FusA-W1P; thus, the leader peptidase, not the lasso cyclase, was intolerant of Pro1.
In addition to resolving biosynthetic ambiguities like core position one tolerance, purified enzymatic reconstitution allows for deeper views into a biosynthetic pathway. In contrast with the E. coli-based expression system, where only mature metabolites are easily extracted and detected, reactions using purified FusB, FusC, and FusE provide straightforward assessments of conversion efficiency for FusA variants. For example, larger residues at core position 1 (e.g. Tyr, Phe, Lys, and Leu) were most efficiently processed. Smaller and charged residues (e.g. His, Glu, and Ala) at position 1 gave comparatively less product by enzymatic reconstitution, which was not discernable by E. coli-expression (Fig. S1). As previously documented with plantazolicin and thiomuracin17,23–25, which derive from two distinct RiPP classes, a greater level of biosynthetic granularity is achieved through enzymatic reconstitution.
Verification of the threaded lasso conformation.
It has remained unanswered if fusilassin prepared enzymatically ex vivo would be in a threaded conformation or in the isobaric “branched-cyclic” state. To address this question, we first performed a MALDI-TOF MS-based hydrogen-deuterium exchange assay on enzymatically prepared, wild-type fusilassin (Fig. S10). Under our H-D exchange conditions, the linear core of FusA was fully deuterated in <1 min while fusilassin prepared enzymatically required ~60 min to obtain the same level of deuteration. To provide additional support for a threaded conformation, we subjected the linear core of FusA and fusilassin to carboxypeptidase Y digestion for 18 h (Fig. S11). Analysis by MS showed the linear core of FusA was completely consumed while the vast majority of the fusilassin peptide remained intact. Indeed, only a single, low intensity peak consistent with proteolytic removal of the C-terminal residue, Ile18, was observed. Lastly, we tested thermal resistance to denaturation of wild-type fusilassin and the W1H and W1L variants. Even after a 2 h, 95 °C heat treatment, all variants exhibited similar resistance to carboxypeptidase Y degradation (Fig. S12). Unfortunately, the poor solubility characteristics and relatively low expression yields of fusilassin by both E. coli expression and enzymatic reconstitution prevented assessment of threadedness by less-sensitive methods, such as NMR.
Altered ring sizes, tail lengths, and biosynthetic insights.
Given the robustness and superior substrate tolerance of the Fus proteins, we sought to determine if FusA variants with expanded and contracted macrolactam rings could be produced. With the exception of the caulosegnin pathway26, which naturally encodes lasso precursor peptides of differing ring sizes, all reported expansion or contraction of the macrolactam ring has abolished biosynthetic processing.27,28 Fusilassin naturally contains a 9-mer macrolactam (Fig. 1A), and readily accepts a 10-mer macrolactam generated by insertion of Ala within the macrocycle (1WYTAAEWGLE10…FI18). Not only was this 10-mer variant easily produced by both E. coli expression and enzymatic reconstitution (Fig. S13), but HRMS-MS/MS confirmed formation of an isopeptide bond at the predicted acceptor site (Fig. S14). Proteolytic treatment of this compound with carboxypeptidase Y supported threadedness (Fig. S15). Contracted fusilassin variants with theoretical 8-mer (1WYTEWGLE8…FI17; ΔA4) and 7-mer (1WYEWGLE7…FI16; ΔT3/A4) macrolactam ring sizes were then prepared and subsequently evaluated for production. While the 8-mer was readily detected from E. coli expression, we note that the same variant was considerably less efficiently produced using purified enzymes (Fig. S13). Continuing the trend, the 7-mer was not be detected by enzymatic reconstitution, and although a mass consistent with the expected product (m/z 2097 Da) was detected from E. coli expression, the intensity was too low to conclusively verify the identity of the product. We attribute the declining efficiency of production to an increased level of steric clashing between residues comprising the fusilassin ring and the C-terminal tail.
The apparent inability of the fusilassin biosynthetic enzymes to produce a reliable 7-mer macrolactam product, whether it be in a threaded (lasso) or unthreaded (branched-cyclic) conformation, suggested that FusC might be incapable of forming the latter. This topic has attracted some debate in the lasso peptide field, especially since the initial report on lassomycin suggested the peptide might naturally possess a branched-cyclic topology.5 A subsequent study from the same group corrected that finding by demonstrating fundamental differences between naturally produced lassomycin and a synthetically prepared branched-cyclic version.29 To date, every reported lasso peptide produced by enzymatic reconstitution, heterologous expression, or native isolation has been in the threaded conformation. To further evaluate the (in)ability of FusC to generate a branched-cyclic peptide, we prepared C-terminal truncations of FusA. The first tested variant contained only the macrolactam ring (1WYTAEWGLE9) and failed to form detectable levels of product under E. coli expression, thus demonstrating the biosynthetic necessity of the loop and/or tail regions (Fig. S15). Truncation of as few as 3 residues from the C-terminus of FusA (ΔRFI18) also failed to generate observable product through E. coli expression. This suggested that the loop region of the FusA core peptide was insufficient for FusC catalysis while the tail region was catalytically necessary. We dissected these findings further using our enzymatic reconstitution platform with Gly-replaced FusA variants at core positions 16–18. All tail variants yielded the expected macrolactam products, albeit formation of the R16G product was impaired relative to the other tested positions (Fig. S15). We postulate that the C-terminal tail of FusA, especially Arg16, is a critical contact point for the lasso cyclase to “pre-fold” the peptide substrate into a lasso-like conformation. Resolving the finer details of this interaction will require extensive additional characterization of the biosynthetic enzymes.
Acceptor specificity and non-cognate lasso peptide formation.
The above data show that the fusilassin biosynthetic enzymes are unusual in that they readily tolerate substitution of the first core position, as well as ring size variants (±1 residue). All previous studies have shown that lasso peptide biosynthetic pathways are intolerant to changes in the macrolactam acceptor site (either Asp or Glu) and location (core positions 7, 8, or 9).27,28 Therefore, we generated a FusA-E9D variant, and found that FusC was also incapable of producing the W1-D9 macrolactam by biosynthetic reconstitution (Fig. 3A).
Figure 3. Acceptor site specificity.
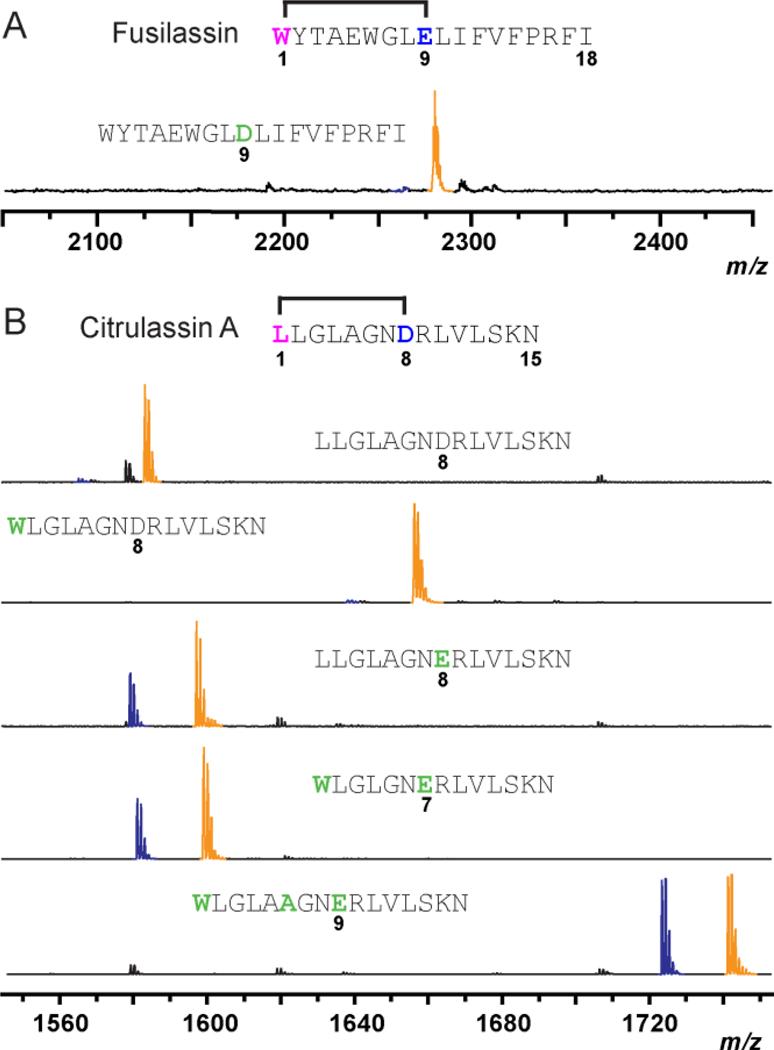
(A) MALDI-TOF-MS of the FusA-E9D variant from enzymatic reconstitution. (B) Evaluation of purified Fus enzymes in processing chimeric substrates bearing wild-type and macrolactam variants of citrulassin A (CitA). Macrolactam-containing products are blue while the linear core (uncyclized) peptides are orange. Substitutions to wild-type FusA in panel A, and CitA in panel B, respectively, are green.
We next applied our new understanding of fusilassin biosynthetic tolerance to the generation of a non-cognate lasso peptide. We generated a chimeric substrate by fusing the leader peptide of FusA (residues −22 thru −1, Fig. 1A) to the core peptide of citrulassin A (CitA), a lasso peptide we characterized previously3 which has no obvious sequence relationship to fusilassin. Notably, CitA possesses a Leu1-Asp8 macrolactam and a core peptide that is 3 residues shorter than that of fusilassin. As expected, purified FusB, FusC, and FusD did not process the chimeric substrate, given the presence of an Asp acceptor (Fig. 3B). The Fus proteins also did not process a Cit-L1W variant, showing the overriding effect of acceptor versus donor site identity. Introduction of D8E to CitA led to a dramatic increase in processing by the Fus proteins. The formation and location of the macrolactam linkage was confirmed by HRMS-MS/MS (Fig. S14). Threadedness of this peptide was supported by recalcitrance to carboxypeptidase Y digestion although the resulting lasso peptide was not stable to prolonged heat treatment (Fig. S16). CitA variants with contracted (W1-E7) and expanded (W1-E9) macrolactam linkages were also readily processed by the Fus proteins (Fig. 3B). The predicted macrolactam for the CitA (W1-E7) macrolactam was demonstrated by HRMS-MS/MS (Fig. S14). Similar to the CitA-D8E variant, we found that the variant bearing an expanded W1-E9 macrolactam ring exhibited resistance to carboxypeptidase Y digestion, which is highly suggestive of a threaded conformation (Fig. S17). Unfortunately, we did not observe product formation from the chimeric substrates using the E. coli expression system. One possible reason could be degradation by endogenous E. coli proteases outcompeted the rate of lasso peptide formation (Fig. S18). Further characterization would be needed to identify the origin of these observations. Nevertheless, our data underscore a remarkable substrate tolerance for the fusilassin pathway, especially with purified proteins.
Genomics-guided characterization of protein-protein interactions.
Given the unprecedented performance of FusB, FusC, and FusE in vitro, we sought to elucidate residues critical to formation of the fusilassin biosynthetic protein complex. Previously, retrospective analysis of RODEO data sets uncovered a conserved YxxP motif in predicted lasso leader peptides.3 Recent work on the lasso peptides paeninodin18 and chaxapeptin19 have confirmed that the YxxP motif functions as the recognition sequence for the RRE, as substitution with Ala caused severe reductions in binding affinity and product formation. To confirm their role in fusilassin biosynthesis, and to investigate other modestly conserved residues within the leader peptide, we prepared a brief panel of Ala-substituted FusA variants. We then employed a fluorescence polarization assay with wild-type FusE as the receptor and a fluorescein-labeled, wild-type FusAleader as a competing ligand. Of the five FusA positions surveyed, only Y(−17)A and P(−14)A were found to decrease affinity for FusE with the Tyr(−17) being critically important (Fig. S19, Table S3).
Since the above data only pertain to the substrate (FusA) side of the interaction, we next sought to probe the RRE-portion (FusE) of this interaction by targeted mutagenesis. To rationally direct our mutagenesis to likely sites of interaction, we employed GREMLIN30, a computational algorithm that predicts interaction sites based on the principle of compensatory mutation. This strategy requires a large amount of diverse sequences, thus necessitating an updated survey of lasso peptide BGCs. We therefore queried RODEO to update the observable lasso peptide genomic space. Our last published survey of this kind was conducted in October 2016.3 Since then, GenBank has grown by 56% and we have made algorithmic improvements that enhance the prediction of leader peptide cleavage sites.15 As of August 2018, RODEO identified 2,993 non-redundant, high-scoring, manually curated lasso precursor peptides in publicly available bacteria and archaeal genomes deposited in GenBank (Supplemental Dataset 1). Our updated RODEO data set provided ample sequence diversity for the application of this algorithm.
Our analysis identified 7 residues of lasso peptide RRE proteins that are under co-evolutionary pressure with the lasso peptide precursors (Fig. S20). As we noted previously3, all sites within the precursor peptide that were under statistically relevant co-evolution with the RRE were confined to the leader peptide. The receptor residues predicted to interact with the leader peptide were overrepresented in the predicted β3 and α3 secondary structures of the RRE. Ala-substitution of four of these positions led to a loss of ~2–fold affinity towards FusAleader. FusE-D74A exhibited a more dramatic reduction (8–fold) (Fig. S21, Table S4). Based on several available crystal structures of RRE proteins, we mapped Asp74 to the start of helix α3. Presumably, Leu78 would reside on the same face of this helix (Fig. 4). In support of this prediction, the FusE-L78A variant displayed a 10–fold reduction in FusAleader affinity.
Figure 4. Residues critical for FusA/E/B ternary complex.
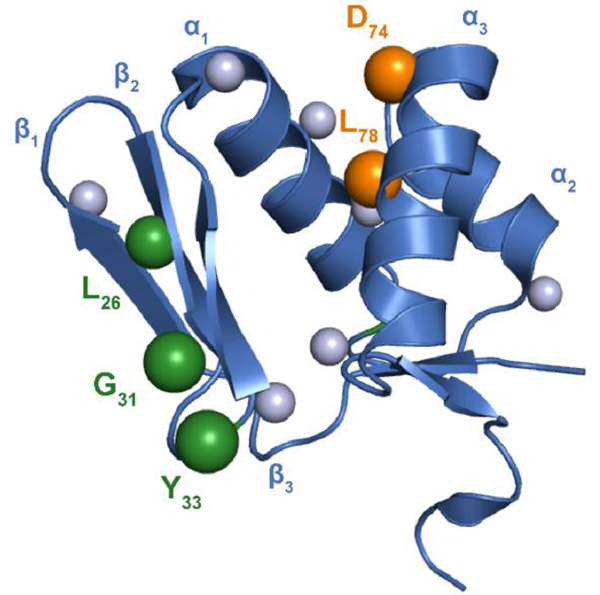
The RRE domain of MccB (residues M1-S86, PDB entry 3H9J, blue) was aligned with FusE. Spheres indicate residues of FusE probed by mutagenesis. Mutagenesis at sites along α3 drastically reduced binding to FusAleader (orange). Ala-substitutions along β2 and β3 did not influence FusAleader binding but did abolish leader peptide cleavage (green). The validity of these predicted protein-protein interaction sites is underscored by the fact mutagenesis elsewhere (gray) did not reduce leader peptide binding or leader peptidase activity.
Sequence analysis of FusE shows that the protein has an abnormally acidic isoelectric point (pI = 4.3). Given that Asp74 was a critical residue for engaging a relatively basic leader peptide (Fig. 1A), we hypothesized that additional acidic residues of FusE may contribute to this interaction. Single Ala-replacement of select residues also caused a ~2–fold loss in leader peptide affinity. It is possible that the difference in pI provides an electrostatic attraction between the RRE and the leader peptide over longer distances. However, once within proximity, we predict that specific hydrophobic interactions, governed by the YxxP motif, will dominate the orientation and specificity of the two interacting proteins. Investigations that further define this interaction are on-going.
Interaction between RRE and leader peptidase.
Sequence analysis of FusB, which collaborates with the RRE (FusE) during leader peptide cleavage, yielded a considerably basic isoelectric point (pI = 10.7). Thus, we propose interaction of these two proteins is also driven, at least initially, by electrostatic attraction. To determine if this phenomenon was more widespread, we calculated the pI’s for all discretely encoded (i.e., non-fused) RRE proteins, leader peptidases, and lasso cyclases identified by RODEO (Fig. S22). While lasso cyclases exhibit neutral pI’s (7.4 ± 1.1), RRE proteins are notably acidic (4.7 ± 1.2) and the leader peptidases are strongly basic (10.5 ± 1.1). Within a given lasso peptide BGC, the difference in pI between the two interacting proteins was considerable (5.7 ± 1.8). This phenomenon is widespread across nearly 2,000 examined lasso peptide BGCs and thus certainly not specific to the fusilassin pathway.
By leveraging the 2018 RODEO-derived dataset, we had a sufficient number of lasso peptide biosynthetic protein sequences to probe co- evolutionary relationships between FusB and FusE in a statistically relevant fashion. GREMLIN analysis on the concatenated protein sequences identified 5–6 residues within FusB and E that were under predicted co-evolutionary pressure (Fig. S20). To evaluate the relative contribution of these residues, we conducted an MS-based leader peptidase cleavage assay that requires both FusB and FusE (Fig. 2A). Under the conditions employed, wild-type FusE orchestrated the efficient delivery of FusA to FusB (Fig. 5). The most statistically probable interaction identified by GREMLIN with FusB involved FusE-Tyr33. Substitution with Ala led to no observed leader peptide cleavage. It is notable that FusE-Y33A has affinity for FusA equal to wild-type FusE (Table S3), which is supported by structure homology arguments as this site does not reside on either the β3 or α3 secondary structure of FusE (Fig. 4).
Figure 5. RRE:Leader peptidase interaction.
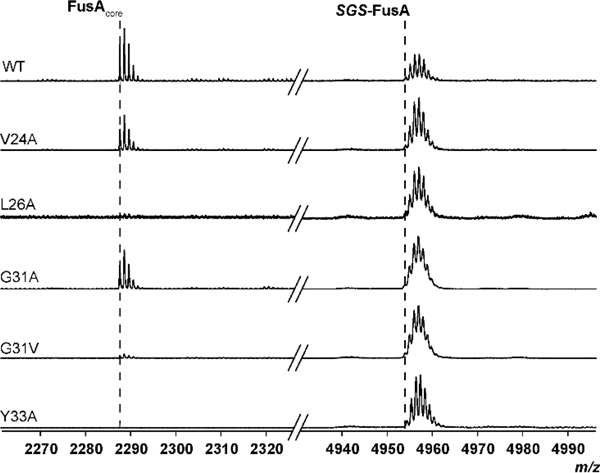
MALDI-TOF-MS was used to monitor leader peptidase activity of FusB and a panel of GREMLIN- and structure-derived FusE variants. Variants demonstrated differing degrees of enzymatic processing, supporting our GREMLIN predictions. SGS refers to the N-terminal Ser-Gly-Ser motif remaining after TEV protease cleavage of MBP-FusA.
The second most strongly FusB-correlated residue of FusE was Gly31, which did not influence leader peptide cleavage upon Ala-substitution. However, replacement of Gly31 with Val nearly abolished leader peptidase activity. This variant only exhibited a minor reduction in leader peptide-binding (2–fold); therefore, we predict it also plays a direct role in FusB-binding (Table S3; Fig. 5). In proximity to the 31GxY33 motif was FusE-Leu26, which we had previously substituted with Ala to evaluate FusAleader-binding (Fig. S21). This variant abolished leader peptidase activity, but the same was not true for the nearby V24A variant, which would theoretically occupy the same face of the β2 strand (Fig. 4).
Bioinformatic insight into lasso cyclases.
The bioinformatically mined lasso cyclases are visualized in the form of maximum likelihood trees and a sequence similarity network (Figs. S23–24). As noted previously, many lasso peptides are found within the actinobacteria, proteobacteria, and firmicutes phyla with minimal evidence of horizontal gene transfer. Other bacterial and archaeal phyla are represented at lower frequencies. Upon viewing the tree color-coded by the identity of the macrolactam acceptor residue (Asp or Glu), we first noted that many clades display a dominant acceptor identity while a few contain a more balanced representation of the two types. Further, Asp- and Glu-specific lasso cyclases are interspersed throughout the tree, meaning that there is no global sequence distinction between enzymes that prefer one acceptor type over the other.
Given the macrolactam size-flexibility of the fusilassin pathway, we conducted a retrospective analysis of the 2018 lasso peptide dataset to determine how many naturally encoded lasso peptides would be 7, 8, or 9-mers. Upon removing ambiguous linkages from the data set, the proportions were found to be 5.5%, 36.6%, and 57.9%, respectively (Fig. S25). Given the acceptor-intolerance of the fusilassin pathway, we also analyzed the same data and found that 29.5% of RODEO-identified, unambiguous lasso core peptides would use Glu as the macrolactam acceptor site and the remaining 69.5% would contain Asp. The plurality (45%) of lasso peptides are predicted to be Asp-containing 9-mers.
Conclusions.
The data presented herein describe the discovery, reconstitution, and initial enzymatic and biophysical characterization of fusilassin, an unusual lasso peptide from a mesothermophilic actinomycete. Our results confirm that the fusilassin pathway can serve as the first lasso peptide biosynthetic platform amenable to detailed mechanistic characterization. The fusilassin enzymes exhibit an unprecedented substrate promiscuity with respect to the identity of the first core position, the size of the macrolactam linkage, and core peptide sequence. We have successfully dissected the role of leader peptide from core processing using purified enzymes and synthetic core peptides, which have delineated the enzymatic preferences of the leader peptidase and lasso cyclase proteins. Our data support an inability of the lasso cyclase to generate the isobaric branched-cyclic peptides, as illustrated by employing a series of truncated precursor peptides, proteolytic degradation studies, and hydrogen-deuterium exchange assays. Moreover, the enzymes described have the exceptional capability to produce non-cognate lasso peptides or simple derivatives thereof, presenting unprecedented access to the ~3,000 RODEO-identified lasso peptides found in prokaryotic genomes. Finally, bioinformatic analysis of this dataset enabled the co-evolutionary and biochemical interrogation of additional sites of interaction between the leader peptide, RRE domain, and leader peptidase. The fusilassin BGC represents the most promising pathway yet described that can be utilized to elucidate molecular-level details of lasso peptide biosynthesis.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants from the David and Lucile Packard Fellowship for Science and Engineering (to D.A.M.) and the Ving & May Lee Discovery Fund from the Department of Chemistry (to D.A.M.). Funds to purchase the Bruker UltrafleXtreme MALDI TOF/TOF mass spectrometer were from the National Institutes of Health (S10 RR027109 A). We acknowledge C. Schwalen, B. Burkhart, and S. Simon for their foundational and technical assistance in cloning and protein purification.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information, including experimental methods and supporting figures, is available free of charge on the ACS Publications website.
The authors declare no competing financial interests.
REFERENCES
- (1).Arnison PG; Bibb MJ; Bierbaum G; Bowers AA; Bugni TS; Bulaj G; Camarero JA; Campopiano DJ; Challis GL; Clardy J; Cotter PD; Craik DJ; Dawson M; Dittmann E; Donadio S; Dorrestein PC; Entian KD; Fischbach MA; Garavelli JS; Göransson U; Gruber CW; Haft DH; Hemscheidt TK; Hertweck C; Hill C; Horswill AR; Jaspars M; Kelly WL; Klinman JP; Kuipers OP; Link AJ; Liu W; Marahiel MA; Mitchell DA; Moll GN; Moore BS; Müller R; Nair SK; Nes IF; Norris GE; Olivera BM; Onaka H; Patchett ML; Piel J; Reaney MJ; Rebuffat S; Ross RP; Sahl HG; Schmidt EW; Selsted ME; Severinov K; Shen B; Sivonen K; Smith L; Stein T; Süssmuth RD; Tagg JR; Tang GL; Truman AW; Vederas JC; Walsh CT; Walton JD; Wenzel SC; Willey JM; van der Donk WA Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep 2012, 30 (1), 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Burkhart BJ; Hudson GA; Dunbar KL; Mitchell DA A Prevalent Peptide-Binding Domain Guides Ribosomal Natural Product Biosynthesis. Nat. Chem. Biol. 2015, 11 (8), 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Tietz JI; Schwalen CJ; Patel PS; Maxson T; Blair PM; Tai H-C; Zakai UI; Mitchell DA A New Genome-Mining Tool Redefines the Lasso Peptide Biosynthetic Landscape. Nat. Chem. Biol. 2017, 13 (5), 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ortega MA; van der Donk WA New Insights into the Biosynthetic Logic of Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products. Cell Chem. Biol. 2016, 23 (1), 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gavrish E; Sit CS; Cao S; Kandror O; Spoering A; Peoples A; Ling L; Fetterman A; Hughes D; Bissell A; et al. Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium Tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem. Biol. 2014, 21 (4), 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Feng Z; Ogasawara Y; Nomura S; Dairi T Biosynthetic Gene Cluster of a D-Tryptophan-Containing Lasso Peptide, MS-271. ChemBioChem., 19 (19), 2045–2048. [DOI] [PubMed] [Google Scholar]
- (7).Allen CD; Chen MY; Trick AY; Le DT; Ferguson AL; Link AJ Thermal Unthreading of the Lasso Peptides Astexin-2 and Astexin-3. ACS Chem. Biol. 2016, 11 (11), 3043–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hegemann JD; Fage CD; Zhu S; Harms K; Leva FSD; Novellino E; Marinelli L; Marahiel MA The Ring Residue Proline 8 Is Crucial for the Thermal Stability of the Lasso Peptide Caulosegnin II. Mol. BioSyst. 2016, 12 (4), 1106–1109. [DOI] [PubMed] [Google Scholar]
- (9).Hegemann JD; Zimmermann M; Xie X; Marahiel MA Lasso Peptides: An Intriguing Class of Bacterial Natural Products. Acc. Chem. Res. 2015, 48 (7), 1909–1919. [DOI] [PubMed] [Google Scholar]
- (10).Hegemann JD; De Simone M; Zimmermann M; Knappe TA; Xie X; Di Leva FS; Marinelli L; Novellino E; Zahler S; Kessler H; et al. Rational Improvement of the Affinity and Selectivity of Integrin Binding of Grafted Lasso Peptides. J. Med. Chem. 2014, 57 (13), 5829–5834. [DOI] [PubMed] [Google Scholar]
- (11).Knappe Thomas A; Manzenrieder Florian; Mas-Moruno Carlos; Linne; Sasse Florenz; Kessler Horst; Xie Xiulan; Marahiel Mohamed A. Introducing Lasso Peptides as Molecular Scaffolds for Drug Design: Engineering of an Integrin Antagonist. Angew. Chem. Int. Ed. 2011, 50 (37), 8714–8717. [DOI] [PubMed] [Google Scholar]
- (12).Naimi S; Zirah S; Hammami R; Fernandez B; Rebuffat S; Fliss I Fate and Biological Activity of the Antimicrobial Lasso Peptide Microcin J25 Under Gastrointestinal Tract Conditions. Front. Microbiol. 2018, 9, 1764–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yamamoto T; Matsui H; Yamaji K; Takahashi T; Øverby A; Nakamura M; Matsumoto A; Nonaka K; Sunazuka T; Ōmura S; et al. Narrow-Spectrum Inhibitors Targeting an Alternative Menaquinone Biosynthetic Pathway of Helicobacter Pylori. J. Inf. Chemother. 2016, 22 (9), 587–592. [DOI] [PubMed] [Google Scholar]
- (14).Lopez FE; Vincent PA; Zenoff AM; Salomón RA; Farías RN Efficacy of Microcin J25 in Biomatrices and in a Mouse Model of Salmonella Infection. J Antimicrob. Chemother. 2007, 59 (4), 676–680. [DOI] [PubMed] [Google Scholar]
- (15).Schwalen CJ; Hudson GA; Kille B; Mitchell DA Bioinformatic Expansion and Discovery of Thiopeptide Antibiotics. J. Am. Chem. Soc. 2018, 140 (30), 9494–9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mahanta N; Liu A; Dong S; Nair SK; Mitchell DA Enzymatic Reconstitution of Ribosomal Peptide Backbone Thioamidation. Proc. Natl. Acad. Sci. U.S.A. 2018, 115 (12), 3030–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hudson GA; Zhang Z; Tietz JI; Mitchell DA; van der Donk WA In Vitro Biosynthesis of the Core Scaffold of the Thiopeptide Thiomuracin. J. Am. Chem. Soc. 2015, 137 (51), 16012–16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).D. Hegemann J; J. Schwalen C; A. Mitchell D; Donk W A. van der Elucidation of the Roles of Conserved Residues in the Biosynthesis of the Lasso Peptide Paeninodin. Chem. Commun. 2018, 54 (65), 9007–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Martin-Gómez H; Linne U; Albericio F; Tulla-Puche J; Hegemann JD Investigation of the Biosynthesis of the Lasso Peptide Chaxapeptin Using an E. Coli-Based Production System. J. Nat. Prod. 2018, 81 (9), 2050–2056. [DOI] [PubMed] [Google Scholar]
- (20).Pavlova O; Mukhopadhyay J; Sineva E; Ebright RH; Severinov K Systematic Structure-Activity Analysis of Microcin J25. J. Biol. Chem. 2008, 283 (37), 25589–25595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Larsen TM; Boehlein SK; Schuster SM; Richards NGJ; Thoden JB; Holden HM; Rayment I Three-Dimensional Structure of Escherichia Coli Asparagine Synthetase B: A Short Journey from Substrate to Product. Biochemistry 1999, 38 (49), 16146–16157. [DOI] [PubMed] [Google Scholar]
- (22).Evnin LB; Vásquez JR; Craik CS Substrate Specificity of Trypsin Investigated by Using a Genetic Selection. Proc. Natl. Acad. Sci. U.S.A. 1990, 87 (17), 6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Deane CD; Burkhart BJ; Blair PM; Tietz JI; Lin A; Mitchell DA In Vitro Biosynthesis and Substrate Tolerance of the Plantazolicin Family of Natural Products. ACS Chem. Biol. 2016, 11 (8), 2232–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Deane CD; Melby JO; Molohon KJ; Susarrey AR; Mitchell DA Engineering Unnatural Variants of Plantazolicin through Codon Reprogramming. ACS Chem. Biol. 2013, 8 (9), 1998–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhang Z; Hudson GA; Mahanta N; Tietz JI; van der Donk WA; Mitchell DA Biosynthetic Timing and Substrate Specificity for the Thiopeptide Thiomuracin. J. Am. Chem. Soc. 2016, 138 (48), 15511–15514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hegemann JD; Zimmermann M; Xie X; Marahiel MA Caulosegnins I–III: A Highly Diverse Group of Lasso Peptides Derived from a Single Biosynthetic Gene Cluster. J. Am. Chem. Soc. 2013, 135 (1), 210–222. [DOI] [PubMed] [Google Scholar]
- (27).Knappe TA; Linne U; Robbel L; Marahiel MA Insights into the Biosynthesis and Stability of the Lasso Peptide Capistruin. Chem. Biol. 2009, 16 (12), 1290–1298. [DOI] [PubMed] [Google Scholar]
- (28).Ducasse R; Yan K-P; Goulard C; Blond A; Li Y; Lescop E; Guittet E; Rebuffat S; Zirah S Sequence Determinants Governing the Topology and Biological Activity of a Lasso Peptide, Microcin J25. ChemBioChem 2012, 13 (3), 371–380. [DOI] [PubMed] [Google Scholar]
- (29).Lear S; Munshi T; S. Hudson A; Hatton C; Clardy J; A. Mosely J; J. Bull T; S. Sit C; L. Cobb S Total Chemical Synthesis of Lassomycin and Lassomycin-Amide. Org. Biomol. Chem. 2016, 14 (19), 4534–4541. [DOI] [PubMed] [Google Scholar]
- (30).Ovchinnikov S; Kamisetty H; Baker D Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. eLife. 2014, 3, e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


