Abstract
Staphylococcus pseudintermedius is commonly associated with colonization or infection in dogs, and was identified as a novel species within the genus Staphylococcus in 2006. Methicillin resistance emerged in S. pseudintermedius during the last decade. We describe here a genomic characterization of the first methicillin-resistant S. pseudintermedius (MRSP) recovered from a human patient in Argentina. The strain was phenotypically identified as MRSP 8510 by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and antimicrobial susceptibility testing. We assessed genetic characterization by mecA PCR, SCCmec (staphylococcal chromosomal cassette) typing, and whole-genome sequencing. MRSP 8510 was phenotypically resistant to six classes of antimicrobial agents, consistent with the genes found in its genome. We concluded that MRSP 8510 was a multidrug-resistant ST1412 isolate. This study highlights the importance of the detection and characterization of pathogens with potential risks of zoonotic transmission to humans, as they may constitute a reservoir of genes associated with antimicrobial resistance.
Keywords: MRSP, Staphylococcus pseudintermedius, MDR
Introduction
The Staphylococcus intermedius group was differentiated in 2006, and is comprised of Staphylococcus intermedius, Staphylococcus pseudintermedius, and Staphylococcus delphini.1 S. pseudintermedius is commonly found as part of the normal skin and nasal flora of healthy dogs and cats, but it can also cause infections ranging from pyoderma and surgical wound infections, to deep infections such as osteomyelitis.2 S. pseudintermedius is a common pathogen in dogs and can occasionally be found in human infection. The first case of endocarditis caused by S. pseudintermedius infection was confirmed by molecular identification and published in 2006.3 Since then, other human infections caused by S. pseudintermedius have been reported, including skin and soft tissue infections, surgical hardware site infections, pneumonia, brain abscesses, and bacteremia.4–6
It is likely that infections caused by S. pseudintermedius in humans are under-reported because of inaccurate identification and often reported instead as Staphylococcus aureus.7 However, the introduction of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems in clinical laboratories has resulted in increased reporting of S. pseudintermedius.8
Over the past decade, methicillin-resistant S. pseudintermedius (MRSP) has emerged in various parts of the world.2 As in S. aureus, methicillin resistance in S. pseudintermedius is mediated by PBP2a, an additional penicillin-binding protein encoded by the mecA gene located in a mobile element of the bacterial chromosome called SCCmec (staphylococcal chromosomal cassette) that can be transferred between different species of staphylococci.
The aim of this study was to perform a genomic characterization of the first isolate of MRSP recovered from a human patient in Argentina and determine whether it was related to the dominant lineages found worldwide.
Methods
Case study
In October 2017, an 86-year-old woman with a history of hypertension, deep vein thrombosis, and chronic venous ulcers was admitted to “Hospital General de Agudos Juan A. Fernández,” Buenos Aires, Argentina, with a three-day history of worsening lower extremity pain. The patient had a history of inferior vena cava filter placement and was anticoagulated with acenocoumarol 2 years prior. The patient was hemodynamically stable, afebrile, and had lower extremity ulcerations associated with poor perfusion, without phlogosis and with cyanosis and lividities in the distal region of the left foot. During hospitalization, the ulcers evolved to dry necrosis of the distal region of the left foot and three compromised toes. In November 2017, the patient had a below the knee amputation. Bone tissue from the patient's left tibia was sent for culture. The surgical wound appeared to be healing; thus, the patient did not receive antibiotic treatment and was discharged from the hospital after 1 week.
Identification and susceptibility testing
The bone culture showed methicillin-resistant S. aureus and MRSP. Species identification was performed using MALDI-TOF MS (Bruker Daltonics Microflex LT, Billerica, MA). Susceptibility testing was performed using Phoenix PMIC-89 Panel, Epicenter V6.41 (Becton Dickinson, Argentina), E-test (BioMerieux, France), and disk diffusion according to manufacturer's recommendations and Clinical Laboratory Standards Institute (CLSI) guidelines. Inducible resistance to clindamycin was performed by D-test.9 The categories of resistant, intermediate, and susceptible classes for each antibiotic were assigned when the applicable breakpoint was available in CLSI documents CLSI M100 28th Edition9 or CLSI VET08 4th Edition.10 Tigecycline was interpreted according to the Food and Drug Administration antimicrobial breakpoints. The MRSP 8510 isolate was received at the National Reference Laboratory on Antimicrobial Resistance INEI-ANLIS “Dr. Carlos G. Malbrán” for molecular studies.
mecA gene PCR
Detection of the mecA gene was performed by PCR as previously described11; S. aureus ATCC 43300 and S. aureus ATCC 29213 were used as the positive and negative controls, respectively.
Genome sequencing
Genomic DNA was extracted using DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA) per manufacturer's instructions; concentration was measured by QubitTM assay (Invitrogen, Carlsbad, CA). Illumina library preparation was carried out by Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA). Hi-seq sequencing was carried out in the Epigenetics and Genomic Laboratory of our affiliate, Weill Cornell Medical College (New York, NY), using an Illumina HiSeq 2000, generating sequencing reads with a coverage of 100 × . Assembly was performed by Velvet and annotation by NCBI Prokaryotic Genome Annotation Pipeline (PGAP). Genome sequence has been deposited at DDBJ/ENA/GenBank under the accession WEIP00000000. The detection of resistance genes was carried out with PATRIC using the available ResFinder (genomicepidemilogy.org) and CARD (Comprehensive Antimicrobial Resistance Database, card.macmaster.ca) databases, and the gene content was compared with the phenotype presented by the isolate.
Identification of SCCmec element
The isolate was first screened for typical SCCmec elements by multiplex PCR.12 S. aureus collection strains were used as controls for SCCmec types I, IA, II, III, IV, and VI (COL, PER34, BK2464, USA100, ANS46, HU25, USA400, and HDE288, respectively); a clinical strain was used as the positive control for SCCmec type V.12 The genome sequence from the isolate was examined in the SCCmecFinder resource13 to determine the SCCmec type. In addition, the SCCmec V (GenBank Id AY894416)14 nucleotide and individual protein sequences were compared by BLAST (Basic Local Alignment Search Tool)15 with the genome of the isolate in PATRIC (Pathosystems Resource Integration Center patricbrc.org).16 A careful examination of the region containing the SCCmec V genes and its flanking regions was conducted using the Proteome Comparison and Compare Region View17 tools found in PATRIC. A broad examination for the presence or absence of the protein families that contain mecA, mecR1, and mecI genes across the MRSP 8510 genome was conducted using PATRIC's Protein Family Sorter.18
Multilocus Sequence Typing analysis
Sequence type was determined using MLST (Multilocus Sequence Typing) software based on the 7-gene MLST scheme that included the genes ack, cpn60, fdh, pta, purA, sar, and tuf.19 The allelic profile of the isolate was compared with allele sequences present in the PubMLST database and submitted to the MLST database curator Vincent Perreten to assign a sequence type.
Results
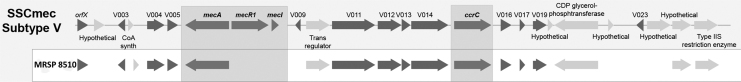
The MRSP 8510 isolate was resistant to penicillin, oxacillin, erythromycin, clindamycin, streptomycin, kanamycin, ciprofloxacin, chloramphenicol, and trimethoprim-sulfamethoxazole, and was susceptible to the other antimicrobial agents tested (Table 1). By using the disk diffusion method, the isolate was shown to be resistant (zone diameter) to oxacillin 1 μg (15 mm), erythromycin 15 μg (6 mm), and clindamycin 2 μg (6 mm), indicating constitutive resistance to macrolides, lincosamides, and streptogramin B (cMLSB). Despite the lack of CLSI-approved interpretative criteria for streptomycin and kanamycin, the isolate showed an inhibition zone of 6 mm with streptomycin 10 μg and kanamycin 30 μg disks, and was considered resistant to both antibiotics. Methicillin resistance was confirmed by mecA gene PCR. SCCmec screening by multiplex PCR showed a pattern that shared two bands with the SCCmec type V element: a 162 bp that corresponded to mecA and a 449 bp that corresponded to ccr complex. However, the pattern differed in the corresponding J1 region band, suggesting that it could be a variant of SCCmec V, and the SCCmec finder tool could not classify this element. Analysis of the genome allowed us to observe that the isolate possessed mecA, but not other genes associated with SCCmec V, including mecR1 and mecI (Fig. 1).
Table 1.
Minimal Inhibitory Concentrations and Antibiotic Resistance Genes in Methicillin-Resistant Staphylococcus pseudintermedius 8510 Strain
| Antibiotics | MICs (μg mL-1) | Interpretive category | Resistance genes from MRSP 8510 genome |
|---|---|---|---|
| Penicillin | >32a | R | blaZ |
| Oxacillin | >2 | Rc | mecA |
| Ceftaroline | 0.5 | Sd | |
| Erythromycin | >4 | R | ermB |
| Clindamycin | >2 | R | — |
| Kanamycin | 6 mmb | NA | aph(3′)-III |
| Streptomycin | 6 mmb | NA | ant(6)-Ia |
| Gentamicin | 4 | S | — |
| Tetracycline | 0.12a | S | — |
| Minocycline | ≤1 | S | — |
| Tigecycline | ≤0.12 | Se | — |
| Trimethoprim-sulfamethoxazole | >2/38 | R | dfrG |
| Chloramphenicol | 64a | R | catA |
| Ciprofloxacin | >2 | R | grlA (Ser80Ile); gyrA (Ser84Leu, Glu88Val)f |
| Vancomycin | ≤0.5 | S | — |
| Teicoplanin | ≤1 | S | — |
| Rifampicin | ≤0.5 | S | — |
| Linezolid | ≤1 | S | — |
| Daptomycin | ≤1 | S | — |
Performed by E-test (BioMerieux).
Performed by disk diffusion.
All the antibiotics were interpreted according to Staphylococcus spp. CLSI breakpoints (M100 28th ed.) except for: coxacillin (Staphylococcus pseudintermedius CLSI breakpoint), dceftaroline (Staphylococcus aureus CLSI breakpoint), etigecycline (Food and Drug Administration breakpoint).
Mutations in the quinolone-resistance determining region of the topoisomerase genes.
—, no gene was detected; CLSI, Clinical Laboratory Standards Institute; MICs, minimal inhibitory concentrations; MRSP, methicillin-resistant S. pseudintermedius; NA, not available; R, resistant; S, susceptible.
FIG. 1.
Comparison of the SCCmec region of the MRSP 8510 with the Staphylococcus aureus TSGH17 (SCCmec V). The MRSP 8510 genome had similar ccr complex and mecA and ccrC genes, but was missing many of the other genes associated with SCCmec V, including mecR1 and mecI. MRSP, methicillin-resistant S. pseudintermedius; SCCmec, staphylococcal chromosomal cassette.
We found by MLST analysis that isolate displayed a new allele combination assigned as ST1412 that has been included into the S. pseudintermedius pubMLST database. Sequencing analysis revealed 2,669,519 bp, 37.2% GC content, and 2,689 predicted coding sequences. In addition to the gene mecA, the isolate carried the β-lactamase gene blaZ, the phosphotransferase gene aph(3′)-III, adenylnucleotidyltransferase gene ant(6)-Ia, the methylase gene ermB, the dihydrofolate reductase gene dfrG, and the chloramphenicol acetyltransferase catA gene (Table 1). The isolate MRSP 8510 was resistant to ciprofloxacin, and mutations in topoisomerase genes gyrA and grlA, Ser84Leu and Glu88Val in the topoisomerase GyrA and Ser80Ile in GrlA were identified. Additional amino acid substitutions were found outside the quinolone-resistance determining region (QRDR) of the topoisomerase genes (Pro5Ser, Asp447Glu, His557Gln, Lys736Asn, and Ile516Thr).
Discussion
In this study, we characterized the first MRSP isolated from a human patient in Argentina. The MRSP was isolated along with a methicillin-resistant S. aureus resistant to erythromycin, with inducible resistance to clindamycin iMLSB, and susceptible to the other antibiotics tested. The MRSA isolate belongs to ST5-SCCmec IV clone,20 which is currently the second most prevalent clone after ST30-SCCmec IV in community isolates in Argentina.
The finding of MRSP in polymicrobial cultures seems to be common.5 The patient reported no contact with dogs or cats before hospital admission, which is consistent with cases reported by other authors.7
Similar to other recent reports,21 our results showed a strong correlation between the resistance phenotype displayed by the isolate and the resistance genes detected.
The SCCmec of S. pseudintermedius displayed some degree of homology to that of S. aureus, but the SCCmec of S. pseudintermedius is sometimes untypeable using SCCmec typing schemes developed for S. aureus, as we observed in our study. The classification of the SCCmec elements in S. pseudintermedius is complex due to the high genetic diversity and the existence of composite cassettes and pseudo-SCCmec elements.22 Several SCCmec elements are reported in MRSP, including SCCmec III (previously described as II-III), which is found in the globally dominant ST71 lineage; SCCmec VT variants; and other newly reported cassettes.22 The SCCmec element present in the isolate MRSP 8510 is largely homologous to SCCmec type V (5C2 and 5), previously called VI or VII of S. aureus but lacking mecR1 and mecI genes. Elements with similar characteristics were found in canine isolates previously studied in Argentina.23
Two of the three mutations identified in topoisomerase genes gyrA and grlA of MRSP 8510, related to ciprofloxacin resistance, were previously described, Ser84Leu and Ser80Ile.21,23–26 Additional amino acid substitutions were found outside the QRDR region of the topoisomerase genes, but their roles in fluoroquinolone resistance have yet to be determined.
The dissemination of MRSP isolates was associated with a limited number of clones,27,28 similar to the situation observed in human S. aureus. CC71, previously described as the epidemic European clone, which is now widespread worldwide. CC258 may be partly replacing CC71, at least in Northern Europe.28,29 CC68, previously described as the epidemic North American clone,30 is frequently reported in this region but also in Europe. CC45 originated in Asia, but is also found in Europe and, to a lesser extent, in North America and CC112 is prevalent in Asia, and it is also found in Europe, Oceania, and North America.28
In a previous study conducted in MRSP isolated from dogs in Argentina, we observed high clonal diversity by MLST (ST339, ST649, ST919, ST920, ST921, and ST922), and the goeBURST analysis of the isolates showed that although they were not related to ST68 or ST71, they were evolutionarily closer to ST68 than to ST71.23 In this study, MRSP 8510 displayed a new allele combination assigned as ST1412, not related to the STs previously found in Argentina.23 ST1412 was not related to the globally disseminated clones mentioned above, nor to the CC551 SCCmec V that emerged in 2015 in Poland and is replacing the previously prevalent ST 71.31 ST1412 is a double locus variant of the ST45, which originated in Asia and was described in Finland, Sweden, the Netherlands, Israel, Thailand, and Australia.2,22,28
Comparing the resistance phenotype of the MRSP 8510 isolate with those of canine origin all were microbial drug resistant, but unlike the MRSP of canine origin the MRSP 8510 was resistant to chloramphenicol. Interestingly, chloramphenicol resistance is strongly associated with CC45 and was recently reported in 95% of ST45 isolates.28
Although it has not yet been determined to what extent S. pseudintermedius contributes to the gene pool of resistance in human pathogens (e.g., S. aureus), animal-derived staphylococci can be the source of determinants of resistance in human S. aureus.32
In conclusion, in this study we reported the characterization of the first multidrug-resistant MRSP isolated from a human case in Argentina. Using MALDI-TOF MS in the clinical laboratory allowed us to correctly identify the S. pseudintermedius species and to clearly define the methicillin resistance phenotype. The transmission of these isolates between dogs or cats and humans represents a potential risk to public health, especially as they could be a source of antibiotic resistance genes.
Acknowledgments
We thank the Epigenomic Core of Weill Cornell Medical College for their WGS Service.
Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported in part from the National Institutes of Health/NIAID grant (1R56AI118756-0A1 to A.E.R.).
References
- 1. Devriese L.A., Hermans K., Baele M., and Haesebrouck F.. 2009. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet. Microbiol. 133:206–207 [DOI] [PubMed] [Google Scholar]
- 2. Gronthal T., Eklund M., Thomson K., Piiparinen H., Sironen T., and Rantala M.. 2017. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J. Antimicrob. Chemother. 72:1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Hoovels L., Vankeerberghen A., Boel A., Van Vaerenbergh K., and De Beenhouwer H.. 2006. First case of Staphylococcus pseudintermedius infection in a human. J. Clin. Microbiol. 44:4609–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Somayaji R., Priyantha M.A, Rubin J.E, and Church D.. 2016. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn. Microbiol. Infect. Dis. 85:471–476 [DOI] [PubMed] [Google Scholar]
- 5. Yarbrough M.L., Lainhart W., and Burnham C.A.. 2018. Epidemiology, clinical characteristics, and antimicrobial susceptibility profiles of human clinical isolates of Staphylococcus intermedius group. J. Clin. Microbiol. 56:e01788–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lozano C., Rezusta A., Ferrer I., Pérez-Laguna V., Zarazaga M., Ruiz-Ripa L., Revillo M.J, and Torres C.. 2017. Staphylococcus pseudintermedius human infection cases in Spain: dog-to-human transmission. Vector Borne Zoonotic Dis. 17:268–270 [DOI] [PubMed] [Google Scholar]
- 7. Viau R., Hujer A.M, Hujer K.M, Bonomo R.A, and Jump R.L.. 2015. Are Staphylococcus intermedius infections in humans cases of mistaken identity? A case series and literature review. Open Forum Infect. Dis. 2:ofv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu M.T., Burnham C.A, Westblade L.F, Dien Bard J., Lawhon S.D, Wallace M.A, Stanley T., Burd E., Hindler J., and Humphries R.M.. 2016. Evaluation of oxacillin and cefoxitin disk and MIC breakpoints for prediction of methicillin resistance in human and veterinary isolates of Staphylococcus intermedius group. J. Clin. Microbiol. 54:535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clinical Laboratory Standards Institute (CLSI). 2018. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical and Laboratory Standards Institute, Wayne, PA, CLSI Document M100-S28 [Google Scholar]
- 10. Clinical and Laboratory Standards Institute (CLSI). 2018. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. VET08 4th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Vannuffel P., Gigi J., Ezzedine H., Vandercam B., Delmee M., Wauters G., and Gala J.L.. 1995. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J. Clin. Microbiol. 33:2864–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milheirico C., Oliveira D.C, and de Lencastre H.. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaya H., Hasman H., Larsen J., Stegger M., Johannesen T.B, Allesøe R.L, Lemvigh C.K, Aarestrup F.M, Lund O., and Larsen A.R.. 2018. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere. 3:e00612–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boyle-Vavra S., Ereshefsky B., Wang C.C, and Daum R.S. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 43:4719–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Altschul S.F. 2005. BLAST algorithm. In Encyclopedia of life sciences. 10.1038/npg.els.0005253 (accessed December27, 2019) [DOI]
- 16. Wattam A.R., Davis J.J, Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M, Disz T., Gabbard J.L, Gerdes S., Henry C.S, Kenyon R.W, Machi D., Mao C., Nordberg E.K, Olsen G.J, Murphy-Olson D.E, Olson R., Overbeek R., Parrello B., Pusch G.D, Shukla M., Vonstein V., Warren A., Xia F., Yoo H., and Stevens R.L.. 2016. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45:D535–D542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Overbeek R., Olson R., Pusch G.D, Olsen G.J, Davis J.J, Disz T., Edwards R.A, Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A.R, Xia F., and Stevens R.. 2013. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42:D206–D214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wattam A.R., Gabbard J.L, Shukla M., and Sobral B.W.. 2014. Comparative genomic analysis at the PATRIC, a bioinformatic resource center, in host-bacteria interactions. Methods Mol. Biol. 1197:287–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Solyman S.M., Black C.C, Duim B., Perreten V., van Duijkeren E., Wagenaar J.A, Eberlein L.C, Sadeghi L.N, Videla R., Bemis D.A, and Kania S.A.. 2013. Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J. Clin. Microbiol. 51:306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Egea A.L., Gagetti P., Lamberghini R., Faccone D., Lucero C., Vindel A., Tosoroni D., Garnero A., Saka H.A, Galas M., Bocco J.L, Corso A., Sola C., and Study Group of S. aureus in Argentina. 2014. New patterns of methicillin-resistant Staphylococcus aureus (MRSA) clones, community-associated MRSA genotypes behave like healthcare-associated MRSA genotypes within hospitals, Argentina. Inter. J. Med. Microbiol. 304:1086–1099 [DOI] [PubMed] [Google Scholar]
- 21. Wegener A., Broens E.M, Zomer A., Spaninks M., Wagenaar J.A, and Duim B.. 2018. Comparative genomics of phenotypic antimicrobial resistances in methicillin-resistant Staphylococcus pseudintermedius of canine origin. Vet. Microbiol. 225:125–131 [DOI] [PubMed] [Google Scholar]
- 22. Worthing K.A., Schwendener S., Perreten V., Saputra S., Coombs G.W, Pang S., Davies M.R, Abraham S., Trott D.J, and Norris J.M.. 2018. Characterization of staphylococcal cassette chromosome mec elements from methicillin-resistant Staphylococcus pseudintermedius infections in Australian animals. mSphere 3:e00491–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gagetti P., Wattam A.R, Giacoboni G., De Paulis A., Bertona E., Corso A., and Rosato A.E.. 2019. Identification and molecular epidemiology of methicillin resistant Staphylococcus pseudintermedius strains isolated from canine clinical samples in Argentina. BMC Vet. Res. 15:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Descloux S., Rossano A., and Perreten V.. 2008. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius. J. Clin. Microbiol. 46:1818–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Onuma K., Tanabe T., and Sato H.. 2012. Antimicrobial resistance of Staphylococcus pseudintermedius isolates from healthy dogs and dogs affected with pyoderma in Japan. Vet. Dermatol. 23:17–22 [DOI] [PubMed] [Google Scholar]
- 26. Gomez-Sanz E., Torres C., Lozano C., Sáenz Y., and Zarazaga M.. 2011. Detection and characterization of methicillin-resistant Staphylococcus pseudintermedius in healthy dogs in La Rioja, Spain. Immunol. Microbiol. Infect. Dis. 34:447–453 [DOI] [PubMed] [Google Scholar]
- 27. McCarthy A.J., Harrison E.M, Stanczak-Mrozek K., Leggett B., Waller A., Holmes M.A, Lloyd D.H, Lindsay J.A, and Loeffler A.. 2015. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 70:997–1007 [DOI] [PubMed] [Google Scholar]
- 28. Pires dos Santos T., Damborg P., Moodley A., and Guardabassi L.. 2016. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: inference of population structure from multilocus sequence typing data. Front. Microbiol. 7:1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duim B., Verstappen K.M, Broens E.M, Laarhoven L.M, van Duijkeren E., Hordijk J., de Heus P., Spaninks M., Timmerman A.J, and Wagenaaret J.A.. 2016. Changes in the population of methicillin-resistant Staphylococcus pseudintermedius and dissemination of antimicrobial-resistant phenotypes in the Netherlands. J. Clin. Microbiol. 54:283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perreten V., Kadlec K., Schwarz S., Grönlund Andersson U., Finn M., Greko C., Moodley A., Kania S.A, Frank L.A, Bemis D.A, Franco A., Iurescia M., Battisti A., Duim B., Wagenaar J.A, van Duijkeren E., Weese J.S, Fitzgerald J.R, Rossano A., and Guardabassi L.. 2010. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J. Antimicrob. Chemother. 65:1145–1154 [DOI] [PubMed] [Google Scholar]
- 31. Kizerwetter-Świda M., Chrobak-Chmiel D., Rzewuska M., and Binek M.. 2017. Changes in the population structure of canine methicillin-resistant Staphylococcus pseudintermedius in Poland. Vet. Microbiol. 208:106–109 [DOI] [PubMed] [Google Scholar]
- 32. Worthing K.A., Brown J., Gerber L., Trott D.J, Abraham S., and Norris J.M.. 2018. Molecular epidemiology of methicillin-resistant staphylococci amongst veterinary personnel, personnel-owned pets, patients and the hospital environment of two companion animal veterinary hospitals. Vet. Microbiol. 223:79–85 [DOI] [PubMed] [Google Scholar]